Abstract
Plants synthesize a chemically diverse range of hormones that regulate growth, development, and responses to environmental stresses. The major classes of plant hormones are specialized metabolites with exquisitely tailored perception and signaling systems, but equally important are the enzymes that control the dose and exposure to the bioactive forms of these molecules. Here, we review new insights into the role of enzyme families, including the SABATH methyltransferases, the methylesterases, the GH3 acyl acid-amido synthetases, and the hormone peptidyl hydrolases, in controlling the biosynthesis and modifications of plant hormones and how these enzymes contribute to the network of chemical signals responsible for plant growth, development, and environmental adaptation.
Keywords: Biosynthesis, Enzyme Structure, Plant Biochemistry, Plant Hormones, Protein Structure
Introduction
Plants produce an array of signaling molecules with essential roles in plant growth and development and control responses to environmental stresses, such as drought, herbivory, and pathogen attack. Many plant growth regulators, or phytohormones, were originally isolated as specialized metabolites with molecular structures that reflect their metabolic origins (Fig. 1A) (1). The chemical diversity of these molecules is linked to their biological functions and in planta effects through various signaling pathways. Over the past decade, efforts to understand the biosynthesis of plant hormones and their associated perception systems have revealed new biochemical pathways and identified the receptors and signaling events for the major classes of these molecules (Table 1) (2–43).
FIGURE 1.
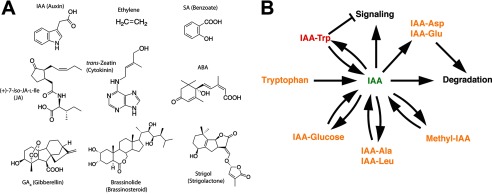
Plant hormones and hormone homeostasis. A, chemical structures of the major classes of plant hormones. B, representative chemical modifications of the auxin IAA. The bioactive (green), inactive (orange), and anti-auxin (red) forms of IAA are shown. These modifications are only some of the possible chemical changes that occur to this molecule.
TABLE 1.
Summary of major plant hormones: biological effects, metabolic precursors, and receptor responses
ET, ethylene; GA, gibberellin; IPP, isopentenyl pyrophosphate; ARR, Arabidopsis response regulator.
| Hormone | Active form | Effects | Precursor | Receptor |
|---|---|---|---|---|
| Auxin | IAA | Cell elongation; apical dominance; tropisms; branching; lateral roots | Tryptophan | AUX/IAA: repressor proteins; TIR1/AFBs: auxin F-box-binding proteins; auxin facilitates binding of AUX/IAA with SCFTIR1/AFB, resulting in degradation of AUX/IAA repressors and altered transcription of auxin-induced genes |
| ET | Ethylene | Flowering/fruit ripening; stress response; seed germination | Methionine | ETR1/ETR2/ERS1/ERS2/EIN4: histidine kinases (dimers); ET represses repressors of ET response, leading to activation of EIN3 and EREBP families of transcription factors |
| JA | JA-Ile | Plant defenses against insect herbivores; root growth inhibition; necrotrophic pathogen responses | α-Linolenic acid | COI1/JAZ co-receptor: COI1 F-box protein; component of the SCFCOI1 E3 ligase complex; targets JAZ proteins for degradation and leads to altered gene expression |
| Cytokinins | trans-Zeatin | Cell division; releases lateral buds from apical dominance; delays senescence; root growth | Adenine | CRE1: similar to histidine kinases; activates histidine kinase activity, initiating a phosphorelay that results in phosphorylation of type B ARRs to induce transcription of type A ARRs |
| Benzoates | SA | Systemic acquired resistance to pathogens; biotrophic pathogen responses | Chorismate | NPR1: transcriptional coactivator; NPR3/ NPR4: CUL3 E3 ligase adaptor proteins |
| ABA | ABA | Stomatal closure; seed maturation, germination, storage, desiccation tolerance; root/shoot growth; leaf senescence | IPP-derived tetraterpene (phytoene) | PYR/PYL/RCAR: START family of ligand-binding proteins; CHLH: cheletase; GTG1/2: G-proteins |
| GAs | GA1, GA4 | Stem elongation; root growth; seed germination; flowering; floral development; fruit growth; stresses | IPP-derived diterpene (geranylgeranyl diphosphate) | GID1: globular protein; GA-GID1 binds DELLA repressors, leading to targeting to the SCF complex and subsequent degradation by the 26 S proteasome |
| Brassinosteroids | Brassinolide | Cell division/elongation in stems/roots; photomorphogenesis; reproductive development; leaf senescence; stress responses | IPP-derived sesquiterpene (farnesyl diphosphate) | BRI1: dual-specificity kinase; dimerizes with BAK1, initiating phosphorelay prevention of phosphorylation of BES1/BZR1 to alter gene expression |
| Strigolactones | Strigol | Branching; leaf senescence; root development; plant-microbe interactions | Carotenoids | D14/DAD2: α/β-hydrolase; putative receptor/co-receptor; initiates SCF-mediated signaling via SCFMAX2 complex |
Although plant development, growth, and environmental responses are all determined by the complex integration of hormone-controlled signaling pathways, changes in cellular concentrations and the chemical structure of a hormone directly affect interaction with cognate receptors to control the duration of activation and potentiation of specific biological effects. The biosynthesis, degradation, and chemical modification of each class of plant hormone contribute to controlling those biological effects. For example, indole-3-acetic acid (IAA),4 the major auxin, triggers hormone responses, whereas modified forms of IAA are used for storage, degradation, or inhibition of auxin signaling (Fig. 1B) (2–5). This minireview provides an overview of plant hormone synthesis; describes new insights into enzymatic modification for controlling plant growth regulators; and highlights how enzymes contribute to the intricate network required for plant growth, development, and environmental adaptations.
Overview of Plant Hormone Biosynthesis
Primary metabolism provides the building blocks of plant growth regulators (Table 1). From a chemical perspective, the molecules that control plant growth are specialized metabolites adapted for interaction with protein receptors to regulate a variety of biological outcomes. Pathways for the biosynthesis of the plant hormones are tightly regulated and integrated to control responses to a diverse array of developmental and environmental inputs.
Amino acid metabolism contributes to the synthesis of ethylene, auxin, and salicylic acid (SA). Production of ethylene, which stimulates fruit ripening and senescence of vegetative tissues, occurs by cyclization of S-adenosyl-l-methionine (AdoMet) into 1-aminocyclopropane-1-carboxylic acid (ACC) by ACC synthase and subsequent oxidation into ethylene by ACC oxidase. Regulation of ACC synthase controls ethylene production (10).
Aromatic amino acids are precursors of auxin and SA synthesis. Originally discovered through their effects on plant growth in light and gravity, auxins, especially the predominant hormone IAA, control a range of processes, such as maintenance of apical dominance, shoot elongation, and root initiation (5, 44). Initially reported as a plant growth regulator in 1926 (45), the biosynthesis of IAA from tryptophan has a long history involving multiple proposed pathways (5); however, new evidence suggests that tryptophan aminotransferase and the YUCCA flavin monooxygenase are the major route to auxin (46, 47). Maintenance of bioactive IAA levels requires a balance of synthesis, storage, degradation, transport, and modification (Fig. 1B) (2–5, 48).
SA plays a critical role in plant responses to biotrophic pathogens, which lead to increased SA levels at infection sites (19, 49). SA synthesis requires chorismate. Isotopic feeding studies suggest that conversion of phenylalanine into cinnamic acid and its subsequent metabolism to SA are one route (50–52). In contrast, genetic studies in Arabidopsis thaliana suggest that the main route for SA synthesis is conversion of chorismate to isochorismate, followed by breakdown to SA and pyruvate (53). Although bacteria metabolize isochorismate to SA and pyruvate using isochorismate-pyruvate lyase (54), a plant homolog of this enzyme remains unidentified.
Metabolites from the lipid and isoprenoid pathways support the synthesis of jasmonates, cytokinins, brassinosteroids, abscisic acid (ABA), and gibberellins. The biosynthesis of these plant hormones generates a wide range of chemical diversity, much of which remains to be explored with respect to their biological effects.
First identified by their ability to inhibit plant growth, jasmonates affect seed germination, fertility, root growth, and responses to pathogens (1, 13, 55). Plants synthesize jasmonic acid (JA) from α-linoleic acid (13, 42, 43). JA synthesis begins in the plastid with conversion of α-linoleic acid to 12-oxo-phytodienoic acid. Following transport of 12-oxo-phytodienoic acid to the peroxisome, a reduction reaction and multiple β-oxidation steps generate (+)-7-iso-JA (i.e. free JA). Conjugation of JA with isoleucine yields the bioactive hormone (+)-7-iso-jasmonoyl-l-isoleucine (JA-Ile) (13, 56, 57).
In plants, cytokinins promote cell division, and their synthesis occurs either de novo or through recycling of tRNAs containing a uridine at the first anti-codon position (58). In the de novo pathway, dephosphorylation of isopentenyl-adenosine-5′-phosphate followed by hydrolysis of the ribose yields trans-zeatin. Addition of chemical moieties at the adenosine amine leads to diverse natural and synthetic cytokinins, such as kinetin (N6-furfuryladenine) and 6-benzylaminopurine (48).
Biosynthesis of brassinosteroids, ABA, strigolactones, and gibberellins extends from isoprenoid metabolism. Synthesis of brassinosteroids shares similar chemistry with mammalian steroid biosynthesis, as assembly of the core scaffold requires squalene synthase, steroid 5α-reductase, and cytochrome P450 monooxygenases for formation of campesterol (31). Additional reactions lead to brassinosteroids with various functional groups (31, 55). Carotenoids are the basis for ABA and strigolactones. For ABA synthesis, epoxidation and processing of zeaxanthin to trans-violaxanthin occur in the plastid. Subsequent oxidative cleavage in the cytosol produces xanthoxin, which is further metabolized to ABA (22). Similarly, strigolactone synthesis involves breakdown of β-carotene; however, the exact pathway remains unclear (38). Gibberellins are a large group of diterpenoid carboxylic acids (27). Produced by plants, as well as fungi and bacteria, their synthesis begins with cyclization of geranylgeranyl diphosphate into ent-kaurene, followed by a series of reactions involving cytochrome P450 monooxygenases and 2-oxoglutarate-dependent dioxygenases (27).
Chemical Modifications
Although the biosynthesis of plant hormones builds from different primary metabolic pathways, these specialized compounds are all modified by common reactions. Modifications of plant hormones mark them for degradation, storage, or activation (Fig. 1B). For example, glycosylated forms of IAA, cytokinins, ABA, JA, and SA are common (19, 48, 55). Glycosylation usually leads to inactive storage forms of plant hormones that can be hydrolyzed for activation, but glycosylation of cytokinins tags these molecules for degradation (55). In addition, methylation, amino acid conjugation, hydroxylation, sulfonation, and carboxylation reactions lead to further tailoring of plant hormones (2, 48, 51, 55, 59).
Substantial progress toward understanding how different families of enzymes modify plant hormones provides a foundation for unraveling the biological consequences of these chemical changes. Because methylation and amino acid conjugation of plant hormones are well studied and the proteins involved in both formation and hydrolysis of these modifications have been structurally and biochemically characterized, the rest of this minireview is focused on the methyltransferases, acyl acid-amido synthetases, and the esterases/hydrolases associated with these modifications.
Plant Hormone Methylation
The methylation and demethylation of plant growth regulators rapidly switch chemical activity (Fig. 2A). Methyl derivatives of many plant growth regulators were isolated even before the parent compound's hormone activity was identified. For example, methyl jasmonate was isolated from Jasminum grandiflorum 2 decades before the role of JA in plant growth was discovered (13, 60). Methylated benzoates are also common across multiple plant species (61–64). Typically, methylated volatiles aid in long-distance communication, and the methylated forms are inactive as hormones and require removal of the modification for effect. Moreover, not all methylated plant hormones were discovered as volatiles. In Arabidopsis, overexpression of a methyltransferase (IAMT1) that converts IAA to methyl-IAA (MeIAA) leads to a curvy leaf phenotype, which is typical of perturbed auxin homeostasis, even though MeIAA has not been identified in vivo (62). Similarly, in planta overexpression of methyltransferases active on gibberellins and JA in vitro results in phenotypes characteristic of low gibberellin and JA levels, respectively (63, 64).
FIGURE 2.
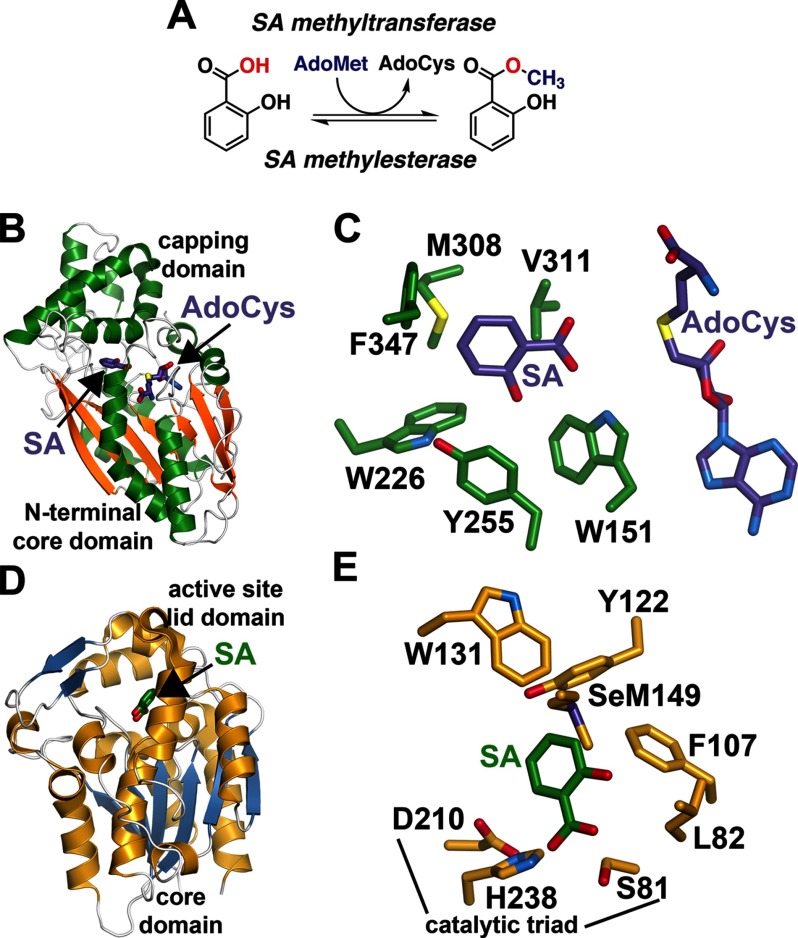
Structural overview of the SABATH and MES families. A, chemical reaction of SA methylation and demethylation. AdoCys, S-adenosylhomocysteine. B, overall structure of the SA methyltransferase from C. breweri (Protein Data Bank code 1M6E) (65) shown as a ribbon diagram. C, close-up view of the SA methyltransferase active site. D, overall structure of the N. tabacum MES SABP2 (Protein Data Bank code 1Y7I) (71). E, active site view of N. tabacum SABP2 showing the positioning of SA and the catalytic triad.
In plants, the SABATH methyltransferases catalyze the AdoMet-dependent addition of methyl groups to a range of molecules (61, 65). Named after the first members of the family to be discovered, i.e. SA methyltransferase, benzoic acid methyltransferase, and theobromine synthase, these enzymes are encoded by large gene families in plants. For example, the genomes of Arabidopsis and rice (Oryza sativa) encode 24 and 41 SABATH methyltransferases, respectively. Typically, SABATH proteins function as O-methyltransferases that target carboxylic acids, but N- and S-methyltransferases have been reported (66, 67). Nearly all of the characterized SABATH methyltransferases display a high degree of substrate specificity, which suggests that members of this family evolved for specialized functions (61). An exception is a dual-specific enzyme in Arabidopsis that methylates benzoic acid and SA (68).
Crystallographic studies of multiple SABATH methyltransferases reveal the molecular basis of substrate specificity and catalysis (65). Following determination of the structure of SA methyltransferase from Clarkia breweri (Fig. 2B) (65), x-ray crystal structures of SABATH proteins involved in caffeine biosynthesis and IAA methylation were determined (66, 69). The structural fold of the SABATH family is an elongated, parallel, seven-stranded β-sheet, which is common to all AdoMet-dependent methyltransferases, with an α-helical domain capping the active site (Fig. 2B). Although the sequences of SABATH methyltransferases are highly divergent, the residues of the AdoMet-binding site are well conserved (65, 66, 69). In a typical SABATH methyltransferase, such as SA methyltransferase (Fig. 2, B and C), apolar residues interact with the adenine ring of AdoMet, and an invariant aspartate forms a bidentate interaction with the hydroxyl groups of the ribose group. Additional protein-ligand contacts with the carboxylate and amine groups of AdoMet anchor this molecule in the active site for catalysis.
Because SABATH proteins methylate chemically diverse molecules, the substrate-binding site is highly variable in sequence and structure. For example, the substrate-binding site of the C. breweri SA methyltransferase (Fig. 2C) consists of multiple apolar residues, with hydrogen bonds between the substrate carboxylic acid and the amine groups of Gln-25 and Trp-151 orienting it toward the AdoMet methyl group (65). In comparison, the substrate-binding site of IAAMT1 utilizes apolar residues for substrate binding (69). Although knowledge of binding residues has aided in the prediction of SABATH methyltransferase function (65, 69), many SABATH proteins with unknown physiological roles remain.
The SABATH methyltransferases catalyze an SN2 reaction using substrate proximity and desolvation effects to drive the reaction (65). In the proposed reaction, the enzyme binds AdoMet and the methyl acceptor substrate to occlude solvent and orient the AdoMet methyl group toward the acceptor site. Thus, residues provided by the SABATH active site provide a three-dimensional scaffold for binding and orientation of the two substrates.
Plant Hormone Demethylation
Demethylation of plant growth regulators by methylesterases (MESs) leads to activation of these molecules for their specific biological function (Fig. 2A) (70). MES proteins are part of the α/β-hydrolase enzyme superfamily and share a Ser-His-Asp triad (71, 72). In plants, the first MES discovered was an SA-binding protein from tobacco, which was proposed to function in methylsalicylic acid (MeSA) perception and signaling (73). Later work showed that this protein hydrolyzed MeSA to SA (71). As with the SABATH family, plants encode multiple MES-like genes. For example, Arabidopsis contains 20 MES-like genes, two of which (A. thaliana (At) MES19 and AtMES20) are likely pseudogenes (72).
Crystallographic analysis of the tobacco (Nicotiana tabacum) MES SABP2 (salicylic acid-binding protein-2) provided the first structural insight into this protein family (71). The overall structure contains a core six-stranded parallel β-sheet surrounded by α-helices with an active site lid domain consisting of a three-stranded antiparallel β-sheet and three α-helices (Fig. 2D). The active site is at the interface of these two domains. The aromatic ring of SA binds to a hydrophobic pocket in the cap domain, and the main domain contains the catalytic triad responsible for hydrolysis of the methyl group (Fig. 2E). Although some of the MES proteins in Arabidopsis contain a non-canonical catalytic triad in which the serine is replaced with an aspartate, this substitution does not compromise esterase activity (70, 72).
Biochemical screening of 15 Arabidopsis MES proteins identified enzymes that hydrolyzed MeIAA, MeSA, methyl jasmonate, and the artificial substrate p-nitrophenyl acetate (70). None of the Arabidopsis MESs were active with methyl gibberellins, and some showed no activity with any compound tested (70). Subsequent studies showed that AtMES16 demethylates chlorophyll catabolites and is important for chlorophyll degradation (74). Further work aimed at examining the expression patterns and quantifying substrate specificities of the plant MES family promises to develop a better understanding of the molecular and physiological roles of these proteins.
Plant Hormone-Amino Acid Conjugation
In plants, amino acid conjugates to IAA and JA dramatically alter the biological roles of these molecules (2–5, 13, 42, 43). Amino acid conjugation of auxins plays a central role in their homeostasis (2). For IAA, the free acid is the biologically active form of the hormone, with amino acid conjugation leading to inactivation (75). The metabolic fate of conjugated IAA depends on which amino acid is attached (Fig. 1B). Conjugation of either alanine or leucine to IAA leads to an inactive but readily hydrolyzed storage form (75). Conjugation of IAA with either aspartate or glutamate leads to hormone degradation (75). IAA-Trp acts as an anti-auxin to inhibit plant growth effects but does not compete with IAA for binding to the TIR1 auxin receptor (76). Typically, modification of IAA negates its plant growth effects. In contrast, amino acid conjugation of JA leads to formation of the biologically active hormone JA-Ile, which binds to the COI1 ubiquitin ligase/JAZ protein co-receptor to elicit its effects (56, 77, 78). Interestingly, JA-Trp also functions as an anti-auxin and suggests possible cross-talk that aids in balancing auxin and jasmonate signaling (77).
In plants, the enzymes that catalyze amino acid conjugation of plant hormones belong to the GH3 (Gretchen Hagen 3) family of acyl acid-amido synthetases (79). The first GH3 gene was identified in 1985 as an early auxin-responsive gene in Glycine max (soybean) (79). As with the SABATH and MES families, multiple genes encode GH3 proteins in each plant. For example, A. thaliana and rice encode 19 and 13 GH3 proteins, respectively (79–81). Genetic and physiological studies of various GH3 proteins indicate a diverse range of biological functions for these enzymes in jasmonate and auxin hormone signaling and for SA-related pathogen responses.
Staswick et al. (82) first characterized the A. thaliana GH3.11 (AtGH3.11/JAR1) protein after identifying the jar1 (jasmonate resistant 1) mutant, which showed defective JA signaling, including reduced male fertility and resistance to exogenous JA treatment. Sequence analysis of AtGH3.11/JAR1 identified low homology with firefly luciferase and the ANL family of adenylating enzymes (83). Named after its founding members (acyl-CoA synthetases, non-ribosomal peptide synthetases, and luciferase), ANL enzymes catalyze a two-step reaction via an adenylated reaction intermediate (83). The GH3 acyl acid-amido synthetases use ATP to adenylate an acyl acid substrate in the first half-reaction. Nucleophilic attack by the amine of an amino acid leads to release of AMP and formation of an amino acid conjugate (84, 85).
Subsequent studies in Arabidopsis and rice identified multiple GH3 proteins with roles in plant growth and development linked to auxin hormone signaling (86–91). Later biochemical analysis of these enzymes demonstrated conjugation of IAA to various amino acids (92). In Arabidopsis, knock-outs of the IAA-utilizing GH3 proteins show little effect on plant growth (86–91), which may result from functional redundancy. Overexpression lines for a few IAA-utilizing GH3 proteins in Arabidopsis all result in a dwarf phenotype along with other traits indicative of altered auxin signaling (86–91). In addition to jasmonate and auxin signaling, Arabidopsis mutants of the AtGH3.12/PBS3 gene display increased disease susceptibility to virulent and avirulent forms of the pathogen Pseudomonas syringae (93). These mutants suggest that amino acid conjugates of SA and related benzoates may function as bioactive inducers of plant pathogen defense responses (94).
Although genetic studies link GH3 proteins to jasmonate, auxin, and SA responses, the biochemical understanding of these proteins was limited to a few examinations of reaction chemistry and substrate specificity (77, 81, 82). New insights into how these proteins modify plant hormones came from the crystal structures of two Arabidopsis GH3 proteins (AtGH3.11/JAR1 and AtGH3.12/PBS3) and a grapevine (Vitis vinifera (Vv)) GH3 protein (VvGH3.1) (95, 96). As mentioned above, AtGH3.11/JAR1 catalyzes formation of JA-Ile, the bioactive jasmonate hormone (77). Although mutants of AtGH3.12/PBS3 display SA-related phenotypes in Arabidopsis, SA is not a substrate of this protein, and the physiologically relevant substrate is not known (94). The grapevine protein (VvGH3.1) conjugates IAA with aspartate (96). These structures define the overall GH3 fold as a large N-terminal domain with a β-barrel and two β-sheets flanked by α-helices and a smaller C-terminal domain consisting of a single four-stranded β-sheet flanked by two α-helices on each side (Fig. 3A). The active site is located at the interdomain interface, and the C-terminal domain is conformationally flexible, with its movement linked to each half-reaction (95).
FIGURE 3.
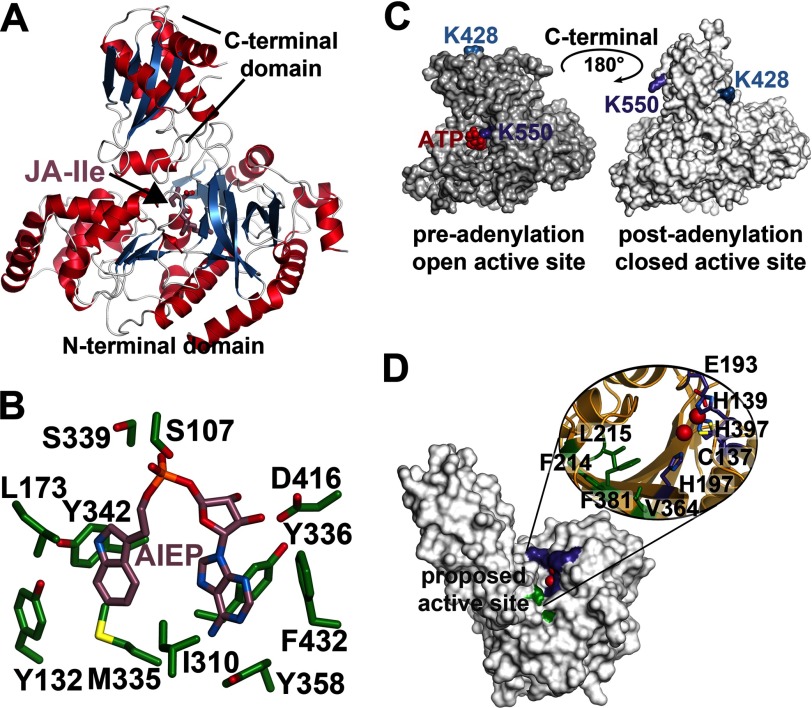
Structural overview of the GH3 acyl-amido synthetases and peptidyl hydrolase ILL. A, overall structure of AtGH3.11/JAR1 (Protein Data Bank code 4EPL) (95). B, active site view of the VvGH3.1 protein in complex with the non-hydrolyzable adenylation intermediate analog adenosine 5′-[2-(1H-indol-3-yl)ethyl]phosphate (Protein Data Bank code 4B2G) (96). C, surface representations of AtGH3.12/PBS3 showing the rotation of the C-terminal domain during the pre-adenylation open active site conformation (Protein Data Bank code 4EWV) (95) and the post-adenylation closed active site conformation (code 4EPM) (95). D, surface view of the AtILL2 peptidyl hydrolase (Protein Data Bank code 1XMB) (99). Residues corresponding to the apolar pocket proposed as the substrate-binding site are colored green, and the residues forming the canonical metal-binding site are colored purple.
The crystal structures reveal how the GH3 enzymes conjugate various amino acids to chemically diverse plant hormones. In each structure, residues of the N-terminal domain form the acyl acid/hormone-binding site. In AtGH3.11/JAR1, JA-Ile is bound in an elongated hydrophobic tunnel, with the oxylipin tail stacking across the surface of a tryptophan (95). The protein was crystallized with a racemic mixture of JA with the final structure including (−)-JA-Ile because the refinement statistics were modestly better with this enantiomer than with (+)-iso-JA-Ile; however, the quality of the ligand density allows modeling of either product. It is possible that the density represents a mixture of product enantiomers. The only polar interaction is a water-mediated hydrogen bond between the JA ketone group and a histidine side chain (95).
In comparison, the hormone-binding site of the IAA-utilizing VvGH3.1 protein reveals extensive changes that provide a different hydrophobic site to accommodate the substrate indole ring and a hydrogen bond between an active site tyrosine hydroxyl group and the amine of the indole ring (Fig. 3B) (96). Surprisingly, there are no interactions with the substrate carboxylate group, but the VvGH3.1 structure was determined with adenosine 5′-[2-(1H-indol-3-yl)ethyl]phosphate, a non-hydrolyzable adenylated IAA analog lacking the IAA carbonyl moiety, so additional interactions may form with the authentic substrate (Fig. 3B). Previous analyses of the IAA-utilizing GH3 proteins demonstrate activity with a variety of different auxins, including phenylacetic acid and naphthaleneacetic acid, which differ in chemical structure from IAA, but the basis for binding these molecules remains unclear (85). Although the physiological substrate of AtGH3.12/PBS3 is not known, the hormone site of this enzyme contains a number of polar residues, suggesting distinct substrate specificity compared with the JA- and IAA-conjugating enzymes.
Across the structures, the nucleotide site is identical to residues conserved in not only GH3 proteins but also the ANL superfamily (83, 95, 96). Three motifs define this site. The first motif is a serine-, threonine-, and glycine-rich P-loop that forms contacts with the α-phosphate; these interactions are essential for catalysis (95). Residues of the second motif (i.e. YGSSE) provide additional binding interactions. The tyrosine contacts the adenosine ring, the second serine hydrogen bonds with the α-phosphate, and the glutamate coordinates binding of the Mg2+ ion required for ATP binding. The third motif provides an aspartate for hydrogen bonding to the ribose hydroxyls of the bound nucleotide. Overall, these features orient ATP for attachment of the hormone carboxylic acid at the α-phosphate in the first half-reaction, leading to an adenylated intermediate (95).
Following adenylation of the acyl acid substrate and pyrophosphate release, rotation of the C-terminal domain by 180° yields an active site that covers the reaction intermediate (Fig. 3C) (95). This rotation prevents either the release or hydrolysis of the reaction intermediate before binding of the amino acid substrate. Rotation of the C-terminal domain also creates a tunnel for the amino acid to enter and orient for nucleophilic attack on the adenylated intermediate (95, 96). Although no crystal structure of a GH3 protein with an amino acid bound is currently available, the placement of the isoleucine group of JA-Ile in AtGH3.11/JAR1 and a bound malate in VvGH3.1 suggests a putative amino acid-binding site (95, 96).
Biochemically, the GH3 proteins catalyze the conjugation of amino acids to acyl acids via an adenylation reaction. Through this simple modification, these proteins regulate levels of active and inactive forms of jasmonates, auxins, and benzoates. The capacity of GH3 proteins to modify multiple related substrates may also allow for changes of metabolic pools across a biosynthetic pathway, in addition to modifying the final hormone product.
Plant Hormone Hydrolases
As with methylated hormones, amino acid-conjugated hormones can be hydrolyzed back to the free hormone and amino acid. The first hydrolase discovered with this activity was identified from a mutant screen of Arabidopsis looking for resistance to IAA-Leu treatment (97). The ILR (IAA-Leu-resistant) protein belongs to the M20 peptidase superfamily, of which there are six other homologs in Arabidopsis (97). Most enzymes in the M20 peptidase family are Zn2+-dependent, but the ILR proteins prefer Mn2+ (98). The crystal structure of an ILR protein from Arabidopsis (i.e. AtILL2) shows the bidomain fold representative of the superfamily (Fig. 3D) (99). The larger domain contains the active site and consists of eight β-strands with α-helical bundles on both sides. The smaller satellite domain adopts an α/β-sandwich topology. In other M20 peptidases, this domain serves as a dimerization interface; however, AtILL2 functions as a monomer (99). Although AtILL2 was crystallized in the absence of metal ions, it contains a canonical histidine-rich metal-binding site (Fig. 3D). Because the crystal structure of AtILL2 is an apoenzyme form, computational ligand docking experiments identified a putative IAA-binding site and suggested that the difference between a leucine and a tyrosine in the site may control access of substrates with smaller versus larger amino acid conjugates, respectively (99). Crystal structures with non-hydrolyzable analogs of the substrates or reactions products along with biochemical analysis of each family member are needed to understand the mechanism of substrate specificity in the plant hormone hydrolases.
Summary
The integration of multiple signaling pathways and hormone responses determines how plants develop and grow; however, the perception of small molecule signals by receptors is only one piece of this biological puzzle. The range of responses controlled by plant hormones requires enzyme action in biosynthetic, storage, degradation, and mobilization pathways and through recognition by cognate receptors (many of which are also enzymes in plants). These processes control the dose and duration of exposure to plant growth regulators.
Recent efforts summarized here provide new molecular insights into how large enzyme families catalyze similar types of modifications on chemically diverse plant growth regulators to alter their biological functions. In addition to the growing biochemical and structural understanding of enzymatic regulation of plant hormones, efforts to elucidate the temporal and spatial expression patterns of hormone-modifying enzymes will provide a physiological context for these reactions. The specialized chemicals that plants use to control their growth are a rich pallet for coloring plant responses to internal and external stimuli.
This work was supported in part by National Science Foundation Grant MCB-1157771.
- IAA
- indole-3-acetic acid
- SA
- salicylic acid
- AdoMet
- S-adenosyl-l-methionine
- ACC
- 1-aminocyclopropane-1-carboxylic acid
- ABA
- abscisic acid
- JA
- jasmonic acid
- JA-Ile
- (+)-7-iso-jasmonoyl-l-isoleucine
- MeIAA
- methyl-IAA
- MES
- methylesterase
- MeSA
- methylsalicylic acid
- At
- A. thaliana
- Vv
- V. vinifera.
REFERENCES
- 1. Santner A., Calderon-Villalobos L. I. A., Estelle M. (2009) Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307 [DOI] [PubMed] [Google Scholar]
- 2. Korasick D. A., Enders T. A., Strader L. C. (2013) Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ludwig-Müller J. (2011) Auxin conjugates: their role for plant development and in the evolution of land plants. J. Exp. Bot. 62, 1757–1773 [DOI] [PubMed] [Google Scholar]
- 4. Ljun K., Hul A. K., Kowalczyk M., Marchant A., Celenza J., Cohen J. D., Sandberg G. (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-4-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 50, 309–332 [DOI] [PubMed] [Google Scholar]
- 5. Woodward A. W., Bartel B. (2005) Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gray W. M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276 [DOI] [PubMed] [Google Scholar]
- 7. Dharmasiri N., Dharmasiri S., Estelle M. (2005) The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445 [DOI] [PubMed] [Google Scholar]
- 8. Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J. S., Jürgens G., Estelle M. (2005) Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 [DOI] [PubMed] [Google Scholar]
- 9. Kepinski S., Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435, 446–451 [DOI] [PubMed] [Google Scholar]
- 10. Wang K. L. C., Li H., Ecker J. R. (2002) Ethylene biosynthesis and signaling networks. Plant Cell 14, S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schaller G. E., Bleecker A. B. (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811 [DOI] [PubMed] [Google Scholar]
- 12. Zhao Q., Guo H. W. (2011) Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol. Plant 4, 626–634 [DOI] [PubMed] [Google Scholar]
- 13. Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 [DOI] [PubMed] [Google Scholar]
- 14. Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S. Y., Howe G. A., Browse J. (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signaling. Nature 448, 661–665 [DOI] [PubMed] [Google Scholar]
- 15. Katsir L., Schilmiller A. L., Staswick P. E., He S. Y., Howe G. A. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U.S.A. 105, 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chini A., Fonseca S., Fernández G., Adie B., Chico J. M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F. M., Ponce M. R., Micol J. L., Solano R. (2007) The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448, 666–671 [DOI] [PubMed] [Google Scholar]
- 17. Hwang I., Sheen J., Müller B. (2012) Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380 [DOI] [PubMed] [Google Scholar]
- 18. Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063 [DOI] [PubMed] [Google Scholar]
- 19. An C., Mou Z. (2011) Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428 [DOI] [PubMed] [Google Scholar]
- 20. Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., Mohan R., Spoel S. H., Tada Y., Zheng N., Dong X. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., De Luca V., Després C. (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647 [DOI] [PubMed] [Google Scholar]
- 22. Nambara E., Marion-Poll A. (2005) Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185 [DOI] [PubMed] [Google Scholar]
- 23. Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J. K., Schroeder J. I., Volkman B. F., Cutler S. R. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pandey S., Nelson D. C., Assmann S. M. (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136, 136–148 [DOI] [PubMed] [Google Scholar]
- 25. Shen Y. Y., Wang X. F., Wu F. Q., Du S. Y., Cao Z., Shang Y., Wang X. L., Peng C. C., Yu X. C., Zhu S. Y., Fan R. C., Xu Y. H., Zhang D. P. (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826 [DOI] [PubMed] [Google Scholar]
- 26. Raghavendra A. S., Gonugunta V. K., Christmann A., Grill E. (2010) ABA perception and signaling. Trends Plant Sci. 15, 395–401 [DOI] [PubMed] [Google Scholar]
- 27. Hedden P., Thomas S. G. (2012) Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25 [DOI] [PubMed] [Google Scholar]
- 28. Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T. Y., Hsing Y. I., Kitano H., Yamaguchi I., Matsuoka M. (2005) Gibberellin insensitive DWARF1 encodes a soluble receptor for gibberellin. Nature 437, 693–698 [DOI] [PubMed] [Google Scholar]
- 29. Nakajima M., Shimada A., Takashi Y., Kim Y. C., Park S. H., Ueguchi-Tanaka M., Suzuki H., Katoh E., Iuchi S., Kobayashi M., Maeda T., Matsuoka M., Yamaguchi I. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46, 880–889 [DOI] [PubMed] [Google Scholar]
- 30. Davière J. M., Achard P. (2013) Gibberellin signaling in plants. Development 140, 1147–1151 [DOI] [PubMed] [Google Scholar]
- 31. Fujioka S., Yokota T. (2003) Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164 [DOI] [PubMed] [Google Scholar]
- 32. He Z., Wang Z. Y., Li J., Zhu Q., Lamb C., Ronald P., Chory J. (2000) Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 28, 2360–2363 [DOI] [PubMed] [Google Scholar]
- 33. Friedrichsen D. M., Joazerio C. A. P., Li J., Hunter T., Chory J. (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 123, 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Z. Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383 [DOI] [PubMed] [Google Scholar]
- 35. Clouse S. D. (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23, 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu J. Y., Sae-Seaw J., Wang Z. Y. (2013) Brassinosteroid signalling. Development 140, 1615–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Humphrey A. J., Beale M. H. (2006) Strigol: biogenesis and physiological activity. Phytochemistry 67, 636–640 [DOI] [PubMed] [Google Scholar]
- 38. Ruyter-Spira C., Al-Babili S., van der Krol S., Bouwmeester H. (2013) The biology of strigolactones. Trends Plant Sci. 18, 72–83 [DOI] [PubMed] [Google Scholar]
- 39. Hamiaux C., Drummond R. S. M., Janssen B. J., Ledger S. E., Cooney J. M., Newcomb R. D., Snowden K. C. (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 [DOI] [PubMed] [Google Scholar]
- 40. Stirnberg P., Furner I. J., Ottoline Leyser H. M. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94 [DOI] [PubMed] [Google Scholar]
- 41. Waters M. T., Nelson D. C., Scaffidi A., Flematti G. R., Sun Y. K., Dixon K. W., Smith S. M. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 [DOI] [PubMed] [Google Scholar]
- 42. Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth, and development. Ann. Bot. 100, 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wasternack C., Hause B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanaka M., Takei K., Kojima M., Sakakibara H., Mori H. (2006) Auxin controls local cytokinin biosynthesis in the nodal stem in apical dominance. Plant J. 45, 1028–1036 [DOI] [PubMed] [Google Scholar]
- 45. Went F. W. (1926) On growth-accelerating substances in the coleoptile of Avena sativa. Proc. Kon. Ned. Akad. Wet. 30, 10–19 [Google Scholar]
- 46. Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Hanada A., Yaeno T., Shirasu K., Yao H., McSteen P., Zhao Y., Hayashi K., Kamiya Y., Kasahara H. (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011) Conversion of tryptophan to indole-3-acetic acid by tryptophan aminotransferases of Arabidopsis and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bajguz A., Piotrowska A. (2009) Conjugates of auxin and cytokinin. Phytochemistry 70, 957–969 [DOI] [PubMed] [Google Scholar]
- 49. Yalpani N., Silverman P., Wilson T. M. A., Kleier D. A., Raskin I. (1991) Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3, 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee H. I., León J., Raskin I. (1995) Biosynthesis and metabolism of salicylic acid. Proc. Natl. Acad. Sci. U.S.A. 92, 4076–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Silverman P., Seskar M., Kanter D., Schweizer P., Metraux J. P., Raskin I. (1995) Salicylic acid in rice biosynthesis, conjugation, and possible role. Plant Physiol. 108, 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yalpani N., Leon J., Lawton M. A., Raskin I. (1993) Pathway of salicylic acid biosynthesis in healthy and virus-inoculated tobacco. Plant Physiol. 103, 315–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565 [DOI] [PubMed] [Google Scholar]
- 54. Serino L., Reimmann C., Baur H., Beyeler M., Visca P., Haas D. (1995) Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 249, 217–228 [DOI] [PubMed] [Google Scholar]
- 55. Piotrowska A., Bajguz A. (2011) Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 72, 2097–2112 [DOI] [PubMed] [Google Scholar]
- 56. Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009) (+)-7-iso-Jasmonoyl-l-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350 [DOI] [PubMed] [Google Scholar]
- 57. Wasternack C., Kombrink E. (2010) Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 5, 63–77 [DOI] [PubMed] [Google Scholar]
- 58. Frébort I., Kowalska M., Hluska T., Frébortová J., Galuszka P. (2011) Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 62, 2431–2452 [DOI] [PubMed] [Google Scholar]
- 59. Koo A. J., Howe G. A. (2012) Catabolism and deactivation of the lipid-derived hormone jasmonyl-isoleucine. Front. Plant Sci. 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Demole E., Lederer E., Mercier D. (1962) Isolation and determination of the structure of methyl jasmonate, a constituent odor characteristic of jasmine oil. Helv. Chim. Acta 45, 675–685 [Google Scholar]
- 61. Pott M. B., Hippauf F., Saschenbrecker S., Chen F., Ross J., Kiefer I., Slusarenko A., Noel J. P., Pichersky E., Effmert U., Piechulla B. (2004) Biochemical and structural characterization of benzenopid carboxyl methyltransferases involved in floral scent production in Stephanotis floribunda and Nicotiana suaveolens. Plant Physiol. 135, 1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L. J. (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17, 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Seo H. S., Song J. T., Cheong J. J., Lee Y. H., Lee Y. W., Hwang I., Lee J. S., Choi Y. D. (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. U.S.A. 98, 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Varbanova M., Yamaguchi S., Yang Y., McKelvey K., Hanada A., Borochov R., Yu F., Jikumaru Y., Ross J., Cortes D., Ma C. J., Noel J. P., Mander L., Shulaev V., Kamiya Y., Rodermel S., Weiss D., Pichersky E. (2007) Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19, 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zubieta C., Ross J. R., Koscheski P., Yang Y., Pichersky E., Noel J. P. (2003) Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15, 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McCarthy A. A., McCarthy J. G. (2007) The structure of two N-methyltransferases from the caffeine biosynthetic pathway. Plant Physiol. 144, 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhao N., Ferrer J. L., Moon H. S., Kapteyn J., Zhuang X., Hasebe M., Stewart C. N., Jr., Gang D. R., Chen F. (2012) A SABATH methyltransferase from the moss Physcomitrella patens catalyzes S-methylation of thiols and has a role in detoxification. Phytochemistry 81, 31–41 [DOI] [PubMed] [Google Scholar]
- 68. Chen F., D'Auria J. C., Tholl D., Ross J. R., Gershenzon J., Noel J. P., Pichersky E. (2003) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36, 577–588 [DOI] [PubMed] [Google Scholar]
- 69. Zhao N., Ferrer J. L., Ross J., Guan J., Yang Y., Pichersky E., Noel J. P., Chen F. (2008) Structural, biochemical, and phylogenetic analyses suggest that indole-3-acetic acid methyltransferase is an evolutionarily ancient member of the SABATH family. Plant Physiol. 146, 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang Y., Xu R., Ma C. J., Vlot A. C., Klessig D. F., Pichersky E. (2008) Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 147, 1034–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Forouhar F., Yang Y., Kumar D., Chen Y., Fridman E., Park S. W., Chiang Y., Acton T. B., Montelione G. T., Pichersky E., Klessig D. F., Tong L. (2005) Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in innate immunity. Proc. Natl. Acad. Sci. U.S.A. 102, 1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vlot A. C., Liu P. P., Cameron R. K., Park S. W., Yang Y., Kumar D., Zhou F., Padukkavidana T., Gustafsson C., Pichersky E., Klessig D. F. (2008) Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 56, 445–456 [DOI] [PubMed] [Google Scholar]
- 73. Kumar D., Klessig D. F. (2003) High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic-acid stimulated lipase activity. Proc. Natl. Acad. Sci. U.S.A. 100, 16101–16106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Christ B., Schelbert S., Aubry S., Süssenbacher I., Müller T., Kräutler B., Hörtensteiner S. (2012) MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol. 158, 628–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. LeClere S., Tellez R., Rampey R. A., Matsuda S. P. T., Bartel B. (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452 [DOI] [PubMed] [Google Scholar]
- 76. Staswick P. E. (2009) The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 150, 1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Staswick P. E., Tiryaki I., Rowe M. L. (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sheard L. B., Tan X., Mao H., Withers J., Ben-Nissan G., Hinds T. R., Kobayashi Y., Hsu F. F., Sharon M., Browse J., He S. Y., Rizo J., Howe G. A., Zheng N. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hagen G., Guilfoyle T. J. (1985) Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 5, 1197–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Okrent R. A., Wildermuth M. C. (2011) Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Mol. Biol. 76, 489–505 [DOI] [PubMed] [Google Scholar]
- 81. Westfall C. S., Herrmann J., Chen Q., Wang S., Jez J. M. (2010) Modulating plant hormones by enzyme action: the GH3 family of acyl acid amido synthetases. Plant Signal. Behav. 5, 1607–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Staswick P. E., Su W., Howell S. H. (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. U.S.A. 89, 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gulick A. M. (2009) Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4, 811–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen Q., Zhang B., Hicks L. M., Wang S., Jez J. M. (2009) A liquid chromatography-tandem mass spectrometry-based assay for indole-3-acetic acid-amido synthetase. Anal. Biochem. 390, 149–154 [DOI] [PubMed] [Google Scholar]
- 85. Chen Q., Westfall C. S., Hicks L. M., Wang S., Jez J. M. (2010) Kinetic basis for the conjugation of auxin by a GH3 family indole-acetic acid-amido synthetase. J. Biol. Chem. 285, 29780–29786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ding X., Cao Y., Huang L., Zhao J., Xu C., Li X., Wang S. (2008) Activation of the indole-3-acetic acid-amido synthetase GH3–8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20, 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang S. W., Li C. H., Cao J., Zhang Y. C., Zhang S. Q., Xia Y. F., Sun D. Y., Sun Y. (2009) Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 151, 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Takase T., Nakazawa M., Ishikawa A., Kawashima M., Ichikawa T., Takahashi N., Shimada H., Manabe K., Matsui M. (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37, 471–483 [DOI] [PubMed] [Google Scholar]
- 89. Nakazawa M., Yabe N., Ichikawa T., Yamamoto Y. Y., Yoshizumi T., Hasunuma K., Matsui M. (2001) DFL1, an axuin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221 [DOI] [PubMed] [Google Scholar]
- 90. Park J. E., Park J. Y., Kim Y. S., Staswick P. E., Jeon J., Yun J., Kim S. Y., Kim J., Lee Y. H., Park C. M. (2007) GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282, 10036–10046 [DOI] [PubMed] [Google Scholar]
- 91. Khan S., Stone J. M. (2007) Arabidopsis thaliana GH3.9 influences primary root growth. Planta 226, 21–34 [DOI] [PubMed] [Google Scholar]
- 92. Staswick P. E., Serban B., Rowe M., Tiryaki I., Maldonado M. T., Maldonado M. C., Suza W. (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nobuta K., Okrent R. A., Stoutemyer M., Rodibaugh N., Kempema L., Wildermuth M. C., Innes R. W. (2007) The GH3 acyl adenylase family member PBS3 regulates salicylic acid-dependent defense responses in Arabidopsis. Plant Physiol. 144, 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Okrent R. A., Brooks M. D., Wildermuth M. C. (2009) Arabidopsis GH3.12 (PBS3) conjugates amino acids to 4-substituted benzoates and is inhibited by salicylate. J. Biol. Chem. 284, 9742–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Westfall C. S., Zubieta C., Herrmann J., Kapp U., Nanao M. H., Jez J. M. (2012) Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 336, 1708–1711 [DOI] [PubMed] [Google Scholar]
- 96. Peat T. S., Böttcher C., Newman J., Lucent D., Cowieson N., Davies C. (2012) Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell 24, 4525–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Bartel B., Fink G. R. (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748 [DOI] [PubMed] [Google Scholar]
- 98. Davies R. T., Goetz D. H., Lasswell J., Anderson M. N, Bartel B. (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bitto E., Bingman C. A., Bittova L., Houston N. L., Boston R. S., Fox B. G., Phillips G. N., Jr. (2009) X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana. Proteins 74, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]