SUMMARY
To use microRNAs to down-regulate mRNA targets, cells must first process these ~22 nt RNAs from primary transcripts (pri-miRNAs). These transcripts form RNA hairpins important for processing, but additional determinants must distinguish pri-miRNAs from the many other hairpin-containing transcripts expressed in each cell. Illustrating the complexity of this recognition, we show that most Caenorhabditis elegans pri-miRNAs lack determinants required for processing in human cells. To find these determinants, we generated >1011 variants of four human pri-miRNAs, sequenced millions that retained function and compared them with the starting variants. Our results confirmed the importance of pairing in the stem and revealed three primary-sequence determinants, including an SRp20-binding motif (CNNC) found downstream of most pri-miRNA hairpins in bilaterian animals but not in nematodes. Adding this and other determinants to C. elegans pri-miRNAs imparted efficient processing in human cells, thereby confirming the importance of primary-sequence determinants for distinguishing pri-miRNAs from other hairpin-containing transcripts.
INTRODUCTION
MicroRNAs (miRNAs) are ~22 nt RNAs that pair to mRNAs to direct post-transcriptional repression (Bartel, 2004). MicroRNAs are processed from hairpin-containing primary transcripts (pri-miRNAs). In the canonical processing pathway of animals, pri-miRNAs are cleaved by the Microprocessor, a protein complex containing an RNase III enzyme Drosha and its cofactor DGCR8/Pasha (Lee et al., 2003; Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004). The liberated portion of the hairpin (the pre-miRNA) is then cleaved by the RNase III enzyme Dicer (Grishok et al., 2001; Hutvagner et al., 2001), leaving two ~22 nt strands that pair to each other with ~2 nt 3′ overhangs (Lee et al., 2003; Lim et al., 2003b). One strand of each duplex is loaded into an Argonaute protein to form the core of the silencing complex, and the other strand is discarded (Khvorova et al., 2003; Schwarz et al., 2003; Liu et al., 2004). Noncanonical pathways also contribute to the miRNA repertoire through the processing of mirtrons (Okamura et al., 2007; Ruby et al., 2007) or other pri-miRNAs that bypass Drosha cleavage (Babiarz et al., 2008), or one pre-miRNA that bypasses Dicer cleavage (Cheloufi et al., 2010; Cifuentes et al., 2010).
A long-standing mystery has been how pri-miRNAs are distinguished from the many other hairpin-containing transcripts for processing as Microprocessor substrates. Determinants of Dicer cleavage are better understood (Zhang et al., 2004; Macrae et al., 2006; Park et al., 2011), as illustrated by both the design (Brummelkamp et al., 2002; Paddison et al., 2002) and prediction (Chung et al., 2011) of Dicer substrates that bypass Drosha processing. For Microprocessor recognition, sequences within 40 nt upstream and 40 nt downstream of the pre-miRNA hairpin are required for ectopic miRNA expression (Chen et al., 2004), which is consistent with both the observation that these flanking sequences tend to pair to each other to extend the stem another turn of the helix beyond the cleavage site (Lim et al., 2003b) and a requirement for this extension and a lack of pairing immediately following it for processing (Han et al., 2006). However, many cellular transcripts have paired regions flanked by single-stranded RNA (ssRNA), and most of these are not Microprocessor substrates. Indeed, attempts to predict canonical miRNA hairpins from genomic sequence yield many thousands of false-positive predictions, which must be eliminated using additional criteria, such as analysis of conservation or experimental evaluation (Lim et al., 2003a; Lim et al., 2003b; Bentwich et al., 2005; Berezikov et al., 2006; Chiang et al., 2010), illustrating a large gap in our understanding of how the Microprocessor distinguishes between authentic substrates and other transcribed hairpins.
Here, we report that transcripts that enter the miRNA pathway in C. elegans failed to do so in human cells. Thus, the definition of a pri-miRNA in one species differs from that in another. To find features that define human pri-miRNAs, we generated >1011 variants of four pri-miRNAs and sequenced millions that were cleaved by the human Microprocessor. Comparison of cleaved and initial variants revealed important sequence and structural features. These features were evolutionarily conserved in non-nematode lineages and sufficient to increase the processing efficiency of C. elegans hairpins in human cells.
RESULTS
Unknown Features Specify Human Pri-miRNAs
To examine whether miRNA processing features are shared across animals, we ectopically expressed a panel of C. elegans, D. melanogaster and human pri-miRNAs in human cells and compared the yields of mature miRNA. Despite variability in the degree of overexpression, presumably reflecting differences in efficiency at various steps of the pathway (Fellmann et al., 2011; Feng et al., 2011), most human miRNAs were efficiently expressed (Figure 1A), as expected (Chiang et al., 2010). Four of nine Drosophila miRNAs also fell within the range observed for human miRNAs. However, the tested C. elegans miRNAs were less efficiently expressed (Figure 1A, p = 1.4 × 10−5, Wilcoxon rank-sum test). Similar results were observed in Drosophila S2 cells (p = 0.024). Thus, most nematode pri-miRNAs lack determinants required for efficient processing in human or insect cells.
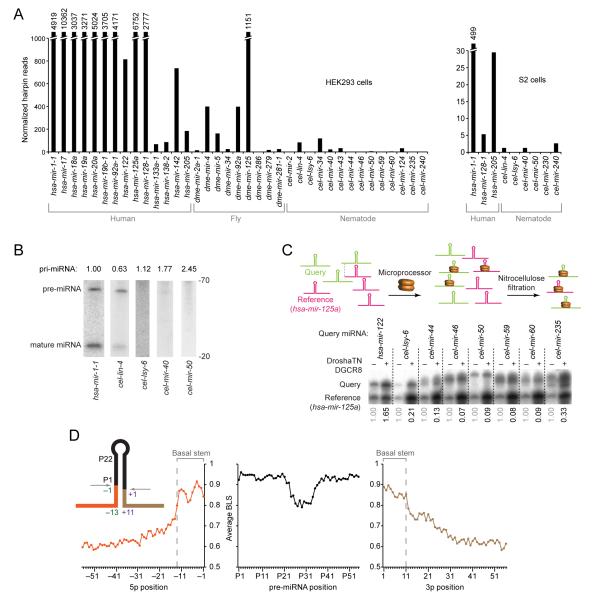
Figure 1. Existence of Unknown Features Specifying Human Pri-miRNAs.
(A) Processing of human, fly, and nematode pri-miRNAs in human cells and Drosophila cells. Cells were transfected with plasmids expressing the indicated pri-miRNA hairpins with ~100 flanking genomic nucleotides on each side of each hairpin (Figure S1A), and total RNA was pooled for small-RNA sequencing. Plotted are small-RNA reads derived from the indicated pri-miRNAs.
(B) Accumulation of pri-miRNA, pre-miRNA and miRNA after expressing the indicated pri-miRNAs in HEK293T cells. Pre-miRNA and mature species were measured by RNA blot of total RNA from cells transfected with plasmids expressing the indicated pri-miRNA (full gel images, including in vitro transcribed cognate positive controls, in Figure S1B). Relative pri-miRNA levels (indicated above the lanes) are from ribonuclease protection assays, normalized to the signals for neomycin phosphotransferase mRNA also expressed from each expression plasmid.
(C) Relative binding of C. elegans and human pri-miRNAs to the Microprocessor. In the competitive binding assay (top, schematic), radiolabeled query pri-miRNA was mixed with the radiolabeled shorter reference pri-miRNA (human mir-125a) and incubated in excess over catalytically impaired Drosha (Drosha-TN) and DGCR8. Bound RNA was filtered on nitrocellulose and eluted for analysis on a denaturing gel. Phosphorimaging (bottom) indicated the relative amounts of input (−) and bound (+) RNAs. Numbers below each lane indicate the ratio of bound query to bound reference pri-miRNAs, normalized to their input ratio.
(D) Nucleotide conservation of human pri-miRNAs conserved to mouse, reported as the average branch-length score (BLS) at each position. Positions are numbered based on the inferred Drosha cleavage site (inset); negative indices are upstream of the 5p Drosha cleavage site, indices with “P” count from the 5′ end of the pre-miRNA, and positive indices are downstream of the 3p Drosha cleavage site.
To isolate the processing defect, we probed for processing intermediates. Consistent with the sequencing results, cel-lin-4 was processed, with detectable pre-miRNA and mature miRNA (Figure 1B). For other C. elegans miRNAs, neither pre-miRNA nor mature miRNA were detected, despite the presence of primary transcripts (Figure 1B and S1B), suggesting that these C. elegans pri-miRNAs were not productively recognized as Microprocessor substrates. To assay directly for Microprocessor binding, we examined binding to catalytically deficient Drosha and DGCR8. Whereas human pri-mir-122 bound the Microprocessor somewhat better than did the reference pri-miRNA (human pri-mir-125a), all seven tested C. elegans pri-miRNAs bound worse (Figure 1C). Thus, most C. elegans pri-miRNAs are missing some of the determinants needed for efficient recognition and processing by the human Microprocessor.
Known features of C. elegans and human pri-miRNAs appear largely similar, as illustrated by the accuracy of an algorithm trained on C. elegans pri-miRNAs in predicting most miRNA genes conserved in mammals and fish (Lim et al., 2003a). Nonetheless, the poor specificity of this algorithm when predicting non-conserved miRNAs suggests that unknown features help define authentic pri-miRNAs. To look for clues regarding these unknown features, we analyzed the conservation of sequence immediately flanking human pre-miRNAs. Residues extending 13 nt upstream of the 5p Drosha cleavage site (i.e., the site corresponding to the 5′ end of the pre-miRNA) and 11 nt downstream of the 3p Drosha cleavage site were conserved above background, consistent with the importance of the ~11 bp basal stem for pri-miRNA processing (Figure 1D). However, the signal beyond the basal stem tailed off rapidly (particularly in the upstream flanking region), suggesting that any determinants in the flanking regions might be either at variable distances from the hairpin or present in only subsets of miRNAs, making them difficult to identify using alignments.
Functional Substrates from Large Libraries of Pri-miRNA Variants
To identify features important for Microprocessor recognition and cleavage, we generated >1011 pri-miRNA variants, sequenced millions that retained function and compared these sequences to those of the initial variants (Figure 2A). This approach resembled classical in vitro selection approaches (Wilson and Szostak, 1999), except we did not perform multiple rounds of selection. Because the starting and the selected pools underwent the same number of transcription, reverse-transcription and amplification steps, any differences between the two pools were subject to neither the compounding effects of multiple rounds nor the confounding effects of amplification biases. Moreover, as with previous analyses of selection results using high-throughput sequencing (Zykovich et al., 2009; Pitt and Ferre-D’Amare, 2010; Slattery et al., 2011), sequencing depth reduced the influence of stochastic sampling. Thus, compared to the results of classical approaches, enrichment or depletion of a residue was a more direct reflection of its contribution to biochemical specificity.
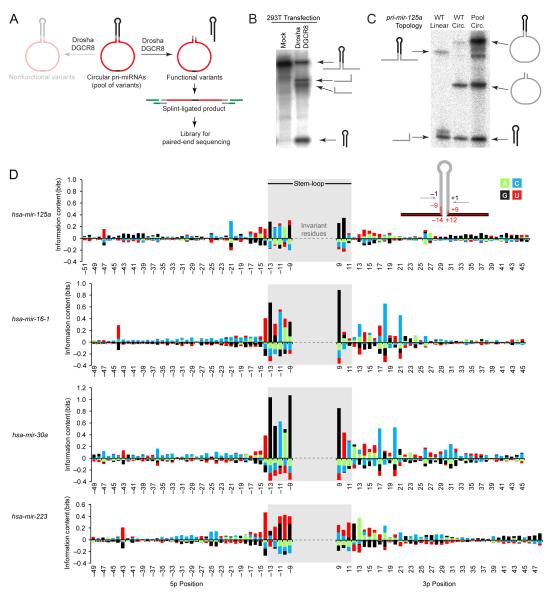
Figure 2.
Selection for functional pri-miRNA variants.
(A) Schematic of the selection. Pri-miRNAs with variable residues (red) flanking the Drosha cleavage site were circularized by ligation and incubated in Microprocessor lysate. Cleaved variants were gel-purified, ligated to adaptors, reverse transcribed, and amplified for high-throughput sequencing.
(B) Cleavage of let-7a in HEK293T whole-cell lysate (mock) and Microprocessor lysate (whole-cell lysate from HEK293T cells transfected with plasmids expressing Drosha and DGCR8). Incubations were 1.5 h. Body-labeled reactants and products were resolved on a denaturing polyacrylamide gel and visualized by phosphorimaging.
(C) Cleavage of linear and circular mir-125a (WT linear and WT circ., respectively) and a pool of circular mir-125a variants (pool). RNAs were incubated for 5 minutes in Microprocessor lysate and analyzed as in (B). The linear RNA was 5′ end-labeled; other RNAs were body-labeled.
(D) Enrichment and depletion at variable residues in functional pri-miRNA variants. At each varied position (inset, red inner line), information content was calculated for each residue (green, cyan, black, and red for A, C, G, and U, respectively).
Four pools of variants were constructed, each based on a different human pri-miRNA (mir-125a, mir-16-1, mir-30a, and mir-223). Residues >8 nt upstream of the 5p Drosha cleavage site and >8 nt downstream of the 3p cleavage site were varied, whereas the remaining hairpin residues were not. At each variable position, 79% of the molecules had the wild-type residue, and the remainder had one of the other three alternatives. As done for self-cleaving ribozymes (Pan and Uhlenbeck, 1992), each variant was circularized so that all of its variable nucleotides resided in a single cleavage product (Figure 2A), thereby enabling a full analysis of sequence interdependencies.
In vitro cleavage reactions were in Microprocessor lysate, i.e., whole-cell lysate from HEK293T cells overexpressing Drosha and DGCR8 to enhance cleavage activity (Figure 2B). At a time in which the lysate cleaved linear and circularized pri-mir-125a to near completion, many pri-mir-125a variants remained uncleaved (Figure 2C), which indicated that some substitutions in the basal stem and flanking regions attenuated Microprocessor cleavage in vitro.
Cleaved variants were purified and sequenced (Figure 2A). At each variant position, the odds of each nucleotide in the cleaved pool were compared to the odds of that nucleotide in the starting pool. These odds ratios were used to calculate the information content of each nucleotide possibility at each variant position—the greater the information content, the more favorable the influence on activity, with positive values indicating beneficial influences and negative values disruptive ones. An advantage of plotting information content is that it reports the relative influence of each nucleotide possibility irrespective of whether it was the wild-type possibility. Because molecular manipulations and computational filtering both selected for cleavage at the wild-type site, nucleotide changes that altered the cleavage site were not distinguished from those that abolished cleavage.
Some positions had substantial enrichment of one or more nucleotide possibilities, with corresponding depletion of others (Figure 2D). When tested in vitro, the results of changing specific residues closely matched those predicted from analysis of sequenced variants (Figure S2A and B). Moreover, the in vitro results predicted the direction and sometimes the magnitude of the effects observed in HEK293T cells (Figure S2C).
Importance of an 11 bp basal stem flanked by at least nine unstructured nucleotides
For all four miRNAs, some of the varied residues with the greatest influence fell within the basal stem (Figure 2D). Covariation matrices listing the odds ratio of each pair of nucleotide identities showed preference for Watson–Crick geometry at each basal pair, with the G:U wobble the most frequently preferred non-Watson–Crick alternative (Figure 3A, S3A). For example, the most favored alternatives to the wild-type C:G pair at positions −11 and +9 of mir-125a are the G:C and U:A pairs, and to a lesser extent the A:U, G:U and U:G pairs (Figure 3A). In fact, Watson–Crick pairing was strongly preferred even if it did not occur in the wild-type sequence. For example, the wild-type A:C pair at positions −12 and +10 of mir-30a was disfavored compared to the four Watson–Crick possibilities (Figure 3A), and the bulged A at position +10 of mir-223 was preferentially incorporated into an alternative continuous helix (Figure S3A–B). Extending these methods to systematically evaluate all pairing possibilities involving all varied positions uncovered no evidence for Watson–Crick pairing outside the basal stem (Figure S3C).
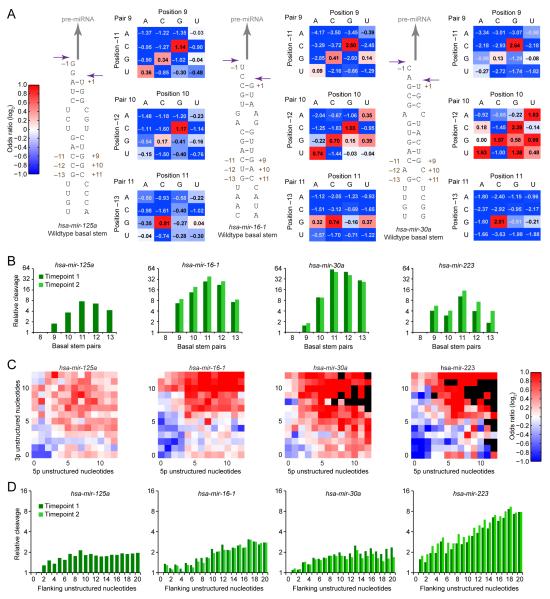
Figure 3.
Basal stem structure in functional pri-miRNA variants.
(A) Predicted basal secondary structures and covariation matrices for mir-125a, mir-16-1, and mir-30a. For each pair of positions, joint nucleotide distributions were tabulated from sequences of the initial and selected pools, and the log odds ratio calculated. Favored and disfavored pairs are colored red and blue, respectively, with color intensity (key) and values indicating magnitudes.
(B) Relative cleavage of variants with different stem lengths. The number of contiguous Watson–Crick pairs was counted, and the relative cleavage calculated, normalized to the 8 bp stem. For selections with two time points, results are shown for both (key).
(C) Enrichment for unstructured nucleotides flanking the basal stem. Predicted folds of variant sequences were generated, and the subset of sequences with wild-type basal stem pairing were classified based on the distance to the nearest consecutive structured nucleotides upstream of position –13 and the nearest consecutive structured nucleotides downstream of position +11. Enrichment (red) and depletion (blue) of unstructured lengths among the selected variants are colored (key), with black indicating that sequencing data were insufficient to calculate enrichment.
(D) Relative cleavage of variants with differing numbers of total unstructured nucleotides flanking the basal stem. Upstream and downstream unstructured lengths predicted in (C) were summed, and the relative cleavage calculated, normalized to zero unstructured nucleotides. For selections with two time points, results are shown for both (key).
Layered on the overall preference for Watson–Crick pairing were primary-sequence preferences specific to each basal pair. For example, at positions −11 and +9 the C:G pair was favored over the other Watson–Crick alternatives. The primary-sequence preference was most acute at the most basal pair, where wobbles or mismatches involving G at –13 were favored over alternative Watson–Crick pairs (Figure 3A). We conclude that primary-sequence features supplement and sometimes supersede structural features important for basal-stem recognition.
The Microprocessor recognizes the junction between the miRNA hairpin and flanking ssRNA to position the active site approximately one helical turn (11 bp of A-form RNA) from the base of the duplex (Han et al., 2006; Yeom et al., 2006). To examine the preferred length of the basal stem, we calculated the relative cleavage efficiencies of different stem-length variants, normalizing to that of an 8 bp stem. Invariant mismatches within symmetric internal loops (e.g., the A:C mismatch at positions −6 and +4 of mir-30a) were assumed to be non-canonical pairs that stacked within the stem to contribute to its length, whereas mismatches at varied positions were assumed to disrupt further pairing and thereby terminate the inferred basal stem. For all four pri-miRNAs, an 11 bp basal stem was optimal (Figure 3B), consistent with the single-turn model. Indeed, an 11 bp basal stem was preferred for mir-223 even though the wild-type sequence was predicted to form a 12 bp stem (Figures 3A and S3A). For most pri-miRNAs, however, the efficiency of the 12-pair stem approached that of the 11-pair stem (Figure 3B). This tolerance of a twelfth pair hinted that other features, such as the G at position –13, help specify the precise site of cleavage.
The single-turn model also posits that the nucleotides immediately flanking the basal stem are unstructured (Han et al., 2006; Yeom et al., 2006). To test this, we used RNAfold (Hofacker and Stadler, 2006) to predict the minimum free-energy structure of each sequenced pri-miRNA variant. For those with predicted wild-type stem pairing, we recorded the number of nucleotides between the base of the stem and the most proximal two consecutive structured residues. Although an imperfect estimate of the size of the unstructured segments flanking the base of the helix, this metric correlated well with cleavage (Figure 3C). Predicted pairing was tolerated in one flank, provided that the other flank contained at least 5–7 unpaired bases, consistent with reports of some cleavage when only one flanking segment is present (Zeng and Cullen, 2005; Han et al., 2006). When summing the flanking unpaired bases from both sides, the optimum plateaued at ~9–18 nt (Figure 3D).
A basal UG motif enhances processing
Among the nucleotides upstream of the stem-loop, the most striking enrichment was for a U at position –14 (Figure 2D). This U immediately preceded the position that, as mentioned above, displayed a strong primary-sequence preference for a G. The U and G at positions –14 and –13 contributed independently; variants with either a U or a G were enriched over variants with neither, and variants with both were even more enriched (Figure 4A). For mir-223, the UG at positions –14 and –13 was preferred (Figure 2D), even though wild-type mir-223 has a UG at positions –15 and –14, respectively. This basal UG motif was also enriched among variants of mir-125a selected for Microprocessor binding rather than cleavage (Figure S4B).
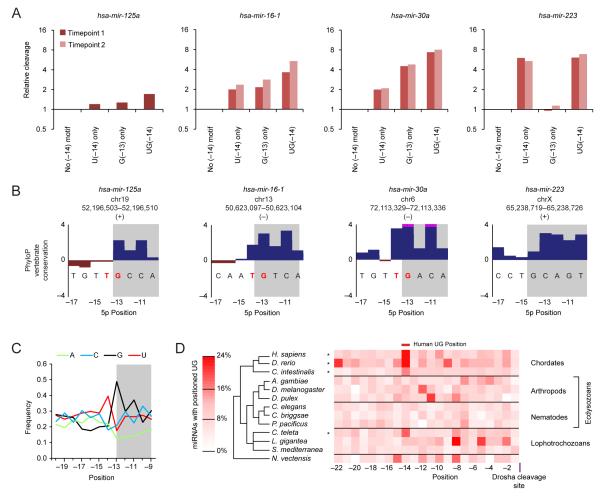
Figure 4.
The basal UG motif.
(A) Relative cleavage of variants with a full UG motif, a partial motif, and no motif. Values were normalized to that of variants with no motif, showing results from two time points, if available (key).
(B) PhyloP conservation across 30 vertebrate species in the region of the basal UG motif (red letters) for the four selected miRNAs. Bars extending beyond the scale of the graph are truncated (pink). Nucleotides predicted to be paired in the wild-type basal stem are shaded.
(C) Frequencies of A, C, G, and U (green, cyan, black, and red, respectively) at the indicated positions of human pri-miRNAs conserved to mouse. Analysis was of 204 pri-miRNAs, each representing a unique paralogous family (Table S2).
(D) Enrichment for the UG dinucleotide in the pri-miRNAs of representative animals with sequenced genomes. UG occurrences were tabulated for the upstream regions of pri-miRNAs aligned on the predicted Drosha cleavage site (Table S2). Species with statistically significant enrichment at position –14 are indicated (asterisks, empirical p-value <10−3).
The basal UG was conserved in vertebrate orthologs of mir-16-1 and mir-30a (Figure 4B). Moreover, the motif was enriched in other mammalian pri-miRNAs, as illustrated by the sequence composition of human pri-miRNAs (Figure 4C). It was also enriched in pri-miRNAs of zebrafish (D. rerio) and tunicate (C. intestinalis) but only sporadically in more distantly related lineages, suggesting that its recognition emerged in a chordate ancestor (Figure 4D).
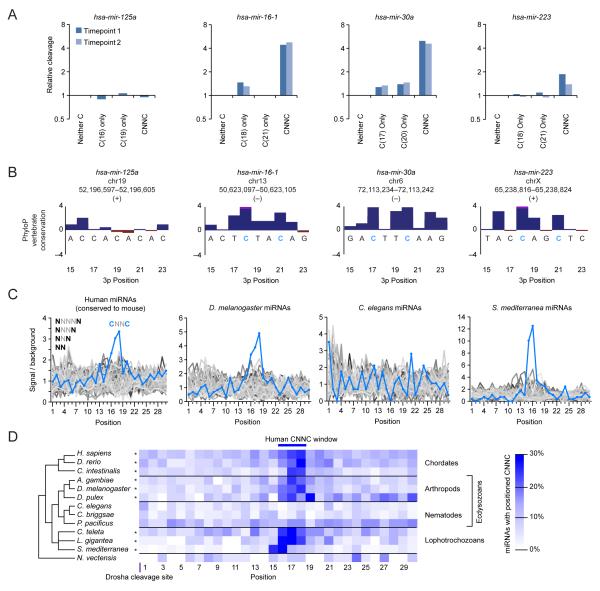
The broadly conserved CNNC motif enhances processing
In mir-16-1, mir-30a, and mir-223 we observed a preference for two C residues, separated by two intervening nucleotides, beginning 17–18 nt downstream of the Drosha cleavage site (Figure 2D). The two C residues of this CNNC motif (N signifies any nucleotide) acted synergistically, in that variants that retained neither C residue were not disfavored much more than those that retained one (Figure 5A). The C residues enriched in the active variants were conserved in vertebrate orthologs of these three pri-miRNAs (Figure 5B).
Figure 5.
The downstream CNNC motif.
(A) Relative cleavage of variants with a full CNNC motif, a partial motif, and no motif. Values were normalized to that of variants with no motif, showing results from two time points, if available (key).
(B) PhyloP conservation across 30 vertebrate species in the region of the downstream CNNC motif (blue letters) for the four selected pri-miRNAs. Bars extending beyond the scale of the graph are truncated (pink).
(C) CNNC enrichment compared to that of 63 other spaced dinucleotide motifs. Occurrences of each motif were tabulated for the downstream regions of pri-miRNAs aligned on the predicted Drosha cleavage site (Table S2). Background expectation was based on the nucleotide composition of pri-miRNA downstream regions in each species.
(D) Enrichment of the CNNC motif in the pri-miRNAs of representative bilaterian animals (Table S2). Species with statistically significant enrichment at positions 16, 17, or 18 are indicated (asterisk, empirical p-value <10−4).
The mir-125a pri-miRNA also had four C residues in the vicinity (positions 16–21), which gave rise to a CNNC at position 16 and the possibility of creating a CNNC at positions 17 or 18 (by changing either A20 or A18 to a C, respectively). However, the CNNC at position 16 was not preferred in the selection, nor were either of the single-nucleotide changes that could create a CNNC (Figures 2D and 5A). Moreover, the position 16 CNNC was not conserved in vertebrate orthologs (Figure 5B). These results indicate that unidentified features present in mir-16-1, mir-30a, and mir-223 but not mir-125a are required for the CNNC to increase processing efficiency.
For the three pri-miRNAs in which the CNNC motif was effective, its position fell in a small window 17–18 nt downstream of the Drosha cleavage site. In variants in which neither wild-type C was present, alternative CNNC motifs were strongly enriched 1–2 nt downstream (Figure S5A), which further indicated that a CNNC motif within a small range of positions can contribute to pri-miRNA recognition.
Of the 64 possible dinucleotide motifs with 0–3 intervening nucleotides, CNNC was the one most highly enriched downstream of the cleavage sites of human pri-miRNAs (Figure 5C). Moreover, enrichment was limited to a small range of positions 16–18 nt downstream of the site, peaking at positions 17 and 18, which matched the positions of the motif within mir-16-1, mir-30a, and mir-223. These results suggest that the CNNC motif enhances processing of many human pri-miRNAs.
Similar analyses of non-mammalian pri-miRNAs indicated strong, position-specific enrichment of the CNNC motif in chordates, arthropods and lophotrochozoans but not in sea anemone (Nematostella vectensis) (Figure 5C–D), suggesting that its recognition emerged with the divergence of bilaterians. Interestingly, enrichment was also absent in nematodes (Figure 5C–D), suggesting an isolated loss in the nematode branch of the ecdysozoans.
Consistent with the results in extracts, mutation of the basal UG and downstream CNNC motifs each reduced accumulation of mature miR-16 and miR-30a in HEK293T cells, with mutation of both reducing accumulation ~4–8-fold relative to wild type (Figure S5B–C). Furthermore, one or both motifs contributed to the accumulation of each of the additional pri-miRNAs tested in cell culture (hsa-mir-28, hsa-mir-129-2, and hsa-mir-193b; Figure S5D–F).
SRp20 binds the CNNC motif and enhances processing
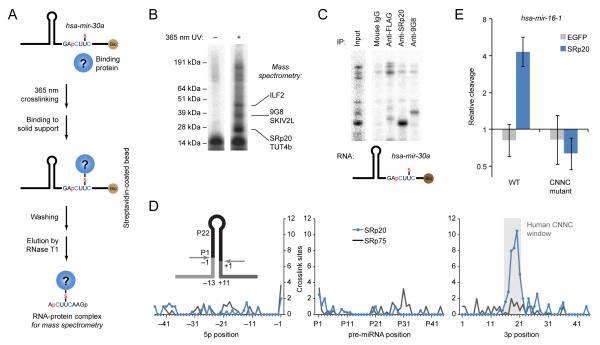
To learn how the CNNC motif is recognized, we used site-specific crosslinking (Wyatt et al., 1992). Proteins that crosslinked to pri-mir-30a RNA with a photoreactive nucleotide (4-thiouridine) placed within the CNNC motif were identified by mass spectrometry (Figure 6A). To guide gel-purification of crosslinked proteins, the procedure was performed in parallel with a radiolabeled pri-miRNA designed to label only proteins that crosslinked in the vicinity of the CNNC (Figure 6A–B). The two strongest candidates were SRp20/SRSF3 and 9G8/SRSF7, closely related proteins implicated in splicing regulation (Zahler et al., 1993; Cavaloc et al., 1994), mRNA export (Huang and Steitz, 2001) and translation initiation (Bedard et al., 2007; Swartz et al., 2007). These proteins both have an RNA-recognition motif (RRM) conserved across bilaterian animals, which recognizes degenerate motifs closely related to the CNNC motif (Heinrichs and Baker, 1995; Cavaloc et al., 1999; Schaal and Maniatis, 1999). NMR studies of this RRM in complex with RNA indicate that the C residues, particularly the first C of the CNNC, are bound in a base-specific manner, with minimal preferences for the two intervening bases (Hargous et al., 2006). Immunopurification of SRp20 and 9G8 confirmed that these two proteins (particularly SRp20) were the ones that most efficiently crosslinked in our assay (Figure 6C).
Figure 6.
Binding and activity of SRp20 at the CNNC motif.
(A) Site-specific crosslinking approach used to identify CNNC-binding proteins. The mir-30a crosslinking substrate contained a photoreactive base in the CNNC motif (4-thiouridine, U–S), a 3′ biotin (Bio), and for some applications, a 32P-labeled phosphate (red p). This substrate was incubated in Microprocessor lysate and irradiated with 365 nm UV light. Crosslinked complexes were captured on streptavidin-coated beads and eluted by RNase T1 digestion.
(B) Proteins within crosslinked RNA–protein complexes. Crosslinked complexes prepared as in (A) were separated on an SDS gel. For each CNNC-crosslinked band, proteins are listed that were identified by mass spectrometry and have known or inferred RNA-binding activity.
(C) Immunoprecipitation of proteins crosslinked to the CNNC motif. After crosslinking as in (A), complexes were enriched using monoclonal antibodies against either FLAG (the tag of the overexpressed Drosha and DGCR8), SRp20 or 9G8, and then resolved on an SDS gel. Input was run on a different region of the same gel for reference.
(D) SRp20 binding downstream of mouse pri-miRNA hairpins in vivo. Sites were obtained by reanalysis of crosslinking data for SRp20 and SRp75 in mouse cells (Anko et al., 2012). Positions are numbered as in Figure 1D. Expected sites of crosslinks to any of the motif nucleotides in the region of motif enrichment (Figure 5D) are shaded (gray).
(E) Enhancement of in vitro pri-miRNA cleavage by SRp20. Wildtype pri-mir-16-1 or pri-mir-16-1 with mutated CNNC were incubated for 3 minutes with immunopurified Microprocessor, supplemented with either FLAG-EGFP or 3X-FLAG-SRp20 purified from HEK293T cells. Reactants and products were resolved on denaturing polyacrylamide gels and quantified by phosphorimaging relative to a buffer-only control (geometric mean ± standard error, n = 3).
To evaluate SRp20 binding in vivo, we analyzed a large dataset of SRp20 crosslinking sites in P19 cells (Anko et al., 2012). Although the published analyses of this dataset focused on sites within pre-mRNAs, we found that many SRp20 sites resided in pri-miRNAs, and, more importantly, that these sites overlapped the region of CNNC enrichment (Figure 6D). This analysis extended our results from in vitro binding to in vivo binding and from one pri-miRNA to many. Some of the crosslinking sites in the CNNC-enriched region were in pri-miRNAs that lacked a CNNC motif, suggesting that SRp20 (and presumably its paralog, 9G8) might play a role even more general than that implied by CNNC conservation and enrichment.
The requirement of SRp20 for cell viability (Jumaa et al., 1999; Jia et al., 2010) confounded attempts to test its function by depleting the protein in cell culture. Therefore, we tested its function in vitro, supplementing immunopurified Microprocessor complex with either immunopurified recombinant SRp20 (Figure S6) or an analogously purified control protein (EGFP). SRp20 enhanced mir-16-1 processing in a CNNC-dependent manner (Figure 6E). Taken together, our results indicate that for many bilaterian miRNAs the CNNC motif is enriched and preferentially conserved because it helps recruit SRp20 (or its homologs), which enhances pri-miRNA recognition and processing.
Loop and apical stem elements can enhance processing
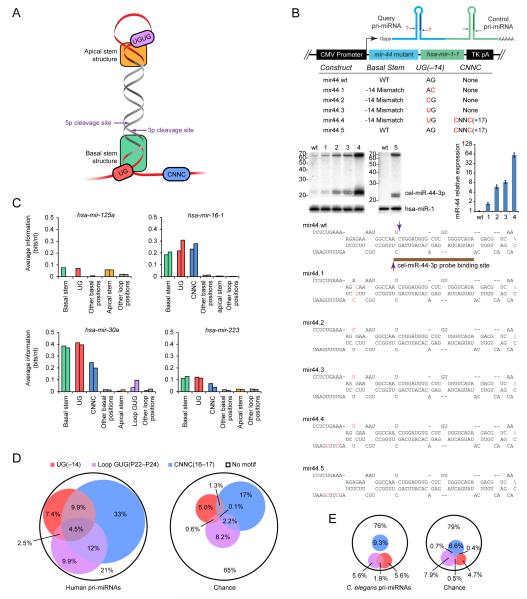
To examine whether additional processing features reside in the loop and apical stem, we extended our approach to those regions (Figure S7A). Pairing at the apical portion of the stem contributed to pri-miRNA recognition and processing for mir-125a and mir-30a but not for mir-16-1 or mir-223 (Figure S7B), consistent with differing conclusions drawn from studies of different miRNAs (Zeng et al., 2005; Han et al., 2006). Primary-sequence preferences were weaker than those observed for basal and flanking residues (Figure S7C). The best candidate for a loop-binding motif was observed only in mir-30a, in which the wild-type UGUG at positions P24–27 was both preferred in the selection (Figure S7D) and conserved in vertebrate orthologs (Figure S7E). Human and zebrafish miRNAs were enriched for UGU or GUG in this region of the loop (empirical p < 10−5 for each species) (Figure S7F), thereby confirming it as the third primary-sequence motif identified in our study (Figure 7A).
Figure 7.
Structural and primary-sequence features important for human pri-miRNA processing.
(A) Summary of human pri-miRNA determinants identified or confirmed in this study.
(B) Processing enhancement from adding human pri-miRNA features to C. elegans mir-44. Changes that introduced the listed features were incorporated into mir-44 within the bicistronic expression vector (top). Secondary structures are shown for mutations predicted to affect the wild-type basal stem (bottom; Drosha cleavage sites, purple arrowheads). After transfection into HEK293T cells, accumulation of miR-44-3p was assessed on RNA blots (middle), with the graph plotting increased miR-44-3p expression normalized to that of the hsa-miR-1 control (geometric mean ± standard error, n = 3). Adding a CNNC to the wild-type sequence (construct mir44.5) enhanced processing ≥20 fold (geometric mean of triplicate experiment), a lower bound set by the wild-type background.
(C) Contributions of individual features to in vitro processing, measured as average information content per nucleotide. If available, results from two time points are shown.
(D) Enrichment of primary-sequence motifs in human pri-miRNAs conserved to mouse (Table S2). Pri-miRNAs were classified based on whether they had the basal UG, the apical GUG or UGU, or the downstream CNNC motif (left). Expectations by chance (right) were estimated based on the nucleotide composition of upstream, pre-miRNA, and downstream regions of human pri-miRNAs for the basal UG, apical GUG or UGU, and CNNC motifs, respectively.
(E) A search for human motifs in C. elegans pri-miRNAs (Table S2). Pri-miRNAs were analyzed as in (D); the smaller diagrams reflect the smaller number of analyzed pri-miRNAs.
Rescue of C. elegans miRNA expression in human cells
The primary-sequence motifs important for mammalian miRNAs were not enriched in the nematode clade, suggesting that their absence might account for the failure of C. elegans pri-miRNAs to be processed in human cells. To test this idea, we added the basal UG and the downstream CNNC motifs to cel-mir-44 in the context of the mir-1 bicistronic vector (Figure 7B). Before adding the motifs, we disrupted the predicted pairing between positions −14 and +12 and substituted the G:C pair at positions −13 and +11 (construct mir44.1). These changes, which were expected to simultaneously enhance processing by shortening the basal stem to its optimal length and inhibit processing by replacing the fortuitous G at position –13, had a marginal net effect on production of mature miR-44 in human cells (Figure 7B). Adding a basal UG enhanced production of mature miR-44 by 5-fold (8-fold over the wild-type), primarily from restoring the G at –13 (Figure 7B). Adding a CNNC 17 nt downstream of the cleavage site (mir44.4) enhanced production another 8-fold, yielding a 64-fold net increase over wild-type (Figure 7B). Similarly, converting the wild-type, asymmetrically bulged stem of cel-mir-50 to a regular, 11-pair stem and adding the UG and CNNC motifs enhanced expression of mature miR-50 by 30-fold (Figure S7G), while adding the motifs to cel-mir-40 enhanced expression of mature miR-40 by 5-fold (Figure S7H). We conclude that primary-sequence motifs discovered in this study help human cells to distinguish pri-miRNAs hairpins from other hairpins and that the absence of these motifs in C. elegans pri-miRNAs helps to explain why human cells do not regard these transcripts as pri-miRNAs.
DISCUSSION
Secondary structure is inadequate on its own to specify pri-miRNA hairpins: primary-sequence features, including the basal UG, the CNNC and the apical GUG motifs, also contribute to efficient processing in human cells (Figure 7A). Complicating the story (and perhaps explaining why these primary-sequence features had not been observed earlier), different pri-miRNAs differentially benefit from the different motifs (Figure 7C). Among human pri-miRNAs, these motifs were nonetheless highly enriched, with 79% of the conserved human miRNAs containing at least one of the three motifs (Figure 7D).
The motifs were not enriched in C. elegans pri-miRNAs (Figure 7E) and when added to the C. elegans pri-miRNAs conferred more efficient processing in mammalian cells (Figure 7B, Figure S7G–H). These experiments also showed the benefit of disrupting pairing normally present at positions −14 and +12 of the C. elegans miRNAs. The presence of pairing that is inhibitory to mammalian processing suggests that measurement from the base of the helix might also differ in nematodes. Thus, despite the many broadly conserved features of miRNAs, some primary-sequence features and some secondary-structure features differ in mammals and nematodes.
About a fifth of human pri-miRNAs lack all three newly identified primary-sequence determinants (Figure 7D). These are attractive subjects for further study, in that the approach implemented here presumably would identify additional unique determinants used by these pri-miRNAs. Other determinants probably also exist at the Microprocessor cleavage site and nearby stem regions, which were inaccessible to our approach as implemented. Indeed, point mutations that disrupt pairing in the middle of the stem dramatically impair processing (Gottwein et al., 2006; Duan et al., 2007; Jazdzewski et al., 2008; Sun et al., 2009), and the SR-domain splicing factor SF2/ASF is reported to enhance the processing of mir-7-1 by binding a motif in the stem near the cleavage site (Wu et al., 2010). Hinting at the possibility of additional primary-sequence preferences within the stem are results from both bacterial RNase III and fungal homologs (Rnt1 and Pac1), which prefer specific base-pair identities near the cleavage site (Lamontagne and Elela, 2004).
The emerging picture is that pri-miRNA recognition is a modular phenomenon in which each module contributes modestly, and each pri-miRNA depends on individual modules to varying degrees. Our results quantify the relative importance of each known module for each pri-miRNA (Figure 7C). Pairing within the basal stem was crucial, as expected (Lim et al., 2003b; Han et al., 2006). In addition, all four miRNAs made use of the basal UG motif, which provided similar information content per nucleotide as did the basal-stem nucleotides. For the three miRNAs that used the CNNC SRp20-binding site, its importance was also comparable to that of the basal stem nucleotides. Compared to the nucleotides within these motifs, other flanking nucleotides contributed very little.
Apical and terminal loop elements were less important than the basal motifs (Figure 7C). We detected significant contributions only in mir-125a, in which the apical stem nucleotides were as important as the basal stem nucleotides, and in mir-30a, in which the loop UGUG motif contributed some information, albeit less than any of the three other features. Together, the features described here explained 61–78% of the information content in the selected sequences. The remaining information content was diffusely distributed among the other partially-randomized positions and might have mostly reflected avoidance of detrimental alternative structures.
Knowledge of biogenesis features will aid in interpreting human mutations. For example, reduced miR-16 expression associated with chronic lymphocytic leukemia (CLL) is typically due to deletions spanning the intron containing mir-15a and mir-16-1 (Calin et al., 2002). However, two of 75 CLL patients studied had tumors that retain the pri-miRNA hairpins and instead carried a germline C>T single-nucleotide polymorphism (SNP) downstream of the mir-16-1 hairpin (Calin et al., 2005). This SNP lowers overexpression of miR-16 in HEK293 cells, and in both patients heterozygosity for the SNP was lost in the leukemic cells (Calin et al., 2005). This SNP corresponds to the first C in the mir-16-1 CNNC, which explains why it lowers miR-16 accumulation and leads to CLL: it affects pri-miRNA processing by disrupting SRp20 recruitment. Discovery of additional features for pri-miRNA recognition and processing might lead to improved diagnostic and therapeutic tools in cancer and other diseases in which miRNAs are dysregulated.
EXPERIMENTAL PROCEDURES
Ectopic Pri-miRNA Expression
Plasmids were derived from pcDNA3.2/V5-DEST and pMT-DEST (Invitrogen) for expression in HEK293 and S2 cells, respectively. Query pri-miRNA sequences and the human pri-mir-1-1 sequence were cloned such that the query pri-miRNAs were transcriptionally fused upstream of mir-1-1. HEK293 and S2 cells were transfected using Lipofectamine 2000 and Cellfectin (Invitrogen), respectively. After 36–48 h, total RNA was extracted, and miRNA expression was assayed by RNA blots, ribonuclease protection assays (Invitrogen), and high-throughput sequencing (Chiang et al., 2010). For additional details including the data analysis pipeline, see Extended Experimental Procedures.
Binding and cleavage assays
To assay binding, T7-transcribed competitor and reference pri-miRNA substrates were radiolabeled and mixed in an equimolar ratio, then incubated with limiting amounts of immunopurified catalytically impaired Microprocessor (Lee and Kim, 2007; Han et al., 2009). RNA–protein complexes were filtered on Immobilon-NC nitrocellulose discs (Whatman), and RNA extracted from the filter was resolved on 5% polyacrylamide gels. To assay cleavage, labeled substrates were incubated with Microprocessor lysate, which was prepared from cells overexpressing Drosha and DGCR8 (Lee and Kim, 2007). After extraction using Tri-Reagent (Ambion), substrates and products were resolved on denaturing 5% polyacrylamide gels. For additional details, see Extended Experimental Procedures.
Synthesis and selection of pri-miRNA variants
Templates for T7 transcription were assembled from oligonucleotides (IDT) synthesized using nucleoside phosphoramidite mixtures designed to introduce variability at specified positions (Table S1). Sequences encoding the HDV self-cleaving ribozyme were appended so that ribozyme cleavage would generate transcripts with defined 3′ ends. Template pools were transcribed using T7 RNA polymerase, and following treatment with TurboDNAse (Ambion) RNA was purified on denaturing polyacrylamide gels. After dephosphorylation of 5′ and 3′ ends using calf intestinal phosphatase (NEB) and T4 polynucleotide kinase (T4 PNK, NEB), followed by 5′ phosphorylation using T4 PNK, transcripts were circularized using T4 RNA ligase 1 (NEB) and gel purified. RNA pools were incubated with Microprocessor lysate, and after gel-purification, cleavage products were ligated to oligonucleotide adaptors, reverse transcribed, amplified, and Illumina sequenced (75 nt paired-end reads). In parallel, the initial pool of RNA was also reverse transcribed, amplified and sequenced. Selections for examining binding or apical stem-loops were similar, except transcripts were not circularized. For additional details including the data analysis pipeline, see Extended Experimental Procedures.
Motif enrichment
Enrichment of a motif within pri-miRNAs of a species was evaluated by comparing to 100,000 cohorts of miRNAs in which the upstream, downstream and pre-miRNA sequences were independently shuffled, preserving dinucleotide frequencies. The numbers of miRNAs that contained a match to the motif in the actual and shuffled cohorts were used to compute an empirical P-value. A list of the representative pri-miRNAs used for analyses is provided (Table S2). For additional details, see Extended Experimental Procedures.
Site-specific crosslinking
The mir-30a pri-miRNA crosslinking substrate was assembled using T4 RNA ligase 2 (NEB) and a DNA splint to join an in vitro transcribed 5′ fragment to a synthetic 3′ fragment containing a 3′-terminal biotin and a 4-thiouridine 3′ within the CNNC motif (Dharmacon). This crosslinking substrate was incubated in Microprocessor lysate and exposed to 1000 mJ of 365 nm UV light in a Stratalinker (Stratagene). For purification of RNA-protein complexes for mass spectrometry, complexes were captured on streptavidin-coated magnetic beads (Invitrogen), washed, and eluted with RNase T1 (Ambion), which cleaves after G. Eluted complexes were either separated on SDS gels and analyzed by HPLC/tandem mass spectrometry, or immunoprecipitated and analyzed by SDS gel. For additional details, see Extended Experimental Procedures.
Supplementary Material
Research Highlights.
C. elegans pri-miRNAs are not processed in human cells, despite similar structure
In vitro selection reveals sequence motifs that help define human pri-miRNAs
The motifs are conserved in pri-miRNAs of other animals but not those of nematodes
Adding the motifs to C. elegans pri-miRNAs rescues their processing in human cells
ACKNOWLEDGEMENTS
We thank D. Shechner, C. Jan, O. Rissland, D. Weinberg, J. Ruby, J. Nam, and V.N. Kim for valuable discussions; O. Rissland for comments on this manuscript; L. Schoenfeld and J. Lassar for technical assistance; J. Stévenin for 9G8 antibody; V.N. Kim and T. Tuschl for plasmids; the Whitehead Institute Genome Technology Core for sequencing; and E. Spooner for mass spectrometry. This work was supported by NIH grants GM067031 and T32GM007753. D.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Accession Numbers Sequencing data have been deposited into the Short Read Archive (SRA051323).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography and References Cited
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26:459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Popielarz M, Fuchs JP, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 2011;21:286–300. doi: 10.1101/gr.113050.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang X, Song Q, Li T, Zeng Y. Drosha processing controls the specificity and efficiency of global microRNA expression. Biochim Biophys Acta. 2011;1809:700–707. doi: 10.1016/j.bbagrm.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cai X, Cullen BR. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J Virol. 2006;80:5321–5326. doi: 10.1128/JVI.02734-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Baker BS. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Li C, McCoy JP, Deng CX, Zheng ZM. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int J Biol Sci. 2010;6:806–826. doi: 10.7150/ijbs.6.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Wei G, Nielsen PJ. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr Biol. 1999;9:899–902. doi: 10.1016/s0960-9822(99)80394-7. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Lamontagne B, Elela SA. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J Biol Chem. 2004;279:2231–2241. doi: 10.1074/jbc.M309324200. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim VN. In vitro and in vivo assays for the activity of Drosha complex. Methods Enzymol. 2007;427:89–106. doi: 10.1016/S0076-6879(07)27005-3. [DOI] [PubMed] [Google Scholar]
- Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003a;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003b;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Uhlenbeck OC. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+ Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JN, Ferre-D’Amare AR. Rapid construction of empirical RNA fitness landscapes. Science. 2010;330:376–379. doi: 10.1126/science.1192001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, Sommer SS, Rossi JJ. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjold ML. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem. 2007;282:19844–19853. doi: 10.1074/jbc.M701660200. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Mol Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt JR, Sontheimer EJ, Steitz JA. Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev. 1992;6:2542–2553. doi: 10.1101/gad.6.12b.2542. [DOI] [PubMed] [Google Scholar]
- Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Stolk JA, Roth MB. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.