Eutrema salsugineum maintains its salt tolerance under very different growth conditions even though its development and metabolism show substantial growth condition-dependent differences.
Abstract
Comparative studies of the stress-tolerant Arabidopsis (Arabidopsis thaliana) halophytic relative, Eutrema salsugineum, have proven a fruitful approach to understanding natural stress tolerance. Here, we performed comparative phenotyping of Arabidopsis and E. salsugineum vegetative development under control and salt-stress conditions, and then compared the metabolic responses of the two species on different growth platforms in a defined leaf developmental stage. Our results reveal both growth platform-dependent and -independent phenotypes and metabolic responses. Leaf emergence was affected in a similar way in both species grown in vitro but the effects observed in Arabidopsis occurred at higher salt concentrations in E. salsugineum. No differences in leaf emergence were observed on soil. A new effect of a salt-mediated reduction in E. salsugineum leaf area was unmasked. On soil, leaf area reduction in E. salsugineum was mainly due to a fall in cell number, whereas both cell number and cell size contributed to the decrease in Arabidopsis leaf area. Common growth platform-independent leaf metabolic signatures such as high raffinose and malate, and low fumarate contents that could reflect core stress tolerance mechanisms, as well as growth platform-dependent metabolic responses were identified. In particular, the in vitro growth platform led to repression of accumulation of many metabolites including sugars, sugar phosphates, and amino acids in E. salsugineum compared with the soil system where these same metabolites accumulated to higher levels in E. salsugineum than in Arabidopsis. The observation that E. salsugineum maintains salt tolerance despite growth platform-specific phenotypes and metabolic responses suggests a considerable degree of phenotypic and metabolic adaptive plasticity in this extremophile.
The study of extremophile plants that survive in harsh environments has gained traction in recent years due to the limitation of using Arabidopsis (Arabidopsis thaliana) for abiotic stress research (Oh et al., 2012). Although Arabidopsis has been the model plant of choice for identifying genes involved in abiotic stress responses due to its unsurpassed genetic and genomic resources, Arabidopsis is a stress-sensitive plant and is unlikely to possess many stress tolerance mechanisms found in naturally stress-tolerant plants. One fruitful approach has been the study of the close Arabidopsis relative, Eutrema salsugineum (formally Thellungiella halophila, renamed Thellungiella salsuginea and now assigned to the genus Eutrema; http://phytozome.net/thellungiella.php#A; Amtmann, 2009; Yang et al., 2013). This species is tolerant to extreme levels of salinity (Inan et al., 2004; Kant et al., 2006), low soil nitrogen (Kant et al., 2008), and high boron levels (Lamdan et al., 2012), and shows similar tolerance to cold and freezing stress as Arabidopsis (Griffith et al., 2007; Lee et al., 2012), reflecting the combination of stresses to which plants living in extreme environments are simultaneously exposed. Importantly, E. salsugineum possesses similar morphology to Arabidopsis as well as many attributes of a plant model system (Amtmann, 2009). These attributes have facilitated many comparative studies of Arabidopsis and E. salsugineum stress responses at the physiological, biochemical, gene expression, and “omics” levels (Amtmann, 2009; Oh et al., 2012; Yang et al., 2013). From such reports, a number of traits are postulated to contribute to E. salsugineum tolerance to abiotic stresses: (1) Differences in regulation between Arabidopsis and E. salsugineum genes that are crucial for stress tolerance have been observed. Thus, expression of the gene encoding the plasma membrane Na+/H+ antiporter SALT OVERLY SENSITIVE1 (SOS1) is constitutively higher in E. salsugineum roots under control conditions compared with Arabidopsis and is induced by salt to a greater extent in E. salsugineum shoots (Taji et al., 2004; Kant et al., 2006). High expression of SOS1 can be correlated with reduced accumulation of Na+ in E. salsugineum shoots. Constitutive down-regulation of E. salsugineum PDH, encoding the Pro catabolic enzyme, Pro dehydrogenase, is correlated with increased levels of Pro under control and salt-stress conditions, whereas constitutive up-regulation of NR2, which encodes nitrate reductase, is associated with increased nitrate uptake under low nitrogen conditions (Kant et al., 2006, 2008). On a global level, E. salsugineum exhibits fewer stress-induced transcripts than Arabidopsis, suggesting that E. salsugineum is “primed” for stress (Taji et al., 2004; Gong et al., 2005). Analysis of the E. salsugineum genome sequence also suggests that biased codon usage may facilitate more efficient translation of proteins related to ion transportation compared with Arabidopsis (Yang et al., 2013). (2) Evaluation of E. salsugineum responses to different stresses using EST libraries and microarrays demonstrates very little overlap between gene expression under the different conditions, suggesting that E. salsugineum is more specific in its stress response than stress-sensitive species (Wong et al., 2005, 2006). (3) Comparison of the recently sequenced E. salsugineum and Eutrema parvulum genomes with the Arabidopsis genome has revealed that gene copy number expansion of stress-related genes and microRNAs appears to have played a role in the evolution of tolerance to extreme environments (Dassanayake et al., 2011; Oh et al., 2012; Wu et al., 2012; Yang et al., 2013). (4) The E. salsugineum genomes possess protein-coding genes that do not share sequence similarity outside the lineage. Thus, 11% of these annotated “lineage-specific” or “taxonomically restricted” genes in the E. parvulum genome show no sequence similarity with any Arabidopsis genes, suggesting the presence of unique stress-response genes (Dassanayake et al., 2011).
With the sequencing of the Eutrema spp. genomes, we are now in a position to take a rigorous systems biology approach to catalog the changes in the response of the E. salsugineum transcriptome, proteome, and metabolome, particularly in comparison with Arabidopsis, and apply powerful computational biology tools to understand how gene, protein, and metabolite networks are wired in E. salsugineum. However, the challenge will remain to accurately associate changes in these molecules with quantifiable developmental and physiological stress adaptation phenotypes and thus allow genotype-to-phenotype predictions to be made. To achieve this goal, deep phenotyping (phenomics) is essential to allow compilation of precise multidimensional, morphological, physiological, and molecular phenotypes of plant responses (Boyes et al., 2001; Granier et al., 2006; Chern et al., 2007; Miyao et al., 2007; Kuromori et al., 2009; Skirycz et al., 2010; Furbank and Tester, 2011). Although Arabidopsis and E. salsugineum possess some fundamental differences in developmental program such as a vernalization requirement for promotion of flowering in E. salsugineum (Bressan et al., 2001), their developmental growth stages from seed to rosette leaves to production of flowering bolts are similar, a feature that facilitates phenotypic comparison between the two species. Yet, no comparative deep phenotyping of E. salsugineum versus Arabidopsis development has been reported. This is particularly important when investigating plant responses to milder stresses, characteristic of many agricultural situations, where stressed and nonstressed plants can look very similar morphologically but deep phenotyping could detect altered developmental progression. Strict definition of each developmental stage and subsequent parallel molecular analysis would then facilitate linking of genotype to phenotype.
A growth-based phenotypic analysis platform for Arabidopsis, based on the BASF, Bayer, Ciba-Geigy, and Hoechst scale of standardized descriptions for growth stages of crops and weeds (Lancashire et al., 1991), was developed by Boyes et al. (2001) and provided a common semantic link between Arabidopsis and crop plants. The entire life cycle of Arabidopsis was divided into growth stages representing developmental landmarks. Early stages of plant development, from radical emergence to appearance of the sixth rosette leaf, were recorded using an in vitro plate system, while growth stages from emergence of two rosette leaves to seed production were observed in soil-grown plants. In this study, our aim was to perform a comparative analysis of Arabidopsis and E. salsugineum vegetative development under control and salt-stress conditions using the Boyes et al. (2001) growth stage system, and then to compare the metabolic responses of the two species between plate and soil growth platforms in a precisely defined leaf developmental stage. Our results reveal growth platform-dependent and -independent phenotypes and metabolic responses that might reflect both core stress tolerance mechanisms and the underlying adaptive plasticity of E. salsugineum.
RESULTS
Plate- and Soil-Based Phenotypic Analysis Platforms
To determine whether the two species exhibited detailed phenotypic differences in growth as an acclimative feature of their response to salt stress, the time of occurrence of plant developmental stages according to a modified in vitro plate-based and soil-based platform of Boyes et al. (2001) was examined (Table I). For the plate-based assay, it was necessary to modify the growth protocol because E. salsugineum was unable to germinate on plates supplemented with NaCl (Inan et al., 2004; Supplemental Fig. S1). Thus, to be able to grow E. salsugineum under salt stress conditions from as early in plant development as possible, it was necessary to germinate E. salsugineum on medium without salt and then to transfer seedlings to salt plates at a later stage. It is possible to transfer seedlings to fresh medium with forceps but not earlier than stage 1 (cotyledons fully open). Hence, to analyze even earlier stages of E. salsugineum development, seeds were first germinated on a silk mesh overlaid upon the control (no salt) medium and then mesh plus seeds could be transferred to salt plates. The mesh size was such that plant roots could penetrate through to the medium, but the seedlings remained on the mesh surface (Supplemental Fig. S2, A and B). Comparison of seedlings transferred to fresh control plates at 12 d after stratification (DAS) with seedlings grown on mesh but not transferred demonstrated that the transfer process did not affect plant growth and development (Supplemental Fig. S2, C and D). No signs of stress, such as leaf yellowing, curling, or reduction in size, were visible and there were no differences in fresh weight or leaf number. Furthermore, with the aid of the mesh, it became possible to transfer seeds when the radicle had just emerged (stage 0.5; Supplemental Fig. S2, E and F), thereby facilitating observation of early stages of plant development.
Table I. Growth stages for in vitro- and soil-based phenotypic analysis.
| Stagea | Description |
|---|---|
| In vitro | |
| 0.5 | Radicle emergence |
| 0.7 | Hypocotyl and cotyledon emergence |
| 1.0 | Cotyledons fully opened |
| 1.02 | 2 rosette leaves > 1 mm in length |
| 1.04 | 4 rosette leaves > 1 mm in length |
| 1.06 | 6 rosette leaves > 1 mm in length |
| Soil | |
| 1.02 | 2 rosette leaves > 1 mm in length |
| 1.04 | 4 rosette leaves > 1 mm in length |
| 1.05 | 5 rosette leaves > 1 mm in length |
| 1.06 | 6 rosette leaves > 1 mm in length |
| 1.07 | 7 rosette leaves > 1 mm in length |
| 1.08 | 8 rosette leaves > 1 mm in length |
| 1.09 | 9 rosette leaves > 1 mm in length |
| 1.10 | 10 rosette leaves > 1 mm in length |
Growth stages based on Boyes et al. (2001).
For phenotypic analysis of the effect of salt stress on Arabidopsis and E. salsugineum development in soil, which is closer to field conditions, seedlings were grown on plates containing Murashige and Skoog (MS) medium and then stage 1 seedlings were transferred to pots with soil. This was the earliest possible time at which seedlings could be moved to soil without damage due to the transfer process. The developmental progression of stages 1.02 to 1.10 (Table I) for control and salt-treated plants was determined. The measurement of leaf development for more rosette leaves was not performed because under our growth room conditions, Arabidopsis started to flower after stage 1.10, whereas E. salsugineum does not flower unless vernalized. The experiment was therefore terminated at stage 1.10.
In Vitro-Grown Arabidopsis and E. salsugineum Exhibit Species-Specific Leaf Emergence Responses to Salt Stress But a Common Response of Leaf Area
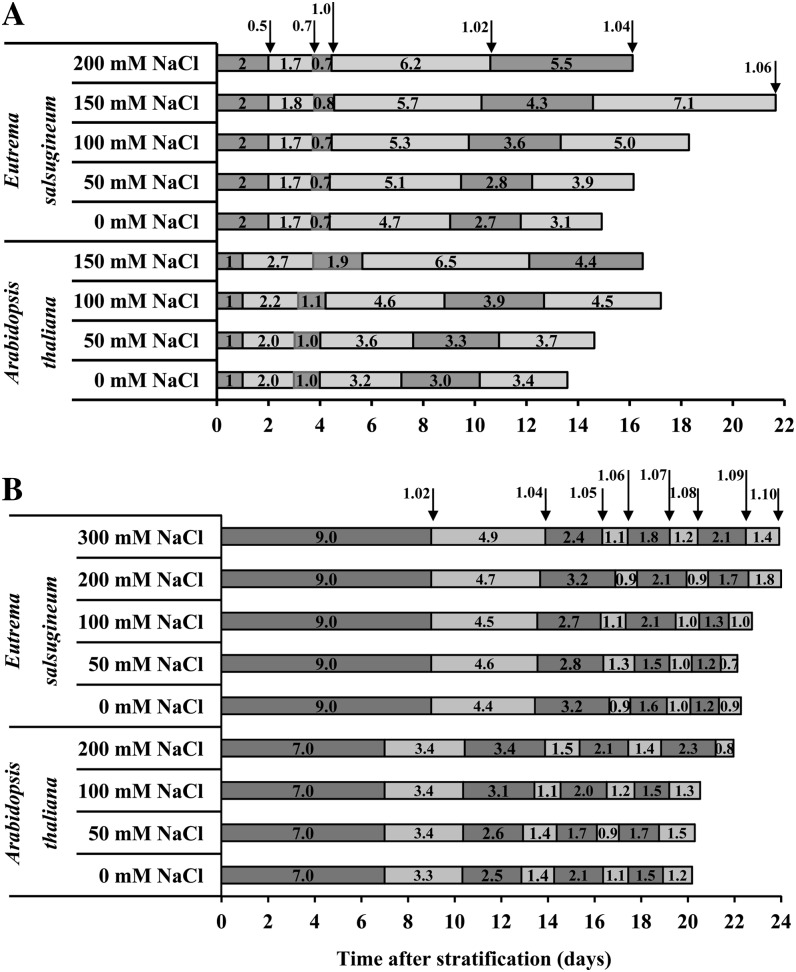
Supplemental Tables S1 and S2 show the cumulative time of occurrence of the developmental stages listed in Table I. sd and coefficient of variation (calculated as the ratio between sd and the average number of days until the stage occurred × 100) between independent biological replicates in which plants were subjected to the same treatment were less than or equal to 12%, indicating highly reproducible progression of development under all treatments. To better visualize the results, the data were depicted in Figure 1 as stacked bar graphs showing the duration of each developmental stage. In in vitro plate-based assays, neither Arabidopsis nor E. salsugineum exhibited significant differences in the cumulative occurrence of the observed developmental stages between control plants and those exposed to mild (50 mm NaCl) salt stress (Fig. 1A; Supplemental Table S1). However, under the 100 mm NaCl treatment, small increases in the duration of each stage of Arabidopsis leaf emergence led to a significant lengthening in the cumulative time taken to reach stage 1.04 and beyond. On the other hand, this level of salt stress had no significant effect on E. salsugineum growth and development compared with control. At more severe stress (150 mm NaCl), Arabidopsis not only exhibited a significant increase in the duration of every developmental stage, but plants only reached stage 1.04, after which no further leaves emerged. In contrast, E. salsugineum exhibited no slowdown in the cumulative time taken to reach stage 1 but developmental progression significantly slowed for each stage from 1.02 and beyond. Moreover, E. salsugineum was able to reach stage 1.06, unlike Arabidopsis. Only E. salsugineum was able to grow at 200 mm NaCl, but only up to stage 1.04, after which no further leaves emerged.
Figure 1.
The effect of salt stress on growth stage progression of in vitro- and soil-grown Arabidopsis and E. salsugineum. A, Seedlings were transferred by mesh at stage 0.5 to fresh control in vitro medium and medium supplemented with the indicated NaCl concentrations. B, Stage 1.0 seedlings were transferred to soil from in vitro plates and subjected to the indicated salt treatments. Salt treatments commenced at stage 1.02 with 50% of the final salt concentration. One hundred percent of each salt level was applied at stage 1.04. Developmental stages are according to Boyes et al. (2001). Boxes represent the time elapsed between the successive growth stages. Junctions between boxes of different shading indicate the occurrence of a growth stage. Data are mean of three independent experiments. Each experiment contained either three or more replicate plates with approximately 15 seedlings per plate (in vitro) or approximately 10 replicate plants (soil).
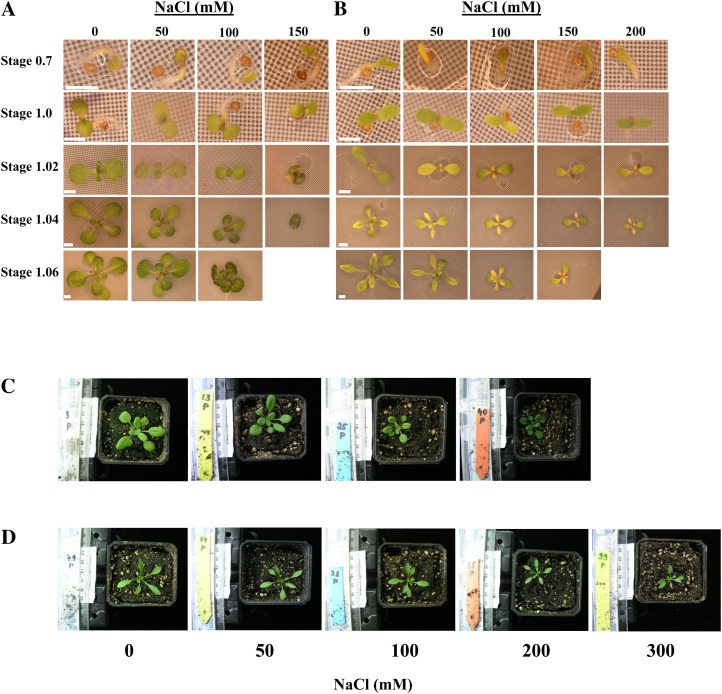
In addition to quantitatively measured parameters, several qualitative observations were recorded that illustrate not only the various growth stages but also the comparative effect of salt stress upon those stages in the two species (Fig. 2). From the earliest stages of plant development, phenotypic differences between control and salt-treated plants were observed. In particular, a reduction in seedling cotyledon/leaf area was visible as early as stage 1 for both plant species under 100 and 150 mm NaCl. At later stages of plant development, symptoms of stress such as leaf curling occurred, which was especially evident for Arabidopsis grown at 150 mm NaCl, where plants did not usually survive past the fourth true leaf. By stage 1.04, E. salsugineum exhibited paling of the true leaf margins even under control conditions, a phenotype that we noted was present in other reports using in vitro-grown E. salsugineum seedlings (Taji et al., 2004; Kant et al., 2008). However, the increased tolerance to salt of E. salsugineum compared with Arabidopsis could still be discerned, particularly under 150 mm NaCl. Here, Arabidopsis exhibited signs of stress-mediated damage and decreased growth rate from the first stage after transfer to salt-containing plates (Figs. 1A and 2A; Supplemental Table S1). In contrast, although there is a reduction in E. salsugineum growth rate at 150 mm NaCl (which occurs at later stages compared with Arabidopsis) and signs of damage (leaf curling and seedling yellowing), E. salsugineum continues to develop new leaves (Figs. 1A and 2B; Supplemental Table S1). As mentioned above, only E. salsugineum could survive on 200 mm NaCl, but did not produce any more true leaves after stage 1.04.
Figure 2.
Arabidopsis and E. salsugineum phenotypes on in vitro medium and on soil (stage 1.10) under control and salt-stress conditions. A, Arabidopsis in vitro. B, E. salsugineum in vitro. C, Arabidopsis soil. D, E. salsugineum soil. White scale bar = 1 mm.
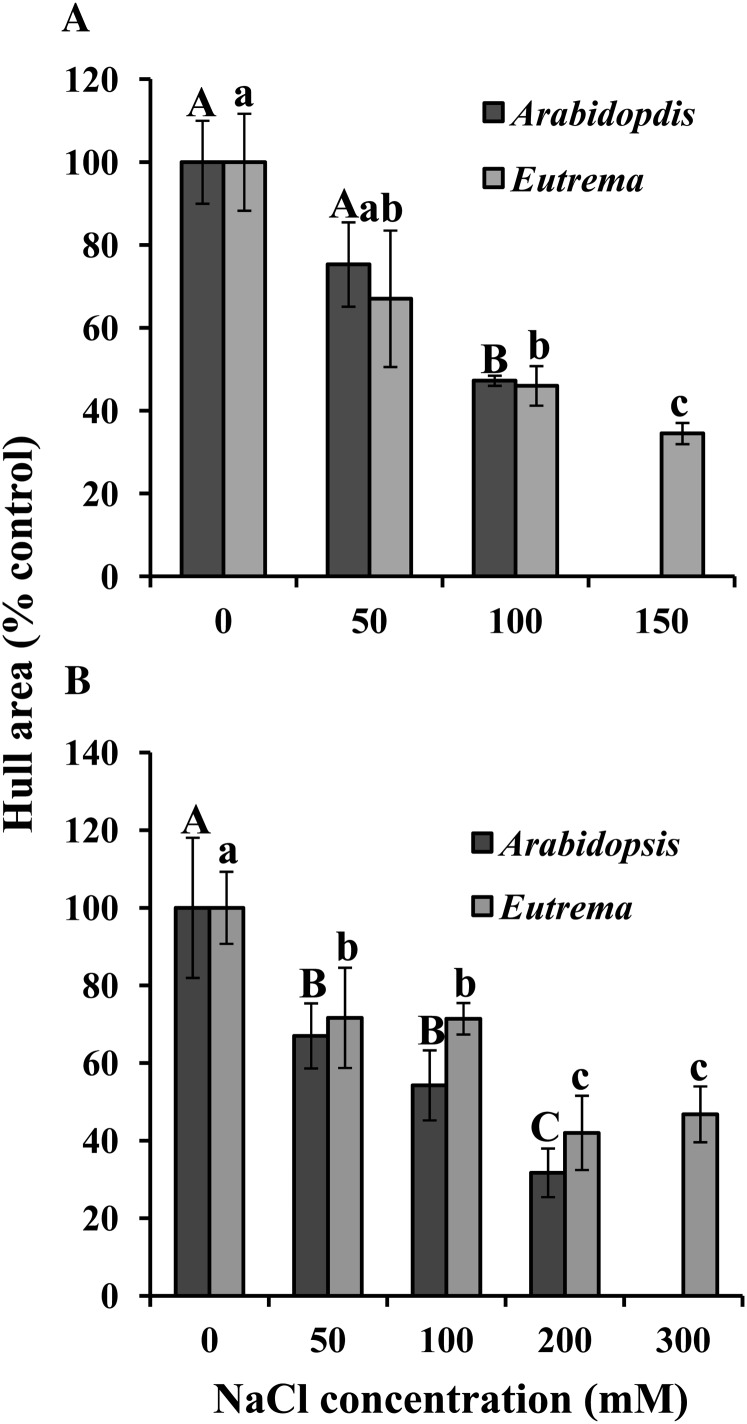
Because the leaves from the in vitro-grown plants were too small to quantitatively measure leaf area, particularly under salt stress, we measured hull area (the area that results from joining the tips of rosette leaves with a line) from digital images of the plants as a parameter of reduction of whole rosette area. Figure 3A and Supplemental Figure S3A show that there is a correlation between the salt concentration and a decrease in plant hull area that is almost equally pronounced in Arabidopsis and E. salsugineum.
Figure 3.
Effect of salt stress on hull area of in vitro- and soil-grown Arabidopsis and E. salsugineum. A, In vitro. B, Soil. Data are mean ± sd (n = 3, in vitro; n = 8, soil). Each replicate in vitro plate contained approximately 15 seedlings. Only E. salsugineum plants could be grown on soil under 300 mm NaCl. Data are representative of similar results from two independent experiments. Bars with different letters (uppercase, Arabidopsis; lowercase, E. salsugineum) indicate significant difference at P ≤ 0.05 (Student’s t test). Each species was statistically analyzed separately between all salt levels.
Soil-Grown Arabidopsis and E. salsugineum Exhibit No Effect of Salt Stress on Leaf Emergence But Display Species-Specific Responses of Leaf Area
In contrast to the in vitro plate experiments, in the soil-based assay there was no significant difference in leaf emergence from stage 1.02 to 1.10 between control and any of the salt treatments in either species (Fig. 1B; Supplemental Table S2). Figure 2, C and D, illustrates the effect of salt stress on the two soil-grown species at developmental stage 1.10. Reduction in leaf and rosette size is clearly visible. Furthermore, there is a darkening in the color of Arabidopsis leaves that is noticeable at 200 mm NaCl, which is not observed in E. salsugineum plants. This darkening is characteristic of anthocyanin production, an oft-used marker of stress that has been shown to accumulate to far lesser extent in E. salsugineum than Arabidopsis under salt stress (Kant et al., 2006).
The stress-induced reduction in plant size was quantitatively measured by determining total rosette leaf area and hull area. Both species exhibited a similar (approximately 30%) reduction in both rosette and hull area, even under 50 mm NaCl (Fig. 3B; Supplemental Fig. S3). In addition, E. salsugineum exhibited a two-step response to salt stress: a decline in leaf area at 50 mm NaCl, no further reduction in size at 100 mm NaCl, and a further 30% decline in size at 200 mm NaCl, with no additional change at 300 mm NaCl. It is not clear whether Arabidopsis exhibited a dose response or a two-step response because only E. salsugineum was able to survive at 300 mm NaCl, and we could therefore not dissect the Arabidopsis response with the additional data point.
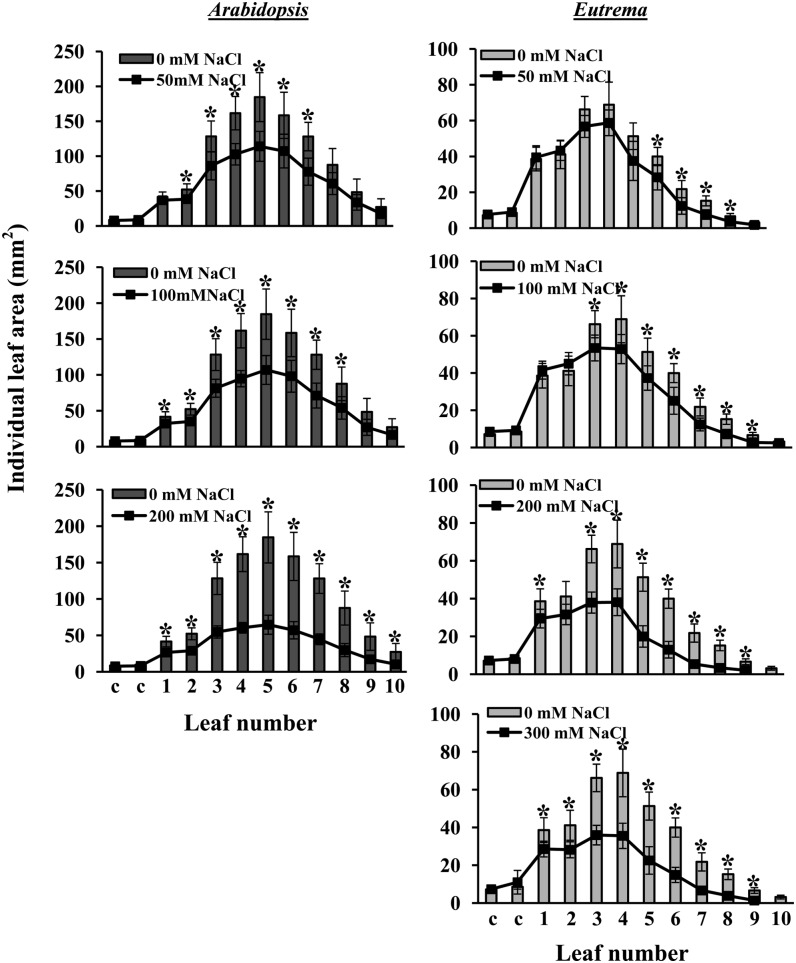
The finding that a reduction in E. salsugineum rosette and hull area occurred at 50 and 100 mm NaCl was surprising because such mild salt stress has not been previously shown to affect E. salsugineum development as leaf area was not assessed as a growth parameter (Inan et al., 2004; Kant et al., 2006; Ghars et al., 2008; Lugan et al., 2010). Therefore, to further examine this reduction in leaf area in detail, the area of each cotyledon and leaf was ascertained when plants of both species reached stage 1.10. All leaves, starting from the fifth leaf onward, emerged after final salt concentration was reached. At the lowest salt concentration, Arabidopsis exhibited a significant reduction in area of the second to seventh leaves, whereas the cotyledons, first true leaf, and eighth to 10th leaves were unaffected (Fig. 4; Supplemental Fig. S4). An even larger reduction in the area of individual leaves was observed at higher salt concentrations and this reduction now included all the true leaves but not the cotyledons. E. salsugineum also exhibited a reduction in the area of individual leaves, but at each salt concentration this decrease was less than that observed in Arabidopsis. Strikingly, in contrast to Arabidopsis, the younger E. salsugineum leaves (sixth to ninth) were affected under mild stress with an effect on older leaves only under higher levels of stress. Similar to Arabidopsis, there was no effect of salt stress on E. salsugineum cotyledons. In agreement with the data for total rosette leaf area and hull area (Fig. 3; Supplemental Fig. S3), E. salsugineum (and possibly Arabidopsis) again exhibited a two-step response: a small and similar reduction in leaf area at 50 and 100 mm NaCl with a further and similar reduction at 200 and 300 mm NaCl. This response can be most clearly seen in Supplemental Figure S4, where all salt treatments are amalgamated in the same graph for each species.
Figure 4.
The effect of salt stress on the area of individual soil-grown Arabidopsis (dark gray bars) and E. salsugineum (light gray bars) leaves. Data are mean ± sd (n = 8). Data are representative of similar results from two independent experiments. Asterisks represent significant difference between control and salt-treated leaf areas (P ≤ 0.01, Student’s t test).
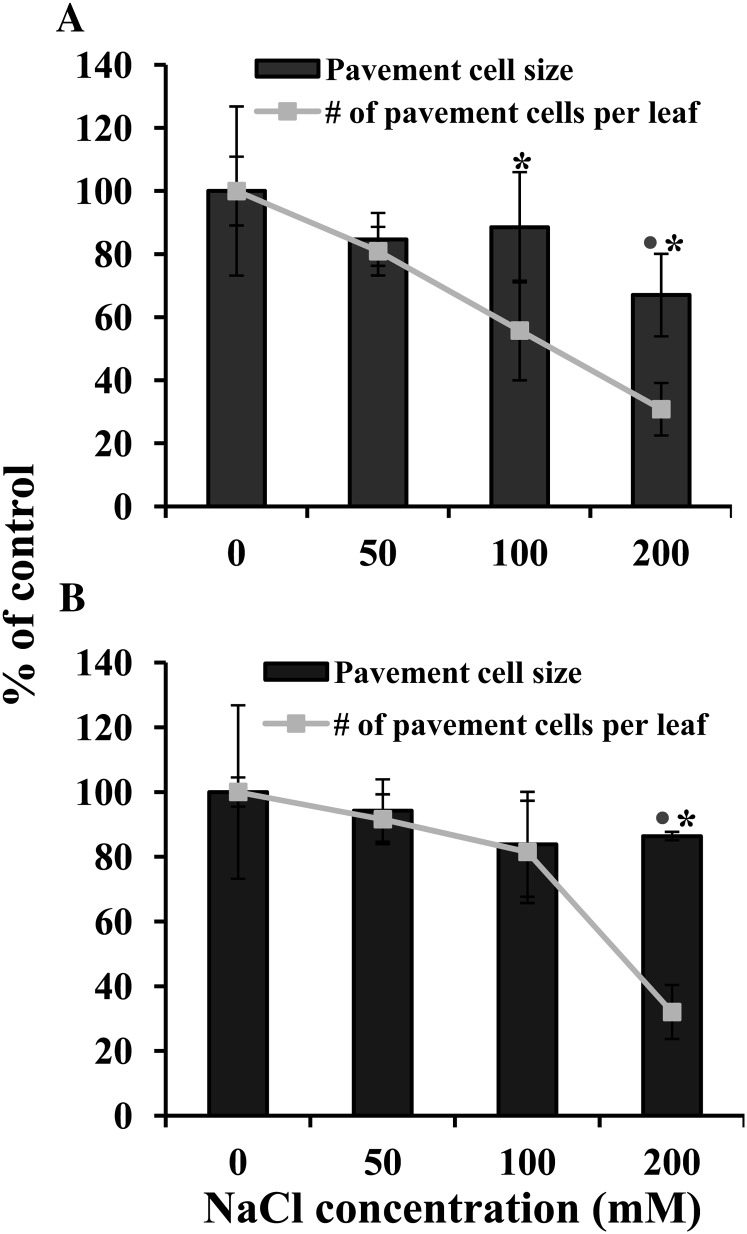
The observed reduction in Arabidopsis and E. salsugineum leaf area could be due to a decrease in cell number, cell size, or a combination of both these parameters (Skirycz et al., 2010). Therefore, to ascertain the cause of the salt-induced reduction in leaf area, we determined pavement cell number and size for the fifth leaf (the first leaf that emerged after 100% of salt treatment), employing the dental resin imprint technique (Weyers and Johansen, 1985; Lawson et al., 1998). No difference in morphology of pavement cells was observed, either due to salt treatment or between the species (Supplemental Fig. S5). Both species exhibited a statistically significant reduction in pavement cell size only at 200 mm NaCl (Fig. 5; Supplemental Table S3). However, whereas Arabidopsis cell size was reduced by about 33%, E. salsugineum only exhibited a 16% reduction. Both species also exhibited a salt-mediated decrease in pavement cell number. Salt stress caused a statistically significant 45% reduction in Arabidopsis cell number at 100 mm NaCl with a further decrease at 200 mm NaCl, reaching a 70% reduction compared with control cell number. E. salsugineum showed a significant 68% fall in cell number only at 200 mm NaCl. Thus, the predominant factor in the salt-mediated reduction in E. salsugineum leaf area (at least at 200 mm NaCl) was a fall in cell number, although a small decrease in cell size also contributed to the overall fall in leaf area. In contrast, a considerable salt-mediated reduction in both cell size and cell number led to a decrease in Arabidopsis leaf area.
Figure 5.
Effect of salt stress on soil-grown Arabidopsis and E. salsugineum cell number and cell size. A, Arabidopsis. B, E. salsugineum. Data are mean (n = 4) ± sd of the area of the fifth leaf from plants at stage 1.10. Fifty to 90 cells from each leaf were examined. Data are representative of two independent experiments. Dots indicate a significant difference in pavement cell size (P ≤ 0.05, Student’s t test) between control and salt treatment. Asterisks indicate a significant difference in number of pavement cells per leaf (P ≤ 0.01; Student’s t test) between control and salt treatment. Values for 100% average pavement cell size ± sd (mm2): Arabidopsis, 1,798.6 ± 196; E. salsugineum, 2,099.4 ± 95. Values for 100% average number of pavement cells per leaf ± sd: Arabidopsis, 166,995 ± 44,757; E. salsugineum, 90,855 ± 2,714. For average cell numbers and cells sizes under salt stress, see Supplemental Table S3.
Global Analysis of Metabolic Profiles of Arabidopsis and E. salsugineum under Control and Salt Stress Conditions Reveals Major Effects of Species, Salt Treatment, and Growth Platform
Our phenotypic analysis revealed both similarities (reduction in leaf area) and differences (leaf emergence) between the effects of salt on in vitro-grown and soil-grown Arabidopsis and E. salsugineum plants. To gain insight into metabolic events underlying the phenotypic changes, we chose well-defined developmental stages and salt treatments that also led to a response in E. salsugineum according to our phenotypic analysis. Thus, for the in vitro experiment, seedlings that had been subjected to 0, 50, 100, or 150 mm NaCl under our in vitro plate conditions were harvested at stage 1.06 (1.04 for Arabidopsis at 150 mm NaCl). For the soil experiment, plants were grown under 0, 100, or 200 mm NaCl until stage 1.10. True leaves 5 and 6, a leaf pair at a very similar stage of development that exhibited the greatest salt-mediated reduction in both species (Fig. 4) and were the first leaves to emerge after final salt concentration was reached, were harvested and pooled. The relative abundance of approximately 50 metabolites was determined for both plant species using standard gas chromatography-mass spectrometry-based protocols (Lisec et al., 2006).
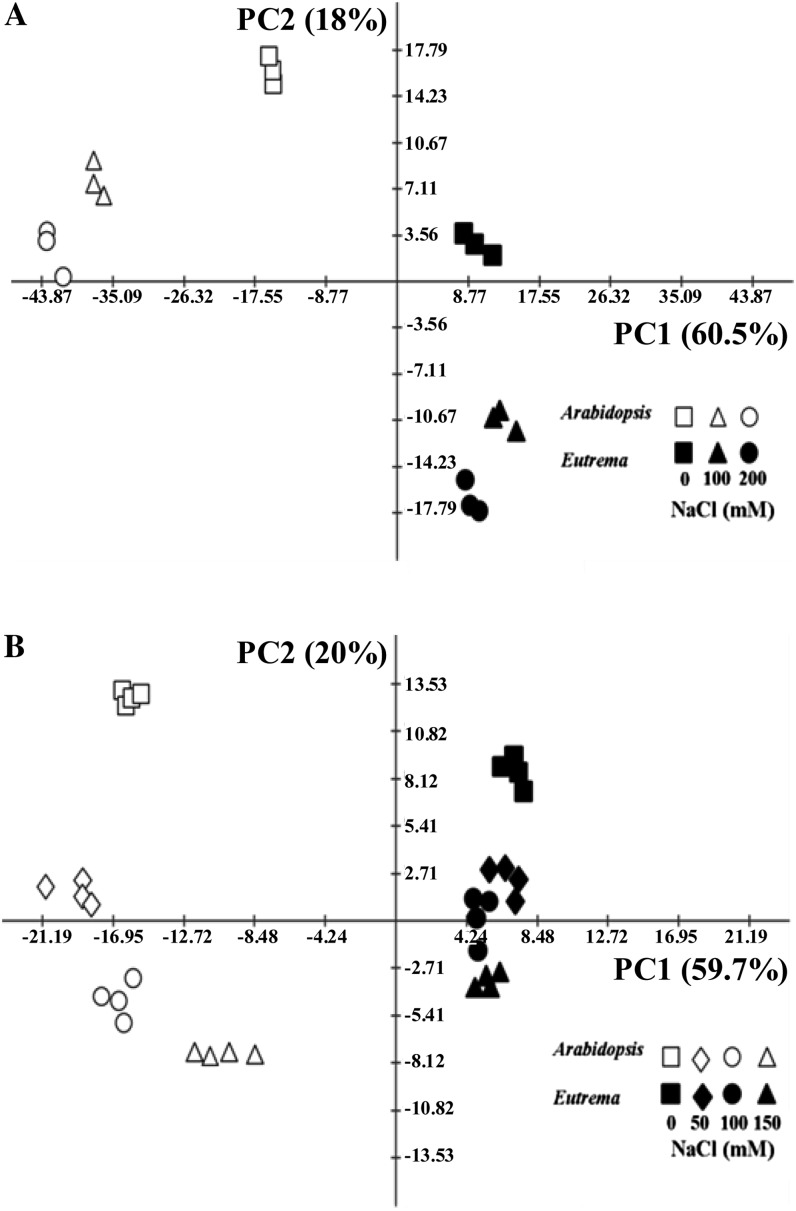
To obtain a global view of the differences between the plant species and across the treatments, principal component analysis (PCA) was employed. Inspection of the first two components, which together account for 78.5% and 79.7% of the total variance within the in vitro and soil data sets, respectively, allowed classification of samples by species and treatments (Fig. 6). For both growth platforms, the first principal component, accounting for 60.5% (in vitro) or 59.7% (soil) of total variance, separated the two species on the basis of their metabolisms. Among the metabolites most affecting the separation between samples on the first component for in vitro-grown seedlings (in decreasing order of an absolute value of eigenvector and confirmed with two-way ANOVA) were Gln, caffeate, raffinose, galactinol, Asn, fumarate, citrate, and malate, and for soil-grown plants were putrescine, Fru, raffinose, caffeate, Gln, Phe, γ-aminobutyric acid (GABA), and Asn (Supplemental Tables S4–S9).
Figure 6.
PCA of metabolic profiles of (A) in vitro-grown (stage 1.06 [stage 1.04 – Arabidopsis 150 mm NaCl]) and (B) soil-grown Arabidopsis and E. salsugineum plants (stage 1.10) under control and salt-stress conditions. Arabidopsis and E. salsugineum are separated along principal component 1 (PC1) while the effect of salt stress is discriminated along principal component 2 (PC2). Variance explained by each component is indicated in brackets. Data are representative of similar results from three independent experiments.
The second principal component, accounting for 18% (plates) or 20% (soil) of the data variance, discriminated samples according to the salt treatment to which the plants were subjected. Among the metabolites that most affected the separation between samples on the second component for in vitro-grown seedlings (in decreasing order of absolute value of eigenvector and confirmed with two-way ANOVA) were Pro, Gly, raffinose, Gln, fumarate, Ala, galactinol and malate, and for soil-grown plants were Pro, citrate, malate, erythronate, ascorbate, Lys, fumarate and putrescine (Supplemental Tables S4–S9). The two-way ANOVA test also showed that 35 out of 46 metabolites (in vitro) and 45 out of 50 metabolites (soil) were significantly different between the species (Supplemental Tables S8 and S9). Among these, 30 and 44 metabolites also had significant species × salt treatment interaction for in vitro and soil experiments, respectively.
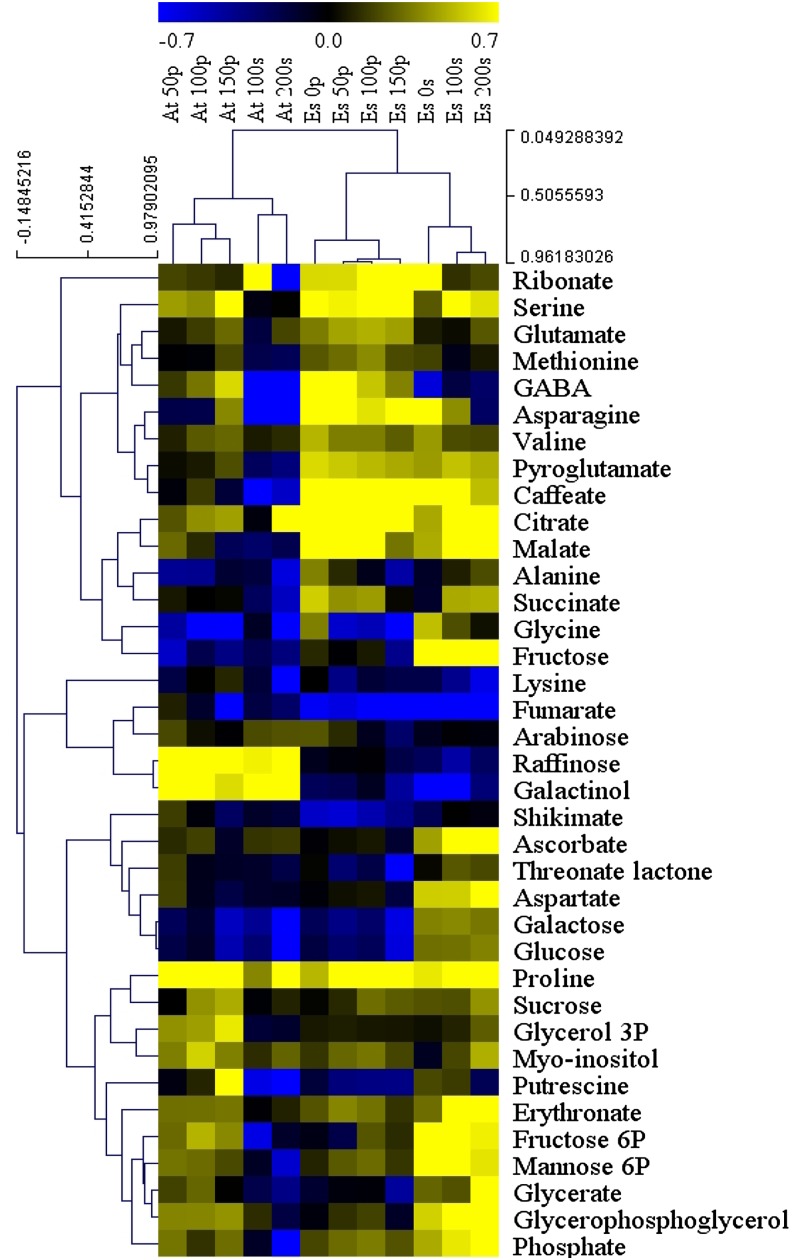
To further explore global effects of salt stress on in vitro and soil-grown plants, metabolic data were expressed as log10 ratios over the Arabidopsis control and then samples were sorted by hierarchical clustering analysis (Fig. 7). Samples clearly clustered according to species first. However, within each species group, clear subclades were observed separated by growth platform. Only within each growth platform clade could the effect of salt treatment be discerned. Thus, salt-mediated metabolic programming in both species was substantially affected by the type of growth medium.
Figure 7.
Hierarchical clustering analysis of metabolites from in vitro- and soil-grown Arabidopsis and E. salsugineum under control and salt-stress conditions. Data are expressed as log10 ratios over Arabidopsis control values. At, Arabidopsis; Es, E. salsugineum; P, plates (in vitro); S, soil. Numbers indicate NaCl concentration.
Common and Growth Platform-Specific Metabolic Responses of Arabidopsis and E. salsugineum under Control and Salt Stress Conditions
To investigate more closely the differences in metabolic responses of the two species on each growth platform, we examined whether the two species exhibited common growth condition-independent and/or condition-specific accumulation of metabolites under control conditions. A few metabolites such as glycerol, myo-inositol, Suc, and Ser showed a statistically significant but marginal difference in comparison with the Arabidopsis control (Fig. 8), and therefore we limited our analysis only to those metabolites exhibiting at least a statistically significant 2-fold change compared with Arabidopsis control plants. A number of metabolites showed common growth platform-independent accumulation, the majority of which displayed higher content in E. salsugineum compared with Arabidopsis (Fig. 8). These included Gly, Val, Thr, Pro, and the nitrogen-rich amino acids Gln and Asn. In addition, the abundance of tricarboxylic acid (TCA) cycle intermediates, citrate and malate, as well as the phenylpropanoid and lignin biosynthesis intermediate, caffeate, was higher in E. salsugineum. Pyro-Glu (5-oxo-Pro) levels were also elevated in E. salsugineum. This metabolite is variously thought to be part of the glutathione recycling pathway, a Glu reservoir and an osmoprotectant (Ohkama-Ohtsu et al., 2008; Kumar and Bachhawat, 2012; Schreiber et al., 2012). On the other hand, levels of the TCA cycle intermediate fumarate, as well as shikimate, a key metabolite involved in aromatic amino acids and phenylpropanoids biosynthesis (Vogt, 2010; Maeda and Dudareva, 2012), were higher in Arabidopsis under control conditions.
Figure 8.
Comparison of metabolites between Arabidopsis and E. salsugineum in vitro- and soil-grown plants under control conditions. Data are representative of similar results from three independent experiments. Asterisks represent significant difference (P ≤ 0.05, Student’s t test) between Arabidopsis and E. salsugineum. Dark gray bars indicate metabolites more abundant in Arabidopsis; light gray bars indicate metabolites more abundant in E. salsugineum.
In contrast to the common metabolic signatures, many metabolites exhibited growth platform-dependent accumulation under control conditions. In particular, sugar and sugar phosphate levels were higher in E. salsugineum compared with Arabidopsis when grown on soil. Thus, Glc and Gal content was higher in E. salsugineum on soil but higher in in vitro-grown Arabidopsis. Glc-6-P, Fru-6-P, and Man-6-P levels were also higher in soil-grown E. salsugineum but on the in vitro platform either exhibited no significant difference between the species or differences were not detected. Glycerophosphoglycerol, the glycerophosphodiester precursor of the sugar alcohol glycerol, showed higher levels in soil-grown E. salsugineum but was not significantly different between the species under in vitro conditions. The amino acid Met was present at higher levels in in vitro-grown E. salsugineum but showed no significant difference between the soil-grown species, whereas Asp content in soil-grown E. salsugineum was higher than in Arabidopsis but under in vitro conditions showed no significant difference between the two species. The nonprotein amino acid GABA exhibited higher levels in E. salsugineum under the in vitro platform but higher levels in Arabidopsis in the soil platform. Other notable growth platform-dependent differences included higher levels of the antioxidant metabolites ascorbate and dehydroascorbate in soil-grown E. salsugineum compared with Arabidopsis but either undetectable (dehydroascorbate) or no significant difference between the species (ascorbate) under in vitro conditions.
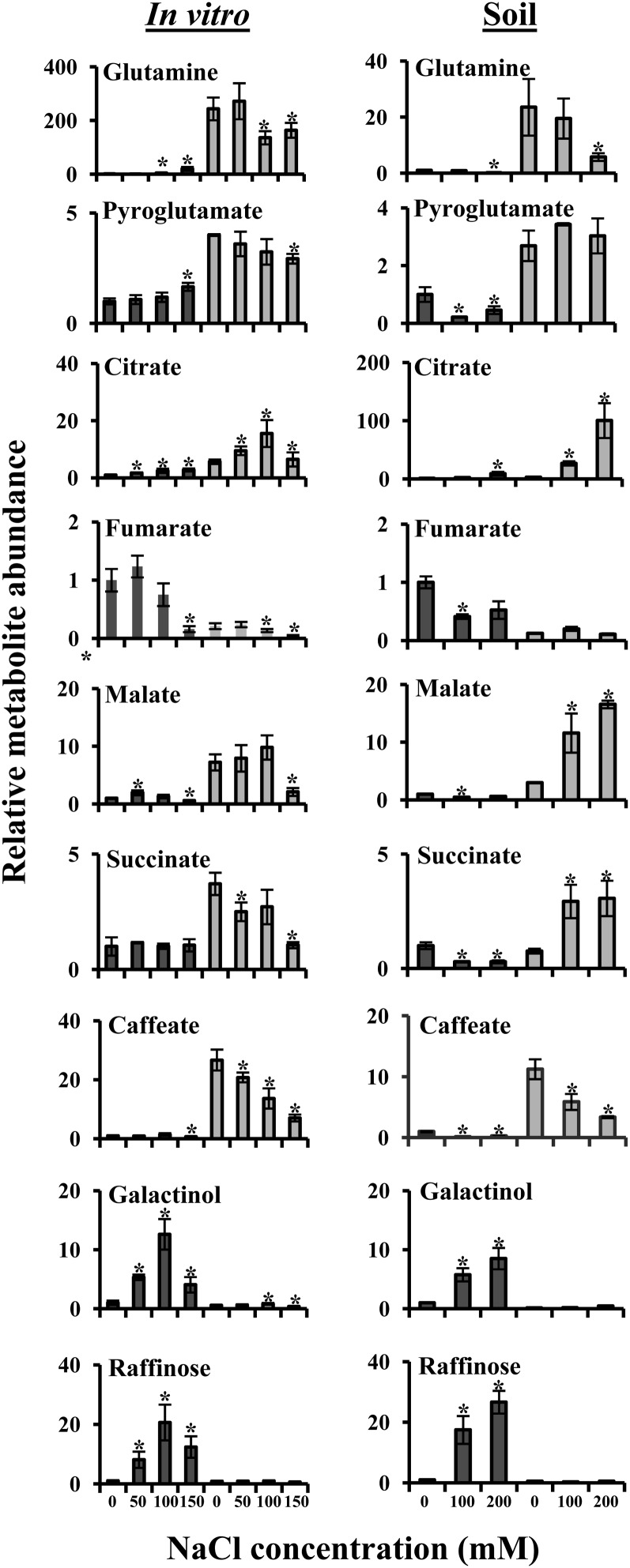
We next examined whether the two species exhibited common growth condition-independent salt stress response signatures. Gln and pyro-Glu exhibited constant high levels in E. salsugineum compared with Arabidopsis, although Gln levels were reduced both in vitro and on soil at higher salt concentrations (Fig. 9). Citrate, succinate, and malate exhibited, in general, high and increasing levels in E. salsugineum compared with constant low levels in Arabidopsis, whereas raffinose and its precursor galactinol displayed high and increasing accumulation in Arabidopsis compared with constitutively low levels in E. salsugineum. Fumarate levels were constantly high in Arabidopsis compared with low levels in E. salsugineum, whereas caffeate exhibited high but decreasing levels in E. salsugineum compared with constitutively low levels in Arabidopsis.
Figure 9.
Growth platform-independent Arabidopsis (dark gray bars) and E. salsugineum (light gray bars) salt response metabolic signatures. Abundance of each metabolite was normalized to the Arabidopsis control level, which was assigned a value of 1. Data are mean (n = 4, in vitro; n = 3, soil) ± sd and are representative of three independent experiments. Asterisks represent significant difference (P ≤ 0.05, Student’s t test) between Arabidopsis or E. salsugineum and their respective controls.
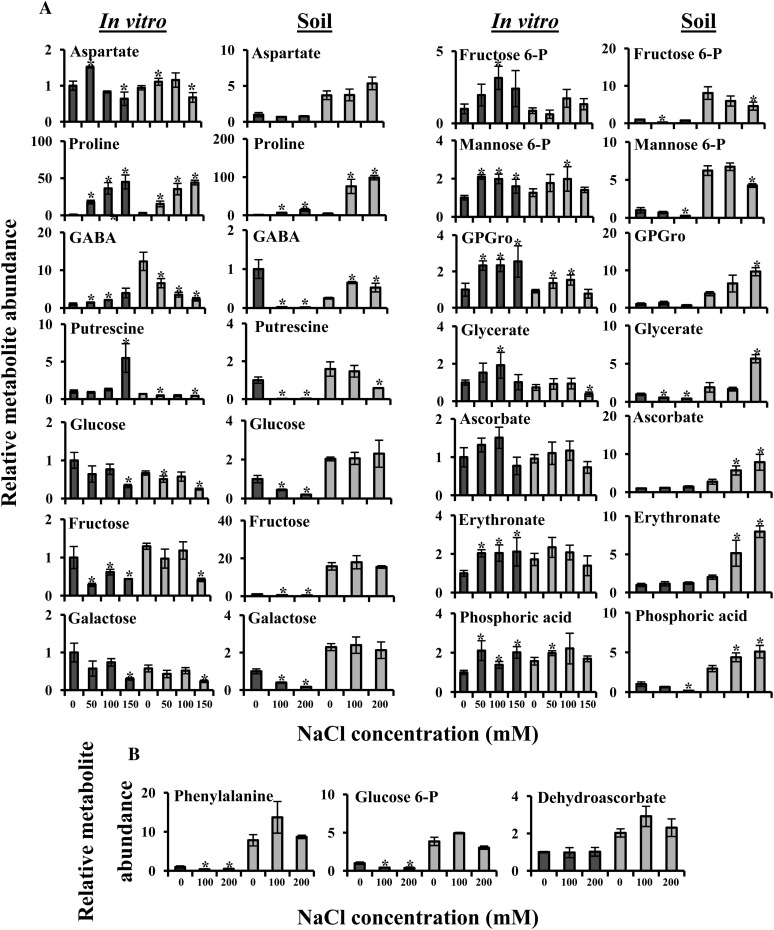
Many metabolites, however, exhibited growth platform-specific responses to salt stress (Fig. 10). Strikingly, a substantial number of metabolites showed generally similar levels in the two in vitro-grown species but high levels in soil-grown E. salsugineum compared with Arabidopsis. These included Asp, Pro, GABA, putrescine, Glc, Fru, Gal, Fru-6-P, Man-6-P, glycerophosphoglycerol, glycerate, ascorbate, erythronate, and inorganic phosphate. In addition, Phe, Glc-6-P, and dehydroascorbate were undetectable in either of the in vitro-grown species but were constitutively high in soil-grown E. salsugineum compared with Arabidopsis. These results suggest that either the in vitro growth conditions repress the accumulation of metabolites in E. salsugineum or that growth on soil represses accumulation of metabolites in Arabidopsis.
Figure 10.
Growth platform-dependent Arabidopsis (dark gray bars) and E. salsugineum (light gray bars) salt response metabolic signatures. A, Metabolites whose accumulation exhibits repression in in vitro-grown compared with soil-grown E. salsugineum. B, Metabolites only detected in soil-grown plants. Abundance of each metabolite was normalized to the Arabidopsis control level, which was assigned a value of 1. Data are mean (n = 4, in vitro; n = 3, soil) ± sd and are representative of three independent experiments. Asterisks represent significant difference (P ≤ 0.05, Student’s t test) between Arabidopsis or E. salsugineum and their respective controls. GPGro, Glycerophosphoglycerol.
DISCUSSION
A Detailed Comparative Phenomics Analysis Unmasks Species-Specific and Experimental Condition-Specific Differences in the Responses of Arabidopsis and E. salsugineum Leaves to Salt Stress
The last decade has seen a growing recognition of the Arabidopsis relative, E. salsugineum, as an important model system for investigating the genetic basis of natural stress tolerance (Amtmann, 2009). The high similarity of E. salsugineum to Arabidopsis in morphology, complementary DNA sequence, and metabolic pathways, yet its dramatic difference in stress tolerance, has allowed fruitful comparative analyses of the stress responses of the two species. Furthermore, a growing body of technical resources has been developed, including ecotype collections (Lee et al., 2012), ESTs (Wong et al., 2005; Zhang et al., 2008), full-length complementary DNA libraries (Du et al., 2008; Taji et al., 2008, 2010), microarrays (Wong et al., 2006), and a genome sequence of both E. salsugineum and E. parvulum (Dassanayake et al., 2011; Wu et al., 2012; Yang et al., 2013).
One aspect lacking in E. salsugineum research has been a detailed phenomics-based study of E. salsugineum growth and development under control and stress conditions. E. salsugineum research has employed a number of growth platforms, including soil in growth rooms, in vitro plate systems, hydroponics, and field conditions (e.g. Volkov et al., 2003; Inan et al., 2004; Kant et al., 2006; Guevara et al., 2012). These systems can lead to vastly different growth phenotypes. For instance, hydroponics yields a very different root phenotype compared with soil (Gibeaut et al., 1997). Both Arabidopsis and E. salsugineum are able to tolerate higher levels of NaCl when grown in soil than when grown in a hydroponic system (Volkov et al., 2003). A recent report demonstrated dramatic differences in growth and development of E. salsugineum (Yukon ecotype) between plants grown in the soil in cabinets and plants grown in native field conditions (Guevara et al., 2012). Cabinet-grown plants produced many basal rosette leaves with a few cauline leaves developing on the main and axillary bolts, whereas in the field only a few or even no rosette leaves were produced. Instead, cauline leaves around the main and axillary bolts were predominant, a phenotype characteristic of vernalized E. salsugineum.
It is thus essential to establish reliable, nondestructive monitoring of E. salsugineum growth and development that would allow detailed, quantitative phenotypes to be linked with genotypic variation in stress tolerance mechanisms, as well as to identify new growth traits that may be masked by conventional growth analyses or manifest as relatively small differences. To begin this task, we employed the growth-based phenotypic analysis platform for Arabidopsis developed by Boyes et al. (2001; Table I) to compare Arabidopsis and E. salsugineum leaf growth and development under control and salt stress conditions using two different growth platforms commonly used in laboratory studies: an in vitro plate system and a pot-based soil system, both of which were placed in the same growth room.
Our results showed that growth responses of both species to salt stress differed depending upon whether plants were grown in vitro or in soil. Leaf emergence was affected in a similar way in both species grown in vitro but the same effects observed in Arabidopsis occurred at higher salt concentrations in E. salsugineum (Fig. 1A; Supplemental Table S1). Thus, mild stress (50 mm NaCl) had no effect on either species, 100 mm NaCl caused a slowdown in Arabidopsis leaf emergence with no effect on E. salsugineum, 150 mm NaCl led to a slowdown in E. salsugineum leaf emergence and inhibition of Arabidopsis leaf emergence after the second true leaf, and 200 mm NaCl led to inhibition of E. salsugineum leaf emergence after the fourth true leaf (Arabidopsis was unable to grow at all on 200 mm NaCl plates). In contrast to our in vitro experiments, leaf emergence of both species was unaffected on soil at all stress levels (only E. salsugineum could grow at 300 mm NaCl; Fig. 1B; Supplemental Table S2). These results are in agreement with research showing that under soil water deficit, plasticity of leaf production was low in 25 Arabidopsis accessions (Aguirrezabal et al., 2006).
Our growth analysis unmasked an effect of salt stress on E. salsugineum development: a reduction in leaf area even under mild salt stress. A salt-mediated reduction in leaf area was observed for Arabidopsis and E. salsugineum in both growth platforms (Figs. 3 and 4; Supplemental Fig. S3). Almost all previous reports measuring the effect of salt stress on E. salsugineum growth have focused on fresh and dry weight parameters (e.g. Inan et al., 2004; Kant et al., 2006; Ghars et al., 2008), which exhibit no reduction in E. salsugineum under mild salt stress (e.g. 50, 75, or 100 mm NaCl). One report did observe a reduction in E. salsugineum leaf area during a screen of salt tolerance of wild Arabidopsis relatives (Orsini et al., 2010), but this was only measured at a higher NaCl concentration (150 mm). One notable difference between growth platforms was that whereas both species exhibited a dose-dependent reduction in leaf area with in vitro-grown plants, on soil, E. salsugineum (and possibly Arabidopsis) displayed a two-step response (Fig. 3; Supplemental Figs. S3 and S4). Thus, there was a 30% reduction in E. salsugineum total rosette and hull area at 50 and 100 mm NaCl with a further reduction to about 50% of the control at 200 and 300 mm NaCl. The physiological basis for such a two-step response is unclear at present but could suggest that E. salsugineum (and perhaps Arabidopsis) may trigger different tolerance mechanisms at certain stress thresholds or that the second step of the response is a symptom of damage.
It is now generally accepted that plants actively reduce their growth in response to stress and that this growth reduction is independent of photosynthesis (Fricke et al., 2006; Hummel et al., 2010; Skirycz and Inzé, 2010; Skirycz et al., 2010, 2011; Baerenfaller et al., 2012). This is particularly true under mild stresses where an adaptive response is probably observed rather than damage characteristic of the extreme stresses often applied in stress studies. Thus, the slowdown in leaf emergence during our in vitro experiments and the leaf area reduction on both growth platforms are likely to be adaptive responses at lower levels of salt stress where symptoms of extreme stress such as leaf curling or anthocyanin production are absent. At higher levels of stress, however, it is possible that the complete cessation of leaf emergence in in vitro-grown Arabidopsis (150 mm NaCl) and E. salsugineum (200 mm NaCl) and the large reduction in leaf area in both species on both growth platforms is not an adaptive response but rather a symptom of stress-induced damage.
In dicotyledon species such as Arabidopsis and E. salsugineum, the final leaf size depends upon the number of founder cells recruited from the shoot apical meristem and the subsequent rates of cell division and expansion of these cells (Beemster et al., 2005). Previous reports have demonstrated that water deficit-mediated reduction in leaf size can be attributed to decreases in both cell number and cell size early in leaf development but that both cell division and expansion rates are adaptable and return to control levels within a few days after leaf initiation, particularly under mild osmotic stress (Aguirrezabal et al., 2006; Skirycz et al., 2010). Our data also showed that salt stress leads to a reduction in cell number and cell size in both species (Fig. 5; Supplemental Table S3). However, the fall in cell size is greater in Arabidopsis than in E. salsugineum under higher salt concentrations. At the highest salt concentration, E. salsugineum cell number reduction is comparable with Arabidopsis, whereas cell size is much less affected in E. salsugineum. This could be due to the ability of E. salsugineum to constitutively maintain a negative leaf osmotic potential in unstressed conditions and to greatly reduce leaf osmotic potential and maintain turgor pressure at high levels of salt stress (Inan et al., 2004), thereby maintaining a greater rate and/or duration of cell expansion under salt stress.
Another phenotype that was revealed by investigating the response to salt stress of each individual leaf showed that whereas only middle to older Arabidopsis leaves were affected under mild stresses, in E. salsugineum it was the younger leaves that showed a reduction in leaf area (Fig. 4). Only as the degree of salt stress increased were other leaves affected sequentially: old-to-young leaves in Arabidopsis and the opposite in E. salsugineum. It is well known that salt stress has different effects on old and young tissues (Vera-Estrella et al., 2005; Yasar et al., 2006; Munns and Tester, 2008; Wang et al., 2012). In rice (Oryza sativa), for instance, young leaves are protected by accumulation of Na+ and Cl– ions in the older leaves possibly due to differential expression of ion transporter genes (Wang et al., 2012). Similarly, in E. salsugineum, the major site of Na+ accumulation occurs in old leaves, followed by young leaves, taproots, and lateral roots (Vera-Estrella et al., 2005). This is perhaps due to high basal and salt-induced expression of the SOS1 Na+/H antiporter (Taji et al., 2004; Vera-Estrella et al., 2005; Kant et al., 2006; Oh et al., 2009, 2010; Taji et al., 2010). Therefore, under very mild salt stress the younger E. salsugineum leaves may not be subjected to the ionic stress component of salt stress but rather perceive the osmotic stress component, a signal that can lead to rapid reduction in leaf growth, as an adaptive, protective response (Skirycz et al., 2010). It should be noted, however, that in their natural habitat E. salsugineum plants are likely exposed to salt throughout their whole life cycle. Nevertheless, even under such conditions it is possible that leaves at various stages of development are affected differently. Overall, our results emphasize the growing realization of the importance of future molecular studies focusing on comparing the response to stress of leaves at different developmental stages. Indeed, it has recently been shown that the transcriptome and metabolome of leaves respond differently to stress depending on whether the leaf is at the proliferating expanding or mature stage of development (Skirycz et al., 2010).
Salt-Mediated Metabolic Reprogramming in Arabidopsis and E. salsugineum Depends on Growth Platform
Our metabolic profiling analysis comparing control and salt-stressed Arabidopsis and E. salsugineum from in vitro- and soil-based growth systems revealed, as expected, many differences in metabolite content and abundance. In agreement with previous reports (Lugan et al., 2010; Guevara et al., 2012), our PCA showed that species was the predominant factor explaining differences in metabolic response to salt stress (Fig. 6). However, hierarchical clustering analysis demonstrated that within each species metabolism could be separated on the basis of growth platform (Fig. 7). Because all tissue in all experiments was harvested at the same time after lights on, differences cannot be explained by large diurnal variations in metabolite content found in Arabidopsis and E. salsugineum (Gibon et al., 2006; Guevara et al., 2012). Thus, although E. salsugineum exhibited greater salt tolerance on both growth systems, its overall metabolic response was dependent upon growth platform. This finding is in agreement with a recent paper showing different metabolic responses between soil-grown E. salsugineum raised in growth cabinets and E. salsugineum plants grown in their native habitat (Guevara et al., 2012).
Common Growth Platform-Independent Metabolic Signatures May Reflect Core Stress Tolerance Mechanisms in E. salsugineum
Although the overall metabolic response of Arabidopsis and E. salsugineum depended upon growth platform, we nevertheless found several metabolites showing common growth platform-independent patterns of abundance under control and salt-stress conditions (Figs. 8 and 9) that might represent core stress tolerance mechanisms in E. salsugineum. Higher malate and citrate levels but constitutively low fumarate, raffinose, and galactinol content in E. salsugineum compared with Arabidopsis were observed. These features are something of a metabolic signature of E. salsugineum and can also be observed under hydroponics (Lugan et al., 2010) and in response to low-nitrogen stress (Kant et al., 2008). A large malate pool in E. salsugineum could allow increased generation of oxaloacetate to provide carbon skeletons for increased amino acid synthesis (Coruzzi and Last, 2000; Siedow and Day, 2000). Under control conditions, E. salsugineum possesses higher levels of Asn, Gln, Gly, Pro, Ser, Thr, and Val, whereas under salt stress Gln and pyro-Glu remain constitutively high. However, continuous export of oxaloacetate could eventually lead to a deficiency in TCA cycle intermediates such as the observed reduced accumulation of fumarate. This could require anaplerotic replenishment of citric acid intermediates such as production of malate in the cytosol from Glc and Fru via phosphoenolpyruvate. Malate can then be transported back into the mitochondria and reenter the citric acid cycle (Dennis and Blakeley, 2000; Siedow and Day, 2000). However, increased levels of Glc and Fru that could provide higher malate accumulation was only observed in soil-grown E. salsugineum. On the other hand, the high malate levels observed in E. salsugineum could arise from reduced malate catabolism or its reduced use as a substrate in other reactions, irrespective of Glc or Fru availability. An alternative way that TCA cycle intermediate levels could be maintained in E. salsugineum might be via the citramalate shunt, whereby malate is converted to pyruvate by malic enzyme, which is then converted to acetyl-CoA to increase citrate levels (Steinhauser et al., 2012).
Raffinose and its precursor galactinol are important osmoprotectants that accumulate during stress in a variety of plant species (Taji et al., 2002; Farrant et al., 2009; dos Santos et al., 2011; Oliver et al., 2011; Saito and Yoshida, 2011; Valluru and Van den Ende, 2011; Bai et al., 2012) and also function to scavenge hydroxyl radicals (Taji et al., 2002; Nishizawa et al., 2008; Nishizawa-Yokoi et al., 2008). It is not clear why galactinol and raffinose do not accumulate in E. salsugineum in response to stress. It is possible that Arabidopsis, which is affected to a far greater extent by salt stress than E. salsugineum, generates raffinose family oligosaccharides as a tolerance mechanism. On the other hand, E. salsugineum may not yet have reached the level of stress requiring raffinose and galactinol production. Alternatively, the absence of raffinose family oligosaccharides in E. salsugineum might be a distinctive trait of this naturally stress-tolerant Brassica species similar to the desiccation-tolerant spike moss Selaginella lepidophylla (Yobi et al., 2012).
Repression of In Vitro-Grown E. salsugineum Metabolite Accumulation Could Reflect Perturbation of Carbon and Nitrogen Balance by Exogenous Suc
The dominant growth platform-specific metabolic difference that we observed was manifested as repression of accumulation of a considerable number of metabolites in E. salsugineum grown in the in vitro system compared with the soil platform (or conversely, repression of metabolites in soil-grown Arabidopsis) under control and salt-stress conditions (Fig. 10). A number of these metabolites are known to be involved in stress responses, including the polyamine putrescine (Takahashi and Kakehi, 2010; Alet et al., 2011; Marco et al., 2011), the antioxidants ascorbate and dehydroascorbate (Huang et al., 2005; Gallie, 2013), GABA (Bouché and Fromm, 2004; Fait et al., 2008; Obata and Fernie, 2012), and Pro (Kavi-Kishor et al., 1995; Hong et al., 2000; Kant et al., 2006; Verbruggen and Hermans, 2008). It is tempting to speculate that the reduction in such a set of stress-tolerance metabolites may account for the paling of leaf margins in in vitro-grown E. salsugineum from stage 1.04 onward even under control conditions (Fig. 2B) and even though E. salsugineum still retained greater stress tolerance than Arabidopsis. Of particular interest were the reduced levels of Pro in control and salt-treated E. salsugineum grown in vitro. Increased levels of Pro, a compatible osmolyte and osmoprotectant, in control and salt-treated E. salsugineum compared with Arabidopsis have been suggested to contribute to E. salsugineum salt tolerance (Inan et al., 2004; Taji et al., 2004; Kant et al., 2006; Lee et al., 2012). However, all of these reports of increased E. salsugineum Pro levels have employed soil-grown plants in growth rooms/cabinets or hydroponics. In contrast, Guevara et al. (2012) reported that E. salsugineum grown on soil but in its native habitat does not exhibit a salt-mediated increase in Pro levels compared with cabinet-grown plants. They further showed that under nitrogen-limiting conditions (characteristic of soils in E. salsugineum’s natural habitat) Pro no longer accumulates in salt-treated, cabinet-grown plants. Instead, there is accumulation of carbohydrates, suggesting an excess of carbon skeletons. Although we did not grow E. salsugineum under low-nitrogen conditions, our in vitro growth medium did contain Suc, which may have increased the carbon:nitrogen ratio in a similar manner to reducing nitrogen levels. Such changes in carbon:nitrogen ratio can have profound effects upon the reciprocal regulation of carbon and nitrogen metabolism by carbon and nitrogen signals (Oliveira et al., 2001; Price et al., 2004). For instance, Suc and Glc have been shown to repress expression of genes involved in nitrogen metabolism, including biosynthesis of amino acids (Lam et al., 1994; Melo-Oliveira et al., 1996; Lam et al., 1998; Oliveira et al., 2001; Price et al., 2004; Sahrawy et al., 2004).
E. salsugineum Maintains Salt Tolerance Despite Growth Platform-Specific Phenotypes and Metabolic Responses
In our phenomics and metabolomics analysis, we have shown both phenotypic and metabolic differences when Arabidopsis and E. salsugineum plants are grown in vitro versus on soil. Despite these differences, E. salsugineum exhibits greater salt tolerance on both growth platforms. It is possible that the existence of core growth platform-independent, common metabolic signatures representing essential stress-response mechanisms in E. salsugineum are sufficient to maintain salt tolerance in this extremophile. Such core signatures might be good targets for discovery of new stress tolerance determinants (e.g. genes and promoters). On the other hand, Guevara et al. (2012) suggest that phenotypic and metabolic adaptive plasticity observed in E. salsugineum between different growth conditions that still facilitate salt tolerance is an inherent trait that allows the flexibility required for an extremophile lifestyle.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Stress Treatments
For plate-based in vitro analysis, Arabidopsis (Arabidopsis thaliana, Columbia-0 ecotype) and Eutrema salsugineum (Shandong ecotype) seeds were surface sterilized with 50% commercial bleach for 5 min, and then rinsed four times with sterile water. Sterilized seeds were suspended in 0.12% agarose and sown on nutrient agar plates containing 0.5 × MS (Murashige and Skoog, 1962) medium, pH 5.7, 0.5 g L−1 MES, 2% (w/v) Suc, and 0.8% (w/v) agar (Duchefa Biochemie). Plates were overlaid with autoclaved mesh (Arta Art Graphic and Office Supplies, Ltd.; http://www.arta-israel.co.il) and 30 seeds were sown uniformly on each plate. Seeds were stratified at 4°C (Arabidopsis, 3 d; E. salsugineum, 7 d) and plates incubated in the growth room (16-h light (130 µmol m−2 s−1)/8-h dark) for 24 (Arabidopsis) or 48 (E. salsugineum) h. Sowing of Arabidopsis and E. salsugineum seeds was staggered so that germination of both species occurred at approximately the same time. Germination of seeds was verified under a binocular microscope, 15 seeds that germinated uniformly were selected, and the rest were removed by forceps. The mesh, along with 15 seeds at the stage of radical emergence, was transferred to fresh 0.5 × MS plates supplemented with various NaCl concentrations. Following stratification and transfer to the growth room, daily measurements were performed until the 1.06 stage of development (Boyes et al., 2001) to evaluate changes in growth of the seedlings. Each day, plates were shuffled so that all plants received equal illumination and to remove shelf position effects.
For verification that transfer by mesh does not affect seedling growth, seeds were sterilized and sown on MS plates. After germination, plates with seeds were randomly divided into two groups and seeds were thinned out. The mesh from the plates of one group was transferred to fresh MS plates while in the second group the mesh was left untouched. Plates were kept in the growth room for 16 d after stratification when plates were photographed with a Canon IXUS 70 digital camera and the fresh weight of pooled plants on each plate was determined. To test the effect of NaCl on germination of Arabidopsis and E. salsugineum, seeds were sterilized and sown (100 seeds per plate) on 0.5 × MS supplemented with one of the following salt concentrations: 0, 50, 100, 150, or 200 mm NaCl. After stratification, plates were incubated in the growth room. Germination was recorded at 7 d after stratification.
For soil-based growth analysis, seeds were surface sterilized, sown on MS plates without mesh, and stratified for 4 d at 4°C before transfer to the growth room. Five-day-old seedlings (cotyledons fully open) were transferred to pots containing Arabidopsis soil growth medium (Weizmann Institute of Science) and irrigated to field capacity with 1 g L−1 of 20-20-20 NPK + micronutrients solution (Haifa chemicals). Flats containing pots were placed in the growth room under the same conditions as for the plate experiments. Flats were covered with plastic domes for 1 to 2 d, which were then gradually removed to allow seedlings to harden.
Salt treatments commenced at stage 1.02 with 50% of the final salt concentration in order to avoid salt shock. One hundred percent of each salt level was applied at stage 1.04. Pots were irrigated with 1 g L−1 of 20-20-20 NPK + micronutrients solution supplemented with 0, 50, 100, 200, or 300 mm NaCl (only E. salsugineum was subjected to 300 mm NaCl). Irrigation was performed by allowing pots to stand in irrigation solution for 30 min and then each pot was irrigated from above with solution of the respective NaCl concentration until leaching occurred to ensure a uniform salt concentration throughout the soil. Evaluations of changes in growth were performed daily until stage 1.10.
Growth Measurements, Cell Size, and Cell Number
For calculation of hull area, plates containing seedlings at stage 1.06 or soil-grown plants at stage 1.10 were photographed with a digital camera (Canon IXUS 70) and acquired images were analyzed using ImageJ software, version 1.43 (http://rsbweb.nih.gov/ij/). For leaf area measurements of soil-grown plants, leaves were detached, placed between two transparency films (Graphic Vision Media), and scanned. Leaf area was calculated with ImageJ software.
For cell size and cell number, adaxial imprints of the surface of the fifth leaf of four soil-grown plants for each treatment at stage 1.10 were made by the dental rubber impression technique (Weyers and Johansen, 1985; Lawson et al., 1998). In brief, the adaxial side of the leaf was covered with silicone impression material (Elite HD+). After the material solidified, the leaf was carefully peeled from the imprint and leaf area was determined using ImageJ software. The imprint was then coated with transparent nail polish and acquired films were mounted on a glass slide in a drop of water, covered with a cover slip, observed, and photographed under a light microscope (Zeiss Axioimager LED microscope with Zeiss Axiocam HRC camera). The photographs were processed in AxioVision 4.6 (Zeiss) and cell area was calculated with ImageJ software. Photographs of 15 areas within each leaf were used for determination of cell area (the areas of at least 50 cells per leaf were used for average cell area calculations). Leaf area and cell area were used to calculate total number of cells per leaf.
Metabolite Profiling and Analysis
For metabolite profiling, plants were harvested at stage 1.06 (stage 1.04 for Arabidopsis in the 150 mm NaCl treatment) for in vitro-grown plants and stage 1.10 for soil-grown plants. In vitro-grown plants from each of four replicate plates were pooled (15 to 20 pooled plants per plate), whereas in the soil experiments leaves 5 and 6 were pooled into three biological replicates (leaves from approximately five plants per pool). Tissue was snap frozen in liquid nitrogen and stored at –80°C until further analysis.
Metabolite extraction was performed according to Lisec et al. (2006) with minor modifications. Briefly, 1 mL of methanol and 42 µL of ribitol (0.2 mg mL−1) were added to approximately 100 mg ground tissue followed by agitation for 15 min. Ribitol was added to the mixture as an internal standard. Samples were centrifuged at 14,000 rpm (Eppendorf microcentrifuge, 5417R) and the supernatant was transferred to a fresh Eppendorf tube. Samples were shaken at room temperature for 15 min, and 536 µL of chloroform and 1,100 µL of water were added to each tube followed by vortexing. All samples were centrifuged at 4,000 rpm (Eppendorf microcentrifuge, 5417R). One hundred microliters of the upper phase was dried in a vacuum concentrator (Eppendorf Concentrator Plus) for further derivatization. In cases where the weight of analyzed samples was other than 100 mg, different volumes of reagents were used but the solvent-to-tissue ratio was conserved.
Derivatization was performed strictly according to the protocol of Lisec et al. (2006). Forty microliters of freshly prepared methoxyaminhydrochloride in pyridine (20 mg mL−1) was added to each sample followed by agitation at 37°C for 2 h. Seventy microliters of N-methyl-N-trifluoroacetamide and 7 µL of alkane mixture prewarmed to 70°C were added to the samples, which were then agitated for 30 min at 37°C followed by transfer to autosampler vials (Thermo Scientific). Separation was carried out on a Thermo Scientific DSQ II GC/MS using a FactorFour Capillary VF-5ms column. Acquired chromatograms and mass spectra were evaluated using Xcalibur (version 2.0.7) software and metabolites were identified and annotated using the Mass Spectral and Retention Time Index libraries available from the Max-Planck Institute for Plant Physiology, Golm, Germany (http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/msri/gmd_msri.html). The level of metabolites was calculated by normalizing the intensity of the peak of each metabolite to the ribitol standard. PCA, ANOVA, Student’s t tests, and hierarchical clustering analysis were performed using the MultiExperiment Viewer 4.8.1 software (Saeed et al., 2003).
Supplemental Material
The following materials are available in the online version of this article.
Supplemental Figure S1. The effect of NaCl on germination of Arabidopsis and E. salsugineum.
Supplemental Figure S2. The effect of transfer by mesh on growth and development of Arabidopsis and E. salsugineum.
Supplemental Figure S3. Actual hull and rosette areas of in vitro- and soil-grown Arabidopsis and E. salsugineum in response to salt stress.
Supplemental Figure S4. The effect of salt stress on the area of individual soil-grown Arabidopsis and E. salsugineum leaves.
Supplemental Figure S5. Microscopic images of adaxial Arabidopsis and E. salsugineum leaf surfaces.
Supplemental Table S1. Time of occurrence of growth stages in control and salt-treated in vitro-grown Arabidopsis and E. salsugineum seedlings.
Supplemental Table S2. Time of occurrence of growth stages in control and salt-treated soil-grown Arabidopsis and E. salsugineum seedlings.
Supplemental Table S3. Average cell numbers and cell sizes of soil-grown Arabidopsis and E. salsugineum plants under control and salt-stress conditions.
Supplemental Table S4. Relative contents of metabolites detected by gas chromatography-quantitative mass spectrometry in polar extracts of in vitro-grown Arabidopsis and E. salsugineum seedlings.
Supplemental Table S5. Relative contents of metabolites detected by gas chromatography-quantitative mass spectrometry in soil-grown leaf polar extracts of Arabidopsis and E. salsugineum plants.
Supplemental Table S6. Absolute metabolite eigenvalues for the first two components of the PCA analysis of in vitro-grown Arabidopsis and E. salsugineum seedlings.
Supplemental Table S7. Absolute metabolite eigenvalues for the first two components of the PCA analysis of soil-grown Arabidopsis and E. salsugineum plants.
Supplemental Table S8. Two-way ANOVA (P < 0.01) of metabolite response of in vitro-grown Arabidopsis and E. salsugineum seedlings.
Supplemental Table S9. Two-way ANOVA (P < 0.01) of metabolite response of soil-grown Arabidopsis and E. salsugineum plants.
Supplementary Material
Acknowledgments
We thank Ruti Shaked, Tania Acuña, and Noga Sikron for excellent technical help.
Glossary
- PCA
principal component analysis
- TCA
tricarboxylic acid
- MS
Murashige and Skoog
- GABA
γ-aminobutyric acid
- DAS
d after stratification
References
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C. (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29: 2216–2227 [DOI] [PubMed] [Google Scholar]
- Alet AI, Sanchez DH, Cuevas JC, Del Valle S, Altabella T, Tiburcio AF, Marco F, Ferrando A, Espasandín FD, González ME, Ruiz OA, Carrasco P. (2011) Putrescine accumulation in Arabidopsis thaliana transgenic lines enhances tolerance to dehydration and freezing stress. Plant Signal Behav 6: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A. (2009) Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol Plant 2: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Massonnet C, Walsh S, Baginsky S, Bühlmann P, Hennig L, Hirsch-Hoffmann M, Howell KA, Kahlau S, Radziejwoski A, et al. (2012) Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol Syst Biol 8: 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Sikron N, Gendler T, Kazachkova Y, Barak S, Grafi G, Khozin-Goldberg I, Fait A. (2012) Ecotypic variability in the metabolic response of seeds to diurnal hydration-dehydration cycles and its relationship to seed vigor. Plant Cell Physiol 53: 38–52 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M. (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Fromm H. (2004) GABA in plants: just a metabolite? Trends Plant Sci 9: 110–115 [DOI] [PubMed] [Google Scholar]
- Bressan RA, Zhang C, Zhang H, Hasegawa PM, Bohnert HJ, Zhu J-K. (2001) Learning from the Arabidopsis experience. The next gene search paradigm. Plant Physiol 127: 1354–1360 [PMC free article] [PubMed] [Google Scholar]
- Chern C-G, Fan M-J, Yu S-M, Hour A-L, Lu P-C, Lin Y-C, Wei F-J, Huang S-C, Chen S, Lai M-H, et al. (2007) A rice phenomics study—phenotype scoring and seed propagation of a T-DNA insertion-induced rice mutant population. Plant Mol Biol 65: 427–438 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Last RL (2000) Amino acids. In RG Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 358-410 [Google Scholar]
- Dassanayake M, Oh D-H, Haas JS, Hernandez A, Hong H, Ali S, Yun D-J, Bressan RA, Zhu J-K, Bohnert HJ, et al. (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis DT, Blakeley SD (2000) Carbohydrate metabolism. In RG Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 630–675 [Google Scholar]
- dos Santos TB, Budzinski IGF, Marur CJ, Petkowicz CLO, Pereira LFP, Vieira LGE. (2011) Expression of three galactinol synthase isoforms in Coffea arabica L. and accumulation of raffinose and stachyose in response to abiotic stresses. Plant Physiol Biochem 49: 441–448 [DOI] [PubMed] [Google Scholar]
- Du J, Huang Y-P, Xi J, Cao M-J, Ni W-S, Chen X, Zhu JK, Oliver DJ, Xiang CB. (2008) Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J 56: 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR. (2008) Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci 13: 14–19 [DOI] [PubMed] [Google Scholar]
- Farrant JM, Lehner A, Cooper K, Wiswedel S. (2009) Desiccation tolerance in the vegetative tissues of the fern Mohria caffrorum is seasonally regulated. Plant J 57: 65–79 [DOI] [PubMed] [Google Scholar]
- Fricke W, Akhiyarova G, Wei W, Alexandersson E, Miller A, Kjellbom PO, Richardson A, Wojciechowski T, Schreiber L, Veselov D, et al. (2006) The short-term growth response to salt of the developing barley leaf. J Exp Bot 57: 1079–1095 [DOI] [PubMed] [Google Scholar]
- Furbank RT, Tester M. (2011) Phenomics—technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16: 635–644 [DOI] [PubMed] [Google Scholar]
- Gallie DR. (2013) The role of L-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J Exp Bot 64: 433–443 [DOI] [PubMed] [Google Scholar]
- Ghars MA, Parre E, Debez A, Bordenave M, Richard L, Leport L, Bouchereau A, Savouré A, Abdelly C. (2008) Comparative salt tolerance analysis between Arabidopsis thaliana and Thellungiella halophila, with special emphasis on K+/Na+ selectivity and proline accumulation. J Plant Physiol 165: 588–599 [DOI] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR. (1997) Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Li P, Ma S, Indu Rupassara S, Bohnert HJ. (2005) Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J 44: 826–839 [DOI] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux J-J, Rolland G, Bouchier-Combaud S, Lebaudy A, et al. (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169: 623–635 [DOI] [PubMed] [Google Scholar]
- Griffith M, Timonin M, Wong ACE, Gray GR, Akhter SR, Saldanha M, Rogers MA, Weretilnyk EA, Moffatt B. (2007) Thellungiella: an Arabidopsis-related model plant adapted to cold temperatures. Plant Cell Environ 30: 529–538 [DOI] [PubMed] [Google Scholar]
- Guevara DR, Champigny MJ, Tattersall A, Dedrick J, Wong CE, Li Y, Labbe A, Ping C-L, Wang Y, Nuin P, et al. (2012) Transcriptomic and metabolomic analysis of Yukon Thellungiella plants grown in cabinets and their natural habitat show phenotypic plasticity. BMC Plant Biol 12: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Lakkineni K, Zhang Z, Verma DPS. (2000) Removal of feedback inhibition of Δ1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol 122: 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, He W, Guo J, Chang X, Su P, Zhang L. (2005) Increased sensitivity to salt stress in an ascorbate-deficient Arabidopsis mutant. J Exp Bot 56: 3041–3049 [DOI] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, Piques M, Rolland G, Dauzat M, Christophe A, Pervent M, Bouteille M, Stitt M, et al. (2010) Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: An integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiol 154: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inan G, Zhang Q, Li P, Wang Z, Cao Z, Zhang H, Zhang C, Quist TM, Goodwin SM, Zhu J, et al. (2004) Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol 135: 1718–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Kant P, Raveh E, Barak S. (2006) Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ 29: 1220–1234 [DOI] [PubMed] [Google Scholar]
- Kant S, Bi Y-M, Weretilnyk E, Barak S, Rothstein SJ. (2008) The Arabidopsis halophytic relative Thellungiella halophila tolerates nitrogen-limiting conditions by maintaining growth, nitrogen uptake, and assimilation. Plant Physiol 147: 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi-Kishor P, Hong Z, Miao GH, Hu C-AA, Verma DPS. (1995) Overexpression of Δ1-pyrroline-5-carboxylate synthetase increases proline production and confers osmotolerance in transgenic plants. Plant Physiol 108: 1387–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Bachhawat AK. (2012) Pyroglutamic acid: throwing light on a lightly studied metabolite. Curr Sci 102: 288–297 [Google Scholar]
- Kuromori T, Takahashi S, Kondou Y, Shinozaki K, Matsui M. (2009) Phenome analysis in plant species using loss-of-function and gain-of-function mutants. Plant Cell Physiol 50: 1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H-M, Peng SS-Y, Coruzzi GM. (1994) Metabolic regulation of the gene encoding glutamine-dependent asparagine synthetase in Arabidopsis thaliana. Plant Physiol 106: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H-M, Hsieh M-H, Coruzzi G. (1998) Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana. Plant J 16: 345–353 [DOI] [PubMed] [Google Scholar]
- Lamdan NL, Attia Z, Moran N, Moshelion M. (2012) The Arabidopsis-related halophyte Thellungiella halophila: boron tolerance via boron complexation with metabolites? Plant Cell Environ 35: 735–746 [DOI] [PubMed] [Google Scholar]
- Lancashire PD, Bleiholder H, van der Boom T, Langeluddeke P, Stauss R, Weber E, Witzenberger A. (1991) A uniform decimal code for growth stages of crops and weeds. Ann Appl Biol 119: 561–601 [Google Scholar]
- Lawson T, James W, Weyers J. (1998) A surrogate measure of stomatal aperture. J Exp Bot 49: 1397–1403 [Google Scholar]
- Lee YP, Babakov A, de Boer B, Zuther E, Hincha DK. (2012) Comparison of freezing tolerance, compatible solutes and polyamines in geographically diverse collections of Thellungiella sp. and Arabidopsis thaliana accessions. BMC Plant Biol 12: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lugan R, Niogret M-F, Leport L, Guégan J-P, Larher FR, Savouré A, Kopka J, Bouchereau A. (2010) Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. Plant J 64: 215–229 [DOI] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. (2012) The shikimate pathway and aromatic amino Acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Marco F, Alcázar R, Tiburcio AF, Carrasco P. (2011) Interactions between polyamines and abiotic stress pathway responses unraveled by transcriptome analysis of polyamine overproducers. OMICS 15: 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo-Oliveira R, Oliveira IC, Coruzzi GM. (1996) Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc Natl Acad Sci USA 93: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Iwasaki Y, Kitano H, Itoh JI, Maekawa M, Murata K, Yatou O, Nagato Y, Hirohiko H. (2007) A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol Biol 63: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Yabuta Y, Shigeoka S. (2008) The contribution of carbohydrates including raffinose family oligosaccharides and sugar alcohols to protection of plant cells from oxidative damage. Plant Signal Behav 3: 1016–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Fernie AR. (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69: 3225–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D-H, Leidi E, Zhang Q, Hwang S-M, Li Y, Quintero FJ, Jiang X, D’Urzo MP, Lee SY, Zhao Y, et al. (2009) Loss of halophytism by interference with SOS1 expression. Plant Physiol 151: 210–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D-H, Dassanayake M, Haas JS, Kropornika A, Wright C, d’Urzo MP, Hong H, Ali S, Hernandez A, Lambert GM, et al. (2010) Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol 154: 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D-H, Dassanayake M, Bohnert HJ, Cheeseman JM. (2012) Life at the extreme: lessons from the genome. Genome Biol 13: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Oikawa A, Zhao P, Xiang C, Saito K, Oliver DJ. (2008) A γ-glutamyl transpeptidase-independent pathway of glutathione catabolism to glutamate via 5-oxoproline in Arabidopsis. Plant Physiol 148: 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver MJ, Guo L, Alexander DC, Ryals JA, Wone BWM, Cushman JC. (2011) A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell 23: 1231–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira IC, Brenner E, Chiu J, Hsieh M-H, Kouranov A, Lam H-M, Shin MJ, Coruzzi G. (2001) Metabolite and light regulation of metabolism in plants: lessons from the study of a single biochemical pathway. Braz J Med Biol Res 34: 567–575 [DOI] [PubMed] [Google Scholar]
- Orsini F, Paino D'Urzo M, Inan G, Serra S, Oh DH, Mickelbart MV, Consiglio F, Li X, Jeong JC, Yun DJ, et al. (2010) A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J Exp Bot 61: 3787–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang J-C. (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16: 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sahrawy M, Avila C, Chueca A, Cánovas FM, López-Gorgé J. (2004) Increased sucrose level and altered nitrogen metabolism in Arabidopsis thaliana transgenic plants expressing antisense chloroplastic fructose-1,6-bisphosphatase. J Exp Bot 55: 2495–2503 [DOI] [PubMed] [Google Scholar]
- Saito M, Yoshida M. (2011) Expression analysis of the gene family associated with raffinose accumulation in rice seedlings under cold stress. J Plant Physiol 168: 2268–2271 [DOI] [PubMed] [Google Scholar]
- Schreiber KJ, Austin RS, Gong Y, Zhang J, Fung P, Wang PW, Guttman DS, Desveaux D. (2012) Forward chemical genetic screens in Arabidopsis identify genes that influence sensitivity to the phytotoxic compound sulfamethoxazole. BMC Plant Biol 12: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN, Day DA (2000) Respiration and photorespiration. In B Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 676–725 [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R, Andriankaja M, Van Aken O, Van Breusegem F, Fernie AR, et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiol 152: 226–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Claeys H, De Bodt S, Oikawa A, Shinoda S, Andriankaja M, Maleux K, Eloy NB, Coppens F, Yoo S-D, et al. (2011) Pause-and-stop: the effects of osmotic stress on cell proliferation during early leaf development in Arabidopsis and a role for ethylene signaling in cell cycle arrest. Plant Cell 23: 1876–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Inzé D. (2010) More from less: plant growth under limited water. Curr Opin Biotechnol 21: 197–203 [DOI] [PubMed] [Google Scholar]
- Steinhauser D, Fernie AR, Araújo WL. (2012) Unusual cyanobacterial TCA cycles: not broken just different. Trends Plant Sci 17: 503–509 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Taji T, Seki M, Satou M, Sakurai T, Kobayashi M, Ishiyama K, Narusaka Y, Narusaka M, Zhu J-K, Shinozaki K. (2004) Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol 135: 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Sakurai T, Mochida K, Ishiwata A, Kurotani A, Totoki Y, Toyoda A, Sakaki Y, Seki M, Ono H, et al. (2008) Large-scale collection and annotation of full-length enriched cDNAs from a model halophyte, Thellungiella halophila. BMC Plant Biol 8: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji T, Komatsu K, Katori T, Kawasaki Y, Sakata Y, Tanaka S, Kobayashi M, Toyoda A, Seki M, Shinozaki K. (2010) Comparative genomic analysis of 1047 completely sequenced cDNAs from an Arabidopsis-related model halophyte, Thellungiella halophila. BMC Plant Biol 10: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Kakehi J. (2010) Polyamines: ubiquitous polycations with unique roles in growth and stress responses. Ann Bot (Lond) 105: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valluru R, Van den Ende W. (2011) Myo-inositol and beyond—emerging networks under stress. Plant Sci 181: 387–400 [DOI] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, García-Ramírez L, Pantoja O. (2005) Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol 139: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. (2008) Proline accumulation in plants: a review. Amino Acids 35: 753–759 [DOI] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Volkov V, Wang B, Dominy PJ, Fricke W, Amtmann A. (2003) Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, possesses effective mechanisms to discriminate between potassium and sodium. Plant Cell Environ 27: 1–14 [Google Scholar]
- Wang H, Zhang M, Guo R, Shi D, Liu B, Lin X, Yang C. (2012) Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol 12: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers JDB, Johansen LG. (1985) Accurate estimation of stomatal aperture from silicone rubber impressions. New Phytol 101: 109–115 [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Whitty BR, Díaz-Camino C, Akhter SR, Brandle JE, Golding GB, Weretilnyk EA, Moffatt BA, Griffith M. (2005) Expressed sequence tags from the Yukon ecotype of Thellungiella reveal that gene expression in response to cold, drought and salinity shows little overlap. Plant Mol Biol 58: 561–574 [DOI] [PubMed] [Google Scholar]
- Wong CE, Li Y, Labbe A, Guevara D, Nuin P, Whitty B, Diaz C, Golding GB, Gray GR, Weretilnyk EA, et al. (2006) Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol 140: 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-J, Zhang Z, Wang J-Y, Oh D-H, Dassanayake M, Liu B, Huang Q, Sun H-X, Xia R, Wu Y, et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Jarvis DE, Chen H, Beilstein MA, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, et al. (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasar F, Uzal O, Tufenkci S, Yildiz K. (2006) Ion accumulation in different organs of green bean genotypes grown under salt stress. Plant Soil Environ 52: 476–480 [Google Scholar]
- Yobi A, Wone BWM, Xu W, Alexander DC, Guo L, Ryals JA, Oliver MJ, Cushman JC. (2012) Comparative metabolic profiling between desiccation-sensitive and desiccation-tolerant species of Selaginella reveals insights into the resurrection trait. Plant J 72: 983–999 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lai J, Sun S, Li Y, Liu Y, Liang L, Chen M, Xie Q. (2008) Comparison analysis of transcripts from the halophyte Thellungiella halophila. J Integr Plant Biol 50: 1327–1335 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.