TIR-NBS and TIR-X proteins are novel plant proteins with activities indicative of roles in plant defense.
Abstract
Toll/interleukin receptor (TIR) domain-containing proteins encoded in the Arabidopsis (Arabidopsis thaliana) genome include the TIR-nucleotide binding site (TN) and TIR-unknown site/domain (TX) families. We investigated the function of these proteins. Transient overexpression of five TX and TN genes in tobacco (Nicotiana benthamiana) induced chlorosis. This induced chlorosis was dependent on ENHANCED DISEASE RESISTANCE1, a dependency conserved in both tobacco and Arabidopsis. Stable overexpression transgenic lines of TX and TN genes in Arabidopsis produced a variety of phenotypes associated with basal innate immune responses; these were correlated with elevated levels of salicylic acid. The TN protein AtTN10 interacted with the chloroplastic protein phosphoglycerate dehydrogenase in a yeast (Saccharomyces cerevisiae) two-hybrid screen; other TX and TN proteins interacted with nucleotide binding-leucine-rich repeat proteins and effector proteins, suggesting that TN proteins might act in guard complexes monitoring pathogen effectors.
Innate immunity is a primary defense mechanism in plants that functions to protect against a variety of biotic stresses (Eitas and Dangl, 2010). The plant basal immune system comprises pattern or pathogen recognition receptors that can recognize a variety of plant pathogens by identifying specific pathogen-associated molecular patterns (PAMPs; Tsuda and Katagiri, 2010). This recognition of PAMPs by plant pattern recognition receptors triggers PAMP-triggered immunity or plant basal immunity (Jones and Dangl, 2006; Zipfel, 2008). Well-known PAMPs or microbe-associated molecular patterns recognized by plants include bacterial flagellin, cold shock proteins, and elongation factor Tu. To suppress PAMP-triggered immunity, plant pathogens secrete an array of virulence factors such as type III effector proteins, while plant resistance (R) proteins function to recognize the effector molecules (Römer et al., 2009; Lewis et al., 2010; Tsuda and Katagiri, 2010; Zhang et al., 2012). Specific recognition of a pathogen effector by a plant R protein triggers a second type of immune response called effector-triggered immunity, resulting in an incompatible reaction (Qi et al., 2011; Sohn et al., 2012; Tahir et al., 2012).
The most commonly known plant R proteins are the nucleotide-binding (NB) site Leucine-rich repeat (LRR) proteins that plants use to detect effector proteins. The NB is often called NB-ARC because of sequence similarities to the human apoptotic protease-activating factor APAF1 and Caenorhabditis elegans homolog CELL DEATH PROTEIN4 (Lukasik and Takken, 2009). Plant NB-LRR proteins often also have, at the N terminus, a Toll/Interleukin-1 receptor (TIR) or coiled coil (CC) domain (Meyers et al., 2003). In animal TIR proteins, this domain is more commonly located at the C terminus and is linked by a transmembrane domain to an N-terminal LRR domain (Torto et al., 2002). In Drosophila spp. and other microbes, a TIR domain has been shown to play an important role in the activation of antifungal immune responses (Jenkins and Mansell, 2010). Toll-like receptors (TLRs) perform an integral role in the activation of antimicrobial responses in many animals (Radhakrishnan and Splitter, 2010).
In plants, two additional TIR-containing protein families, TIR-NB site (TN) and TIR-unknown/random (TX), were identified, which are distinct from the longer TIR-NB-LRR (TNL) R protein homologs (Meyers et al., 2002). TN proteins contain TIR and NBS domains but lack LRRs, while TX proteins lack both NBS and LRR domains, yet often have a small and variable C-terminal domain (Meyers et al., 2002). In the Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) genome, there are 30 TX genes and 21 TN genes (Meyers et al., 2003). The crystal structure of a TIR domain from an Arabidopsis TN protein (At1g72930/NP_177436) contains a compact globular fold resembling the mammalian (TLR1 and MYELOID DIFFERENTIATION PRIMARY RESPONSE GENE88 [MYD88]) and bacterial TIR domain proteins (Chan et al., 2010). Although plant TIR domains share less than 20% sequence identity with the human TLR domains, the structures of the TIR domain in plants, mammalian TLRs, and bacterial TIR domain proteins have strong similarity (Chan et al., 2010).
A high proportion of TX and TN genes were previously reported to be in complex clusters with TNL genes; these clusters were found to be duplicated to multiple locations in the genome (Meyers et al., 2002). The existence of genetically linked pairs or sets of genes such as RESISTANCE TO PERONOSPORA PARASITICA2A (RPP2A)-RPP2B, RESISTANCE TO PSEUDOMONAS SYRINGAE1 (RPS1)-RPS4, LEAF RUST RESISTANCE GENE10 (LR10)-RESISTANCE GENE ANALOGUE2 (RGA2), RICE BLAST RESISTANCE GENE AT PIK LOCUS1 (PIKM-1)-TS-PIKM2-TS, and RICE BLAST RESISTANCE GENE AT PI LOCUS1 (PI5-1)-PI5-2 in the genomes and their role in disease resistance suggests that these linked genes are required to effect a defense response in plants (Eitas and Dangl, 2010). The genomic pairing of the TNL genes with TX or TN genes suggests a role of the tightly linked TN protein in the function of its cognate TNL protein or proteins (and vice versa).
The specific direct or indirect interaction between an R gene and a corresponding avirulence (Avr) gene in the characterized pairs of interaction resulted in an immune response in the form of localized programmed cell death, called the hypersensitive response (HR; Burch-Smith et al., 2007; Caplan et al., 2008). The recognition of avirulence proteins from pathogens by the cognate R proteins induces a cascade of changes that increases the levels of salicylic acid (SA), jasmonic acid (JA), phenyl ammonium lyase, and systemin (Liu et al., 2010). The production of several of these biochemical signals is known to trigger multiple convergent ‘R’-gene signaling pathways, leading to programmed cell death and further changes in gene expression patterns (Vlot et al., 2008a, 2008b). Structure function analysis of Arabidopsis R proteins RPS4 (Zhang et al., 2004) and RPP1A (Michael Weaver et al., 2006) have shown that TIR and NBS domains of the proteins without the LRR domain (TNL truncations) could be sufficient to induce HR. Studies using overexpression of plant R genes (particularly the truncated TNL genes) suggest that the TIR and NBS domains by themselves might be sufficient to induce HR and to initiate plant defense responses (Michael Weaver et al., 2006; Swiderski et al., 2009).
In this study, we present experimental and computational data that are collectively consistent with a role for Arabidopsis TX and TN proteins in plant defenses. For example, the ability of the TX and TN genes to induce HR responses upon transient expression is dependent on ENHANCED DISEASE RESISTANCE1 (EDS1). This EDS1 dependency in induced HR was demonstrated in both tobacco (Nicotiana benthamiana) and in Arabidopsis. Stable transgenic overexpression in Arabidopsis of TX and TN genes resulted in a variety of phenotypes involved with basal innate immune responses that are dependent on SA. We also demonstrated the interaction of TX and TN proteins with plant pathogenic elicitor proteins and other plant signal transduction proteins.
RESULTS
Phylogeny and Conservation of TX and TN Proteins
Based on a phylogenetic analysis of the TIR protein domain within the Arabidopsis Col-0 genome, TX proteins are distinct from TN proteins (Supplemental Fig. S1; Meyers et al., 2002). This is consistent with additional features distinct from TNL plant R proteins such as a lack of NBS domain sequences and or the presence of variable domains at their C terminus. The N-terminal TIR domain is conserved among all three groups. The conserved motifs in the NBS region of the TN protein sequences were shown to be consistent with motifs in the NBS region of the TNL proteins, except for the total absence of a conserved domain within the NBS known as RNBS-A motifs (Meyers et al., 2002). Though TX and TN proteins are diversified in their C-terminal domains, they are more closely related and conserved in their TIR domain regions compared with TNL proteins. Two TN proteins, AtTN17 and AtTN21 (Supplemental Fig. S1 contains a key to the shorthand identifiers), uniquely possess unusual domains at their N and C termini, and thus they have been identified as “XTNX” proteins (Meyers et al., 2002). The phylogenetic analysis demonstrated that these two proteins are outliers in the TX and TN phylogeny, consistent with our prior analysis (Meyers et al., 2002). The X domain in the TX and XTNX proteins was comprised of a variety of diverse sequences ranging in length from 28 to 235 amino acids that were either unique or similar to other TX proteins. Among other variations in their structures, TX and TN genes differ substantially in the number of introns, with TN genes (16 out of 21) comprised of two exons with a single intron, while the TX genes typically included more than one intron, which are more often intertwined into X (unknown) domains (Supplemental Fig. S1).
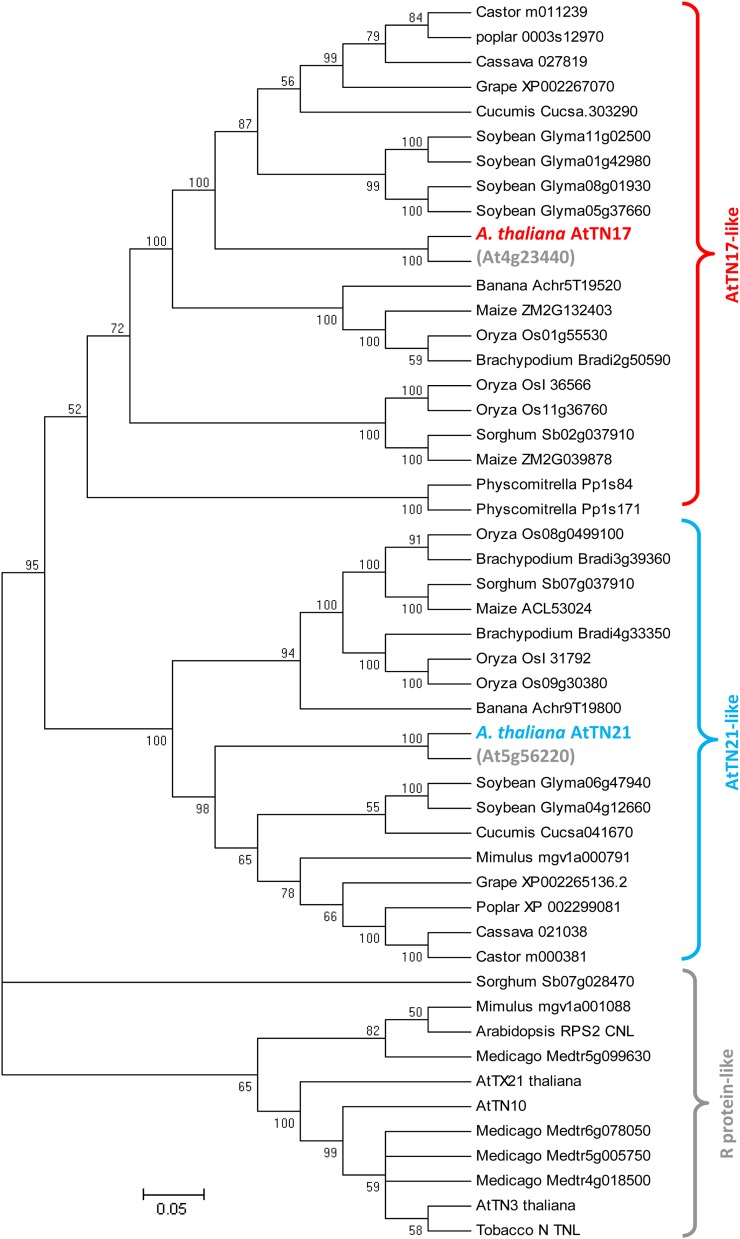
We next examined the conservation of the TX, TN, and XTNX proteins using plant genomic sequences from GenBank. To identify homologs, we used Arabidopsis TN (AtTN3, AtTN10, and AtTN11) and TX (AtTX21) protein sequences plus the two outlying XTNX proteins AtTN17 and AtTN21 for this analysis. Five of the TN proteins yielded protein homologs in a BLAST search against sequences from monocots, basal angiosperms, and magnoliids (Supplemental Table S1). Arabidopsis coiled coil-NBS-LRR (CNL) and TNL reference proteins such as RESISTANCE TO PSEUDOMONAS SYRINGAE1 (RPM1) and RPP1 displayed a broad range of homology to all the members identified from lower monocots and gymnosperms (Supplemental Table S1). Thirty-five different homologs of AtTN17 and AtTN21 were found in rice (Oryza sativa), grape (Vitis vinifera), soybean (Glycine max), poplar (Populus spp.), Sorghum spp., Physcomitrella spp., castor bean (Ricinus communis), maize (Zea mays), cassava (Manihot esculenta Crantz.), Cucumis spp., papaya (Carica papaya), Mimulus spp., Brachypodium spp., and banana (Musa spp.; Fig. 1), indicating that these two proteins are conserved among diverse plant species. These matches exceeded 60% identity for 41 of the proteins, demonstrating that the XTNX clade is a highly conserved group across all plants in which homologs were found. In rice, three homologs (LOC_Os01g55530, LOC_Os08g38970, and LOC_Os09g30380) of the Arabidopsis XTNX proteins AtTN21 or AtTN17 were identified. Similarly, in banana (a nongrass monocot), two homologs (Achr9T19800_001 and Achr5T19520_001) of the Arabidopsis XTNX proteins AtTN21 or AtTN17 were identified. The rice XTNX homologs LOC_Os08g38970 and LOC_Os01g55530 are moderately expressed (Supplemental Table S2). The two XTNX proteins were found to have highest percentage identity (60%–70%) among basal angiosperms (Amborella trichopoda, Aristolochia fimbriata, and Nuphar advena), monocots (Asparagus officinalis and Musa acuminata), and magnoliids (Persea americana, Saruma henryi, and Liriodendron tulipifera). The other TN proteins that have homologs in Amborella trichopoda, Nuphar advena, Liriodendron tulipifera, and M. acuminata were AtTN3, AtTN10, and AtTN11 (Supplemental Table S1). In banana, a single additional TX protein (GSMUA_AChr9G24500_001, 59% identity to At1g61105/AtTX9 and At1g52900/AtTX3) was identified, the first TIR-encoding gene other than a XTNX identified in a monocot. This banana protein has 55% to 60% overall identity to the Arabidopsis TIR protein (TIR motif from TX and TNL proteins).
Figure 1.
Phylogeny and conservation of TX and TN protein domains. A neighbor-joining phylogenetic tree shows the homology of TN sequences across plants. Arabidopsis TN proteins AtTN21 and AtTN17 (highlighted in color) were used as template sequences to compare against plant protein databases. The evolutionary history was inferred using the neighbor-joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved 50 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1,556 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
Expression of TX and TN Genes in Arabidopsis
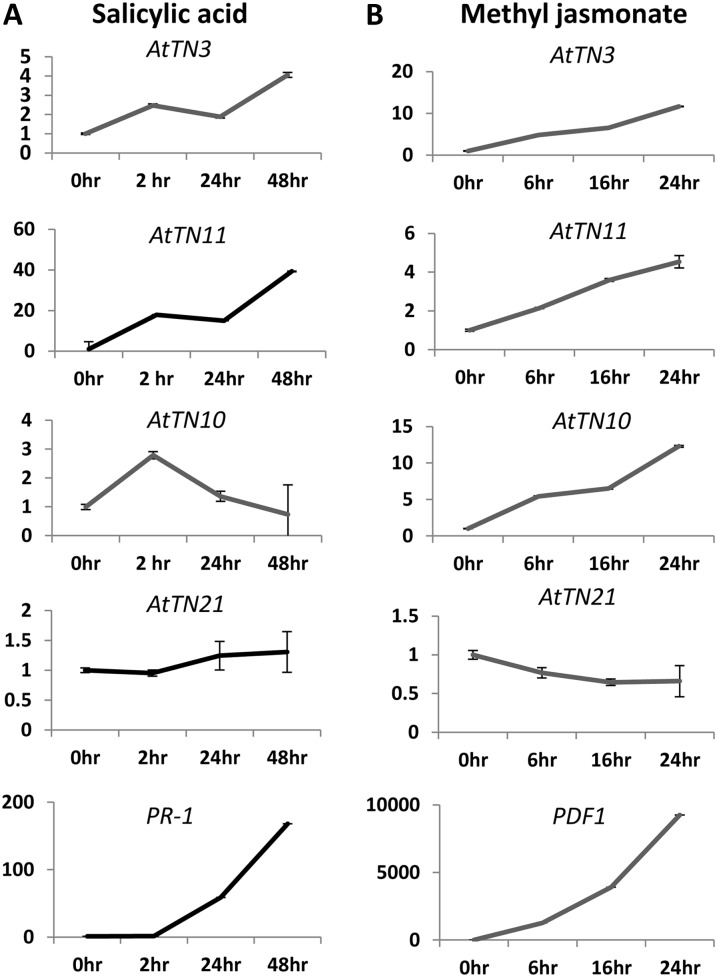
The pattern of expression of TX and TN genes may provide clues to their cellular roles. Our prior work demonstrated expression of TX and TN genes in EXPRESSED SEQUENCE TAGS (EST) or massively parallel signature sequences data sets (Meyers et al., 2002). With the availability of much larger data sets in Arabidopsis, such as microarray or Illumina-based sequencing-by-synthesis (SBS) data sets, we examined expression of the TX and TN genes under a variety of conditions, including biotic stress based on microarray data sets available from Genevestigator (Supplemental Fig. S2). TX and TN genes in Arabidopsis were found to be induced under biotic stress conditions as expressed in microarray data sets (Supplemental Fig. S2). Some of the TN genes (AtTN10, AtTN11, AtTN2, and AtTN3) showed a higher fold induction in their expression under biotic stress treatments, including infection by Blumeria graminis, Erysiphe cichoracearum, and Pseudomonas syringae infection (Supplemental Fig. S2). The expression of TX and TN genes in 17 SBS libraries of Arabidopsis tissues was also analyzed (Table I; Supplemental Table S3). AtTN10 and AtTN13 had the highest and most ubiquitous expression. After SA treatments, five of the TN genes (AtTN1, AtTN7, AtTN10, AtTN11, and AtTN13) were induced by at least 10-fold compared with their expression in the leaf libraries either at 4 or 52 h after treatment (Supplemental Table S3). We checked by quantitative reverse transcription (RT)-PCR the expression levels of four different TN genes (AtTN3, AtTN10, AtTN11, and AtTN21) in SA-treated plant tissues (Fig. 2). Among the TN genes tested, AtTN3 and AtTN11 displayed an elevated expression at 48 h compared with the zero-hour time points after SA treatments, which are in agreement with the massively parallel signature sequences data (Fig. 2A; Supplemental Table S3). The highly conserved Arabidopsis gene AtTN21 was found to have relatively stable expression at different time points, as the gene expression level was relatively unaffected by SA treatments (Fig. 2A). Quantitative RT-PCR measurements also recapitulated SA-induced expression of the genes AtTN10 and AtTN11 found in public databases such as Genevestigator and public SBS libraries (Fig. 2; Supplemental Table S3). The marker gene PATHOGENESIS-RELATED PROTEIN1 (PR-1) (control) was substantially induced with the exogenous application of SA (Fig. 2A). This induction suggests that TX and TN genes might play a role in SA-mediated defense signaling or responses. In agreement with the microarray data, following the methyl jasmonate (MeJA) treatment, the genes AtTN3, AtTN10, and AtTN11 were increasingly induced from 6 to 24 h after treatment (Fig. 2B; Supplemental Fig. S2). Expression levels were 10- to 15-fold higher in AtTN3 and AtTN11 compared with its basal levels in Col-0 following the treatment of 10 µm MeJA treatments (Fig. 2B). The marker gene PATHOGEN-INDUCIBLE PLANT DEFENSIN1.2 (PDF1.2; the control) was substantially induced with the exogenous application of MeJA (Fig. 2B). Thus, these TN genes are responsive to external application of JA and could act in JA-dependent plant pathways. In summary, the expression patterns of several TX and TN genes are consistent with roles in or downstream of plant defense pathways.
Table I. Expression of TX and TN genes in Arabidopsis, detected in public SBS data sets.
| TX/TN Gene | Annotation | Signature Abundancea | No. of Libraries in which Gene Is Expressedb | Leaf Control | SA Induction (52 h) |
|---|---|---|---|---|---|
| Transcripts per million | |||||
| At1g17610 | AtTN1 | 4–103 | 3 | 4 | 103 |
| At1g47370 | AtTX1 | 3–19 | 5 | 0 | 0 |
| At1g65390 | AtTX10 | 2–6 | 4 | 2 | 0 |
| At1g66090 | AtTN3 | 1–21 | 5 | 0 | 1 |
| At1g72850 | AtTN4 | 1–25 | 10 | 4 | 7 |
| At1g72870 | AtTN5 | 1–8 | 6 | 0 | 0 |
| At1g72890 | AtTN6 | 2–191 | 14 | 20 | 13 |
| At1g72900 | AtTN7 | 3–1,382 | 10 | 199 | 805 |
| At1g72910 | AtTN8 | 9–15 | 2 | 15 | 9 |
| At1g72920 | AtTN9 | 2–26 | 6 | 0 | 0 |
| At1g72930 | AtTN10 | 1–1,499 | 17 | 369 | 1,499 |
| At1g72940 | AtTN11 | 3–215 | 15 | 93 | 117 |
| At1g72950 | AtTN12 | 2–30 | 7 | 0 | 0 |
| At2g03300 | AtTX12 | 0 | 0 | 0 | 0 |
| At2g20145 | AtTX13 | 2–79 | 7 | 4 | 12 |
| At2g32140 | AtTX14 | 4 | 1 | 4 | 0 |
| At3g04210 | AtTN13 | 6–2,008 | 14 | 654 | 2,008 |
| At4g04110 | AtTN14 | 0 | 0 | 0 | 0 |
| At4g09420 | AtTN15 | 2–76 | 6 | 2 | 0 |
| At4g11340 | AtTX15 | 0 | 0 | 0 | 0 |
| At4g16990 | AtTN16 | 1–476 | 8 | 90 | 20 |
| At4g19920 | AtTX17 | 4–112 | 4 | 0 | 0 |
| At4g23440 | AtTN17 | 1–58 | 15 | 38 | 1 |
| At4g23513 | AtTX21 | 15–132 | 3 | 0 | 0 |
| At5g44920 | AtTX25 | 1–695 | 14 | 0 | 30 |
| At5g45000 | AtTX26 | 0 | 0 | 0 | 0 |
| At5g45070 | AtTX27 | 8–150 | 3 | 0 | 0 |
| At5g45080 | AtTX28 | 22–32 | 2 | 0 | 0 |
| At5g45090 | AtTX29 | 2–4 | 2 | 0 | 0 |
| At5g45220 | AtTX30 | 12–13 | 3 | 0 | 0 |
| At5g46480 | AtTN19 | 0 | 0 | 0 | 0 |
| At5g48780 | AtTN20 | 1–5 | 5 | 0 | 0 |
| At5g56220 | AtTN21 | 9–49 | 14 | 9 | 0 |
Ranges in different libraries indicated in transcripts per million representing maximum and minimum levels. bThe number with detectable expression out of the 17 mRNA libraries available at http://mpss.udel.edu/at.
Figure 2.
Expression of TN and TX genes in Arabidopsis Col-0 plants after SA and MeJA treatments. The values on the y axis represent fold change of expression at corresponding time intervals on the x axis. A, Expression of TN and TX genes that were chosen based on SBS data from SA-treated libraries, measured by real-time PCR following SA-treatments. The PR-1 marker gene known to be induced by SA was used as a control (bottom). B, Expression level of TN and TX genes as measured by real-time PCR experiment following MeJA treatments. The PDF1 marker gene known to be induced by MeJA was used as a control (bottom).
Transient Overexpression of the TX and TN Genes in Tobacco Results in Cell Death Responses
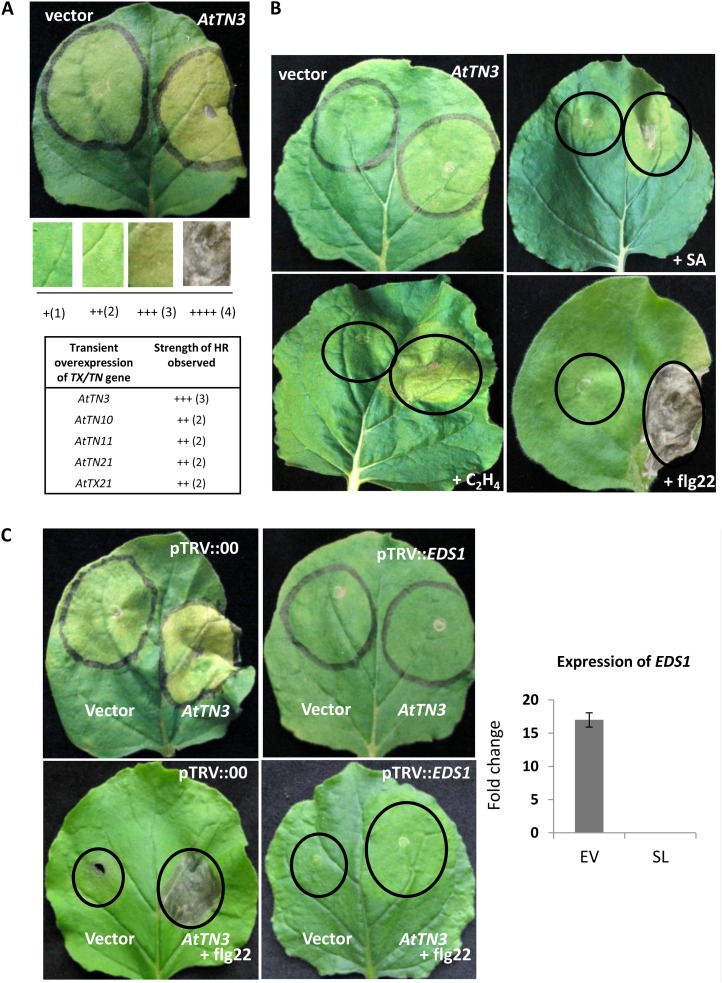
We next sought to investigate the potential function of these genes using transient assays, knowing that transient overexpression of R genes may induce spontaneous HR (Swiderski et al., 2009). Transient Agrobacterium tumefaciens-mediated overexpression of Arabidopsis TX and TN proteins in tobacco resulted in HR, a cell death phenotype, for five (AtTN3, AtTN10, AtTN11, AtTN21, and AtTX21) out of 15 TX and TN genes tested (Supplemental Table S4). HR cell death was scored based on a relative scoring method using a system of multiple plus signs as the strength increases (Fig. 3A). The cell death phenotype observed for AtTN3 (+++ = 3) was stronger over the other four transient overexpression events (AtTN10, AtTN11, AtTN21, and AtTX21) that resulted in a mild HR response (++ = 2) in tobacco (Fig. 3A). The other 10 TX and TN genes tested with transient overexpression in tobacco did not display any visible phenotypes (Supplemental Table S5). AtTN3 induced a more robust cell death response (++++ = 4) when transiently overexpressed together with the exogenous addition of the elicitors SA or ethephon (Fig. 3B). This enhanced cell death was even more apparent when AtTN3 was transiently overexpressed in conjunction with exogenously applied flagellin (flg22) peptide. The enhanced cell death effect observed by transient overexpression of AtTN3 with the flg22 peptide is possibly a synergistic mechanism in tobacco (Fig. 3B). There was no enhancement effect of these exogenously applied elicitors on the ability of the other 14 TX or TN genes to induce cell death in tobacco leaves. Therefore, some members of the TX and TN protein family can trigger cell death via ectopic expression, similar to previous reports that used N-terminal domains of the TNL genes RPS4 and RPP1A (Zhang et al., 2004; Michael Weaver et al., 2006).
Figure 3.
Transient overexpression of Arabidopsis TN and TX genes in tobacco results in partial cell death response. A, Transient overexpression of AtTN3 in tobacco results in cell death. Visual scoring of HR responses and HR strength score table (++++ = 4; +++ = 3; ++ = 2; and + = 1) indicate the strength of HR observed with the transient overexpression of other TX and TN genes compared with the TN gene AtTN3. B, Transient overexpression of AtTN3 along with elicitors SA, ethephon, and flagellin results in increased cell death responses in tobacco. C, Transient overexpression of AtTN3 in an eds1-silenced plant along with or without flagellin resulted in a decreased HR in tobacco. Expression of EDS1 was checked in tobacco plants in both empty vector transformed lines (EV) and EDS1-silenced lines (SL) by quantitative RT-PCR.
Dependency of TX and TN Genes on EDS1 to Induce Cell Death in Tobacco
Plant TIR domain-containing proteins are reported to signal via pathway that includes EDS1 and PHYTOALEXIN DEFICIENT4 (PAD4) (Nishimura and Dangl, 2010). To investigate the importance of EDS1 downstream of TX and TN proteins, transient overexpression of TX and TN genes was tested in tobacco transiently silenced for EDS1 by viral-induced gene silencing (VIGS). Transient overexpression of the gene AtTN3 induced partial cell death of the leaf tissue in the plants inoculated with the empty viral vector. However, AtTN3-induced partial cell death was lost in the lines silenced for EDS1 (Fig. 3C).
Because flg22 can induce defense response genes and trigger responses to plant pathogenic bacteria (Zipfel et al., 2004; Zipfel, 2008), transient overexpression of AtTN3 gene was tested along with flg22 in both EDS1-silenced and nonsilenced plants. In the EDS1 nonsilenced plant, coinfiltration of AtTN3 and flg22 was able to cause cell death (Fig. 3C). However, in the EDS1-silenced plants, cell death was abolished after coinfiltration (Fig. 3C). Quantitative real-time PCR analysis of both the empty VIGS vector-transformed and EDS1-silencing plants showed that expression of the EDS1 gene was greater than 15-fold higher in the nonsilenced versus silenced plants (Fig. 3C). The results were replicated over five different subsets of plants. Thus, signaling by AtTN3 that results in cell death in transient overexpression experiments appears dependent on EDS1.
Role of TX and TN Genes in Plant Defense Responses
Induction of plant defense responses occurs upon overexpression of R genes (such as Suppressor of NONEXPRESSOR OF PR GENES1 (NPR1) Constitutive1 [SNC1]) in Arabidopsis (Li et al., 2010). Consistent with this, A. tumefaciens-mediated transient overexpression of five TN and TX genes, AtTN3, AtTN10, AtTN11, AtTN21, and AtTX21, in tobacco triggered cell death. Encouraged by the transient studies in tobacco described above, we assessed the functions of TX and TN genes in Arabidopsis. Initially, knockouts of the TX and TN genes were screened for visible phenotypes and for susceptibility to plant pathogens (Albugo candida, Tobacco Mosaic Virus (TMV), avirulent P. syringae, and Hyaloperonospora parasitica). No visible phenotype was observed for the transfer DNA lines analyzed (data not shown). Lack of an obvious phenotype in Arabidopsis knockouts prompted us to generate stable overexpression lines for TX and TN genes in Arabidopsis. We generated transgenic lines of Arabidopsis Col-0 with stable overexpression of TN and TX genes (AtTN1 to AtTN4, AtTN10, AtTN11, AtTN13, AtTN15, AtTN17 to AtTN19, AtTN21, AtTX6 to AtTX10, AtTX14, AtTX16, AtTX21, AtTX24, AtTX26, and pK2GW7 as a control) to observe their phenotypic effects (Supplemental Table S6). One of the TN genes, AtTN3, did not yield any transgenics from transformation into a Col-0 background, potentially due to deleterious effects of the overexpressed transgene. However, we did obtain overexpression lines for AtTN3 in an Arabidopsis eds1 background.
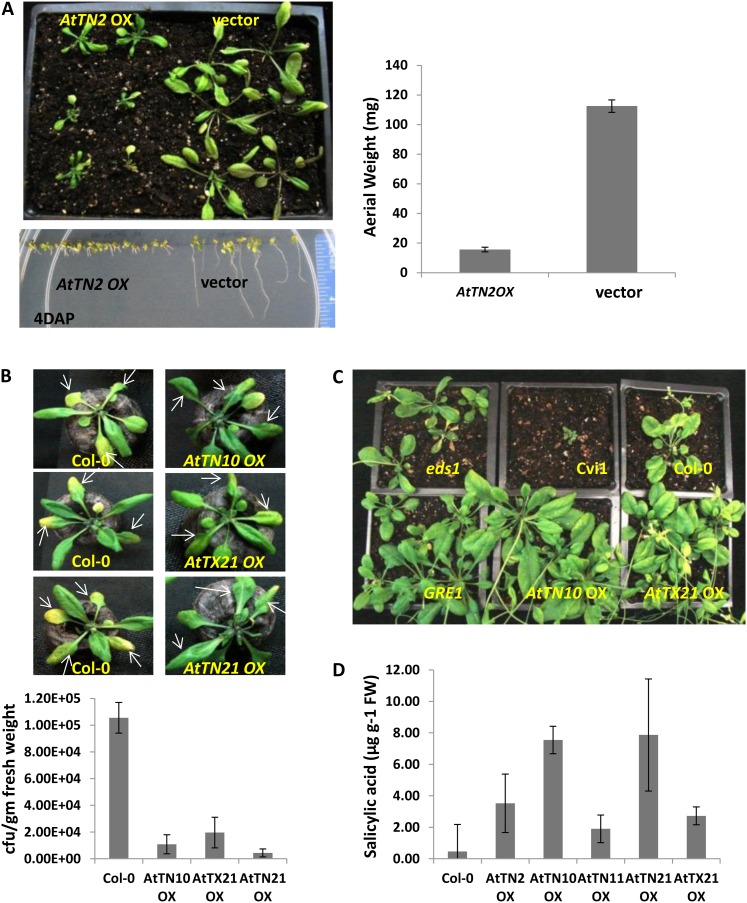
All of the overexpression lines, except for AtTN3, in Col-0 background were analyzed for developmental phenotypes and tested for response phenotypes to a set of bacterial and fungal plant pathogens. Out of 22 different overexpression stable transgenic lines developed for different TX and TN genes (Supplemental Table S6), one of the overexpression transgenics, AtTN2, displayed stunted growth and developmental defects, both on soil and in plates, and had less aerial weight (Fig. 4A), consistent with phenotypes for AtTN2 overexpression published by other labs (Li et al., 2009). Lines overexpressing AtTN10, AtTN21, and AtTX21 displayed reduced disease symptoms along with lower bacterial titers compared with the wild type, when challenged with virulent bacterial plant pathogen Pseudomonas syringae pv tomato DC3000 (Pst DC3000; Fig. 4B).
Figure 4.
Stable transgenics of TN and TX genes in Arabidopsis Col-0 plants display plant defense responses. A, Stunted phenotype of overexpression (OX) transgenics of AtTN2 gene. B, TX and TN overexpression transgenics showing resistance to virulent bacterial pathogen Pst DC3000. Bacterial colony count numbers expressed as colony forming units (CFUs) per gram fresh weight of leaf infected shows lower colony forming units in overexpression transgenics AtTN10 OX, AtTN21 OX, and AtTX21 OX. C, TN and TX overexpression transgenics showing resistance to F. oxysporum strain O-685. D, HPLC analysis of stable TX and TN overexpression transgenics measuring free SA levels in plants.
Because overexpression of NPR1 in Arabidopsis can confer broad-spectrum resistance to the necrotrophic fungal pathogen Fusarium oxysporum (Parkhi et al., 2010), we investigated whether TX- and TN-overexpressing transgenic lines might similarly affect resistance to F. oxysporum. The roots of the stable overexpression lines (AtTN10, AtTN11, AtTN21, and AtTX21) were inoculated with F. oxysporum strain O-685. AtTN10 and AtTX21 lines showed increased resistance to the pathogen (Fig. 4C). Upon further analysis of the lines, they showed an increase in SA levels (free SA levels) compared with the control samples (Fig. 4D). Increased SA levels (free SA) could potentially prime systemic acquired resistance (SAR) and other defense responses leading to resistance in these transgenics. We concluded that stable overexpression in Arabidopsis of several TX and TN genes resulted in increased resistance to bacterial (Pst DC3000) and fungal (F. oxysporum strain O-685) pathogens.
Subcellular Localization and Interactions of TX and TN Proteins with Effector Proteins Indicate Their Role in Plant Defense Signal Transduction Pathway
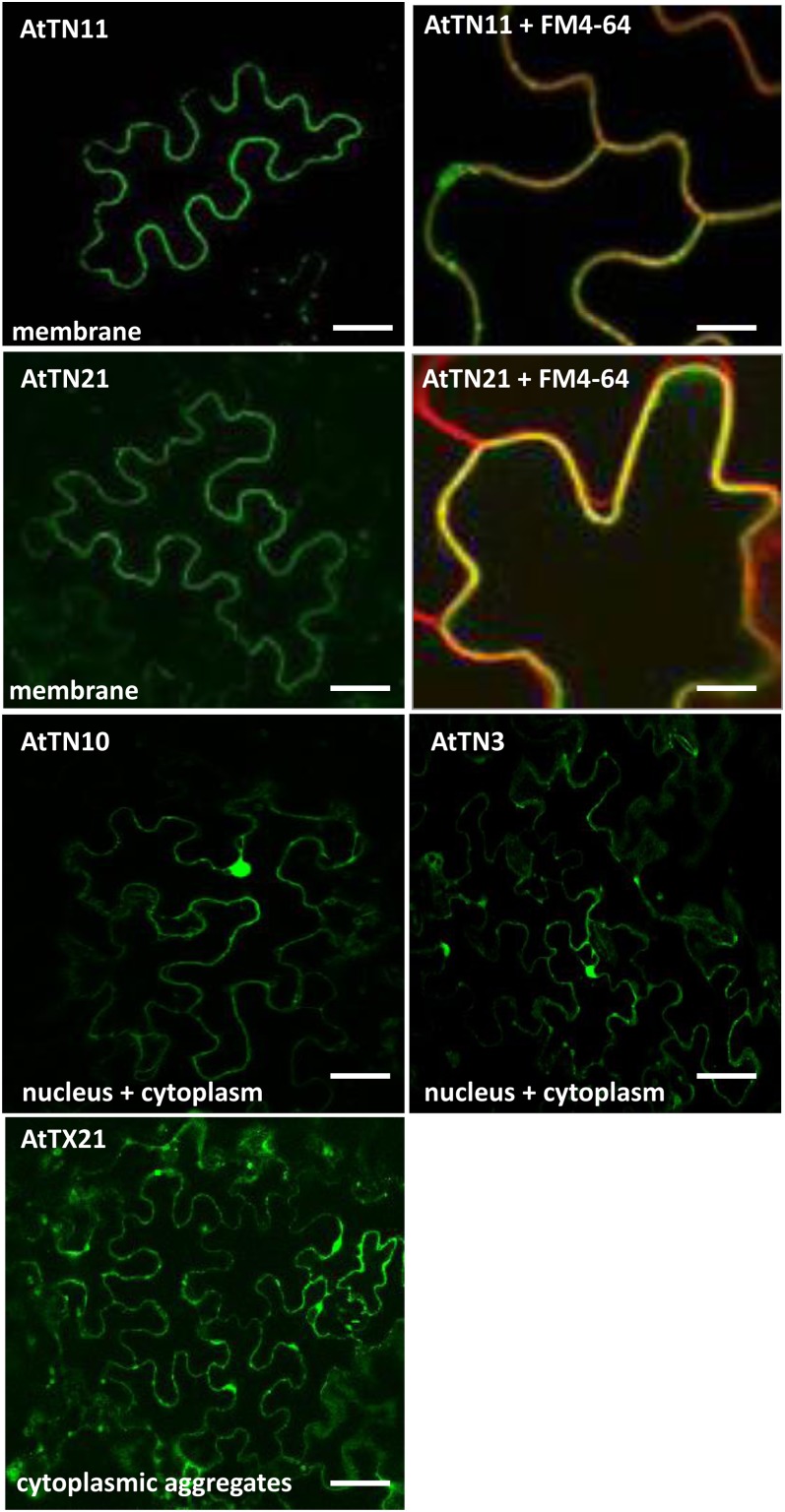
We next examined the subcellular localization of TX and TN proteins by transient expression as GFP fusion proteins under the 35S promoter in tobacco. AtTN3 and AtTN10 both demonstrated nuclear and cytoplasmic localization, while AtTN11 and AtTN21 were plasma membrane localized (Fig. 5A). The expression of the fusion proteins AtTN3 and AtTN10 was measured using anti-GFP antibody in a Western blot, and they both were found to have predicted protein fusion sizes, which were 48 and 76 kD, respectively (Supplemental Fig. S3). The membrane localization of AtTN11 and AtTN21 was confirmed using FM4-64 FX staining (Fig. 5A). The transient overexpression of AtTX21 resulted in the formation of cytoplasmic aggregates (Fig. 5A). Thus, TN and TX proteins were localized to distinct, diverse cellular locations, suggesting diverse roles for these proteins.
Figure 5.
Transient overexpression of TN and TX proteins in tobacco leaves. AtTN11 and AtTN21 proteins show membrane localization, observed through FM4-64 staining in the insets. AtTN11 and AtTN21 proteins are showed in green and FM4-64 staining in red. The colocalization of FM4-64 and Arabidopsis membrane-localizing TN proteins is shown in yellow. AtTN10 and AtTN3 show nuclear and cytoplasmic localization pattern in tobacco. The localization of AtTN10 and AtTN10 is shown in green. Transient overexpression of AtTX21 shows cytoplasmic aggregates and cytoplasmic localization. The localization of AtTX21 was shown in green. Bars = 20 µm.
To identify potential signal transduction proteins and effector proteins that could interact with TX and TN proteins, given that R proteins are known to interact with plant signal transduction proteins (Eitas and Dangl, 2010), we performed a yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) experiment with known and candidate effector targets, components of NB-LRR proteins. A subset of the TX and TN proteins (AtTN1, AtTN3, AtTN10, AtTN11, AtTN15, AtTN21, AtTX6, AtTX8, AtTX16, and AtTX21) was chosen based on their phenotypes in earlier experiments (transient and overexpression studies). The representative class of TX and TN proteins screened against plant pathogenic effectors displayed interactions with bacterial, fungal, and nematode effector proteins (Table II). Based on the number of interactions with TX and TN proteins and the strength of interactions, mapping was done to describe the interactions along with their strength index (Supplemental Fig. S4). Among the TX and TN proteins that were chosen as baits, AtTN21 has the highest number of interactions with candidate elicitor proteins from P. syringae, Ralstonia solanacearum, Bremia lactucae, and Hyaloperonospora arabidopsidis. Among other notable interactions, few plant proteins (AtEDS1 and Arabidopsis serine/threonine Protein Kinase PBS1; AtPBS1) that are involved in disease resistance pathways were found to interact with AtTN21. Similarly, AtTN15 was found to interact with effectors from P. syringae, R. solanacearum, B. lactucae, and H. arabidopsidis as well as plant NBS-LRR proteins (Supplemental Fig. S4; Table II). Two of the TN proteins, AtTN3 and AtTN21, interacted with components of NB-LRR proteins, At3g14470 and At5g60320, respectively. The Arabidopsis protein At3g14470 is an RPP13-like NB-LRR protein, while At5g60320 is a lectin kinase-like protein. Therefore, TN and TX proteins may function with other proteins involved in plant disease resistance pathways.
Table II. TN and TX proteins interacted with a diverse set of elicitors and NBS-LRR proteins in a Y2H screen.
| TX/TN Protein | Effectors and Signal Transduction Proteins Identified as Interacting |
|---|---|
| AtTN1 | Protein kinase (Pto)66 (AvrPto) of P. syringae and Ral011t (R. solanacearum), Magnaporthe oryzae (Mor003) |
| AtTN3 | Pto42 (HopY), Brl028 (B. Lactucae), At3g14470 (NB-LRR) |
| AtTN6 | No interactions detected |
| AtTN7 | Ral019 (HopF2; R. solanacearum) |
| AtTN10 | Rbp001 (root knot nematode effector), Urf004 |
| AtTN11 | Hpp001 (H. parasitica) |
| AtTN13 | Pto42 (HopY), Rbp001 (nematode effector) |
| AtTN15 | Psy(2), Pto(8), Ral(4), Rbp(3), Pvx001, Pph(2), Urf003, Urf004; Brl011, Pma005 |
| Plant proteins: At1g59124, AtACT, Arabidopsis Jasmonate response1, At1g61190, At5g05400, At1g31540 | |
| AtTN21 | P. syringae: Psy(2), Pto(5), Pph(3) |
| R. solanacearum: Ral(6) | |
| Pseudomonas flourescens: Pfl003, B. lactucae: Brl(6), H. parasitica: Hpp001, Hpp021 | |
| Nematode: Rbp001, Rbp002, Mor003; Pvx001 | |
| Plant proteins: At5g63020 (NB-LRR), At1g12210, At4g16950, At4g36150, At5g63020, AtACT, AtEDS1, AtPBS1, AtTGA1, AtTGA2, AtTGA3, AtTGA7, AtMPK4, LeCMPG1, AtTN21 | |
| AtTX6 | No interactions detected |
| AtTX8 | Ral019 (HopF2; R. solanacearum) |
| AtTX16 | (Psy022, psy023), Pto(4), Ral025, Pph022, Mor003; plant proteins: At3g14470, AtACT, AtTGA3, Arabidopsis MAP Kinase4, LePto |
| AtTX21 | Pto66 (AvrPto) of P. syringae |
Arabidopsis TN Protein AtTN10 Interacts in the Cytoplasm with a Chloroplastic Protein, 3-Phosphoglycerate Dehydrogenase
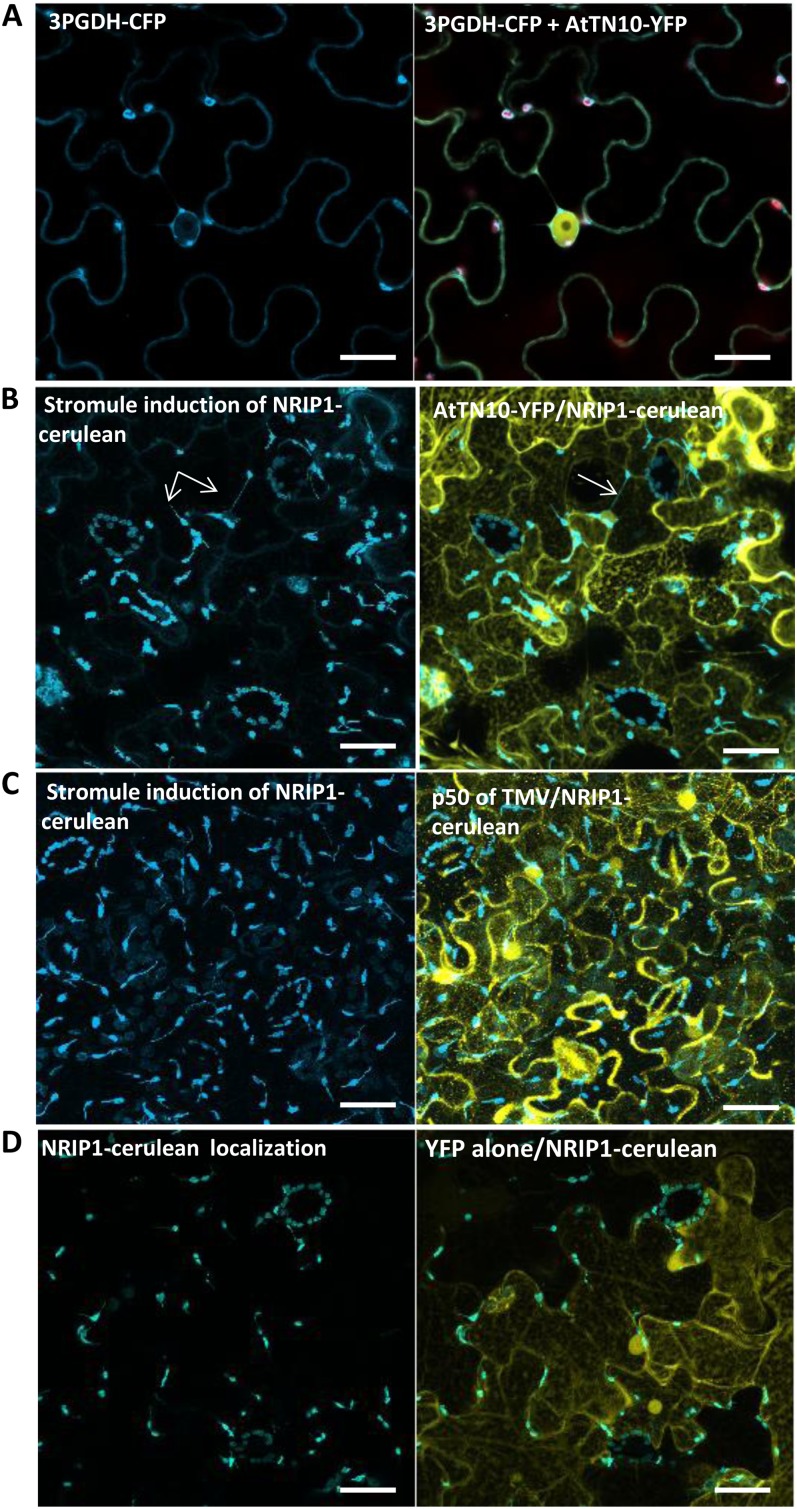
Next, we sought to expend the set of cellular partners with which TX or TN proteins might function. The TN protein AtTN10 was selected for analysis as a bait protein for screening a Y2H library of prey proteins constructed from mRNA derived from several growth stages of Arabidopsis. Strong interactions were found between AtTN10 and 3-phosphoglycerate dehydrogenase (3PGDH, At1g17745), a chloroplastic protein. We used a colocalization assay to assess their interaction in vivo. When transiently coexpressed in epidermal cells of tobacco, the two proteins AtTN10 and 3PGDH both colocalized in the cytoplasm (Fig. 6A). Additional control experiments that involved the localization of AtTN10 with cyan fluorescent protein (CFP) nonfusion protein helped us to confirm the nuclear exclusion interaction of 3PGDH with AtTN10 (Fig. 6A; Supplemental Fig. S5). The sequence comparison of 3PGDH with orthologous proteins from other plant species shows the highly conserved nature of the protein (Supplemental Fig. S6). A search of available expression data from Genevestigator showed the induced expression (more than 2.5 times) of the gene 3PGDH in Col-0 plants infected by plant pathogens such as P. syringae, B. graminis, Botrytis cinerea, and Alternaria brassicola. Transient overexpression of 3PGDH-tagged CFP fusion protein was found to localize in the subcellular compartments of chloroplasts along with cytoplasm (Fig. 6A).
Figure 6.
Arabidopsis TN protein AtTN10 interacts with 3PGDH in the cytoplasm. A, AtTN10 transiently cooverexpressed with 3PGDH in tobacco leaves results in cytoplasmic localization. B, AtTN10 induces strong stromule formation, as observed by the NRIP1 in chloroplasts when AtTN10 is transiently overexpressed in NRIP1 transgenic background. C, p50 protein of TMV induces strong stromule formation in NRIP1 background. D, Negative control showing YFP protein alone transiently overexpressed in NRIP1. Stromule induction is shown by white arrows. Bars = 20 µm.
To understand whether AtTN10 might be playing a role as an R protein, we transiently overexpressed AtTN10 in a transgenic background with N RECEPTOR-INTERACTING PROTEIN1 (NRIP1) cerulean fusion protein (which fluoresces blue). The TNL protein N of tobacco was shown to interact with p50 domain of TMV through its TIR domain (Burch-Smith et al., 2007). NRIP1 was recently identified to interact with both the TIR domain of N and the p50 domain of TMV (Caplan et al., 2008). NRIP1 transgenics were previously generated by expressing NRIP1 tagged with C-terminal cerulean under the control of the NRIP1 genomic promoter (Caplan et al., 2008), and NRIP1 has been shown to localize in the chloroplasts (Caplan et al., 2008). Transient overexpression of AtTN10 in the NRIP1 background resulted in a strong stromule induction of NRIP1 (Fig. 6B), comparable to the stromule induction by p50 protein of TMV in NRIP1 (Fig. 6C). Stromule induction in the NRIP1 background appeared to require AtTN10, as the nonfusion yellow fluorescent protein (YFP) control protein did not induce this phenotype (Fig. 6D). Therefore, AtTN10 overexpression can induce a cellular phenotype (stromule formation) similar to a TMV-triggered immune response.
DISCUSSION
Plant TX- and TN-encoding genes appear to be largely restricted to dicot genomes that parallel the taxonomic distribution of the related TNL R genes. TN proteins were found to be conserved among many diverse plant genomes, including gymnosperms, basal angiosperms, monocots, and magnoliids. Two divergent XTNX proteins are highly conserved, apparently in many flowering plants, suggesting an ancient and important role of these proteins. The presence of a single TN-like TIR gene in the recently sequenced M. acuminata genome (a nongrass monocot) indicates that TIR genes have been largely but not completely eliminated from monocot genomes (XTNX genes excluded); the banana TX gene may be a vestige of larger numbers of such genes in earlier plants. The specific functions of the larger set of TX and TN plant proteins remain unclear, but we present evidence consistent with their function in defense against pathogens (Supplemental Table S7). The expression of few of the TN genes in the publicly available microarray databases and the SBS databases under different biotic treatments suggests the possibility of their role in plant defenses. Furthermore, the effect of exogenously supplied SA resulted in the induction of two TN genes, AtTN3 and AtTN11 in Arabidopsis Col-0 plants. Interestingly, similar effect of induction was observed with three Arabidopsis TN genes, AtTN3, AtTN10, and AtTN11, when MeJA was applied. Evidence from previous reports suggests a reciprocal antagonism between two pathways, SA and JA (Thaler et al., 2012). The transcriptional induction of two TN genes AtTN3 and AtTN11 in both the applications of SA and JA could be due to their position at the convergence of these two pathways or might be due to their interference with the SA-JA cross talk regulators (Verhage et al., 2010).
Overexpression of TX and TN Genes Activates Defense in Both Tobacco (Transient Expression) and Arabidopsis (Stable Transgenic Lines)
We used reverse genetics approaches to study the functions of genes encoding TX and TN proteins in Arabidopsis. Initially, knockouts of the TX and TN genes were screened for phenotypes, such as loss of resistance or susceptibility to plant pathogens. Lack of an obvious phenotype in Arabidopsis knockouts prompted us to generate transient and stable overexpression lines for TX and TN genes in tobacco and Arabidopsis. Out of 15 TX and TN genes tested for transient overexpression in tobacco, four of the TN genes and one TX gene induced partial cell death responses. Stable overexpression of TN and TX genes in Arabidopsis resulted in phenotypes consistent with plant defense responses, including a lethal phenotype (by AtTN3), similar to that observed in the leaves of tobacco. Induction of a localized cell death has been demonstrated previously in tobacco when an R gene is transiently overexpressed in the absence of a corresponding Avr protein (Swiderski et al., 2009). Cell death induced in response to the transient overexpression of TNL genes was shown in the cases of RPP1A and RPS4 (Zhang et al., 2004; Swiderski et al., 2009). Thus, the TIR domain is in common between the TX, TN, and TNL proteins and may have a shared roll in triggering cell death.
Stable transgenic overexpression lines of AtTN10, AtTX21, and AtTN21 resulted in resistance to the bacterial plant pathogen Pst DC3000 and, in AtTN10 and AtTX21, resistance to the necrotrophic fungal pathogen F. oxysporum. AtTN2, which caused a stunted phenotype upon stable overexpression, has also been identified as lesion cell death gene4 (LCD4) in a transgenic experiment (Li et al., 2009). The same gene was also identified as a modifier of SNC1, which is negatively regulated by BONZAI1 (BON1) and BON3 (Li et al., 2009). In Arabidopsis, the BON1/copine1 (CPN1) gene in association with BON2 or BON3 has been shown to negatively regulate cell death and defense responses (Yang et al., 2006). We found that stable AtTN2-expressing transgenic lines displayed a stunted phenotype and had high free SA levels. Reduced plant fitness has also been observed from priming of SA-related defense responses upon R gene overexpression in Arabidopsis (including RPM1, Pseudomonas resistance and fenthion sensitivity (Prf), SNC1, and ACCELERATED CELL DEATH6 (ACD6); Li et al., 2010; Todesco et al., 2010). Thus, the stable transgenic lines that we obtained overexpressing TX and TN proteins also displayed phenotypes that were phenocopies of overexpression lines for genes involved in plant defenses.
The Dependency of TX and TN Genes on EDS1, a Key Component in the Plant Defense Signaling Pathway
EDS1, PAD4, and NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1) transduce signals in plants upon recognition of plant pathogens (Eitas and Dangl, 2010; Nishimura and Dangl, 2010; Bhattacharjee et al., 2011). We tested the hypothesis that TX and TN proteins might have a similar requirement for EDS1 signaling by transient overexpression of TX and TN genes in tobacco plants silenced for EDS1. As in similar experiments with RPS4 and RPP1A (TNL genes; Zhang et al., 2004; Swiderski et al., 2009), cell death caused by the transient overexpression of AtTN3 gene was dependent on EDS1. In Arabidopsis, we were unable to generate any AtTN3 transgenics in a Col-0 background (data not shown), while we were able to develop these in an eds1 background (Supplemental Table S6). The synergistic cell death effect observed by the transient overexpression of AtTN3 and flg22 peptide in tobacco was also found to be dependent on EDS1 (Fig. 3C).
The dependency of AtTN3 on EDS1 in heterologous tobacco system demonstrates the requirement of EDS1 for plant defense responses by AtTN3.
Interaction of TX and TN Proteins with Effectors and Plant Defense Signal Transduction Proteins
We tested the interaction of TX and TN with plant signal transduction proteins and pathogen effector proteins using the Y2H system. The protein interaction study resulted in a diverse set of interactions between TX or TN proteins with the candidate effectors tested. The interacting proteins identified through the Y2H experiment (Table II; Fig. 5) suggests the possible role of the TN or TX proteins in recognizing the plant pathogenic effectors either directly or indirectly. One of the XTNX proteins, AtTN21, interacted with several of the candidate effector proteins from at least four different plant pathogens. Intriguingly, AtTN21 was also shown to interact with other plant proteins, including PBS1, a signaling component of CC-NBS-LRR protein RPS5. The interactions of AtTN21 with plant proteins such as PBS1 or effector proteins might have to do with a role as an adapter protein. This is particularly interesting given the level of conservation observed for AtTN21 with homologs in grass and other nongrass genomes. We also searched for interactions of the TX and TN proteins with a broader set of Arabidopsis proteins, and AtTN10 was found to interact with a chloroplastic protein, 3PGDH. More interestingly, colocalization experiments performed by fluorescence tagging of AtTN10 and 3PGDH resulted in both the proteins colocalizing with each other, suggesting that the interaction might occur in the cytoplasm rather than the chloroplasts. Bacterial and viral pathogens are known to target chloroplasts or chloroplastic proteins such as NRIP1 for degradation, reducing host defenses (Caplan et al., 2008). Transient overexpression of 3PGDH in tobacco leaves showed a chloroplastic and cytoplasmic localization of the protein, whereas expression of AtTN10 in an NRIP1 transgenic background strongly induced chloroplastic stromule formation. The formation of stromules is induced by the TMV p50 protein during HR in tobacco (Caplan et al., 2008). AtTN10-induced stromules formed in the absence of effector proteins suggests that AtTN10 could function in responses similar to those induced by plant R proteins. It is possible that the 3PGDH protein could be the “guardee,” and AtTN10 might play a role as an adapter for this protein, allowing for degradation or modification (Supplemental Fig. S7).
Arabidopsis TX and TN proteins were previously hypothesized to function as adapter proteins in plants similar to MyD88 and MyD88-adapter-like (Mal) adapter proteins in mammalian and Drosophila spp. immune responses (Meyers et al., 2002). Here, some of the experimental data reveals a possibility of such a role for TX and TN proteins in the plant defense against plant pathogens (Supplemental Fig. S7). In the model, we hypothesize that the two classes of TN and TX proteins may function as adapter proteins in plant defense signaling (Supplemental Fig. S7). The interaction of TN proteins with NB-LRR plant proteins and with proteins that are involved in the plant defense response pathways strengthens the hypothesis of these proteins acting as adapters in conjunction with TNL proteins.
CONCLUSION
We have presented several lines of evidence consistent with some TN and TX proteins functioning in plant defenses. A small number of TN proteins are well conserved from gymnosperms to flowering plants, and a larger set of TIR-containing proteins are found in dicots. A subset of TX and TN genes were induced in SA-treated Arabidopsis tissues, and TX and TN overexpression in tobacco and Arabidopsis resulted in phenotypes that were consistent with a role in disease responses. The ability of overexpression of TX and TN proteins to induce cell death in tobacco was dependent on EDS1. Other published data suggests that more than one NB-LRR protein can function together in the recognition of pathogens (Eitas and Dangl, 2010). The presence of the TNL::TX or TN::TNL protein fusion pairs in Arabidopsis genome (Meyers et al., 2002) is consistent with a cofunctional role of TX or TN proteins together with TNL proteins. Thus, the TX and TN proteins could function together with TNL proteins to facilitate pathogen recognition or downstream signaling (Supplemental Fig. S6), and the XTNX may have other nondefense roles in monocots and other plants.
Though the specific functions of the majority of the TX and TN plant proteins remain unclear, our experimental evidence suggests the possibility for a role of TX and TN proteins in plant defense system. The TX and TN genes were diverse in their expression patterns, as observed with a variety of biotic stress treatments. Very few TN and TX genes were induced with the exogenous application of SA, suggesting that not all proteins might be involved similarly in a single pathway. The induction of two TN genes, AtTN3 and AtTN11, by the application of both SA and JA indicates the possibility of their role in the SA-JA cross talk. Consistent with their expression patterns, the TX and TN proteins also show diversity in their localization patterns, as observed with fluorescence tagging the proteins at their C terminus. The diversity in their localization patterns in the plant cell and their observed in vitro Y2H interactions with elicitor proteins and components of NBS-LRR proteins suggest a functional diversity of these TX and TN proteins. The presence of the XTNX protein orthologs (AtTN21 and AtTN17) in other plant genomes at a high rate of conservation signifies that these proteins are not lost in evolution and might play an important role.
MATERIALS AND METHODS
Bioinformatics Analysis
The complete set of protein sequences for TX and TN proteins was obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org) based on published lists (Meyers et al., 2002). The conservation of the TN and TX genes and their encoded proteins from other plant genomes was assessed using Jalview from Phytozome (http://www.phytozome.net). Arabidopsis (Arabidopsis thaliana) TN proteins AtTN21 and AtTN17 were used as reference sequences for comparison to plant protein databases. In the resulting sequence set and alignment, all ambiguous positions were removed for each sequence pair. There were a total of 1,556 positions in the final data set. A bootstrap consensus tree was inferred from 1,000 replicates using the neighbor-joining method (Saitou and Nei, 1987). Branches corresponding to partitions reproduced in less than 50% of the bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using a Poisson correction method and are in the units of the number of amino acid substitutions per site. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011).
Expression analysis of TX and TN genes used the Genevestigator microarray data sets (https://www.genevestigator.com) and public SBS data sets for biotic and abiotic stress responses (http://mpss.udel.edu/at_sbs). The expression of the Arabidopsis TX and TN genes was also presented as the sum of abundance of their expression in each of the available libraries. Assessment of posttranslational modifications was performed using Expasy (http://expasy.org/tools).
Plant Growth Conditions
Arabidopsis plants (Col-0) were grown in controlled environmental growth chambers. Soil-grown plants were grown in the growth chambers at 21°C in 72% relative humidity under 16-h days. Tobacco (Nicotiana benthamiana) plants were grown in controlled environmental growth chambers with a controlled temperature of 24°C and under 16-h days. Tobacco plants were used at the 3-week stage for inoculations and Agrobacterium tumefaciens infiltrations.
RNA Isolation and cDNA Preparation
Plant RNA was isolated using TRIzol Reagent (Invitrogen), based on the manufacturer’s recommendations. Following RNA isolation, complementary DNA (cDNA) was made from the total RNA using the SuperScript III RT kit for RT-PCR from Invitrogen according to the manufacturer’s recommendations. RT was performed with SuperScript III reverse transcriptase followed by PCR amplification of the cDNA.
Real-Time PCR Analysis
Quantitative real-time PCR was carried out using 2X QuantiTect SYBR Green PCR Master Mix on an Applied Biosystems Fast System 7500 cycler. The PCR conditions were followed according to the manufacturer’s recommendations (Qiagen). The PCR amplification protocol consisted of a 10-min denaturation step followed by 40 cycles of 15 s at 95°C, 30 s at 54°C, and 30 s at 72°C. The primers for the amplification of the TX and TN genes were used as listed (Supplemental Table S8).
SA and JA Treatments
SA solution of 100 µm was applied as foliar spray on Arabidopsis plants at the 3-week stage. One milliliter of 50 µm MeJA solution was applied as a volatile in a sealed 100-mL Magenta jar.
Plasmid Construction
The TX and TN genes were amplified from either cDNA or genomic DNA of Arabidopsis (Col-0). The primers for the amplification of the genes were designed manually based on the annotations in The Arabidopsis Information Resource database (Supplemental Table S9). Most cloning procedures utilized the Gateway cloning technology (Invitrogen). The amplified PCR products were cloned into pDONR207 using the BP clonase (Invitrogen). The resulting sequences were then transferred into pK2GW7 or pEarleyGate 100 series vectors (pEG101, pEG102, and pEG103) using LR clonase (Invitrogen). The inserts were then cloned into plant binary vectors pCB302-rfB and pEarleyGate 100 series vectors (used as a destination vectors), using the protocol from Invitrogen. The genes cloned in pK2GW7 and pEarleyGate 100 series expression vectors were driven by the 35S promoter. The nonredundant sets of clones in the final binary expression vectors pK2GW7 and pEarleyGate 100 series vectors were sequenced to confirm that they were identical to the corresponding clones in pDONR207.
Transient and Stable Transgenic Experiments
A. tumefaciens strain GV3101 was transformed with variants of pK2GW7 or pEarleyGate 100 series vectors using electroporation. Transient assays of tobacco were performed as described previously (Bernal et al., 2005). For all procedures, A. tumefaciens GV3101 containing pK2GW7 clones was grown in Luria-Bertani medium containing gentamycin at 50 µg mL–1 and rifampicin at 30 µg mL–1. A. tumefaciens GV3101 containing pEarleyGate 100 series clones was grown in Luria-Bertani medium containing kanamycin at 50 µg mL–1 and rifampicin at 30 µg mL–1. The resuspended A. tumefaciens cells were used to inoculate the leaves of tobacco on the abaxial surface of the leaves. Imaging under confocal microscope was done 42 h postinfiltration. Similar assays were performed in tobacco plants silenced for EDS1 gene. EDS1 in tobacco plants was silenced using VIGS, and AtTN3-dependent cell death was transiently induced in those plants as described previously (Swiderski et al., 2009).
The A. tumefaciens-mediated plant transformation of Arabidopsis was done using the floral-dip method (Clough and Bent, 1998). The T1 generation plants were screened on Murashige and Skoog plates with kanamycin. A minimum of six independent transgenics were tested per gene.
Pathogen Response Assays
For the fungal response assay, a root-dip inoculation method was followed. Ten-day-old seedlings of Arabidopsis wild-type Col-0 plants and the stable overexpression transgenic lines of TX and TN were grown in the soil. The mutant plants of eds-1 and ecotypes Greenville and Cape Verde Island, ecotypes with known resistance to Fusarium oxysporum, were grown as controls. The roots were dipped in the fungal conidial suspension of concentration of 106 conidia mL–1 for one minute, and the seedlings were transferred to autoclaved soil. The plants were allowed to grow for another 3 weeks, and the plants were scored based on the number of plants dead or surviving.
For the bacterial response assay, Arabidopsis Col-0 plants and stable overexpression transgenic lines were grown until 3 weeks on soil pellets. Leaves of the plants were infiltrated with the virulent Pseudomonas syringae pv tomato DC3000 bacteria with an optical density at 600 nm value of 0.002 (approximately 1 × 106 colony forming units mL–1). The leaves were left until dry on the surface, and the tray was covered with a dome until the completion of the experiment. Observations were recorded at 24, 48, and 72 h posttreatment. Bacterial growth analysis was performed as described previously (Melotto et al., 2006). Bacterial colonies were counted at each time point with at least three technical replicates.
HPLC Estimation of SA Levels
Leaf samples from Arabidopsis plants were weighed and then ground in liquid nitrogen; 400 µL of 80% (v/v) methanol was added, and samples were incubated overnight at 4°C with continuous agitation. The samples were centrifuged twice at 14,000 rpm, and the pellet was discarded. The supernatant was fractionated by HPLC with a 0% (v/v) to 100% (v/v) linear gradient of methanol in 2 mm formic acid, 30 µL total volume, and 60-min run time. Free SA levels in the plants were estimated using a C 18 column. The data were tabulated as microgram per gram of fresh weight.
Confocal Imaging
Live plant imaging was performed on a Zeiss LSM510 confocal microscope using a 40X C-Apochromat (NA = 1.2) water immersion objective lens. Tissue samples were cut from leaves of tobacco at approximately 42 h postinoculation (Caplan et al., 2008). The 458- and 514-nm laser lines of a 25-mW argon laser with appropriate emission filters were used to image cyan fluorescence and yellow florescence, respectively. Membrane localization of the TN protein was confirmed using FM4-64 staining as an additional marker.
Y2H Experiments
Pairwise Y2H experiments were performed using the Matchmaker GAL4 Two-Hybrid System according to the manufacturer’s recommendations (Clontech). TX and TN bait proteins were expressed as a fusion to the yeast transcription activator protein Gal4 (GAL4) DNA-binding domain, and libraries of prey proteins (pathogen elicitor proteins, plant signal transduction proteins, and NB-LRR proteins) were expressed as fusions to the GAL4 activation domain. The interactions were scored for their ability to grow on selective media lacking His and adenine for a double selection. Matings were replicated four times. The interactions were mapped using Cytoscape (Smoot et al., 2011).
The Y2H experiments on AtTN10 were performed by screening against cDNA prey libraries by Dualsystems using their DUALhybrid screening system. The cDNA library for target prey libraries was constructed using whole plant of Arabidopsis (equal mix of roots, leaves, and inflorescence) at 3-week stage. The complexity of the library was 2.7 × 107 independent clones with an average insert size of 1.8 kb.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Phylogeny and conservation of TX and TN protein domains.
Supplemental Figure S2. Expression levels of TN and TX genes vary under biotic stresses.
Supplemental Figure S3. Western blots of the TN proteins AtTN10 and AtTN3 using anti-GFP antibodies confirm protein expression and localization in the nucleus and cytoplasm.
Supplemental Figure S4. Yeast two hybrid interactions of TX and TN proteins with candidate elicitor protein library.
Supplemental Figure S5. Colocalization of YFP with 3PGDH-CFP or AtTN10-YFP with CFP resulted in cytoplasmic localization.
Supplemental Figure S6. Arabidopsis 3PGDH protein At1g17745 is well conserved among other plant genomes.
Supplemental Figure S7. Model for the function of Arabidopsis TN and TX proteins.
Supplemental Table S1. Primers designed for the amplification of transcript abundance through the use of real time PCR.
Supplemental Table S2. Primers designed for the amplification of TX and TN transcripts for cloning and generation of pENTRY gateway vectors.
Supplemental Table S3. Expression data from rice orthologs of Arabidopsis TIR-NBS genes.
Supplemental Table S4. Arabidopsis TX and TN proteins used for HR assays on N. benthamiana.
Supplemental Table S5. Phylogeny and conservation of TN protein domains among monocots and other basal angiosperms.
Supplemental Table S6. Expression of TN and TX genes from public MPSS database libraries.
Supplemental Table S7. Stable transgenic overexpression lines generated in Arabidopsis ecotype Col-0.
Supplemental Table S8. Salk T-DNA knockout lines for TX and TN genes.
Supplemental Table S9. Summary of TX and TN phenotypes.
Supplementary Material
Acknowledgments
We thank Luis Williams and Huaqin Xu for technical and database support.
Glossary
- PAMP
pathogen-associated molecular pattern
- Col-0
ecotype Columbia
- HR
hypersensitive response
- SA
salicylic acid
- JA
jasmonic acid
- MeJA
methyl jasmonate
- SBS
sequencing-by-synthesis
- RT
reverse transcription
- VIGS
viral-induced gene silencing
- Pst DC3000
Pseudomonas syringae pv tomato DC3000
- Y2H
yeast two-hybrid
- cDNA
complementary DNA
References
- Bernal AJ, Pan Q, Pollack J, Rose L, Kozik A, Willits N, Luo Y, Guittet M, Kochetkova E, Michelmore RW. (2005) Functional analysis of the plant disease resistance gene Pto using DNA shuffling. J Biol Chem 280: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Halane MK, Kim SH, Gassmann W. (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Mamillapalli P, Burch-Smith TM, Czymmek K, Dinesh-Kumar SP. (2008) Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Mukasa T, Santelli E, Low LY, Pascual J. (2010) The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci 19: 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. (2010) NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins KA, Mansell A. (2010) TIR-containing adaptors in Toll-like receptor signalling. Cytokine 49: 237–244 [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Wu R, Guttman DS, Desveaux D. (2010) Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet 6: e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pennington BO, Hua J. (2009) Multiple R-like genes are negatively regulated by BON1 and BON3 in Arabidopsis. Mol Plant Microbe Interact 22: 840–848 [DOI] [PubMed] [Google Scholar]
- Li Y, Tessaro MJ, Li X, Zhang Y. (2010) Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol 153: 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PP, Bhattacharjee S, Klessig DF, Moffett P. (2010) Systemic acquired resistance is induced by R gene-mediated responses independent of cell death. Mol Plant Pathol 11: 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasik E, Takken FL. (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Morgante M, Michelmore RW. (2002) TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J 32: 77–92 [DOI] [PubMed] [Google Scholar]
- Michael Weaver L, Swiderski MR, Li Y, Jones JD. (2006) The Arabidopsis thaliana TIR-NB-LRR R-protein, RPP1A; protein localization and constitutive activation of defence by truncated alleles in tobacco and Arabidopsis. Plant J 47: 829–840 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Dangl JL. (2010) Arabidopsis and the plant immune system. Plant J 61: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhi V, Kumar V, Campbell LM, Bell AA, Shah J, Rathore KS. (2010) Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res 19: 959–975 [DOI] [PubMed] [Google Scholar]
- Qi Y, Tsuda K, Glazebrook J, Katagiri F. (2011) Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol 12: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan GK, Splitter GA. (2010) Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem Biophys Res Commun 397: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer P, Recht S, Lahaye T. (2009) A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci USA 106: 20526–20531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Hughes RK, Piquerez SJ, Jones JD, Banfield MJ. (2012) Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc Natl Acad Sci USA 109: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Birker D, Jones JD. (2009) The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact 22: 157–165 [DOI] [PubMed] [Google Scholar]
- Tahir J, Watanabe M, Jing HC, Hunter D, Tohge T, Nunes-Nesi A, Brotman Y, Fernie AR, Hoefgen R, Dijkwel P. (2013) Activation of R-mediated innate immunity and disease susceptibility is affected by different mutations in a cytosolic O-acetylserine (thiol) lyase in Arabidopsis. Plant J 73: 118–130 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17: 260–270 [DOI] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Hu TT, Traw MB, Horton M, Epple P, Kuhns C, Sureshkumar S, Schwartz C, Lanz C, et al. (2010) Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 465: 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto TA, Rauser L, Kamoun S. (2002) The pipg1 gene of the oomycete Phytophthora infestans encodes a fungal-like endopolygalacturonase. Curr Genet 40: 385–390 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F. (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- Verhage A, van Wees SC, Pieterse CM. (2010) Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol 154: 536–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Klessig DF, Park SW. (2008a) Systemic acquired resistance: the elusive signal(s). Curr Opin Plant Biol 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Vlot AC, Liu PP, Cameron RK, Park SW, Yang Y, Kumar D, Zhou F, Padukkavidana T, Gustafsson C, Pichersky E, et al. (2008b) Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J 56: 445–456 [DOI] [PubMed] [Google Scholar]
- Yang S, Yang H, Grisafi P, Sanchatjate S, Fink GR, Sun Q, Hua J. (2006) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45: 166–179 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dorey S, Swiderski M, Jones JD. (2004) Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J 40: 213–224 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y. (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20: 10–16 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.