GhTCP14 is a dual-function transcription factor able to positively or negatively regulate expression of auxin response and transporter genes, and it may act as a crucial regulator in auxin-mediated differentiation and elongation of cotton fiber cells.
Abstract
Plant-specific TEOSINTE-BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors play crucial roles in development, but their functional mechanisms remain largely unknown. Here, we characterized the cellular functions of the class I TCP transcription factor GhTCP14 from upland cotton (Gossypium hirsutum). GhTCP14 is expressed predominantly in fiber cells, especially at the initiation and elongation stages of development, and its expression increased in response to exogenous auxin. Induced heterologous overexpression of GhTCP14 in Arabidopsis (Arabidopsis thaliana) enhanced initiation and elongation of trichomes and root hairs. In addition, root gravitropism was severely affected, similar to mutant of the auxin efflux carrier PIN-FORMED2 (PIN2) gene. Examination of auxin distribution in GhTCP14-expressing Arabidopsis by observation of auxin-responsive reporters revealed substantial alterations in auxin distribution in sepal trichomes and root cortical regions. Consistent with these changes, expression of the auxin uptake carrier AUXIN1 (AUX1) was up-regulated and PIN2 expression was down-regulated in the GhTCP14-expressing plants. The association of GhTCP14 with auxin responses was also evidenced by the enhanced expression of auxin response gene IAA3, a gene in the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) family. Electrophoretic mobility shift assays showed that GhTCP14 bound the promoters of PIN2, IAA3, and AUX1, and transactivation assays indicated that GhTCP14 had transcription activation activity. Taken together, these results demonstrate that GhTCP14 is a dual-function transcription factor able to positively or negatively regulate expression of auxin response and transporter genes, thus potentially acting as a crucial regulator in auxin-mediated differentiation and elongation of cotton fiber cells.
TEOSINTE-BRANCHED1/CYCLOIDEA/PCF (TCPs) constitute a large group of plant-specific genes found in various species, ranging from lower to higher plants, with 24 TCPs in Arabidopsis (Arabidopsis thaliana) and more than 20 TCPs in rice (Oryza sativa; Cubas, 2002; Navaud et al., 2007; Yao et al., 2007). TCP proteins have a TCP domain that contains a helix-loop-helix (bHLH) motif (Martín-Trillo and Cubas, 2010). Based on sequence variation in the TCP domains, these proteins can be divided into two subfamilies, class I (TCP-P) and class II (TCP-C) TCPs (Cubas et al., 1999; Kosugi and Ohashi, 2002; Navaud et al., 2007). A number of TCP proteins have been characterized as functional transcription factors, acting either as activators or repressors, and the members of the two TCP subclasses bind to different consensus DNA elements (Cubas et al., 1999; Kosugi and Ohashi, 2002; Trémousaygue et al., 2003).
Accumulating evidence demonstrates that TCP transcription factors play important roles in plant growth and development (Cubas et al., 1999; Li et al., 2005; Hervé et al., 2009). In particular, great progress has been made in functional characterization of the class II TCPs in Arabidopsis. These transcription factors play important roles in leaf, flower, shoot morphogenesis, and hormone biosynthesis (Damerval et al., 2007; Koyama et al., 2007; Schommer et al., 2008; Nag et al., 2009; Guo et al., 2010). The cellular functions of class I TCPs are less well explored, but several previous studies indicated that they are involved in a number of cellular or developmental processes. For example, Arabidopsis AtTCP14 and AtTCP16 participate in regulation of seed germination and pollen development, respectively (Takeda et al., 2006; Tatematsu et al., 2008a), and AtTCP20 and AtTCP15 are involved in the regulation of cell division, expansion, and differentiation (Hervé et al., 2009; Kieffer et al., 2011). In addition, several lines of evidence have demonstrated that the functions of class I TCP proteins are associated with phytohormone signaling, such as auxin, jasmonic acid (JA), and cytokinin (Kosugi and Ohashi, 2002; Danisman et al., 2012; Steiner et al., 2012; Uberti-Manassero et al., 2012). Based on transcriptome analysis, Kosugi and Ohashi (2002) proposed that class I TCP proteins are likely to be involved in up-regulation of auxin-induced genes. This notion is further supported by a recent study of Uberti-Manassero et al. (2012), which showed that AtTCP15 regulates the expression of boundary-specific genes, presumably through alteration of auxin homeostasis. Although these studies represent important achievements in understanding the roles of this family of proteins, elucidation of the functions of class I TCP proteins is still limited (Li et al., 2012).
Dynamic changes in auxin distributions in plant cells or tissues trigger various differentiation and developmental processes (Benjamins and Scheres, 2008; Vanneste and Friml, 2009). Polar auxin transport, which is regulated by auxin efflux carriers such as PIN-FORMED2 (PIN2) and PIN-FORMED3 (PIN3) and auxin influx carriers such as AUXIN1 (AUX1), is essential for dynamic auxin distribution (Leyser, 2005). The functions of the auxin transporters are well characterized in root gravity response of Arabidopsis plant (Sabatini et al., 1999; Sun et al., 2011). Coordinated activities of PINs and AUX1 proteins led to asymmetric auxin distribution in root apex and resulted in alteration of the growth orientation (Bennett et al., 1996; Sabatini et al., 1999; Swarup and Péret, 2012). Auxin transporters also act as key modulators in root hair formation (Ganguly et al., 2010). It has been shown that AUX1 promoted root hair initiation by stimulating auxin influx to specific sites (Rahman et al., 2002; Jones et al., 2009), and overexpression of AUX1 or PIN2 was found to promote or block the root hair elongation (Cho et al., 2007; Ganguly et al., 2010). In addition to regulating the auxin transporter genes, auxin also induces the expression of genes in the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) family, including auxin response gene IAA3, which plays important roles in auxin-mediated regulation of root growth, gravitropism, and lateral root formation (Tian and Reed, 1999; Tian et al., 2002; Knox et al., 2003).
The cotton (Gossypium hirsutum) fiber is a highly elongated seed hair and is considered to be a unique system for studying the mechanisms of plant cell expansion (Kim and Triplett, 2001; Qin and Zhu, 2011). Like Arabidopsis trichomes, cotton fibers are derived from single epidermal cells (Basra and Malik, 1984). Regulation of cotton fiber development are attributed to multiple phytohormone signaling processes, of which the auxin signaling was shown to be important for fiber initiation and elongation (Samuel Yang et al., 2006; Shi et al., 2006; Gou et al., 2007). Several transcriptome analyses revealed the enriched transcripts of auxin response factors during fiber initiation and fast-elongation stages (Gou et al., 2007; Liu et al., 2012), and external application of IAA promoted fiber initiation (Gialvalis and Seagull, 2001). Recently, Zhang et al. (2011) showed that overexpression of the key IAA biosynthesis gene iaaM led to enhanced initiation of fiber cells on the ovule epidermal layer and increased fiber length. All these results pointed out the important roles of auxin signaling in cotton fiber development. However, the upstream factors that control the distribution of auxin signaling in cotton fibers remain unknown.
In this study, we isolated a fiber-specific TCP gene (designated GhTCP14) from upland cotton. GhTCP14 is expressed primarily in developing fiber cells during the initiation and elongation stages. As Arabidopsis has been employed successfully as a model system for functional characterization of several cotton fiber-specific genes (Wang et al., 2004; Guan et al., 2008, 2011; Shangguan et al., 2008), we therefore investigated the effect of GhTCP14 on trichome development in Arabidopsis. We show that ectopic expression of GhTCP14 in Arabidopsis promoted the differentiation and elongation of trichome and root hair cells; moreover, overexpression of GhTCP14 resulted in alteration of auxin distribution and altered expression levels of auxin-related genes such as AUX1, PIN2, and IAA3. We found that GhTCP14 bound directly to the promoters of these genes in vitro and in vivo. We also found misregulation of these genes in elongation-deficient fibers of the Ligon lintless1 (Li1) cotton mutant, in which GhTCP14 was down-regulated. Our results provide important insights on the function of class I TCP transcription factors in the development of epidermal cells such as Arabidopsis trichomes and cotton fibers.
RESULTS
Identification and Sequence Analysis of GhTCP14
A cotton fiber-specific complementary DNA (cDNA) library was constructed by suppression subtractive hybridization using cDNA prepared from 9-DPA fibers (“tester”) and cotton leaves (“driver”; Gao et al., 2007). A cDNA clone that encodes a putative TCP-domain protein was found highly expressed in cotton fiber. The full-length cDNA of this clone was obtained by 5′ and 3′ RACE PCR. It encodes a predicted 395 amino acid protein that contains a bHLH-type DNA-binding domain at its N-terminal region. Phylogenetic analysis of the putative protein with TCP proteins from Arabidopsis, rice, snapdragon (Antirrhinum majus), sea-island cotton (Gossypium barbadense), and corn (Zea mays) showed that it belongs to the class I TCP protein group and has the greatest similarity to AtTCP14 (Supplemental Fig. S1); thus, this gene was designated as GhTCP14 (accession no. AF165924). Domain analysis showed that the bHLH domain of GhTCP14 is highly similar to those of other TCP proteins (Supplemental Fig. S1). Motif analysis revealed a nuclear localization signal at residues 237 to 240 (RKKR) of GhTCP14 protein.
GhTCP14 Protein Localizes to the Nucleus
To examine the subcellular localization of GhTCP14, a GFP reporter gene was fused in frame to the GhTCP14 coding region under the control of the 35S promoter to produce a GhTCP14-GFP fusion protein in transgenic Arabidopsis plants. As seen in Supplemental Figure S2, GhTCP14-GFP fusion proteins were detected predominantly in the nuclei, consistent with sequence-based prediction of the subcellular localization and putative transcription factor activity of GhTCP14.
GhTCP14 Possesses DNA-Binding and Transactivation Activities
The amino acid sequence and subcellular distribution of GhTCP14 pointed to a potential function as a TCP transcription factor. To identify sequences preferentially bound by GhTCP14, we conducted a random binding site selection analysis using recombinant GhTCP14. Binding assays indicated that GhTCP14 can specifically bind to two sequences, TGGGTCCCACAT and TTGTGGGCCCCT (Supplemental Fig. S3). This exhibited a perfect match with the consensus TCP DNA-binding specificity of class I (GGNCCCAC) and class II (G[T/C]GGNCCC), indicating that GhTCP14 could bind to two types of TCP binding sites.
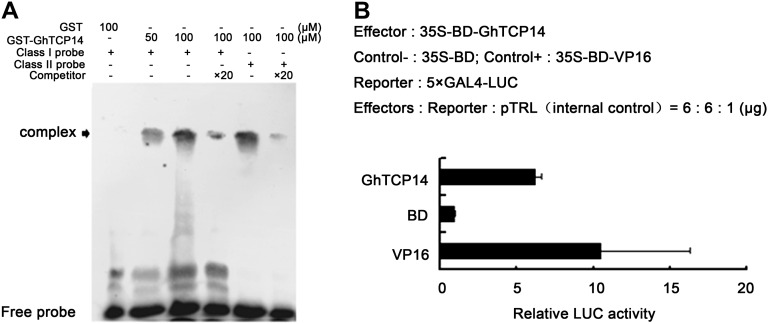
Electrophoretic mobility shift assay (EMSA) was conducted to verify the binding of these DNA sequences. Bacterially expressed glutathione S-transferase (GST)-GhTCP14 proteins were purified. Double repeats of sequences TGGGTCCCACAT and TTGTGGGCCCCT were annealed, labeled, and incubated with purified GST-GhTCP14 fusion proteins. GhTCP14 formed a complex with the labeled probes, and the signal was dramatically decreased by addition of unlabeled DNA probe (competitor; Fig. 1A), indicating that GhTCP14 could bind specifically to the two sequences in vitro.
Figure 1.
DNA-binding specificity and transcriptional activation activity of GhTCP14. A, Gel-shift assay. GhTCP14 proteins were incubated with biotin-labeled probes (class I probe: 2×TGGGTCCCACAT and class II probe: 2×TTGTGGGCCCCT) in the presence or absence of 20-fold excess of nonlabeled competitor. Arrow indicates the position of protein-DNA complexes. B, Transcriptional activation ability of GhTCP14 in Arabidopsis protoplasts. The GAL4 DNA-binding domain (BD) and BD-VP16 were used as negative or positive control, respectively. Error bars represent the sd of three biological replicates.
The ability of GhTCP14 to activate transcription was examined using a dual-luciferase reporter assay system in Arabidopsis protoplasts (Ohta et al., 2001). The coding sequence of GhTCP14 was fused to the DNA sequence encoding yeast transcriptional activator GAL4 DNA-binding domain to generate the effector plasmid pBD-GhTCP14. GhTCP14 activated the reporter (Fig. 1B), indicating that GhTCP14 has transcription activation activity in vivo.
Phenotypes of Transgenic Arabidopsis Plants Overexpressing GhTCP14
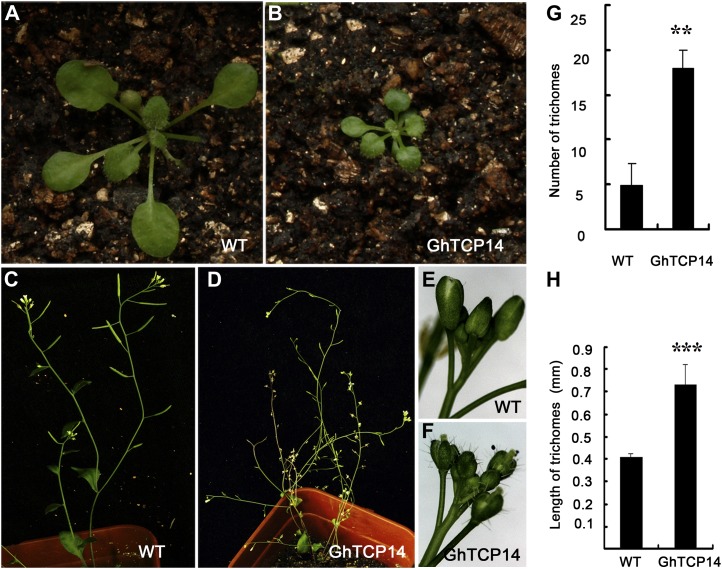
To examine GhTCP14 function, we generated transgenic Arabidopsis plants to bypass the difficulties and delays of cotton transformation. The GhTCP14 cDNA was subcloned into the pPZP111 expression vector under the control of the Cauliflower mosaic virus 35S promoter, and more than 20 Arabidopsis transgenic lines were obtained. Severe phenotypic alterations were observed with the transgenic plants. For example, nine lines showed strong inhibition of root development; roots were barely developed, and as a result, most of these plants did not survive. The remaining lines had pleiotropic phenotypes. Some plants had highly elongated trichomes and denser trichomes on sepals (Fig. 2, E–H) but displayed dwarf growth (Fig. 2, A and B). In addition, these plants showed delayed flowering, and a large portion of the siliques were aborted with few seeds set (Fig. 2, C and D).
Figure 2.
Phenotypes of transgenic Arabidopsis plants overexpressing GhTCP14 under 35S promoter. A and B, Three-week-old wild-type (A) and transgenic (B) seedlings. C and D, Six-week-old wild-type (C) and transgenic (D) plants. E and F, Trichomes on sepals of wild-type (E) and transgenic (F) plants. G and H, Quantitative analyses of the number (G) and length (H) of sepal trichomes. Error bars represent the sd of three biological replicates (n = 30 each). Asterisks denote values statistically different from the corresponding wild-type controls (** P < 0.01, *** P < 0.001). WT, Wild type. [See online article for color version of this figure.]
Induced Expression of GhTCP14 Increases the Density and Length of Root Hairs and Trichomes and Affects Gravitropism
Because constitutive overexpression of GhTCP14 severely affected the survival and fertility of the transgenic plants, it was difficult to assess the effect of GhTCP14 overexpression on the plants. As constitutive expression of GhTCP14 under the 35S promoter is lethal to the transgenic plants, we therefore employed an induced expression strategy to clarify the effect of GhTCP14. To this end, GhTCP14 was placed under the control of the estrogen-inducible promoter pER8 (Zuo et al., 2000; Wang et al., 2009), and the construct was transformed into Arabidopsis plants. To investigate the effect of GhTCP14 expression on plant growth and development, T3 homozygote seeds of three lines were germinated on medium with dimethyl sulfoxide (DMSO) as the solvent-only negative control, or with 10 µm β-estradiol, to induce transgene expression. No phenotype was observed in transgenic plants grown on medium containing DMSO. In the presence of β-estradiol, however, all three lines had similar, strong developmental abnormalities. When seeds were germinated and grown on induction medium, the plants were dwarf and lost the gravitropism response (Fig. 3, A and B). The cotyledons were unable to expand and appeared light green (Fig. 3, A and B). Induction was also carried out on 4-d-old plants in which both root and shoot are differentiated. Seeds were grown on Murashige and Skoog (MS) medium for 4 d, and the seedlings were then transferred to media in the presence or absence of β-estradiol and grown for 7 d. A striking phenotypic change was observed with the roots of the transgenic plants. The root appeared as callus-like tissue and turned green when expression of the transgene was induced (Fig. 3, C and D). Also, induced expression of GhTCP14 significantly increased the density and length of root hairs (Fig. 3, E and F). To find out whether GhTCP14 can also affect trichomes, we used 10 µm β-estradiol to spray the wild-type and transgenic plants once per day. Following this treatment, the density and length of trichomes on both stem and sepal were increased in transgenic plants compared with wild-type plants (Fig. 3, G–K)
Figure 3.
Phenotypes of pER8:GhTCP14 transgenic Arabidopsis plants. A and B, Seeds directly sown on noninducing (A) or inducing (B) medium with 10 µm estradiol and grown for 5 d. C and D, Four-day-old seedlings transferred to noninducing (C) or inducing (D) medium and grown for 7 d. E and F, Root hairs of plants grown under noninducing (E) or inducing (F) conditions. G to J, Trichomes on stems (G and H) and sepals (I and J) of plants grown under noninducing (G and I) or inducing (H and J) conditions. K, Quantitative analysis of trichome length. Error bars represent the sd of three biological repeats (n = 30 each). Asterisks denote values statistically different from the corresponding wild-type controls (** P < 0.01, *** P < 0.001). NI, Noninduced; I, induced. Bars = 1 mm. [See online article for color version of this figure.]
Induced Expression of GhTCP14 Alters Auxin Levels and Distribution in Arabidopsis Roots and Sepal Trichomes
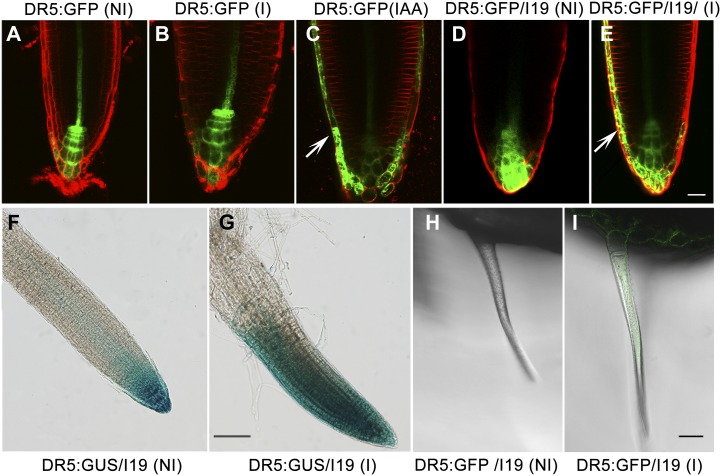
Some phenotypes of the GhTCP14-expressing transgenic plants (loss of gravitropism, increased root hair density and length) resembled those of plants in which auxin content or distribution was abnormal. This prompted us to investigate if auxin distribution was affected in the GhTCP14-expressing plants. To monitor auxin distribution patterns in the transgenic plants, we introduced the auxin-responsive reporter constructs DR5:GFP and DR5:GUS into the pER8:GhTCP14 plants. DR5 is a synthetic auxin-responsive promoter with increased auxin responsiveness (Ulmasov et al., 1997). We observed that the activity of the DR5 promoter was obvious in the roots. In the GhTCP14-induced root tips, the expression of DR5:GFP (Fig. 4E) and DR5:GUS (Fig. 4G) exhibited dramatic increase compared with that of noninduced plants (Fig. 4, D and F) and plants without GhTCP14 (Fig. 4, A and B), indicating that auxin levels were substantially increased in lateral root cap cells. In addition, the distribution of signals in the lateral root cap cells was asymmetric. This staining pattern appeared similar to that of wild-type plants treated with exogenous auxin (IAA; Fig. 4C). This may cause the loss of gravitropism observed in the GhTCP14-expressing plants (Fig. 3B). Interestingly, GFP could also be visualized in the sepal trichomes and epidermal cells of the DR5:GFP plants when GhTCP14 expression was induced (Fig. 4I). By contrast, the corresponding cells in the noninduced plants showed no visible fluorescence (Fig. 4H).
Figure 4.
Effects of GhTCP14 on the expression of auxin-responsive reporter genes and auxin distribution. A to C, Root tips of DR5:GFP plants grown on noninducing medium (A), inducing medium (B), or medium containing 1 µm IAA (C). D and E, Root tips of DR5:GFP × pER8:GhTCP14 transgenic plants grown on noninducing (D) or inducing (E) medium. F and G, Root tips of DR5:GUS × pER8:GhTCP14 plants grown on noninducing (F) or inducing (G) medium. H and I, Sepal trichomes of DR5:GFP × pER8:GhTCP14 plants grown on noninducing (H) or inducing (I) medium. Arrows show fluorescent signals in the lateral root cap cells. NI, Noninduced; I, induced; I19, pER8:GhTCP14. Bars = 25 µm (A–E), 30 µm (F and G), and 100 µm (H and I). [See online article for color version of this figure.]
Induced Expression of GhTCP14 Affects Transcription of Genes Involved in Auxin Responses
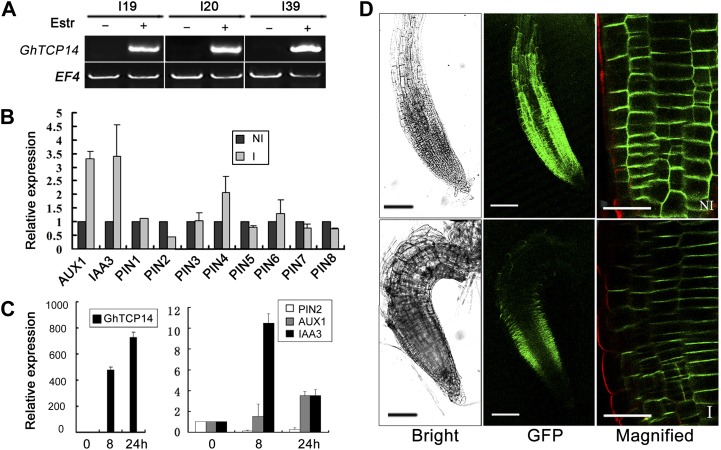
The changed auxin content and distribution in pER8:GhTCP14 transgenic plants indicated that overexpression of the GhTCP14 transcription factor may change the expression of auxin-related genes. Thus, we next used qualitative reverse transcription (RT)-PCR to measure the expression of genes involved in auxin homeostasis, including auxin synthesis, auxin-mediated gene expression, and auxin distribution. In the induced transgenic plants the expression of AUX1, which encodes an auxin uptake carrier, was increased (Fig. 5, B and C). By contrast, expression of PIN2, which encodes an auxin efflux transporter, was down-regulated, but PIN1, PIN3, PIN4, PIN5, PIN6, PIN7, and PIN8 were not significantly changed (Fig. 5, B and C). In addition, expression of IAA3, which encodes an auxin-responsive protein, increased significantly (Fig. 5, B and C). These results indicated that ectopic expression of GhTCP14 (Fig. 5A) alters the expression of both auxin transporter and auxin-responsive genes (Fig. 5, B and C). To verify this effect, we examined GFP expression from pPIN2:PIN2-GFP in pER8:GhTCP14 transgenic plants (Fig. 5D). In the GhTCP14-induced root tip, the brightness of PIN2-GFP signal was dramatically lowered compared with that of noninduced plants (Fig. 5D), indicating the reduced activity of PIN2 promoter.
Figure 5.
Effects of GhTCP14 on the expression of auxin-related genes. A, Induction of GhTCP14 expression in pER8:GhTCP14 transgenic lines (I19, I20, and I39) analyzed by RT-PCR. The elongation factor gene (EF4) was used as an internal control. B, Quantitative RT-PCR analysis of the expression of genes related to auxin transport or auxin response in pER8:GhTCP14 plants grown on noninducing (NI) or inducing (I) medium. C, Time curse expressions of GhTCP14 (left) and some auxin-related genes (right). Line I19 plants were grown in liquid medium and harvested at 8 or 24 h after supplementation with 10 µm estradiol. For B and C, Actin2 was used as an internal control. Error bars represent the sd of three biological replicates, and value of the noninduced plants was set to 1. D, Expression of PIN2-GFP under the PIN2 promoter in root tips of pER8:GhTCP14 plants grown on noninducing (top) or inducing (bottom) medium. Bars = 100 µm (left and middle) and 20 µm (right). [See online article for color version of this figure.]
The GhTCP14 Transcription Factor Binds Directly to a Specific Motif in Target Promoters
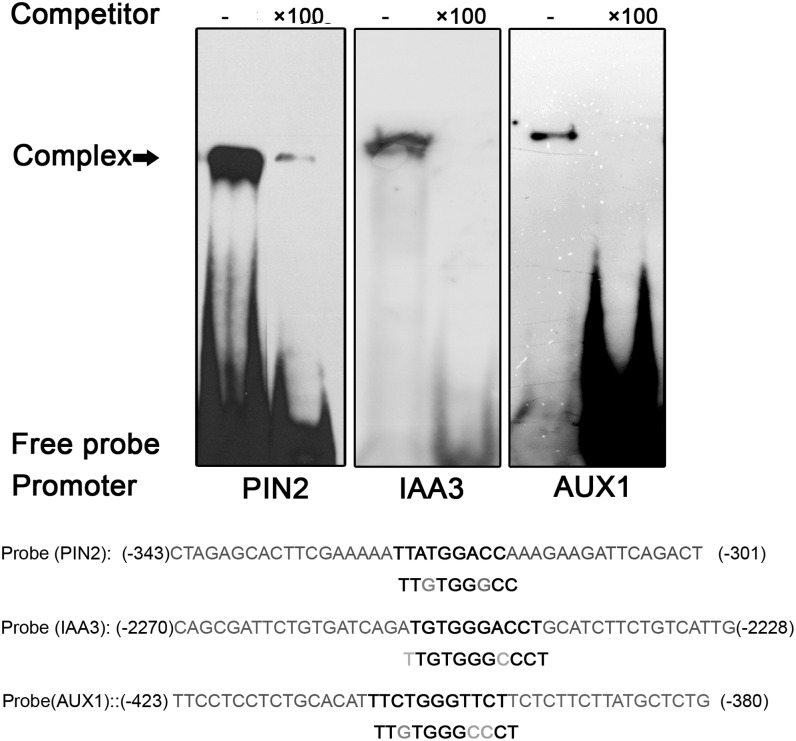
The up- or down-regulated expression of auxin-related genes implied that GhTCP14 may regulate their expression by acting as a transcription factor. The IAA3 promoter contains TCP binding sites (Koyama et al., 2010). Also, sequence analysis indicated that PIN2 and AUX1 promoters contain putative TCP binding elements. To determine whether the transcription of these genes could be affected by GhTCP14 binding directly to their promoters, we conducted EMSA with AUX1, IAA3, and PIN2 promoter sequences and found that GhTCP14 can bind to the promoters of the three genes (Fig. 6).
Figure 6.
Binding of GhTCP14 to AUX1, PIN2, and IAA3 promoter sequences. EMSA analysis was conducted with the recombinant GhTCP14 proteins and DNA sequences containing the promoter region of PIN2, IAA3, and AUX1 genes, respectively. The promoter sequences used in EMSA are shown in the bottom portion of the figure. Numbers indicate positions relative to the translation start site of the corresponding gene. Putative TCP binding sequences are in bold.
Expression of GhTCP14 in Cotton
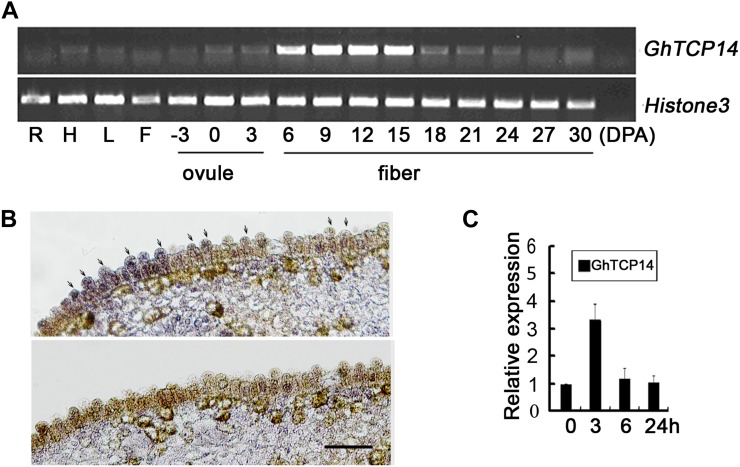
To gain insights into the native functions of GhTCP14 in cotton, we analyzed its expression pattern. RT-PCR analysis showed that GhTCP14 was expressed most strongly in developing fiber cells. GhTCP14 transcript levels were high in the rapid fiber elongation stage (6–15 DPA), and the expression level was rather low in all other tissues examined (Fig. 7A). Because it is technically unfeasible to separate the fiber initials from the ovules at early developmental stages, in situ hybridization was conducted to examine the expression of GhTCP14 in 0-DPA ovules when cotton fibers initiate from the ovule epidermal cells. GhTCP14 transcripts were also abundant in the fiber initials (Fig. 7B). These results showed that GhTCP14 expression is associated with fiber initiation and elongation.
Figure 7.
Expression of GhTCP14 in cotton. A, Transcript profile of GhTCP14 in cotton. R, Root; H, hypcotyl; L, leaf; F, flower. The developmental stages of ovule and fiber are indicated as DPA. The Histone3 gene was used as an internal control. B, In situ hybridization of GhTCP14 transcripts in the ovule (0 DPA) of cotton with antisense (top) and sense (bottom) probes. Arrows show hybridization signals in fiber initials. Bar = 100 µm. C, Expression of GhTCP14 induced by IAA. Quantitative RT-PCR analysis was conducted with the total RNAs extracted from 1-DPA cotton ovules with fiber initials. The value of untreated ovules was set to 1. Error bars represent the sd of three biological repeats. [See online article for color version of this figure.]
The fiber-specific expression feature and the effect of GhTCP14 on auxin signaling observed in the transgenic Arabidopsis plants likely reflects a native role of GhTCP14 in cotton fiber differentiation and elongation through auxin-mediated signaling. To assess this role of GhTCP14, we analyzed the impact of IAA on its expression. As shown in Figure 7C, when 1-DPA cotton ovules were treated with 1 µm IAA for 3, 6, and 24 h, GhTCP14 expression was induced shortly in about 3 h, and its transcription was increased 3-fold, showing that the expression of GhTCP14 was up-regulated by auxin at an early stage of fiber development.
GhAUX1, GhPIN2, and GhIAA3 May Be Direct Targets of GhTCP14 during Cotton Fiber Development
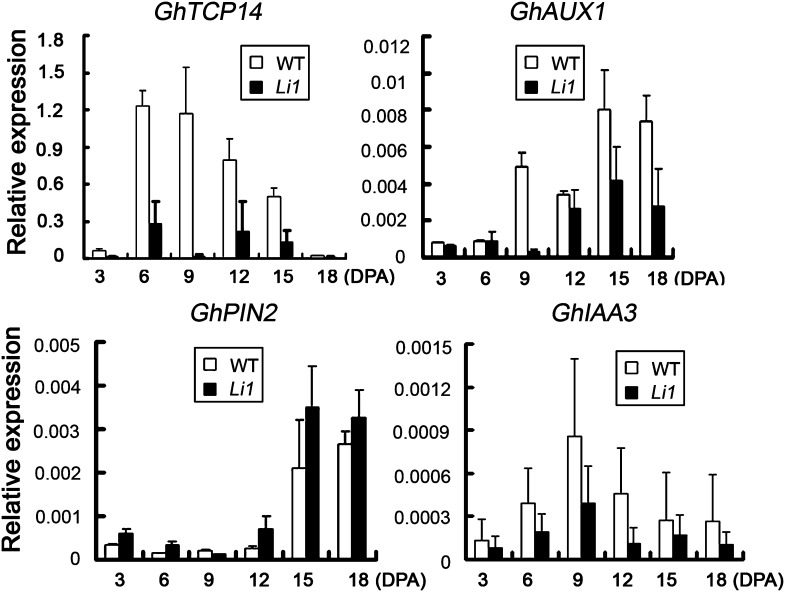
To confirm the association of GhTCP14 expression with fiber elongation, we analyzed its expression in the fibers of Li1 cotton mutant. Li1 was originally discovered by Griffee and Ligon (1929), and subsequent studies demonstrated that it is a dominant monogenic mutant with a defect in fiber elongation and plant growth. GhTCP14 transcript levels were much lower in the short fiber cells compared with the wild-type control during the fast elongation stage of fiber development at 6 to 15 DPA (Fig. 8). Furthermore, we conducted quantitative RT-PCR analysis to see if the expression of the auxin-related genes AUX1, PIN2, and IAA3 was coregulated with down-regulation of GhTCP14 in Li1 fibers. Due to the lack of complete genome sequence of G. hirsutum, Arabidopsis AUX1, PIN2, and IAA3 protein sequences were used to search against the Gossypium spp. database (tblasn), and an AUX1-like gene (GhAUX1, CD486543.1), a PIN2-like gene (GhPIN2, AY148428), and an IAA3-like gene (GhIAA3, HQ452481.1) were subsequently cloned. Then, we examined the expression of these auxin-related genes in Li1 and wild-type cotton fibers. Consistent with our hypothesis, the expression of GhAUX1 and GhIAA3 was reduced along with the down-regulation of GhTCP14 in Li1 fibers. By contrast, the expression of GhPIN2 increased with the repression of GhTCP14 in these fiber cells (Fig. 8). These results suggested that GhTCP14 might regulate cotton fiber development by direct modulation of auxin-related genes, including GhAUX1, GhPIN2, and GhIAA3.
Figure 8.
Expression of GhTCP14, GhAUX1, GhPIN2, and GhIAA3 genes in the wild type and Li1 mutant. Quantitative RT-PCR analysis was conducted with the total RNAs extracted from ovules (3 DPA) and fiber cells (6–18 DPA). The Histone3 gene was used as the internal control. Error bars represent the sd of three biological replicates.
DISCUSSION
A complex group of transcription factors functions in either promoting or inhibiting leaf trichome initiation and development in Arabidopsis. These include the MYB transcription factors, the bHLH transcription factors, WD repeat proteins, and homeodomain proteins (Tominaga-Wada et al., 2011). Likewise, transcription factors participating in cotton fiber development have been reported by several studies. For example, a few MYB transcription factors were found to play important roles in cotton fiber initiation (Cedroni et al., 2003; Suo et al., 2003; Wang et al., 2004; Pu et al., 2008; Machado et al., 2009; Guan et al., 2011; Walford et al., 2011). In addition, it was recently reported that GbTCP (from sea-island cotton), which shares 47% protein sequence identity with GhTCP14, is involved in cotton fiber development, possibly through JA signaling (Hao et al., 2012). Here, our data indicated that GhTCP14 may be another important TCP transcription factor acting in the regulation of epidermal cell differentiation and development, and more importantly, we found that it may function through auxin pathways.
GhTCP14 Is a Dual-Functional Transcription Factor
It has been suggested that TCP proteins may function either as transcriptional activators or repressors, depending on their interactions with other proteins (Hervé et al., 2009). As the GhTCP14-overexpressing plants phenocopied auxin distribution mutants such as pin2, we analyzed the expression of PIN2 and other auxin-related genes in the transgenic plants and found that the expression of several auxin-related genes was either increased or decreased. Sequence analyses indicated that the promoters of these genes contain putative TCP-binding element to which GhTCP14 was able to bind, and dual-luciferase reporter assays showed that GhTCP14 possesses transcription factor activity. These results demonstrated that GhTCP14 acts as a dual functional transcription factor in the regulation of the expression of certain auxin-related genes. Although transcriptional activator or repressor activity has been reported for TCP proteins, to our best knowledge, this is the first report of the dual function of a TCP protein. The distinctive activities of GhTCP14 may depend on its interaction with other proteins (Hervé et al., 2009).
Recently, Koyama et al. (2010) reported that expression of PIN1, PIN5, and PIN6 and PIN3, PIN4, and PIN7 was either up- or down-regulated, respectively, by overexpression of the class II family member TCP3 chimeric repressor gene (TCP3SRDX), but PIN2 expression was not affected in the transgenic plants. In our study, we found that PIN2 expression was down-regulated, while expression levels of other PIN genes were not changed in GhTCP14-expressing transgenic plants. Combining these data, it is apparent that TCP transcription factors regulate the expression of PIN genes. In addition, our results suggested that the class I and II subclasses may have divergent functions in transcriptional regulation of the subgroups of PINs.
Overexpression of GhTCP14 Altered Auxin Patterning
Several TCPs have been shown to function in leaf and flower development or embryogenesis via auxin-signaling pathways (Koyama et al., 2010; Uberti-Manassero et al., 2012). In our study, overexpression of GhTCP14 altered expression of PIN2, AUX1, and IAA3 and also produced phenotypic changes that resembled those of auxin-related mutants (Müller et al., 1998; Marchant et al., 1999; Ganguly et al., 2010). In particular, the numbers and lengths of the root hairs and trichomes were changed. In the root tip, the auxin distribution was substantially altered, and the lateral epidermal cells accumulated higher levels of auxin than other cells. In the trichomes, auxin accumulated more abundantly at the base of transgenic cells, but the signal was almost undetectable in the corresponding cells of the control plants. These results indicate that overexpression of GhTCP14 affected distribution and homeostasis of auxin, which is required for the differentiation and elongation of the root hair and trichome, both of which differentiate from epidermal cells. Previously, molecular genetic analyses using Arabidopsis mutants demonstrated that the differentiation of both types of cells is regulated by similar molecular mechanisms (Tominaga-Wada et al., 2011). Here, our results provide novel evidence to show that the development of root hairs and trichomes might be controlled by a common auxin-mediated mechanism involving TCP transcription factor(s).
In addition to alterations in epidermal cell development, the association of GhTCP14 function with auxin signaling is further evidenced by the gravitropism defect of the transgenic plants. Under inducing conditions, GhTCP14-overexpressing plants showed a severe defect in root gravitropism, which appeared similar to that of plants with elevated IAA levels, observed by Müller et al. (1998), who showed that down-regulation of the auxin transporter gene PIN2 led to an auxin-hypersensitive phenotype, including a severe gravitropism defect.
Regulation of Auxin-Mediated Trichome Development by TCPs
Compared with the root hairs, the information on auxin-regulated trichome development is limited, although evidence showing the roles of gibberellins, JA, salicylic acid, and cytokinins in trichome initiation and growth is accumulating (Traw and Bergelson, 2003; Steiner et al., 2012). Recently, it was reported that the sepals of Arabidopsis tcp14 tcp15 double mutants were glabrous, but several trichomes grow on each sepal of the wild type and the tcp14 or tcp15 single mutant (Steiner et al., 2012). Here, we showed that ectopic overexpression of GhTCP14 affected the differentiation and elongation of sepal and stem trichomes. Also, these phenotypic changes were coupled with a change in the auxin level at the base of the trichomes, implying that the TCP transcription factor participated in trichome development through auxin patterning. Interestingly, although the development of the sepal and stem trichomes and root hairs were both affected by induced GhTCP14 expression, we did not see an obvious alteration in leaf trichome development in the transgenic plants, suggesting that the leaf trichomes may be under control of distinct mechanism(s). In line with our observation, Steiner et al. (2012) reported that the Arabidopsis tcp14 tcp15 double mutant lacked sepal trichomes, whereas the leaf trichomes developed normally.
It has been established that auxin and cytokinin interact to control many aspects of plant growth and development (Moubayidin et al., 2009; Müller and Leyser, 2011). Previously, Steiner et al. (2012) showed that Arabidopsis TCP14 and TCP15 acted with SPINDLY to promote cell proliferation through cytokinin signaling in leaves and flowers. Here, our results demonstrate that GhTCP14 regulated epidermal cell initiation and elongation through auxin signaling in roots and trichomes. Together, these results indicated that class I TCP transcription factors are involved in both auxin and cytokinin signaling and perhaps play roles in the cross talk between these two hormones.
The Possible Roles of TCP Transcription Factors in Developing Cotton Fibers
Cotton fibers differentiate from the epidermal layer of the ovule. Usually, only 30% of epidermal cells differentiate into fiber cells. Several transcriptome analyses have indicated that auxin signaling is important for fiber cell initiation and elongation (Gou et al., 2007; Liu et al., 2012). Accordingly, elevated expression of the key auxin synthesis gene iaaM was found to promote fiber cell differentiation and elongation, improving fiber yield and quality (Zhang et al., 2011). All these studies pointed out the importance of auxin signaling in fiber development and also raised the question of what mechanism(s) regulate these auxin-related processes. In this work, we found that (1) GhTCP14 is specifically expressed at the fiber initiation and elongation stages; (2) the expression of GhTCP14 was up-regulated by auxin at the early stage of fiber development; (3) ectopic expression of GhTCP14 in Arabidopsis led to alterations in auxin levels and distribution, which affected root and trichome epidermal cell initiation and elongation; and (4) in the Li1 short-fiber mutant, expression of GhTCP14 was down-regulated, accompanied by decreased expression of GhAUX1 and GhIAA3 and increased expression of GhPIN2. Based on these results, we speculate that GhTCP14 may be an upstream regulator of auxin-mediated fiber initiation and elongation, functioning in transcriptional regulation of these auxin-related genes. Future work should further elucidate the functions of GhTCP14 in cotton plants.
MATERIALS AND METHODS
Plant Materials and Treatments
The cotton variety Texas-Maker-1 (TM-1; the genetic standard of Gossypium hirsutum) and Li1 mutant were used in this study. Seeds were sown on agar plates containing MS medium (Murashige and Skoog, 1962) and grown under tissue culture conditions for 2 weeks. Roots, hypocotyls, and leaves were collected from the seedlings. Flowers and fibers were harvested from field-grown cotton plants. Fibers were removed from the ovules at 6 to 30 DPA. The collected materials were immediately frozen in liquid nitrogen and stored at –80°C
Seeds of Arabidopsis (Arabidopsis thaliana ecotype Columbia) were germinated on agar plates containing MS salts, 1% Suc (w/v), and 0.7% (w/v) agar, pH 5.7. After vernalization at 4°C for at least 2 d, seedlings were grown in an illuminated growth chamber at 23°C. The induction of GhTCP14 gene was carried out by two means: (1) direct germination of seeds on medium supplemented with 10 µm β-estradiol or the solvent DMSO (used for mock treatment) and (2) transplant of the 4-d-old seedlings to medium containing β-estradiol or DMSO with continuing growth for 7 d.
Gene Cloning and RNA Analysis
The cDNAs of cotton genes were amplified by RT-PCR from total RNAs of cotton root, hypocotyl, leaf, flower, ovules, and fiber cells according to the ultracentrifugation method (John and Crow, 1992). The cDNAs of Arabidopsis genes were amplified by RT-PCR from total RNA extracted from the seedlings using TRIzol reagent (Invitrogen). In each case, 2 µg of total RNA was used for reverse transcription. Quantitative RT-PCR assays were performed by using SYBR Green Real-Time PCR Master Mix (Toyobo) and the DNA Engine Option 2 Real-Time PCR Detection System (MJ Research). The primers used in RT-PCR and quantitative RT-PCR are listed in Supplemental Table S1.
Sequence Alignment, Domain, and Phylogenetic Analyses
The protein sequences were aligned by DNAMAN 6.0, the conserved domains were predicted at the National Center for Biotechnology Information, and phylogenetic analysis was performed with ClustalX version 1.83 (Thompson et al., 1997) and MEGA4 (Tamura et al., 2007) by the neighbor-joining method. Nuclear localization signal was predicted with WoLF PSORT.
Plasmid Construction and Plant Transformation
Full-length GhTCP14 cDNA was amplified using the BD SMART RACE cDNA Amplification Kit. An EcoR1-Sal1 fragment of GhTCP14 cDNA was cloned into the pGEX-6p-1 vector for production of GST-GhTCP14 proteins. To construct 35S:GhTCP14 and pER8:GhTCP14 plasmids, GhTCP14 open reading frame was amplified and inserted into plant expression vectors pPZP111 or pER8. Both constructs were introduced into Arabidopsis by the Agrobacterium tumefaciens floral dip procedure (Clough and Bent, 1998), and transgenic plants were selected using 50 mg mL–1 kanamycin. Homozygous T3 pER8:GhTCP14 plants were cross fertilized with plants carrying DR5:GFP, DR5:GUS, or PIN2:PIN2-GFP to generate DR5:GFP × pER8:GhTCP14, DR5:GUS × pER8:GhTCP14, and PIN2:PIN2-GFP × pER8:GhTCP14 plants.
Subcellular Localization of GhTCP14-GFP Fusion Proteins
The GhTCP14 open reading frame was fused to the GFP coding sequence and cloned into pPZP111 vector under the control of the Cauliflower mosaic virus 35S promoter. Transgenic Arabidopsis plants were generated by floral dip method and selected using 50 mg mL–1 kanamycin. Leaves of GhTCP14-GFP transgenic plants were stained with 4, 6-diamidino-2-phenylindole (Roche), and the signals were visualized with a confocal laser microscope (Leica TCS SP5).
EMSA
GST-GhTCP14 fusion proteins were expressed in Escherichia coli strain BL21 and then purified using glutathione-Sepharose 4B according to the manufacturer’s instructions (Amersham Pharmacia Biotech). Random binding site selection analysis (Nørby et al., 1992) was conducted using recombinant GhTCP14, and EMSAs were carried out using biotin-labeled probes and the Pierce LightShift Chemiluminescent EMSA kit (Thermo). The binding reaction was carried out in 20 µL reaction mixture containing indicated GST-GhTCP14 proteins and 20 nmol of synthetic biotin-labeled DNA probe in the presence or absence of nonlabeled competitor. The reaction mixtures were incubated at room temperature for 30 min and then applied onto a 6% native polyacrylamide gel in 0.5× Tris-borate/EDTA buffer. The labeled probes were detected according to the instructions provided with the EMSA kit.
Protoplast Assay of Transcription Factor Activity
The dual-luciferase reporter assay (Ohta et al., 2001) was conducted to assess the transcription factor activity of GhTCP14 in Arabidopsis protoplasts. Reporter plasmid and plasmid pPTRL were kindly provided by Masaru Ohme-Takagi. The GAL4 reporter plasmid contains the firefly luciferase gene, driven by the minimal TATA region of the 35S promoter with five GAL4 binding elements upstream. The Renilla luciferase gene driven by the 35S promoter was used as an internal control. For construction of effector plasmid, the coding region of GhTCP14 was cloned into the pRT-BD vector to generate 35S-BD-GhTCP14. The effector, reporter, and internal control plasmids were cotransfected into Arabidopsis protoplasts by PEG transformation. After culturing for 16 h, luciferase assays were performed with the Promega dual-luciferase reporter assay system and the GloMax 20-20 luminometer (Promega).
GUS Assay
GUS activity was analyzed by histochemical staining using a solution containing 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide; 10 mm phosphate buffer, pH 7.0; 0.05 mm potassium ferrocyanide; 0.05 mm potassium ferricyanide; 2 mm EDTA; and 0.1% (v/v) Triton X-100. Arabidopsis seedlings were incubated in staining solution at 37°C for 6 h in the dark, followed by washing with 3:1 (v/v) ethanol:acetic acid solution before observation. Samples were examined under an Olympus BX51 microscope.
In Situ Hybridization
The 3′-untranslated region fragment of GhTCP14 cDNA was PCR amplified (primers used are listed in Supplemental Table S1) and subcloned into the pMD-18T Simple vector (Takara). The plasmid was used for probe preparation according to the manual of DIG RNA Labeling Kit (SP6/T7; Roche). Cotton ovules were collected at 0 DPA and embedded in paraffin. GhTCP14 mRNAs were detected with a digoxigenin-labeled riboprobe using the method described by Marrison and Leech (1994). The hybridized probes were detected using the anti-DIG-alkaline phosphatase conjugate and 5-Bromo-4-chloro-3-indolyl phosphate/Nitro blue tetrazolium as substrates. Sections were visualized under an Olympus BX51 microscope.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF165924.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogenetic analysis of TCP proteins and bHLH domain comparison.
Supplemental Figure S2. Nuclear localization of GhTCP14.
Supplemental Figure S3. Random binding site selection analysis using recombinant GhTCP14.
Supplemental Table S1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Tong-Lin Mao and Guo-Hua Mi (China Agricultural University) for generously providing seeds of DR5:GFP and DR5:GUS plants and Abidur Rahman (Iwate University) for kindly providing pPIN2:PIN2-GFP.
Glossary
- cDNA
complementary DNA
- EMSA
electrophoretic mobility shift assay
- DMSO
dimethyl sulfoxide
- MS
Murashige and Skoog
- RT
reverse transcription
- EMSA
electrophoretic mobility shift assay
References
- Basra AS, Malik CP. (1984) Development of the cotton fibre. Int Rev Cytol 87: 65–113 [Google Scholar]
- Benjamins R, Scheres B. (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA. (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273: 948–950 [DOI] [PubMed] [Google Scholar]
- Cedroni ML, Cronn RC, Adams KL, Wilkins TA, Wendel JF. (2003) Evolution and expression of MYB genes in diploid and polyploid cotton. Plant Mol Biol 51: 313–325 [DOI] [PubMed] [Google Scholar]
- Cho M, Lee SH, Cho HT. (2007) P-glycoprotein4 displays auxin efflux transporter-like action in Arabidopsis root hair cells and tobacco cells. Plant Cell 19: 3930–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cubas P (2002) Role of TCP genes in the evolution of morphological characters in angiosperms. In JA Hawkins, QCB Cronk, RM Bateman, eds, Developmental Genetics and Plant Evolution. CRC Press, London, pp 247–266 [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Damerval C, Le Guilloux M, Jager M, Charon C. (2007) Diversity and evolution of CYCLOIDEA-like TCP genes in relation to flower development in Papaveraceae. Plant Physiol 143: 759–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Lee SH, Cho M, Lee OR, Yoo H, Cho HT. (2010) Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells. Plant Physiol 153: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Zhao PM, Wang J, Wang HY, Wu XM, Xia GX. (2007) Identification of genes preferentially expressed in cotton fibers: a possible role of calcium signaling in cotton fiber elongation. Plant Sci 173: 61–69 [Google Scholar]
- Gialvalis S, Seagull RW. (2001) Plant hormones alter fiber initiation in unfertilized, cultured ovules of Gossypium hirsutum. J Cotton Sci 5: 252–258 [Google Scholar]
- Gou JY, Wang LJ, Chen SP, Hu WL, Chen XY. (2007) Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res 17: 422–434 [DOI] [PubMed] [Google Scholar]
- Griffee F, Ligon L. (1929) Occurrence of “lintless” cotton plants and the inheritance of the character “lintless. J Am Soc Agron 21: 711–717 [Google Scholar]
- Guan XY, Lee JJ, Pang MX, Shi XL, Stelly DM, Chen ZJ. (2011) Activation of Arabidopsis seed hair development by cotton fiber-related genes. PLoS ONE 6: e21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XY, Li QJ, Shan CM, Wang S, Mao YB, Wang LJ, Chen XY. (2008) The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol Plant 134: 174–182 [DOI] [PubMed] [Google Scholar]
- Guo ZX, Fujioka S, Blancaflor EB, Miao S, Gou XP, Li J. (2010) TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22: 1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J, Tu L, Hu H, Tan J, Deng F, Tang W, Nie Y, Zhang X. (2012) GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J Exp Bot 63: 6267–6281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C, Dabos P, Bardet C, Jauneau A, Auriac MC, Ramboer A, Lacout F, Tremousaygue D. (2009) In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol 149: 1462–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John ME, Crow LJ. (1992) Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc Natl Acad Sci USA 89: 5769–5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HMO, Grierson CS. (2009) Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol 11: 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B. (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. (2001) Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361–1366 [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O. (2003) AXR3 and SHY2 interact to regulate root hair development. Development 130: 5769–5777 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. (2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. (2010) TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. (2005) Auxin distribution and plant pattern formation: how many angels can dance on the point of PIN? Cell 121: 819–822 [DOI] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZY, Li B, Dong AW. (2012) The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol Plant 5: 270–280 [DOI] [PubMed] [Google Scholar]
- Liu K, Sun J, Yao L, Yuan Y. (2012) Transcriptome analysis reveals critical genes and key pathways for early cotton fiber elongation in Ligon lintless-1 mutant. Genomics 100: 42–50 [DOI] [PubMed] [Google Scholar]
- Machado A, Wu YR, Yang YM, Llewellyn DJ, Dennis ES. (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59: 52–62 [DOI] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Müller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrison JL, Leech RM. (1994) The subcellular and intra-organelle recognition of nuclear and chloroplast transcripts in developing leaf-cells. Plant J 6: 605–614 [Google Scholar]
- Martín-Trillo M, Cubas P. (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sabatini S. (2009) Cytokinin-auxin crosstalk. Trends Plant Sci 14: 557–562 [DOI] [PubMed] [Google Scholar]
- Müller A, Guan C, Gälweiler L, Tänzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. (1998) AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J 17: 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller D, Leyser O. (2011) Auxin, cytokinin and the control of shoot branching. Ann Bot (Lond) 107: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nag A, King S, Jack T. (2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaud O, Dabos P, Carnus E, Tremousaygue D, Hervé C. (2007) TCP transcription factors predate the emergence of land plants. J Mol Evol 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Nørby PL, Pallisgaard N, Pedersen FS, Jørgensen P. (1992) Determination of recognition-sequences for DNA-binding proteins by a polymerase chain reaction assisted binding site selection method (BSS) using nitrocellulose immobilized DNA binding protein. Nucleic Acids Res 20: 6317–6321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L, Li Q, Fan XP, Yang WC, Xue YB. (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Zhu YX. (2011) How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol 14: 106–111 [DOI] [PubMed] [Google Scholar]
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S. (2002) Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol 130: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Samuel Yang S, Cheung F, Lee JJ, Ha M, Wei NE, Sze SH, Stelly DM, Thaxton P, Triplett B, Town CD, et al. (2006) Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47: 761–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan XX, Xu B, Yu ZX, Wang LJ, Chen XY. (2008) Promoter of a cotton fibre MYB gene functional in trichomes of Arabidopsis and glandular trichomes of tobacco. J Exp Bot 59: 3533–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18: 651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. (2012) The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chen Q, Qi L, Jiang H, Li S, Xu Y, Liu F, Zhou W, Pan J, Li X, et al. (2011) Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol 191: 360–375 [DOI] [PubMed] [Google Scholar]
- Suo JF, Liang XE, Pu L, Zhang YS, Xue YB. (2003) Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim Biophys Acta 1630: 25–34 [DOI] [PubMed] [Google Scholar]
- Swarup R, Péret B. (2012) AUX/LAX family of auxin influx carriers—an overview. Front Plant Sci 3: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Amano K, Ohto MA, Nakamura K, Sato S, Kato T, Tabata S, Ueguchi C. (2006) RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol Biol 61: 165–177 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Nakabayashi K, Kamiya Y, Nambara E. (2008a) Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J 53: 42–52 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW. (1999) Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711–721 [DOI] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14: 301–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Ishida T, Wada T. (2011) New insights into the mechanism of development of Arabidopsis root hairs and trichomes. Int Rev Cell Mol Biol 286: 67–106 [DOI] [PubMed] [Google Scholar]
- Traw MB, Bergelson J. (2003) Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol 133: 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trémousaygue D, Garnier L, Bardet C, Dabos P, Hervé C, Lescure B. (2003) Internal telomeric repeats and ‘TCP domain’ protein-binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J 33: 957–966 [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH. (2012) The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63: 809–823 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Walford SA, Wu Y, Llewellyn DJ, Dennis ES. (2011) GhMYB25-like: a key factor in early cotton fibre development. Plant J 65: 785–797 [DOI] [PubMed] [Google Scholar]
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY. (2004) Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16: 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Niu QW, Teng C, Li C, Mu J, Chua NH, Zuo J. (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res 19: 224–235 [DOI] [PubMed] [Google Scholar]
- Yao X, Ma H, Wang J, Zhang D. (2007) Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J Integr Plant Biol 49: 885–897 [Google Scholar]
- Zhang M, Zheng X, Song S, Zeng Q, Hou L, Li D, Zhao J, Wei Y, Li X, Luo M, et al. (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29: 453–458 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Chua NH. (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in plants. Plant J 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.