Abstract
Hookworms, parasitic nematodes that infect nearly one billion people worldwide, are a major cause of anemia and malnutrition. We hypothesize that hookworms actively manipulate the host immune response through the production of specific molecules designed to facilitate infection by larval stages and adult worm survival within the intestine. A full-length cDNA encoding a secreted orthologue of the human cytokine, Macro-phage Migration Inhibitory Factor (MIF) has been cloned from the hookworm Ancylostoma ceylanicum. Elucidation of the three-dimensional crystal structure of recombinant AceMIF (rAceMIF) revealed an overall structural homology with significant differences in the tautomerase sites of the human and hookworm proteins. The relative bioactivities of human and hookworm MIF proteins were compared using in vitro assays of tautomerase activity, macrophage migration, and binding to MIF receptor CD74. The activity of rAceMIF was not inhibited by the ligand ISO-1, which was previously determined to be an inhibitor of the catalytic site of human MIF. These data define unique immunological, structural, and functional characteristics of AceMIF, thereby establishing the potential for selectively inhibiting the hookworm cytokine as a means of reducing parasite survival and disease pathogenesis.
Hookworms are blood feeding intestinal nematodes that currently infect more than 700 million people in developing countries (1). The hookworm life cycle begins when eggs excreted in the feces of an infected individual hatch in soil and undergo successive molts to the infectious L3 stage. After contacting the skin of a permissive host, larvae migrate to the pulmonary vasculature, traverse alveolar capillaries, ascend the respiratory tree and are swallowed. Hookworms molt to the adult stage in the intestine, where they attach to the mucosal surface and feed on blood and tissue. Chronic blood and serum protein loss attributable to hookworm infection is associated with anemia, malnutrition, and growth/developmental delay, resulting in the loss of tens of millions of disability adjusted life-years annually (2).
There is no clear evidence of sterile immunity in humans following naturally acquired infection, suggesting that hookworms may modulate the host immune response, perhaps during tissue migration, and/or while attached to the intestinal mucosa. The fact that adult hookworms can survive within a single human host for many years (3) further suggests that these worms are capable of evading or dampening host immune responses that might kill parasites and/or trigger expulsion. Of the immunomodulatory activities that have been identified from the hookworms Ancylostoma or Necator, none has yet been shown to play a definitive role in the pathogenesis of infection or intestinal disease (4–7).
Macrophage migration inhibitory factor (MIF)4 is a pro-inflammatory cytokine first identified as a product of activated T cells, and subsequently demonstrated to have diverse biological functions (8). Mammalian MIF inhibits the random migration of macrophages and can act as a counter regulator of the immunosuppressive and anti-inflammatory effects of endogenous glucocorticoids (9). MIF promotes cytokine (TNF-α, IL-1β, IL-6, IL-8, and IL-12) production by macrophages, triggers proliferation of T cells, and induces the release of nitric oxide, matrix metalloproteases, COX-2, and prostaglandin E2 (10–12). The molecular mechanism of action of MIF involves sustained activation of the ERK-1/2 family of mitogen-activated protein kinases (MAPK), which leads to increased levels of phospholipase A2 and arachidonic acid, the precursors of proinflammatory prostaglandins and leukotrienes (13, 14). Infectious diseases in which host and/or pathogen derived MIF is thought to play an important role include tuberculosis, malaria, filariasis, taeniasis, and schistosomiasis (15–21).
We report here the molecular cloning of a cDNA corresponding to an orthologue of MIF that is secreted by host-dwelling stages of the hookworm parasite Ancylostoma ceylanicum. In vitro characterization confirms that the recombinant A. ceylanicum MIF (rAceMIF) is an active tautomerase and lymphocyte chemoattractant, similar to the human orthologue. Unlike human MIF, however, AceMIF is not inhibited by the small molecule ligand ISO-1, and the three-dimensional crystal structure reveals functionally relevant differences between the hookworm and human proteins. Together, these data establish a structural basis for the development of pathogen-specific MIF inhibitors as potential treatments for infectious diseases, including hookworm.
Experimental Procedures
Parasites
The life cycle of A. ceylanicum was maintained in Syrian golden hamsters (Mesocricetus auratus)(22) under protocols approved by the Yale Animal Care and Use committee. Soluble hookworm protein extracts (HEX) were prepared from A. ceylanicum by grinding adult worms in a tissue homogenizer, followed by centrifugation at 10,000 × g. Protein preparations of eggs/first stage (L1) A. ceylanicum larvae, third stage (L3) larvae and adult hookworm excretory/secretory (ES) proteins were prepared as described (23, 24).
Cloning of the AceMIF cDNA
A partial sequence of the AceMIF gene was initially identified in the A. ceylanicum EST data base available through the Nematode EST project (25). The 451-bp EST sequence (GenBank™ accession no. BM131124) corresponds to a 98-amino acid fragment of the mature AceMIF protein. To clone the cDNA corresponding to the AceMIF gene, A. ceylanicum RNA was extracted from adult worms in the presence of TRIzol reagent (Invitrogen) following manufacturer's protocol. First-strand cDNA was prepared with oligo(dT) primer and Superscript II reverse transcriptase as previously described (24). The full-length AceMIF cDNA was subsequently amplified from adult A. ceylanicum cDNAbyRT-PCR using a gene-specific 3′-primer and a 5′-primer corresponding to the nematode spliced leader sequence (SL-1) (26). Alignment of the various MIF clones was conducted using MegAlign software from DNASTAR, Inc. and BLAST analysis was conducted using the NCBI BLAST utility.
Expression and Purification of Recombinant AceMIF
The cDNA corresponding to the mature AceMIF protein was direc-tionally cloned into the pET11b expression plasmid vector as described (27). The protein was expressed by transforming Escherichia coli BL21-Gold (DE3) competent cells, and individual colonies containing the rAceMIF cDNA were identified through screening by PCR. Expression of recombinant AceMIF protein was induced by adding isopropyl-1-thio-β-D-galacto-pyranoside to a culture of E. coli containing the expression plasmid, followed by shaking at 37 °C for 3 h. The rAceMIF protein was purified from soluble cell lysates using sequential anion exchange, cation exchange, and size exclusion chromatography following procedures described previously for mammalian MIF (27). The protein concentration was determined using the BCA kit (Pierce), and the mass of the purified rAceMIF was determined using electrospray mass spectrometry as described (26).
Generation of a Polyclonal a-AceMIFIgG and Immunodetection of AceMIF Protein
A single New Zealand White rabbit was immunized with 700 μg of rAceMIF in Freund's Complete adjuvant. The rabbit was subsequently boosted twice at 3-week intervals with 700 μg of protein in Freund's incomplete adjuvant. Serum was collected after the third vaccination, and the IgG purified using affinity chromatography (26, 28).
The presence of native AceMIF in soluble protein extracts of A. ceylanicum eggs/L1 larvae, L3 larvae, adult worms, as well as pooled adult worm ES proteins was assessed by immunoblot (23). Nitrocellulose membranes containing various native hookworm protein preparations were probed with the rabbit α-rAceMIF IgG, followed by a secondary horseradish peroxidase-conjugated goat α-rabbit IgG and chemiluminescent detection. A similar method was used to perform immunoblots of rAceMIF and recombinant human MIF (rhMIF) using a monoclonal antibody that recognizes the human protein (29).
X-ray Crystal Structure of rAceMIF
For crystallographic studies, rAceMIF was expressed in BL21(DE3)E. coli cells transformed with the pET-11b expression vector containing the AceMIF cDNA and purified using anion and cation exchange chromatography as described above. Se-methionine containing rAceMIF was prepared for multiple-wavelength anomalous diffraction (MAD) phasing according to a previously reported method (30) with minor modifications. The rAceMIF was initially crystallized in 2.0 m ammonium sulfate, 0.1 m sodium citrate pH 5.6, and 0.2 m potassium sodium tartrate (Crystal Screen2 14 from Hampton Research) at 20 °C using the hanging drop method. Se-methionine rAceMIF was crystallized in 4% of isopropyl alcohol and 2.2–2.3 m ammonium sulfate. X-ray diffraction data were collected at the X29 beam line in Brookhaven National Laboratory. All crystals were soaked in 20% glycerol in the crystallization solution for cryoprotection under liquid nitrogen during data collection. Native crystal diffraction data were collected at 1.1 Å by 1° oscillation for 180°. Diffraction images were indexed, integrated, and scaled using HKL2000 (31). The space group was confirmed by examining images of the processed data in reciprocal space using HKLVIEW (31). Crystallographic statistics for data collection, processing, and refinement are summarized in Table 1. Selenium sites and phases were determined using ShelxD and E (32) via HKL2MAP (33). Model building was initiated by ArpWarp (34) and finalized by manual fitting using Xfit (35). The model was refined using CNS (36) and Refmac (37). Solvent molecules were added at the last stage of model refinement.
Table 1. Crystallographic statistics for structure of AceMIF.
| Native | SeMet | |||
|---|---|---|---|---|
| Data collection | ||||
| Space group | P6322 | P6322 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 115.2, 115.2, 199.2 | 115.4, 115.4, 199.3 | ||
| a, b, g (°) | 90, 90, 120 | 90, 90, 120 | ||
|
|
||||
| Peak | Inflection | Remote | ||
|
|
||||
| Wavelength | 1.1 | 0.9791 | 0.9793 | 0.9565 |
| Resolution (Å) | 1.6 | 2.5 | 2.5 | 2.5 |
| Rsym or Rmerge | 0.116 | 13.6 | 12.8 | 14.4 |
| I/ σI | 3.8 | 7.9 | 6.1 | 5.6 |
| Completeness (%) | 100 | 99.98 | 99.98 | 99.96 |
| Redundancy | 14.5 | 19.9 | 34.0 | 35.3 |
|
| ||||
| Refinement | ||||
| Resolution (Å) | 1.6 | |||
| No. reflections | 88,238 | |||
| Rwork/Rfree | 0.199/0.209 | |||
|
| ||||
| No. atoms | ||||
| Protein | 3648 | |||
| Ligand/ion | 20 | |||
| Water | 267 | |||
|
| ||||
| B-factors | ||||
| Protein | 19.8 | |||
| Ligand/ion | 9.6 | |||
| Water | 28.2 | |||
|
| ||||
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.014 | |||
| Bond angles (°) | 1.495 | |||
Structure Analysis and Visualization
Dihedral angles and bond lengths of the refined rAceMIF structure were evaluated using ProCheck (38). Crystal packing and the global structure of the protein were analyzed using SPOCK. Hydrogen bonds and hydrophobic interactions were analyzed using LigPlot (39) with the integrated hydrogen bond analysis program HBPlus (40). The structural figures in this manuscript were generated using PyMol (41) and SPOCK.
In Vitro Characterization of rAceMIFActivity
Tautomerase activity was assessed using the substrate p-hydroxyphenylpyruvate (HPP) prepared as described (42). The substrate and buffer were mixed in 96-well plates, followed by addition of purified protein (rAceMIF or rhMIF). Tautomerase activity was deter- mined by measuring the increase in absorbance at 306 nm (A306) over 30–90 s. The inhibitory effect of ISO-1 (Calbiochem) (43, 44) was determined by preincubating rAceMIF or rhMIF with inhibitor prior to addition of HPP. The raw data were processed using Prism4 from GraphPad to obtain the kinetic parameters. The molar extinction coefficient of the enol form of HPP (∊ = 11,400 m−1) was determined experimentally (42).
The chemoattractant activities of rAceMIF and rhMIF were measured using human peripheral blood monocytes (PBMCs) isolated from whole blood by centrifugation on Histopaque-1077 (Sigma) (45, 46). The cells were washed in RPMI 1640, diluted to 1 × 106 cells/ml and analyzed immediately. These assays were carried out in 24-well tissue culture plates utilizing 8.0-μm cell culture inserts (Falcon). Washed cells were placed in the cell culture insert with or without recombinant MIF, followed by incubation for 3 h at 37 °C, 5% CO2. Experiments were also carried out by incubating PMBCs with recombinant MIF proteins (AceMIF or hMIF) in the presence of ISO-1. Cells that migrated through the membrane were fixed in methanol and stained with Geimsa, followed by counting under light microscopy. Results are expressed as the mean number of cells counted per high power field for each of two replicates ± S.E.
Analysis of Binding of rAceMIF to CD74
Binding to the MIF receptor (CD74) was characterized by coating wells of a 96-well plate with recombinant, soluble CD74 ectodomain (sCD7473–232) as described (47). Biotinylated human MIF (2 ng/μl) (Roche Applied Sciences) was added together in triplicate wells with increasing concentrations of purified (non-biotinylated) recombinant human MIF (rhMIF) (27), heat-denatured rhMIF, rAceMIF, heat-denatured rAceMIF, or a neutralizing, α-CD74 monoclonal antibody (clone LN2) (47). The bound biotinylated rhMIF was detected by adding streptavid in-conjugated alkaline phosphatase (R&D), followed by detection of the alkaline phosphatase with p-nitrophenyl phosphate (Sigma). Absorbance at 405 nm was plotted as percent A405 relative to wells containing biotinylated human MIF alone. Each plot represents at least three independently performed assays, and each data point depicts a S.E. ≤10%.
The real time binding interaction of rAceMIF with CD74 was measured by surface plasmon resonance using a BIAcore 2000 optical biosensor (BIAcore AB, Sweden) (47). The CM5 sensor chips and the BIA Evaluation software were obtained MIF receptor (sCD7473–232) was immobilized according to a standardized method using the Biacore Amine Coupling Kit. The immobilized CM5 chips were equilibrated in HEPES (pH 8.0) at 20 μl/min, and the ligand (rAceMIF) was introduced at five serial dilutions in BIAcore buffer (1 mM dithiothreitol, 2.5 mM MgCl2, 20 mM HEPES, 1 mM EDTA, 150 mM NaCl, 0.005% P20) at a flow rate of 20 μl/min. Binding was measured at 25 °C for 5 min, followed by 15 min of dissociation. After regeneration with 1 m NaCl/50 mM NaOH, the process was repeated 3 times for each dilution sample. Sensorgram response data were analyzed in the BIA evaluation kinetics package and the equilibrium binding constants calculated.
Results
Cloning and Expression of the Ace-MIF cDNA
A partial nucleotide sequence with homology to human and nematode MIF genes was identified from the adult A. ceylanicum EST data base (GenBank accession no. BM131124) (25). A full-length AceMIF cDNA was subsequently amplified from adult A. ceylanicum cDNA by RT-PCR using a 5′-primer corresponding to the nematode spliced leader (SL-1) (28, 48) and a 3′ -primer that was derived from the original EST sequence. The translated amino acid sequence predicted by the AceMIF cDNA corresponds to a mature protein that is 119 amino acids in length, with a predicted mass of 12,993 Da and pI of 7.92. Similar to other MIF orthologues (49), the AceMIF cDNA contained no secretory signal sequence following the initiating methionine codon.
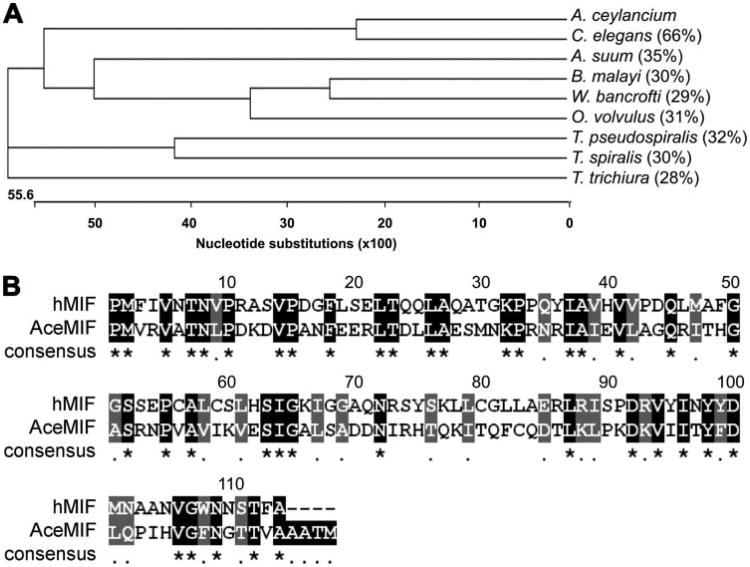
Analysis of the translated amino acid sequence of AceMIF reveals that the hookworm protein shares between 28–35% sequence identity and 53–63% similarity with MIF orthologues from other parasitic nematodes (Fig. 1A). Alignment of the deduced amino acid sequences from nine parasitic nematode MIF proteins indicates that there are 40 highly conserved amino acids, including 17 that are invariant among all nine sequences. By comparison, AceMIF is 33% identical and 53% similar to the human MIF amino acid sequence (GenBank™ accession no. AAP36881) (Fig. 1B).
Figure 1.
A, cladogram of MIF nucleotide sequences from nematodes constructed using the neighbor joining method. The length of each pair of branches represents the MIF sequence divergence, while the units at the bottom of the tree indicate the number of nucleotide substitution events. Numbers in parentheses signify the percent amino acid sequence identity of each compared with AceMIF as determined using the BLASTP algorithm. GenBank™ accession numbers of sequences above are as follows: Brugia malayi: CAC70155; Onchocerca volvulus: AAK66563; Ascaris suum: BAD24819; Trichinella spiralis: AAL12629; Trichinella pseudospiralis: AAL12630; Wuchereria bancrofti: O44786; Trichuris trichiura: CAB46355; Caenorhabditis elegans: NP_506003. A. ceylancium, Ancylostoma ceylancium. B, amino acid sequence alignment of hookworm (AceMIF) and human MIF (hMIF) proteins. Identical residues are shaded in black and indicated in the consensus line with an asterisk (*); conserved residues are shaded in gray and indicated in the consensus line with a period (.).
The cDNA corresponding to the mature AceMIF coding sequence was expressed in E. coli and purified as described previously for mammalian MIF (27). Analysis of the purified rAceMIF protein using electrospray mass spectrometry (48) revealed a mass of 12,996 Da (data not shown), which is within 0.02% of the predicted mass of the translated amino acid sequence (12,993 Da).
Stage-specific Expression of rAceMIF and Comparison of Immunoreactivity with Human MIF
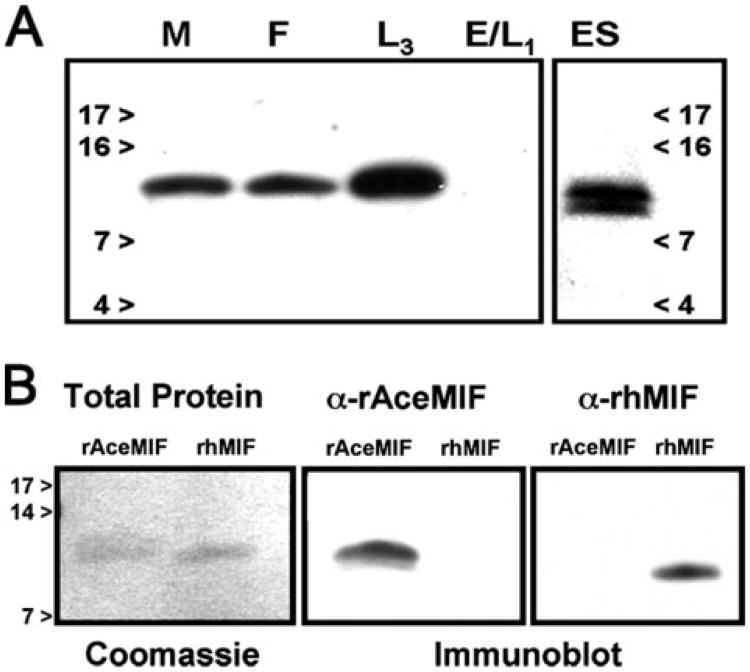
Probing of immunoblots with the α-rAceMIF IgG revealed recognition of a single protein band at a mass of ∼13 kDa present in soluble protein extracts of adult male, female, and L3A. ceylanicum worm (Fig. 2). In contrast, no signal was observed in the soluble protein preparation of eggs and newly hatched (L1) larvae, suggesting that the translation of AceMIF protein is developmentally regulated in vivo, with expression initiated between the L1 and L3 stage. Immunoblots of pooled adult worm ES proteins identified a protein doublet, with the higher MW species at the estimated size of the full-length AceMIF (12,993 Da), and a smaller species likely representing a breakdown product. The immunoblot data suggest that the native protein does not undergo significant post-translational modification, given the close approximate size of the protein band compared with the predicted size of AceMIF.
Figure 2.
A, life cycle stage specificity of AceMIF. Soluble protein extracts from A. ceylanicum life cycle stages (adult Males (M) or Females (F), L3 larvae (L3), Eggs/LI (E/L1), and pooled adult hookworm ES proteins (ES)) were probed by immunoblot using a polyclonal α-rAceMIF IgG. Numbers refer to mass (kDa) of molecular weight markers. B, immunoblots of rAceMIF and rhMIF probed with antibodies raised against the two proteins demonstrate the specificity of each IgG for its respective immunogen.
To determine whether there are any cross reacting epitopes shared by the recombinant human MIF (rhMIF) and rAceMIF proteins, immunoblot experiments were carried out using specific antisera (Fig. 2B). The α-rhMIF IgG only recognizes the human protein, while the α-rAceMIF IgG recognizes only the hookworm recombinant, confirming that the epitope recognized by the α-rhMIF is not present in the hookworm protein. Moreover, the fact that the polyclonal α-rAceMIF IgG does not recognize the human homologue, despite 33% amino acid sequence identity and 53% similarity, suggests that the two recombinants share few immunoreactive epitopes. In control blots, preimmune IgG did not react with either protein (not shown).
Three-dimensional Crystal Structure of rAceMIF and Analysis of the Active Site
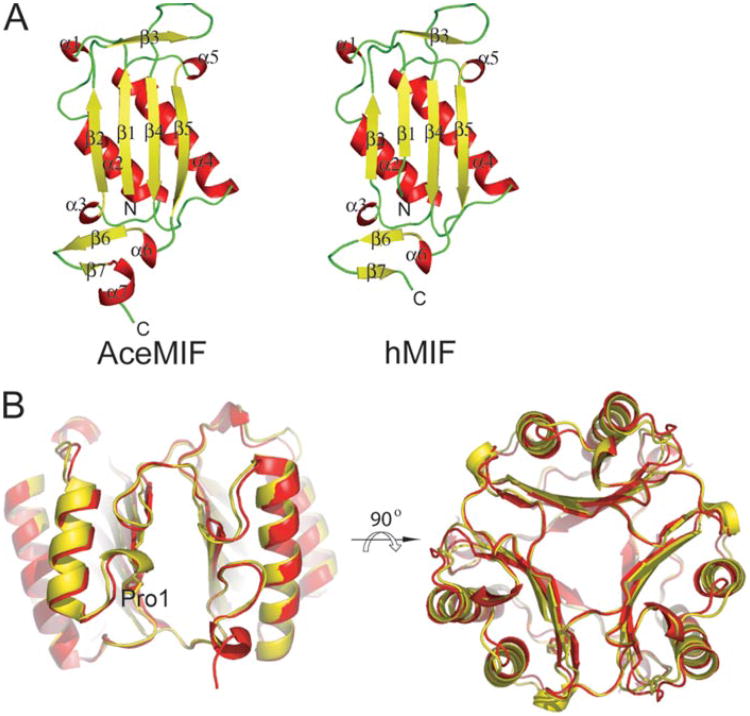
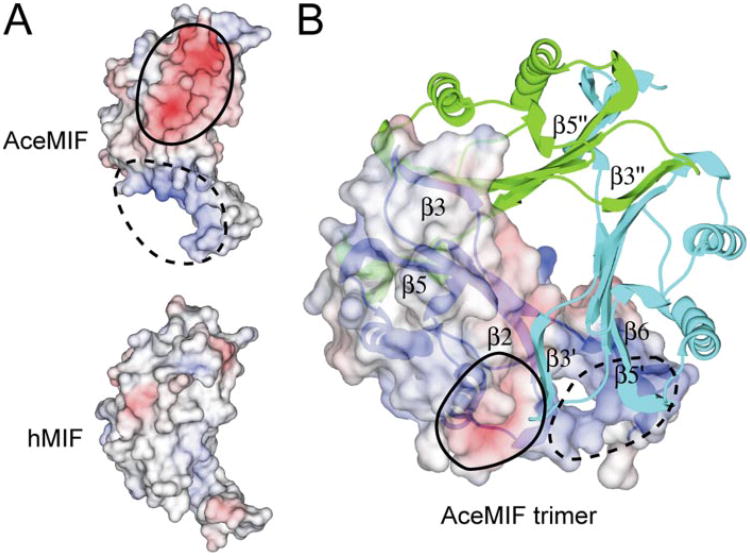
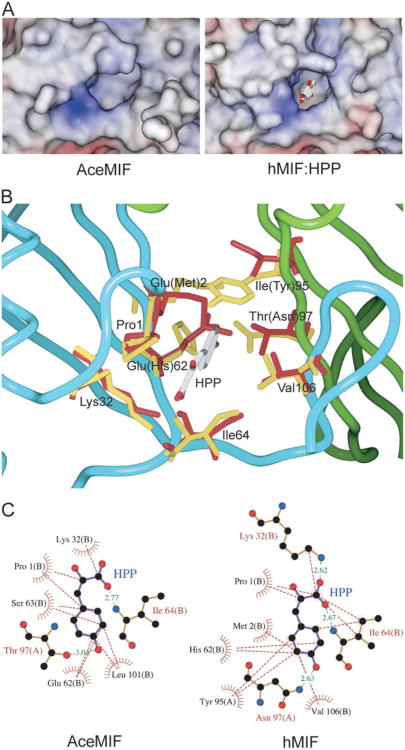
The structure of rAceMIF, as defined by the crystallography data (Table 1), consists of four protomers in the asymmetric unit (Fig. 3). Three of the protomers form a trimer, and the last monomer forms a trimer with two other protomers from adjacent asymmetric units. As expected, the topology is very similar to human MIF (Fig. 3). However, despite the backbone similarity, the protein surface and electrostatic potential of AceMIF are distinct (Fig. 4A). Also, the interface between subunits of the AceMIF trimer is highly charged compared with human MIF (Fig. 4B), making it unlikely that the two MIF molecules form heterotrimers in solution.
Figure 3. Topological comparison of AceMIF and hMIF.
A, backbone ribbon diagrams of AceMIF and hMIF. Secondary structural elements are represented in red (α-helix),yellow(β-strand), and green (random coil). B, trimers of the two MIFs are superimposed. Human MIF is represented in yellow and AceMIF in red. The approximate location of the active site is between two subunits and indicated by the designation Pro1.
Figure 4. Electrostatic surface potential of AceMIF and hMIF.
The negatively charged surface potential is colored in red and the positive potential in blue. These potentials are circled with solid or dashed lines in each of the panels. A, electrostatic potentials of AceMIF or hMIF are depicted on the surface of each protomer. B, charged surface potential for one monomer is shown in the context of the trimer, with the other subunits drawn as ribbon diagrams. β-Strandsof theunsurfaced subunits are labeled with single and double primes.
Analysis of the structure of AceMIF shows that the active site of the hookworm protein is wider than that of hMIF (Fig. 5A). The active site of AceMIF shares a group of identical amino acids with hMIF (Pro1, Lys32, Ile64, Val106), which include residues that interact with the substrate hydroxyphenylpyruvate (HPP) (Fig. 5B). However, the presence of three unique active site residues likely contributes to the decreased HPP tautomerase activity of AceMIF relative to hMIF (Table 2), as well as to differences in the inhibition of hMIF and AceMIF by ISO-1 (see below). A threonine in AceMIF replaces asparagine at residue 97 of hMIF, which directly interacts with the hydroxyl group of HPP in hMIF (Fig. 5C, right) (50). In the AceMIF modeled with substrate based on the hMIF:HPP complex, Thr97 shows weakened hydrogen bonding compared with Asn97 in hMIF (Fig. 5C, left). Another potentially significant amino acid change in AceMIF is Glu62, which is a His in hMIF. His62 of hMIF is one of the five residues involved in hydrophobic contacts with phenol ring of HPP (Fig. 5C, right). The aliphatic carbon atoms of Glu62 of AceMIF still maintain hydrophobic contact (Fig. 5C, left), but with a different part of the HPP phenol ring. The hydroxy-phenyl group of hMIF Tyr95 is perpendicular to the phenol ring of HPP to form an aromatic-aromatic interaction (50). Residue Ile95 in AceMIF, which corresponds to a tyrosine in hMIF (Fig. 5B), cannot contribute to an aromatic-aromatic interaction with the substrate, and thereby diminishes the interaction with HPP (Fig. 5C, left).
Figure 5. Comparative views of the active sites of hMIF and AceMIF.
A, view into the cavity of the active sites of AceMIF and hMIF. The figure shows the substrate HPP bound in the active site of hMIF (50), which is displayed in stick representation. B, superposition of the active sites of AceMIF and hMIF, with HPP bound to hMIF. Residues within hydrogen-bonding distance to HPP are colored in red (AceMIF) or yellow (hMIF). Residue codes correspond to AceMIF. Residues in parentheses correspond to hMIF. Two adjacent protomers (colored in cyan and green) in the trimer form the active site, and there are three active sites in each trimer. C, hydrogen bonds and hydrophobic interactions between HPP and interacting residues in a model of AceMIF:HPP and the crystal structure of hMIF:HPP (50), respectively, are shown. The residues involved in hydrophobic interactions with HPP are drawn as a spiked hemisphere. Distances for hydrogen bonds are in green dashed lines with their distance in Angstrom units.
Table 2. Steady state kinetic values for the HPP tautomerase activity of rhMIF and rAceMIF.
Although the Vmax of rAceMIF is 3.4-fold less than that of rhMIF, the substrate specificity index (kcat/Km) of AceMIF is 1.7-fold higher than that of rhMIF. This difference is largely due to the 6-fold decrease in the Km value for AceMIF relative to the human protein.
| rhMIF | rAceMIF | |
|---|---|---|
| Vmax (μM s−1) | 3.85 ± 0.10 | 1.13 ± 0.02 |
| Km (mM) | 1.20 ± 0.06 | 0.21 ± 0.02 |
| kcat(s−1) | 38.4 | 11.2 |
| kcat/ Km(mM−1s−1) | 32.0 | 53.3 |
| kcat/kcat,hMIF | 1.0 | 0.3 |
| (kcat/Km)/(kcat/Km,hMIF) | 1.0 | 1.7 |
rAceMIF Is Not Inhibited by ISO-1
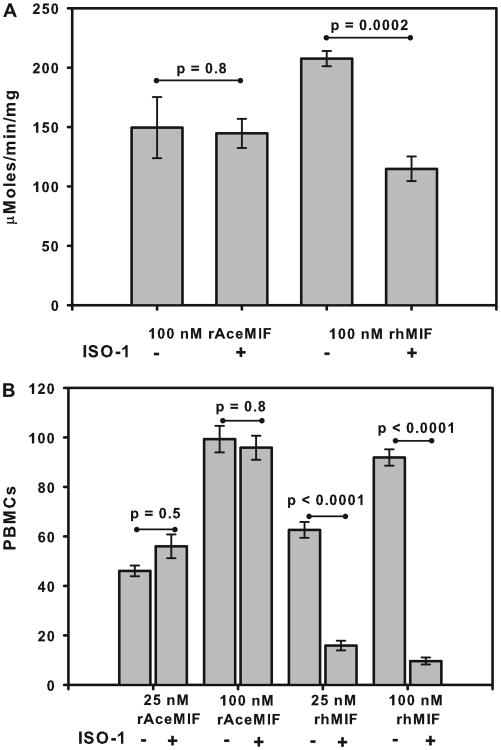
In light of differences within the active sites of the hookworm and human MIF orthologues revealed by the crystal structures, we investigated whether rAceMIF might also be sensitive in vitro to the inhibitor of rhMIF, ISO-1 (44, 51). As shown in Fig. 6A, preincubation with a 10-fold molar excess of ISO-1 reduced the tautomerase activity of rhMIF by 45%, from a baseline value of 208 ± 6 μmol/min/mg to 115 ± 10 μMoles/min/mg (p = 0.002). In contrast, a similar excess of ISO-1 had no effect on the tautomerase activity of rAceMIF (150 ± 26 μmol/min/mg versus 145 ± 12 μmol/min/mg; p = 0.8). Of note, rAceMIF demonstrated comparable tautomerase activity to values previously reported for the filarial nematode Brugia malayiMIF-1 (106.8 μmol/min/mg) andMIF-2 (517.2 μmol/min/ mg) (45, 52), and was significantly more active in vitro than the MIF orthologue from Trichinella spiralis (3.9 μmol/min/mg) (45).
Figure 6. Selective inhibition of rhMIF by ISO-1.
A, effect of a 10-fold excess of the inhibitor ISO-1 on the activity of rAceMIF and rhMIF using an in vitro tautomerase assay. Results are presented as the average ± S.E. of data from three replicate wells. B, effect of ISO-1 (10-fold excess) on the chemo attractant activity of rAceMIF and rhMIF using a chamber assay. Results are expressed as the mean number of cells counted per high power field for each of two replicates ± S.E.
We also compared the response of rAceMIF to the human MIF inhibitor ISO-1 using a cell migration assay. As shown in Fig. 6B, rAceMIF promotes the migration of human PBMCs across a membrane in a concentration-dependent manner, with similar activity to rhMIF. As with the tautomerase assay, addition of a 10-fold molar excess of ISO-1 failed to inhibit the chemoattractant activity of rAceMIF. In contrast, inhibition of rhMIF-mediated migration of human PBMCs was observed in the presence of ISO-1, with a nearly 4-fold reduction in chemoattractant activity noted at 25 nM rhMIF (p < 0.0001), and a 9-fold reduction at 100 nM (p < 0.0001).
rAceMIF Binds to the Human MIF Receptor CD74
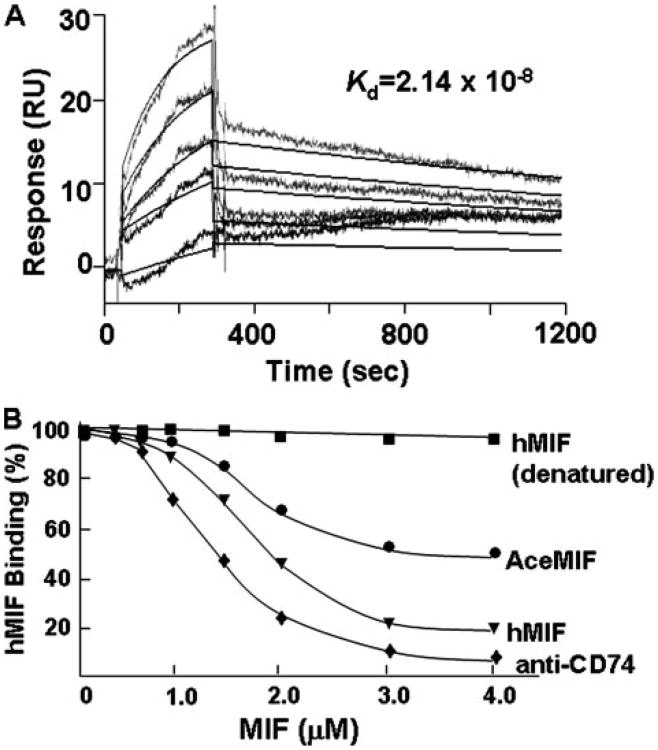
MIF activates cells by engaging its cell surface receptor CD74, and a high affinity interaction between hMIF and the CD74 ectodomain (CD7473–232, sCD74) has previously been demonstrated (47). We determined the equilibrium dissociation constant for the binding of rAceMIF to sCD74 by BIAcore surface plasmon resonance, a technique that measures real-time binding interactions by changes in the refractive index of a biospecific surface. Analysis of the binding interaction between AceMIF and sCD74 showed an equilibrium dissociation constant (Kd) of 2.14 × 10−8m (Fig. 7A). This contrasts with a previously determined Kd for the binding of human MIF with sCD74 of 9.0 × 10−9m(47). We also examined the potential interaction between AceMIF and the MIF receptor by studying the ability of rAceMIF to compete with hMIF for binding to immobilized sCD74 (47). As shown in Fig. 7B, rAceMIF inhibited the binding of hMIF to sCD74, but not to the level of inhibition observed if non-biotinylated hMIF was used as competitor.
Figure 7.
A, representative biosensorgrams showing the interaction between sCD74 (sCD7473-232) and increasing concentrations of rAceMIF sensor chips, as measured by surface plasmon resonance (see “Experimental Procedures” for details). Data from biosensorgrams were used to derive the equilibrium dissociation constant (Kd) shown for the interaction between rAceMIF to sCD74. B, comparative binding of rAceMIF and rhMIF to the human MIF receptor ectodomain (sCD74) in an in vitro, capture assay employing immobilized sCD74 and biotinylated-MIF as competitor. Heat denatured rAceMIF showed the same lack of binding as denatured rhMIF (data not shown).
Discussion
MIF is a pro-inflammatory mammalian cytokine produced by numerous cell types, including T cells, eosinophils, fibroblasts, monocytes, and macrophages (53, 54). Human MIF (hMIF) mediates many of its inflammatory properties by interacting with the CD74-CD44 receptor complex, modulating ERK1/2 MAPK-specific pathways to effect downstream cell signaling cascades associated with pro-inflammatory, Th1 biased responses (13). Experiments utilizing mif−/− knock-out mice, anti-MIF monoclonal antibodies, and the small molecule inhibitor ISO-1 to block the activity of MIF in vivo highlight the potential of MIF as a therapeutic target. Interestingly, the role of host MIF in mediating a protective immune response to microbial pathogens has also been characterized in model systems of bacterial and parasitic diseases (15,18,19, 43, 55-57).
Orthologues of MIF have been identified in numerous invertebrate species, including nematodes, protozoa, and ticks (17, 45, 46, 58–62). Among parasitic nematodes, MIF orthologues have been described in filarial worms, including Brugia, Wuchereria, and Onchocerca, as well as Trichinella, Ascaris, and Trichuris. It has been proposed that MIF homologues may play a role in immune evasion strategies of parasitic helminths by altering macrophage influx, immune cell activation and ultimately, host cytokine production (46). In filarial infections, the B. malayi MIF-1 (BmMIF-1) has been shown to upregulate the expression of eosinophil chemoattractant factor in macrophages, which augments eosinophil recruitment (17).
We report here the molecular cloning of a hookworm MIF cDNA (AceMIF) from the bloodfeeding hookworm A. ceylanicum, based on a partial sequence first identified in the Nema-tode.net hookworm EST data base (25). Although most nematode MIFs, including AceMIF, share limited primary amino acid sequence homology (Fig. 1A), their crystal structures exhibit a high degree of similarity (52, 60). The presence of related sequences in such disparate phyla suggests either a highly conserved function of MIF, or that individual species have modified the MIF protein scaffold to meet divergent physiologic needs.
A recombinant AceMIF (rAceMIF) was expressed in E. coli, and the hookworm life stage specificity was investigated using a polyclonal α-rAceMIF IgG. We first demonstrated by immunoblot that native AceMIF is present in infectious third stage (L3) larvae, adult worm extracts, and ES proteins, with none detected in eggs or newly hatched (L1) larvae (Fig. 2). These studies confirm that native AceMIF is present exclusively in those stages of the parasite that must confront host immune responses, either during tissue migration (L3) or intestinal attachment (adults). In contrast, eggs and L1 larvae are primarily environmental, i.e. soil-dwelling, stages of Ancylostoma, which do not interact directly with the host. Of note, a number of potential hookworm virulence factors have been identified in ES, including anticoagulants, platelet inhibitors, hemoglobin-degrading proteases, and inhibitors of host pancreatic enzymes (7). The fact that AceMIF is secreted by the adult hookworm at the site of intestinal attachment suggests a potential role in modulating the host immune response.
We also used immunoblotting to confirm that the recombinant human and hookworm MIF proteins, which share 33% amino acid sequence identity and 53% similarity (Fig. 1B), are distinct in terms of immunoreactive epitopes. Our observation that a polyclonal α-rAceMIF IgG recognizes both native and recombinant hookworm proteins, yet fails to react with the human orthologue, establishes an essential proof of concept for ongoing studies aimed at generating neutralizing antibodies that target AceMIF without interfering with the functions of the host-derived MIF. As a likely immunomodulatory virulence factor, AceMIF thus represents a viable target for the development of a vaccine that would impair the ability of hookworms to modulate the host immune response to infection.
The three-dimensional structure of rAceMIF was solved using x-ray crystallography (Fig. 3). Although the global topology of AceMIF is similar to that of hMIF, the active site of the hookworm molecule contains three potentially significant amino acid substitutions. These amino acid substitutions, along with a somewhat larger opening into the active site (Fig. 5), might confer greater flexibility within the active site of AceMIF relative to the human orthologue. These structural findings are also consistent with differences in the steady-state kinetics of rAceMIF tautomerase activity (Table 2).
As shown in Fig. 6, AceMIF is resistant to the inhibitory activity of the small molecule ISO-1, which has previously been shown to bind to the tautomerase active site and block the bioactivities of murine and human MIFs (43, 44, 51, 63). This is the first demonstration that ISO-1 can be used in a species-specific fashion to target host MIF activity. The observation that rAceMIF is resistant to ISO-1 establishes a biochemical basis for the development of specific inhibitors of pathogen-derived MIF orthologues. Ultimately, we anticipate that a small molecule inhibitor that selectively targets AceMIF in vivo would significantly alter the pathogenesis of hookworm disease by blocking an essential parasite virulence factor.
We also demonstrate that rAceMIF is capable of binding to the previously characterized cell surface mammalian MIF receptor, CD74 (8, 47) (Fig. 7). Using surface plasmon resonance, the equilibrium dissociation constant (Kd) for the Ace-MIF-CD74 interaction was measured at 2.1 × 10−8m, which is 4.5-fold lower than that previously reported for hMIF (9.0 × 10−9m)(47). Interestingly, the fact that an excess of rAceMIF was only partially effective at displacing hMIF from CD74 using the solid phase binding assay is consistent with these data, but additionally suggests that the two proteins may bind to the receptor by distinct mechanisms. Whether AceMIF modulates MIF receptor and target cell function by inducing activation, or by engaging the receptor in a non-productive fashion, will be of interest to examine in the context of the hookworm pathogenesis.
These data offer the first evidence that a pathogen-derived MIF binds to the host receptor, further suggesting that AceMIF may play a role in modulating the host inflammatory response, perhaps during skin penetration and tissue migration by infectious L3, as well as during intestinal attachment by adult parasites. In fact, despite repeated exposure to skin penetrating larvae, individuals in hookworm endemic areas fail to develop sterile immunity, and it has long been theorized that the parasites must effectively dampen the host immune response. Not surprisingly, it has been shown both in humans and animal models that hookworm infection is associated with varying degrees of immunosuppression, manifested by impaired lymphocyte responses to both hookworm and heterologous antigens (64). To date, however, no specific parasite factor has yet been proven in vivo to modulate host responses in the setting of hookworm infection. Because host MIF is constitutively expressed in the skin (65) and intestinal epithelium (57), we hypothesize that the hookworm orthologue AceMIF exerts its biological effect through a direct interaction with the MIF receptor CD74 at these two critical sites. In light of the data presented here, as well as that from other parasite systems in which MIF is thought to play a role, it is quite possible that AceMIF mediates hookworm pathogenesis through its immunomodulatory activity. Ultimately, however, defining an essential role for AceMIF in hookworm pathogenesis will require neutralization of its activity in vivo, either through chemotherapeutic or vaccination strategies.
In summary, we report here evidence that hookworm-secreted AceMIF is both a functional tautomerase and capable of binding to the human cell surface MIF receptor. We hypothesize that AceMIF functions in vivo as a parasite virulence factor by modulating the host immune response to skin penetrating larvae and adult worms attached to the intestine. Furthermore, because of its unique immunoreactive, structural, and functional properties, AceMIF represents a viable target for novel drug and/or vaccine based strategies aimed at limiting the ability of hookworms to infect and survive within the mammalian host, thereby reducing the global health burden of this major parasitic disease.
Acknowledgments
We thank Richard Bungiro and Blaise Dondji for thoughtful suggestions during the course of this work and Courtney McDonald for assistance with the receptor binding studies.
Footnotes
This work was supported by National Institutes of Health Grants AI065029 (to E. L.), AI42310 (to R. B.), AI51306 (to R. B.), AI007404 (to J. J. V.), and AI058980 (to M. C.).
The abbreviations used are: MIF, macrophage migration inhibitory factor; HPP, p-hydroxyphenylpyruvate; ES, excretory/secretory; ISO-1, (S,R)-3-(4-hydroxyphenyl)-4,5-dihydro-5-isoxazole acetic acid methyl ester; IL, interleukin; R.m.s., root mean-squared; EST, expressed sequence tag
References
- 1.de Silva NR, Brooker S, Hotez PJ, Montresor A, Engels D, Savioli L. Trends Parasitol. 2003;19:547–551. doi: 10.1016/j.pt.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Stephenson LS, Latham MC, Ottesen EA. Parasitology. 2000;121(suppl.):S23–S38. doi: 10.1017/s0031182000006491. [DOI] [PubMed] [Google Scholar]
- 3.Beaver PC. Am J Trop Med Hyg. 1988;39:369–372. doi: 10.4269/ajtmh.1988.39.369. [DOI] [PubMed] [Google Scholar]
- 4.Moyle M, Foster DL, McGrath DE, Brown SM, Laroche Y, De Meutter J, Stanssens P, Bogowitz CA, Fried VA, Ely JA, Soule HR, Vlasuk GP. J Biol Chem. 1994;269:10008–10015. [PubMed] [Google Scholar]
- 5.Culley FJ, Brown A, Conroy DM, Sabroe I, Pritchard DI, Williams TJ. J Immunol. 2000;165:6447–6453. doi: 10.4049/jimmunol.165.11.6447. [DOI] [PubMed] [Google Scholar]
- 6.Kasper G, Brown A, Eberl M, Vallar L, Kieffer N, Berry C, Girdwood K, Eggleton P, Quinnell R, Pritchard DI. Parasite Immunol. 2001;23:141–152. doi: 10.1046/j.1365-3024.2001.00366.x. [DOI] [PubMed] [Google Scholar]
- 7.Jones BF, Cappello M. Drug Disc Today. 2004;1:217–222. [Google Scholar]
- 8.Leng L, Bucala R. Cell Res. 2006;16:162–168. doi: 10.1038/sj.cr.7310022. [DOI] [PubMed] [Google Scholar]
- 9.Aeberli D, Leech M, Morand EF. Rheumatology (Oxford) 2006;45:937–943. doi: 10.1093/rheumatology/kel142. [DOI] [PubMed] [Google Scholar]
- 10.Sampey AV, Hall PH, Mitchell RA, Metz CN, Morand EF. Arthritis Rheum. 2001;44:1273–1280. doi: 10.1002/1529-0131(200106)44:6<1273::AID-ART219>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell RA, Metz CN, Peng T, Bucala R. J Biol Chem. 1999;274:18100–18106. doi: 10.1074/jbc.274.25.18100. [DOI] [PubMed] [Google Scholar]
- 12.Onodera S, Nishihira J, Iwabuchi K, Koyama Y, Yoshida K, Tanaka S, Minami A. J Biol Chem. 2002;277:7865–7874. doi: 10.1074/jbc.M106020200. [DOI] [PubMed] [Google Scholar]
- 13.Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lue H, Kapurniotu A, Fingerle-Rowson G, Roger T, Leng L, Thiele M, Calandra T, Bucala R, Bernhagen J. Cell Signal. 2006;18:688–703. doi: 10.1016/j.cellsig.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez-Sosa M, Rosas LE, David JR, Bojalil R, Satoskar AR, Terrazas LI. Infect Immun. 2003;71:1247–1254. doi: 10.1128/IAI.71.3.1247-1254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stavitsky AB, Metz C, Liu S, Xianli J, Bucala R. Parasite Immunol. 2003;25:369–374. doi: 10.1046/j.1365-3024.2003.00641.x. [DOI] [PubMed] [Google Scholar]
- 17.Falcone FH, Loke P, Zang X, MacDonald AS, Maizels RM, Allen JE. J Immunol. 2001;167:5348–5354. doi: 10.4049/jimmunol.167.9.5348. [DOI] [PubMed] [Google Scholar]
- 18.McDevitt MA, Xie J, Shanmugasundaram G, Griffith J, Liu A, McDonald C, Thuma P, Gordeuk VR, Metz CN, Mitchell R, Keefer J, David J, Leng L, Bucala R. J Exp Med. 2006;203:1185–1196. doi: 10.1084/jem.20052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oddo M, Calandra T, Bucala R, Meylan PR. Infect Immun. 2005;73:3783–3786. doi: 10.1128/IAI.73.6.3783-3786.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell RA, Bucala R. Semin Cancer Biol. 2000;10:359–366. doi: 10.1006/scbi.2000.0328. [DOI] [PubMed] [Google Scholar]
- 21.Bacher M, Meinhardt A, Lan HY, Mu W, Metz CN, Chesney JA, Calandra T, Gemsa D, Donnelly T, Atkins RC, Bucala R. Am J Pathol. 1997;150:235–246. [PMC free article] [PubMed] [Google Scholar]
- 22.Bungiro RD, Jr, Greene J, Kruglov E, Cappello M. J Infect Dis. 2001;183:1380–1387. doi: 10.1086/319867. [DOI] [PubMed] [Google Scholar]
- 23.Brown AC, Harrison LM, Kapulkin W, Jones BF, Sinha A, Savage A, Villalon N, Cappello M. Mol Biochem Parasitol. 2007;151:141. doi: 10.1016/j.molbiopara.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milstone AM, Harrison LM, Bungiro RD, Kuzmic P, Cappello M. J Biol Chem. 2000;275:29391–29399. doi: 10.1074/jbc.M002715200. [DOI] [PubMed] [Google Scholar]
- 25.Wylie T, Martin JC, Dante M, Mitreva MD, Clifton SW, Chinwalla A, Waterston RH, Wilson RK, McCarter JP. Nucleic Acids Res. 2004;32:D423–D426. doi: 10.1093/nar/gkh010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bungiro RD, Jr, Solis CV, Harrison LM, Cappello M. Infect Immun. 2004;72:2203–2213. doi: 10.1128/IAI.72.4.2203-2213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernhagen J, Mitchell RA, Calandra T, Voelter W, Cerami A, Bucala R. Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 28.Del Valle A, Jones BF, Harrison LM, Chadderdon RC, Cappello M. Mol Biochem Parasitol. 2003;129:167–177. doi: 10.1016/s0166-6851(03)00121-x. [DOI] [PubMed] [Google Scholar]
- 29.Donnelly SC, Haslett C, Reid PT, Grant IS, Wallace WA, Metz CN, Bruce LJ, Bucala R. Nat Med. 1997;3:320–323. doi: 10.1038/nm0397-320. [DOI] [PubMed] [Google Scholar]
- 30.Sun HW, Swope M, Cinquina C, Bedarkar S, Bernhagen J, Bucala R, Lolis E. Protein Eng. 1996;9:631–635. doi: 10.1093/protein/9.8.631. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Schneider TR, Sheldrick GM. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 33.Pape T, Schneider TR. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 34.Lamzin VS, Wilson KS. Acta Crystallogr D Biol Crystallogr. 1993;49:129–147. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 35.McRee DE. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 39.Wallace AC, Laskowski RA, Thornton JM. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 40.McDonald IK, Thornton JM. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. Drug Discov Today. 2005;10:213–217. doi: 10.1016/S1359-6446(04)03363-X. [DOI] [PubMed] [Google Scholar]
- 42.Lin EC, Pitt BM, Civen M, Knox WE. J Biol Chem. 1958;233:668–673. [PubMed] [Google Scholar]
- 43.Al-Abed Y, Dabideen D, Aljabari B, Valster A, Messmer D, Ochani M, Tanovic M, Ochani K, Bacher M, Nicoletti F, Metz C, Pavlov VA, Miller EJ, Tracey KJ. J Biol Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 44.Lubetsky JB, Dios A, Han J, Aljabari B, Ruzsicska B, Mitchell R, Lolis E, Al-Abed Y. J Biol Chem. 2002;277:24976–24982. doi: 10.1074/jbc.M203220200. [DOI] [PubMed] [Google Scholar]
- 45.Tan TH, Edgerton SA, Kumari R, McAlister MS, Roe SM, Nagl S, Pearl LH, Selkirk ME, Bianco AE, Totty NF, Engwerda C, Gray CA, Meyer DJ, Rowe SM. Biochem J. 2001;357:373–383. doi: 10.1042/0264-6021:3570373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pastrana DV, Raghavan N, FitzGerald P, Eisinger SW, Metz C, Bucala R, Schleimer RP, Bickel C, Scott AL. Infect Im-mun. 1998;66:5955–5963. doi: 10.1128/iai.66.12.5955-5963.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bungiro RD, Harrison LM, Cappello M. Mol Biochem Parasitol. 2002;119:147–151. doi: 10.1016/s0166-6851(01)00408-x. [DOI] [PubMed] [Google Scholar]
- 49.Flieger O, Engling A, Bucala R, Lue H, Nickel W, Bernhagen J. FEBS Lett. 2003;551:78–86. doi: 10.1016/s0014-5793(03)00900-1. [DOI] [PubMed] [Google Scholar]
- 50.Lubetsky JB, Swope M, Dealwis C, Blake P, Lolis E. Biochemistry. 1999;38:7346–7354. doi: 10.1021/bi990306m. [DOI] [PubMed] [Google Scholar]
- 51.Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. J Immunol. 2006;177:8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- 52.Zang X, Taylor P, Wang JM, Meyer DJ, Scott AL, Walkinshaw MD, Maizels RM. J Biol Chem. 2002;277:44261–44267. doi: 10.1074/jbc.M204655200. [DOI] [PubMed] [Google Scholar]
- 53.Calandra T, Roger T. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lue H, Kleemann R, Calandra T, Roger T, Bernhagen J. Microbes Infect. 2002;4:449–460. doi: 10.1016/s1286-4579(02)01560-5. [DOI] [PubMed] [Google Scholar]
- 55.Satoskar AR, Bozza M, Rodriguez Sosa M, Lin G, David JR. Infect Immun. 2001;69:906–911. doi: 10.1128/IAI.69.2.906-911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calandra T. Scand J Infect Dis. 2003;35:573–576. doi: 10.1080/00365540310016277. [DOI] [PubMed] [Google Scholar]
- 57.Maaser C, Eckmann L, Paesold G, Kim HS, Kagnoff MF. Gastroenterology. 2002;122:667–680. doi: 10.1053/gast.2002.31891. [DOI] [PubMed] [Google Scholar]
- 58.Miska KB, Fetterer RH, Lillehoj HS, Jenkins MC, Allen PC, Harper SB. Mol Biochem Parasitol. 2006;151:173–183. doi: 10.1016/j.molbiopara.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 59.Marson AL, Tarr DE, Scott AL. Gene (Amst) 2001;278:53–62. doi: 10.1016/s0378-1119(01)00706-5. [DOI] [PubMed] [Google Scholar]
- 60.Wu Z, Boonmars T, Nagano I, Nakada T, Takahashi Y. J Parasitol. 2003;89:507–515. doi: 10.1645/0022-3395(2003)089[0507:MEACOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 61.Augustijn KD, Kleemann R, Thompson J, Kooistra T, Crawford CE, Reece SE, Pain A, Siebum AH, Janse CJ, Waters AP. Infect Immun. 2007;75:1116–1128. doi: 10.1128/IAI.00902-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umemiya R, Hatta T, Liao M, Tanaka M, Zhou J, Inoue N, Fujisaki K. Exp Parasitol. 2007;115:135. doi: 10.1016/j.exppara.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Cheng KF, Al-Abed Y. Bioorg Med Chem Lett. 2006;16:3376–3379. doi: 10.1016/j.bmcl.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 64.Geiger SM, Massara CL, Bethony J, Soboslay PT, Correa-Oliveira R. Clin Exp Immunol. 2004;136:334–340. doi: 10.1111/j.1365-2249.2004.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shimizu T, Ohkawara A, Nishihira J, Sakamoto W. FEBS Lett. 1996;381:199. doi: 10.1016/0014-5793(96)00120-2. [DOI] [PubMed] [Google Scholar]