Background: Fibroblast growth factor 14 (FGF14) binds to and regulates the voltage-gated Na+ (Nav) channel.
Results: Inhibition of glycogen synthase kinase 3 (GSK3) modifies FGF14/Nav channel interaction, with effects on Na+ currents and subcellular distribution of the FGF14·Nav channel complex.
Conclusion: The FGF14·Nav channel complex is a new target of GSK3.
Significance: We provide evidence for modulation of Nav channels by GSK3 through FGF14.
Keywords: Ion Channels, Neurons, Phosphorylation, Protein/Protein Interactions, Sodium Channels, FGF14, GSK3, Excitability
Abstract
The FGF14 protein controls biophysical properties and subcellular distribution of neuronal voltage-gated Na+ (Nav) channels through direct binding to the channel C terminus. To gain insights into the dynamic regulation of this protein/protein interaction complex, we employed the split luciferase complementation assay to screen a small molecule library of kinase inhibitors against the FGF14·Nav1.6 channel complex and identified inhibitors of GSK3 as hits. Through a combination of a luminescence-based counter-screening, co-immunoprecipitation, patch clamp electrophysiology, and quantitative confocal immunofluorescence, we demonstrate that inhibition of GSK3 reduces the assembly of the FGF14·Nav channel complex, modifies FGF14-dependent regulation of Na+ currents, and induces dissociation and subcellular redistribution of the native FGF14·Nav channel complex in hippocampal neurons. These results further emphasize the role of FGF14 as a critical component of the Nav channel macromolecular complex, providing evidence for a novel GSK3-dependent signaling pathway that might control excitability through specific protein/protein interactions.
Introduction
The intracellular FGFs (FGF11–14, also known as fibroblast growth factor homologous factors) are relevant components of the macromolecular complex of Nav3 channels (Nav1.1–1.9) (1–13). Among this group of molecules, FGF14 stands out for its potent, specific, and diverse modulatory actions on Nav channels in the CNS, features that have been attributed to its unique nonconserved N-terminal domain (4–7, 14). A number of in vitro experiments and studies in heterologous cells have examined the relative binding affinities of FGF14 to various Nav isoforms and evaluated the functional effects of FGF14 expression on Na+ currents, especially the ones encoded by the neuronal Nav1.2 and Nav1.6 isoforms (6). It has been shown that through high affinity binding to the intracellular C terminus of the α subunit of Nav channels (7), FGF14 potently modulates amplitude and voltage dependence of Na+ currents, producing functional outcomes on Na+ currents of magnitude and direction that depend upon the channel isoform and the cell background (4–6).
As part of the macromolecular complex of proteins at the axonal initial segment (AIS), FGF14 also serves to cluster Nav channels at the AIS, preserving action potential firing (5). Evidence comes from studies in hippocampal neurons where expression of a dominant negative FGF14 mutant protein suppresses Na+ currents and reduces expression and clustering of Nav channels at the AIS, leading to impairment of neuronal excitability (5). Consistent with these findings are the animal studies in fgf14−/− mice showing that genetic deletion of fgf14 leads to decreased excitability (12, 13) and impaired synaptic transmission (15), resulting in complex motor and cognitive deficits (16, 17). Finally, expression of the naturally occurring FGF14F145S missense mutation in humans results in ataxia, severe mental retardation, and neurodegeneration (18, 19), indicating an evolutionary conserved role of FGF14.
Altogether these studies provide compelling evidence for FGF14 as a complex multivalent molecule vital for brain function. However, the intracellular signaling mechanisms regulating the assembly and stability of this protein/protein interaction complex remain largely unknown. We posited that identifying cellular pathways upstream of the FGF14·Nav channel complex would provide new fundamental knowledge on the mechanisms decoding intracellular signaling into functional outputs, advancing our understanding of the molecular determinants of neuronal excitability in the normal and diseased brain.
Toward this end, we have applied the LCA (14) to screen a library of kinase inhibitors against the FGF14·Nav1.6 C-tail complex and identified GSK3 inhibitors as hits. Through a combination of LCA validations, co-immunoprecipitation, patch clamp electrophysiology, and quantitative confocal immunofluorescence, we demonstrate that pharmacological inhibition of GSK3 reduces the assembly of the FGF14·Nav channel complex, modifies FGF14-dependent modulation of Na+ currents, and induces subcellular redistribution of the native FGF14·Nav channel complex in hippocampal neurons. Collectively, these results add a new layer of complexity to the FGF14-dependent regulation of Nav channels, providing a potential link between the multifaceted GSK3 intracellular pathway (20) and neuronal excitability through specific molecular interactions.
EXPERIMENTAL PROCEDURES
Plasmids
The CLuc-FGF14, CD4-Nav1.6-NLuc, and Fgf14–6xmyc (Fgf14-1b-6xmyc) fusion constructs and the pcDNA3.1 vector (Invitrogen) were engineered and characterized as described previously (4, 5, 14). The corresponding gene ID numbers are as follows: NM_175929.2 (human FGF14-1b), NT_009759.16 (human CD4), and NM_014191.3 (human Nav1.6). The plasmid pGL3 expressing full-length Firefly (Photinus pyralis) luciferase was a gift from Dr. P. Sarkar (Dept. of Neurology, University of Texas Medical Branch).
Cell Culture and Transient Transfections
All reagents were purchased from Sigma unless noted otherwise. HEK-293 cells and stably expressing human Nav1.6 (HEK-Nav1.6 cells) were the gifts from Dr. Enzo Wanke (Universita di Milano-Bicocca, Italy). HEK-293 cells stably expressing rat Nav1.2 (HEK-Nav1.2 cells) and HEK-Nav1.6 were maintained in medium composed of equal volumes of DMEM and F-12 (Invitrogen) supplemented with 0.05% glucose, 0.5 mm pyruvate, 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 500 μg/ml G418 (Invitrogen) for selection of Nav1.6 and Nav1.2 stably transfected cells, and incubated at 37 °C with 5% CO2 (6, 21). Cells were transfected according to manufacturer's instructions at 90–100% confluency using Lipofectamine 2000 (Invitrogen).
Bioluminescence Assays and Kinase Inhibitor Library Screening
HEK-293 cells were seeded in 24-well CELLSTAR® tissue culture plates (Greiner Bio-One, Monroe, NC) at 4.5 × 105 cells per well and co-transfected within 24 h post-plating with either CLuc-FGF14 and CD4-Nav1.6-NLuc (1 μg per construct/well) or with CLuc-FGF14 and pcDNA3.1 (1 μg per construct/well); 48 h post-transfection cells were trypsinized for 5 min at 37 °C and seeded in white clear-bottom CELLSTAR® μClear® 96-well plates (Greiner Bio-One) at ∼105 cells/well in 200 μl of medium; 72 h post-transfection the culture medium was replaced with 100 μl of serum-free, phenol red-free DMEM/F-12 medium (Invitrogen). The chemical library used for the screening includes 385 protein kinase inhibitors obtained from the John S. Dunn Gulf Coast Consortium for Chemical Genomics (Houston, TX). Test compounds dissolved in DMSO at 6 mm (stock solution) were automatically dispensed into the 96-well plates at 30 μm final concentration in the medium (final concentration of DMSO 0.5%) using a Tecan Freedom EVO 200 liquid handling system (Tecan US, Durham, NC). Treated cells were incubated for 1 h at 37 °C, at which time the reaction substrate d-luciferin, dissolved in serum-free phenol red-free DMEM/F-12 medium (final concentration 0.75 mg/ml), was automatically dispensed into the 96-well plates with a SynergyTM H4 Multi-Mode Microplate Reader (BioTek, Winooski, VT). Following 15 min of incubation, luminescence values per well (relative luminescence units, integration time of 1 s) were acquired with the SynergyTM H4 Microplate Reader. The cells were maintained at 37 °C throughout the screening. Luminescence values expressed as percent average signal intensity from two independent screenings were normalized to per plate control (CLuc-FGF14 + CD4-Nav1.6-NLuc treated with 0.5% DMSO). Control experiments were performed as described above to test the influence of compounds on the luciferase enzymatic activity, except that cells were transfected with pGL3 (1 μg per well) expressing full-length Photinus luciferase instead of the LCA constructs.
For all other LCA experiments, the final concentration of DMSO was maintained at 0.5% unless stated otherwise, and selective kinase inhibitors were added manually in the culture medium (1–50 μm final concentration). Luminescence values per well (relative luminescence units, integration time 0.5 s) were acquired at 2-min intervals for 20–30 min using the SynergyTM H4 Microplate Reader. Signal intensity for each well was calculated as an average value of peak luminescence derived from three adjacent time points and expressed as percent control. Dose-response curves and IC50 values were obtained by fitting the data with a nonlinear regression (14).
Chemicals
d-Luciferin was purchased from Gold Biotechnology (St. Louis, MO), prepared as a 30 mg/ml stock solution in PBS, and further diluted at 1.5 mg/ml (intermediate solution); GSK3 inhibitor XIII (EMD Chemicals San Diego) and CHIR99021 (CT99021; SelleckBio, Houston, TX) were dissolved in DMSO (20 mm stock solution), aliquoted, and stored at −20 °C.
Measurement of Cell Viability
The influence of compounds on cell viability was measured in transiently transfected HEK-293 cells expressing full-length Photinus luciferase using CytoTox-GloTM cytotoxicity assay (Promega, Madison, WI), according to the manufacturer's instructions.
Western Blotting
Transfected HEK-293 (or HEK-Nav1.2) cells treated for 1 h at 37 °C with kinase inhibitors (or DMSO) were washed with phosphate-buffered saline (PBS) and lysed in buffer containing (in mm) 20 Tris-HCl, 150 NaCl, 1% Nonidet P-40. Protease inhibitor mixture for mammalian cells and tissue extract (Sigma) or protease inhibitor mixture Set 3 (Calbiochem) was added immediately before cell lysis. Cell extracts were collected and sonicated for 20 s and then centrifuged at 4 ° at 15,000 × g for 15 min. Supernatant was mixed with 4× sample buffer containing 50 mm tris(2-carboxyethyl)phosphine, and mixtures were heated for 10 min at 65 °C and resolved on 4–15% polyacrylamide gels (Bio-Rad). Resolved proteins were transferred to PVDF membranes (Millipore, Bedford, MA) for 1.5–2 h at 4 °C, 75 V and blocked in TBS with 3% nonfat dry milk and 0.1% Tween 20. Membranes were then incubated in blocking buffer containing goat polyclonal anti-luciferase polyclonal antibody (Promega, Madison, WI), rabbit polyclonal anti-calnexin (1:1000; Cell Signaling Technology, Danvers, MA), mouse monoclonal anti-GSK3 (1:1000; Millipore, Bedford, MA), mouse monoclonal anti-Myc (1:1000; 9E10 clone Santa Cruz Biotechnology) or mouse monoclonal anti-PanNav channel (1:1000; Sigma) antibody overnight. Washed membranes were incubated with donkey anti-goat, goat anti-rabbit, or goat anti-mouse HRP antibody (1:5000–10,000; Vector Laboratories, Burlingame, CA) and detected with ECL Advance Western blotting Detection kit (GE Healthcare); protein bands were visualized using FluorChem® HD2 System and analyzed with AlphaView 3.1 software (ProteinSimple, Santa Clara, CA).
siRNA Gene Silencing of GSK3
To selectively silence GSK3, siRNA against GSK3 α/β was obtained from Cell Signaling (siRNA 6301) along with an unrelated siRNA used as negative control (Ambion 16706). For knockdown experiments, HEK-293 cells were plated in 6- or 24-well plates (50% confluency) and incubated 1 day later with 10 μl of siRNA or negative control siRNA (both from a 10 μm stock) using RNAiMAX (Invitrogen) at a ratio of 10:5 (μl/μl) according to the manufacturer's instructions. Two days later, cells were replated in either 24- or 96-well plates and transfected with a mixture of cDNA plasmid and with a second pulse of siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Forty eight h later, LCA and/or Western blot analyses were performed to confirm reduction in the complementation of CLuc-FGF14 and CD4 Nav1.6-C-tail-NLuc and silencing of GSK3.
Immunoprecipitations
Immunoprecipitations from HEK-Nav1.2 cells were essentially as described previously (5). For immunoprecipitations from rat brain, crude membrane extract was prepared from adult rat brains and homogenized as described previously (22) in a 0.3 m sucrose, 10 mm sodium phosphate monobasic homogenization buffer, 1 mm EDTA, pH 7.4, containing the following protease inhibitors: leupeptin (1 μg/ml), aprotinin (1 μg/ml), pepstatin (1 μg/ml), and PMSF (1 mm). The homogenate was then centrifuged for 10 min (3000 × g) at 4 °C to remove nuclear debris, and the supernatant was centrifuged for 90 min (45,000 × g) at 4 °C; the resulting pellet (crude membrane extract) was resolubilized in the homogenization buffer described above. Crude membrane extract protein concentrations were measured by BCA (Pierce). Cell membrane extract was solubilized at 1:10 (∼1 mg/ml) in a lysis buffer consisting of: 1% Triton X-100, 0.15 m NaCl, 1 mm EDTA, 10 mm sodium azide, 10 mm Tris-HCl, pH 8.0, 2 mm NaF, BSA (1 mg/ml), aprotinin (1.5 μg/ml), antipain (10 μg/ml), leupeptin (10 μg/ml), benzamidine (0.1 mg/ml), and PMSF (1 mm) on a tube rotator for 30 min at 4 °C (23). Next, the samples were centrifuged (16,000 × g) for 30 min at 4 °C to remove the insoluble fraction. Monoclonal antibody mouse anti-PanNav (mAb K58/35, Sigma) at 2.5 μg/ml was added to the soluble fraction and incubated on a tube rotator at 4 °C overnight. Then the antibody·Nav channel complex was immobilized with 250 μl of slurry of protein-A/G magnetic beads (Pierce) prewashed three times in lysis buffer at 4 °C for 2 h on a tube rotator. Following incubation, beads were washed six times in 1 ml of lysis buffer without BSA and eluted with 4% acetic acid by incubation at 40 °C for 15 min. Immunoprecipitated protein concentrations were measured with Nanodrop (Thermo Scientific, Wilmington, DE).
Electrophysiology
Transfected cells were dissociated and re-plated at low density ∼12 h post-transfection. Recordings were performed at room temperature (20–22 °C) 12–18 h post-transfection using an Axopatch 200A amplifier (Molecular Devices). Borosilicate glass pipettes with resistance of 3–6 megohms were made using a Narishige PP-83 vertical Micropipette Puller (Narishige International Inc.). The recording solutions were as follows: extracellular (mm): 140 NaCl, 3 KCl, 1 MgCl2, 1 CaCl2, 10 HEPES, 10 glucose, pH 7.3; intracellular: 130 CH3O3SCs, 1 EGTA, 10 NaCl, 10 HEPES, pH 7.3. Membrane capacitance and series resistance were estimated by the dial settings on the Axopatch 200A amplifier (Molecular Devices). Capacitive transients and series resistance were compensated electronically by 70–80%. Data were acquired at 20 kHz and filtered at 5 kHz prior to digitization and storage. All experimental parameters were controlled by Clampex 7 software (Molecular Devices) and interfaced to the electrophysiological equipment using a Digidata 1200 analog-digital interface (Molecular Devices). Voltage-dependent inward currents for HEK-Nav1.6 and HEK-Nav1.2 cells were evoked by depolarizations to test potentials between −100 mV (Nav1.6) or −60 mV (Nav1.2) and +60 mV from a holding potential of −70 mV followed by a voltage pre-step pulse of −120 mV (Nav1.6) or −90 mV (Nav1.2). Steady-state (fast) inactivation of Nav channels was measured with a paired-pulse protocol. From the holding potential, cells were stepped to varying test potentials between −120 mv (Nav1.6) or −110 mV (Nav1.2) and +20 mV (pre-pulse) prior to a test pulse to −20 mV (Nav1.6) or −10 mV (Nav1.2).
Electrophysiology Data Analysis
Current densities were obtained by dividing Na+ current (INa) amplitude by membrane capacitance. Current-voltage relationships were generated by plotting current density as a function of the holding potential. Conductance (GNa) as calculated by Equation 1,
where INa is the current amplitude at voltage Vm, and Erev is the Na+ reversal potential.
Steady-state activation curves were derived by plotting normalized GNa as a function of test potential and fitted using the Boltzmann Equation 2,
where GNa,max is the maximum conductance; Va is the membrane potential of half-maximal activation; Em is the membrane voltage, and k is the slope factor. For steady-state inactivation, normalized current amplitude (INa/INa, max) at the test potential was plotted as a function of prepulse potential (Vm) and fitted using the Boltzmann Equation 3,
where Vh is the potential of half-maximal inactivation, and k is the slope factor.
Data analysis was performed using Clampfit9 software (Molecular Devices) and Origin 8.6 software (OriginLab, Northampton, MA).
Mice
Fgf14−/− mice (17) were maintained on an inbred C57/BL6J background (greater than 10 generations of backcrossing to C57/BL6J). All genotypes described were confirmed by PCR analysis (Charles River).
Immunohistochemistry
Fgf14−/− and fgf14+/+ mice (P30) and rat pups (P11) derived from timed pregnant Sprague-Dawley rat females (Charles River, Wilmington, MA) were sacrificed through isoflurane exposure followed by decapitation. Whole brains were removed, frozen in liquid nitrogen vapor, and stored at −80 °C until sectioning. Coronal sections (5–10 μm) were serially cut using a crytostat, mounted on Superfrost® glass microscope slides (Fisher Scientific, Waltham, MA), and left to dry overnight at room temperature. Sections were then fixed in fresh acetone at −20 °C for 8–10 min and dried at room temperature for 30 min, as described previously (24). Samples were washed three times with TBS, blocked for 1 h in TBS containing 0.3% Triton X-100 and 3% normal goat serum, and incubated overnight at 4 °C with the following primary antibodies: mouse anti-FGF14 (monoclonal 1:100; Sigma) and rabbit anti-PanNav (polyclonal 1:100; Sigma) dissolved in TBS containing 1% normal goat serum and 0.3% Triton X-100. Samples were then washed and incubated for 1 h with the following secondary antibodies at a 1:250 dilution as follows: Alexa 488-conjugated goat anti-mouse, Alexa 568-conjugated goat anti-rabbit, and Alexa 647-conjugated goat anti-chicken (Molecular Probes, Eugene, OR). In the experiments involving mouse tissue, 4′,6′-diamino-2-phenylindole·2HCl nuclear staining was performed according to the manufacturer's instruction (Santa Cruz Biotechnology). Samples were then mounted on glass slides with Prolong Gold anti-fade reagent (Invitrogen). Confocal images were then acquired with a Zeiss LSM-510 Meta confocal microscope with a ×20 air objective (0.75 numerical aperture), a ×40 (0.95 correction) air objective, and a C-Apochromat ×63 (1.2-watt corr) water objective. Multitrack acquisition was performed with excitation lines at 488 nm for Alexa 488, 543 nm for Alexa 568, and 364 nm for DAPI. Respective emission filters were band pass 505–530 nm, band pass 560–615 nm low pass 650, and 385–470 nm. The optical slices were 1 μm (mice) and 2 μm (rats); for rat brain Z-stacks were collected at z-steps of 1 μm with a frame size of 1024 × 1024, pixel time of 1.26 μs (×20 objective) or 1.60 μs (×40 objective), pixel size 0.22 × 0.22 μm, and an 8-frame Kallman-averaging. For mouse brain, Z-stacks were collected with ×63 water objective at z-steps of 0.5 μm with a frame size of 512 × 512, pixel time of 3.2 μs, pixel size 0.40 × 0.40 μm (0.7× zoom), 0.14 × 0.14 μm (2× zoom), and a 4-frame Kallman-averaging.
Primary Hippocampal Cultures
Banker's style hippocampal neuron cultures (25) were prepared from embryonic day 18 (E18) rat embryos using previously described methods (5). Briefly, following trituration through a Pasteur pipette, neurons were plated at low density (1–5 × 105 cells/dish) on poly-l-lysine-coated coverslips in 60-mm culture dishes in minimum Eagle's medium supplemented with 10% horse serum. After 2–4 h, coverslips (containing neurons) were inverted and placed over a glial feeder layer in serum-free minimum Eagle's medium with 0.1% ovalbumin and 1 mm pyruvate (N2.1 media; Invitrogen) separated by ∼1 mm wax dot spacers. To prevent the overgrowth of the glia, cultures were treated with cytosine arabinoside (5 μm; Calbiochem) at day 3 in vitro (DIV).
Immunocytochemistry
Hippocampal neurons (DIV14) were treated with either kinase inhibitors or DMSO (0.5%) for 24 h and then fixed in fresh 4% paraformaldehyde and 4% sucrose in phosphate-buffered saline (PBS) for 15 min. Following permeabilization with 0.25% Triton X-100 and blocking with 10% BSA for 30 min at 37 °C, neurons were incubated overnight at room temperature with the following primary antibodies: mouse anti-FGF14 (monoclonal 1:100; Sigma), rabbit anti-PanNav (1:100; Sigma), and chicken anti-MAP2 (polyclonal 1:25000; Covance, Princeton, NJ) diluted in PBS containing 3% BSA. In some experiments, a rabbit anti-MAP2 (polyclonal 1:10,000; Millipore) was used in combination with a chicken anti-β-IV-spectrin (1:25,000, gift from Dr. M. Komada (Tokyo Institute of Technology, Tokyo, Japan)). Neurons were then washed three times in PBS and incubated for 45 min at 37 °C with appropriate secondary antibodies as described for brain tissue staining. Coverslips were then washed six times with PBS and mounted on glass slides with Prolong Gold anti-fade reagent.
Image Analysis
Confocal images were acquired with a Zeiss LSM-510 Meta confocal microscope with a ×63 oil immersion objective (1.4 NA). Multitrack acquisition was done with excitation lines at 488 nm for Alexa 488, 543 nm for Alexa 568, and 633 nm for Alexa 647. Respective emission filters were band pass 505–530 nm, band pass 560–615 nm, and low pass 650. Optical slices were 2 μm for the first dataset and 0.8 μm for the second dataset; Z-stacks were collected at z-steps of 1 μm for the first dataset and 0.4 μm for the second dataset with a frame size of 512 × 512, pixel time of 2.51 μs, pixel size 0.28 × 0.28 μm, and a four-frame Kallman-averaging. All acquisition parameters, including photomultiplier gain and offset, were kept constant throughout each set of experiments. Acquired Z-stacks were sum-projected using MetaMorph 7.1.3.0 (Molecular Devices, Sunnyvale, CA), and pixel intensity values from the resulting stacked TIFF images were analyzed with ImageJ (National Institutes of Health). A region of interest (ROI) corresponding to a line of 6 pixels (1.68 μm) in width and 72 pixels (20 μm, first set) or 150 pixels (42 μm, second set) in length was highlighted on an overlay image of MAP2, PanNav channel, and FGF14 staining along the MAP2-negative and FGF14- and PanNav channel-positive processes. The “starting point” for the ROI was the point of reduced MAP2 staining intensity that corresponds to the AIS as described previously (4–7). A similar ROI was created for the dendrites on the Alexa 647 images corresponding to the MAP2 staining. The dendrites were defined as MAP2 positive processes. For each analyzed cell, the ROI for both the AIS and dendrites was then transferred to the Alexa 488 and Alexa 568 images and used for quantification of the FGF14 and the PanNav channel fluorescence intensity values, respectively. Total fluorescence intensity of immunolabeled PanNav channels and FGF14 at the AIS and in the dendrites was measured from cells in each set of experiments. The axonal-dendritic ratio was defined as the ratio of total fluorescence intensity in the AIS divided by the total fluorescence intensity in the dendrite within the ROI of the same size and was used as an index of polarized distribution of FGF14 and the Nav channel. The red/green ratio was defined as the total fluorescence intensity of the red channel divided by the total fluorescence intensity of the green channel in either AIS or dendrites and represented the level of co-localization/association of the Nav channel with FGF14. The intensity of off-cell background fluorescence was determined by measuring an area free of cells but close to the ROI and was numerically subtracted at the end of the analysis prior to fluorescence quantification. To combine data from independent sets of experiments, total fluorescence intensity values were normalized to the mean value of the control (DMSO-treated neurons) of that set. Intensity profiles of individual traces for each experimental group were generated by plotting pixel intensity values along the distance. Data were tabulated and analyzed with Excel, Origin 8.6, and SigmaStat (Jendel Corp., San Rafael, CA). Linear filters available in Adobe Photoshop were applied to images for illustration purpose only.
Statistical Analysis
Results were expressed as means ± S.E. The statistical significance of observed differences among groups was determined by Student's t test and one-way ANOVA or the corresponding nonparametric tests, Mann-Whitney rank sum, and Kruskal-Wallis tests, respectively, based on the distribution of the samples underlying the populations. A p < 0.05 was regarded as statistically significant. Bonferroni or Dunnett's tests were used for ANOVA post hoc analysis.
RESULTS
Kinase Inhibitor Screening of the FGF14·Nav Channel Complex
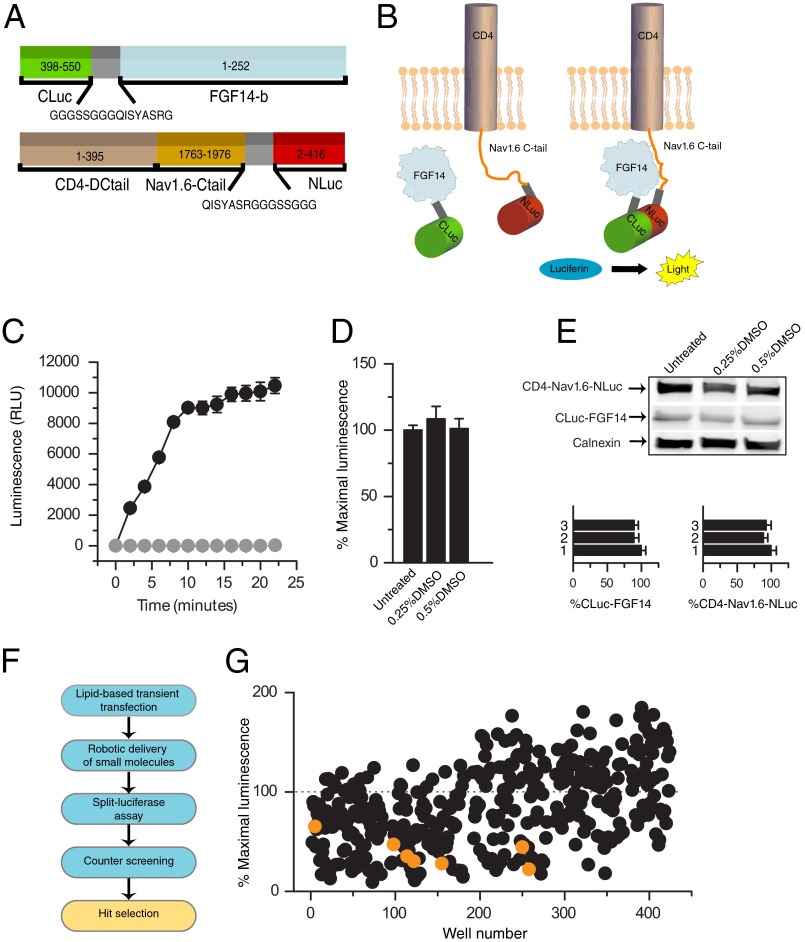
In previous studies, we introduced the LCA (26) to detect the assembly of FGF14 with the C-terminal tail of the Nav1.6 channel in live cells (14). In this assay, the C-terminal and the N-terminal fragments of the P. pyralis luciferase are fused, respectively, to FGF14, CLuc-FGF14, and a chimera expressing CD4 fused to the Nav1.6 C-tail, CD4-Nav1.6-NLuc (Fig. 1A). Upon transient co-expression of the two constructs in HEK-293 cells, the FGF14·Nav1.6 C-tail complex formation is detected upon the addition of the substrate d-luciferin (Fig. 1, B and C). To determine the applicability of LCA for pharmacological screenings, we tested its sensitivity to dimethyl sulfoxide (DMSO) commonly used as a vehicle for small molecule inhibitors. We found that neither the CLuc-FGF14·CD4-Nav1.6-NLuc complex formation (Fig. 1, C and D, n = 3, p = 0.22, one-way ANOVA) nor the relative protein expression levels of the two constructs are significantly affected by DMSO (Fig. 1E, n = 3, p = 0.70 for Cluc-FGF14, p = 0.89 for CD4-Nav1.6-NLuc, one-way ANOVA). These features combined with the assay sensitivity, reliability, and favorable signal-to-noise ratio characterized in previous studies (14) prompted us to conclude that LCA would be suitable for small molecule screenings.
FIGURE 1.
Bioluminescence screening of a chemical library of kinase inhibitors against the FGF14·Nav1.6 channel complex. A, constructs expressing the CLuc (amino acids 398–550) and NLuc (amino acids 2–415) fragments of firefly P. pyralis luciferase were, respectively, fused to full-length FGF14-1b (amino acids 1–252) and a chimera of the transmembrane protein CD4 (amino acids 1–395) and the C-terminal tail of Nav1.6 (amino acids 1763–1976). A flexible linker (gray) spaces the FGF14 and CD4-Nav1.6 cDNA from the two halves of luciferase. B, spontaneous association of FGF14 and Nav1.6 C-tail brings in close proximity the two halves of luciferase leading to reconstitution of the luciferase enzymatic activity and light production in the presence of the substrate d-luciferin (right); reprinted with permission from Assay and Drug Development; 10/2, published by Mary Ann Liebert, Inc., New Rochelle, NY. C, HEK-293 cells transiently transfected with either CLuc-FGF14 plus CD4-Nav1.6-NLuc (black circles) or CLuc-FGF14 alone (gray circles) were incubated with 0.5% DMSO for 1 h. The assembly of the FGF14·Nav1.6 C-tail complex is detected as luminescence (relative luminescence units, RLU) upon the addition of the d-luciferin (0.75 mg/ml) substrate at time 0; data are mean ± S.E. from quadruplicate wells from one representative experiment; CLuc-FGF14 alone serves as background. D, low concentrations of DMSO have no significant effect on the split luciferase reporter activity. Bars represent % maximal luminescence (normalized to untreated control) measured upon complementation of the Cluc-FGF14·CD4-Nav1.6-NLuc complex following 1 h of incubation of transfected cells with the indicated concentrations of DMSO; data are mean ± S.E. from quadruplicate wells from three independent experiments; the % maximal luminescence produced was comparable across conditions (no statistically significant difference between the conditions as determined by one-way ANOVA, F(2,6) = 1.51, p = 0.22). E, Western blots of whole-cell extracts (equal amount of protein per lane) from cells co-transfected with CLuc-FGF14 and CD4-Nav1.6-NLuc following the indicated treatments were probed with a polyclonal anti-luciferase antibody. Immunodetection of calnexin (rabbit polyclonal antibody) is used as loading control. At the bottom, densitometry analysis of CLuc-FGF14 (left) and CD4-Nav1.6-NLuc (right) in the presence of 0.25% DMSO (2) or 0.5% DMSO (3) normalized to untreated control (1) is shown; the expression levels of either CLuc-FGF14 or CD4-Nav1.6-NLuc (normalized to calnexin) in conditions 1 and 2 were comparable with untreated control (n = 3 for all groups; one-way ANOVA, F(2,6) = 0.39, p = 0.70 for CLuc-FGF14, and F(2,6) = 0.12, p = 0.89 for CD4-Nav1.6-NLuc); data are means ± S.E. F, scheme depicting the workflow of compound screening using LCA, counter-screening of compounds against intact Photinus luciferase and hit selection is shown. G, compound library screening of 385 kinase inhibitors against the CLuc-FGF14·CD4-Nav1.6·NLuc protein complex. Results for each compound are reported as average maximal luminescence response from the two independent screenings normalized to per plate controls; kinase inhibitors that target GSK3 are highlighted (orange). Few compounds produced a signal that was >200% and are omitted from the plot.
To gain insights into how the FGF14·Nav channel complex may be regulated by intracellular signaling cascades, we screened a small molecule library of 385 kinase inhibitors against the CLuc-FGF14·CD4-Nav1.6-NLuc complex, seeking pathways that could ultimately be relevant for neuronal excitability. Using the protocol illustrated in Fig. 1F, compounds were screened twice at a nominal test concentration of 30 μm (delivered in DMSO, 0.5% final concentration). Among the potential hits, our screen identified seven structurally heterogeneous compounds targeting GSK3, producing a significant reduction in the LCA signal compared with control (change in signal >2 S.D. of the mean in control; Fig. 1G, orange dots). These compounds, including indirubin-3′-monoxime, kenpaullone, and the GSK3 inhibitors XIII, XII, X, IX, and VIII (Fig. 2A), were then run through a counter-screening to account for potential interference of test compounds with the luciferase enzymatic activity (27, 28), and three were declared hits (Fig. 2B, black dots). Studies have shown that some GSK3 inhibitors can also block the GSK3 closely related cyclin-dependent kinases (29). However, neither roscovitine nor roscovitine S-isomer, two pan-cyclin-dependent kinase inhibitors included in our library, were effective against the CLuc-FGF14·CD4-Nav1.6-NLuc channel complex (79 ± 23% S.D., n = 4, and 98 ± 23% S.D., n = 2, respectively), providing further evidence for a GSK3-specific effect on our target. Identification of GSK3 inhibitors as hits was of particular significance due to the documented role of GSK3 in regulating neuronal polarity (30), synaptic plasticity (31), and ion channels (20) and considering its relevance as a therapeutic target against brain disorders (32, 33).
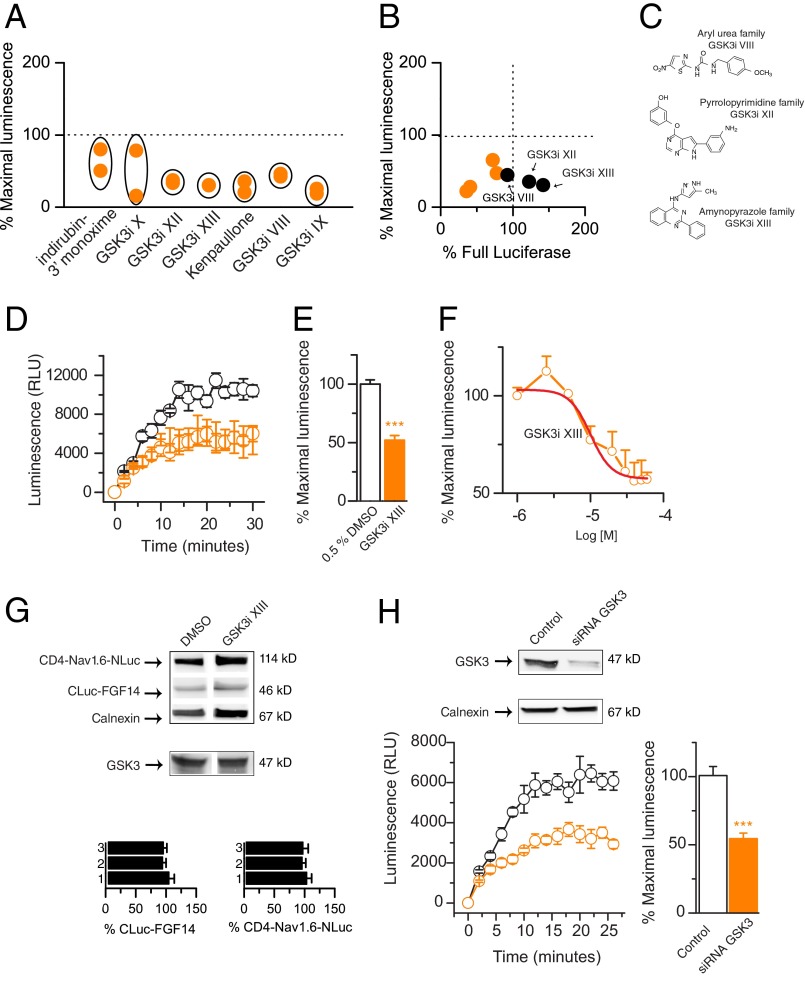
FIGURE 2.
GSK3 inhibitors reduce the FGF14·Nav1.6 channel complex formation. A, CLuc-FGF14·CD4-Nav1.6-NLuc complex formation is reduced upon exposure of GSK3 inhibitors. Results for the indicated compounds are reported as maximal luminescence response normalized to per plate controls from two independent rounds of library screening. B, scatter plot illustrates the effect of indicated compounds on the split luciferase reporter versus intrinsic enzymatic activity of the full-length Photinus luciferase; data are reported as averages from two independent experiments. GSK3 inhibitors (GSK3i) VIII, XII, and XIII decreased the reporter activity without inhibiting full-length luciferase and were declared hits (black dots). Compounds in the lower left quadrant interfere with full-length luciferase activity and were considered potential false positives. The chemical structure of the hit compounds is shown (C). D, complementation of CLuc-FGF14 and CD4-Nav1.6-NLuc detected as luminescence is decreased upon treatment with 30 μm GSK3 inhibitor XIII (orange circles) compared with DMSO-treated control (black circles); data are mean ± S.E., representing quadruplicates from one representative experiment. E, bar graph representing % maximal luminescence shows a reduction of the CLuc-FGF14·CD4-Nav1.6-NLuc complex assembly after 1 h of incubation with 30 μm GSK3 inhibitor XIII compared with control (0.5% DMSO). Data are mean ± S.E. (n = 4); ***, p < 0.001 (Student's t test). F, concentration-dependent inhibition of the luminescence signal by GSK3 inhibitor XIII. Data are mean ± S.E. (n = 4). The relative IC50 value calculated by fitting the nonlinear regression equation (red line) is 10 ± 0.25 μm (adjusted R2 = 0.97). G, Western blots of lysates from cells co-transfected with CLuc-FGF14 and CD4-Nav1.6-NLuc following the indicated treatments were probed with a polyclonal anti-luciferase antibody. Immunodetection of calnexin (rabbit polyclonal antibody) is used as loading control. The presence of GSK3 in HEK-293 cells is confirmed by detection with a mouse monoclonal anti-GSK3 antibody (middle). The expression levels of either CLuc-FGF14 or CD4-Nav1.6-NLuc (normalized to calnexin) in conditions 2 and 3 were comparable with untreated control (n = 3; one-way ANOVA: F(2,6) = 0.39, p = 0.70 for CLuc-FGF14 and F(2,6) = 0.12, p = 0.89 for CD4-Nav1.6-NLuc); data are means ± S.E. At the bottom, Western blots of lysate from cells transfected with unrelated siRNA (control) or GSK3 siRNA confirms specific silencing of GSK3. The calnexin antibody is used to control for loading and siRNA specificity. H, complementation of CLuc-FGF14 and CD4-Nav1.6-NLuc detected as luminescence is decreased upon GSK3 silencing (orange circles) compared with control (black circles); data are mean ± S.E., representing quadruplicates from one representative experiment. Bar graph representing % maximal luminescence shows a reduction of the CLuc-FGF14·CD4-Nav1.6-NLuc complex assembly upon GSK3 silencing compared with control. Data are mean ± S.E. (n = 4 independent experiments, each with four replicates); ***, p < 0.001 (Student's t test).
We followed up with the aminopyrazole GSK3 inhibitor XIII (Fig. 2, C and D; pubChem number 649766), a potent cell-permeable inhibitor used in cell culture studies (27, 28), because of its robust effect (reduction of the LCA signal to 30 ± 0.1% S.D. compared with DMSO control, n = 2, see Fig. 2A), and low toxicity profile (∼151.6% cell viability compared with DMSO control; data not shown). We confirmed with additional LCA experiments that GSK3 inhibitor XIII significantly reduced the CLuc-FGF14·CD4-Nav1.6-NLuc complex formation (Fig. 2, D and E; 48 ± 3.7%, n = 4, compared with DMSO control, n = 4, p < 0.001, Student's t test) with an apparent IC50 of 10 ± 0.25 μm (Fig. 2E, n = 4) without affecting the protein expression levels of either the LCA constructs or endogenous GSK3 (Fig. 2G).
GSK3 expression was then reduced using validated siRNA (Fig. 2G). As illustrated in Fig. 2H, the assembly of the CLuc-FGF14·CD4-Nav1.6-NLuc complex upon GSK3 silencing was significantly reduced compared with siRNA negative control (p < 0.001, n = 4, Student's t test) to a level comparable with GSK3 inhibitor XIII (Fig. 2D). Taken together, these data demonstrate that our strategy allowed the discovery of a signaling pathway identifying the FGF14·Nav channel complex as a new target of GSK3, a highly relevant multifaceted kinase with a central role in the normal and diseased brain (20, 33–35).
FGF14-dependent Regulation of Nav1.6 Currents Is Modified by GSK3 Inhibitors
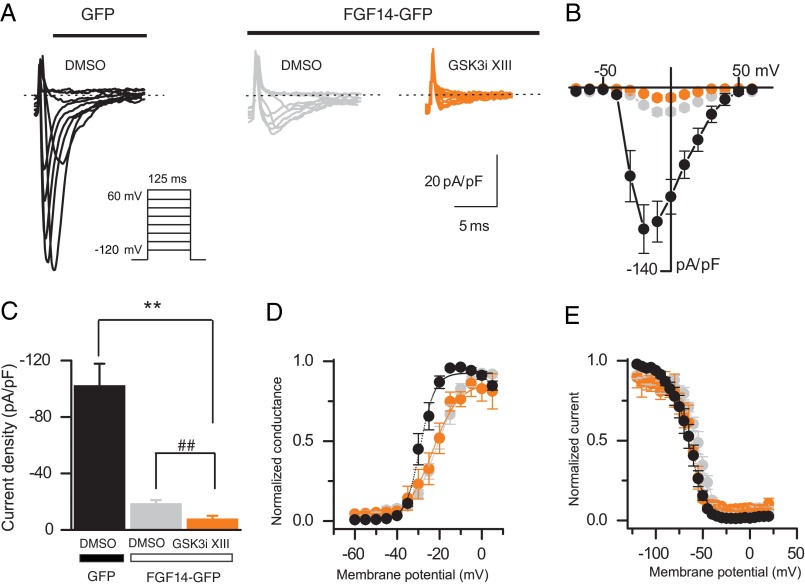
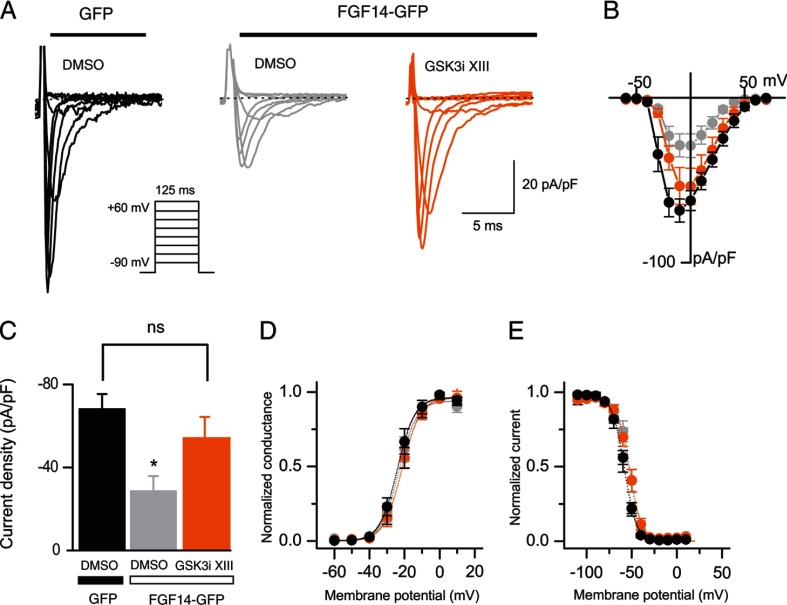
Previous studies have shown that FGF14 regulates amplitude and voltage dependence of Nav-encoded Na+ currents in cell lines (4, 6). To determine whether pharmacological inhibition of GSK3 affected FGF14-dependent modulation of Na+ currents, HEK-Nav1.6 cells were transiently transfected with Gfp or Fgf14-Gfp and treated with either DMSO (0.15% final concentration, control group) or GSK3 inhibitor XIII (30 μm, final concentration) 30–60 min prior to the experiments. In agreement with previous studies, we found that HEK-Nav1.6 cells expressing FGF14-GFP show significantly lower Na+ current (INa) amplitudes than cells expressing GFP (−18.7 ± 2.4 pA/pF, n = 13, for FGF14-GFP-expressing cells, −102.3 ± 15.6 pA/pF, n = 10, for GFP-expressing cells, F(2,31) = 37.36, p < 0.0001, one-way ANOVA, post hoc Bonferroni, see Fig. 3, A–C). Notably, GSK3 inhibitor XIII induces a further decrease in the level of Na+ current amplitude (−7.8 ± 2.2 pA/pF, n = 11, for FGF14-GFP-expressing cells treated with GSK3 inhibitor XIII compared with FGF14-GFP-expressing cells, p < 0.01, unpaired t test, see Fig. 3C). As expected from previous studies (6), analysis of voltage dependence of INa activation and steady-state inactivation revealed significant changes in the kinetics of Na+ currents in the cells expressing FGF14-GFP compared with control (Fig. 3, D and E, and Table 1). Interestingly, treatment with GSK3 inhibitor XIII rescues the depolarizing shift of the steady-state inactivation induced by FGF14-GFP expression (approximately +10 mV) back to control levels (F(2,22) = 6.68, p < 0.0054, one-way ANOVA, post hoc Dunnett's test; Fig. 3, D and E, and Table 1). Thus, pharmacological inhibition of GSK3 modifies regulation of Na+ current amplitude and kinetics by FGF14, providing functional relevance to our hypothesis.
FIGURE 3.
Pharmacological inhibition of GSK3 modifies functional modulation of Nav1.6 currents by FGF14. A, representative traces of voltage-gated Na+ currents (INa) recorded from HEK-Nav1.6 cells transiently expressing GFP or FGF14-GFP in response to voltage steps from −120 mV to +60 mV from a holding potential of −70 mV (inset). Only selected current traces in response to voltage steps are shown. GFP-expressing cells were treated with 0.15% DMSO (black traces), whereas FGF14-GFP-expressing cells were treated either with 0.15% DMSO (gray traces) or with 30 μm GSK3 XIII inhibitor (GSK3i XIII; orange traces). B, current-voltage relationships of INa from the experimental groups described in A. C, bar graphs representing peak current densities measured in individual cells HEK-Nav1.6 cells expressing GFP (treated with 0.15% DMSO; black bar), FGF14 (treated with 0.15% DMSO; gray bar), or FGF14 (treated with 30 μm GSK3 inhibitor XIII; orange bar). Data are mean ± S.E. Pharmacological inhibition of GSK3 in cells expressing FGF14-GFP (orange bar) suppresses peak current densities in comparison with DMSO-treated control (**, p < 0.01, one-way ANOVA, post hoc Bonferroni) and FGF14-GFP-expressing cells (##, p < 0.01, unpaired t test). D and E, voltage dependences of INa activation (D) and steady-state inactivation (E) were measured as described under “Experimental Procedures” and means ± S.E. values are plotted as a function of the membrane potential. The activation and inactivation data were fitted with the Boltzmann function as described under “Experimental Procedures.” The fitted parameters are provided in Table 1.
TABLE 1.
Voltage-gated Na+ currents in HEK-Nav1.6 cells
| Condition | Peak density | Activation V½ | kact | Inactivation V½ | kinact |
|---|---|---|---|---|---|
| pA/pF | mV | mV | mV | mV | |
| GFP (DMSO control) | −102.26 ± 15.6 (10) | −28.1 ± 1.2 (10) | 2.8 ± 0.4 (10) | −64.5 ± 2.8 (10) | −7.64 ± 0.5 (10) |
| FGF14 (DMSO control) | −18.7 ± 2.4 (13)a | −20.3 ± 1.4 (9)a | 6.1 ± 0.7 (9)a | −53.6 ± 2.0 (9)b | −7.2 ± 0.5 (9) |
| FGF14 (GSK3 inhibitor XIII) | −7.8 ± 2.2 (11) a,c | −21.6 ± 2.8 (6)a | 5.1 ± 0.7 (6)a | −60.2 ± 2 (4) | −8.1 ± 0.9 (4) |
a p < 0.01, one-way ANOVA, post hoc Dunnett's, and/or Bonferroni compared with GFP (DMSO control); data are mean ± S.E.
b p < 0.05.
c p < 0.01, unpaired t tests.
GSK3 Inhibition Reduces the Assembly of the FGF14·Nav1.2 Channel Complex and Controls FGF14-dependent Modulation of Nav1.2 Currents
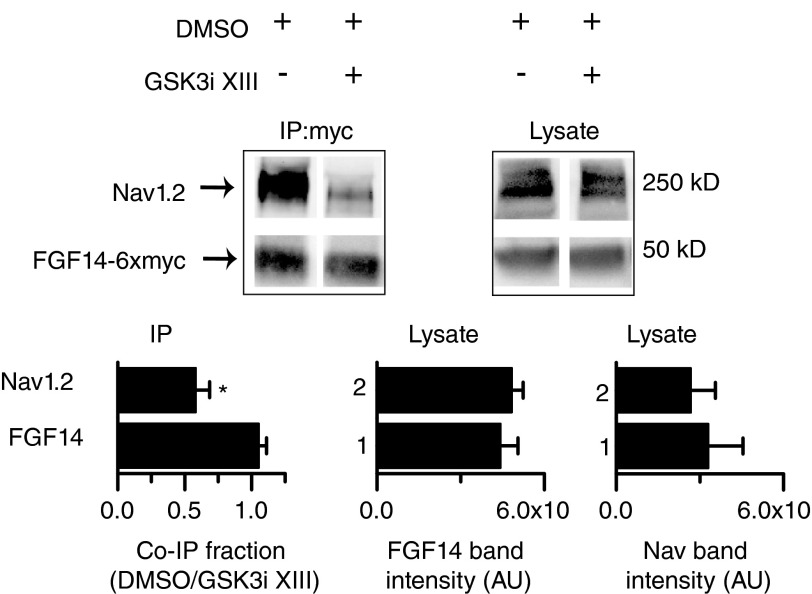
To extend our studies to another neuronal Nav channel isoform regulated by FGF14 (6), HEK-Nav1.2 cells were transiently transfected with Fgf14–6xmyc and treated with either GSK3 inhibitor XIII (30 μm) or DMSO (0.5% final concentration) for 2 h prior to harvest, and immunoprecipitations were performed with anti-Myc-agarose beads. As illustrated in Fig. 4, the fraction of Nav1.2 co-immunoprecipitating with FGF14-6×myc was significantly lower (n = 4, p < 0.05, Student's t test) in cells treated with GSK3 inhibitor XIII compared with DMSO control. Importantly, the levels of FGF14-6×myc and Nav1.2 expression in the cell lysate were similar in cells treated with GSK3 inhibitor XIII compared with DMSO (Fig. 4), indicating that pharmacological inhibition of GSK3 is not likely to affect stability and/or degradation of FGF14 and Nav1.2 channel, but rather they decrease binding between the two interacting proteins.
FIGURE 4.
Pharmacological inhibition of GSK3 reduces the assembly of FGF14 and the Nav1.2 channel. Western blot analysis of co-immunoprecipitation (IP:myc) and cell lysate showing the effect of 30 μm GSK3 inhibitor XIII on the FGF14–6×myc·Nav1.2 channel complex. Densitometric ratios between Nav1.2 or FGF14–6×myc in the co-immunoprecipitated (co-IP) fraction from inhibitor-treated and control cells are shown. GSK3 inhibitor XIII treatment reduces the co-immunoprecipitated fraction of Nav1.2 without affecting FGF14–6×myc (n = 4; *, p < 0.05, Student's t test). The levels of FGF14–6×myc and Nav1.2 protein in the lysate were unchanged (n = 4; p = 0.6 and p = 0.9, respectively, Student's t test). AU, arbitrary units.
Similarly to the experiments illustrated in Fig. 3, whole-cell patch recordings were performed in HEK-Nav1.2 cells transiently transfected with Gfp or Fgf14-1b-Gfp and treated with either DMSO or GSK3 inhibitor XIII. In agreement with previous studies, we found that HEK-Nav1.2 cells expressing FGF14-GFP show significantly lower Na+ current amplitudes than cells expressing GFP (−28.83 ± 7 pA/pF, n = 11, for FGF14-GFP-expressing cells, −68.37 ± 7.03 pA/pF, n = 13, for GFP-expressing cells, F(2,37) = 4.06, p < 0.05, one-way ANOVA, post hoc Dunnett's, see Fig. 5, A–C). Pretreatment with GSK3 inhibitor XIII antagonized this FGF14-mediated phenotype, rescuing the level of Na+ current amplitude back to control levels (−53.42 ± 11.14 pA/pF, n = 16, for FGF14-1b-GFP-expressing cells treated with GSK3 inhibitor XIII compared with −62.4 ± 5.9 pA/pF, n = 13, for GFP-expressing control cells; F(2,37) = 3.3, p > 0.05, one-way ANOVA, post hoc Dunnett's, see Fig. 5C), although no significant changes of Nav1.2 kinetics were found across conditions (Fig. 5, D and E, and Table 2). The lack of effect of FGF14 on the kinetics of Nav1.2 is in disagreement with previous studies (6) and might be attributable to an effect of DMSO.4 Thus, GSK3 exerts a modulatory action on the FGF14·Nav channel complex that depends upon the Nav channel isoform, revealing an additional layer of complexity underlying this newly discovered pathway.
FIGURE 5.
Pharmacological inhibition of GSK3 prevents functional modulation of Nav1.2 currents by FGF14. A, representative traces of voltage-gated Na+ currents (INa) recorded from HEK-Nav1.2 cells transiently expressing GFP or FGF14-GFP in response to voltage steps from −60 mV to +60 mV from a holding potential of −90 mV (inset). Only selected current traces in response to voltage steps are shown. GFP-expressing cells were treated with 0.15% DMSO (black traces), whereas FGF14-GFP-expressing cells were treated either with 0.15% DMSO (gray traces) or with 30 μm GSK3 XIII inhibitor (GSK3i XIII; orange traces). B, current-voltage relationships of INa from the experimental groups described in A. C, bar graphs representing peak current densities measured in individual HEK-Nav1.2 cells expressing GFP treated with 0.15% DMSO (black bar), FGF14 treated with 0.15% DMSO (gray bar), or FGF14 treated with 30 μm GSK3 inhibitor XIII (orange bar). Data are mean ± S.E. Cells expressing FGF14-GFP treated with DMSO exhibit peak current densities of significantly lower amplitude than the corresponding cells expressing GFP (*, p < 0.05, one-way ANOVA, post hoc Dunnett's). Pharmacological inhibition of GSK3 in cells expressing FGF14-GFP produces peak current densities indistinguishable from GFP-expressing control cells (p = 0.3, one-way ANOVA, post hoc Dunnett's). ns, non-significant. D and E, voltage dependences of INa activation (D) and steady-state inactivation (E) were measured as described under “Experimental Procedures,” and means ± S.E. values are plotted as a function of the membrane potential. The activation and inactivation data were fitted with the Boltzmann function as described under “Experimental Procedures.” The fitted parameters are provided in Table 2.
TABLE 2.
Voltage-gated Na+ currents in HEK-Nav1.2 cells
| Condition | Peak density | Activation V½ | kact | Inactivation V½ | kinact |
|---|---|---|---|---|---|
| pA/pF | mV | mV | mV | mV | |
| GFP (DMSO control) | −68.4 ± 7 (13) | −23.4 ± 2 (8) | 4.46 ± 0.3 (8) | −56.6 ± 1.3 (13) | −6.2 ± 0.3 (13) |
| FGF 14 (DMSO control) | −29.0 ± 7 (11)a | −22.5 ± 2 (8) | 4.1 ± 0.5 (8) | −53.0 ± 2.2 (8) | −5.8 ± 0.6 (8) |
| FGF 14 (GSK3 inhibitor XIII) | −53.7 ± 11 (16) | −21.0 ± 1.6 (13) | 4.4 ± 0.3 (13) | −53.1 ± 2.4 (14) | −5.2 ± 0.3 (14) |
a p < 0.05, one-way-ANOVA, post hoc Dunnett's, compared with GFP (DMSO control); data are mean ± S.E.
FGF14 and Nav Channels Form a Complex in Native Tissue
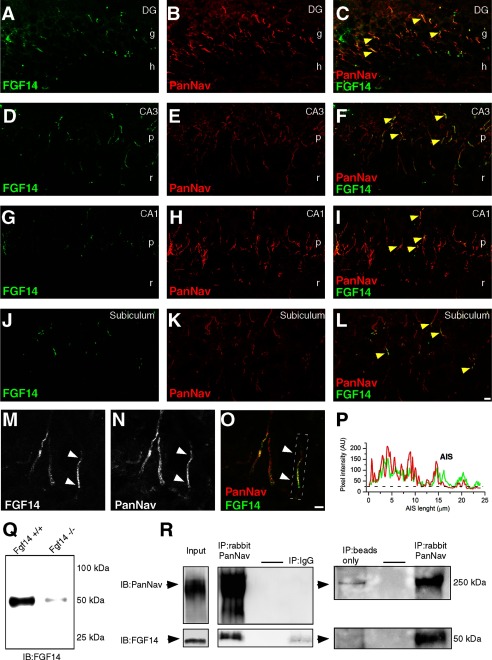
The role of neuronal FGF14 has been evaluated in neuronal cultures (4, 5, 15) and in animal models using genetic manipulations (12, 13, 15, 16). Yet studies of the distribution of FGF14 expression in native tissue are still missing. To gain insights into the expression pattern and subcellular distribution of native FGF14 and Nav channels, rat brain slices were fixed and probed with a rabbit antibody against PanNav channels and a mouse monoclonal antibody against FGF14 (Fig. 6, A–O). In the hippocampus, confocal analysis revealed a strong co-localization of FGF14 and Nav channels at the AIS, especially in the CA3 region and in the dentate gyrus (Fig. 6, A–F and M–P); a similar co-localization pattern, albeit in fewer cells, was observed in the CA1 region and in the subiculum (Fig. 6, G–I and J–L, respectively).
FIGURE 6.
Distribution and isolation of the native FGF14·Nav channel complex from brain tissue. A–P, confocal images of principal neurons in the dentate gyrus (DG), CA1 and CA3 regions of the rat hippocampus, and subiculum stained with a mouse monoclonal anti-FGF14 antibody, visualized with an Alexa 488-conjugated secondary antibody (A, C, D, F, G, I, J, L, M, and P) and a rabbit anti-PanNav α subunit antibody visualized with an Alexa 568-conjugated secondary antibody (B, C, E, F, H, I, K, L, N, and P). Arrows in M–P point to areas of co-localization of FGF14 and Nav channels along the AIS of a CA3 pyramidal neuron. Overlay image of the green (FGF14) and red (PanNav channels) is shown (C, F, I, L, and P). The boxed region in P highlights a representative AIS with pixel intensity profiles of both channels shown on the side (Q). Scale bar, 10 μm; g = granule cell layer; h = hilus; p = stratum pyramidalis; r = stratum radiatum. R, representative Western blot of brain homogenate obtained from fgf14+/+ and fgf14−/− adult mice (equal amount of proteins per lane). S, co-immunoprecipitation of FGF14 and the Nav channel from native tissue. Crude membrane extract (Input; 1:25 dilute) from whole adult rat brain was used for immunoprecipitation with a rabbit PanNav channel antibody (IP:PanNav), IgG (rabbit anti-luciferase, control), or blank (beads only). Western blots of input and co-immunoprecipitated fraction using a mouse anti-PanNav (IB:PanNav) and a mouse anti-FGF14 (IB:FGF14) antibodies are shown. Representative blot is from n = 4 independent experiments.
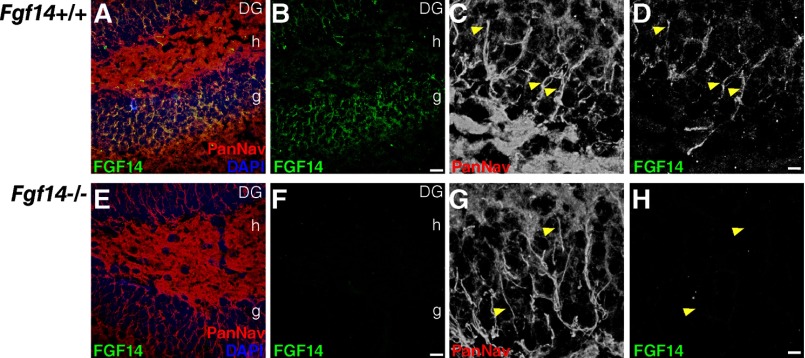
We further confirmed that FGF14 and Nav channels form a complex in the mammalian brain using co-immunoprecipitation from whole brain tissue followed by Western blot analysis (Fig. 6, Q and R). In addition to Western blot analysis (Fig. 6Q), the anti-FGF14 antibody was validated with immunofluorescence labeling of brain tissue of fgf14−/− mice (Fig. 7). Altogether, these results provide further evidence for FGF14 as an integral component of the Nav channel complex at the AIS (5) with distinct regional brain localization, supporting specialized functions for this protein complex.
FIGURE 7.
Validation of the mouse monoclonal anti-FGF14 antibody. Confocal images of the dentate gyrus (DG) in fgf14+/+ and fgf14−/− mice showing immunolabeling with a mouse monoclonal anti-FGF14 antibody, visualized with an Alexa 488-conjugated secondary antibody (A, B, D, E, F, and H) and a rabbit anti-PanNav α subunit antibody visualized with an Alexa 568-conjugated secondary antibody (A, C, E, and G). Overlay images of the green (FGF14), red (PanNav channels), and blue (DAPI nuclear staining) are shown (A and E). Higher resolution images of dentate granule cells are shown in gray scale (C, D, G, and H). Arrows point to representative AIS labeled with anti-FGF14 and anti-PanNav α subunit antibodies. Scale bar, 15 μm (A, B, E, and F) and 5 μm (C, D, G, and H); g = granule cell layer; h = hilus.
GSK3 Inhibition Induces Subcellular Redistribution of the FGF14·Nav Complex in Neurons
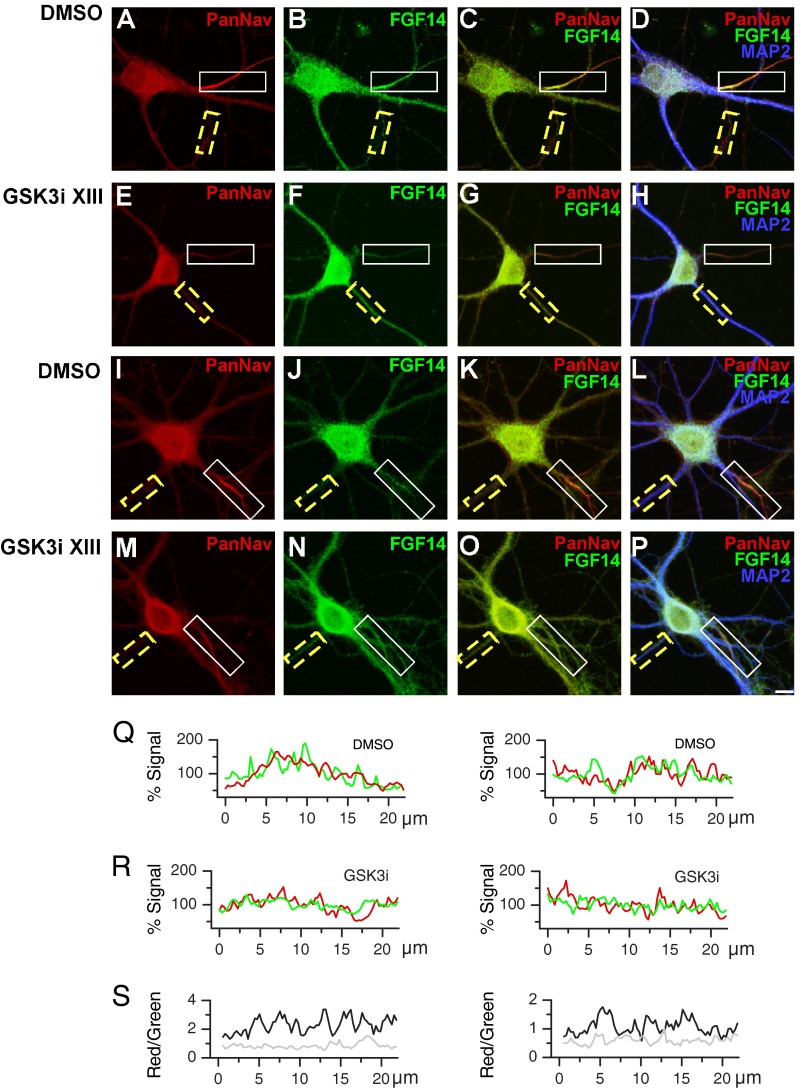
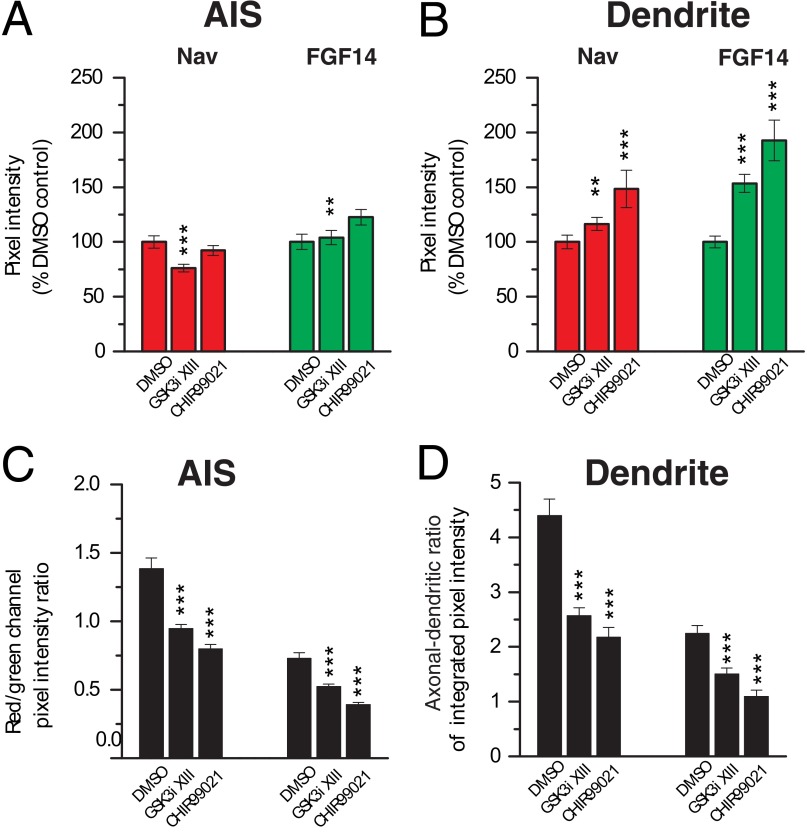
To determine the impact of the GSK3 signaling on the stability and subcellular distribution of the native FGF14·Nav channel complex, primary hippocampal neurons (DIV14) were exposed to GSK3 inhibitor XIII (30 μm) or vehicle (0.5% DMSO) for 24 h and processed for confocal immunofluorescence analysis. Neurons were fixed and triple-labeled with a mouse monoclonal antibody against FGF14, rabbit polyclonal antibody against PanNav, and a chicken antibody against the somato-dendritic marker MAP2 (Fig. 8, A–P). In controls, FGF14 and Nav channels are found highly expressed and clustered at the AIS (5) and expressed at low level in the dendrites (Fig. 8, A–D and I–L). Following the inhibitor treatment, a significant loss in the FGF14 and Nav channel axonal polarity and co-localization was observed as evident from individual confocal images (Fig. 8, E–H and M–P), fluorescence intensity profiles, and red/green signal ratios (Fig. 8, Q and R and S, respectively). Quantitative immunofluorescence analysis of confocal stacks over a large population of cells from two independent cultures (n = 58 and 63 analyzed cells per category, respectively) revealed a prominent subcellular re-distribution of the FGF14·Nav channel complex from the AIS to dendrites upon pharmacological inhibition of GSK3 (Fig. 9). We observed a significant loss of Nav channel immunofluorescence at the AIS (p < 0.001, Student's t test, Fig. 9A), although its content in the dendrites increased (p < 0.05, Mann-Whitney test, see Fig. 9B). Conversely, FGF14 expression increased significantly in the dendrites (p < 0.001, Student's t test, see Fig. 9B), while remaining overall unchanged in the AIS (p = 0.522, Mann-Whitney test, see Fig. 9A). These changes had a significant impact on the degree of association of the Nav channel with the FGF14 protein, as indicated by a prominent reduction in the red/green ratio at both the AIS and in the dendrites upon exposure to GSK3 inhibitor XIII (Figs. 8S and 9C). Furthermore, a drastic decrease in the axonal-dendritic ratio of both the Nav channels and FGF14 was observed (Fig. 9D; p < 0.001, Student's t test for both groups).
FIGURE 8.
Pharmacological inhibition of GSK3 leads to a redistribution of the FGF14·Nav channel complex in hippocampal neurons. A–P, confocal images of DIV14 primary hippocampal neurons following 24 h of exposure to 0.5% DMSO (A–D and I–L) or 30 μm GSK3 inhibitor XIII (GSK3i XIII; E–H, and M–P), labeled with a rabbit anti-Nav α subunit-specific antibody, PanNav, visualized with an Alexa 568-conjugated secondary antibody (red; A, E, I, and M), a mouse monoclonal anti-FGF14 antibody, visualized with an Alexa 488-conjugated secondary antibody (green; B, F, J, and N), and a chicken anti-MAP2 antibody, visualized with an Alexa 647-conjugated secondary antibody (blue; D, H, L, and P). Co-localization of PanNav and FGF14 is shown in red and green, respectively (C, G, K, and O). Overlay images of PanNav, FGF14 and MAP2 are shown in red, green, and blue channels, respectively (D, H, L, and P). The boxed regions highlight the AIS (white line) and dendrites (yellow dotted line). Scale bar, 10 μm. Q–S, representative examples of Nav α subunit (red) and FGF14 (green) immunofluorescence intensity along the AIS and dendrite regions in individual neurons treated with DMSO (Q) or GSK3 inhibitor XIII (GSK3i XIII; R) illustrated in I–L and M–P, respectively. Fluorescence intensity values along AIS and dendrites are expressed as % mean fluorescence intensity in the corresponding process. S illustrates the level of co-localization of the Nav α subunit and FGF14 along the AIS and dendrites in neurons illustrated in I–L and M–P, expressed as the ratio between red and green fluorescence intensity (neuron exposed to DMSO, black; neuron exposed to GSK3 inhibitor XIII, gray). Note the marked decrease of the red/green ratio in both the AIS and dendrites of the neurons treated with the GSK3 inhibitor compared with the DMSO-treated control.
FIGURE 9.
Immunofluorescence quantification of pharmacologically induced redistribution of the FGF14·Nav channel complex in hippocampal neurons. A and B, summary bar graphs showing pixel intensity values from confocal images corresponding to FGF14 and the Nav α channel at the AIS (A) and dendrites (B) following 24 h of treatment with 0.5% DMSO, 30 μm GSK3 inhibitor XIII (GSK3i XIII) or 3 μm CHIR99021. Values are normalized to the average pixel intensity in the DMSO-treated control. A, treatment with GSK3 inhibitor XIII (n = 59) causes a 23.8 ± 3.5% reduction of Nav channel in the AIS compared with the control (n = 63; ***, p < 0.001, Student's t test) without changing the level of FGF14 (p = 0.522, Mann-Whitney test); CHIR99021 treatment results in a slight (7.73 ± 4.6%) but not statistically significant decrease of the Nav channel content (left) in the AIS (n = 20; p = 0.921, Mann-Whitney test) and a 22.6 ± 7.0% increase of FGF14 (right) (**, p < 0.01, Mann-Whitney test). B, GSK3 inhibition causes an increase of Nav channel (left) in the dendrites both in the case of GSK3 inhibitor XIII (16.5 ± 5.8%; *, p < 0.05, Student's t test) and CHIR99021 (48.5 ± 17.2%; ***, p < 0.001, Mann-Whitney test) treatment; FGF14 (right) is also significantly increased in the dendrites in the presence of both GSK3 inhibitor XIII (53.5 ± 8.2%; ***, p < 0.001, Student's t test) and CHIR99021 (92.7 ± 18.5%; ***, p < 0.001, Student's t test). C, data from A and B are expressed as red/green ratio (C) (for the AIS p < 0.001 both in the GSK3 inhibitor XIII group and the CHIR99021 group; for the dendrites p < 0.001 in both groups; Student's t test) and axo-dendritic ratio of FGF14 and the Nav channel (D). Ratios between the red (Nav channel) and green (FGF14) pixel intensity values of the confocal images reflect the relative abundance of these proteins in the AIS (left) and dendrites (right). The red/green ratio in the AIS is significantly reduced after incubation with GSK3 inhibitor XIII (0.95 ± 0.03; ***, p < 0.001, Student's t test) and CHIR99021 (0.80 ± 0.03; ***, p < 0.001, Student's t test) compared with the DMSO control (1.38 ± 0.08). D, axo-dendritic ratio of FGF14 and the Nav channel is significantly decreased after incubation with GSK3 inhibitor XIII (2.57 ± 0.14 compared with 4.40 ± 0.30 in the control for Nav channels and 1.51 ± 0.11 compared with 2.25 ± 0.14 in the control for FGF14; ***, p < 0.001, Student's t test) and CHIR99021 (2.17 ± 0.18 for Nav channels, 1.09 ± 0.12 for FGF14; ***, p < 0.001, Student's t test) compared with the DMSO control.
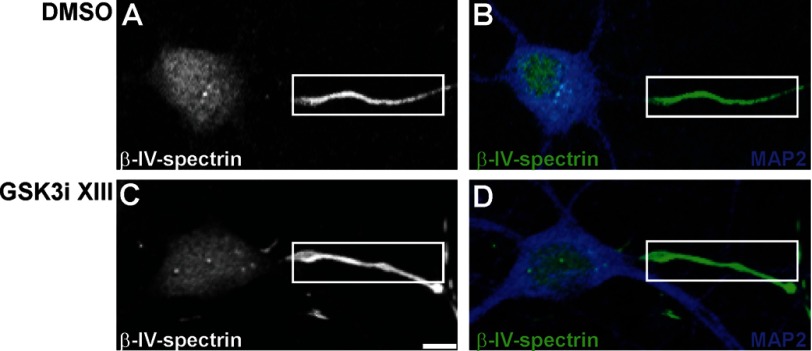
To confirm the observed phenotype, we extended our analysis to another GSK3 inhibitor, CHIR99021 (CT99021) (36, 37). Treatment with 3 μm CHIR99021 induced a phenotype very similar to the one observed with the GSK3 inhibitor XIII, except that the level of reduction of Nav channels at the AIS was not statistically significant (Fig. 9A, n = 20, p = 0.921, Mann-Whitney test), although the FGF14 content significantly increased (Fig. 9B, n = 20, p < 0.01, Mann-Whitney test), probably reflecting slightly different pharmacological properties of the two compounds. Importantly, the resulting reduction in the axonal-dendritic polarity and in the level of co-localization of FGF14 and Nav channels at the AIS and in dendrites was even more pronounced with CHIR99021 treatment (Fig. 9, C and D). Notably, the subcellular distribution of β-IV-spectrin, a component of the AIS (38), was unaffected by the treatment with GSK3 inhibitor XIII (Fig. 10) or CHIR99021 (data not shown), indicating a specific effect of GSK3 inhibition on the FGF14·Nav channel complex.
FIGURE 10.
Immunofluorescence staining of β-IV-spectrin in the presence of GSK3 inhibitor XIII. A–D, representative confocal images of DIV14 primary hippocampal neurons following 24 h of exposure to 0.5% DMSO (A and B) or 30 μm GSK3 inhibitor XIII (GSK3i XIII; C and D), labeled with a chicken anti-β-IV-spectrin antibody, visualized with an Alexa 488-conjugated secondary antibody (gray or green, A–D), and a rabbit anti-MAP2 antibody visualized with an Alexa 647-conjugated secondary antibody (blue; B and D). The distribution of β-IV-spectrin, a component of the AIS, is unaffected by the treatment with GSK3 inhibitor XIII. Overlay images of β-IV-spectrin and MAP2 are shown in green and blue channels, respectively (B and D). The boxed region highlights the AIS (white line). Scale bar, 6 μm.
DISCUSSION
In this study we applied a combination of bioluminescence screenings, co-immunoprecipitation, patch clamp electrophysiology, and quantitative immunofluorescence to identify a link between the FGF14·Nav channel complex and GSK3. This kinase has well documented roles in neuronal polarity (30) and synaptic plasticity (20, 31), and it has been found dysfunctional in a plethora of brain disorders (33–35). Collectively, our results provide consistent evidence for a modulation of interaction between the Nav channel and its multivalent regulatory protein, FGF14, upon inhibition of GSK3.
Our discovery was driven by the results of a bioluminescence-based screening of kinase inhibitors, implementing the LCA (26). Building on previous studies (14), we designed a screening platform incorporating counter-screening and additional validation steps that improved assay specificity and performance. We envision our approach as a novel tool to survey protein/protein interactions in the broad context of signaling networks allowing the identification of new regulatory pathways relevant for ion channels.
One of the key advantages of screening a chemical library as opposed to genetically silencing kinases (e.g. with RNAi knockdown) is that it provides the opportunity to easily screen transiently transfected cells. The limitation of this approach arises from off-target activity and cell toxicity exhibited by many kinase inhibitors (39). Several lines of evidence argue against nonspecific effects of GSK3 inhibitors on the FGF14·Nav complex. First, all compounds declared as potential hits that had GSK3 as their primary target (seven compounds) were structurally heterogeneous, yet consistently reduced the assembly of the FGF14·Nav channel complex, indicating conserved activity across different families of compounds. Second, inhibitors of cyclin-dependent kinases, such as roscovitine, did not significantly affect the FGF14·Nav channel complex, ruling out the most common source of side effects of GSK3 inhibitors (29). Third, the reduction of the FGF14·Nav1.6 C-tail complex assembly was unlikely ascribed to protein degradation (Fig. 2G) or cell toxicity (∼151.6% cell viability compared with DMSO control; data not shown). Fourth, the reduction in the FGF14·Nav channel complex assembly was reproducible by silencing GSK3 with siRNA (Fig. 2H).
Our electrophysiological experiments highlight the functional relevance of our study and reveal a new layer of complexity underlying the mechanism of regulation of Nav channels by FGF14 through GSK3. Previous studies have reported that expression of FGF14 in heterologous cells leads to inhibition of peak amplitude of Na+ currents encoded by Nav1.6 and Nav1.2 channel isoforms (6) and that these changes are likely to result from modulation in the number of cell surface available channels (4, 5). Importantly, although GSK3 inhibitors act synergistically with FGF14 further suppressing Nav1.6 current, they antagonize FGF14 modulation of Nav1.2 rescuing Na+ current amplitudes back to control levels. In contrast, GSK3 inhibitors antagonize the action of FGF14 on Nav1.6 kinetics (rescuing V½ of steady-state inactivation back to control levels, see Fig. 3D and Table 1), with no detectable effects on Nav1.2 biophysical properties (Fig. 5, D and E, and Table 2). Although future studies are needed to identify the molecular nature of these differences, these results clearly demonstrate Nav isoform-specific differences in the effect exerted by GSK3 on the FGF14·Nav channel complex. One can speculate that GSK3 inhibition might favor alternative binding modalities of FGF14 to the Nav1.6 channel (for example through tethering of the FGF14 N-terminal tail with other Nav1.6 intracellular domains) resulting in an almost complete block of Na+ currents and a rescue of the channel kinetics (steady-state inactivation). Alternatively, GSK3 inhibitors might simply decrease or destabilize the interaction of FGF14 with the Nav1.2 channel, opposing the FGF14-mediated suppression of Nav1.2 currents. Given the specialized subcellular segregation (40) and the diverse properties of Nav1.2 and Nav1.6 (6), these intricate effects of GSK3 on the FGF14·Nav channel complex are expected to greatly impact neuronal excitability.
At present, it remains unknown whether FGF14 or Nav channels are the primary target of GSK3 or whether the action of GSK3 on the FGF14·Nav channel complex requires any intermediate proteins/signaling molecules. Inspection of the primary amino acid sequence reveals a number of predicted GSK3 phosphorylation motifs throughout FGF14 and in the intracellular domains of Nav channels, but further studies are required to validate any of these Ser/Thr sites as GSK3 substrate(s).
To further validate the significance of our newly identified signaling mechanism, we carried out a detailed analysis of confocal images showing that pharmacological inhibition of GSK3 leads to decreased co-localization and subcellular redistribution of the native FGF14 and Nav channel complex, leading to reversal of its axonal-dendritic polarity in hippocampal neurons (Figs. 8 and 9). This effect was largely determined by a reduction of the Nav channel expression at the AIS with a concomitant increase in the dendrites and an increase of FGF14 expression mostly observed in the dendrites. Slight differences in the effect of CHIR99021 versus GSK3 inhibitor XIII were observed especially for the level of FGF14 at the AIS, but they did not affect the net result on the protein complex association (red/green ratio) or reversal of axonal-dendritic polarity. Diminished association and loss of axonal-dendritic polarity of FGF14 and Nav channels might be mechanistically linked. FGF14 is rich in potential motifs critical for cellular trafficking, and the Nav channel C-tail contains endocytic di-leucine motifs that are required for its axonal compartmentalization (41). Although speculative, uncoupling of FGF14 from the Nav channel complex upon GSK3 inhibition could (i) directly impair cellular trafficking of Nav channels, leading to their dendritic accumulation and loss in axonal polarity, and (ii) prevent the accessibility of the channel C-tail di-leucine motifs, blocking channel endocytosis in the dendrites and limiting its compartmentalization in the axon. Notably, under our experimental conditions, the subcellular distribution of β-IV-spectrin, a resident component of the AIS (38), was unchanged upon GSK3 inhibition (Fig. 10), suggesting a specific action of GSK3 on the FGF14·Nav channel complex. However, future studies are required to examine our results in the light of the role of GSK3 in establishing and maintaining neuronal polarity (30, 42–45).
Overall, these results further emphasize the significance of FGF14 and provide evidence for a novel GSK3-dependent signaling pathway that acts at the level of the Nav channel macromolecular complex through specific protein/protein interactions, advancing our understanding of the molecular determinants of excitability in the normal and diseased brain.
Acknowledgments
We thank Dr. Enzo Wanke (Dipartimento di Biotecnologie e Bioscienze, Università di Milano-Bicocca) for providing the HEK-293 cells stably expressing human Nav1.6; Dr. Sarkar Partha (Department of Neurology, University of Texas Medical Branch) for providing the Firefly full-length luciferase; Adriana Paulucci for critical insights on the confocal image acquisition and analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants MH095995 from NIMH (to F. L.) and 5UC7A107008304 from NIAID (to N. B.) and by Clinical and Translational Science Award UL1TR000071 from NCATS (to F. L.). This work was also supported by a Research starter grant from the PhRMA Foundation (to F. L.).
M. Nenov, T. F. James, and F. Laezza, unpublished observations.
- Nav
- voltage-gated Na+
- LCA
- luciferase complementation assay
- ROI
- region of interest
- ANOVA
- analysis of variance
- AIS
- axonal initial segment
- DIV
- days in vitro
- pF
- picofarad.
REFERENCES
- 1. Dover K., Solinas S., D'Angelo E., Goldfarb M. (2010) Long-term inactivation particle for voltage-gated sodium channels. J. Physiol. 588, 3695–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang C., Chung B. C., Yan H., Lee S. Y., Pitt G. S. (2012) Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure 20, 1167–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wei E. Q., Barnett A. S., Pitt G. S., Hennessey J. A. (2011) Fibroblast growth factor homologous factors in the heart: a potential locus for cardiac arrhythmias. Trends Cardiovasc. Med. 21, 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lou J. Y., Laezza F., Gerber B. R., Xiao M., Yamada K. A., Hartmann H., Craig A. M., Nerbonne J. M., Ornitz D. M. (2005) Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J. Physiol. 569, 179–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Laezza F., Gerber B. R., Lou J. Y., Kozel M. A., Hartman H., Craig A. M., Ornitz D. M., Nerbonne J. M. (2007) The FGF14(F145S) mutation disrupts the interaction of FGF14 with voltage-gated Na+ channels and impairs neuronal excitability. J. Neurosci. 27, 12033–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laezza F., Lampert A., Kozel M. A., Gerber B. R., Rush A. M., Nerbonne J. M., Waxman S. G., Dib-Hajj S. D., Ornitz D. M. (2009) FGF14 N-terminal splice variants differentially modulate Nav1.2- and Nav1.6-encoded sodium channels. Mol. Cell. Neurosci. 42, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goetz R., Dover K., Laezza F., Shtraizent N., Huang X., Tchetchik D., Eliseenkova A. V., Xu C. F., Neubert T. A., Ornitz D. M., Goldfarb M., Mohammadi M. (2009) Crystal structure of a fibroblast growth factor homologous factor (FHF) defines a conserved surface on FHFs for binding and modulation of voltage-gated sodium channels. J. Biol. Chem. 284, 17883–17896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang C., Wang C., Hoch E. G., Pitt G. S. (2011) Identification of novel interaction sites that determine specificity between fibroblast growth factor homologous factors and voltage-gated sodium channels. J. Biol. Chem. 286, 24253–24263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wittmack E. K., Rush A. M., Craner M. J., Goldfarb M., Waxman S. G., Dib-Hajj S. D. (2004) Fibroblast growth factor homologous factor 2B: association with Nav1.6 and selective colocalization at nodes of Ranvier of dorsal root axons. J. Neurosci. 24, 6765–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Cj, Dib-Hajj S. D., Waxman S. G. (2001) Fibroblast growth factor homologous factor 1B binds to the C terminus of the tetrodotoxin-resistant sodium channel rNav1.9a (NaN). J. Biol. Chem. 276, 18925–18933 [DOI] [PubMed] [Google Scholar]
- 11. Liu C. J., Dib-Hajj S. D., Renganathan M., Cummins T. R., Waxman S. G. (2003) Modulation of the cardiac sodium channel Nav1.5 by fibroblast growth factor homologous factor 1B. J. Biol. Chem. 278, 1029–1036 [DOI] [PubMed] [Google Scholar]
- 12. Goldfarb M., Schoorlemmer J., Williams A., Diwakar S., Wang Q., Huang X., Giza J., Tchetchik D., Kelley K., Vega A., Matthews G., Rossi P., Ornitz D. M., D'Angelo E. (2007) Fibroblast growth factor homologous factors control neuronal excitability through modulation of voltage-gated sodium channels. Neuron 55, 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shakkottai V. G., Xiao M., Xu L., Wong M., Nerbonne J. M., Ornitz D. M., Yamada K. A. (2009) FGF14 regulates the intrinsic excitability of cerebellar Purkinje neurons. Neurobiol. Dis. 33, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shavkunov A., Panova N., Prasai A., Veselenak R., Bourne N., Stoilova-McPhie S., Laezza F. (2012) Bioluminescence methodology for the detection of protein/protein interactions within the voltage-gated sodium channel macromolecular complex. Assay Drug Dev. Technol. 10, 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiao M., Xu L., Laezza F., Yamada K., Feng S., Ornitz D.M. (2007) Impaired hippocampal synaptic transmission and plasticity in mice lacking fibroblast growth factor 14. Mol. Cell. Neurosci. 34, 366–377 [DOI] [PubMed] [Google Scholar]
- 16. Wozniak D. F., Xiao M., Xu L., Yamada K. A., Ornitz D. M. (2007) Impaired spatial learning and defective θ burst induced LTP in mice lacking fibroblast growth factor 14. Neurobiol. Dis. 1, 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Q., Bardgett M. E., Wong M., Wozniak D. F., Lou J., McNeil B. D., Chen C., Nardi A., Reid D. C., Yamada K., Ornitz D. M. (2002) Ataxia and paroxysmal dyskinesia in mice lacking axonally transported FGF14. Neuron 35, 25–38 [DOI] [PubMed] [Google Scholar]
- 18. Brusse E., de Koning I., Maat-Kievit A., Oostra B. A., Heutink P., van Swieten J. C. (2006) Spinocerebellar ataxia associated with a mutation in the fibroblast growth factor 14 gene (SCA27): A new phenotype. Mov. Disord. 21, 396–401 [DOI] [PubMed] [Google Scholar]
- 19. van Swieten J. C., Brusse E., de Graaf B. M., Krieger E., van de Graaf R., de Koning I., Maat-Kievit A., Leegwater P., Dooijes D., Oostra B. A., Heutink P. (2003) A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia (corrected). Am. J. Hum. Genet. 72, 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wildburger N. C., Laezza F. (2012) Control of neuronal ion channel function by glycogen synthase kinase-3: new prospective for an old kinase. Front. Mol. Neurosci. 5, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schiavon E., Sacco T., Cassulini R. R., Gurrola G., Tempia F., Possani L. D., Wanke E. (2006) Resurgent current and voltage sensor trapping enhanced activation by a β-scorpion toxin solely in Nav1.6 channel. Significance in mice Purkinje neurons. J. Biol. Chem. 281, 20326–20337 [DOI] [PubMed] [Google Scholar]
- 22. Trimmer J. S. (1991) Immunological identification and characterization of a delayed rectifier K+ channel polypeptide in rat brain. Proc. Natl. Acad. Sci. U.S.A. 88, 10764–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Berendt F. J., Park K. S., Trimmer J. S. (2010) Multisite phosphorylation of voltage-gated sodium channel α subunits from rat brain. J. Proteome Res. 9, 1976–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liao Y., Deprez L., Maljevic S., Pitsch J., Claes L., Hristova D., Jordanova A., Ala-Mello S., Bellan-Koch A., Blazevic D., Schubert S., Thomas E. A., Petrou S., Becker A. J., De Jonghe P., Lerche H. (2010) Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain 133, 1403–1414 [DOI] [PubMed] [Google Scholar]
- 25. Goslin K., Asmussen H., Banker G. (1998) in Culturing Nerve Cells (Banker G., Goslin K., eds) pp. 339–370, MIT Press, Cambridge, MA [Google Scholar]
- 26. Luker K. E., Smith M. C., Luker G. D., Gammon S. T., Piwnica-Worms H., Piwnica-Worms D. (2004) Kinetics of regulated protein/protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. U.S.A. 101, 12288–12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schütz S. V., Cronauer M. V., Rinnab L. (2010) Inhibition of glycogen synthase kinase-3β promotes nuclear export of the androgen receptor through a CRM1-dependent mechanism in prostate cancer cell lines. J. Cell. Biochem. 109, 1192–1200 [DOI] [PubMed] [Google Scholar]
- 28. Rinnab L., Schütz S. V., Diesch J., Schmid E., Küfer R., Hautmann R. E., Spindler K. D., Cronauer M. V. (2008) Inhibition of glycogen synthase kinase-3 in androgen-responsive prostate cancer cell lines: are GSK inhibitors therapeutically useful? Neoplasia 10, 624–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eldar-Finkelman H., Martinez A. (2011) GSK-3 inhibitors: preclinical and clinical focus on CNS. Front. Mol. Neurosci. 4, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim Y. T., Hur E. M., Snider W. D., Zhou F. Q. (2011) Role of GSK3 signaling in neuronal morphogenesis. Front. Mol. Neurosci. 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peineau S., Taghibiglou C., Bradley C., Wong T. P., Liu L., Lu J., Lo E., Wu D., Saule E., Bouschet T., Matthews P., Isaac J. T., Bortolotto Z. A., Wang Y. T., Collingridge G. L. (2007) LTP inhibits LTD in the hippocampus via regulation of GSK3β. Neuron 53, 703–717 [DOI] [PubMed] [Google Scholar]
- 32. Hernández F., Gómez de Barreda E., Fuster-Matanzo A., Lucas J. J., Avila J. (2010) GSK3: a possible link between β-amyloid peptide and tau protein. Exp. Neurol. 223, 322–325 [DOI] [PubMed] [Google Scholar]
- 33. Jope R. S., Roh M. S. (2006) Glycogen synthase kinase-3 (GSK3) in psychiatric diseases and therapeutic interventions. Curr. Drug Targets 7, 1421–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Polter A. M., Yang S., Jope R. S., Li X. (2012) Functional significance of glycogen synthase kinase-3 regulation by serotonin. Cell. Signal. 24, 265–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X., Jope R. S. (2010) Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology 35, 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clayton E. L., Sue N., Smillie K. J., O'Leary T., Bache N., Cheung G., Cole A. R., Wyllie D. J., Sutherland C., Robinson P. J., Cousin M. A. (2010) Dynamin I phosphorylation by GSK3 controls activity-dependent bulk endocytosis of synaptic vesicles. Nat. Neurosci. 13, 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang M., Villaescusa J. C., Luo S. X., Guitarte C., Lei S., Miyamoto Y., Taketo M. M., Arenas E., Huang E. J. (2010) Interactions of Wnt/β-catenin signaling and sonic hedgehog regulate the neurogenesis of ventral midbrain dopamine neurons. J. Neurosci. 30, 9280–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Komada M., Soriano P. (2002) βIV-spectrin regulates sodium channel clustering through ankyrin-G at axon initial segments and nodes of Ranvier. J. Cell Biol. 156, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anastassiadis T., Deacon S. W., Devarajan K., Ma H., Peterson J. R. (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 29, 1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van Wart A., Trimmer J. S., Matthews G. (2007) Polarized distribution of ion channels within microdomains of the axon initial segment. J. Comp. Neurol. 500, 339–352 [DOI] [PubMed] [Google Scholar]
- 41. Garrido J. J., Fernandes F., Giraud P., Mouret I., Pasqualini E., Fache M. P., Jullien F., Dargent B. (2001) Identification of an axonal determinant in the C terminus of the sodium channel Nav1.2. EMBO J. 20, 5950–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leroy K., Brion J. P. (1999) Developmental expression and localization of glycogen synthase kinase-3β in rat brain. J. Chem. Neuroanat. 16, 279–293 [DOI] [PubMed] [Google Scholar]
- 43. Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. (2005) GSK-3β regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 120, 137–149 [DOI] [PubMed] [Google Scholar]
- 44. Kim W. Y., Snider W. D. (2011) Functions of GSK-3 signaling in development of the nervous system. Front. Mol. Neurosci. 4, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Namekata K., Harada C., Guo X., Kimura A., Kittaka D., Watanabe H., Harada T. (2012) Dock3 stimulates axonal outgrowth via GSK-3β-mediated microtubule assembly. J. Neurosci. 32, 264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]