Abstract
Despite significant attractiveness of anti-sense oligonucleotide/RNAi technology, its clinical application has been precluded by a lack of methods for targeted delivery and transduction of primary immune cells in vivo. Here, we devised a chemokine CCL17-based strategy (TARC-arp) that transiently silences expression of genes in immune cells by delivering inhibitory oligonucleotides via their chemokine receptors. In modeling studies using mice with established 4T1.2 breast cancer, we show that IL10 produced by CCR4+ cells, in particular FoxP3+ regulatory T cells (Tregs), plays an important role in lung metastasis. As such, TARC-arp-mediated silencing of IL-10 or FoxP3 in CCR4+ Tregs is sufficient to block lung metastasis. Thus, we provide a simple solution that circumvents the problems of RNAi use in vivo, indicating that a disease outcome can be successfully controlled by delivering inhibitory oligonucleotides with chemokines to inactivate a selective subset of immune cells.
Keywords: Chemokine, Tregs, Metastasis, siRNA delivery, IL10, FoxP3
INTRODUCTION
Despite their small numbers (less than 5% of circulating CD4+ T cells), regulatory T cells (Tregs) play a key role in controlling peripheral tolerance to self and inflammatory responses [1]. To date, a number of different Tregs are known to exist, such as thymic natural naïve (nnTregs), mature and memory CD25+CD4+ Tregs [2–4], extrathymically generated adaptive Tregs [5,6], and IL-10 producing murine effector-memory Tregs (TREM) [7] and human Tr1 cells [8]. Tregs mostly utilize a cell contact-dependent regulatory process involving FasL, galectin-1/βGBP [9,10] and perforin/granzymes [11,12], although soluble factors such as IL-10 and TGFβ are shown to be used by TREM, Tr1 and ICOS+FoxP3+ Tregs [8,13]. In vivo, Tregs appear to exhibit non-redundant functions tailored for a specific tissue and inflammatory setting [6,14]; and the choice of a particular Treg subset or regulatory pathway seems to depend on the type or stage of disease as well as the strength and/or type of stimulation. While natural Tregs prevent systemic and tissue-specific autoimmunity, extrathymically generated Tregs control Th2 responses to microbiota at mucosal interfaces [6] and inflammation at environmental interfaces (the skin, lung and colon) by producing IL10 [14,15] upon activation with antigens presented by DCs [15].
Tissue infiltration/homing of Tregs is mostly controlled by differentially expressed chemokine receptors. For example, while CXCR4 and CCR7-expressing nnTregs and other Tregs migrate into bone marrow and secondary lymphoid organs [2,16], CCR6 and CCR4 are used for homing of TREM cells and other Tregs into the skin and lungs [7,17]. As such, the presence of CCR4+ Tregs is often a sign of bad disease outcome in human and mice with tumors [18,19]. We recently found that metastasis (the stage that is presumably most vulnerable to immune attack) of murine 4T1.2 breast cancer depends on CCR4+ Tregs infiltrated into the lungs to protect metastasizing cancer cells from NK cells [20]. Despite its highly aggressive nature, 4T1.2 cancer could not metastasize into the lungs in the absence of Tregs, for example, in NOD/SCID mice deficient in T and B cells [20] or if Tregs are depleted in immunocompetent WT mice using CD25 or CCR4 - targeting antibodies and toxins [20,21]. Overall, Treg inactivation is shown to inhibit cancer escape in various murine tumor models, although their therapeutic depletion seldom provides clinical benefits or augments immune responses to cancer vaccines in humans [21,22]. The main problem appears to be lack of strategies that only and specifically inactivate Tregs. A widely used method of depletion with anti-CD25 antibody can also non-specifically inactivate other potentially beneficial immune effector cells.
Here, to circumvent this problem, we wanted to employ anti-sense oligonucleotide/RNAi silencing technology, a potent and specific strategy for inactivation of gene expression of cells in vitro. Since its therapeutic use is limited or even precluded by lack of reliable and simple ways to transduce primary immune cells in vivo, we created chemokine CCL17 (TARC-arp) modified to bind oligonucleotides by linking it with a 15 amino acid fragment of a single-stranded DNA/RNA – binding (RBD) portion of the capsid antigen of HBV. The idea was to utilize TARC-arp as a vehicle for oligonucleotide delivery into CCR4+ Tregs, as we reported for exogenous antigens efficiently internalized into cell cytosol if fused with chemokines [23,24]. We show that TARC-arp can indeed transduce CCR4-expressing CD4+ T cells and Tregs with siRNA transiently silencing the genes of interest. In mice with established 4T1.2 breast cancer (a widely used model of regulatory immune cell-mediated cancer escape [25]), we found that IL10 produced from CCR4+ Tregs plays a non-redundant supporting role in lung metastasis. As such, TARC-arp abrogates lung metastasis by efficiently and specifically delivering siRNA and silencing the genes of interest, such as IL10 or FoxP3, in CCR4+ Tregs. Overall, considering the simplicity and potency of TARC-arp, we propose that chemokine-based gene silencing can be therapeutically used to modulate immune cells and improve outcome of diseases, such as by inactivating Tregs to block cancer escape and metastasis.
MATERIALS AND METHODS
Chemicals and reagents were from Sigma (St. Louis, MO), unless specified otherwise. Antisense and sense oligonucleotides to mouse IL10 gene (TGAGATCTGCAATGCA and GCCAGTCAGTAAGAGCAG, respectively) were purchased from The Midland Certified Reagent Company (Midland, TX); Ambion in vivo siRNA to mouse IL10 (s68180), FoxP3 (A, s73597 and B, s73595) and control siRNA (in vivo ready) were from Ambion Products (Austin, TX). The following antibodies were used: anti-mouse CD4-FITC, anti-mouse CD25-PE (Biolegend) and anti-mouse Foxp3-APC (eBioscience); rabbit anti-mouse CCR4 Ab (Capralogics, Biolegend); anti-human CCL17 Ab (Abcam, ab9816), Fc blocker (anti-CD16/32; BD Biosciences).
Chemoarp production
TARC-arp and RANTES-arp (collectively named chemoarp) encode mature sequences of human chemokines CCL17 (NM_002987) and CCL5 (NM_002985) fused in frame with hypothetical single DNA/RNA-binding domain (RBD) of HBcAg of HBV [26] (Arya et al., Patent is pending). TARC-FN was created from TARC-arp by replacing RBD with irrelevant peptide of the same size. Coding sequences of chemoarps were cloned using XhoI and NotI restriction sites under signal sequence of yeast α-factor into pPIC9 plasmid (Invitrogen). All constructs were verified by DNA sequencing (Keck DNA Sequencing Lab, New Haven, CT). Chemoarps were produced using methanol-inducible Pichia expression kit (Invitrogen) in Pichia pastoris GS115 following manufacturer’s instructions. Briefly, after one week of methanol induction, culture chemoarp-containing supernatants were collected by centrifugation at 3000–5000g and filtered through 0.25 μ filter. Then, chemoarp was purified using SP-Sepharose™ Fast Flow and Heparin-HP trap column chromatography (GE Healthcare) with Fast performance liquid chromatography (FPLC) (Bio-Rad BioLogic Duoflow). Chemoarp was eluted using NaCl gradient elution in 20 mM Na-phosphate buffer, pH8.0. Chemoarp-containing fractions were combined and dialysed against PBS in dialysis chambers with 3000 cutoff (Pierce, Thermo Fisher Scientific Inc.). Purity of proteins was (>95%), as verified by Coomassie Blue staining and western blotting with respective antibodies.
Cells and mice
The 4T1 mouse mammary carcinoma cells (CRL-2539), human acute T-lymphoblastic leukemia cell lines CCRF-CEM (CEM, CCL-19) were from the American Type Culture Collection, Rockville, MD); 4T1.2 is a single cell subclone of 4T1 cells and a gift from Dr. Robin L. Anderson (Peter McCallum Cancer Center, Australia). Cells were cultured in RPMI 1640 (Invitrogen Corporation, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum, HEPES-Sodium Pyruvate, non-essential amino acids, 2-Mercaptoethanol, L-glutamine, and Penicillin-Streptomycin (complete RPMI). Mouse CD3+ T cells were isolated from naïve mouse spleen using T cell enrichment columns (R&D Systems, Minneapolis, MN). To generate non-Tregs (purity >99.5 %), CD4+ T cells were isolated by CD4 negative selection kit (Miltenyi Biotec Inc) and separated from CD25+ cells using CD25 Microbead kit (Miltenyi Biotec Inc). The CD25+CD4+ cells were used as Tregs. The lung mononuclear cells were isolated using Ficoll density separation after digesting lungs with collagenase/DNase/elastase mixture (Sigma).
In vitro siRNA manipulations
siRNA binding was evaluated by incubating 2 pmol siRNA with serial dilutions of TARC-arp in PBS on ice for 15 min. Upon binding with TARC-arp, siRNA losses ability to be separated by electrophoresis in 2% ethidium bromide stained agarose gel in TAE buffer (Invitrogen). To evaluate siRNA uptake, 4T1 cells (20,000 cells/well) were treated with 20 pmol of Invitrogen’s BLOCK-iT™ Alexa Fluor® Red Fluorescent Oligo (Invitrogen) complexed with TARC-arp for 18 hours at 37°C. After washing 3 times in PBS to remove free siRNA, fluorescence was visualized using a Zeiss Axiovert 200 microscope (Carl Zeiss, Heidelberg, Germany). Images were processed using NIH ImageJ software. Lipofectamine-2000 (Invitrogen) – mediated siRNA transfection was used as the positive control. Viability of 4T1.2 cells was tested using WST-1 assay (Roche) following manufacturer’s instructions. In brief, titrated amounts of TARC-arp (5–25 μg/ml, with or without siRNA) were in vitro cultured with 20,000 cells in triplicate 96-well plates in cRPMI for 24, 48 and 72h.
To inhibit gene expression, indicated amounts of siRNA and TARC-arp were mixed in PBS and pre-incubated on ice for 15–30 min prior adding onto cells. For example, 6 μg/ml TARC-arp complexed with 18 μg/ml siRNA was incubated with freshly isolate 50,000–100,000 CD4+ T cells/50 μl/well in serum-free RPMI for 1h at 37°C. Then, after presumable internalization of the complex, the medium was replaced with complete RPMI. Cells were harvested after 24, 48, 72 and 96 hours of incubation at 37°C to test expression of targeted genes using FACS staining with respective antibodies against mouse and human FoxP3 (eBioscience, San Diego, CA), CD4 and CCR4 (Biolegend, San Diego, CA). Conditioned media of cells were tested for CCL17 expression using ELISA kit (MAB529, R&D, Minneapolis USA) and IL6 by capturing with anti-mouse IL-6 antibody and detecting with biotin-conjugated anti-IL6 antibody and HRP-Streptavidin (eBioscience). Chemotaxis of cells was assessed in a 48-well microchemotaxis chamber (NeuroProbe) with a 10-μm polycarbonate filter (Osmonics) coated with rat tail collagen type I (BD Bioscience) or using 24-well tissue transwell plate (5-μm pore size, Costar; Corning Life Science, Acton, MA), as previously described [20].
In vivo manipulations
Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (NIH Publication No. 86-23, 1985). All experiments were performed using 4–8 weeks old female BALB/C mice and BALB/C background mice with IL10 deficiency (C.129P2(B6)-Il10tm1Cgn/J; stock 004333; The Jackson Laboratory, Bar Harbor, ME) or CCR4 deficient mice (the gift from Dr. Hogaboam, C.M. (University of Michigan Medical School, Ann Arbor, MI) in a pathogen-free environment at the National Institute on Aging Animal Facility, Baltimore, MD. Mice were challenged s.c. into the fourth mammary gland (pad) with 5 × 104 of tumor cells. After 28 days of tumor challenge, primary tumor was excised and weighed and the lungs were analyzed for metastasis by ex vivo injecting India ink through the trachea, which was destained in Fekete’s solution to count tumor nodules. To test the in vivo efficacy of TARC-arp (40 μg) complexed with siRNA (20 μg), tumor-bearing mice were intravenously injected via the tail vein at days 3, 6, 9, 13, 15, 18 and 21 post tumor challenge. The role of IL10 – expressing Tregs were assessed by adoptively transferring splenic Tregs (106 Tregs) purified from naïve WT BALB/C or IL-10 KO mice. Tregs were i.v. injected at days −1 and 5 post tumor challenge into tumor-bearing IL10 KO mice.
Statistical Analysis
The results are presented as the mean of triplicates ± SD of at least three experiments. Differences were tested using Student’s t test and a 2 sided p-value less than 0.05 was considered statistically significant.
RESULTS
CCL17-arp inactivates gene expression of CCR4+ cells by delivering anti-sense oligonucleotides
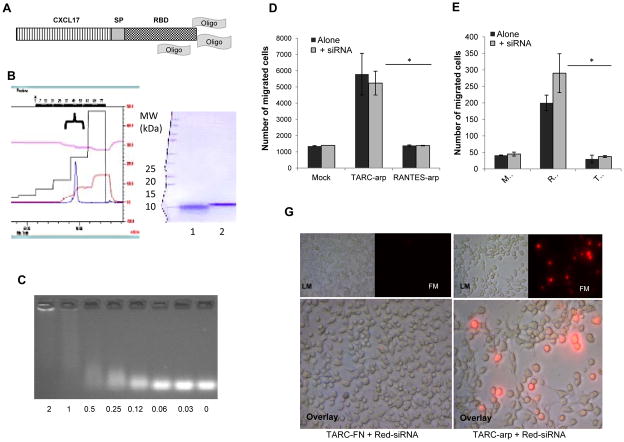
Immunotherapeutic utilization of RNAi and antisense technology is hampered by its inefficient transduction of primary immune cells. To circumvent this problem we generated chimeric CCL17 protein (TARC-arp, Fig. 1A) fused with RNA-binding domain of HBV (to enable binding of short DNA or RNA oligonucleotides). When expressed and purified from culture supernatant of Pichia pastoris as monomeric protein (>95% purity, Fig. 1B), TARC-arp could efficiently bind siRNA in physiological buffers (Fig. 1C). Importantly regardless of the presence or absence of siRNA, TARC-arp retained chemotactic property of parental CCL17, as it specifically induced chemotaxis of CCR4-expressing CEM cells (Fig. 1D). Cells that did not express CCR4, such as control CCR5+ Jurkat cells (Fig. 1E), did not migrate towards TARC-arp. Conversely, a different CCL5-based formulation (RANTES-arp) was only chemotactic for CCR5+ cells (Fig. 1E), but not CCR4+ cells (Fig. 1D).
FIGURE 1.
(A) Schema of TARC-arp. CCL17 was fused with an RNA-binding fragment (RBD) to enable binding of oligonucleotides to deliver into the cytosol of Tregs through CCR4-mediated internalization. CCL17 and RBD were separated with a (Gly4Ser)3 spacer fragment (SP) to enable their unhindered functions and binding of oligonucleotides (Oligo) in physiological buffers. (B) TARC-arp was purified from culture supernatant using SP-Sepharose™ Fast Flow and Heparin-HP trap FPLC chromatography. It was eluted as a single peak with 1M NaCl in 20 mM Na-phosphate buffer, pH 8.0 (left panel) with >95% purity (as judged by Coomasie brilliant blue staining, right panel). Compared with control chemokine (1), TARC-arp migrates slower in 4–20% PAAG gel due to RBD (2, right panel). (C) TARC-arp binding with free siRNA was assessed by the loss/delay of siRNA migration in 2% agarose gel electrophoresis in TAE buffer. Shown, titration results of TARC-arp mixed with a constant amount of siRNA (μg/ml) incubated for 15 min in PBS. Binding of siRNA does not affect chemotaxis of TARC-arp (D) and RANTES-arp (E). Y-axis shows the number of migrated CCR4+ CEM cells (D) and CCR5+ Jurkat cells (E) ± SEM treated with 100 ng/ml TARC-arp or control RANTES-arp alone or complexed with siRNA. (G) TARC-arp delivers Alexa Fluor® Red Fluorescent oligonucleotide (Red-siRNA) into 4T1 cancer cells that had about 30% CCR4+ cells. Control cells were treated with Red-siRNA alone (not shown) or mixed with TARC-FN. After 18 h incubation at 37°C, the cells were washed with PBS and visualized under normal light (LM, top panel) and 555 nm wavelength excitation (FM, top panels, and overlay, lower panels) using a fluorescent microscope. Shown representative of triplicate experiments reproduced at least three times. *P<0.05 is for TARC-arp vs. RANTES-arp.
Since siRNA-bound TARC-arp retained cognate activity of CCL17, TARC-arp would also be expected to deliver/internalize siRNA into CCR4-expressing cells, as we previously reported for exogenous antigens fused with chemokines [23,24]. To test this possibility, we treated 4T1 cells (20000 cells/well) with TARC-arp (1000 pmol) coupled with Alexa Fluor® Red Fluorescent oligonucleotide (Red-siRNA, 20 pmol). After 18 hours of incubation at 37°C, the cells were extensively washed with PBS to remove free Red-siRNA and their fluorescence was assessed. In concordance with our previous report that about 30% of 4T1 cells express CCR4 [20], we detected a comparable and significant proportion of cells fluorescent after the treatment with TARC-arp + Red-siRNA (Fig. 1G). In contrast, control cells treated with Red-siRNA free (data not shown) or coupled with TARC-FN protein, which had RBD replaced with an irrelevant peptide (Fig. 1G), did not have significant fluorescence. Hence, TARC-arp can indeed transduce CCR4+ cells with oligonucleotides.
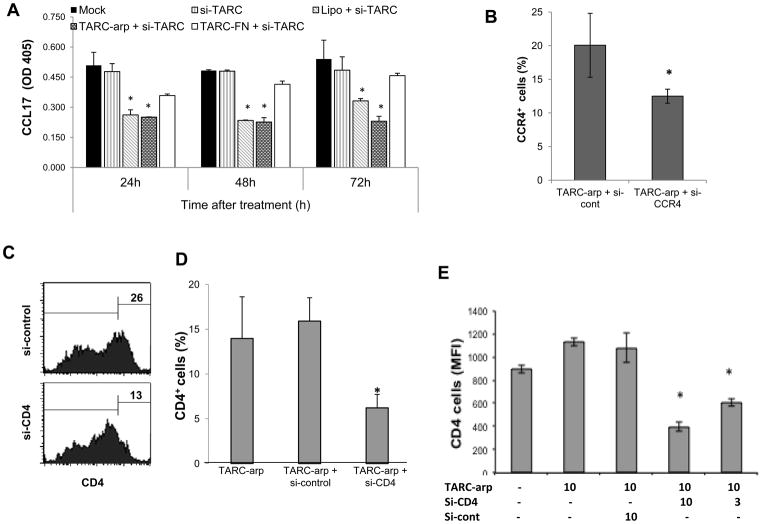
To test whether the transduction also leads to gene silencing, we treated 4T1 cells with siRNA to CCL17 (si-TARC, 1000 pmol) coupled with TARC-arp or control TARC-FN. The gene silencing was assessed by measuring production of CCL17 in conditioned medium (CM). TARC-arp + si-TARC did not affect viability of 4T1 cells (data not shown), but significantly decreased CCL17 in CM (Fig. 2A). The reduction was transient, as it was no longer detectable after four days of incubation (Fig. 2A and data not shown). In contrast, no tangible reduction of CCL17 was detected in CM of control cells treated with TARC-FN + si-TARC (Fig. 2A). The extent of the inhibition was comparable with the use of lipofectamine (Lipo + si-TARC, Fig. 2A), a widely used strategy for silencing genes in cell lines in vitro, but not suitable for in vivo applications due to the inability to transduce primary immune cells.
FIGURE 2.
(A – E) TARC-arp + si-RNA specifically inhibits expression of targeted genes, such as CCL17 in the culture medium of adherent 4T1 cells (A) and CCR4 (%, B) and CD4 (C–E) on the surface of human (C,D) and murine (E) CD4+ T cells. In (E), numbers show concentration of TARC-arp and si-RNA (μg/ml) used to treat 5×105 cells. TARC-arp + siRNA did not affect viability of 4T1.2 cells (data not shown). Y-axis shows % of CCR4+ (B) and CD4+ (D,E) cells ± SEM gated in CD4+ cells. Control cells were treated mock (PBS), or TARC-arp alone or mixed with control siRNA (A–E), or siRNA transduced using lipofectamine-2000 (A). Shown in (A), ELISA of conditioned media of cells after 24, 48 and 72 h post treatment. Histograms (C) show % of CD4+ cells of representative data shown in (D). *P<0.05 is for comparisons between with TARC-arp + si-cont. All experiments were reproduced at least three times, each sample in triplicate.
Next, TARC-arp was tested using primary CD4+ T cells purified from human peripheral blood and murine spleens, which usually contain about 20% circulating CCR4+ cells [10]. Expression of CCR4 (Fig. 2B) and CD4 (Fig. 2C–E) on the surface of CD4+ T cells was significantly reduced when the cells were treated with TARC-arp coupled with siRNA to CCR4 or CD4 genes (TARC-arp + si-CCR4 and TARC-arp + si-CD4), respectively. The inhibition was specific, as expression of CCR4 and CD4 was not changed in cells treated with TARC-arp alone or coupled with irrelevant siRNA (TARC-arp + si-cont, Fig. 2B–E). Although the oligonucleotide transduction may also non-specifically affect function and viability of primary immune cells, we did not detect any alteration of cell viability even after repeated treatment (data not shown). In summary, TARC-arp allows to specifically and transiently inhibit gene expression in primary CCR4+ T cells by delivering siRNA.
Transient in vivo inactivation of FoxP3 expression in CCR4+ Tregs abrogates lung metastasis of mice with breast cancer
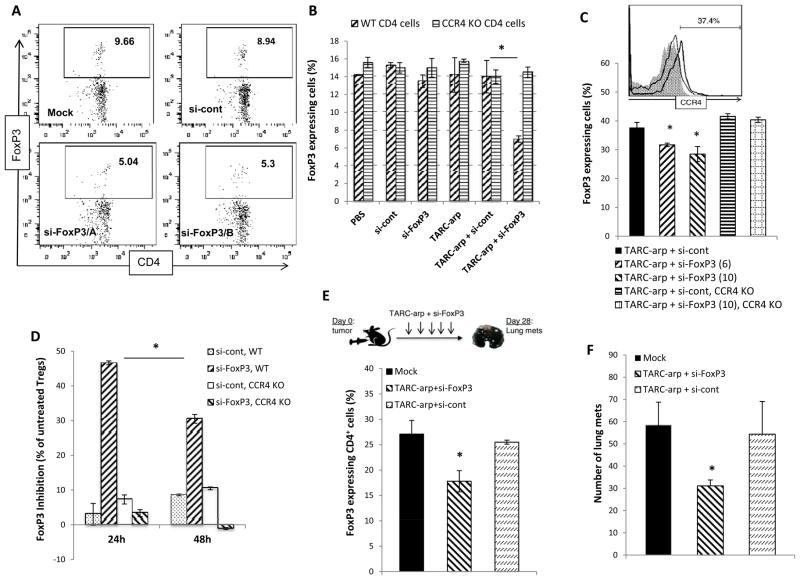
Since CCR4-expressing FoxP3+ Tregs are required for cancer escape and metastasis [19,20], the process can probably be blocked by siRNA-mediated silencing of FoxP3, a key Treg-restricted gene [27]. First, we evaluated ability of TARC-arp to inactivate FoxP3 expression of CCR4+ Tregs in vitro using murine splenic CD4+ T cells. Although CCR4+ Tregs usually represent about 5–10% cells [10], we clearly detected significant reduction of FoxP3-expressing cells (Tregs, Fig. 3A,B) within CD4+ T cells treated with TARC-arp coupled with two different si-RNA to FoxP3 (si-FoxP3/A and si-FoxP3/B). The inhibition was specific for si-FoxP3, as control treatments with saline (mock) or with TARC-arp + conrol siRNA failed to affect FoxP3 expression (Fig. 3A,B). Importantly, TARC-arp + si-FoxP3 also failed to silence FoxP3 expression if CD4+ T cells did not express CCR4 due to genetic knock out (CCR4 KO, Fig. 3B), indicating that the process required CCR4 expression. To further confirm these results, Tregs were purified from splenic CD4+ T cells using anti-CD25 Ab, which usually yields > 80% CD25+ Tregs with about 30–40% expressing CCR4 (Fig. 3C) [10]. Treatment of Tregs with TARC-arp + si-FoxP3, but not TARC-arp + si-cont, also caused significant (up to 50%, Fig. 3D) and in a dose-dependent reduction of FoxP3 expression (p<0.05, Fig. 3C,D). Importantly, in the absence of CCR4 expression, TARC-arp + si-FoxP3 failed to reduce FoxP3 expression in Tregs (CCR4 KO, Fig. 3C,D), suggesting that TARC-arp also required CCR4-dependent internalization process to deliver siRNA into Tregs.
FIGURE 3.
In vitro, TARC-arp efficiently delivers siRNA to FoxP3 (si-FoxP3) and silences expression of FoxP3 in murine splenic CD4+ T cells (A,B). In (A), numbers indicate % FoxP3+ cells within the depicted gate in a representative dot plot of CD4+ cells stained for FoxP3 (intracellular) after the following treatments: Mock, or TARC-arp mixed with control siRNA (si-cont) or two different si-FoxP3s (A and B, lower panels). The FoxP3 silencing ability of TARC-arp is lost in murine CD4+ T cells (B) and purified Tregs (CD25+CD4+ T cells, C,D) deficient in CCR4 (CCR4 KO). Y-axis shows % of FoxP3+ within CD4+ cells ± SEM (B,C) and % ± SEM of FoxP3 inhibition compared with mock-treated Tregs (D) of triplicate experiments. Histogram (C) shows expression of CCR4 on Tregs from WT and CCR4 KO mice. (E) To test in vivo efficacy of TARC-arp delivery, 5–10 per group BALB/c mice challenged with 4T1.2 cancer in the mammary gland (day 0) were i.v. injected with 40 μg TARC-arp + 20 μg siRNA at days 3, 6, 9, 13, 15, 18 and 21. All mice were euthanized at day 28 to assess FoxP3-expression in peripheral blood Tregs (gated on CD4 cells, E) and to count metastatic foci in the lungs (F). Y-axis shows % of FoxP3+ (gated on CD4 cells, E) or number of metastatic foci in the lungs (F) ± SEM of four-five mice per group experiment. *P<0.05 is for comparisons between TARC-arp + si-FoxP3 and TARC-arp + si-cont (B, C, E, F) or for % of inhibition FoxP3 between 24h and 48h incubations with TARC-arp + si-FoxP3 (D). Shown are data independently reproduced at least three times (B, C, E, F) and twice (D).
Although the strategy only yields transient reduction of FoxP3 in Tregs with maximum effects detected at 24 h post treatment (Fig. 3D), we hypothesized that TARC-arp/si-FoxP3 may also enable us to inactivate in vivo expression of FoxP3 in natural CCR4+ Tregs promoting cancer metastasis. To study this possibility, we used BALB/c mice s.c. challenged with 4T1.2 breast cancer cells in the mammary gland, a model that requires CCR4+ FoxP3+ Tregs for successful lung metastasis [20]. Although genetic deficiency in FoxP3 causes fatal autoimmune disorders in mice and humans [27], its transient inactivation in CCR4+ Tregs in tumor-bearing mice remains unknown. We treated tumor-bearing mice intravenously seven times with 40 μg TARC-arp coupled with 20 μg si-FoxP3 (Fig. 3E). Control mice treated with TARC-arp + control siRNA did not affect FoxP3 expression in Tregs in the peripheral blood (Fig. 3E) nor lung metastasis of 4T1.2 cells (Fig. 3F), as expected. In contrast, we detected significantly reduced numbers of FoxP3-expressing CD4+ cells in the peripheral blood (Fig. 3E), which was associated with decreased numbers of metastatic foci in the lungs of mice treated with TARC-arp + si-FoxP3 (p<0.05, Fig. 3F). Overall, TARC-arp + si-FoxP3 significantly reduced lung metastasis (p<0.05) in all five independently reproduced experiments involving 5–10 per group mice. Thus, confirming the importance of CCR4+Tregs in lung metastasis of 4T1.2 cancer cells [20], our data clearly indicate that their metastasis - supporting activity can be efficiently blocked by transient and exclusive inactivation of FoxP3 in natural Tregs of tumor-bearing mice.
In vivo inactivation of IL10 in CCR4+ cells abrogates lung metastasis by enhancing CD8+ T cell infiltration in the lungs of tumor-bearing mice
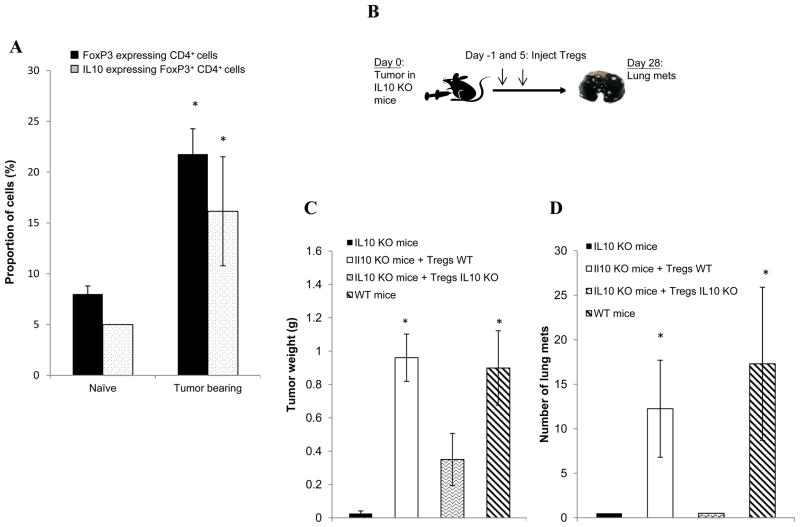
Next, to further confirm the in vivo efficacy of TARC-arp strategy, we also tested therapeutic benefit of inactivation of IL10. We recently reported that IL10 produced by Th2-type CD4+ T cells is only needed for primary tumor progression, but not essential for lung metastasis of 4T1.2 cancer cells [28]. Despite this, lung metastasis in this model appears to require IL10 expressed by CCR4+ Tregs, as the inability of cancer to metastasize in the absence of Tregs can be reversed by adoptive transfer of WT, but not IL10-deficient splenocytes [20,29]. To test this possibility, first we quantified numbers of IL10+ Tregs in peripheral blood of 4T1.2 cancer-bearing mice that is known to cause FoxP3+ Treg increase [20,29]. Indeed, compared with naïve mice, tumor-bearing mice contained significantly increased amounts of both IL10+ Tregs and FoxP3+ Tregs (p<0.05, Fig. 4A). Next, to prove the importance of IL10+ Tregs in lung metastasis, we implanted 4T1.2 cancer cells into the mammary glands of WT or congenic IL10-deficient (IL10 KO) mice (Fig. 4B). Compared with WT mice, IL10 KO mice poorly supported 4T1.2 cancer growth (Fig. 4C) and metastasis (Fig. 4D). However, the inability of cancer to progress and metastasize in IL10 KO mice was completely reversed if IL10 KO mice were adoptively transferred with congeneic WT Tregs (Fig. 4C,D). In contrast, IL10 KO mice replenished with Tregs from IL10 KO mice only slightly enhanced tumor progression (Fig. 4C), but still remained free of lung metastasis (Fig. 4D). Thus, 4T1.2 cancer progression and metastasis also require IL10 expressed by Tregs.
FIGURE 4.
(A) Compared with naïve mice, peripheral blood of 4T1 tumor-bearing mice contains elevated proportions of IL10 and FoxP3–expressing CD4+ T cells. Numbers represent % of IL10+ and FoxP3+ within (gated) CD4+ cells, as compared with isotype control IgG. (B) IL10 KO mice were i.v injected with PBS or adoptively transferred with 5×106 Tregs (CD25+CD4+) purified from naïve IL10 KO or WT BALB/C mice one day before and 5 days after s.c. challenge with 4T1.2 cancer cells (day 0). At day 28, tumor growth (C) and metastasis in the lungs (D) were compared with ones in control BALB/c mice (WT mice) challenged with 4T1.2 cells in a side-by-side experiment. Y-axis shows weight (g) of primary tumor (C) and number of metastatic foci in the lungs (D) ± SEM of five mice per group experiment. All results were reproduced at least twice. *P<0.05 is for comparisons between naïve and tumor-bearing mice (A) and groups injected with WT and IL10 KO Tregs (C, D).
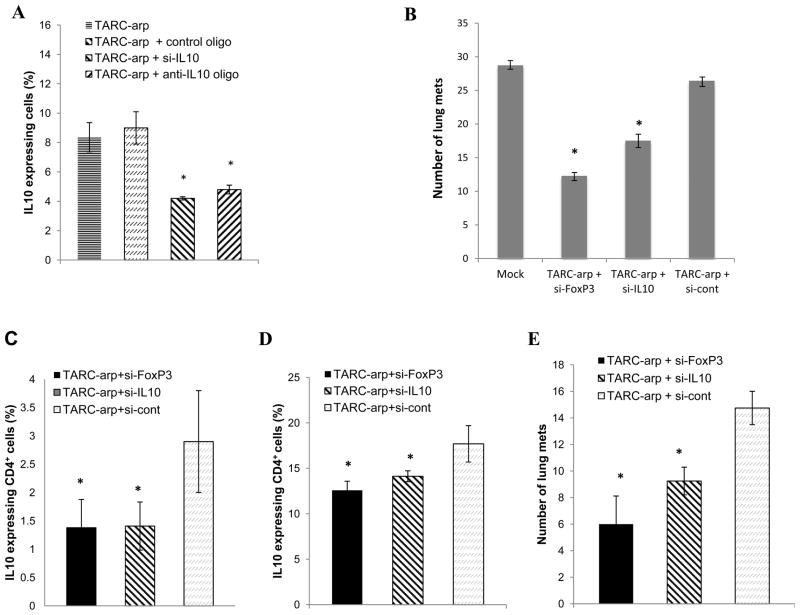
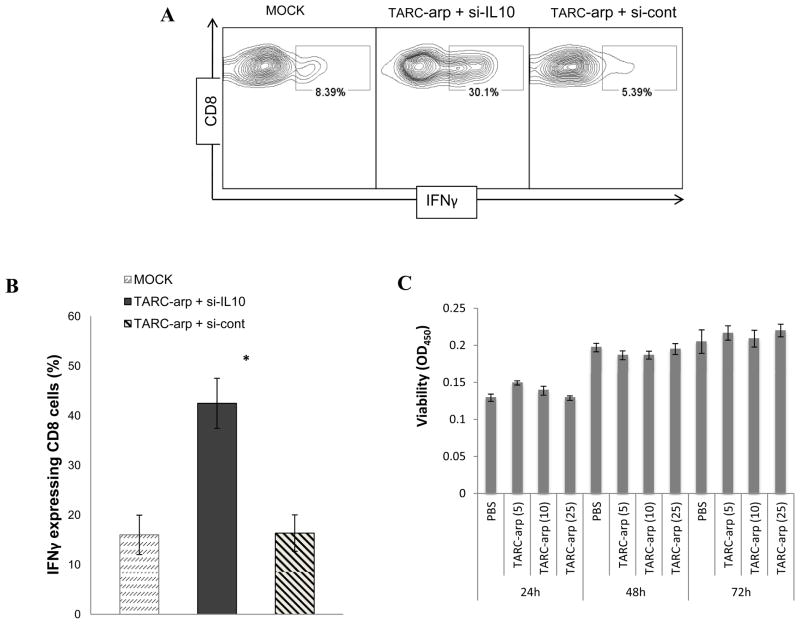
Since the majority of metastasis-promoting Tregs express CCR4 [20], lung metastasis may be controlled by silencing their IL10 expression with TARC-arp. First, we tested whether TARC-arp coupled with IL10-specific siRNA (si-IL10) or anti-sense DNA oligonucleotide (anti-IL10 oligo) can in vitro inhibit IL10 expression from purified murine Tregs (CD25+ CD4+ T cells). As shown in Fig. 5A, the proportion of IL10-expressing Tregs was significantly reduced by treatment with TARC-arp + si-IL10 or TAC-arp + anti-IL10 oligo, but not TARC-arp + control scrambled oligo. Next, to test therapeutic benefit of IL10 inhibition in CCR4+ Tregs in vivo, we treated 4T1.2 cancer-bearing mice with TARC-arp + si-IL10 or TARC-arp+si-FoxP3 in a side-by-experiment. While all control mice (mock-treated or injected with TARC-arp + si-cont, Fig. 5B) succumbed to massive metastasis, both TARC-arp + si-IL10 and TARC-arp + si-FoxP3 significantly abrogated lung metastasis (Fig. 5B). Thus, considering the essential importance of Tregs in the metastasis of 4T1.2 cells, we speculated that the two si-RNAs probably targeted the same type of Tregs. To confirm this possibility, we repeated the experiment to compare lung metastasis and expression of IL10 in Tregs. Indeed, both TARC-arp + si-IL10 and TARC-arp + si-FoxP3, but not TARC-arp + si-cont, comparably reduced numbers of IL-10+ Tregs isolated from peripheral blood (Fig. 5C) and the lungs (Fig. 5D), which was associated with the inhibition of lung metastasis (Fig. 5E). Considering the importance of IL10 in immunoregulation, the loss/reduction of IL10-expressing Tregs is expected to release immunosuppression and to activate antitumor effector immune responses. In support, we found drastically increased IFNγ-producing CD8+ T cells (key antitumor cells associated with a favorable clinical outcome in cancer patients [30,31]) in the lungs of tumor-bearing mice treated with TARC-arp + si-IL10 (Fig. 6A,B). Control mice treated with TARC-arp + si-cont succumbed to massive metastasis (Fig. 3F and 5B,E) and did not reduce the numbers of IL10+ Tregs (Fig. 5C,D) nor elevate proportion of IFNγ-expressing CD8+ T cells in the lungs (Fig. 6A,B). Taken together, as for FoxP3 inactivation, these results indicate that TARC-arp/si-IL10 can also provide anti-metastatic benefit in mice with aggressive and metastatic breast cancer by transiently down-regulating IL10 expression in CCR4+ cells, particularly Tregs. Although a focus of a different study, these results suggest that IL10 from Tregs plays a non-redundant role in lung metastasis, as in the same model we previously reported that the IL10-producing Th2-type CD4+ T cells were only needed for primary tumor progression, but not for lung metastasis [28].
FIGURE 5.
(A) siRNA (si-IL10) or anti-sense oligonucleotide (anti-IL10 oligo) reduce the proportion of IL10-expressing Tregs, if delivered with TARC-arp. Control treatment was TARC-arp complexed with sense oligo to IL10. Shown, intracellular staining for IL10 within CD4+ T cells ± SEM of triplicate experiment. (B) Lung metastasis of 4T1.2 cancer cells is abrogated by treating tumor-bearing mice with TARC-arp + si-FoxP3 or + si-IL10. Y-axis shows # of metastatic foci in the lungs ± SEM of four BALB/c mice per group experiment reproduced trice. In an independent experiment (C–E), the proportion (% ± SEM) of IL10+ Tregs (gated in CD4) was reduced in the peripheral blood (C) and lungs (D) of 4T1.2 cancer-bearing mice treated with TARC-arp + si-IL10 and TARC-arp + si-FoxP3, which was associated with the inhibition of lung metastasis (E). Mice were treated with TARC-arp and si-RNA as in Fig. 3E. *P<0.05 is for comparisons between TARC-arp + si-IL10/si-FoxP3 and TARC-arp + si-cont.
FIGURE 6.
The reduction of IL10+FoxP3+ Tregs (Fig. 5C,D) and lung metastasis (Fig. 5E) is associated with the increased % of IFNγ-expressing CD8+ T cells (A and B) in the lungs of tumor bearing mice. Numbers in (A) show % of IFNγ in within CD8+ T cells (smaller gate) of a representative result summarized in (B) ± SEM. *P<0.05 is for comparisons between TARC-arp + si-IL10 and TARC-arp + si-cont, and was independently reproduced twice. (C) TARC-arp is not cytotoxic for 4T1.2 cancer cells. Y-axis shows viability of 4T1.2 cells (OD450 ± SEM in WST-1 assay, Roche) incubated for 24 h, 48 h and 72 h with titrated amounts of TARC-arp (5–25 μg/ml).
DISCUSSION
Here, we report the development of a simple chemokine-based therapeutic anti-sense oligonucleotide strategy for a targeted transient inactivation of key genes in immune cells to control disease outcome. We demonstrate that expression of genes of interest in primary immune cells can be transiently silenced using chimeric chemokine CCL17 (TARC-arp) modified to bind short DNA and RNA oligonucleotides. Based on our modeling studies in mice with breast cancer, we think that this strategy has significant practical and clinical value. First of all, it successfully circumvents a stumbling block of therapeutic utilization of de facto potent anti-sense oligonucleotide/RNAi technology - the inability to transduce primary immune cells in vivo. Unlike the viral vector-based shRNA technology that is mostly used to substitute the lack of non-cytotoxic methods of transduction of primary immune cells, we show that TARC-arp efficiently silences genes in primary CD4+ T cells and Tregs by delivering siRNA both in vitro and in vivo. The specificity of the strategy is controlled at several levels: by differential expression of CCR4 and the uniqueness of a targeted gene, such as FoxP3. Unlike the antibody-based approach [32], the chemokine-based method is versatile and easy to generate in large quantities at any research facility without the need for a specific antibody. Since chemokine receptors recycle back to the cell surface, TARC-arp can also be applied repeatedly without detectable cytotoxicity (data not shown), if prolonged gene silencing is needed.
In BALB/c mice with 4T1.2 breast cancer, a widely utilized model that imitates human breast cancer escape and metastasis [33], we demonstrate that the targeted silencing of FoxP3 in CCR4+ Tregs is sufficient to reduce their metastasis-promoting functions and to block lung metastasis. Although CCR4 is also expressed on about 30% of 4T1.2 cancer cells [20], we do not think that TARC-arp directly affected them, as TARC-arp alone or complexed with control si-RNA did not affect in vitro viability of 4T1.2 cells (Fig. 6C, Fig. 2A and data not shown) nor reduced metastasis in mice (TARC-arp + si-cont, Fig. 3F, 5B, 5E). Confirming the data in mice with a genetic deficiency of FoxP3, which completely abrogates the regulatory activity of Tregs causing fatal autoimmune disorders in mice and humans [27], our data clearly indicate that transient inactivation of FoxP3 in CCR4+ Tregs with TARC-arp/si-FoxP3 is also sufficient to block metastasis presumably via inactivation of their regulatory functions. This is despite the fact that a number of regulatory and suppressive immune cells also participate in 4T1 cancer escape and lung metastasis [29,34,35]. The anti-metastatic activity of TARC-arp + si-FoxP3/si-IL10 underscores the importance of CCR4+ Tregs and further confirms our previous findings that the absence and dysfunction of Tregs preclude lung metastasis of 4T1.2/4T1 cancer cells [20,28,29].
The simplicity of the TARC-arp use not only enabled us to elucidate the role of CCR4+ Treg subsets in lung metastasis, but also to demonstrate the importance of IL10-producing CCR4+ Tregs. Although various immune cells produce IL10 to mediate immune regulation, our recent data indicate that in the same tumor model the IL10-producing Th2-polarized CD4+ T cells do not control metastasis [28]. Instead, they were mostly required for progression of cancer at the primary site, in the mammary glands [28]. However, our data shown here indicate that IL10 produced by CCR4+FoxP3+ Tregs plays a non-redundant and essential role in lung metastasis. First, 4T1.2 cancer did not metastasize in congenic IL10 KO mice unless reconstituted with WT Tregs, but not IL10 KO Tregs. Second, the TARC-arp-mediated silencing of IL10 in CCR4+ Tregs was sufficient to block metastasis in tumor-bearing WT mice. Although the molecular mechanism of IL10 produced from CCR4+ Tregs in lung metastasis is not known and is the subject of a different study, it is tempting to speculate that this is to function as IL10 –producing Tregs that specifically regulate inflammation at environmental interfaces [14,15]. Alternatively, IL10 may be involved in modulation of NK cell activity [36], NK cell-mediated cancer killing [37], or to abrogate cross-talk between NK cells and DCs [38], thus, disabling anti-tumor protective functions of effector immune cells. It is also plausible that the metastasis-promoting role of IL10 is to modify target NK cells to make them susceptible for the CCR4+ Treg-mediated killing with βGBP [20]. On the other hand, as in IL10 deficient mice that are protected from ultraviolet-induced carcinogenesis due to Th1-type skewed responses and antitumor CD8+ T cells [39], we also detected significant increase in IFNγ-producing CD8+ T cells in the lungs and the secondary lymphoid organs of tumor-bearing mice treated with TARC-arp + si-IL10. The infiltration of CD8+ T cells is usually associated with antitumor clearance in mice and humans and is a sign of a favorable clinical outcome in cancer patients [30,31]. In concordance, in TARC-arp + si-IL10 –treated mice, the increase in IFNγ-producing CD8+ T cells in the lungs was associated with reduced ability of 4T1.2 cancer to metastasize into the lungs. Thus, an additional benefit of silencing of IL10 in CCR4+ Tregs is to disable their regulation and free antitumor effector adaptive immune cells, such as IFNγ-producing CD8+ T cells. Confirming this, Tregs utilize IL10 to inhibit IFNγ production and proliferation of CD8+ T cells [40] or modulate CD4+ helper T cells [41] and DCs [19,27]. Overall, in cases such as a very aggressive and difficult to treat murine 4T1 breast cancer where CCR4+ Tregs promote lung metastasis [20], TARC-arp allows to transiently silence essential genes in Tregs and thereby abrogate lung metastasis. Thus, the use of TARC-arp + siRNA alone or in combination with other treatment modalities may also help controlling metastasis in cancer patients, a primary cause of bad disease outcome.
Acknowledgments
We are grateful to Drs. Edward Goetzl (UCSF) and Dan Longo, Ana Lustig and Rachel Munk (NIA/NIH) for helpful comments and suggestions; Karen Madara (NIA/NIH) for providing human blood samples. This research was supported by the Intramural Research Program of the National Institute on Aging, NIH.
Footnotes
Conflict of Interest: This work is fully executed by employees of US government. The authors declare no conflict of interest.
References
- 1.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 2.Valmori D, Merlo A, Souleimanian NE, Hesdorffer CS, Ayyoub M. A peripheral circulating compartment of natural naive CD4 Tregs. J Clin Invest. 2005;115:1953–1962. doi: 10.1172/JCI23963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubtsov YP, Niec RE, Josefowicz S, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonuleit H, Schmitt E, Stassen M, et al. Identification and functional characterization of human CD4(+)CD25(+) T cells with regulatory properties isolated from peripheral blood. J Exp Med. 2001;193:1285–1294. doi: 10.1084/jem.193.11.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bluestone JA, Abbas AK. Natural versus adaptive regulatory T cells. Nat Rev Immunol. 2003;3:253–257. doi: 10.1038/nri1032. [DOI] [PubMed] [Google Scholar]
- 6.Josefowicz SZ, Niec RE, Kim HY, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleinewietfeld M, Puentes F, Borsellino G, et al. CCR6 expression defines regulatory effector/memory-like cells within the CD25(+)CD4+ T-cell subset. Blood. 2005;105:2877–2886. doi: 10.1182/blood-2004-07-2505. [DOI] [PubMed] [Google Scholar]
- 8.Levings MK, Roncarolo MG. T-regulatory 1 cells: a novel subset of CD4 T cells with immunoregulatory properties. J Allergy Clin Immunol. 2000;106:S109–S112. doi: 10.1067/mai.2000.106635. [DOI] [PubMed] [Google Scholar]
- 9.Baatar D, Olkhanud PB, Wells V, et al. Tregs utilize beta-galactoside-binding protein to transiently inhibit PI3K/p21ras activity of human CD8+ T cells to block their TCR-mediated ERK activity and proliferation. Brain Behav Immun. 2009;23:1028–1037. doi: 10.1016/j.bbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baatar D, Olkhanud P, Sumitomo K, et al. Human Peripheral Blood T Regulatory Cells (Tregs), Functionally Primed CCR4+ Tregs and Unprimed CCR4- Tregs, Regulate Effector T Cells Using Fas L. J Immunol. 2007;178:4891–4900. doi: 10.4049/jimmunol.178.8.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossman WJ, Verbsky JW, Barchet W, et al. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 2004;21:589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. Cutting edge: contact-mediated suppression by CD4+CD25+ regulatory cells involves a granzyme B-dependent, perforin-independent mechanism. J Immunol. 2005;174:1783–1786. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, Hanabuchi S, Wang YH, et al. Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity. 2008;28:870–880. doi: 10.1016/j.immuni.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rubtsov YP, Rasmussen JP, Chi EY, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 15.McLachlan JB, Catron DM, Moon JJ, Jenkins MK. Dendritic cell antigen presentation drives simultaneous cytokine production by effector and regulatory T cells in inflamed skin. Immunity. 2009;30:277–288. doi: 10.1016/j.immuni.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou L, Barnett B, Safah H, et al. Bone marrow is a reservoir for CD4+CD25+ regulatory T cells that traffic through CXCL12/CXCR4 signals. Cancer Res. 2004;64:8451–8455. doi: 10.1158/0008-5472.CAN-04-1987. [DOI] [PubMed] [Google Scholar]
- 17.Iellem A, Mariani M, Lang R, et al. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2001;194:847–853. doi: 10.1084/jem.194.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gobert M, Treilleux I, Bendriss-Vermare N, et al. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer research. 2009;69:2000–2009. doi: 10.1158/0008-5472.CAN-08-2360. [DOI] [PubMed] [Google Scholar]
- 19.Curiel TJ, Coukos G, Zou L, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 20.Olkhanud PB, Baatar D, Bodogai M, et al. Breast cancer lung metastasis requires expression of chemokine receptor CCR4 and regulatory T cells. Cancer Res. 2009;69:5996–6004. doi: 10.1158/0008-5472.CAN-08-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrne WL, Mills KH, Lederer JA, O’Sullivan GC. Targeting regulatory T cells in cancer. Cancer research. 2011;71:6915–6920. doi: 10.1158/0008-5472.CAN-11-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powell DJ, Jr, Felipe-Silva A, Merino MJ, et al. Administration of a CD25-directed immunotoxin, LMB-2, to patients with metastatic melanoma induces a selective partial reduction in regulatory T cells in vivo. Journal of immunology. 2007;179:4919–4928. doi: 10.4049/jimmunol.179.7.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiavo R, Baatar D, Olkhanud P, et al. Chemokine receptor targeting efficiently directs antigens to MHC class I pathways and elicits antigen-specific CD8+ T-cell responses. Blood. 2006;107:4597–4605. doi: 10.1182/blood-2005-08-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baatar D, Olkhanud P, Newton D, Sumitomo K, Biragyn A. CCR4-expressing T cell tumors can be specifically controlled via delivery of toxins to chemokine receptors. J Immunol. 2007;179:1996–2004. doi: 10.4049/jimmunol.179.3.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lelekakis M, Moseley JM, Martin TJ, et al. A novel orthotopic model of breast cancer metastasis to bone. Clin Exp Metastasis. 1999;17:163–170. doi: 10.1023/a:1006689719505. [DOI] [PubMed] [Google Scholar]
- 26.Le Pogam S, Chua PK, Newman M, Shih C. Exposure of RNA templates and encapsidation of spliced viral RNA are influenced by the arginine-rich domain of human hepatitis B virus core antigen (HBcAg 165–173) J Virol. 2005;79:1871–1887. doi: 10.1128/JVI.79.3.1871-1887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 28.Olkhanud PB, Rochman Y, Bodogai M, et al. Thymic stromal lymphopoietin is a key mediator of breast cancer progression. Journal of immunology. 2011;186:5656–5662. doi: 10.4049/jimmunol.1100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olkhanud PB, Damdinsuren B, Bodogai M, et al. Tumor-evoked regulatory B cells promote breast cancer metastasis by converting resting CD4 T cells to T-regulatory cells. Cancer research. 2011;71:3505–3515. doi: 10.1158/0008-5472.CAN-10-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 31.Pages F, Berger A, Camus M, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 32.Song E, Zhu P, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nature Biotechnology. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 33.Heppner GH, Miller FR, Shekhar PM. Nontransgenic models of breast cancer. Breast Cancer Res. 2000;2:331–334. doi: 10.1186/bcr77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha P, Clements VK, Ostrand-Rosenberg S. Interleukin-13-regulated M2 macrophages in combination with myeloid suppressor cells block immune surveillance against metastasis. Cancer Res. 2005;65:11743–11751. doi: 10.1158/0008-5472.CAN-05-0045. [DOI] [PubMed] [Google Scholar]
- 35.Bunt SK, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. 2006;176:284–290. doi: 10.4049/jimmunol.176.1.284. [DOI] [PubMed] [Google Scholar]
- 36.Barao I, Hanash AM, Hallett W, et al. Suppression of natural killer cell-mediated bone marrow cell rejection by CD4+CD25+ regulatory T cells. Proc Natl Acad Sci USA. 2006;103:5460–5465. doi: 10.1073/pnas.0509249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth MJ, Teng MW, Swann J, et al. CD4+CD25+ T regulatory cells suppress NK cell-mediated immunotherapy of cancer. J Immunol. 2006;176:1582–1587. doi: 10.4049/jimmunol.176.3.1582. [DOI] [PubMed] [Google Scholar]
- 38.Terme M, Chaput N, Combadiere B, et al. Regulatory T cells control dendritic cell/NK cell cross-talk in lymph nodes at the steady state by inhibiting CD4+ self-reactive T cells. J Immunol. 2008;180:4679–4686. doi: 10.4049/jimmunol.180.7.4679. [DOI] [PubMed] [Google Scholar]
- 39.Loser K, Apelt J, Voskort M, et al. IL-10 controls ultraviolet-induced carcinogenesis in mice. Journal of immunology. 2007;179:365–371. doi: 10.4049/jimmunol.179.1.365. [DOI] [PubMed] [Google Scholar]
- 40.Endharti AT, Rifa’I M, Shi Z, et al. Cutting edge: CD8+CD122+ regulatory T cells produce IL-10 to suppress IFN-gamma production and proliferation of CD8+ T cells. J Immunol. 2005;175:7093–7097. doi: 10.4049/jimmunol.175.11.7093. [DOI] [PubMed] [Google Scholar]
- 41.Annacker O, Pimenta-Araujo R, Burlen-Defranoux O, et al. CD25+ CD4+ T cells regulate the expansion of peripheral CD4 T cells through the production of IL-10. J Immunol. 2001;166:3008–3018. doi: 10.4049/jimmunol.166.5.3008. [DOI] [PubMed] [Google Scholar]