Cooperative interactions between the sensor domain and the molecular switch domain of plant immune receptors are structurally defined.
Abstract
Many plant and animal immune receptors have a modular nucleotide-binding-leucine-rich repeat (NB-LRR) architecture in which a nucleotide-binding switch domain, NB-ARC, is tethered to a LRR sensor domain. The cooperation between the switch and sensor domains, which regulates the activation of these proteins, is poorly understood. Here, we report structural determinants governing the interaction between the NB-ARC and LRR in the highly homologous plant immune receptors Gpa2 and Rx1, which recognize the potato cyst nematode Globodera pallida and Potato virus X, respectively. Systematic shuffling of polymorphic sites between Gpa2 and Rx1 showed that a minimal region in the ARC2 and N-terminal repeats of the LRR domain coordinate the activation state of the protein. We identified two closely spaced amino acid residues in this region of the ARC2 (positions 401 and 403) that distinguish between autoactivation and effector-triggered activation. Furthermore, a highly acidic loop region in the ARC2 domain and basic patches in the N-terminal end of the LRR domain were demonstrated to be required for the physical interaction between the ARC2 and LRR. The NB-ARC and LRR domains dissociate upon effector-dependent activation, and the complementary-charged regions are predicted to mediate a fast reassociation, enabling multiple rounds of activation. Finally, we present a mechanistic model showing how the ARC2, NB, and N-terminal half of the LRR form a clamp, which regulates the dissociation and reassociation of the switch and sensor domains in NB-LRR proteins.
Resistance (R) proteins play a central role in the recognition-based immune system of plants. Unlike vertebrates, plants lack an adaptive immune system with highly specialized immune cells. Instead, they rely on an innate immune system in which each cell is autonomous. Two types of immune receptors can be distinguished in plants, pathogen-associated molecular patterns recognition receptors that detect conserved molecular patterns in plant pathogens and intracellular R proteins that recognize specific effectors employed by pathogens as modifiers of host metabolism or defense mechanisms (Jones and Dangl, 2006). Effector-triggered activation of R proteins leads to an array of protective responses, often culminating in programmed cell death at the site of infection (Greenberg and Yao, 2004), thereby preventing further ingress of the pathogen. Pathogens have evolved mechanisms to evade recognition by R proteins and to regain their virulence (Dodds and Rathjen, 2010). This continuous coevolutionary process between host and pathogen has resulted in a reservoir of highly diverse R proteins in plants, enabling them to counteract a wide range of pathogens and pests.
The most common class of R proteins consists of nucleotide-binding (NB)-leucine-rich repeat (LRR) proteins with a tripartite domain architecture, which roughly corresponds to an N-terminal response domain (a coiled coil [CC] or Toll/Interleukin-1 receptor [TIR] domain) involved in downstream signaling, a central molecular switch domain (the NB domain present in the mammalian apoptosis regulator Apaf1, plant R proteins, and the Caenorhabditis elegans apoptosis regulator CED4 [NB-ARC]), and a C-terminal sensor domain (the LRR domain). The NB-ARC domain is an extended nucleotide-binding domain that plant immune receptors share with metazoan apoptosis regulators and immune receptors such as Apaf1, CED4, and nucleotide-binding oligomerization domain (NOD-like) receptors (NLRs) and belongs to the STAND (signal transduction ATPases with numerous domains) family of nucleoside triphosphatase domains (van der Biezen and Jones, 1998; Leipe et al., 2004; Albrecht and Takken, 2006; Maekawa et al., 2011b). The overall modular architecture of metazoan STAND nucleoside triphosphatase is similar to that of NB-LRR plant immune receptors, but the domains flanking the NB-ARC domain often differ. In NLRs, for example, several N-terminal domains can be found, including caspase-recruiting domains and Pyrin domains (Proell et al., 2008). In the mammalian protein Apaf1, the sensor involved in cytochrome c detection consists of C-terminal WD40 repeats (Zou et al., 1997).
In plant NB-LRR resistance proteins, the recognition of a pathogen effector via the LRR domain is thought to switch the conformation of the protein from a closed, autoinhibited “off” state into an open, active “on” state (Lukasik and Takken, 2009). The activation of NB-LRR proteins is most likely a multistep process in which the NB-ARC domain plays a central role. The three subdomains of the NB-ARC, the NB, ARC1, and ARC2, collectively form a nucleotide-binding pocket that adopts different conformations depending on the bound nucleotide. This mechanism seems to be conserved between proteins from organisms as distant as bacteria, metazoans, and plants (Rairdan and Moffett, 2007; Danot et al., 2009; Takken and Tameling, 2009). The conformational change coincides with the exchange of bound ADP for ATP in the NB-ARC, probably stabilizing the active conformation (Tameling et al., 2006; Ade et al., 2007). Hydrolysis of the bound ATP is hypothesized to return the domains to their inactive state. The exact mechanism by which elicitor recognition via the LRR leads to a conformational change of the NB-ARC and the subsequent activation of immune signaling pathways is not clear.
Previous studies have shown that the CC/TIR, NB-ARC, and LRR domains in plant immune receptors interact and cooperate with each other in an interdependent manner (Moffett et al., 2002; Leister et al., 2005; Ade et al., 2007; Rairdan et al., 2008). From these data, a picture emerges in which the LRR domain is not only involved in pathogen recognition, but also plays a role in maintaining an autoinhibited resting state in the absence of pathogens via its interactions with the other domains (Bendahmane et al., 2002; Hwang and Williamson, 2003; Ade et al., 2007; Qi et al., 2012). A similar role as regulatory domain has been found for the sensor domains of other NLRs, such as the mammalian Apaf1 (Hu et al., 1998). For the potato (Solanum tuberosum) immune receptor Rx1, a model plant NB-LRR protein, it has been shown that the LRR cooperates with the ARC subdomains in retaining the inactive state of the protein. The deletion of the ARC and LRR domains leads to a constitutive activity of the NB (Bendahmane et al., 2002; Rairdan et al., 2008). In addition, it was demonstrated that the elicitor, the Potato virus X (PVX) coat protein, modifies the interdomain interactions in Rx1 (Moffett et al., 2002; Rairdan et al., 2008). Sequence exchanges between Rx1 and the highly homologous nematode resistance protein Gpa2 (88% amino acid identity) resulted in incompatibilities between the domains that give rise to inappropriate activation of cell death responses (Rairdan and Moffett, 2006), indicating that the cooperation between the sensor and switch domains depends on an interaction fine tuned by intramolecular coevolution. In this light, it is interesting to note that a functional ortholog of Rx1, Rx2 from Solanum acaule, is almost identical to Rx1 in its LRR region but displays a higher similarity to Gpa2 in stretches of its CC-NB-ARC sequence (Bendahmane et al., 2000).
The aim of our study was to pinpoint the molecular determinants controlling the switch between the resting and activation state of NB-LRR proteins. The incompatibility between the ARC and LRR domains of Rx1 and Gpa2 was used as a guideline to dissect the molecular and structural determinants involved in the cooperation between the switch (NB-ARC) and sensor (LRR) domain. An extensive exchange of polymorphic residues between these two homologous NB-LRR proteins resulted in the identification of a minimal fragment of 68 amino acid residues in the ARC2 domain and the first LRR repeats as being crucial for proper activation of Gpa2 and Rx1. Within this minimal region, we identified two amino acids that, despite their proximity in the amino acid sequence, differentiate between elicitor-dependent (position 401) and independent activation (position 403). However, structural modeling of the domains shows that the residue at position 403 operates at the interface of the ARC2 and N-terminal part of the LRR domain, while residue 401 mapped at the interface between the ARC2 and NB domain. Furthermore, an acidic loop region in the ARC2 domain and complementary-charged basic patches in the N-terminal half of the LRR domain are shown to be required for the physical interaction between these domains. We demonstrate that the binding between the CC- NB-ARC and LRR domains is disrupted upon elicitor-dependent activation and that the complementary-charged residues are predicted to facilitate reassociation. Two independent docking simulations of the NB-ARC and LRR domain indicate that the LRR domain binds to the NB-ARC domain at the surface formed by the interaction of the ARC2 and NB subdomains. We present a mechanistic model in which the first repeats of the LRR, the ARC2 subdomain, and the NB form a clamp, which governs the shuttling between a closed, autoinhibited “off” state and an open, active “on” state of the resistance protein. Finally, we discuss the consequences of the functional constraints imposed by the interface of the NB, ARC2, and LRR domain for the generation of novel resistance specificities via evolutionary processes and genetic engineering.
RESULTS
A Minimal Region in the ARC2 of Gpa2 Determines the Incompatibility with the LRR Domain of Rx1
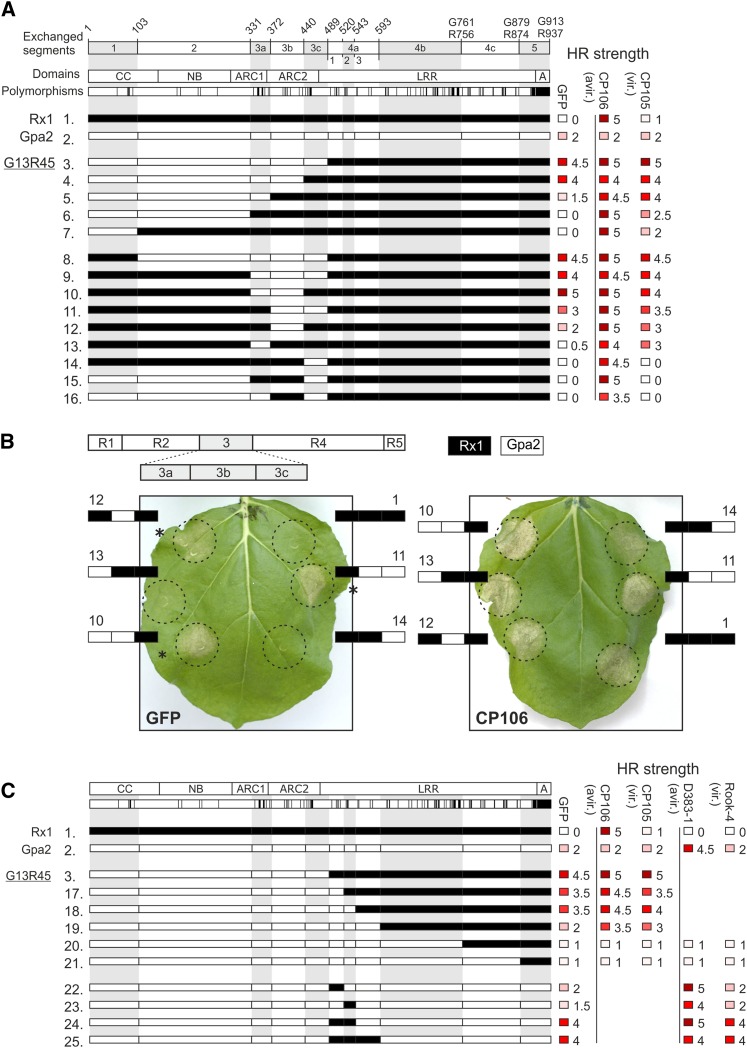
To identify the amino acids in Gpa2 that caused the incompatibility with the LRR of Rx1 (Rairdan and Moffett, 2006), we systematically replaced parts of the CC-NB-ARC of Gpa2 with the corresponding Rx1 sequences in a constitutively active chimera of Rx1 and Gpa2 (G13R45 construct [Fig. 1A, constructs 3–7]). Constitutive (elicitor-independent) activation was assessed by transiently coexpressing the constructs with GFP as control in Nicotiana benthamiana plants and scoring the hypersensitive response. Elicitor-dependent activation was assessed by coexpressing the PVX coat protein to activate Rx1 (eliciting CP106 and noneliciting CP105) and variants of the RanBPM-like effector (RBP1) of the potato cyst nematode Globodera pallida to activate Gpa2 (i.e. eliciting RBP1 variant D383-1 and noneliciting RBP1 variant Rook4; Sacco et al., 2009). The strength of the observed hypersensitive response was ranked from 0 to 5 (no hypersensitive response to full hypersensitive response; Supplemental Fig. S1), and scores of at least three independent experiments with four replicates each were averaged. All tested chimeric constructs are untagged; however, the constructs that displayed a loss of function were recloned with an N-terminal c-Myc-tag to assess their stability via an immunoblot (Supplemental Fig. S2).
Figure 1.
Phenotypes of sequence exchange constructs exploring the incompatibility between the Gpa2 NB-ARC and Rx1 LRR. A, The top row shows sequence fragments exchanged between Gpa2 and Rx1 with the positions of the breakpoints in the amino acid sequence given above. If the numbering of the positions differ between Rx1 and Gpa2, both numbers are given. The second row shows the domain architecture with the CC, NB, ARC1, and ARC2 subdomain and the LRR domain. In the third row, the amino acid positions polymorphic between Rx1 and Gpa2 are indicated. The Rx1 sequence is depicted in white and the Gpa2 sequence is depicted in black in the chimeric constructs. The constructs 1 to 16 are created to delineate the Gpa2 NB-ARC segment incompatible with the Rx1 LRR underlying the constitutive activity of G13R45. On the right-hand side, the hypersensitive response (HR) phenotype of these constructs is given for coexpression with GFP (elicitor-independent activity), CP106 (elicitor-dependent activity), and CP105 from the Rx1-breaking strain PVXHB to test recognition specificity. The strength of the hypersensitive response phenotypes after 7 d are given by a scale from 0 (white = no hypersensitive response) to 5 (dark red = full necrosis), averaged over at least three repetitions and rounded to 0.0 or 0.5. B, Phenotypes of the constructs in which fragments of the ARC domain in the Rx1 background are replaced by Gpa2 sequences. The ARC fragments (3a, 3b, and 3c) are shown in white (Rx1) or black (Gpa2) to depict the composition of the chimeric construct. The numbering corresponds to the numbering of the constructs in Figure 1A. Coexpression with GFP shows autoactive response. Coexpression with PVX CP106 shows the elicitor-dependent activation. A minimal incompatible domain (G3b, amino acid positions 372–440) was identified in which seven residues differ between Rx1 and Gpa2 (construct 12). C, Overview of the sequence exchange constructs aimed at defining the LRR region responsible for the observed incompatibility between the Gpa2 CC-NB-ARC and Rx1 LRR (G13R45). Like Figure 1A, the observed autoactive and specific cell death responses are indicated, and the responses to RBP D383-1 (an elicitor of Gpa2) and RBP Rook-4 from a Gpa2-breaking Globodera pallida population are included.
Replacing the C-terminal part of the ARC2 (fragment 3c) resulted in a slight reduction of both autoactivity and elicitor-dependent activity (Fig. 1A, construct 4). This fragment contains seven of the 14 polymorphic amino acid positions in the ARC2. Replacing the complete ARC2 domain of Gpa2 (fragment 3b and 3c) with the Rx1 sequence (Fig. 1A, construct 5) strongly reduced the autoactive response, while the elicitor-dependent response became only slightly weaker. Exchanging both ARC1 and ARC2 of Gpa2 in G13R45 (Fig. 1A, construct 6) resulted in a complete loss of autoactivity and similar responses to CP106 as Rx1.
Replacing only the ARC1 and ARC2 domains of Rx1 with the corresponding Gpa2 sequence (Fig. 1, A and B, construct 9) resulted in autoactivity indistinguishable from that seen for G13R45. Replacing the single fragments (3a–3c) of the ARC domain in an Rx1 background by Gpa2 sequences only led to constitutive activity when the N-terminal half of the ARC2 (fragment 3b) was also replaced (Fig. 1, A and B, constructs 12–14). However, combining fragment G3b of the ARC2 with either the N-terminal- or C-terminal-flanking Gpa2 fragments (G3ab and G3bc) resulted in a level of constitutive activity comparable to that seen when the full ARC domain derived from Gpa2 was introduced (Fig. 1, A and B, constructs 10 and 11). The construct in which only G3b is replaced with R3b in the G13R45 background shows no autoactivity at all (Fig. 1A, construct 16).
In conclusion, these data show that the N-terminal part of the ARC2 region (fragment 3b; aa372-440) has a central role in the cooperation of the NB-ARC with the LRR domain.
The First Two N-Terminal Repeats in the LRR Region of Rx1 Are Incompatible with the ARC2 of Gpa2
The incompatibility between the ARC2 of Gpa2 and the LRR of Rx1 depends not only on differences between the Gpa2 and Rx1 in the ARC2 domain, but on polymorphisms in the LRR as well. To assess whether we could delineate a region in the LRR of Rx1 responsible for the incompatibility with the ARC2 of Gpa2, we stepwise replaced fragments of the LRR of Rx1 in the constitutively active G13R45 construct (Fig. 1C, construct 3) with sequences of Gpa2 from the junction between the ARC2 and LRR toward the C terminus (Fig. 1C, constructs 17–21). A loss of the autoactive response was observed when the first three repeats of the LRR were replaced by the Gpa2 sequence (Fig. 1C, constructs 17–19). These constructs could still be activated by the Rx1 elicitor CP106, giving a consistently stronger response when coexpressed with the PVX coat protein than when coexpressed with GFP. However, replacing repeats 4 to 9 and 10 to 14 of the LRR domain of Rx1 with the corresponding Gpa2 sequences did not enable a specific activation by either the viral or the nematode elicitors (Fig. 1C, constructs 20 and 21). MYC-tagged versions of constructs 19 to 21 could be detected by western blotting at levels similar to those of Gpa2 and Rx1, demonstrating that any loss of function cannot be attributed to a loss of protein stability (Supplemental Fig. S2).
Because exchanging the fragment encompassing the first three repeats of the LRR of Rx1 by the corresponding Gpa2 sequence abolished the autoactivity observed for G13R45 (Fig. 1C, construct 3), we further explored the role of this region (4a) of the LRR in the interdomain incompatibility. Segments 4a1 to 4a3, consisting of individual Leu-rich repeats and combinations thereof, were replaced by their Rx1 counterparts in Gpa2 (Fig. 1C, constructs 22–25). Replacing only the first or only the second Leu-rich repeat in the LRR domain of Gpa2 with the corresponding Rx1 sequences did not result in constructs that were phenotypically different from wild-type Gpa2 (Fig. 1C, constructs 22 and 23). Exchanging both the first and second repeat of the LRR domain in Gpa2 with the Rx1 sequence on the other hand resulted in a strongly autoactive protein (Fig. 1C, construct 24). No further increase in the autoactivity was observed when the third repeat from Rx1 was replaced in addition to the first two (Fig. 1C, construct 25). Based on these data, we conclude that the amino acid sequence divergence in the first two N-terminal repeats of the LRR region of Rx1 causes this domain region to be incompatible with the ARC2 of Gpa2.
Combining the ARC2 of Gpa2 with the LRR of Gpa2 Restores Elicitor-Dependent Activity
Unlike the strongly autoactive G13R45, the reciprocal construct R13G45 (Fig. 2A, construct 26), in which we combined the CC-NB-ARC of Rx1 and the LRR of Gpa2, shows a loss of elicitor-dependent activation. The weak elicitor-independent activation displayed upon overexpression of Gpa2 is also not observed for R13G45. MYC-tagged versions of Gpa2 and R13G45 were detected at similar levels on western blots of total leaf extracts, showing that the loss of autoactivity was not due to instability of the R13G45 protein (Supplemental Fig. S2). Based on our previous results, we hypothesized that replacing the region that determines incompatibility between the CC-NB-ARC of Rx1 and the LRR of Gpa2 in R13G45 with the corresponding Gpa2 sequence would lead to a reconstitution of the Gpa2-specific elicitor-dependent activation. Replacing only the C-terminal half of the ARC2 of Rx1 in R13G45 with the corresponding sequences of Gpa2 resulted in a weak RBP1-dependent activation (Fig. 2A, construct 27). Replacing the complete ARC2 (fragments 3b and 3c) resulted in an intermediate cell death response to RBP1(Fig. 2A, construct 28). When both ARC1 and ARC2 of Rx1 were exchanged for their Gpa2 counterparts, the resulting elicitor-dependent and elicitor-independent responses became indistinguishable from the full-length Gpa2 protein (Fig. 2A, construct 30). Thus, when an increasingly large segment of the CC-NB-ARC of Rx1 in R13G45 was replaced with the corresponding Gpa2 sequence, we observed a gradual regain of elicitor-dependent activation in response to the elicitor RBP1 (Fig. 2A, constructs 27–32). The stability of MYC-tagged versions of constructs 26 to 32 was confirmed by western blotting (Supplemental Fig. S2).
Figure 2.
Exploring the incompatibility of the CC-NB-ARC of Rx1 with the Gpa2 LRR in chimeric constructs. A, Constructs 26 to 36 explore the domain incompatibility between the CC-NB-ARC of Rx1 and the LRR of Gpa2, which is characterized by a loss of elicitor-dependent activation in construct R13G45. Elicitor-dependent activation (after coexpression with RBP D383-1) and elicitor-independent activation (the response observed after coexpression with GFP) are indicated on a scale from 0 to 5 (no response to full necrosis).
The C-terminal half of the ARC2 domain of Gpa2 (fragment 3c) in the background of the Gpa2 protein can be replaced with the corresponding Rx1 sequence without a complete loss of RBP1-induced activation (Fig. 2A, construct 33). The exchange of the N-terminal half of the ARC2 in Gpa2 (fragment 3b) with the Rx1 sequence resulted in loss of elicitor-dependent activation and the weak autoactivation typically associated with overexpression of full-length Gpa2 (Fig. 2A, construct 35). MYC-tagged versions of constructs 33 to 36 could be shown to be expressed to similar levels as Gpa2 or Rx1 on western blot (Supplemental Fig. S2). Overall, our data support a model in which parts of the ARC2 subdomain and the first N-terminal repeats of the LRR domain form a regulatory unit within Gpa2 and Rx1. In the absence of the elicitor, these domain regions keep the resistance protein in an autoinhibited state, while in the presence of the elicitor, they cooperate in the activation of the R protein.
Two Residues that Are Polymorphic between Gpa2 and Rx1 Distinguish between Elicitor-Independent and Elicitor-Dependent Activation
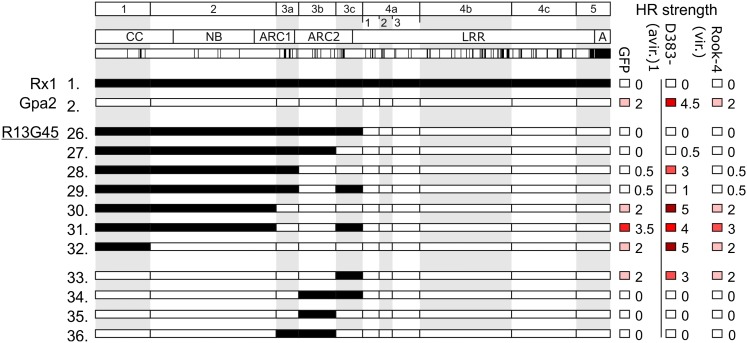
Based on our Rx1/Gpa2 sequence exchanges, we concluded that among the seven polymorphic residues in the ARC2 region from amino acid positions 372 to 440 (fragment 3b) of Gpa2 and Rx1, there are residues that play an important role in the functional interactions between the NB-ARC and LRR domain (Fig. 3A). Position 401 in the ARC2 domain of Rx1 harbors a Gln, while Gpa2 has an Arg at this position. We hypothesized that a large and positively charged residue at this position could have a major impact on the conformation of the NB-ARC domain. Interestingly, in a previous study, the mutagenesis of the residues at the adjacent positions 399 and 400 in Rx1 has been shown to cause the constitutive activation of Rx1 (Bendahmane et al., 2002). Therefore, this position was selected for further analysis. The second notable difference between Gpa2 and Rx1 occurs at position 403, where Rx1 carries a Ser and Gpa2 a Tyr. Here, we hypothesized that the difference in characteristics between the small Ser (van der Waals volume 73 Å3) and the bulky aromatic Tyr (141 Å3) might have contributed to the observed incompatibility after exchanging this sequence fragment between Rx1 and Gpa2. In addition, position 403 is one of only two positions (403 and 405) in which Rx2 differs from Gpa2 in the ARC2 between amino acid positions 372 to 440 (Bendahmane et al., 2000). Like Rx1, Rx2 carries a Ser at position 403, which makes the Tyr at this position unique for Gpa2 and interesting for further testing.
Figure 3.
The role of the polymorphic ARC2 positions 401 and 403 in elicitor-dependent and elicitor-independent activation. A, Alignment of the amino acid sequences of Gpa2 and Rx1 in sequence exchange fragments 3a, 3b, and 3c. The predicted secondary structure is shown above the sequences. Bars represent α-helices, and arrows denote β-strands. Polymorphic positions are highlighted by gray boxes. Positions 401 and 403 are indicated by stars. The identities of these positions were exchanged between Gpa2 and Rx1 via targeted mutagenesis and introduced in constructs either as single substitutions or as a pair. B, Agroinfiltration assay of Gpa2 or the autoactive Gpa2/Rx1 construct G13R45 and mutant versions thereof. The Gpa2 variants were coexpressed with its elicitor RBP1 from Globodera pallida. The G13R45 variants were expressed in the absence of an elicitor to assess the elicitor-independent cell-death response. C, The effect of exchanging the residues at positions 401 and 403 between Rx1 and Gpa2 on the CC-NB-ARC/LRR interaction. In c-Myc-tagged versions of the CC-NB-ARC domains of Gpa2 and Rx1, the residues at positions 401 (Gpa2 R and Rx1 Q) and 403 (Gpa2 Y and Rx1 S) were exchanged. All possible combinations of Gpa2 and Rx1 CC-NB-ARC and LRR domains were tested for interaction in an anti-c-Myc coimmunoprecipitation assay. The immunoblots (bottom) show the coimmunoprecipitated 4×HA-tagged LRR domains. The LRR of Gpa2 binding the CC-NB-ARC R401Q of Gpa2 is indicated by a chevron. D, Elicitor-dependent weakening of the CC-NB-ARC/LRR interaction. The CC-NB-ARC and LRR domains of either Gpa2 or Rx1 were coexpressed in the absence and presence of their elicitors (G. pallida RBP1 and PVX CP106, respectively). Protein was extracted 32 hours post-infiltration, and the c-Myc-tagged CC-NB-ARC domains were immunoprecipitated. E, Interaction of the wild-type and R401Q version of the CC-NB-ARC of Gpa2, with the FLAG-tagged LRR in the presence of the coexpressed elicitor (G. pallida RBP1). The c-Myc-tagged CC-NB-ARC domains were immunoprecipitated. [See online article for color version of this figure.]
We exchanged the individual amino acids at positions 401 and 403 between the ARC2 sequence of Gpa2 and Rx1 to study the effect on elicitor-dependent and elicitor-independent activation of Gpa2, Rx1, and the constitutively active G13R45 (Fig. 3A). These substitutions were introduced individually or paired, resulting in nine full-length constructs: G13R45 (R401Q, Y403S, and RY/QS), Gpa2 (R401Q, Y403S, and RY/QS), and Rx1 (Q401R, S403Y, and QS/RY). The resulting constructs were tested for elicitor-dependent activation and elicitor-independent autoactivation in agroinfiltration assays (Fig. 3B). The mutations at aa positions 401 and 403 resulted in distinct phenotypes. Both Gpa2 R401Q and Gpa2 RY/QS showed a reduced elicitor-dependent cell death response, which was not seen for Gpa2 Y403S (Fig. 3B). G13R45 Y403S and G13R45 RY/QS, on the other hand, showed a reduction in the autoactive response, whereas G13R45 R401Q did not (Fig. 3B). However, neither the individual Q401R or S403Y substitutions nor the combination of the two in Rx1 resulted in autoactivation, as observed for the Rx1 in which fragment 3b was exchanged for the Gpa2 sequence (Fig. 1A, construct 12; data not shown). The substitutions in Rx1 did not result in a change in the strength of the elicitor-dependent hypersensitive response (not shown) or in a significant change in the PVX resistance mediated by Rx1 in a transient resistance assay (Fig. 3B). Apparently, from the seven polymorphic residues in fragment 3b, other residues in addition to those at positions 401 and 403 need to be exchanged to reproduce the elicitor-independent activation observed for construct 12 (Fig. 1A).
To determine if the observed phenotypes of the 401/403 substitutions are linked to the physical interaction of the CC-NB-ARC and LRR, we performed coimmunoprecipitation experiments with coexpressed CC-NB-ARC and LRR constructs (Fig. 3C). All combinations of the CC-NB-ARC wild-type and mutant variants with the wild-type LRR domains of Rx1 and Gpa2 were tested in this experiment. The substitutions at position 403 did not lead to a reduction or increase of the physical interaction between the CC-NB-ARC and LRR, compared with the interaction of the wild-type constructs. Gpa2 CC-NB-ARC R401Q, however, showed a consistently higher affinity for the LRR domain of Gpa2 and as well as the LRR of Rx1, compared with the wild-type CC-NB-ARC of Gpa2 or the CC-NB-ARC of Gpa2 with Y403S substitution (Fig. 3C). We noticed in several experiments that the coexpression of Gpa2 CC-NB-ARC-LRR R401Q led to a slightly higher accumulation of the wild-type Gpa2 LRR domain, which indicates that the LRR is stabilized by the increased interaction with the CC-NB-ARC R401Q. However, we observed that the wild-type Gpa2 LRR coimmunoprecipitated at higher levels with CC-NB-ARC R401Q, even when its levels in the input material were not slightly elevated.
The CC-NB-ARC and LRR of Rx1 dissociate in the presence of the PVX coat protein in plants in which expression of the co-chaperone SGT1 (Suppressor of G2 allele of SKP1) is silenced, in which Rx1 cannot initiate a cell death response (Moffett et al., 2002), indicating that the dissociation of these domains is part of the activation mechanism. This earlier finding led us to hypothesize that the reduction in elicitor-dependent activation of the R401Q mutant of Gpa2 (Fig. 3B) could be caused by a stronger association of the CC-NB-ARC and LRR domains. We tested the effect of the elicitor on the interaction between the CC-NB-ARC and the LRR for both Gpa2 and Rx1 in nonsilenced N. benthamiana plants to determine if both R proteins respond to the elicitor in a similar way (Fig. 3D). Coexpression of the elicitor leads to a cell death response in approximately 40 to 50 h after agroinfiltration of Rx1 or Gpa2. To avoid the cell death, the leaves were harvested 30 h post agroinfiltration. For both Gpa2 and Rx1, the presence of the elicitor led to a reduction of the level of the LRR construct that was pulled down with the CC-NB-ARC (Fig. 3D). The Gpa2 CC-NB-ARC R401Q retained a stronger LRR interaction, even when coexpressed with the elicitor (Fig. 3E). If the dissociation of the CC-NB-ARC and LRR is essential for the elicitor-dependent activation of Gpa2 and Rx1, the higher affinity between CC-NB-ARC and the LRR domain resulting from the R401Q substitution in ARC2 may reduce the elicitor-dependent activation of Gpa2.
In Silico Modeling of the NB-ARC and LRR Domains of Rx1 and Gpa2 Reveals Two Novel Structural Elements
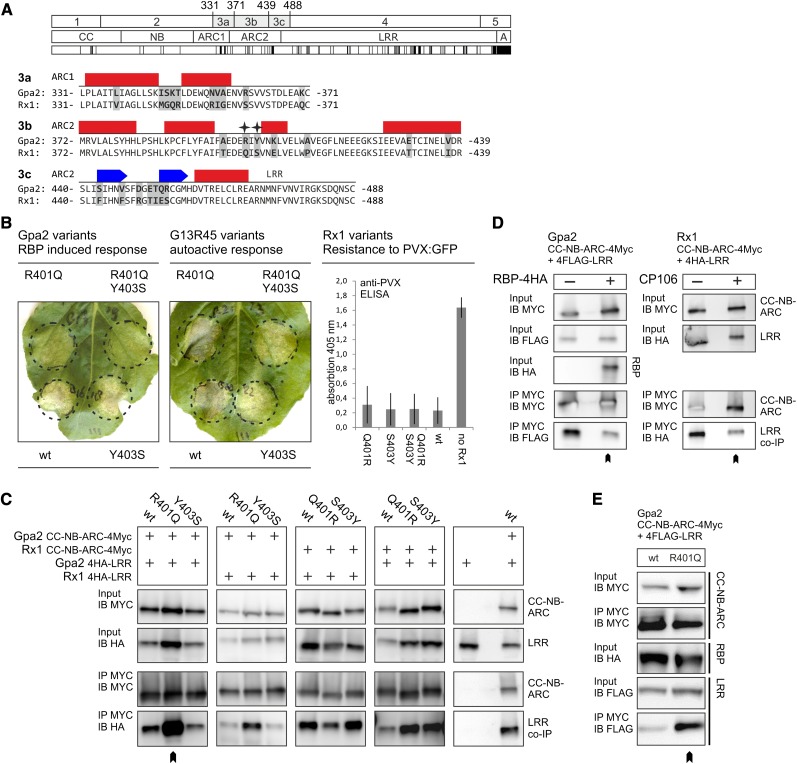
As the polymorphisms between Gpa2 and Rx1 in the N-terminal repeats of the LRR and the ARC2 affect interdomain interaction and cooperation, we hypothesized that the relevant residues should be surface exposed. To test this, remote homology models of the NB-ARC and LRR three-dimensional structure were generated. The crystal structure of the ADP-bound human apoptosis regulator Apaf1 (Protein Data Bank [PDB] entry 1z6t; Riedl et al., 2005) was selected as a template to model the NB-ARC of Gpa2 based on similarity between the Gpa2 and Apaf1 amino acid sequence and predicted secondary structure, as exemplified by a high BLAST e-value (1e–5) and the best score in the fold recognition using Phyre with a confidence value of 100% (2.1e–21). Due to the high sequence similarity of Gpa2 and Rx1, the overall predicted structure of their domains is highly similar. Therefore, the structure prediction model of the NB-ARC of Gpa2 is presented here as an example.
The amino acid sequences of Gpa2 and Apaf1 were aligned, while taking into account the secondary structure predictions, hydrophobicity, contact propensity profiles, and conserved critical amino acids (Supplemental Fig. S3). From the five subdomains present in the Apaf1 structure (1z6t), only the nucleotide-binding domain, helical domain I, and winged-helix domain have counterparts in Gpa2 (the NB, ARC1, and ARC2, respectively). Although the sequence identity of Gpa2 and Apaf1 is only 19.3%, the similarity (49.1%) is sufficient to generate a three-dimensional model by remote homology modeling. The regions with conserved sequences were modeled by coordinate transfer. In the variable regions, the side chains were reconstructed, and loop regions were generated randomly and filtered for steric constraints, followed by successive rounds of simulated annealing and energy minimization, which led to an increase of the overall model accuracy from 3.80 Å to 2.58 Å according to MetaMQAP estimation of root-mean-square deviation (RMSD). The amino acids forming the nucleotide-binding pocket in ADP-bound Apaf1 have a perfect match in the Gpa2 NB-ARC structure (Apaf1/Gpa2): K160/K176 (P-loop motif, NB), D244/D245 (Walker B motif, NB), P321/P332 (GxP motif, ARC1), and H438/H459 (MHD motif, ARC2). In the ADP-bound state, the NB, ARC1, and ARC2 subdomains each contribute to the ADP-binding pocket, forming a compact globular structure around the nucleotide with the N-terminal NB subdomain interacting with the C-terminal ARC2 subdomain. Interestingly, a loop, absent in the Apaf1 structure, protrudes from the ARC2 subdomain of Gpa2 and Rx1 in the model. This loop protrudes the surface of the predicted ARC2 structure and contains most of a remarkable acidic cluster of nine Glu residues between positions 409 to 435 (ELWAV EGFLN EEEGK SIEEV AETCI NE).
A composite three-dimensional structural model was build for the LRR domain of Gpa2. No overall matching template for the LRR domain of Gpa2 and Rx1 was available in the PDB, mainly due to the irregularity of the Leu-rich repeats (Supplemental Fig. S4) and low global sequence identities. Therefore, rather than use a unique global template, we decided to use the best local repeat templates instead and build a composite LRR model by a special remote homology procedure known as the Optimized Joined Fragment Remote Homology modeling (OJFRHM) method (Sela et al., 2012; see “Materials and Methods”). LRR segments were modeled after the best local templates: repeats 1–4 and 8 and 9, rna1p (PDB entry 1yrg), repeats 5 to 7, decorin (PDB entry 1xku), and repeats 10 to 15, Toll-like receptor3 (PDB entry 2a0z). The overall template was built by joining the local templates into the global LRR architecture using the strict constraints imposed by the hydrogen bond network of the LRR motif and maximizing the atomic contacts of the interface between local templates. The model (Fig. 4A) was refined by repeated rounds of energy minimization resulting in a fine-tuning of the slight overall curvature and twist of the LRR structure. Iterative local refinement led to an increase of the overall model accuracy from 3.98 to 3.41 Å according to MetaMQAP root-mean-square deviation estimation, with higher scores, up to a RMSD of 1.0 Å, within the LxxLxLxxN/C motif region and in the lateral and concave regions of the surface.
Figure 4.
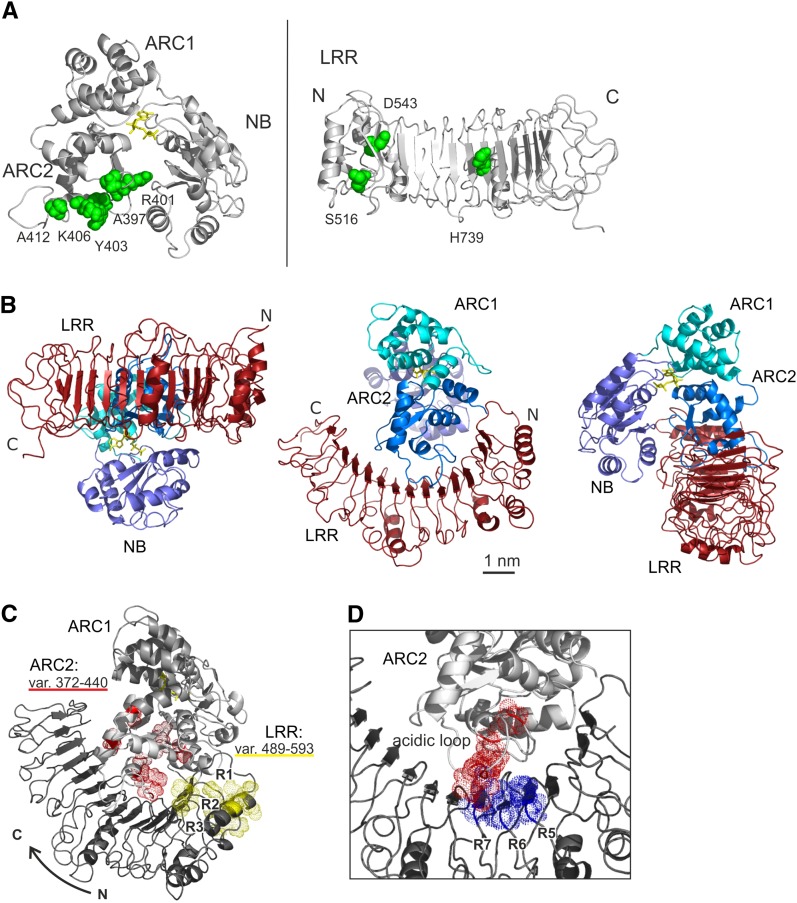
Three-dimensional models of the structure of the NB-ARC and LRR domains of Gpa2. A, The structure of the NB-ARC of Gpa2 created by remote homology modeling after the corresponding Apaf1 crystal structure is shown on the left. The three subdomains (NB, ARC1, and ARC2) are provided in different shades of blue. In the structure, the main residues from each of the subdomains contributing to the nucleotide-binding pocket are shown in red in the structure (NB: K176 [P loop] and D245 [Walker B]; ARC1: P332 [GxP motif]; and ARC2: H459 [MHD motif]). The ADP molecule is shown in yellow. The predicted three-dimensional structure model for the LRR domain of Gpa2 as based on the OJFRHM method (see “Materials and Methods”) is shown on the right. The origin of the template segments used in the modeling study are indicated by colors on the LRR structure. The matching sequence stretches in the templates and the LRR of Gpa2 are given below. B, The positions of the Gpa2/Rx1 polymorphisms in the various sequence exchange fragments (Fig.1) are shown in the NB-ARC and LRR models. The van der Waals radius of the Gpa2 residues matching the polymorphisms in the minimal incompatible regions in the ARC2 (3b) and the N-terminal repeats of the LRR (4a) is indicated by a dotted surface. An alignment of the amino acid segments 4a1 to 4a3 is shown below, with the amino acids that differ between Gpa2 and Rx1 indicated in the colors used in the model of the LRR structure. The β-strands in repeats 1 to 3 (R1–R3) are indicated by a gray box above the sequence. C, The surface electropotential of the Gpa2 NB-ARC and LRR domains. The surface charge (negative in red, positive in blue, and neutral in white) was mapped on the van der Waals surface of the NB-ARC and LRR domains.
In LRR repeat 9 (amino acid positions 734–740 in Gpa2), sequence polymorphisms were observed in the LxxLxLxxN/C motif of Rx1 (Supplemental Fig. S5A), which are predicted to have an impact on the local structure. The amino acids present in repeat 9 in Gpa2 follow the canonical LRR motif, whereas the amino acids present in Rx1 are highly unlikely to form a regular LRR repeat structure, as they introduce polar and charged residues to the hydrophobic core of the LRR structure (Supplemental Fig. S5A). The fact that the local sequence of Rx1 shows a high contact-forming propensity and no sign of intrinsic disorder suggests that the region corresponding to repeat 9 is not an unstructured linker, but forms a compact protruding structure separating the two halves of the LRR (Supplemental Fig. S5B).
The Majority of the Polymorphic Residues Involved in the NB-ARC/ LRR Interactions Are Predicted to be Surface Exposed
To determine the spatial distribution of the amino acids varying between Rx1 and Gpa2 in the ARC2 and N-terminal end of the LRR, they were mapped onto our three-dimensional models (Fig. 4B). The majority of the seven polymorphic positions in the ARC2 region essential for NB-ARC/LRR cooperation (3b in Fig. 1A; Fig. 3A) are surface exposed in our model, which is consistent with a role in domain interactions. This segment of the ARC2 borders the groove where the NB and ARC2 interact and close around the nucleotide-binding pocket (Fig. 4B, polymorphic residues depicted in red). Even though positions 401 and 403 are only one amino acid apart in the protein sequence, their positions in the NB-ARC structure are different. In our model, residue 401 lies in the interface between the ARC2 and the NB subdomains. The residue at position 403 is placed on the outer surface of the ARC2, but close to the interface of the NB and ARC2 (Fig. 4B). Their proximity to the nucleotide-binding pocket (401) and the interface between the NB and ARC2 could account for the observed effects on elicitor-dependent or independent activation (Fig. 3).
Out of 10 polymorphisms in the N-terminal repeats of the LRR responsible for the LRR/ARC2 incompatibility (4a in Fig. 1; Fig. 4B), six mapped to one lateral-side C terminus to the LxxLxL motif and the adjacent positions on the convex side of the LRR (Fig. 4B). Only two polymorphisms in exchange segment 4a1 (E507 and V511) are present in the model and are located on the concave surface (Fig. 4B). Three polymorphisms in this segment could not be mapped on the predicted structure because they lay outside the sequence that was used to model the LRR domain. So, a small set of amino acid residues polymorphic between Gpa2 and Rx1 on the variable side of the LRR structure appears to be important in the cooperation between the N-terminal repeats of the LRR and the ARC2.
The LRR Domain of Rx1 and Gpa2 Can Be Divided into Two Subunits Based on Their Charge Distribution Patterns
The prediction of an acidic loop in the ARC2 prompted us to map the surface electropotential on the three-dimensional models of the NB-ARC and LRR domains to be able to evaluate its functional significance, for example in interdomain interactions. The NB-ARC has a mostly negative surface, with a few small positive spots on the NB and ARC1 (Fig. 4C). The ARC2 has a highly negative potential, to which the concentration of Glu residues in the acidic loop makes an important contribution. In contrast to this mostly uniform charge distribution on the NB-ARC domain, the charge distribution of the LRR domain is highly segregated over the horseshoe-like structure, neatly separating the surface into a predominantly positively charged N-terminal region (repeats 1–8) and a second half that is predominantly negatively charged (repeats 10–14; Fig. 4C).
Such a strict segregation in charge distribution might be indicative of functional differences between the two halves. Interestingly, this charge distribution pattern corresponds with the division of the LRR of Rx1 in two subunits by the structural motif at the position of LRR repeat 9. Furthermore, a high local concentration of positively charged residues, distant in sequence but contiguous on the surface, is formed on one lateral side of the N-terminal half of the LRR structure in repeats 5 to 7 by K604, R628, K629, K650, K675, and K676.
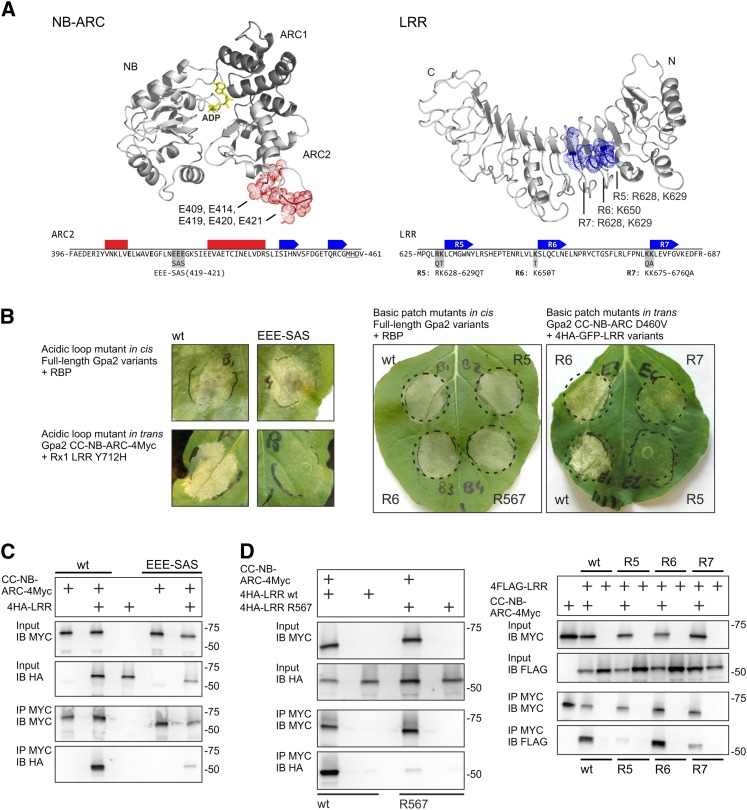
An Acidic Loop on the ARC2 and a Cluster of Basic Residues on the N-Terminal Half of the LRR Are Required for the Interaction between the CC-NB-ARC and LRR
To assess the putative functional relevance of the basic patch in the LRR and the acidic loop in the ARC2 subdomain (Fig. 5A) in the interaction between the CC-NB-ARC and the LRR domains, Gpa2 constructs were made in which residues in these charged patches were altered to noncharged residues via site-directed mutagenesis (Fig. 5A). The effect of these substitutions on the functioning of Gpa2 was tested both in full-length constructs (in cis) and in coexpressed CC-NB-ARC and LRR domains (in trans). Because functionality in trans depends fully on the interaction between the domains, it was expected that mutations influencing this domain interaction would have a greater impact on the functionality of in trans coexpressed domains than on functionality of the domains expressed in cis.
Figure 5.
Charged residues in a basic patch and acidic loop are required for the CC-NB-ARC/LRR interaction. A, Targeted mutagenesis of the acidic loop in the NB-ARC and the basic patch in the LRR. The residues forming the acidic loop in the NB-ARC structure and the location of the residues forming a basic patch on the LRR are indicated on the modeled structures. The positions of the mutations in the ARC2 and LRR are shown in the Gpa2 sequence below. The predicted local secondary structure is indicated by bars (α-helices) or arrows (β-strands). In the LRR, the repeat numbers (R5–R7) are given. B, In cis and in trans functionality of the acidic loop and basic patch mutants of Gpa2. The in cis functionality was tested by coexpressing full-length Gpa2 carrying the mutations with the Globodera pallida elicitor RBP1. The in trans functionality was tested by coexpressing the mutated CC-NB-ARC or LRR domains with complementary domains carrying interaction-dependent autoactivating mutations (Rx1 LRR Y712H or Gpa2 CC-NB-ARC D460V). C, Immunoprecipitation assay to test the role of the acidic loop in the ARC2 domain in the CC-NB-ARC/LRR interaction. Wild type (wt) and mutant (EEE419-421SAS) versions of the CC-NB-ARC of Gpa2 were tested for their ability to coimmunoprecipitate the Gpa2 LRR. D, Immunoprecipitations of the CC-NB-ARC of Gpa2 with the basic patch mutant versions of the LRR. LRR versions were tested in which either the full basic patch was mutated (R567) or basic residues from single repeats were mutated (R5, R6, and R7). [See online article for color version of this figure.]
Three Glu residues (419-EEE-421) in Gpa2, which form the core of the acidic loop, were replaced by smaller uncharged amino acids (EEE-SAS). The role of the basic patch in the LRR was assessed by a series of mutations altering the basic residues in LRR repeat 5 (R5: R628Q/K629T), repeat 6 (R6: K650T), and repeat 7 (R7: K675Q/K676A) to uncharged residues. In the final constructs, these mutations were either combined (R567) or introduced per repeat (R5, R6, and R7). In the full-length Gpa2, none of the basic patch or acidic loop mutations caused a clear loss of function; even Gpa2 R567 showed a cell death response similar to the wild type when coexpressed with the elicitor (Fig. 5B).
Unlike the domains of Rx1, the CC-NB-ARC and LRR of Gpa2 show only a very low level of elicitor-dependent cell death induction in trans. To obtain a more robust response, we applied domain combinations that give an interaction-dependent autoactive response in trans. The LRR of Rx1 harboring the Y712H mutation, which is able to activate the wild-type Gpa2 and Rx1 CC-NB-ARC in trans in an elicitor-independent manner (Rairdan and Moffett, 2006), could not activate the CC-NB-ARC EEE-SAS in trans (Fig. 5B). The constructs of the LRR of Gpa2, in which the basic residues in repeats 5 or 7 were replaced, could no longer provide elicitor-independent transactivation in combination with the Gpa2 CC-NB-ARC D460V constructs. The K650T mutation in repeat 6 did not affect the in trans activation of the CC-NB-ARC D460V construct by the LRR of Gpa2 (Fig. 5B).
To test the effect of the mutations in the basic patch of the LRR or the acidic loop in the ARC2 domain on the physical association between the CC-NB-ARC and LRR domains of Gpa2, coimmunoprecipitations were performed with the epitope-tagged domains transiently coexpressed in N. benthamiana. The mutations in the acidic loop (EEE-SAS) strongly reduced the physical association of the CC-NB-ARC of Gpa2 with the wild-type Gpa2 LRR (Fig. 5C). The mutations in LRR repeats 5 and 7 (R5: R628Q/K629T and R7: K675Q/K676A) disrupted the physical association between the LRR and the CC-NB-ARC domains of Gpa2, whereas the K650T mutation in repeat 6 did not affect this interaction (Fig. 5D). No difference in protein stability between the mutated and the wild-type products was detected on western blots (Fig. 5, C and D). Interestingly, the acidic loop mutations (EEE-SAS) and the basic patch mutations in repeats 5 and 7, which led to a loss of function in the transactivation assay, also resulted in a loss of interaction between the CC-NB-ARC and LRR in the immunoprecipitations. The K650T substitution in repeat 6, on the other hand, did not affect the in trans activation of the domains and did not affect the physical interaction, as shown by the immunoprecipitation (Fig. 5D). These results indicated that the cluster of acidic residues on the ARC2 surface and the clustered basic residues on the LRR surface are both involved in the physical interaction between the CC-NB-ARC and LRR. In addition, it shows that local reduction of the surface charge affects both the physical association and the in trans functionality of the CC-NB-ARC and LRR domains.
The LRR Is Predicted to Bind the NB-ARC at the Surface Formed by the Interaction of the ARC2 and NB Subdomains
Two independent docking simulations of the individual three-dimensional models of the NB-ARC and LRR domains were performed to gain insight in the overall structure of Gpa2. The docking software platform HADDOCK allows the definition of a priori ambiguous interaction restraints to guide the docking by fuzzily constraining the distance between residues suspected or known to be involved in the interaction (Dominguez et al., 2003; see “Materials and Methods”).
In the first docking, we based the interaction restraint for the NB-ARC on the surface-exposed polymorphic residues in N-terminal part of the ARC2 (amino acid positions 372–440: A397, R401, Y403, K406, and A412; Fig. 6A), which were shown by sequence exchange experiments to be involved in the cooperation with the LRR (Fig. 1). The positions of three known autoactivating mutations in the LRR of Rx1 were used to create an interaction restraint for the LRR (mutations S516H, D543E, and H739K; Bendahmane et al., 2002; Farnham and Baulcombe, 2006; Fig. 6A).
Figure 6.
Analysis of the interface between the NB-ARC and LRR via a simulated docking. A, Residues forming the basis for the ambiguous interaction restraints applied in the HADDOCK docking computations for the NB-ARC and LRR of Gpa2, indicated in green on the NB-ARC and LRR structures. In the ARC2 domain, the surface-exposed residues that are poly-morphic between Gpa2 and Rx1 in the region between amino acid 372 and 440 were used. In the LRR, the positions of three known autoactivating mutations were used to define the ambiguous interaction constraint. B, The predicted NB-ARC/LRR complex in Gpa2 resulting from the HADDOCK docking simulation viewed from three sides to show the relative positions of the LRR and NB-ARC subdomains. C, View of the NB-ARC/LRR docking model with the residues polymorphic between Gpa2 and Rx1 in the ARC2 regions 372 to 440 highlighted in red and those in the first three repeats of the LRR (amino acid positions 489–593) highlighted in yellow. D, The main residues of the basic patch on the conserved side of the LRR and of the acidic loop on the ARC2 domain are indicated in blue (basic) and red (acidic), showing their proximity in the NB-ARC/LRR docking model.
Both the ARC2 and the NB are predicted to have an interface with the LRR (Fig. 6B). The interface between the NB-ARC and LRR is formed by the concave surface of the N-terminal half of the LRR and by the outer surface of the NB-ARC at the position where the ARC2 and NB interact with each other in the ADP-bound conformation. The C terminus of the ARC2 and the N terminus of the LRR, which are connected by a linker in the full-length protein, are oriented toward each other in the predicted docking. The ARC1 is positioned at the opposite side of the NB-ARC and does not contact the LRR in this configuration. The interface formed by the LRR, ARC2, and NB displaced 2,203 Å2 from the solvent accessible area, which falls within the normal range for protein-protein interactions. Many of the surface-exposed residues polymorphic between Rx1 and Gpa2, which were not used as input for the docking simulation, were also found in the interface formed by the ARC2 and N-terminal end of the LRR domain (Fig. 6C).
Interestingly, the docking model brings the acidic loop on the ARC2 in close contact with the cluster of basic residues on the LRR, both of which were shown to be crucial in the interaction between the CC-NB-ARC and the LRR (Fig. 6D). The basic patch on the conserved side of LRR repeats 5 to 7 (R628 and K629 in repeat 5, K650 in repeat 6, and K675 and K676 in repeat 7) and the acidic loop region in the ARC2 domain (amino acid positions 409–435) are positioned sufficiently close to each other in space that the attraction between their complementary charges would strengthen the affinity of the domain interaction (Fig. 6D).
In the second docking simulation, the interaction restraints were based on the positions of acidic loop on the ARC2 surface and of the basic patch on the LRR. The domain docking resulting from the second docking simulation was almost identical to the first docking model. Both models varied from each other within 1.83 Å RMSD at the level of the interface and within 2.57 Å RMSD at the level of all Cα atoms of the NB-ARC and LRR domains (Supplemental Fig. S6). The fact that both docking simulations resulted in highly similar domain configurations is remarkable, because in contrast to the interaction restraints used in the first docking, the clusters of charged residues used in the second docking simulation are not located at center but at the edge of the predicted interface, which makes the surface areas probed during the docking simulation extremely large. It is noted that both docking simulations predict that surface-exposed hydrophobic residues contact each other. A relatively large hydrophobic interface is observed between the ARC2 and LRR, and a smaller interface is observed between the NB and LRR. The ARC2 residues I395, A397, W411, A412, and V413 contact V511, C513, V541, and L546 on the concave surface of the N-terminal half of the LRR. The smaller hydrophobic interface between the NB and the variable side of the LRR is formed by the NB residues L211, L212, and L215 contacting F569, P608, and W634 on the LRR.
Amino acid position 403, for which the Y403S substitution specifically reduced the autoactivity of the chimeric construct G13R45 (Fig. 3), is located in the hydrophobic interface between ARC2 and LRR in both docking models. As mentioned previously (Fig. 4B), the residue at position 401, which has a role in elicitor-dependent activation and for which the R401Q substitution strengthens the interaction between the CC-NB-ARC and the LRR of Gpa2, is located in the interface between the NB and ARC2 and not directly in the interface of the ARC2 and the LRR. This residue might indirectly affect the interaction of the CC-NB-ARC with the LRR via the NB, for example via the small hydrophobic interface formed by the NB and LRR.
Overall, the two docking models point to a NB-ARC-LRR domain configuration wherein the interface between the LRR sensor and NB-ARC switch is formed by interactions of the N-terminal half of the LRR with mainly the ARC2 and, to a lesser extent, the NB.
DISCUSSION
Our results demonstrate that residues polymorphic between Rx1 and Gpa2 and complementary-charged residues present in a small region of the ARC2 and the first N-terminal repeats of the LRR play a role in the physical association and cooperation of these domains. From the three-dimensional structural models of the NB-ARC and LRR of Rx1 and Gpa2, we inferred that the majority of these residues are likely located at the surface of the domains, consistent with their role in the physical interdomain interaction. Collectively, our data support a model in which complementary-charged patches and surface-exposed Gpa2/Rx1 polymorphic residues contribute to the formation of a close interface between the ARC2 and N-terminal end of the LRR when the NB-LRR protein is in an ADP-bound resting state. Disruption of this interface compromises the molecular switch function, forcing the NB-LRR protein into an autoactive “on” state or into an unresponsive “off” state. As discussed below, our data indicate that different forces may play a role in the dissociation and reassociation of the switch and sensor domain. The electrostatic forces mediated by the complementary-charged residues are predicted to mediate a fast reassociation of the NB-ARC and LRR domain, enabling multiple rounds of activation. Other intramolecular interactions are assumed to operate at different steps in the conformational switch, for example by influencing the interface between the NB and the ARC2, and may regulate in this way the sensitivity for effector-triggered activation.
In this study, we show that the first repeats in the N-terminal end of the LRR domain of Gpa2 and Rx1 are sufficient to keep these NB-LRR immune receptors in an inactive state by cooperating with a small region in the ARC2 domain. The C-terminal end of the LRR domain is the main determinant of the recognition of the cognate elicitors, demonstrating a dual role of the LRR domain in NB-LRR protein functioning. Remarkably, a division of the LRR domain into two subunits was observed when mapping the electrostatic potential on the surface of the LRR domain. Whereas the N-terminal half of the LRR is predicted to have a positively charged surface, the C-terminal half is predicted to have a predominantly acidic surface. It will be interesting to see how this charge distribution pattern relates to the distinct functional roles of the LRR subunits in the regulation of NB-LRR protein activity and pathogen recognition. The presence of a novel structural element at repeat 9 in the LRR domain of Rx1 correlates with the observed segregation in electrostatic potential, providing additional support for a division of the LRR domain into two modules with distinct functions.
Interestingly, incompatibility between the ARC2 and the LRR of Rx1 and Gpa2 results in two distinct phenotypes despite their high sequence similarity (88%). When the LRR of Rx1 is combined with the ARC2 of Gpa2, an autoactive cell death response resulted, whereas a loss of activation was observed when the LRR of Gpa2 was combined with the ARC2 of Rx1. These regions in Gpa2 and Rx1 only differ in a small number of amino acid residues, showing that NB-LRR protein functioning is easily compromised by disruption of the interface between the ARC2 and N-terminal end of the LRR. From this, we concluded that the ARC2 and N-terminal end of the LRR cooperate as a functional complex. In the resistance protein RPS5 from Arabidopsis (Arabidopsis thaliana), the N-terminal four repeats of the LRR are sufficient to keep the protein in an autoinhibited state (Qi et al., 2012). In Gpa2 and Rx1, the role of the N-terminal repeats of the LRR seem to have a positive role in the activation of the protein; the deletion of the LRR does not result in autoactivation of the CC-NB-ARC and the autoactivating effect of D460V mutation in the ARC2 MHD motif depends on the presence of the LRR (Moffett et al., 2002).
The incompatibility of the ARC2 and N-terminal LRR repeats of Rx1 and Gpa2 shows de facto the effect of the reversal of the coevolutionary fine-tuning between functional domains. This is supported by the observation that the N-terminal end of the LRR in Rx1 and Gpa2 is more conserved than the C-terminal region, consistent with a dual role of this domain in NB-LRR protein functioning. In the conserved N-terminal repeats of the LRR, several autoactivating mutations have been identified in Rx1 (Bendahmane et al., 2002; Farnham and Baulcombe, 2006), and in the resistance protein RPS5, a mutation at this location negatively affected the functioning of RPS5 itself and other resistance proteins (Warren et al., 1998). From the results of the Gpa2/Rx1 sequence exchange study, we conclude that as long as the cooperating N-terminal LRR regions and the ARC2 domains are retained as a compatible unit, the C-terminal region of the LRR responsible for recognition can be exchanged and still yield a functional protein.
We showed, without silencing the Suppressor of G2 allele of skp1 (SGT1), that for both Rx1 and Gpa2, the interaction between the CC-NB-ARC and LRR is partially disrupted in the presence of their respective elicitors. More interestingly, we found one position (amino acid position 401) in the ARC2 domain of Gpa2 that results in an increased binding between the CC-NB-ARC and the LRR domain when we replaced the Arg present in Gpa2 with the Gln of Rx1 (R401Q). Enhancing the physical association between the CC-NB-ARC and LRR domains correlated with a reduced elicitor-dependent activation of Gpa2. In the three-dimensional model of the NB-ARC, position 401 is located on the ARC2, close to the interface with the NB, which suggests that the observed difference in the interdomain interactions could either be caused by a direct disruption of the interface between the ARC2 and LRR or indirectly because of its influence on the interface between the ARC2 and NB, and thus the overall structure of the NB-ARC. Apparently, it is more difficult for the Gpa2 R401Q to respond to the elicitor RBP1, indicating that the (temporary) disruption of the interaction between the CC-NB-ARC and LRR domains in the presence of the elicitor is essential for the transition of the NB-LRR protein from a closed ADP-bound resting state into an open ATP-bound activated state. Whether the disruption is achieved by direct binding of the Rx1 and Gpa2 elicitor to the NB-LRR protein or by elicitor-induced modifications of the accessory protein Ran GTPase-Activating Protein 2 (RanGAP2) (Sacco et al., 2007, 2009; Tameling and Baulcombe, 2007) needs further investigation.
Remarkably, we found a second position in the ARC2 domain of Gpa2 (amino acid position 403) that resulted in a reduced elicitor-independent activation when the Tyr in Gpa2 was replaced with the Ser of Rx1. However, this reduction in the autoactivity of the G13R45 chimera did not correlate with a change in the binding of the CC-NB-ARC and LRR or in a change in elicitor-dependent activation. This indicates that the autoactive construct mimics an activated state of the protein that follows after the elicitor-triggered release of the CC-NB-ARC/LRR interaction. These results demonstrate that the activation of NB-LRR proteins such as Rx1 and Gpa2 is a multistep process. Earlier, it was demonstrated that an autoactivating D460V mutation in the ARC2 MHD motif does not disrupt the CC-NB-ARC/LRR interaction (in a P-loop mutant background), and neither does the P-loop mutation itself, but that in both cases, the presence of the Rx1 elicitor could still cause a decrease in the CC-NB-ARC/LRR binding (Rairdan and Moffett, 2006). Similarly, the interaction between the SD-NB-ARC and LRR of the tomato resistance protein Mi1.2 against Meloidogyne incognita, is not disrupted by autoactivating mutations (van Ooijen et al., 2008). Now, we can conclude that the dissociation of the LRR upon elicitor recognition appears to be an initial step in the elicitor-dependent activation of the NB-LRR protein and that mutations that strengthen the interaction of the LRR with the other domains suppress elicitor-dependent activation.
Here, we further show that conserved complementary-charged amino acid residues in the ARC2 and N-terminal end of the LRR contribute to the complex formation between the CC-NB-ARC and LRR domain of Rx1 and Gpa2. Replacing the charged residues present at the predicted ARC2 and LRR interface with noncharged residues resulted in a loss of autoactivation in the in trans complementation assays and in a disruption of the binding between the two domains in coimmunoprecipitation assays. From these results, we conclude that complementary-charged regions in the ARC2 and LRR domain are involved in the establishment of an electrostatic interaction between the two separate domains and their assembly into a functional protein. Remarkably, replacing the charged residues in the full-length Gpa2 protein by noncharged residues did not change the elicitor-dependent activation response. We therefore concluded that the mutations are not sufficient to disrupt the intramolecular interactions in the full-length protein and secondly, that these residues are not directly involved in signaling. Electrostatic forces have been shown to accelerate the initial binding steps in the formation of protein complexes (Sheinerman et al., 2000; Godderz et al., 2003; Ivanova and Lu, 2008), and the complementary-charged residues we predict to be located at the edge of the ARC2 and LRR interface could be envisioned to play such a role in the interdomain interactions of the CC-NB-ARC and LRR. After the elicitor-mediated dissociation of the CC-NB-ARC and LRR domains, as shown in this study, a fast reassociation is expected to take place that enables the NB-LRR protein to be activated multiple times (Rairdan and Moffett, 2006). The electrostatic forces between the ARC2 and LRR could enhance this reassociation and thereby the transition of the protein thorough the activation cycle. Sequence comparison revealed that the residues forming the complementary-charged surface regions in the ARC2 and LRR of Gpa2 and Rx1 are conserved in the NB-LRR plant immune receptors Bs2 conferring resistance against bacterial spot disease in pepper (Capsicum annuum; Tai et al., 1999; Mazourek et al., 2009), RPS5 conferring resistance against Pseudomonas syringae in Arabidopsis (Warren et al., 1998), and MLA conferring resistance against polymorphic barley mildew A in barley (Hordeum vulgare; Halterman et al., 2001). In a recent model of the LRR of the resistance protein Lr10 , a cluster of basic residues was identified at the same surface region as at the LRR of Gpa2 and Rx1 (Sela et al., 2012). The conserved nature of these structural elements indicates that electrostatic interactions present a common characteristic of NB-LRR proteins important for the physical association between the NB-ARC and LRR domains when switching “on” and “off.”
In the docking simulations, a relatively large hydrophobic interface is predicted between the ARC2 and the LRR, and a smaller interface is predicted between the NB and the LRR. Preliminary data suggests that various hydrophobic residues play a role in the interaction. A change in the NB-ARC conformation, and thus the relative positions of the ARC2 and NB, is expected to affect at least one of the predicted hydrophobic interfaces of the NB-ARC with the LRR. In this way, a substitution that affects the interaction between the NB and ARC2 subdomains, such as R401Q, might indirectly influence the interface of the NB-ARC and LRR. However, it is important to consider the fact that the CC domain is also required for the domain configuration in the full-length protein and requires both the NB-ARC and LRR for its intramolecular interaction (Moffett et al., 2002; Rairdan et al., 2008).
The recent elucidation of the structures of the CC domain of MLA and the TIR domain of L6, a resistance protein against flax rust, both shown to form homodimers, has led to renewed attention for the role of self-association in the functioning of R proteins (Bernoux et al., 2011; Maekawa et al., 2011a). Multimerization via the NB-ARC domain after activation is a common theme in proteins belonging to the STAND clade (Danot et al., 2009). For R proteins, there is no clear evidence that the NB-ARC can mediate multimerization, and maybe this role has shifted to their N-terminal domains. However, there is no indication that multimerization plays a role in the functioning of Rx1 (Moffett et al., 2002; Sacco et al., 2007). In our experiments, we have not been able to show multimerization of full-length Gpa2, Rx1, or their individual domains (NB, NB-ARC, CC-NB-ARC, LRR, and CC; Supplemental Fig. S7; E.J. Slootweg, unpublished results). This does not exclude the possibility that multimerization plays a role during the activation or signaling of Gpa2, but if self-association occurs, it does so at a much lower affinity than the affinity at which the domains interact with each other.
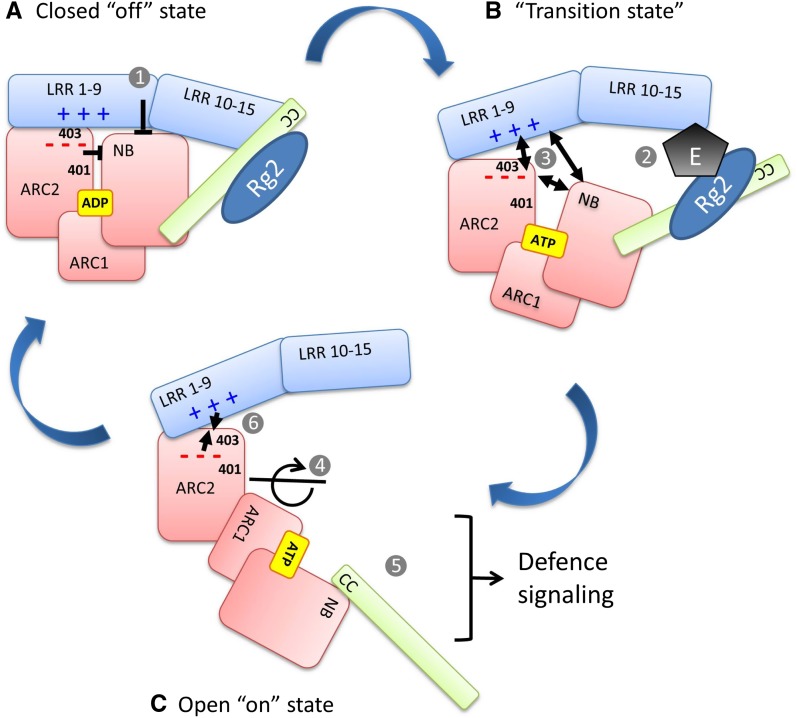
Combining our results with recent insights in the functioning of NB-LRR proteins, we can sketch an activation mechanism (Fig. 7), in which recognition of the pathogen elicitor by the C-terminal end of the LRR results in dissociation of the ARC2 and the N-terminal end of the LRR. This allows the protein to switch from a closed, autoinhibited resting state into an open, signaling-competent state (Fig. 7, A–C). In the closed ADP-bound state, the NB, ARC1, and ARC2 of Rx1 and Gpa2 are predicted to form a compact globular structure around the nucleotide. The horseshoe-like LRR domain docks onto this structure, keeping the NB-ARC in an inactive ADP-bound conformation (Fig. 7A, 1). The sequence exchange experiments show that these interactions are fine tuned and readily disrupted. The direct or indirect recognition of the elicitor leads to a dissociation of the switch and sensor domains (Fig. 3, D and E; Fig. 7B, 2). Residue R401 is predicted to be located in the interface of the ARC2 and NB, but influences the dissociation of the CC-NB-ARC from the LRR and the elicitor-dependent activation. The docking model predicts that both the ARC2 and the NB bind the LRR. A change in their relative position will affect their interface with the LRR (Fig. 7B, 3). The recently resolved crystal structure of the autoinhibited murine Apaf1 shows that the WD40 sensor domain in Apaf1 functions like a clamp by interacting with both the NB and helical domain II (ARC3, absent in NB-LRR R proteins; Reubold et al., 2011). Binding of cytochrome c changes the overall structure of the WD40 sensor and breaks its interaction with the NB, enabling or even forcing the NB and ARC1 to rotate to the open, ATP-bound conformation (Fig. 7C, 4).
Figure 7.
A mechanistic model of the activation of the NB-LRR proteins Gpa2 and Rx1. This model illustrates the conformational changes underlying the switch between the autoinhibited “off” state (A) and the active-signaling “on” state (C) of the NB-LRR immune receptors Gpa2 and Rx1. The autoinhibited state (A) is stabilized via interactions of the ADP-bound NB and ARC2 with each other and their interface with the LRR. The differential effect of the residues at positions 401 and 403 on elicitor-dependent and elicitor-independent activation is due to their presence in two distinct interfaces. B, Disruption of these domain interfaces by the elicitor (E), potentially facilitated by the presence of a guarded host protein, RanGAP2 (Rg2), at the N-terminal domain (CC), and the exchange of ADP for ATP when the protein is in a transition state allows the NB-ARC1 unit to rotate away from the ARC2 and expose surfaces required for signaling (C). The complementary-charged surfaces of the ARC2 and LRR (– – – and + + +) could accelerate the reassociation of the ARC2 and LRR when the protein returns to the resting state. The numbers in the figure are referred to in the discussion. [See online article for color version of this figure.]
In a similar way, upon dissociation of the Gpa2 CC-NB-ARC/LRR complex, the conformation of the nucleotide-binding pocket will change, enabling the exchange of ADP for ATP. Interestingly, the ARC2 residue connecting the NB and ARC2 in the autoinhibited Apaf1 via a salt bridge (D392) is conserved in Gpa2 and Rx1 (D399) and is positioned close to amino acid 401. Mutations at this position (D399V and E400K) have been shown to result in constitutive activity (Bendahmane et al., 2002).
To gain insight into the effect the conformational changes of the NB-ARC have on the overall structure of Gpa2 or Rx1, a three-dimensional model was made for the NB-ARC domain of Rx1 using the ATP-bound crystal structure of the Caenorhabditis elegans death protein CED4 as a template (Yan et al., 2005). For the transition from the resting state to the activation state, the orientation of the NB and ARC1 in respect to the ARC2 has to change dramatically (Supplemental Fig. S8; Fig. 7C, 4). The reorientation of the NB-ARC1 and ARC2 implies a repositioning of the CC and LRR domain, as they are physically connected to the NB and ARC2, respectively. Eventually, a change of the overall conformation will lead to the exposure of initially shielded surfaces of signaling-competent domains (Fig. 7C, 5). In case of Rx1, it was shown that the NB domain itself can initiate a cell death response in the absence of the other domains (Rairdan et al., 2008), but for several other NB-LRR R proteins, the N-terminal CC or TIR domain has signaling capabilities (Swiderski et al., 2009; Bernoux et al., 2011; Collier et al., 2011; Maekawa et al., 2011a). The electrostatic attraction between the conserved clusters of charged residues on the ARC2 and LRR is predicted to facilitate the reassociation of the domains when the R protein completes the activation cycle and returns to its resting state (Fig. 7C, 6).
Our data indicate that the clamp formed by the ARC2, NB, and N-terminal half of the LRR imposes strong functional constraints for the generation of novel resistance genes via evolutionary process, but also through genetic engineering. The autoactivity and loss-of-function phenotypes that were observed in the chimeric R protein constructs (Figs. 1 and 2) show that when Gpa2 and Rx1 diverged, interactions between cooperating domains were maintained through coevolution. As a consequence, the occurrence of such fine-tuned domain cooperations constrains the possibilities for reshuffling sequences within NB-LRR-encoding R gene clusters, an important mechanism in the generation of novel recognition specificities (Cooley et al., 2000; Leister, 2004; Meyers et al., 2005). Random sequence exchange between homologous R genes, either by natural or artificial recombination events, will readily result in the generation of genes that encode for nonfunctional or constitutively active NB-LRR proteins. Surprisingly, for Rx1 and Gpa2, this constraint was limited to the ARC2 and the N-terminal repeats of the LRR. As long as those segments were left intact as a functional unit, the C-terminal segments of the LRR, which determine the recognition specificity, could be exchanged without the loss of proper functioning. These results show that understanding the structural elements involved in controlling the activation of NB-LRR plant immune receptors is a prerequisite for eventually engineering novel functional R genes with custom recognition specificities.
MATERIALS AND METHODS
Protein Structure Analysis
Secondary Structure and Domain Organization
Rx1 and Gpa2 were profiled for motifs, patterns, and the following properties: intrinsic disorder, hydrophobicity, accessibility, contact-forming tendency, secondary structure state propensities, turn, and interdomain linker location. To increase the prediction reliability, several methods were used for every type of analysis. Domain delineation was based on the assessment of intrinsic disorder and contact-forming propensities, secondary structure prediction, fold recognition methods, sequence alignments, detection of LRR motifs, and domain linker prediction (Miyazaki et al., 2002). Intrinsic disorder was assessed with DisEMBL (Linding et al., 2003), IUPRED (Dosztányi et al., 2005), and DISOPRED (Ward et al., 2004). Secondary structure prediction was performed with GOR IV (Garnier et al., 1996), Jpred (Cuff et al., 1998), nnPredict (McClelland and Rumelhart, 1988), Porter (Pollastri and McLysaght, 2005), PSA (Stultz et al., 1993), PSIPRED (Jones, 1999), SOPMA (Geourjon and Deléage, 1995), and SCRATCH software, which also returned contact and accessibility propensities (Cheng et al., 2005). Turns were identified with BETATPRED2 (Kaur and Raghava, 2003) and COUDES (Fuchs and Alix, 2005). Fold recognition was performed with Phyre (Bennett-Lovsey et al., 2008; Kelley and Sternberg, 2009). Multiple sequence alignments were performed using ClustalW (Thompson et al., 1994).
Molecular Modeling
Automated fold recognition and sequence homology were assessed with Phyre and BLAST (1 and 2). Sequence-conserved regions were modeled by coordinate transfer. In sequence-variable regions, the side chains were reconstructed and iteratively optimized, while insertion loops were generated randomly and filtered for steric constraints, followed by successive rounds of simulated annealing and energy minimization. Molecular modeling, including side-chain reconstruction, constrained ab initio loop generation, and local and global simulation annealing for relieving steric conflicts, as well as the model structure analysis were performed using InsightII (Accelrys).
OJFRHM of the Irregular Gpa2 LRR Domain
For modeling the LRR domain in Rx1/Gpa2 proteins, a special procedure was designed, called the OJFRHM method. This involves five main steps. (1) The development and structural analysis of a comprehensive database of LRR templates named the Structural Assessment of LRR Motif database (SALRRM-DB). (2) The identification and locking of LRR repeats along the target sequence. False positive LRR motifs were eliminated by retaining the motifs best matching those resulting from the sequence analysis of LRR families from SMART databases, LRR_CC, LRR_RI, LRR_TYP, LRR LRRNT, LRR_SD22, LRR_BAC, and LRRCT (Schultz et al., 1998), and the structural analysis of LRR motif distribution in SALRRM-DB, including LRR repeat length, volume, secondary structure, and interrepeat contact analysis. (3) The search of best-matching local templates in SALRRM-DB. (4) The assemblage of the overall template framework by joining the local templates in a single structure by imposing the specific LRR hydrogen bond pattern at their borders and optimizing the repeat contacts (Fig. 3A). (5) The generation of the three-dimensional model of the target sequence by using established remote homology modeling procedures. The SALRRM-DB was generated by an exhaustive search in PDB with software specially designed to detect LRR signatures in protein three-dimensional structures. In this way, over 100 LRR domains were identified in the PDB. These were clustered in the SALRRM-DB in 30 nonredundant protein domain subclasses belonging to proteins from a variety of taxa, primarily with recognition functions. The nonredundant number of LRR repeats found in the LRR domains of SALRRM-DB is 255.
Internal Model Assessment
Model quality assessment was performed iteratively using MetaMQAP (Pawlowski et al., 2008). Stability tests were done by molecular dynamics simulations with NAMD (Phillips et al., 2005) using a Charmm27 force field on a high-performance computing facility. Solvent accessibility was computed with the NACCESS program (Hubbard and Thornton, 1993). Electrostatic potential was calculated using both Poisson-Boltzmann and Coulomb methods.
Docking studies between LRR and NB-ARC were performed with HADDOCK in conjunction with CNS due to the facilities this module has in using constraints and its high score obtained in the CAPRI evaluation (Dominguez et al., 2003; van Dijk et al., 2005; de Vries et al., 2007). All the docking was performed according to the following protocol. (1) Ten-thousand complexes were generated during a rigid-body minimization step. (2) The best 200 complexes based on a HADDOCK-specific score function (the enthalpic term) were then retained for the semiflexible simulated annealing step. (3) The 200 complexes were subjected to explicit solvent refinement. (4) Complexes derived from the previous step were clustered based on RMSD to compute the entropic term. Each cluster was analyzed in terms of score average. In described docking, the most abundant cluster had the best score average. The complex with the best score from the most abundant cluster was retained for further analysis.
DNA Constructs
Construction of Expression Vectors
All constructs used in transient expression assays were cloned in the pBINPLUS binary vector system (van Engelen et al., 1995). The transcription of the constructs was controlled by the Cauliflower mosaic virus 35S promoter and A. tumefaciens terminator of nopaline synthase (Tnos) sequences. Constructs harboring the Gpa2 elicitors RBP-D383-1, RBP-Rook6 (eliciting), and RBP-Rook4 (noneliciting) are described elsewhere (Sacco et al., 2009).
Domain Swaps
To construct the chimeric sequences, synthetic gene fragments were introduced in the Gpa2 and Rx1 sequences. Thereby, unwanted restriction sites were removed and new unique sites were added. Care was taken not to change the encoded amino acid sequence. The introns in the Gpa2 and Rx1 sequences were retained. The following changes were made in the Gpa2 sequence (A in ATG is 1): T1179C, A1182G, T1228C, A1230C, T1320C, C1321T, C1335T, T1377C, G1383C, A1498T, G1499C, C1501A, G1503A, T1554G, C1563T, G1596C, A1559G, A1623G, C1659A, A1695C, T1696A, C1697G, T1698C, T1749C, A1782T, A1812G, C1818T, T1833C, G1857C, G2052A, T2172A, T2175C, A2244T, T2247G, C2379A, T2403C, C2406G, A2439G, T2568G, and A2571G. In the Rx1 sequence, the following nucleotides were altered: A990G, T1179C, A1182G, T1228C, A1230C, T1377C, G1383C, A1498T, G1499C, C1501A, G1503A, T1554G, A1584G, G196C, A1599G, A1623G, T1749C, A1782T, A1812G, G1887T, A1971G, A2106G, T2172A, T2175C, T2244G, A2313G, T2388C, C2391G, C2394T, A2424G, T2553G, and A2556G.
Functionality of the synthetic constructs was found to be identical to the original Gpa2 and Rx1 constructs in agroinfiltration assays.
Sequences were exchanged at the restriction sites indicated in Figure 1A. For the exchanges at amino acid positions 489 (ApaLI), 593 (ClaI), Gpa2 761/Rx1 756 (BspEI), and Gpa2 879/Rx1 874 (EcoRI), the original Gpa2 and Rx1 sequences were used.
Site-Directed Mutagenesis
Site-directed mutagenesis was attained by inserting small sections of synthetic DNA with specific codons. The amino acids 419-EEE in the acidic loop were replaced by SAS via the change of the coding sequence (1255–2263) from GAAGAAGAG to AGCGCTTCT. The mutation R401Q was made by changing 1201-CGG to CAG and Y403S by changing 1207-TAT to TCT. In the targeted mutagenesis of the Gpa2 basic patch, the following changes were made in the Gpa2 coding sequence: Gpa2 R5 (RK628-629QT) was made by changing 1882-AGGAAG into CAGACT, Gpa2 R6 (K650T) was made by changing 1948AAA into ACA, and Gpa2 R7 (KK675-676-QA) was made by changing 2023AAGAAG into CAGGCT. In the Rx1 coding sequence, the mutation Q401R was made by changing 1201-CAG to CGG, and the mutation S403Y was made by changing 1207-TCT to TAT. The mutation Y712H was created by changing the corresponding codon from TAT to CAT.
All DNA constructs were sequenced.
Protein Tags
Multimeric c-Myc (EQKLISEEDL) and hemagglutinin (HA; YPYDVPDYA) tags were fused to the C terminus of CC-NB-ARC constructs via a NotI site, creating a short linker consisting of three alanines. Fusions to the N terminus of the LRR were made through an NcoI site overlapping the start codon in this construct, with a short linker in between the 4×HA tag and the LRR sequence. A subset of the full-length chimeric Rx1/Gpa2 constructs was tagged with a single c-Myc tag at their N terminus.
Plant Expression
For agroinfiltration experiments, Agrobacterium tumefaciens strain pMOG101 was cultured in 1 L YEB medium (5 g beef extract, 1 g yeast [Saccharomyces cerevisiae] extract, 5 g bactopeptone, 5 g Suc, and 2 mL MgSO4) with the appropriate antibiotics as described earlier (Van der Hoorn et al., 2000). After growing overnight at 28°C, the bacteria was pelleted by centrifugation, resuspended in 1 L infiltration medium (5 g Murashige and Skoog salts, 1.95 g MES, and 20 g Suc, pH 5.6, with NaOH and 200 μm acetosyringone), and incubated at room temperature for 2 h. For agroinfiltration, the bacterial suspensions were diluted to final concentrations between optical density at 600 nm (OD600) of 0.2 and 1.0. Leaves were infiltrated from 6-week-old Nicotiana benthamiana plants grown in the greenhouse at 20°C and in 16 h of light. Each combination was tested in at least three independent experiments with four replicates each. The strength of observed hypersensitive responses was ranked at a scale of 0 to 5, from no visible signs of hypersensitive response to full necrosis. Hypersensitive responses were scored after 2 and 7 d after infiltration. For protein extraction, leaves were harvested 2 d after infiltration. Transient PVX resistance assays were performed by coinfiltrating A. tumefaciens carrying the Rx1 constructs (OD600 of 0.05) with A. tumefaciens carrying a PVX:GFP amplicon (OD600 of 0.002). At 5 d post infiltration, the leaves were harvested, and an extract (24 mg leave material in 250 μL 50 mm phosphate buffer, pH 7) was incubated in anti-PVX coat protein antibody-coated plates. After stringent washing, the presence of PVX was detected using alkalic phosphatase-conjugated anti-PVX antibodies (Prime Diagnostics). Phosphatase-mediated turnover of the substrate para-nitrophenylphosphate into para-nitrophenol was measured in a photospectrometer via the absorption at 405 nm.
Protein Methods
Total protein extract of N. benthamiana leaves transiently expressing the construct was made by grinding leaf material in protein extraction buffer (50 mm Tris-HCl, pH 7.5, 10% (v/v) glycerol, 150 mm NaCl, 1 mm EDTA, 2% (w/v) polyvinylpolypyrrolidone, 0.4 mg mL–1 Pefabloc SC plus [Roche], and 5 mm dithiothreitol) on ice. For immunoprecipitation, the total protein extract was first passed over a Sephadex G-25 column. The protein extract was precleared with rabbit-IgG agarose (40 μL slurry mL–1 protein extract). After preclearing, the protein extract was mixed with 25 μL anti-Myc agarose beads (Sigma) or anti-HA agarose beads (Roche) and incubated for 2 h at 4°C. After washing six times (washing buffer: protein extraction buffer with 0.15% Igepal CA-630), the beads were resuspended in Laemmli buffer, and the bound protein was separated by SDS-PAGE and blotted on polyvinylidene difluoride membrane. For immunodetection, the following antibodies were used: 9E10 anti-Myc (Sigma), 3F10 anti-HA (Roche), and ab290 anti-GFP (Abcam). Peroxidase activity was visualized using Thermo Scientific SuperSignal West Femto or Dura substrate and imaging the luminescence with G:BOX gel documentation system (Syngene).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Scoring scale used to classify the strength of the cell death response.
Supplemental Figure S2. Western blot to assess the stability of the chimeric Rx1/Gpa2 constructs that display complete or partial loss of function.
Supplemental Figure S3. Alignment of the sequences of Gpa2 and Apaf-1 encompassing the NB, ARC1 and ARC2 domains.
Supplemental Figure S4. The position of the leucin-rich repeat motifs in the Gpa2 amino acid sequence.
Supplemental Figure S5. Predicted structure of the irregular repeat 9 in the LRR of Rx1.
Supplemental Figure S6. Comparison of two independent simulations of the docking of the Gpa2 NB-ARC and LRR domains.
Supplemental Figure S7. Immunoprecipitation experiments to detect (homo-)multimerisation of the CC-NB-ARC and LRR domains of Gpa2 and Rx1.
Supplemental Figure S8. Comparison of the predicted structures of the NB-ARC in the ADP- and ATP-bound conformation.
Supplementary Material
Glossary
- PDB
Protein Data Bank
- OJFRHM
Optimized Joined Fragment Remote Homology modeling
- RMSD
root mean square deviation
- SALRRM-DB
Structural Assessment of LRR Motif database
- HA
hemagglutinin
- OD600
optical density at 600 nm
- PVX
Potato virus X
- NB-LRR
nucleotide-binding-leucine-rich repeat
References
- Ade J, DeYoung BJ, Golstein C, Innes RW. (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M, Takken FLW. (2006) Update on the domain architectures of NLRs and R proteins. Biochem Biophys Res Commun 339: 459–462 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Farnham G, Moffett P, Baulcombe DC. (2002) Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J 32: 195–204 [DOI] [PubMed] [Google Scholar]
- Bendahmane A, Querci M, Kanyuka K, Baulcombe DC. (2000) Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J 21: 73–81 [DOI] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. (2008) Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70: 611–625 [DOI] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN. (2011) Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Randall AZ, Sweredoski MJ, Baldi P. (2005) SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res 33: W72 –W76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P. (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Cooley MB, Pathirana S, Wu HJ, Kachroo P, Klessig DF. (2000) Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics 14: 892–893 [DOI] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. (2009) Wheel of life, wheel of death: a mechanistic insight into signaling by STAND proteins. Structure 17: 172–182 [DOI] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. (2007) HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins 69: 726–733 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737 [DOI] [PubMed] [Google Scholar]
- Dosztányi Z, Csizmok V, Tompa P, Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21: 3433–3434 [DOI] [PubMed] [Google Scholar]
- Farnham G, Baulcombe DC. (2006) Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc Natl Acad Sci USA 103: 18828–18833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs PF, Alix AJ. (2005) High accuracy prediction of beta-turns and their types using propensities and multiple alignments. Proteins 59: 828–839 [DOI] [PubMed] [Google Scholar]
- Garnier J, Gibrat JF, Robson B. (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol 266: 540–553 [DOI] [PubMed] [Google Scholar]
- Geourjon C, Deléage G. (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11: 681–684 [DOI] [PubMed] [Google Scholar]
- Godderz LJ, Rahman NS, Risinger GM, Arbuckle JL, Rodgers KK. (2003) Self-association and conformational properties of RAG1: implications for formation of the V(D)J recombinase. Nucleic Acids Res 31: 2014–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT, Yao N. (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6: 201–211 [DOI] [PubMed] [Google Scholar]
- Halterman D, Zhou F, Wei F, Wise RP, Schulze-Lefert P. (2001) The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J 25: 335–348 [DOI] [PubMed] [Google Scholar]
- Hu Y, Ding L, Spencer DM, Núñez G. (1998) WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J Biol Chem 273: 33489–33494 [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. (1993) NACCESS. In Department of Biochemistry and Molecular Biology. University College, London [Google Scholar]
- Hwang CF, Williamson VM. (2003) Leucine-rich repeat-mediated intramolecular interactions in nematode recognition and cell death signaling by the tomato resistance protein Mi. Plant J 34: 585–593 [DOI] [PubMed] [Google Scholar]
- Ivanova E, Lu H. (2008) Allosteric and electrostatic protein-protein interactions regulate the assembly of the heterohexameric Tim9-Tim10 complex. J Mol Biol 379: 609–616 [DOI] [PubMed] [Google Scholar]
- Jones DT. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kaur H, Raghava GP. (2003) Prediction of β-turns in proteins from multiple alignment using neural network. Protein Sci 12: 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. (2009) Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. (2004) STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 343: 1–28 [DOI] [PubMed] [Google Scholar]
- Leister D. (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20: 116–122 [DOI] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. (2003) Protein disorder prediction: implications for structural proteomics. Structure 11: 1453–1459 [DOI] [PubMed] [Google Scholar]
- Lukasik E, Takken FLW. (2009) STANDing strong, resistance proteins instigators of plant defence. Curr Opin Plant Biol 12: 427–436 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen Q-H, Micluta MA, Somssich IE, et al. (2011a) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. (2011b) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12: 817–826 [DOI] [PubMed] [Google Scholar]
- Mazourek M, Cirulli ET, Collier SM, Landry LG, Kang BC, Quirin EA, Bradeen JM, Moffett P, Jahn MM. (2009) The fractionated orthology of Bs2 and Rx/Gpa2 supports shared synteny of disease resistance in the Solanaceae. Genetics 182: 1351–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland JL, Rumelhart DE. (1988) Explorations in Parallel Distributed Processing, Vol 3 MIT Press, Cambridge, MA, pp 318–362 [Google Scholar]
- Meyers BC, Kaushik S, Nandety RS. (2005) Evolving disease resistance genes. Curr Opin Plant Biol 8: 129–134 [DOI] [PubMed] [Google Scholar]
- Miyazaki S, Kuroda Y, Yokoyama S. (2002) Characterization and prediction of linker sequences of multi-domain proteins by a neural network. J Struct Funct Genomics 2: 37–51 [DOI] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC. (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski M, Gajda MJ, Matlak R, Bujnicki JM. (2008) MetaMQAP: a meta-server for the quality assessment of protein models. BMC Bioinformatics 9: 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26: 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollastri G, McLysaght A. (2005) Porter: a new, accurate server for protein secondary structure prediction. Bioinformatics 21: 1719–1720 [DOI] [PubMed] [Google Scholar]
- Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. (2008) The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS ONE 3: e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, DeYoung BJ, Innes RW. (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158: 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan G, Moffett P. (2007) Brothers in arms? Common and contrasting themes in pathogen perception by plant NB-LRR and animal NACHT-LRR proteins. Microbes Infect 9: 677–686 [DOI] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. (2008) The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S. (2011) Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis. Structure 19: 1074–1083 [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Li WY, Chao Y, Schwarzenbacher R, Shi YG. (2005) Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature 434: 926–933 [DOI] [PubMed] [Google Scholar]
- Sacco MA, Koropacka K, Grenier E, Jaubert MJ, Blanchard A, Goverse A, Smant G, Moffett P. (2009) The cyst nematode SPRYSEC protein RBP-1 elicits Gpa2- and RanGAP2-dependent plant cell death. PLoS Pathog 5: e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco MA, Mansoor S, Moffett P. (2007) A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J 52: 82–93 [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela H, Spiridon LN, Petrescu AJ, Akerman M, Mandel-Gutfreund Y, Nevo E, Loutre C, Keller B, Schulman AH, Fahima T. (2012) Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine-rich repeat (LRR) domains. Mol Plant Pathol 13: 276–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheinerman FB, Norel R, Honig B. (2000) Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol 10: 153–159 [DOI] [PubMed] [Google Scholar]
- Stultz CM, White JV, Smith TF. (1993) Structural analysis based on state-space modeling. Protein Sci 2: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Birker D, Jones JD. (2009) The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact 22: 157–165 [DOI] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ. (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 96: 14153–14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FLW, Tameling WIL. (2009) To nibble at plant resistance proteins. Science 324: 744–746 [DOI] [PubMed] [Google Scholar]
- Tameling WIL, Baulcombe DC. (2007) Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to potato virus X. Plant Cell 19: 1682–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WIL, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJC, Takken FLW. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG. (1998) The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8: R226–R227 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RAL, Laurent F, Roth R, De Wit PJ. (2000) Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol Plant Microbe Interact 13: 439–446 [DOI] [PubMed] [Google Scholar]
- van Dijk AD, de Vries SJ, Dominguez C, Chen H, Zhou HX, Bonvin AM. (2005) Data-driven docking: HADDOCK’s adventures in CAPRI. Proteins 60: 232–238 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ. (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288–290 [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Albrecht M, Cornelissen BJC, Takken FLW. (2008) Transcomplementation, but not physical association of the CC-NB-ARC and LRR domains of tomato R protein Mi-1.2 is altered by mutations in the ARC2 subdomain. Mol Plant 1: 401–410 [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337: 635–645 [DOI] [PubMed] [Google Scholar]
- Warren RF, Henk A, Mowery P, Holub E, Innes RW. (1998) A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Chai JJ, Lee ES, Gu LC, Liu Q, He JQ, Wu JW, Kokel D, Li HL, Hao Q, et al. (2005) Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature 437: 831–837 [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90: 405–413 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.