Abstract
Phylogenetic analyses of small viral K+ channels suggests that they did not originate from their hosts, but instead could be the source of the postulated pore precursor in the evolution of K+ channels.
Cells communicate among themselves by electrical activity. Sophisticated membrane-embedded proteins, called ion channels, catalyze rapid, selective, and regulated ion fluxes across membranes (Hille, 2001). The resulting membrane currents are responsible for neuronal activity and the systemic propagation of electrical signals in animals. The activity of some channels is important for muscle movement in animals or growth in plants; other channels sense the concentration of physiological signals and modulate key processes in all kinds of eukaryotic cells. Among the many diverse ion channels in higher organisms, K+ channels are among the most important. One feature of K+ channels is that they conduct K+ ions much better than slightly smaller Na+ ions (Hille, 2001). The selective transport of K+ is involved in many physiological functions, including homeostasis of the membrane potential and the repolarization of the action potential in excitable cells. Because of a universal requirement for selective K+ fluxes across membranes, K+ channels are present in plasma membranes of all cell types in animals and plants. K+ channels also exist in organellar membranes, including mitochondria, chloroplasts, and endoplasmic reticula.
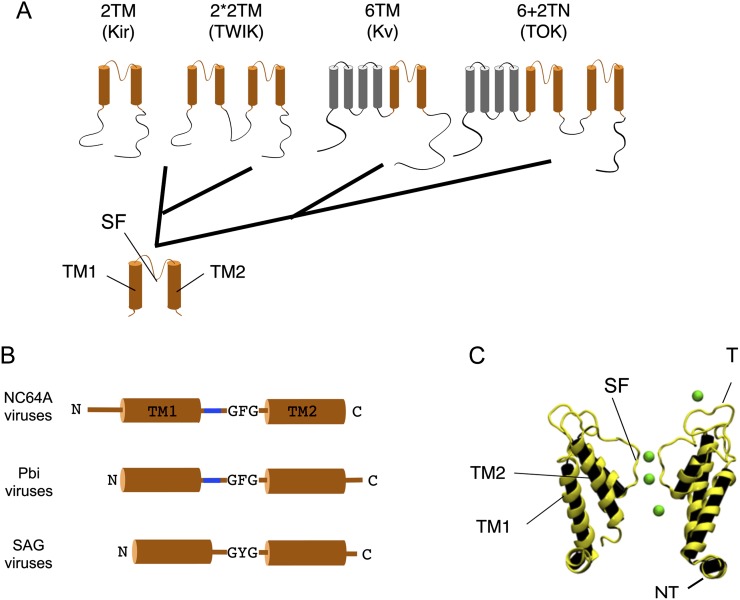
The evolution of K+ channels has been traced back to the prokaryotic world, where K+ channel proteins are common in eubacteria and archea (Jan and Jan, 1992; Derst and Karschin, 1998; Anderson and Greenberg, 2001; Martinac et al., 2008). Phylogenetic studies and structure/function properties of channel proteins have led to the hypothesis that the complex K+ channels in animals and plants evolved from a simple bacterial precursor channel (Anderson and Greenberg, 2001). It is postulated that the precursor channel protein resembled the pore module of modern K+ channels (Fig. 1A). The pore module, which consists of two transmembrane domains connected by a pore helix containing the canonical filter sequence of K+ channels, assembles into a functional tetramer. The central pore, which is created by oligomerization of the monomers, is the conduction pathway for the ions (Doyle et al., 1998). A pore module with this overall architecture exists in all K+ channels, and more complex K+ channels are believed to have started with this pore module (Wei et al., 1996; Anderson and Greenberg, 2001). There is genetic evidence that the plethora of K+ channels results from several major genetic events, such as gene fusions and gene duplications (Anderson and Greenberg, 2001). Furthermore, minor genetic changes involving many single base mutations and deletions also contributed to the evolution of the diversity of K+ channels. In this brief review, we suggest that a viral-encoded K+ protein might be the evolutionary ancestor of all K+ channel proteins.
Figure 1.
Hypothetical phylogenetic relationship within K+ channels. A, Common to all structural classes of K+ channels is a pore module (orange) containing the selectivity filter (SF) located between two transmembrane domains (TM1 and TM2). The different structural classes presumably evolved from such a two transmembrane (2TM) precursor. The Kir channels with the 2TM architecture could be the direct descendants of this precursor. The tandem architecture of the TWIK channels, with the tandem 2TM structure (2*2TM) structure, probably occurred after a gene duplication of the pore module. The fusion of the precursor channel with a voltage sensor domain (gray) could be the origin of the 6TM architecture, which is typical for Kv channels. Gene fusion of the precursor to the latter could explain the structure of the TOK channel in which the 2TM motive and the 6TM motive are combined (6+2TM). B, Cartoon of Kcvs from different chloroviruses (left). The structures differ in the length of their cytosolic N and C termini, the presence/absence of a turret (blue), and the amino acid sequence of their selectivity filter, GFG versus GYG. C, Predicted structure of viral K+ channel Kcv (courtesy of Stefan M. Kast, Technische Universität-Dortmund). The image shows a snapshot from a molecular dynamic simulation including K+ ions (green). For clarity, only two of the four protein subunits are illustrated. The nomenclature for the description of the domains in C corresponds to that in A; also, the location of the turret (T) and the cytosolic N terminus (NT) is indicated.
VIRUSES AND THE ORIGIN OF LIFE
To address the question of the origin of the precursor of K+ channels, it is informative to mention a few recent ideas on evolution and the discovery of K+ channel proteins coded by some viruses. Viruses are often ignored in the evolution of prokaryotes and eukaryotes because they were only believed to be on the receiving end of gene fluxes. Their origin and evolution was believed to be a progressive development in which they recruited genes from their hosts and escaped as particles from cells (Moreira and López-García, 2009). However, the relatively recent discovery of viruses with double-stranded DNA (dsDNA) genomes in the 300- to 1,200-kb size range has led to alternative hypotheses. Some of these giant viruses encode more genes than many bacteria. The size of these viruses and their genetic complexity has fostered the idea that these large DNA viruses might even represent a fourth domain of life (Raoult et al., 2004; Boyer et al., 2010; Legendre et al., 2012; Nasir et al., 2012). One version of this hypothesis is that these viruses were descendants of more complex, autonomous organisms that lost the capacity to replicate by themselves and consequently became parasitic (Raoult et al., 2004; Legendre et al., 2012). This view of virus evolution implies that viruses are not only receptors of genes, but that they could also donate genes to their host(s). In fact, there is accumulating data that support the idea that some viruses are donors of genes and that gene transfer from viruses to prokaryotes and eukaryotes may have occurred very early in evolution. An even more radical idea, the virus-first hypothesis, suggests that some viruses may have existed before prokaryotes and eukaryotes (Forterre, 2001; Villarreal, 2005). In this primordial world, replicating RNA molecules may have preceded cellular life.
It should be noted, however, that the origin of large dsDNA viruses is controversial, and some evolutionary biologists have suggested that instead of starting out as large viruses, they evolved from a smaller virus or viruses (Filée et al., 2007, 2008, 2009). The colorful debate among virologists and evolutionary biologists on the origin of these large dsDNA viruses will continue for some time, and it is possible that none of the current theories completely explains the origin of these viruses. It is not our intent in this review to resolve the controversy about the evolutionary origin of large dsDNA viruses but to explore the possibility that some large dsDNA viruses that infect eukaryotic algae encode a K+ channel protein that might be the evolutionary ancestor of all K+ channel proteins.
CHANNEL AND TRANSPORTER GENE PRODUCTS CODED BY ALGAL VIRUSES
Viruses in the family Phycodnaviridae encompass a diverse and rapidly expanding collection of large, icosahedral dsDNA viruses that infect algae. These lytic and lysogenic viruses have genomes ranging from 160 to 560 kb (Wilson et al., 2009). The family consists of six genera based initially on host range and supported by sequence comparisons. The family is monophyletic with branches for each genus; however, the phycodnaviruses have evolutionary roots that connect them with several other families of large DNA viruses, referred to as nucleocytoplasmic large DNA viruses, including the huge mimiviruses that infect amoeba. Phycodnaviruses are found in both terrestrial and marine aquatic environments throughout the world. They are thought to play dynamic, albeit largely undocumented, roles in regulating algal communities, such as the termination of algal blooms, which has implications in global geochemical cycling and weather patterns (Brussaard, 2004; Wilson et al., 2009).
One genera in the phycodnaviruses is the Chloroviruses. Chloroviruses are large icosahedral (190 nm in external diameter) viruses that have an internal lipid membrane (Van Etten and Dunigan, 2012). The chloroviruses infect symbiotic chlorella, often called zoochlorellae, which are associated with either the protozoan Paramecium bursaria, the coelenterate Hydra viridis, or the heliozoon Acanthocystis turfacea. The zoochlorellae are resistant to virus infection in the symbiotic state. Fortunately, some zoochlorellae can be grown independently of their hosts in the laboratory, permitting plaque assay of the viruses and synchronous infection of their hosts. Three such zoochlorellae are Chlorella NC64A (recently renamed Chlorella variabilis, and its viruses are called NC64A viruses), Chlorella SAG 3.83 (renamed Chlorella heliozoae, and its viruses are called SAG viruses), and Chlorella Pbi (renamed Micratinium conductrix, and its viruses are called Pbi viruses).
Currently, the sequences of 41 chloroviruses are in the public databases. Collectively, these 41 viruses encode 632 putative proteins, of which, any one virus encodes 319 to 416 proteins (Jeanniard et al., 2013). Some chloroviruses encode functional aquaglyceroporins (Gazzarrini et al., 2006), K+/H+ transporters (Greiner et al., 2011), and Ca++-transporting ATPases (Bonza et al., 2010). Relevant to this discussion, 39 of the 41 chloroviruses have genes predicted to encode K+ channel proteins (Jeanniard et al., 2013). One peculiarity of the chlorovirus K+ channel proteins is that their monomers are small. The most extensively studied channel, K+ chlorella virus (Kcv), from the prototype chlorovirus PBCV-1 (named KcvPBCV-1), consists of 94 amino acid residues (Plugge et al., 2000). The monomer of a related chlorovirus, ATCV-1 K+ channel (KcvATCV-1), is only 82 amino acid residues (Gazzarrini et al., 2009), which is the smallest known K+ channel protein. All of these small proteins have the predicted architecture of a K+ channel pore module with two transmembrane domains, which are linked by a pore helix, including a selectivity filter that is present in all K+ channels. However, the predicted structures of these 39 chlorovirus-encoded Kcvs indicate that they can be separated into one of three structural groups, depending on their host (Fig. 1B). The NC64A viruses have a short N-terminal tail; the Pbi viruses instead have a short C-terminal cytoplasmic tail. The SAG viruses encode the smallest proteins and lack an extracellular turret domain (Gazzarrini et al., 2009).
One or more representative Kcvs from each of these three groups has been expressed in heterologous cells, including Xenopus laevis oocytes (Plugge et al., 2000) and mammalian cells (Moroni et al., 2002), and shown to produce K+ selective currents. They can also complement yeast (Saccharomyces cerevisiae) cells that are defective in K+ uptake (Balss et al., 2008; Chatelain et al., 2009).
Like all K+ channel proteins, the viral monomers assemble into stable tetramers with a central pore (Fig. 1C; Pagliuca et al., 2007; Chatelain et al., 2009; Tayefeh et al., 2009). As expected from their predicted structures, many studies in the past 15 years have demonstrated that the chlorovirus channels have the basic properties of a K+ channel; they are ion selective, exhibit gating, and are sensitive to typical K+ channel blockers (Thiel et al., 2011). With the combination of structural simplicity and basic functionality, these viral proteins have features expected for a primordial precursor of modern K+ channels.
Viruses from several other genera in the phycodnavirus family have been sequenced, and members from two of them encode K+ channel proteins, including viruses that infect the filamentous brown alga Ectocarpus siliculosus (Delaroque et al., 2001) and viruses that infect some of the smallest known algae (the Prasinophytes), Ostreococcus (Derelle et al., 2008; Weynberg et al., 2009), Micromonas (Moreau et al., 2010), and Bathycoccus spp. (Moreau et al., 2010). To our knowledge, K+ channel-encoding proteins have only been reported in DNA viruses that infect algae.
ORIGIN OF CHLOROVIRUS K+ CHANNELS
As noted above, a common assumption is that viruses are molecular pirates, which acquire genes from their hosts (Sinkovics et al., 1998; Christie and Dokland, 2012). While this view of viruses as mere “pick pockets” (Moreira and López-García, 2009) can explain the origin of some viral genes (Murphy, 1994), it does not explain the origin of all virus genes, including the virus-encoded K+ channel-encoding genes. If the chlorovirus K+ channel genes were acquired from their algal host, one would expect that coevolution occurred between the virus and host K+ channel-encoded genes. This assumption of coevolution was recently tested experimentally because the genomes of two algal virus hosts, C. variabilis (Blanc et al., 2010) and E. siliculosus (Cock et al., 2010), have been sequenced and annotated. The coccal green alga Chlorella spp. and the filamentous brown alga Ectocarpus spp. are evolutionary very distant (Yoon et al., 2004). However, they are hosts for phycodnaviruses PBCV-1 and Esv-1, respectively. The viruses, which infect these two algae, each have genes encoding K+ channels. Comprehensive phylogenetic analyses of the viral and host K+ channel genes indicate that the viral K+ channel genes are more closely related to each other than to the K+ channel genes coded by their hosts, i.e. there is no evidence of coevolution between the viral K+ channels and the host K+ channels (Hamacher et al., 2012). These data strongly argue against molecular piracy as the origin of the viral K+ channels, at least from their current hosts.
This conclusion is supported by a recent comprehensive bioinformatics analysis of 35 newly sequenced chloroviruses, plus six previously sequenced chloroviruses (Jeanniard et al., 2013). This analysis included 14 NC64A viruses, 13 SAG viruses, and 14 Pbi viruses. The viruses contained 319 to 416 putative protein-encoding genes, of which 155 were present in all the viruses and were considered to be “core genes.” More than one-half of the chlorovirus gene families are predicted to have a potential recent origin (after chlorovirus divergence), among which a significant portion show compositional evidence for horizontal gene transfer. Surprisingly, only a few of these “recently acquired” gene products had close homologs in databases, raising the question of the true donor organism(s). We speculate that the donor(s) might be uncharacterized viruses. Of all the virus PBCV-1-encoded genes, there were only seven families of genes that were exchanged between the current algal host and PBCV-1, and of these seven, four of them were probably transferred from the virus to the alga (Jeanniard et al., 2013).
Because these data indicate that the algal viral K+ channels did not originate from their current hosts, it is reasonable to ask if the genes encoding the K+ channel proteins could have been exchanged with other organisms or viruses. We compiled a representative collection of pro- and eukaryotic K+ channel proteins from the literature and BLAST searches in public sequence databases.
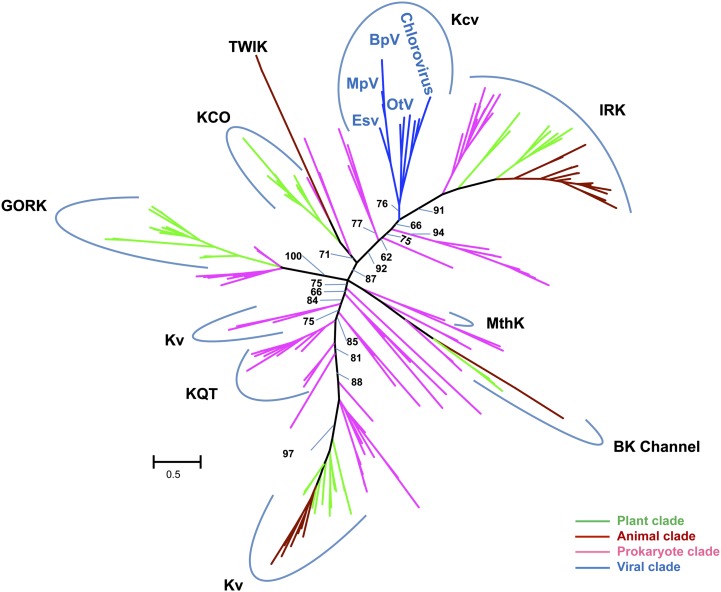
The overall level of sequence conservation outside the selectivity filter was very low between K+ channel families. Therefore, we used structural information to guide us in the construction of the multiple alignment of K+ channels using the 3D-Coffee/Expresso program (Armougom et al., 2006). The 3D-Coffee alignment was further corrected manually, and poorly aligned regions outside the selectivity filter and pore helix were discarded. The resulting alignment, containing 80 amino acid positions, of which 32 had gaps in one or more sequences, was used in phylogenetic reconstructions using the maximum-likelihood (ML), neighbor-joining (NJ), maximum-parsimony, and Bayesian methods. Figure 2 shows the unrooted ML tree. NJ reconstruction produced approximately the same clades, but the relative branching order of the different channel families differed between the ML and NJ phylogenetic trees (Supplemental Fig. S1). The MP and MB methods produced unresolved phylogenetic trees containing a high number of multifurcating nodes and were therefore not analyzed further. The discrepancy between the four reconstruction methods and the associated low bootstrap support for many interior branches in the ML and NJ trees reflect the high divergence and poor phylogenetic signal in the K+ channel alignment. Bacteria and eukaryotes were often mixed in various reconstructed clades, suggesting that the diversification of major K+ channel families predates the split between bacteria and eukaryotes. However, we urge caution in interpreting the phylogenetic trees due to the overall lack of robustness.
Figure 2.
Unrooted ML tree constructed with the pore modules of representative K+ channels from viruses (blue), prokaryotes (purple), plants (green), and animals (brown). The phylogenetic reconstruction was performed using the PhyML program and LS+Γ substitution model (Guindon et al., 2010). Support for the branches was estimated using the approximate-likelihood ratio test and 500 bootstrap replicates. We only show support values (approximate-likelihood ratio test/bootstrap format) for important interior branches separating the main K channel families. The scale bar for branch length is expressed as average number of substitutions per site. K+ channel families include a calcium-gated potassium channel from Methanobacterium thermoautotrophicum (MthK), Kvs, KQT-like subfamily channels (KQT for K+ channels related to long QT syndrome), big potassium channels (BK channel), outwardly rectifying K+ potassium channels (GORK), inwardly rectifying K+ channels (IRK), potassium channel openers (KCO), two-pore domain potassium channels (TWIK), and viral potassium channels (Kcv).
Nevertheless, the three phylogenetic reconstruction methods grouped the K+ channels encoded by the phycodnaviruses in a separate clade, including chloroviruses, EsV, and prasinoviruses Ostreococcus tauri virus 4 (OtV4), OtV6, Micromonas pusilla virus 1 (MpV1), MpV2, Bathycoccus prasinos virus 1 (BbV1), and BbV2. Taken together, the phylogenetic analyses neither support nor refute the hypothesis that the viral channels are the evolutionary ancestors of more complex K+ channels. However, the data also do not support the canonical view that K+ channels originate from bacteria. Consequently, one hypothesis is as plausible as the other.
At this point, we return to the idea that a comparative analysis of diverse K+ channels suggests that eukarya and prokarya obtained their K+ channels from a precursor, which provided the common pore-forming segment (Anderson and Greenberg, 2001). Without any robust data that support a different scenario, it is reasonable to suggest that the small chlorovirus-encoded channels provided the ancestor pore unit. With their small size, their functional simplicity, and their self-assembly into tetramers, these Kcv proteins have all the hallmarks expected for such a pore precursor unit (Thiel et al., 2011). In addition, the fact that these large DNA viruses might predate the separation of the three main branches of cellular life, namely bacteria, archaea, and eukarya (Raoult et al., 2004; Boyer et al., 2010; Legendre et al., 2012; Nasir et al., 2012), is consistent with the idea that viral Kcv channels could be the common ancestor of more complex K+ channels.
In this context, it is interesting to note that the aforementioned bioinformatics analyses of 41 chloroviruses (Jeanniard et al., 2013) suggest that four genes were horizontally transferred from the PBCV-1 virus to its host, C. variabilis. Furthermore, some phycodnaviruses like EsV-1 are lysogenic and incorporate their genomes into the host genome during their replication cycle (Müller et al., 1998). Such a process could link virus-driven gene emergence to host survival and evolution.
VIRAL K+ CHANNELS ARE IDEAL FOR BUILDING MORE COMPLEX AND MODULAR K+ CHANNELS
As mentioned above, K+ channels are modular proteins. They have a central pore module surrounded by auxiliary domains, which perceive external stimuli and translate the stimuli into channel activity. This concept is supported by experimental evidence. For example, a bacterial voltage-gated potassium (Kv) channel can still function after separation of the pore unit from its voltage sensor (Santos et al., 2008). Further support for the modular composition of channels is that regulatory domains such as voltage sensors (VSs), cyclic nucleotide-binding domains, or regulatory PAS domains, which control K+ channels, also occur in proteins with completely different functions. One example is a VS domain in bacterial Kv channels that is part of a voltage-regulated phosphatase in the sea squirt Ciona intestinalis (Murata et al., 2005). A key prediction for the modularity of K+ channels is that the complex arrangement of Kv channels resulted from a simple gene fusion. However, experimental proof for this concept has been difficult to obtain because attempts to exchange entire membrane modules between K+ channels have been unsuccessful (Caprini et al., 2001). Only small portions of the two domains, e.g. the “paddle” motif of the VS domain and the P loop with most of the transmembrane domains of the pore, could be exchanged between K+ channels (Lu et al., 2001; Alabi et al., 2007). These results led to the conclusion that membrane modules have coevolved so intimately in Kv proteins that it is impossible to exchange them entirely without losing the function of the chimeric channels.
At this point, we return to the concept that chlorovirus-encoded ion channels might be the precursor of modern K+ channels. Recent experiments indicate that the viral K+ channel Kcv can be fused to the VS domain of the aforementioned phosphatase from C. intestinalis and produce a functional channel (Arrigoni et al., 2013). Different forms of this chimera with various size linkers between the two proteins resulted in K+ channels with the functional features of a canonical K+ outward rectifier, e.g. the voltage-dependent conductance generated by modern Kv channels (Timpe et al., 1988; Gaymard et al., 1998). The results of these experiments support the hypothesis that complex K+ channels could have evolved from a single genetic event by the fusion of two unrelated genes. Unlike the channel pores of modern pro- and eukaryotic K+ channels, which do not tolerate such fusions, the chlorovirus Kcv channel is sufficiently simple and robust to tolerate fusion with another domain without losing function.
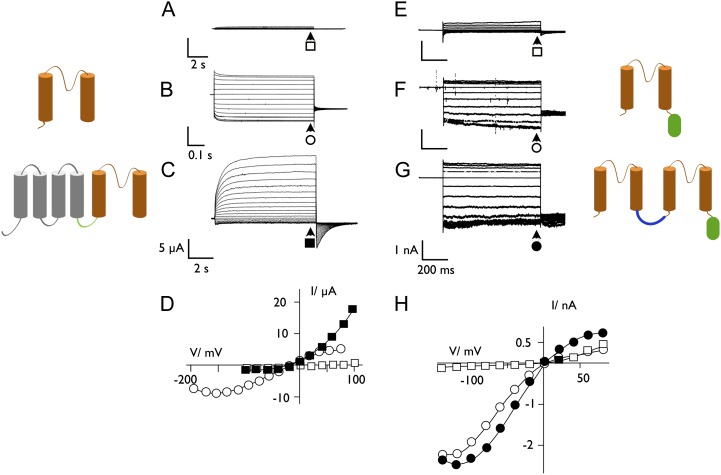
Additional experiments indicate that the viral Kcv channels have features required to be a precursor of more complex K+ channels. One class of K+ channels is composed of two coupled pore modules (Ketchum et al., 1995). Two of these tandem pore modules assemble into a dimer to form a functional K+ channel (Fig. 1A). We created this type of channel by fusing the genes of two chlorovirus Kcv channels. Expression of these tandem genes in mammalian Human Embryonic Kidney (HEK)293 cells produced a functional K+ channel (Fig. 3). The results of this experiment established that the viral Kcv channels tolerate fusion of two monomers like modern tandem K+ channels and produce a functional channel.
Figure 3.
The viral K+ channel Kcv is a suitable element for constructing complex channels as they occur in nature. A to D, Synthetic 6 transmembrane domain (TM) channel from a Kcv linked to the VS domain from C. intestinalis. When expressed in Xenopus spp. oocytes, the VS domain alone generates no current (A). The Kcv channel (2 TM subunit in orange on the left) causes a quasi-ohmic conductance in oocytes (B). When the VS domain (4 TM subunit in gray) and Kcv are linked, the synthetic channel has the features of a K+ outward rectifier with a slow activation at positive voltages (C). In this case, the two units are linked (green line) via the short N terminus of Kcv (Fig. 1B). Currents were measured as reported by Arrigoni et al. (2013). E to H, Synthetic 2*2TM channel from a tandem of Kcv subunits generates an active K+ channel. Exemplary currents measured in a nontransfected HEK293 cell (E), in a cell expressing the Kcv monomer (F), and in a cell expressing a tandem version of Kcv (G). A sketch of the structure of the respective channel subunits is shown on the right. For a tandem channel, two subunits are linked (blue line) by six Gly; a GFP (green oval) is fused to the C terminus. Currents were measured as described in Hertel et al. (2010). The steady-state current/voltage (I/V) relation of measurements in A to C and E and F are plotted in D and H, respectively. Currents were collected at the end of the voltage pulse, indicated by arrows. The symbols on the current traces correspond to the symbols in the respective I/V relations (data in A–D are from Cristina Arrigoni, University of Milan; data in E–H are from Jenifer Hewing, Technische Universität-Darmstadt).
These experiments indicate that viral K+ channels have the appropriate structural features expected for a simple precursor of more complex K+ channels. Furthermore, the viral channels are sufficiently robust to tolerate a bottom-up type evolution in which a complex structure with new functional properties evolves by simple genetic events such as gene fusion or gene duplication.
Virus-encoded K+ channels can also be engineered to change their intracellular location, i.e. differentially sorted. For example, expression of the chlorovirus Kcv channels in yeast cells transverse through the endoplasmic reticulum and are targeted to the plasma membrane. By contrast, the aforementioned E. siliculosus-infecting EsV virus-encoded K+ channel is targeted to the mitochondria. Chimeras formed between these two ion channels established that one could change their subcellular location by altering the length of their transmembrane domain 2 (Balss et al., 2008).
One can also ask about the origin of other domains in complex modern channels and determine if there is evidence of viral precursors. For example, some K+ channels contain PAS domains (Morais Cabral et al., 1998) or ankyrin repeat domains (Ehrhardt et al., 1997). The available databases indicate that some viruses, including chloroviruses, have genes that encode proteins similar to ankyrin repeats or PAS domains (Iyer et al., 2006). Genes encoding the VS domain of Kv channels or cyclic nucleotide-binding domains have not been found in viral genomes. However, when the sequence of the S4 domain of the Shaker channel, i.e. the VS domain, is BLASTed against the databank of virus genomes, a partial match with a chlorovirus protein appears and the matching area contains the critical charged amino acids required for voltage sensing (Fig. 4). The fact that BLAST searches do not uncover an entire VS domain in virus genomes does not mean that they do not exist. The huge gene pool of viruses in aquatic systems (Suttle, 2005, 2007) has only recently been discovered, and one might expect that genes for other K+ channel domains will be discovered in viral genomes.
Figure 4.
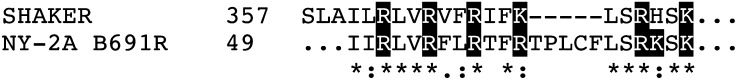
Alignment of voltage-sensing S4 element from Shaker K+ channel with a protein (B619R) of unknown function from chlorovirus New York 2A (NY-2A). The critical charged amino acids are shown in red. Note the large degree of homology in this domain.
ADDITIONAL VIRUSES CODE FOR ION CHANNEL PROTEINS
The idea that K+ channels may have originated in viruses implies that other viruses might encode channel proteins. Recent reports suggest that this occurs. The K+ channels in the chlorella viruses are just one example of a collection of viral proteins with channel function; these proteins have been given the name viroporins (Carrasco, 1995; Nieva et al., 2012; Schindler and Fischer, 2012). Since the seminal discovery of the M2 protein and its role as an H+ channel in the influenza A virus (Pinto et al., 1992), there are an increasing number of reports that viruses from various families encode proteins with ion channel function. Most of the viral viroporins share no or only weak similarity with proteins from prokaryotes and eukaryotes (Hsu et al., 2004; Schindler and Fischer, 2012); however, most of them have a monomer length of about 100 amino acids (Fischer and Sansom, 2002). This is probably about the minimum size for an ion channel protein. From either experimental structures or from structural predictions, the transmembrane domains of all these viral channels are α-helices. Variable numbers of monomers assemble in a membrane to form oligomers with a central pore, which serves as an ion-conductive pathway (Schindler and Fischer, 2012). In this sense, the viral channel proteins, including the viral K+ channels, are designed with the same architectural principle as channels from prokaryotes and eukaryotes, where assembly of different monomers creates a central conducting pore. The general message from this comparison is that the collection of viral channels exhibits multiple attempts to form a stable channel using the smallest molecular fold possible to achieve this goal.
Experimental characterization of the viral channels reveals that many of them are rather nonselective and only select between anions or cations (Wang et al., 2011). In the case of channels, with these functional features, this may be sufficient for virus infection and replication. In other cases, however, where the proteins require a more sophisticated function, viral channels have a higher degree of selectivity. The M2 channel from influenza A for example has a high selectivity for H+ (Pinto et al., 1997). This selectivity is required in the infection process (Wang et al., 2011) and guaranteed by the structure of the pore lining (Pinto et al., 1997). This finding can be interpreted as evidence that a selection pressure shapes the structure/function correlates of a viral channel in the absence of coevolution with the host.
The variability of sequences furthermore implies that channel-forming proteins developed independently in different viruses. The immense power of viruses for gain of functional genes is best described in the case of influenza A. A plus-one frame shift in a gene for an RNA polymerase (PB1) (Chen et al., 2001) is sufficient to generate a second overlapping open reading frame (F2) for a gene product, the PB1-F2 protein, with channel function (Henkel et al. 2010).
VIRAL CHANNEL EVOLUTION IS DRIVEN BY SELECTION PRESSURE
The suggestion that viruses participated in the evolution of ion channels requires one to address the question whether these proteins have a functional role in the viral life cycles and whether this role would create selection pressure. After the discovery of virus-encoded channel proteins, many of the channels were tested to determine their role(s) in the viral life cycle. In those cases where it has been examined, it turned out that functional viral channels, including the viral K+ channels, benefited viral replication (Takeda et al., 2002; Hsu et al., 2004; Greiner et al., 2009). Their functional importance in the viral life cycle was further substantiated by experiments in which channel-encoding genes were deleted or mutated in a viral genome. Typically, these procedures did not prevent viral replication, but they reduced formation of viral progeny and viral pathogenicity, thus reducing the competitiveness and fitness of the virus (Takeda et al., 2002; Gonzalez and Carrasco, 2003). The conclusion from these experiments is that there was strong selection pressure on the evolution of viral channels.
The function of the chlorovirus-encoded K+ channels is reasonably well understood, and its function can be illustrated by a brief description of the prototype virus PBCV-1 infection process. PBCV-1 attaches to the cell wall of its host via a spike structure that is present at one of the icosahedral vertices of the virus particles (Cherrier et al., 2009; Zhang et al., 2011). Attachment is followed by localized degradation of the host wall by a virus-packaged enzyme or enzymes (Meints et al., 1984). During this process, PBCV-1 undergoes a structural change that allows its internal membrane to fuse with the host membrane (Zhang et al., 2011). This membrane fusion results in activation of Kcv channels located in the PBCV-1 internal membrane (Thiel et al., 2010; G. Romani, A. Piotrowski, S. Hillmer, S. Gazzarrini, J. Gurnon, J. L. Van Etten, A. Moroni, G. Thiel, and B. Hertel, unpublished data) that leads to the rapid depolarization of the host plasma membrane (Frohns et al., 2006). Potassium ions are released rapidly from the infected cells (Neupärtl et al., 2008), and this lowers the turgor pressure of the cell, which is hypothesized to aid the transfer of DNA from the virus particle into the host. Virus infection is inhibited by the same compounds that block channel function in heterologous systems (Mehmel et al., 2003; Frohns et al., 2006). Other experiments indicate that depolarization of the host plasma membrane by the Kcv channel also limits infection of the host to a single virus (Greiner et al., 2009). In competition experiments, one chlorovirus species outcompetes another one, and this outcome is correlated with the speed with which the virus depolarizes the host plasma membrane.
CONCLUSION
Comparative analysis of diverse K+ channels suggests that eukarya and prokarya obtained their K+ channels from a common precursor, which provided the common pore-forming segment (Anderson and Greenberg, 2001). The small size of the Kcv viral channels, which is basically the pore-forming segment, their functional simplicity, their self-assembly into tetramers, and their robustness (Thiel et al., 2011) suggest that they are primitive and therefore could predate more complex K+ channel proteins. Also, two examples are provided in this review of how complex K+ channels can be formed by a single genetic event involving the Kcv channel. Currently, phylogenetic algorithms are unable to identify the origin of modern K+ channels. One potential explanation could be that all K+ channels, including those from viruses, evolved from a common but now extinct form of life. If this occurred, we have to assume that the evolution of the viral channels did not progress further. An alternative explanation is that K+ channels evolved from viruses. This view is consistent with the fact that some large DNA viruses might predate the separation of the three main branches of cellular life, namely bacteria, archea, and eukarya (Boyer et al., 2010; Legendre et al., 2012; Nasir et al., 2012). In this context, it is interesting to note that the aforementioned bioinformatics analysis of multiple chloroviruses (Jeanniard et al., 2013) provides evidence that some genes are transferred from the virus to the host. Also, some large DNA viruses such as the algal virus EsV-1 are lysogenic and incorporate their genomes into the host genome (Müller et al., 1998). Such a process could link virus-driven gene emergence to host survival and evolution.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Unrooted neighbor joining tree for K+ channels pore modules.
Supplementary Material
Glossary
- Kcv
K+ chlorella virus
- ML
maximum-likelihood
- NJ
neighbor-joining
- Kv
voltage-gated potassium
- dsDNA
double-stranded DNA
- VS
voltage sensor
- VSs
voltage sensors
- PAS
definition to come
References
- Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. (2007) Portability of paddle motif function and pharmacology in voltage sensors. Nature 450: 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PAV, Greenberg RM. (2001) Phylogeny of ion channels: clues to structure and function. Comp Biochem Physiol B Biochem Mol Biol 129: 17–28 [DOI] [PubMed] [Google Scholar]
- Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. (2006) Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res 34: W604–W608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni C, Schroeder I, Romani G, Van Etten JL, Thiel G, Moroni A. (2013) The voltage-sensing domain of a phosphatase gates the pore of a potassium channel. J Gen Physiol 141: 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balss J, Papatheodorou P, Mehmel M, Baumeister D, Hertel B, Delaroque N, Chatelain FC, Minor DL, Jr, Van Etten JL, Rassow J, et al. (2008) Transmembrane domain length of viral K+ channels is a signal for mitochondria targeting. Proc Natl Acad Sci USA 105: 12313–12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, et al. (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22: 2943–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonza MC, Martin H, Kang M, Lewis G, Greiner T, Giacometti S, Van Etten JL, De Michelis MI, Thiel G, Moroni A. (2010) A functional calcium-transporting ATPase encoded by chlorella viruses. J Gen Virol 91: 2620–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Madoui MA, Gimenez G, La Scola B, Raoult D. (2010) Phylogenetic and phyletic studies of informational genes in genomes highlight existence of a 4 domain of life including giant viruses. PLoS ONE 5: e15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CP. (2004) Viral control of phytoplankton populations—a review. J Eukaryot Microbiol 51: 125–138 [DOI] [PubMed] [Google Scholar]
- Caprini M, Ferroni S, Planells-Cases R, Rueda J, Rapisarda C, Ferrer-Montiel A, Montal M. (2001) Structural compatibility between the putative voltage sensor of voltage-gated K+ channels and the prokaryotic KcsA channel. J Biol Chem 276: 21070–21076 [DOI] [PubMed] [Google Scholar]
- Carrasco L. (1995) Modification of membrane permeability by animal viruses. Adv Virus Res 45: 61–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelain FC, Gazzarrini S, Fujiwara Y, Arrigoni C, Domigan C, Ferrara G, Pantoja C, Thiel G, Moroni A, Minor DL., Jr (2009) Selection of inhibitor-resistant viral potassium channels identifies a selectivity filter site that affects barium and amantadine block. PLoS ONE 4: e7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, et al. (2001) A novel influenza A virus mitochondrial protein that induces cell death. Nat Med 7: 1306–1312 [DOI] [PubMed] [Google Scholar]
- Cherrier MV, Kostyuchenko VA, Xiao C, Bowman VD, Battisti AJ, Yan X, Chipman PR, Baker TS, Van Etten JL, Rossmann MG. (2009) An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc Natl Acad Sci USA 106: 11085–11089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie GE, Dokland T. (2012) Pirates of the Caudovirales. Virology 434: 210–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouzé P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, et al. (2010) The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621 [DOI] [PubMed] [Google Scholar]
- Delaroque N, Müller DG, Bothe G, Pohl T, Knippers R, Boland W. (2001) The complete DNA sequence of the Ectocarpus siliculosus Virus EsV-1 genome. Virology 287: 112–132 [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Escande ML, Eychenié S, Cooke R, Piganeau G, Desdevises Y, Bellec L, Moreau H, Grimsley N. (2008) Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS ONE 3: e2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derst C, Karschin A. (1998) Evolutionary link between prokaryotic and eukaryotic K+ channels. J Exp Biol 201: 2791–2799 [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280: 69–77 [DOI] [PubMed] [Google Scholar]
- Ehrhardt T, Zimmermann S, Müller-Röber B. (1997) Association of plant K+(in) channels is mediated by conserved C-termini and does not affect subunit assembly. FEBS Lett 409: 166–170 [DOI] [PubMed] [Google Scholar]
- Filée J. (2009) Lateral gene transfer, lineage-specific gene expansion and the evolution of nucleo cytoplasmic large DNA viruses. J Invertebr Pathol 101: 169–171 [DOI] [PubMed] [Google Scholar]
- Filée J, Pouget N, Chandler M. (2008) Phylogenetic evidence for extensive lateral acquisition of cellular genes by nucleocytoplasmic large DNA viruses. BMC Evol Biol 8: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filée J, Siguier P, Chandler M. (2007) I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet 23: 10–15 [DOI] [PubMed] [Google Scholar]
- Fischer WB, Sansom MS. (2002) Viral ion channels: structure and function. Biochim Biophys Acta 1561: 27–45 [DOI] [PubMed] [Google Scholar]
- Forterre P. (2001) Genomic and early cellular evolution. The origin of the DNA world. C R Acad Sci III 24: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Frohns F, Käsmann A, Kramer D, Schäfer B, Mehmel M, Kang M, Van Etten JL, Gazzarrini S, Moroni A, Thiel G. (2006) Potassium ion channels of Chlorella viruses cause rapid depolarization of host cells during infection. J Virol 80: 2437–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H. (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94: 647–655 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Kang M, Abenavoli A, Romani G, Olivari C, Gaslini D, Ferrara G, van Etten JL, Kreim M, Kast SM, et al. (2009) Chlorella virus ATCV-1 encodes a functional potassium channel of 82 amino acids. Biochem J 420: 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, Kang M, Epimashko S, Van Etten JL, Dainty J, Thiel G, Moroni A. (2006) Chlorella virus MT325 encodes water and potassium channels that interact synergistically. Proc Natl Acad Sci USA 103: 5355–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Carrasco L. (2003) Viroporins. FEBS Lett 552: 28–34 [DOI] [PubMed] [Google Scholar]
- Greiner T, Frohns F, Kang M, Van Etten JL, Käsmann A, Moroni A, Hertel B, Thiel G. (2009) Chlorella viruses prevent multiple infections by depolarizing the host membrane. J Gen Virol 90: 2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner T, Ramos J, Alvarez MC, Gurnon JR, Kang M, Van Etten JL, Moroni A, Thiel G. (2011) Functional HAK/KUP/KT-like potassium transporter encoded by chlorella viruses. Plant J 68: 977–986 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Hamacher K, Greiner T, Ogata H, Van Etten JL, Gebhardt M, Villarreal LP, Cosentino C, Moroni A, Thiel G. (2012) Phycodnavirus potassium ion channel proteins question the virus molecular piracy hypothesis. PLoS ONE 7: e38826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel M, Mitzner D, Henklein P, Meyer-Almes F-J, Moroni A, Difrancesco ML, Henkes LM, Kreim M, Kast SM, Schubert U, et al. (2010) The proapoptotic influenza A virus protein PB1-F2 forms a nonselective ion channel. PLoS ONE 5: e11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel B, Tayefeh S, Kloss T, Hewing J, Gebhardt M, Baumeister D, Moroni A, Thiel G, Kast SM. (2010) Salt bridges in the miniature viral channel Kcv are important for function. Eur Biophys J 39: 1057–1068 [DOI] [PubMed] [Google Scholar]
- Hille B (2001) Ion Channels of Excitable Membranes. Sinauer, Sunderland, MA [Google Scholar]
- Hsu K, Seharaseyon J, Dong P, Bour S, Marbán E. (2004) Mutual functional destruction of HIV-1 Vpu and host TASK-1 channel. Mol Cell 14: 259–267 [DOI] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. (2006) Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res 117: 156–184 [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. (1992) Tracing the roots of ion channels. Cell 69: 715–718 [DOI] [PubMed] [Google Scholar]
- Jeanniard A, Dunigan DD, Gurnon JR, Agarkova IV, Kang M, Vitek J, Duncan G, McClung OW, Larsen M, Claverie JM, et al. (2013) Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics 14: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum KA, Joiner WJ, Sellers AJ, Kaczmarek LK, Goldstein SAN. (1995) A new family of outwardly rectifying potassium channel proteins with two pore domains in tandem. Nature 376: 690–695 [DOI] [PubMed] [Google Scholar]
- Legendre M, Arslan D, Abergel C, Claverie JM. (2012) Genomics of megavirus and the elusive fourth domain of life. Commun Integr Biol 5: 102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Klem AM, Ramu Y. (2001) Ion conduction pore is conserved among potassium channels. Nature 413: 809–813 [DOI] [PubMed] [Google Scholar]
- Martinac B, Saimi Y, Kung C. (2008) Ion channels in microbes. Physiol Rev 88: 1449–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehmel M, Rothermel M, Meckel T, Van Etten JL, Moroni A, Thiel G. (2003) Possible function for virus encoded K+ channel Kcv in the replication of chlorella virus PBCV-1. FEBS Lett 552: 7–11 [DOI] [PubMed] [Google Scholar]
- Meints RH, Lee K, Burbank DE, Van Etten JL. (1984) Infection of a Chlorella-like alga with the virus, PBCV-1: ultrastructural studies. Virology 138: 341–346 [DOI] [PubMed] [Google Scholar]
- Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. (1998) Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell 95: 649–655 [DOI] [PubMed] [Google Scholar]
- Moreau H, Piganeau G, Desdevises Y, Cooke R, Derelle E, Grimsley N. (2010) Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J Virol 84: 12555–12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, López-García P. (2009) Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol 7: 306–311 [DOI] [PubMed] [Google Scholar]
- Moroni A, Viscomi C, Sangiorgio V, Pagliuca C, Meckel T, Horvath F, Gazzarrini S, Valbuzzi P, Van Etten JL, DiFrancesco D, et al. (2002) The short N-terminus is required for functional expression of the virus-encoded miniature K+ channel Kcv. FEBS Lett 530: 65–69 [DOI] [PubMed] [Google Scholar]
- Müller DG, Kapp M, Knippers R (1998) Viruses in marine brown algae. In K Maramorosch, FA, Murphy, AJ, Shatkin, eds, Advances in Virus Research, Vol 50. Academic Press, New York, pp 49–67 [DOI] [PubMed] [Google Scholar]
- Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. (2005) Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Murphy PM. (1994) Molecular piracy of chemokine receptors by herpesviruses. Infect Agents Dis 3: 137–154 [PubMed] [Google Scholar]
- Nasir A, Kim KM, Caetano-Anolles G. (2012) Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol Biol 12: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupärtl M, Meyer C, Woll I, Frohns F, Kang M, Van Etten JL, Kramer D, Hertel B, Moroni A, Thiel G. (2008) Chlorella viruses evoke a rapid release of K+ from host cells during the early phase of infection. Virology 372: 340–348 [DOI] [PubMed] [Google Scholar]
- Nieva JL, Madan V, Carrasco L. (2012) Viroporins: structure and biological functions. Nat Rev Microbiol 10: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca C, Goetze TA, Wagner R, Thiel G, Moroni A, Parcej D. (2007) Molecular properties of Kcv, a virus encoded K+ channel. Biochemistry 46: 1079–1090 [DOI] [PubMed] [Google Scholar]
- Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. (1997) A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA 94: 11301–11306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ, Lamb RA. (1992) Influenza virus M2 protein has ion channel activity. Cell 69: 517–528 [DOI] [PubMed] [Google Scholar]
- Plugge BS, Gazzarrini S, Nelson M, Cerana R, Van Etten JL, Derst C, DiFrancesco D, Moroni A, Thiel G. (2000) A potassium channel protein encoded by chlorella virus PBCV-1. Science 287: 1641–1644 [DOI] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. (2004) The 1.2-megabase genome sequence of Mimivirus. Science 306: 1344–1350 [DOI] [PubMed] [Google Scholar]
- Santos JS, Grigoriev SM, Montal M. (2008) Molecular template for a voltage sensor in a novel K+ channel. III. Functional reconstitution of a sensorless pore module from a prokaryotic Kv channel. J Gen Physiol 132: 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Fischer WB. (2012) Sequence alignment of viral channel proteins with cellular ion channels. J Comput Biol 19: 1060–1072 [DOI] [PubMed] [Google Scholar]
- Sinkovics J, Horvath J, Horak A. (1998) The origin and evolution of viruses (a review). Acta Microbiol Immunol Hung 45: 349–390 [PubMed] [Google Scholar]
- Suttle CA. (2005) Viruses in the sea. Nature 437: 356–361 [DOI] [PubMed] [Google Scholar]
- Suttle CA. (2007) Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5: 801–812 [DOI] [PubMed] [Google Scholar]
- Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. (2002) Influenza a virus M2 ion channel activity is essential for efficient replication in tissue culture. J Virol 76: 1391–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayefeh S, Kloss T, Kreim M, Gebhardt M, Baumeister D, Hertel B, Richter C, Schwalbe H, Moroni A, Thiel G, et al. (2009) Model development for the viral Kcv potassium channel. Biophys J 96: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, Baumeister D, Schroeder I, Kast SM, Van Etten JL, Moroni A. (2011) Minimal art: or why small viral K+ channels are good tools for understanding basic structure and function relations. Biochim Biophys Acta 1808: 580–588 [DOI] [PubMed] [Google Scholar]
- Thiel G, Moroni A, Dunigan DD, Van Etten JL (2010) Initial events associated with virus PBCV-1 infection of Chlorella NC64A. In U Lüttge, W Beyschlag, B Büdel, eds, Progress in Botany, Vol 71. Springer-Verlag, Berlin, pp 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpe LC, Schwarz TL, Tempel BL, Papazian DM, Jan YN, Jan LY. (1988) Expression of functional potassium channels from Shaker cDNA in Xenopus oocytes. Nature 331: 143–145 [DOI] [PubMed] [Google Scholar]
- Van Etten JL, Dunigan DD. (2012) Chloroviruses: not your everyday plant virus. Trends Plant Sci 17: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal LP (2005) Viruses and the Evolution of Life. American Society for Microbiology Press, Washington, DC [Google Scholar]
- Wang K, Xie S, Sun B. (2011) Viral proteins function as ion channels. Biochim Biophys Acta 1808: 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei A, Jegla T, Salkoff L. (1996) Eight potassium channel families revealed by the C. elegans genome project. Neuropharmacology 35: 805–829 [DOI] [PubMed] [Google Scholar]
- Weynberg KD, Allen MJ, Ashelford K, Scanlan DJ, Wilson WH. (2009) From small hosts come big viruses: the complete genome of a second Ostreococcus tauri virus, OtV-1. Environ Microbiol 11: 2821–2839 [DOI] [PubMed] [Google Scholar]
- Wilson WH, Van Etten JL, Allen MJ. (2009) The Phycodnaviridae: the story of how tiny giants rule the world. Curr Top Microbiol Immunol 328: 1–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21: 809–818 [DOI] [PubMed] [Google Scholar]
- Zhang X, Xiang Y, Dunigan DD, Klose T, Chipman PR, Van Etten JL, Rossmann MG. (2011) Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc Natl Acad Sci USA 108: 14837–14842 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.