Background: CAO domain structure has changed during evolution.
Results: Micromonas CAO is split into two proteins, both of which are required for chlorophyll b synthesis.
Conclusion: Establishment of a new light-harvesting system has been accompanied by changes in the CAO structure during evolution.
Significance: This is the first report showing the relationship between chlorophyll metabolism and structures of the light-harvesting system.
Keywords: Chloroplast, Enzymes, Evolution, Photosynthesis, Photosynthetic Pigments, CAO, Chlorophyll, Photosystem
Abstract
Chlorophyll b is found in photosynthetic prokaryotes and primary and secondary endosymbionts, although their light-harvesting systems are quite different. Chlorophyll b is synthesized from chlorophyll a by chlorophyllide a oxygenase (CAO), which is a Rieske-mononuclear iron oxygenase. Comparison of the amino acid sequences of CAO among photosynthetic organisms elucidated changes in the domain structures of CAO during evolution. However, the evolutionary relationship between the light-harvesting system and the domain structure of CAO remains unclear. To elucidate this relationship, we investigated the CAO structure and the pigment composition of chlorophyll-protein complexes in the prasinophyte Micromonas. The Micromonas CAO is composed of two genes, MpCAO1 and MpCAO2, that possess Rieske and mononuclear iron-binding motifs, respectively. Only when both genes were introduced into the chlorophyll b-less Arabidopsis mutant (ch1-1) was chlorophyll b accumulated, indicating that cooperation between the two subunits is required to synthesize chlorophyll b. Although Micromonas has a characteristic light-harvesting system in which chlorophyll b is incorporated into the core antennas of reaction centers, chlorophyll b was also incorporated into the core antennas of reaction centers of the Arabidopsis transformants that contained the two Micromonas CAO proteins. Based on these results, we discuss the evolutionary relationship between the structures of CAO and light-harvesting systems.
Introduction
Photosynthesis consists of two processes as follows: one involves harvesting light energy and producing ATP and NADPH, and the second involves the fixation of CO2 to sugar using ATP and NADPH. Light energy is harvested by photosynthetic pigments such as chlorophyll, carotenoid, and phycobilin (1, 2). Most oxygenic photosynthetic organisms contain chlorophyll a, which is involved in harvesting light energy in antenna systems and in driving electron transfer in the reaction centers (3), although Prochlorococcus contains 8-vinyl-chlorophyll a, and Acaryochloris contains chlorophyll d in their reaction centers. In addition to chlorophyll a, some photosynthetic organisms have an additional pigment that exists in the peripheral antenna complex and plays an important role in absorbing a broad light spectrum for photosynthesis (4). For example, cyanobacteria and red algae contain phycobilins, which form a large complex called the phycobilisome (5). Fucoxanthin is a major pigment in brown algae and exists in fucoxanthin-chlorophyll-protein complexes (6). The acquisition of these pigments has played an essential role in environmental adaptation by establishing new light-harvesting systems.
Chlorophyll b is widely distributed in prokaryotes and primary and secondary endosymbionts (7). However, the light-harvesting systems are quite different among these organisms, although they all contain chlorophyll b as a major light-harvesting pigment. In Prochlorococcus, 8-vinyl-chlorophyll b binds to prochlorophyte chlorophyll-binding protein (Pcb)2 (8) and forms the photosystem (PS)I-Pcb and PSII-Pcb supercomplexes (9). Pcb belongs to the six-transmembrane helix family, which includes CP43 and CP47 and the iron-stress-induced protein A (IsiA) (10). In land plants and most green algae, chlorophyll b binds to Lhc proteins containing three transmembrane helices that associate with the core antennas of reaction centers of PSI and PSII (11–13). In the prasinophyte order Mamiellales, the content of chlorophyll b in the light-harvesting system is extremely high (14) compared with land plants and green algae; however, the distribution of chlorophyll b in their light-harvesting system has not been well studied. In addition, prasinophyte photosystems contain the prasinophyte-specific light-harvesting chlorophyll a/b-protein complex (LHC) type designated LHCP (14), which binds chlorophyll a, chlorophyll b, and chlorophyll c-like pigments and other carotenoids.
Chlorophyll b is synthesized from chlorophyll a by chlorophyllide a oxygenase (CAO) (15). CAO catalyzes two successive oxygenation reactions, converting a methyl group at the C7 position to a formyl group via a hydroxymethyl group to generate chlorophyll b (16). CAO is not structurally related to bciD, which is involved in the C7-methyl oxidation (formylation) in bacteriochlorophyll e synthesis (17). CAO is a unique enzyme, and all the chlorophyll b-containing organisms, without exception, have CAO. However, the structure of CAO is variable among photosynthetic organisms, in contrast to other chlorophyll metabolic enzymes whose structures are largely conserved (18). Eukaryotic CAOs, except for that of Mamiellales, consist of three domains as follows: the A, B, and C domains in order from the N terminus (18). The A domain controls the accumulation of the Arabidopsis CAO (AtCAO) protein in response to the presence of chlorophyll b (19, 20). The C domain is the catalytic domain, which contains a Rieske center and a mononuclear iron-binding motif. The B domain might serve as a linker between the A and C domains. The prokaryotic Prochlorothrix hollandica CAO (PhCAO) has high sequence similarity to higher plant CAOs, but it only contains the sequence corresponding to the catalytic domain and has no regulatory domain (21). Prochlorococcus CAOs also lack a regulatory domain and have low sequence homology to other CAOs, although they contain Rieske and mononuclear iron-binding motifs (22). The low sequence similarity is partly because Prochlorococcus uses 8-vinyl-chlorophyll instead of chlorophyll and therefore had to adapt to fit 8-vinyl-chlorophyll. Mamiellales has a unique CAO that is quite different from others. The complete sequence of the genomes of the Mamiellales Micromonas and Ostreococcus revealed the presence of two genes that have sequence similarity to CAO (23). Interestingly, these two genes conserve either the Rieske center or mononuclear iron-binding motif, suggesting that these two gene products cooperate in the synthesis of chlorophyll b, although no experimental evidence for this had not been demonstrated so far.
As mentioned previously, the evolution of the light-harvesting systems of chlorophyll b-containing organisms has been accompanied by changes in the CAO structure. In this study, we examined the light-harvesting systems of Micromonas by BN-PAGE and native-green gel followed by pigment analysis and found that Micromonas has a characteristic light-harvesting system in which chlorophyll b is incorporated into the core antennas of reaction centers. When both Micromonas CAO genes were simultaneously introduced into the Arabidopsis chlorophyll b-less (ch1-1) mutant, chlorophyll b was incorporated into the core antennas of reaction centers, indicating that the light-harvesting systems of the transgenic plant shared a common structure with that of Micromonas. Based on these results, we discuss the co-evolution of CAO and the light-harvesting systems.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Arabidopsis thaliana (Columbia ecotype) was grown in a chamber equipped with white fluorescent lamps under a 16-h photoperiod at a light intensity of 70 μmol photons m−2 s−1 at 23 °C. The Arabidopsis ch1-1 mutant (24) was used as the chlorophyll b-less plant in this study. Micromonas pusilla (CCMP1545) was inoculated into 100 ml of liquid L1 medium (25) in a 300-ml Erlenmeyer flask. All culture experiments with M. pusilla were performed at 23 °C under a 16-h photoperiod of 15 μmol photons m−2 s−1. Artificial genes encoding MpCAO1 (MicpuC2 16022) and MpCAO2 (MicpuC2 60555) were designed to optimize their codon usage for Arabidopsis (Fig. 3). To construct an MpCAO1-FLAG vector, the full coding region for MpCAO (FeS) was fused with a transit peptide of AtCAO (1–168 bp) and a C-terminal FLAG tag (GCCTCGTCAGTGATAAAACGAGAAGACTACAA) and was subcloned into the pGreenII vector. To construct an MpCAO2-HA vector, the full coding region for MpCAO2 was fused with a transit peptide of AtCAO (1–168 bp) and a C-terminal HA tag (GCCTCGTCAGTGATAAAACGAGAAGACTACAA) and was subcloned into the pGreenII vector. The resulting plasmids were introduced into ch1-1 plants via Agrobacterium tumefaciens-mediated transformation. We named these mutant lines MpCAO1 and MpCAO2, respectively. Primary transformants were selected on agar plates containing 50 mg/liter kanamycin or 20 mg/liter hygromycin, respectively. To construct a PhCAO vector, the full coding region for a P. hollandica CAO gene (PhCAO) was fused with a transit peptide of AtCAO (1–168 bp) and subcloned into the pGreenII vector (21). To construct a BCFLAG vector, the coding region for the B and C domains of AtCAO (Pro171–Gly535) was fused with a transit peptide of AtCAO (1–168 bp) and subcloned into the pGreenII vector. In addition, we generated double mutants by co-transformation (MpCAO1+MpCAO2–1) and by genetic crossing (MpCAO1+MpCAO2–2). Confirmation of genotypes was performed by PCR. Genomic DNA was extracted as described in the previous report (26). The primers used for amplification of MpCAO1 and MpCAO2 are as follows: MpCAO1Fw, 5′-ATGAGATGTGACGCTGAAGGA-3′; MpCAO1Rv, 5′-TTACTTATCATCGTCATCTTT-3′; MpCAO2Fw, 5′-ATGGCTCCAGAAGTATCTTCC-3′; and MpCAO2Rv, 5′-TTATGCGTAATCAGGAACATC-3′. The PCR products were separated on ethidium bromide-stained 2.0% agarose gels.
FIGURE 3.
Sequences of synthetic genes of MpCAO1 and MpCAO2.
Expression and Purification of Recombinant AtCAO
The coding region of AtCAO lacking its transit peptide was amplified by PCR (KOD FX NEO DNA polymerase; Toyobo, Japan) using the following primers: 5′-TAGGCATATGGAGCTCTTGTTTGATGTGGAGGATCCTA-3′ (the underlined region is an engineered SacI site) and 5′-CCTATCTAGACTGCAGTTAGCCGGAGAAAGGTAGTTTA-3′ (the underlined region is an engineered PstI site) and cloned into the SacI/PstI sites of the pCold ProS2 vector (Takara, Japan). The recombinant protein obtained from this plasmid possesses a hexahistidine tag at the N terminus. The expression plasmid was introduced into Escherichia coli C41 cells (Avidis SA, France). The transformed cells were grown at 37 °C in 1 liter of Luria-Bertani medium containing ampicillin (50 mg/liter) and 0.1 mm ammonium ferric citrate until the absorbance at 600 nm reached 1.5 (27). The expression of the recombinant AtCAO gene was induced with 0.3 mm isopropyl β-d-thiogalactopyranoside for 18 h at 18 °C. After incubation, the culture was harvested by centrifugation, and the collected cells were resuspended in buffer (20 mm imidazole, 500 mm NaCl, and 20 mm Tris-HCl (pH 7.9)) and disrupted by sonication. Cells were then centrifuged at 21,600 × g for 5 min at 4 °C to remove cell debris. The soluble fraction containing the recombinant protein was purified with a nickel column (His-Trap HP kit; GE Healthcare), according to the manufacturer's protocol. The eluent fractions were concentrated to 0.05 ml by a centrifugal filter device (Amicon Ultra-15, 30,000 MWCO; Millipore) and used for two-dimensional BN/SDS-PAGE.
Isolation of Thylakoid Membranes
Arabidopsis thylakoid membranes (wild type, MpCAO1+ MpCAO2, BCFLAG, and PhCAO) were prepared from the leaves of 5-week-old plants as described previously (28). Micromonas thylakoid membranes were prepared from 10-day-old cells. Cells were harvested by centrifugation at 2,500 × g in growth phase and suspended in 50 mm Tricine/NaOH (pH 8.0). The cells were broken with glass beads (100 μm in diameter) at 4 °C using a Mini-Bead Beater (Waken B Tech, Japan). After removal of the beads and unbroken cells, the supernatant was centrifuged at 21,600 × g for 5 min at 4 °C. The green pellets were washed with 5 mm EDTA-2Na three times and used as Micromonas thylakoid membranes.
Native Green Gel Electrophoresis
Thylakoid membranes of wild-type and MpCAO1 Arabidopsis and M. pusilla CCMP1545 were solubilized in 1.0% SDS at 4 °C and centrifuged at 21,600 × g for 5 min at 4 °C. The supernatants were applied onto a 8% polyacrylamide disk gel (5 mm in diameter) containing 0.1% SDS. Electrophoresis was carried out at 4 °C for ∼1.5 h at 0.5 mA/tube according to the method of Anderson et al. (29), provided that the upper and lower reservoir buffer contained 0.1% SDS.
One-dimensional SDS-PAGE
The leaf (15 mg flesh weight) was solubilized by SDS extraction buffer (30) and separated onto slab gels containing 14% polyacrylamide using the Laemmli buffer system (31). For the immunoblot analysis of PsaA/PsaB (CP1), the leaves were separated on 14% polyacrylamide gels that contained 4 m urea.
Two-dimensional SDS/SDS-PAGE
The total leaf proteins were extracted from the leaf (100 mg flesh weight) of 25-day-old plants (wild type, MpCAO1+MpCAO2) using LDS sample buffer (63 mm Tris-HCl (pH 6.8), 10% glycerol, and 0.5% LDS). After centrifugation at 18,800 × g for 5 min at 4 °C, the supernatants were loaded onto an SDS-polyacrylamide gel (14% acrylamide gel) at 4 °C for 4 h at 6 mA using the Laemmli buffer system (31). For the second dimension using SDS-PAGE, the SDS gel strips were soaked for 1 h at room temperature in a solution that contained 1% (w/v) SDS and 1% (v/v) 2-mercaptoethanol and run on a second dimension SDS-polyacrylamide gel (14% polyacrylamide gels that contained 4 m urea) using the Laemmli buffer system (31). After the second dimension SDS-PAGE, gels were stained with the Pierce Silver Stain kit for mass spectrometry (Thermo Fisher Scientific, Japan) according to instructions.
Two-dimensional BN/SDS-PAGE
BN-PAGE was performed essentially according to the method described by Takabayashi et al. (28). Briefly, the thylakoid membrane proteins (which corresponded to 5 μg of Chl) of Arabidopsis wild-type, MpCAO1+MpCAO2, BCFLAG, and PhCAO plants or the purified recombinant AtCAO proteins (which corresponded to 4 μg of protein) were suspended in ice-cold resuspension buffer A that contained 50 mm imidazole-HCl (pH 7.0), 20% glycerol, 5 mm 6-aminocaproic acid, and 1 mm EDTA-2Na and were then solubilized with 1% (w/v) β-dodecyl maltoside on ice for 5 min. For the second dimension using SDS-PAGE, the BN gel strips were soaked for 1 h at 37 °C in a solution that contained 1% (w/v) SDS and 1% (v/v) 2-mercaptoethanol and run on a second dimension SDS-polyacrylamide gel (14% polyacrylamide) using the Laemmli buffer system (31).
Immunoblot Analysis
Immunoblot analysis was performed essentially according to the method described by Takabayashi et al. (28). The transferred proteins were detected with an anti-AtCAO rabbit primary antibody (19), an anti-PsaA/PsaB (CP1) antibody (32), an anti-CP47 antibody (Agrisera, Sweden), an anti-Lhcb2 antibody (Agrisera, Sweden), an anti-FLAG antibody (Sigma), an anti-HA antibody (Funakoshi, Japan), and Penta-His HRP conjugate kit (Qiagen, Japan).
Co-immunoprecipitation
The thylakoid membrane proteins (which corresponded to 5 μg of Chl) of Arabidopsis wild-type and MpCAO1+MpCAO2 plants were suspended in ice-cold resuspension buffer A and solubilized with 1% (w/v) β-dodecyl maltoside on ice for 5 min. Extracts were incubated with anti-DYKDDDDK beads (Wako, Japan) and anti-HA beads (Wako, Japan) for 8 h at 4 °C. Supernatants were discarded, and beads were washed with buffer A three times. Bound complexes were eluted with SDS extraction buffer for 10 min at 100 °C. After centrifugation at 21,600 × g for 5 min at 4 °C, the supernatants were separated on slab gels containing 14% polyacrylamide.
Pigment Determination
Leaves were weighed and pulverized in acetone using the ShakeMaster grinding apparatus (BioMedical Science, Japan), and the extracts were centrifuged at 21,600 × g for 5 min at 4 °C. The supernatant was loaded onto a Symmetry C8 column (4.6 × 150 mm; Waters, Japan) as described previously (33). The elution profiles were monitored by measuring the absorbance at 650 nm using a diode array detector (SPD-M20A, Shimadzu, Japan), and the pigments were identified by their retention times and spectral patterns. Pigment quantification was performed using the areas of the peaks, described previously (34). All reported Chl quantities were the means of three biologically independent samples.
Multiple Alignment of Amino Acid Sequences
The amino acid sequences listed below were retrieved from GenBankTM (www.ncbi.nlm.nih.gov) and Joint Genome Initiative (jgi.doe.gov). The amino acid sequences were aligned with the ClustalW program (35) via the BioEdit program (36).
Accession Numbers
The amino acid sequences used in this work are as follows. GenBankTM identification numbers: A. thaliana (At), AtCAO,15219408; Chlamydomonas reinhardtii (Cr), CrCAO,159463890; P. hollandica (Ph), PhCAO, 38488612; M. pusilla (Mp) protein IDs are from the Joint Genome Initiative: MpCAO (Fe-S), 16022 and MpCAO (Fe), 60555.
RESULTS
Diversity of CAO Domain Structure in Chlorophyll b-containing Organisms
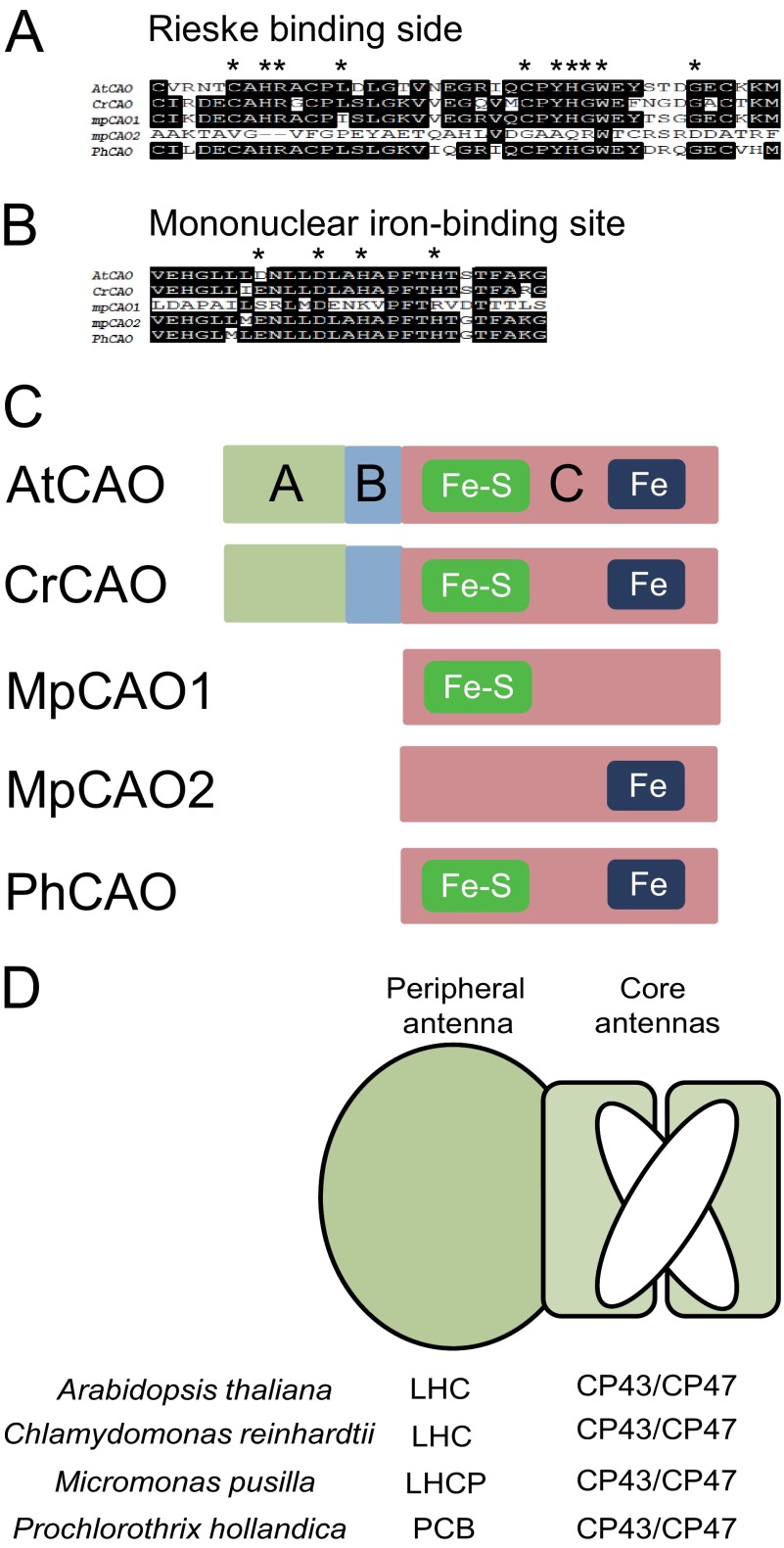
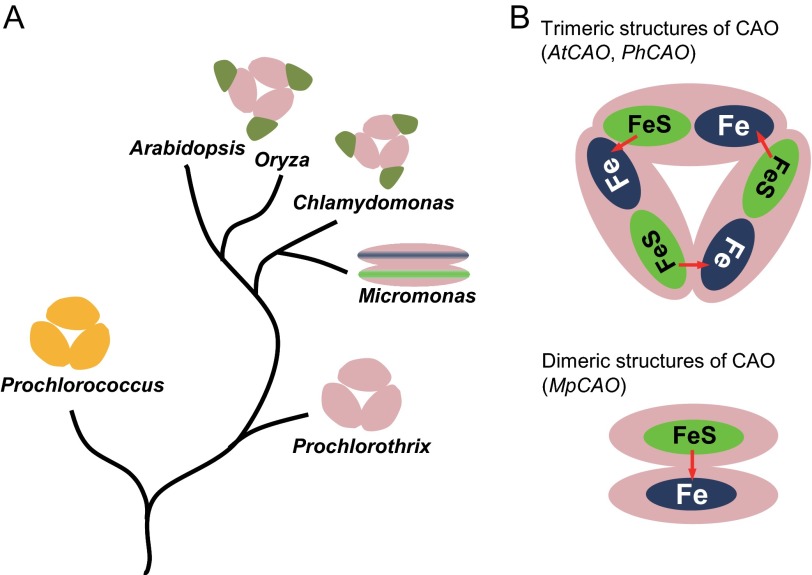
CAOs of vascular plants and most of the green algae have the same domain structure, which consists of the regulatory domain (A domain), catalytic domain (C domain), and the linker (B domain) (Fig. 1C). The amino acid sequence of the B domain is quite different, likely because this domain has no function besides linking the A and C domains (18). Sequence variations are observed in the A domain between vascular plants and the green algae Chlamydomonas (Fig. 2), likely due to different regulatory mechanisms of chlorophyll b synthesis. Prochlorophyte and Mamiellales CAOs have no sequence corresponding to the regulatory and linker domains (Fig. 1C). The amino acid sequences of the catalytic domains of CAOs are highly conserved except for the Prochlorococcus (22) and Mamiellales CAOs (Fig. 2). In Micromonas, CAO is composed of two subunits, one (MpCAO1) that contains only a Rieske center motif and another (MpCAO2) that contains only a mononuclear iron-binding motif (Fig. 1, A and B).
FIGURE 1.
Comparison of the domain structure of the CAO protein among photosynthetic organisms. A, comparison of amino acid sequences of the Rieske center in CAO proteins. B, comparison of amino acid sequences of the mononuclear iron-binding motif in CAO proteins. In both panels, identical residues are shown in white type on a black background. C, domain structure of CAO proteins. Rieske center (Fe-S) and mononuclear iron (Fe)-binding motif are in the C domain. AtCAO, A. thaliana CAO; CrCAO, C. reinhardtii CAO; MpCAO1, M. pusilla CAO (Fe-S); MpCAO2, M. pusilla CAO (Fe); PhCAO, P. hollandica CAO. D, schematic drawing of core/peripheral antenna complexes of Arabidopsis, Chlamydomonas, Micromonas, and Prochlorothrix.
FIGURE 2.
Multiple amino acid sequence alignment of CAO proteins. Identical residues are shown in white type on a black background. The AtCAO sequences corresponding to the A domain, B domain, and C domain are shown. Asterisks and closed squares show binding sites of Rieske center and mononuclear iron-binding motif, respectively.
Cooperation between MpCAO1 and MpCAO2 in Chlorophyll b Synthesis
CAO, which contains both Rieske and mononuclear iron-binding motifs, is not found in the Micromonas or Ostreococcus genomes, and MpCAO1 and MpCAO2 are the only genes that have sequence similarity to CAO in these organisms. This suggests that MpCAO1 and MpCAO2 are responsible for chlorophyll b synthesis in these organisms, although the structure of these proteins is largely different from other CAOs. Introduction of a gene to chlorophyll b-less organisms or mutants is a convenient method to determine whether the gene in question is responsible for chlorophyll b synthesis or not (21, 37). However, the GC content of the Micromonas genes is high (25), and the gene is not thought to be efficiently translated in Arabidopsis. Therefore, the codons of MpCAO1 and MpCAO2 were optimized for expression in Arabidopsis (Fig. 3) and fused with FLAG and HA tags, respectively.
The synthetic DNAs encoding these proteins were expressed under the control of the cauliflower mosaic virus 35S promoter in the Arabidopsis ch1-1 mutant, which lacks chlorophyll b. PCR experiments showed that the MpCAO1 and MpCAO2 genes were successfully introduced into the Arabidopsis ch1-1 mutant (Fig. 4B). Fig. 4A shows the plants 25 days after germination. The ch1-1 mutant plants were small compared with the WT and exhibited a pale green color due to the lack of chlorophyll b. The transgenic plants with either MpCAO1 or MpCAO2 exhibited the same phenotype as the ch1-1 mutant. When both MpCAO1 and MpCAO2 genes were introduced into the ch1-1 mutant, the size and the color of the transgenic plants became similar to that of the WT. Next, we examined the accumulation of the transgene products by immunoblotting using antibodies against the FLAG (MpCAO1) or HA (MpCAO2) tags (Fig. 4C). As shown in Fig. 4, the MpCAO1 and MpCAO2 proteins accumulated in the transgenic plants harboring MpCAO1 or MpCAO2 genes, respectively. Neither MpCAO1 nor MpCAO2 was detected in the WT and the ch1-1 mutant plants, and two lines of MpCAO1+MpCAO2 plants accumulated both proteins.
FIGURE 4.
Generation of MpCAO overexpression of Arabidopsis lines. A, phenotypes of wild-type and transgenic lines. 1, wild type; 2, MpCAO1+MpCAO2–1; 3, MpCAO1+MpCAO2–2; 4, ch1-1; 5, MpCAO1; and 6, MpCAO2. B, confirmation of transgenic lines by PCR. C, confirmation of transgenic lines by immunoblot analysis. The MpCAO1-FLAG and MpCAO2-HA proteins were detected using anti-FLAG or anti-HA antibodies, respectively. Lane 1, wild type; lane 2, ch1-1; lane 3, MpCAO1; lane 4, MpCAO2; lane 5, MpCAO1+MpCAO2–1; lane 6, MpCAO1+MpCAO2–2.
Next, we determined the chlorophyll content of the transgenic plants by HPLC (Table 1). The chlorophyll a/b ratio of the WT was 3.03, whereas chlorophyll b was not detected in the ch1-1 mutant as reported previously. The chlorophyll content of ch1-1 was lower than WT due to the deficiency in LHCII (28), which is a major chlorophyll-binding complex. The chlorophyll content of the transgenic plants that have either MpCAO1 or MpCAO2 was similar to that of the ch1-1 mutant, indicating that neither MpCAO1 nor MpCAO2 alone can synthesize chlorophyll b. In contrast, a large amount of chlorophyll b accumulated when MpCAO1 and MpCAO2 were co-introduced. These results clearly indicate that cooperation of MpCAO1 and MpCAO2 is indispensable for the synthesis of chlorophyll b. It should be noted that the chlorophyll a/b ratio of the MpCAO1+MpCAO2 plants was significantly lower than that of the WT. It was reported that in Arabidopsis, the chlorophyll a/b ratio does not significantly change even when full-length AtCAO is overexpressed because the A domain strictly regulates chlorophyll b synthesis by a negative feedback mechanism. Therefore, the expression of Micromonas MpCAO1 and MpCAO2 might not be properly regulated in Arabidopsis.
TABLE 1.
Chlorophyll content of wild-type and transgenic Arabidopsis plants
The value given is the mean ± S.D. of three experiments. ND stands for not detected; FW is flesh weight.
| Sample | Chl a | Chl b | Total Chl | Chl a/b |
|---|---|---|---|---|
| μg/g FW | ||||
| WT | 1053.1 ± 47.8 | 347.8 ± 14.9 | 1400.9 ± 62.1 | 3.03 ± 0.04 |
| ch1-1 | 760.6 ± 67.6 | ND | 760.6 ± 67.6 | ND |
| MpCAO1 | 671.4 ± 16.4 | ND | 671.4 ± 16.4 | ND |
| MpCAO2 | 650.1 ± 37.8 | ND | 650.1 ± 37.8 | ND |
| MpCAO1+MpCAO2-1 | 737.9 ± 68.9 | 445.5 ± 30.5 | 1182.5 ± 68.9 | 1.65 ± 0.03 |
| MpCAO1+MpCAO2-2 | 747.6 ± 69.9 | 490.0 ± 31.0 | 1237.6 ± 69.9 | 1.53 ± 0.09 |
Subunit Structures of CAO
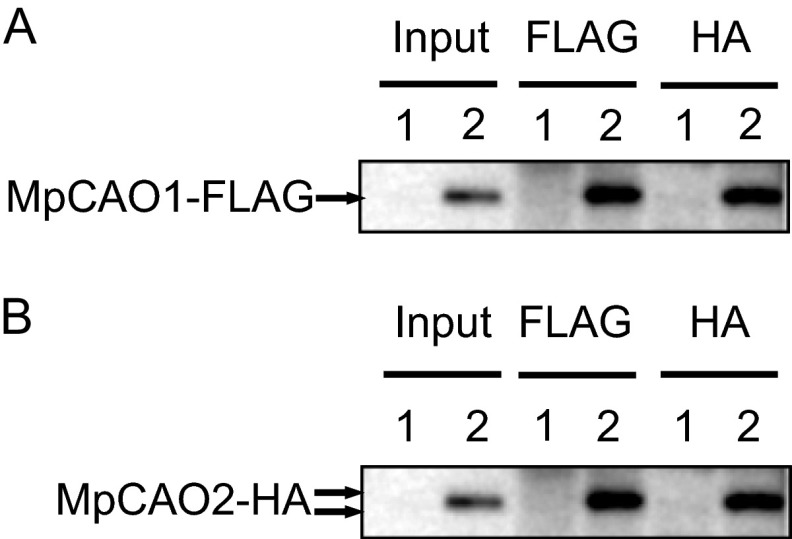
As part of the catalytic activity of CAO, an electron must be transferred to a mononuclear iron from a Rieske center. For efficient electron transfer, it is reasonable to assume that the two subunits, MpCAO1 and MpCAO2, form a complex. To examine this possibility, we carried out a co-immunoprecipitation experiment using anti-FLAG or anti-HA antibodies. MpCAO1-FLAG and MpCAO2-HA were co-expressed in the ch1-1 mutant, and the thylakoid membranes of the transgenic plants were isolated. Solubilized thylakoid membranes were subjected to co-immunoprecipitation with anti-FLAG antibodies. Co-precipitated proteins were eluted and subjected to immunoblot analysis. As shown in Fig. 5A, MpCAO2-HA co-precipitated with the anti-FLAG antibody, indicating that MpCAO2 interacted with MpCAO1. The same result was obtained when MpCAO2 was precipitated by the anti-HA antibody (Fig. 5B). These experiments showed that MpCAO1 and MpCAO2 form a complex.
FIGURE 5.
Co-immunoprecipitation of MpCAO1-FLAG and MpCAO2-HA proteins. The solubilized thylakoid membrane proteins from wild type or MpCAO1+MpCAO2 plants were co-immunoprecipitated with the anti-FLAG antibody (FLAG) or anti-HA antibody (HA) and subjected to SDS-PAGE. Detection of MpCAO1-FLAG and MpCAO2-HA proteins was performed by immunoblot analysis with anti-FLAG antibody (A) or anti-HA antibody (B), respectively. Lane 1, wild type; lane 2, MpCAO1+MpCAO2.
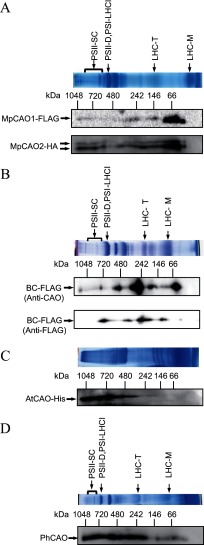
Next, we examined whether MpCAO1 and MpCAO2 form a heterodimer by BN-PAGE followed by immunoblot analysis. Thylakoid membranes were isolated from MpCAO1+MpCAO2 plants and subjected to BN-PAGE followed by immunoblot analysis using anti FLAG or anti-HA tag antibodies. Both MpCAO1 and MpCAO2 bands were found at ∼40 kDa, corresponding to monomeric MpCAO1 and MpCAO2 (Fig. 6A). In addition to these monomeric bands, an 80-kDa band was observed with both anti-FLAG and -HA antibodies (Fig. 6A), suggesting that an MpCAO1 and MpCAO2 heterodimer was formed. Interestingly, higher molecular weight bands than the heterodimer were found, and the immunoblot profile with anti-FLAG tag antibodies was almost the same as that with anti-HA tag (Fig. 6A).
FIGURE 6.
Analysis of the oligomeric states of the CAO proteins. A, two-dimensional BN/SDS-PAGE followed by immunoblotting indicated that MpCAO1 and MpCAO2 proteins accumulated in monomeric, heterodimeric (86 kDa), and higher molecular weight forms in MpCAO1+MpCAO2 plant. B, two-dimensional BN/SDS-PAGE followed by immunoblotting indicated that BCFLAG proteins accumulated in monomeric (43 kDa), trimeric (126 kDa), and higher molecular weight forms in the BCFLAG overexpression plants. The BCFLAG proteins were detected using an anti-FLAG antibody or anti-CAO antibody. C, two-dimensional BN/SDS-PAGE followed by immunoblotting indicated that recombinant AtCAO protein formed higher molecular weight protein complexes in E. coli. The AtCAO proteins were detected using an anti-His antibody. D, two-dimensional BN/SDS-PAGE followed by immunoblotting indicated that PhCAO formed monomeric (41 kDa), trimeric (123 kDa, and higher molecular weight complexes. The PhCAO proteins were detected using an anti-CAO antibody.
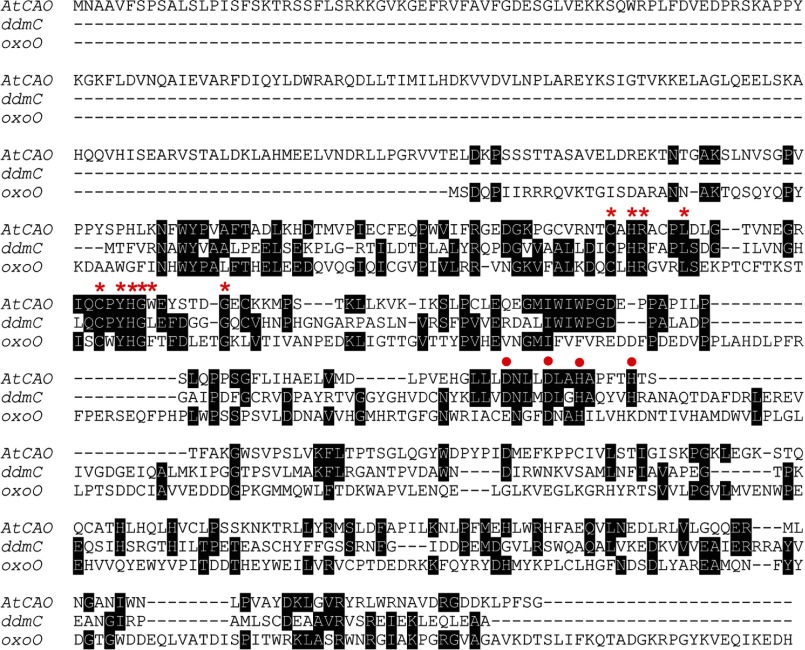
We then examined the oligomeric state of Arabidopsis CAO in chloroplasts. Because full-length AtCAO (ABC) is under the detectable level of immunoblotting as we reported previously (19), we expressed the AtCAO protein without the A domain fused with a FLAG tag (BCFLAG) in the Arabidopsis ch1-1 mutant. The thylakoid membranes were solubilized and analyzed by two-dimensional BN/SDS-PAGE followed by immunoblotting. Almost the same profiles were observed with the anti-CAO and -FLAG antibodies (Fig. 6B). The smallest band was ∼50 kDa, which likely represents monomeric BCFLAG protein (42 kDa) (Fig. 6B). In addition to the monomeric form, higher molecular weight bands were observed on the gel, estimated as ∼160, 240–300, 500, and 750 kDa (Fig. 6B). CAO has sequence homology to dicamba monooxygenase and 2-oxoquinoline 8-monooxygenase (Fig. 7), which contain a Rieske-mononuclear iron oxygenase motif also present in CAO. The crystal structure of the oxygenase component of dicamba monooxygenase and 2-oxoquinoline 8-monooxygenase revealed a homotrimer and showed a ring-shaped, symmetric arrangement (39). Considering this report, it is reasonable to assume that the 160-kDa band corresponds to a trimeric form of AtCAO (BCFLAG). If this assumption is true, then the 240–300-, 500-, and 750-kDa bands probably correspond to a 2 × 3-mer, 3 × 3-mer, and 5 × 3-mer, respectively.
FIGURE 7.
Multiple sequence alignment of AtCAO, dicamba monooxygenases (ddmC), and 2-oxoquinoline 8-monooxygenase (oxoO) proteins. Identical residues are shown in white type on a black background. Red asterisks and red circles show binding sites of Rieske center and mononuclear iron-binding motif, respectively.
The experimental results shown in Fig. 6B do not necessarily exclude the possibility that full-length CAO does not form the same oligomeric structures with the A domain-deleted AtCAO or that AtCAO assembled with other proteins to form a large complex. To examine this possibility, we expressed full-length AtCAO fused with FLAG (AtCAO-FLAG) in E. coli. The soluble fraction of the E. coli lysate was subjected to BN-PAGE followed by immunoblotting (Fig. 6C). A large complex and trimeric forms were observed as in the experiments with transgenic plants (Fig. 6B). These results clearly show that full-length CAO is capable of forming a homotrimer and larger complexes.
P. hollandica is a cyanobacterium that contains chlorophyll b. Prochlorothrix CAO (PhCAO) has no regulatory or linker domains and contains only the catalytic domain. To examine whether PhCAO forms trimeric and multimeric complexes as AtCAO does, we expressed PhCAO in the Arabidopsis ch1-1 mutant and examined the oligomeric state of PhCAO (Fig. 6D). A large amount of the PhCAO protein was detected between 170 and 1,200 kDa in addition to the trimeric and monomeric PhCAO (Fig. 6D).
Pigment Composition of Chlorophyll Protein Complexes of Micromonas and the Arabidopsis Transformants Harboring Micromonas CAO
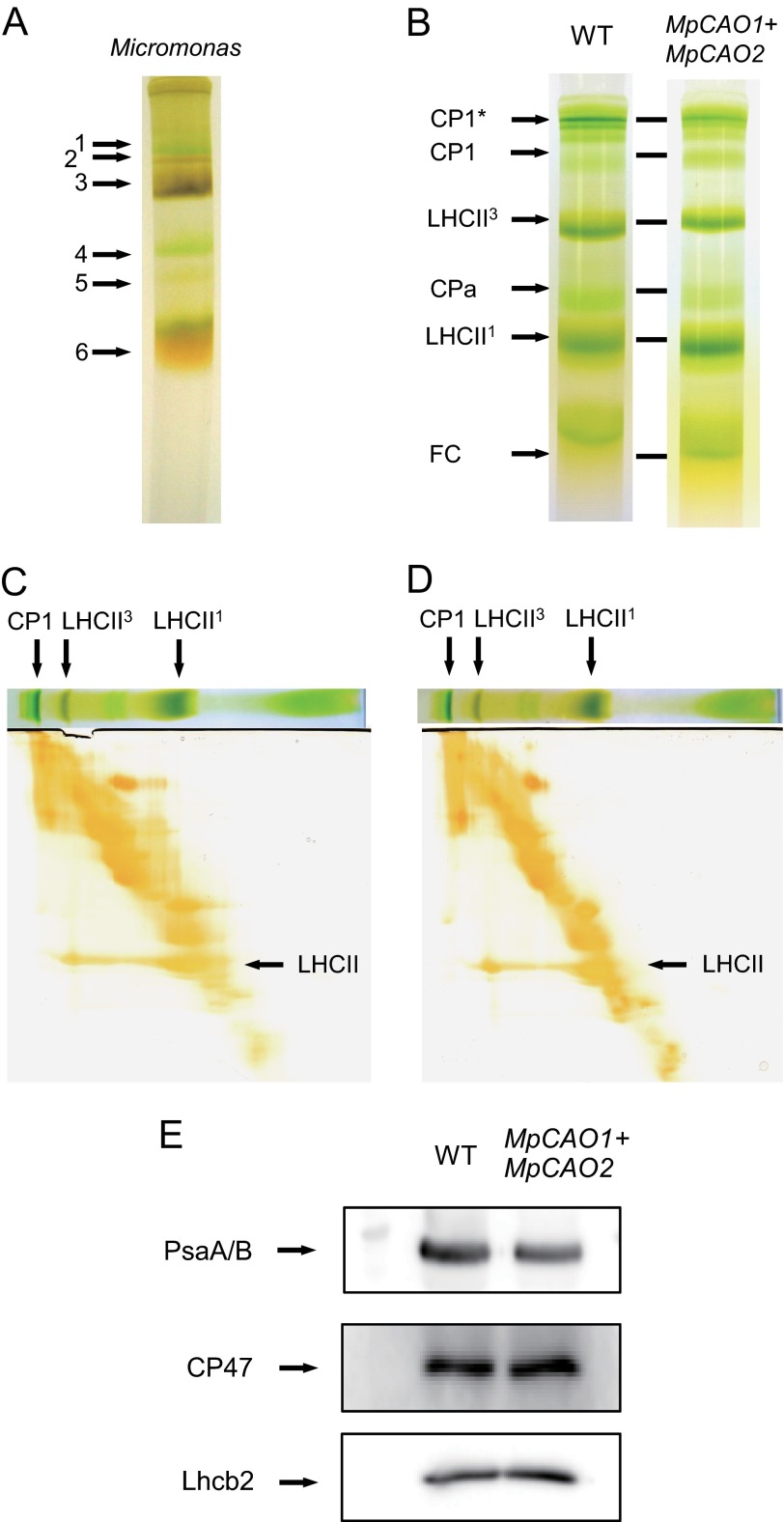
In green plants, it is reported that chlorophyll b exists in peripheral antennas such as LHCI and LHCII. However, when the A domain deleted-CAO (B and C domains only) is expressed in Arabidopsis, chlorophyll b was distributed not only in the peripheral antenna but also in the inner antenna of both photosystems (40), indicating the close relationship between the CAO structure and the distribution of chlorophyll b among various light-harvesting complexes. To examine whether a different structure of the light-harvesting system is constructed in Micromonas due to the characteristics of its CAO, Micromonas chlorophyll-protein complexes were separated by native-green gel (Fig. 8A). Six green bands were resolved. Bands 1 and 4 are the core complexes of photosystem I and II, respectively. Bands 3 and 5 are the trimeric and monomeric LHCII, respectively. The level of monomeric LHCII was very low, indicating that the LHCII trimer is more stable compared with that of other green plants. The separation profile of these bands was very close to that of Arabidopsis and other green plants except for the LHCII band between the PSI core antennas and LHC trimer. Band 6 is a free pigment band. Next, pigments were extracted from these bands and analyzed by HPLC. The chlorophyll a/b ratios of the trimeric and monomeric LHCII bands were 0.98 and 1.38 (Table 2), respectively, which is consistent with that of higher plants. 8-Vinyl-protochlorophyllide and prasinoxanthin are the characteristic pigments of Micromonas. These pigments were found only in LHCII and not in the core antennas of reaction centers. In contrast, β-carotene is predominantly found in the core antennas of both photosystems. The chlorophyll a/b ratios of the core antennas of PSI and PSII were 3.22 and 5.73, respectively, which are low values compared with those of other photosynthetic organisms, indicating that chlorophyll b is significantly incorporated into core antennas of reaction centers in Micromonas. One of the reasons for the distribution of chlorophyll b in the core antennas of reaction centers might be the different structure of CAO. To examine this possibility, we introduced Micromonas MpCAO1 and MpCAO2 into the Arabidopsis ch1-1 mutant and analyzed the pigment composition of the green bands (Fig. 8B). The chlorophyll a/b ratios of the core antennas of PSI and PSII of wild type Arabidopsis plants were 21.35 and 13.09 (Table 3), respectively, indicating that these core antennas of reaction centers consist primarily of chlorophyll a, and the level of chlorophyll b is very low. In contrast, the ratios of the transgenic plants with MpCAO1 and MpCAO2 were 3.43 and 3.50, respectively. This indicates that chlorophyll b is incorporated into core antennas of reaction centers when AtCAO is substituted with Micromonas CAO, suggesting a strong evolutionary relationship between the CAO structure and light-harvesting systems.
FIGURE 8.
Separation of chlorophyll-protein complexes. Thylakoid membranes of M. pusilla (A), A. thaliana (WT) (B, C, and E), and MpCAO1+MpCAO2 plants (B, D, and E) were prepared. Chlorophyll-protein complexes of those plants were separated by native green gel electrophoresis (A and B). The identity of the chlorophyll-protein complexes corresponding to the bands are presented on the left side of the gel. CP1*, CP1 (PsaA/B)-LHCI complexes; CPa, core complexes of PSII (CP43/CP47); LHCII1 and LHCII3, monomeric and trimeric LHCII, respectively; FC, free Chl. In addition, chlorophyll-protein complexes of WT and MpCAO1+MpCAO2 plants were also analyzed by two-dimensional SDS/SDS-PAGE, followed by silver staining (C and D). The amounts of PsaA/B, CP47, and Lhcb2 proteins were estimated by SDS-PAGE, followed by immunoblot analysis with their specific antibodies (E).
TABLE 2.
Chlorophyll a/b ratios and other pigments of chlorophyll-protein complexes in Micromonas
Chlorophyll-protein complexes were separated by native green gel electrophoresis and were extracted from the gel. Their chlorophyll a/b ratios and other pigments were determined by HPLC analysis. The band numbers correspond to Fig. 8A. Chl, Chlorophyll; β-car, β-carotene; DVP*, 8-vinyl-protochlorophyllide; Pra*, prasinoxanthin; ND, not detected.
| Band no. | Chl a/b | β-car*/Chl a | DVP*/Chl a | Pra*/Chl a |
|---|---|---|---|---|
| 1 | 3.22 | 0.26 | ND | ND |
| 2 | 1.03 | 0.03 | 0.61 | 0.91 |
| 3 | 0.98 | 0.01 | 0.69 | 0.77 |
| 4 | 5.73 | 0.48 | ND | ND |
| 5 | 1.38 | 0.22 | ND | 2.17 |
| 6 | 1.66 | 0.48 | ND | 4.73 |
TABLE 3.
Chlorophyll a/b ratios of chlorophyll-protein complexes in WT and MpCAO1+MpCAO2 plants
Chlorophyll-protein complexes were separated by native green gel electrophoresis and were extracted from the gel. CP1 is a heterodimer of PsaA/PsaB. Their chlorophyll a/b ratios were determined by HPLC analysis. Chl, chlorophyll.
| Chl-protein complexes | Wild type | MpCAO1+MpCAO2 |
|---|---|---|
| CP1-LHCI complexes (CP1*) | 8.75 | 3.25 |
| CP1 | 21.35 | 3.43 |
| Trimeric LHCII (LHCII3) | 1.49 | 0.90 |
| Core complexes of PSII (Cpa) | 13.09 | 3.50 |
| Monomeric LHCII (LHCII1) | 1.57 | 0.86 |
| Free Chls | 1.82 | 0.79 |
Although the green band of CP1 (heterodimer of PsaA/PsaB) was reported to contain only the core antennas of PSI (41), co-migration of LHC on this region cannot be completely excluded because the CP1-LHCI band was close to CP1 on the gel (Fig. 8B). To purify the core antennas of PSI without LHC, thylakoid membranes were solubilized in 0.5% SDS and separated by Tris/glycine SDS-PAGE for 4 h. The CP1 band was clearly separated from trimeric LHC with this electrophoresis, and the CP1-LHCI bands disappeared (Fig. 8, C and D). Second-dimensional SDS-PAGE visualized with silver staining clearly shows that CP1 bands did not contain LHC in both wild type and MpCAO1+MpCAO2 plants, indicating that PsaA/PsaB was purified by this method. Chlorophyll a/b ratio of WT CP1 band was 19.30, which is consistent with the previous report (37). Chlorophyll a/b ratio of the CP1 band of the MpCAO1+MpCAO2 plant was 4.46, indicating that chlorophyll b was incorporated into the core antennas of PSI. The incorporation of chlorophyll b into the core antennas of reaction centers was also observed in the BC-Arabidopsis plants in which A domain of CAO is deleted (40). It should be noted that LHCII amounts were slightly high and PsaA/B amounts were slightly low in the MpCAO1+MpCAO2 plant compared with wild type (Fig. 8E). This is consistent with the idea that the level of LHCII is regulated by the amount of chlorophyll b.
DISCUSSION
Oligomeric Structure of CAO
Dicamba monooxygenase and 2-oxoquinoline monooxygenase are Rieske-mononuclear iron oxygenases (39). Their crystal structures show a symmetric trimer and ring-like structure. In the trimeric form, an electron is transferred from the Rieske center to the mononuclear iron in the neighboring subunit, where the catalytic reaction takes place. It is reasonable to speculate that CAO has the same oligomeric structure and electron transfer chain because CAO is a Rieske-mononuclear iron oxygenase and has sequence similarity to dicamba monooxygenase and 2-oxoquinoline monooxygenase. BN-PAGE also indicated that both AtCAO and PhCAO form a trimer and larger complexes (Figs. 6 and 7). Interestingly, CAO in Arabidopsis formed complexes larger than the trimer. Recombinant CAO protein expressed in E. coli also showed the same electrophoretic profile on the BN-gel, indicating that AtCAO and PhCAO proteins form trimers and multimers. It should be noted that neither AtCAO nor PhCAO associated with LHCII or PSII to form large complexes because these complexes did not co-migrate with the CAO protein on the BN gels, suggesting that the trimeric CAO, a unit structure of CAO, assembles into a large complex by itself. Although it is still unclear whether phytylation or formylation is the last step of chlorophyll b biosynthesis (44), CAO is one of the enzymes that catalyze the last part of chlorophyll synthesis. This suggests the involvement of CAO in the formation of LHC. One hypothesis is that a large complex contributes to supplying sufficient chlorophyll b for the formation of LHC, which requires ∼7 chlorophyll b molecules. The other hypothesis is that a large complex is an inactive form. At present, the biochemical importance of this large complex is not clear. Further study is necessary to clarify the importance of the large complex.
Subunit Structure of Micromonas CAO
Micromonas CAO is composed of two proteins, which contain either Rieske- (MpCAO1) or mononuclear iron (MpCAO2)-binding motifs. Neither of these proteins can synthesize chlorophyll b by itself. Only when the two proteins were simultaneously expressed was chlorophyll b accumulated. Co-immunoprecipitation and BN-PAGE followed by immunoblotting showed that the MpCAO1 and MpCAO2 proteins form a heterodimer. As far as we know, the Rieske-mononuclear iron oxygenase forms a homotrimer of c3 symmetry (45–47). The Micromonas CAO may be the first example of a Rieske-mononuclear iron oxygenase that forms a heterodimer. Based on the present results and previous report of the Rieske-mononuclear iron oxygenase (39), the electron transfer route between the CAO subunits is proposed in Fig. 9. In the trimeric form of CAO, Rieske is reduced from ferredoxin, and then the electron is transferred to a mononuclear iron in the neighboring subunit, converting chlorophyll a into chlorophyll b. In the Micromonas CAO, ferredoxin reduces the Rieske center in MpCAO1, which transfers electron to the mononuclear iron in MpCAO2 where chlorophyll b is synthesized.
FIGURE 9.
Evolution of CAO structure. A, prochlorophyte CAOs have only the C domain (catalytic domain) and form a trimer. CAOs in green algae and land plants have A, B, and C domains and form a trimer. Mamiellales Micromonas CAOs are composed of two proteins with either a Rieske (Fe-S)- or mononuclear iron (Fe)-binding motifs. Neither protein has an A or B domain. They interact with each other to form a dimer. B, in each case, electrons must be transferred to a mononuclear iron (Fe) from a Rieske center (Fe-S) across the protein-protein interface.
Evolution of CAOs and Light-harvesting Systems
It has been reported that chlorophyll metabolism is closely related to the formation and degradation of chlorophyll-protein complexes. Direct interaction between light-harvesting complexes and a chlorophyll metabolic enzyme was reported for chlorophyll b reductase, which converts chlorophyll b to 7-hydroxymethyl chlorophyll a in the chlorophyll cycle (48). Chlorophyll b reductase directly catalyzes the reduction of chlorophyll b in the LHC, which is the first step of LHCII degradation (27). For this reason, LHC is never degraded in the cbr mutant (38). When CAO was overexpressed in Arabidopsis, the LHC level increased (42). Interestingly, when the A domain-deleted CAO was introduced into the ch1-1 mutant, an unusual light-harvesting system was constructed, and chlorophyll b was incorporated not only into the LHC but also into the core antennas of both photosystems (21, 43). These results lead us to speculate that the evolution of CAO is closely related to that of the light-harvesting systems.
The evolution of CAOs and light-harvesting systems is summarized in Fig. 9. Prochlorophyte CAOs consist of only catalytic domains, and they have no sequence corresponding to the A and B domains. In these organisms, chlorophyll b does not exist in the LHC but is found in Pcb (10), which has no sequence similarity to LHC but is similar to core antennas (CP43/CP47) of photosystem II. The Prochlorothrix CAO has high sequence similarity to the C domain of higher plants, but Prochlorococcus CAO has less similarity (22). This may be at least partly due to the different chlorophyll species used by each enzyme. Prochlorococcus use 8-vinyl-chlorophyll, whereas other oxygenic organisms use chlorophyll. In eukaryotes, photosynthetic organisms acquired an LHC with a three-membrane helix, and most chlorophyll b is incorporated into the LHC. At this stage of evolution, CAO acquired the A domain, which might be important for preferential incorporation of chlorophyll b into the LHC as well as the regulation of chlorophyll synthesis. This hypothesis is supported by the finding that when full-length CAO (with A, B, and C domains) was substituted by the A domain-deleted CAO, chlorophyll b accumulates in excess and is incorporated into the core antennas of reaction centers (21). The A domain is conserved in land plants and green algae in which LHCII is a major light-harvesting complex, and chlorophyll b does not exist in the core antennas of reaction centers. However, in prasinophytes, the A domain was lost, and CAO separated into MpCAO1 and MpCAO2 over the course of evolution. Acquisition of the two separate genes encoding CAO might not be an evolutionarily difficult process because an electron is transferred from Rieske and mononuclear iron between two neighboring CAO, which was already the case in the trimeric CAO.
Interestingly, the light-harvesting system of Micromonas also drastically changed, although chlorophyll b is a major light-harvesting pigment as in other green algae and land plants. The chlorophyll a/b ratio of Micromonas is low, and chlorophyll b is incorporated into core antennas of reaction centers. This characteristic organization of the light-harvesting system of Micromonas might be at least partly related to the changes in the CAO structure. This hypothesis is supported by experiments showing that when the Micromonas CAO was introduced into Arabidopsis, the chlorophyll a/b ratio became low, and chlorophyll b was incorporated into the core antennas of reaction centers, which is similar to light-harvesting systems of Micromonas.
Acknowledgments
We thank Dr. Hisashi Ito (Hokkaido University) for technical assistance, and we also thank Dr. Ryouichi Tanaka (Hokkaido University) for helpful discussions.
This work was supported by Grant-in-aid for Young Scientists 23770035 (to A. Takabayashi) and by Scientific Research 24370017 (to A. Tanaka) from the Japan Society for the Promotion of Science.
This article was selected as a Paper of the Week.
- Pcb
- prochlorophyte chlorophyll-binding protein
- CAO
- chlorophyllide a oxygenase
- PS
- photosystem
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- LHC
- light-harvesting complex
- AtCAO
- Arabidopsis CAO
- BN
- blue native
- Chl
- chlorophyll.
REFERENCES
- 1. Neilson J. A., Durnford D. G. (2010) Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth. Res. 106, 57–71 [DOI] [PubMed] [Google Scholar]
- 2. Green B. R., Durnford D. G. (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 685–714 [DOI] [PubMed] [Google Scholar]
- 3. Renger T., Schlodder E. (2010) Primary photophysical processes in photosystem II: bridging the gap between crystal structure and optical spectra. Chemphyschem 11, 1141–1153 [DOI] [PubMed] [Google Scholar]
- 4. Chen M., Schliep M., Willows R. D., Cai Z. L., Neilan B. A., Scheer H. (2010) A red-shifted chlorophyll. Science 329, 1318–1319 [DOI] [PubMed] [Google Scholar]
- 5. Adir N. (2005) Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth. Res. 85, 15–32 [DOI] [PubMed] [Google Scholar]
- 6. Lepetit B., Volke D., Szabó M., Hoffmann R., Garab G., Wilhelm C., Goss R. (2007) Spectroscopic and molecular characterization of the oligomeric antenna of the diatom Phaeodactylum tricornutum. Biochemistry 46, 9813–9822 [DOI] [PubMed] [Google Scholar]
- 7. Tomitani A., Okada K., Miyashita H., Matthijs H. C., Ohno T., Tanaka A. (1999) Chlorophyll b and phycobilins in the common ancestor of cyanobacteria and chloroplasts. Nature 400, 159–162 [DOI] [PubMed] [Google Scholar]
- 8. Garczarek L., Hess W. R., Holtzendorff J., van der Staay G. W., Partensky F. (2000) Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc. Natl. Acad. Sci. U.S.A. 97, 4098–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bibby T. S., Mary I., Nield J., Partensky F., Barber J. (2003) Low-light-adapted Prochlorococcus species possess specific antennae for each photosystem. Nature 424, 1051–1054 [DOI] [PubMed] [Google Scholar]
- 10. van der Staay G. W., Yurkova N., Green B. R. (1998) The 38-kDa chlorophyll a/b protein of the prokaryote Prochlorothrix hollandica is encoded by a divergent pcb gene. Plant Mol. Biol. 36, 709–716 [DOI] [PubMed] [Google Scholar]
- 11. Kouřil R., Dekker J. P., Boekema E. J. (2012) Supramolecular organization of photosystem II in green plants. Biochim. Biophys. Acta 1817, 2–12 [DOI] [PubMed] [Google Scholar]
- 12. Schmid V. H. (2008) Light-harvesting complexes of vascular plants. Cell. Mol. Life Sci. 65, 3619–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koziol A. G., Borza T., Ishida K., Keeling P., Lee R. W., Durnford D. G. (2007) Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol. 143, 1802–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Six C., Worden A. Z., Rodríguez F., Moreau H., Partensky F. (2005) New insights into the nature and phylogeny of prasinophyte antenna proteins: Ostreococcus tauri, a case study. Mol. Biol. Evol. 22, 2217–2230 [DOI] [PubMed] [Google Scholar]
- 15. Tanaka A., Ito H., Tanaka R., Tanaka N. K., Yoshida K., Okada K. (1998) Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. U.S.A. 95, 12719–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ito H., Ohtsuka T., Tanaka A. (1996) Conversion of chlorophyll b to chlorophyll a via 7-hydroxymethyl chlorophyll. J. Biol. Chem. 271, 1475–1479 [DOI] [PubMed] [Google Scholar]
- 17. Harada J., Mizoguchi T., Satoh S., Tsukatani Y., Yokono M., Noguchi M., Tanaka A., Tamiaki H. (2013) Specific gene bciD for C7-methyl oxidation in bacteriochlorophyll e biosynthesis of brown-colored green sulfur bacteria. PLoS One 8, e60026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagata N., Satoh S., Tanaka R., Tanaka A. (2004) Domain structures of chlorophyllide a oxygenase of green plants and Prochlorothrix hollandica in relation to catalytic functions. Planta 218, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 19. Yamasato A., Nagata N., Tanaka R., Tanaka A. (2005) The N-terminal domain of chlorophyllide a oxygenase confers protein instability in response to chlorophyll B accumulation in Arabidopsis. Plant Cell 17, 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakuraba Y., Tanaka R., Yamasato A., Tanaka A. (2009) Determination of a chloroplast degron in the regulatory domain of chlorophyllide a oxygenase. J. Biol. Chem. 284, 36689–36699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hirashima M., Satoh S., Tanaka R., Tanaka A. (2006) Pigment shuffling in antenna systems achieved by expressing prokaryotic chlorophyllide a oxygenase in Arabidopsis. J. Biol. Chem. 281, 15385–15393 [DOI] [PubMed] [Google Scholar]
- 22. Satoh S., Tanaka A. (2006) Identification of chlorophyllide a oxygenase in the Prochlorococcus genome by a comparative genomic approach. Plant Cell Physiol. 47, 1622–1629 [DOI] [PubMed] [Google Scholar]
- 23. Tanaka R., Ito H., Tanaka A. (2010) in Regulation and Functions of the Chlorophyll Cycle in The Chloroplast: Basics and Applications (Rebeiz C. A., Benning C., Bohnert H. J., Daniell H., Hoober J. K., Lichtenthaler H. K., Portis A. R., Tripathy B. C., eds) pp. 55–77, Springer, The Netherlands [Google Scholar]
- 24. Oster U., Tanaka R., Tanaka A., Rüdiger W. (2000) Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 21, 305–310 [DOI] [PubMed] [Google Scholar]
- 25. Worden A. Z., Lee J. H., Mock T., Rouzé P., Simmons M. P., Aerts A. L., Allen A. E., Cuvelier M. L., Derelle E., Everett M. V., Foulon E., Grimwood J., Gundlach H., Henrissat B., Napoli C., McDonald S. M., Parker M. S., Rombauts S., Salamov A., Von Dassow P., Badger J. H., Coutinho P. M., Demir E., Dubchak I., Gentemann C., Eikrem W., Gready J. E., John U., Lanier W., Lindquist E. A., Lucas S., Mayer K. F., Moreau H., Not F., Otillar R., Panaud O., Pangilinan J., Paulsen I., Piegu B., Poliakov A., Robbens S., Schmutz J., Toulza E., Wyss T., Zelensky A., Zhou K., Armbrust E. V., Bhattacharya D., Goodenough U. W., Van de Peer Y., Grigoriev I. V. (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272 [DOI] [PubMed] [Google Scholar]
- 26. Edwards K., Johnstone C., Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimoda Y., Ito H., Tanaka A. (2012) Conversion of chlorophyll b to chlorophyll a precedes magnesium dechelation for protection against necrosis in Arabidopsis. Plant J. 72, 501–511 [DOI] [PubMed] [Google Scholar]
- 28. Takabayashi A., Kurihara K., Kuwano M., Kasahara Y., Tanaka R., Tanaka A. (2011) The oligomeric states of the photosystems and the light-harvesting complexes in the Chl b-less mutant. Plant Cell Physiol. 52, 2103–2114 [DOI] [PubMed] [Google Scholar]
- 29. Anderson J. M., Waldron J. C., Thorne S. W. (1978) Chlorophyll–protein complexes of spinach and barley thylakoids: Spectral characterization of six complexes resolved by an improved electrophoretic procedure. FEBS Lett. 92, 227–233 [Google Scholar]
- 30. Tsugama D., Liu S. K., Takano T. (2011) A rapid chemical method for lysing Arabidopsis cells for protein analysis. Plant Methods 7:22, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 32. Tanaka A., Yamamoto Y., Tsuji H. (1991) Formation of chlorophyll-protein complexes during greening. 2. Redistribution of chlorophyll among apoproteins. Plant Cell Physiol. 32, 195–204 [Google Scholar]
- 33. Zapata M., Rodriguez F., Garrido J. L. (2000) Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 195, 29–45 [Google Scholar]
- 34. Tanaka R., Rothbart M., Oka S., Takabayashi A., Takahashi K., Shibata M., Myouga F., Motohashi R., Shinozaki K., Grimm B., Tanaka A. (2010) LIL3, a light-harvesting-like protein, plays an essential role in chlorophyll and tocopherol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 16721–16725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hall T. A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 37. Satoh S., Ikeuchi M., Mimuro M., Tanaka A. (2001) Chlorophyll b expressed in Cyanobacteria functions as a light-harvesting antenna in photosystem I through flexibility of the proteins. J. Biol. Chem. 276, 4293–4297 [DOI] [PubMed] [Google Scholar]
- 38. Horie Y., Ito H., Kusaba M., Tanaka R., Tanaka A. (2009) Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 284, 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martins B. M., Svetlitchnaia T., Dobbek H. (2005) 2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure 13, 817–824 [DOI] [PubMed] [Google Scholar]
- 40. Sakuraba Y., Yokono M., Akimoto S., Tanaka R., Tanaka A. (2010) Deregulated chlorophyll b synthesis reduces the energy transfer rate between photosynthetic pigments and induces photodamage in Arabidopsis thaliana. Plant Cell Physiol. 51, 1055–1065 [DOI] [PubMed] [Google Scholar]
- 41. Anderson J. M. (1980) P-700 content and polypeptide profile of chlorophyll-protein complexes of spinach and barley thylakoids. Biochim. Biophys. Acta 591, 113–126 [DOI] [PubMed] [Google Scholar]
- 42. Tanaka R., Tanaka A. (2005) Effects of chlorophyllide a oxygenase overexpression on light acclimation in Arabidopsis thaliana. Photosynth. Res. 85, 327–340 [DOI] [PubMed] [Google Scholar]
- 43. Sakuraba Y., Balazadeh S., Tanaka R., Mueller-Roeber B., Tanaka A. (2012) Overproduction of chl B retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol. 53, 505–517 [DOI] [PubMed] [Google Scholar]
- 44. Tanaka R., Tanaka A. (2011) Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes. Biochim. Biophys. Acta 1807, 968–976 [DOI] [PubMed] [Google Scholar]
- 45. D'Ordine R. L., Rydel T. J., Storek M. J., Sturman E. J., Moshiri F., Bartlett R. K., Brown G. R., Eilers R. J., Dart C., Qi Y., Flasinski S., Franklin S. J. (2009) Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation. J. Mol. Biol. 392, 481–497 [DOI] [PubMed] [Google Scholar]
- 46. Jakoncic J., Jouanneau Y., Meyer C., Stojanoff V. (2007) The crystal structure of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1. FEBS J. 274, 2470–2481 [DOI] [PubMed] [Google Scholar]
- 47. Ferraro D. J., Gakhar L., Ramaswamy S. (2005) Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338, 175–190 [DOI] [PubMed] [Google Scholar]
- 48. Kusaba M., Ito H., Morita R., Iida S., Sato Y., Fujimoto M., Kawasaki S., Tanaka R., Hirochika H., Nishimura M., Tanaka A. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]