Background: Selenium is incorporated into selenoproteins as the amino acid, selenocysteine.
Results: Dietary selenium supplementation increases ribosome density downstream of selenocysteine-encoding UGA codons.
Conclusion: Dietary selenium levels differentially regulate selenoprotein expression by controlling the rate-limiting step of selenocysteine incorporation.
Significance: The mechanisms by which dietary selenium can affect the readout of the genetic code and selenoprotein expression have been illuminated.
Keywords: Selenium, Selenocysteine, Selenoprotein, Transfer RNA (tRNA), Translation Control, Recoding, Ribosome Profiling
Abstract
Incorporation of selenium into ∼25 mammalian selenoproteins occurs by translational recoding whereby in-frame UGA codons are redefined to encode the selenium containing amino acid, selenocysteine (Sec). Here we applied ribosome profiling to examine the effect of dietary selenium levels on the translational mechanisms controlling selenoprotein synthesis in mouse liver. Dietary selenium levels were shown to control gene-specific selenoprotein expression primarily at the translation level by differential regulation of UGA redefinition and Sec incorporation efficiency, although effects on translation initiation and mRNA abundance were also observed. Direct evidence is presented that increasing dietary selenium causes a vast increase in ribosome density downstream of UGA-Sec codons for a subset of selenoprotein mRNAs and that the selenium-dependent effects on Sec incorporation efficiency are mediated in part by the degree of Sec-tRNA[Ser]Sec Um34 methylation. Furthermore, we find evidence for translation in the 5′-UTRs for a subset of selenoproteins and for ribosome pausing near the UGA-Sec codon in those mRNAs encoding the selenoproteins most affected by selenium availability. These data illustrate how dietary levels of the trace element selenium can alter the readout of the genetic code to affect the expression of an entire class of proteins.
Introduction
Selenium is a naturally occurring trace element that is essential for human health in small amounts but toxic at high levels. Human diseases associated with extreme selenium deficiency have been identified in geographic regions that are very low in selenium soil content, e.g. Keshan disease, a regional cardiomyopathy, which is endemic in regions of China (1). In regions of high selenium soil content, selenium toxicity can manifest as a condition called selenosis that is characterized by gastrointestinal upset, hair loss, fatigue, mood changes, and nerve damage (2, 3). Although disease symptoms resulting from severe selenium deficiency or intoxication are rare, there is strong evidence that less-overt variations in selenium status have multifaceted health effects including the risk of cancer, diabetes, skeletal muscle, and cardiac disease as well as maintenance of proper immune function (4).
One well established role of selenium is its incorporation into ∼25 mammalian selenoproteins as the amino acid, selenocysteine (Sec)2 (5, 6). This is accomplished by an expansion of the normal rules of decoding to allow UGA codons to encode Sec in addition to its normal role in terminating translation. Ribosome reprogramming and the process of UGA-Sec codon redefinition is orchestrated by cis-acting Sec insertion sequence (SECIS) elements that reside in the 3′-UTR of selenoprotein mRNAs (7–10), as well as trans-acting factors including the SECIS-binding protein, SBP2 (11), and the Sec-specific elongation factor, eEFSec (12, 13), which ultimately delivers Sec-tRNA[Ser]Sec to the ribosome (4).
In the context of selenoproteins, Sec mediates reductive/oxidation reactions on a number of substrates important for cellular redox homeostasis, thyroid hormone metabolism, protein folding, and disulfide formation or isomerization (4). It is also known that selenium availability can have a profound effect on selenoprotein levels with synthesis of certain essential housekeeping selenoproteins, thioredoxin reductase 1 (Txnrd1) and glutathione peroxidase 4 (Gpx4), for example, being more resistant to selenium limitation than stress-related selenoproteins, such as glutathione peroxidase 1 (Gpx1) (14–16). The “hierarchy” of differential selenoprotein expression in response to selenium availability and the specific substrates and biochemical pathways affected likely account for the diversity of health effects associated with dietary selenium intake.
Despite the identification of key components of selenocysteine synthesis and incorporation, the mechanism by which dietary selenium affects selenoprotein synthesis remains controversial. One commonly invoked mechanism involves the nonsense-mediated decay (NMD) pathway in which, under selenium limiting conditions, the UGA-Sec codon in nonessential selenoprotein messages is recognized as a premature termination codon causing RNA degradation. For example, the reduction of Gpx1 mRNA levels under selenium deficiency has been shown to be due to post-transcriptional mRNA turnover rather than reduced transcription (17) by induction of the NMD pathway (18, 19), whereas Gpx4 mRNA levels are unaffected and resistant to NMD. Other explanations implicate changes in Sec insertion efficiency during translation due to differences in SECIS elements and their ability to recruit the Sec incorporation machinery (20–22). Also, it is known that selenium levels directly correlate with the degree of Um34 methylation of Sec-tRNA[Ser]Sec wherein the Sec-tRNA[Ser]Sec population consists of two isoforms, the non-Um34-containing isoform, 5-methoxycarbonylmethyluridine (mcm5U), and the Um34-containing isoform, 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um) (23). Based on the observation that the mcm5Um isoform is required for efficient expression of the stress-related selenoproteins but not for the housekeeping selenoproteins, it has been proposed that differential utilization of the two Sec-tRNA[Ser]Sec isoforms during translation of selenoprotein mRNAs could account for selenium regulation of selenoprotein synthesis (24–26). Together these studies suggest that biosynthesis of selenoproteins is a complex process that may involve multiple levels of control; however, the degree of which each process may contribute to selenium-dependent regulation is unknown.
To directly examine the mechanisms of selenium-dependent regulation of selenoprotein expression in vivo, we have applied ribosome profiling, RNA-Seq, and traditional biochemical approaches to examine selenoprotein synthesis in liver. Using these approaches, we have defined the changes in mRNA abundance and translational activity that occur in response to altered dietary selenium intake and expression of a mutant Sec-tRNA[Ser]Sec that prevents Um34 synthesis. Quantitative mapping of ribosome distribution at nucleotide resolution on selenoprotein mRNAs revealed that translational control of selenoprotein synthesis occurred primarily due to changes in Sec incorporation efficiency. Furthermore, translation of Gpx1, Sepx1, Sepw1, and Selh mRNAs requires the Sec-tRNA[Ser]Sec mcm5Um isoform for efficient UGA-Sec decoding and involves an excess of ribosome protection upstream or at the UGA-Sec codon. Finally, we find an excess of ribosome-protected fragments upstream of the initiation codons for Selh, Sephs2, Txnrd1, and Selt. These studies provide a unique view of selenoprotein translational control and UGA redefinition efficiency in mammalian tissue and its regulation by dietary selenium intake.
EXPERIMENTAL PROCEDURES
Lysate Preparation
Three-week-old male WT and TrspA37G mice in a FVB/N background were fed diets supplemented with 0, 0.1, and 2.0 ppm selenium diets for 6 weeks before euthanasia. All animal work was approved under National Institutes of Health Institutional Animal Care and Use Committees protocol BRL-002. Livers were rapidly excised, cut into four equal parts, and immediately frozen in liquid nitrogen. For each experiment, 100 mg of frozen tissue was placed in a 2-ml tube containing a ¼-inch-diameter stainless steel ball with 1.5 ml of pre-chilled lysis buffer (10 mm Tris-Cl (pH 7.5), 300 mm KCl, 10 mm MgCl2, 200 μg/ml cycloheximide (Sigma), 1 mm DTT, and 1% Triton X-100). Samples were homogenized by agitation at full speed for 45 s in the Mini-Beadbeater-8 (Biospec Products, Inc.).
Ribonuclease Digestion and Total RNA Preparation
To minimize biological variation, total RNA for RNA-Seq analysis and ribosome-protected fragments were prepared from the same lysate. 300 μl of crude lysate were immediately transferred to 2 ml of TRIzol reagent (Invitrogen), and total RNA was extracted according to the manufacturer's specification. The remaining lysate was incubated with 500 units of RNase I (Invitrogen) for 30 min at 25 °C. Monosomes were isolated by centrifugation at 48,000 rpm for 2 h in a TL100 ultracentrifugation (Beckman Coulter). Ribonuclease-resistant RNA fragments were isolated from the pellet by TRIzol extraction and electrophoresed on a 15% polyacrylamide, 8 m urea gel. The region of the gel containing 26–36-nucleotide-size RNA fragments was excised, and RNA was isolated by passive elution before library construction.
Ribosome Profiling and Library Construction
Gel-purified ribosome footprints and hydrolyzed total RNA fragments were treated with 5 units of Antarctic phosphatase (New England Biolabs) in the presence of 40 units of RNase inhibitor (Ambion) for 30 min at 37 °C followed by 5 min at 65 °C to deactivate the enzyme. Fragments were then treated with 20 units of polynucleotide kinase (New England Biolabs) for 60 min at 37 °C and purified using RNeasy MinElute columns (Qiagen) according to the manufacturer's recommendations for small RNA. RPF and total RNA libraries were built using the TruSeq Small RNA Sample preparation kit (Illumina) according to the manufacturer's specifications with rRNA subtraction using Ribo-Zero (Epicenter). Limited PCR was used to enrich for ligated fragments.
Illumina Sequencing and Processing of Reads
Library aliquots were run on an Agilent 2100 DNA high sensitivity bioanalyzer chip to validate the library size range. RPF and total RNA libraries were subjected to 50 cycles of single-end sequencing on an Illumina HiSeq 2000 instrument. Adapter sequences were removed using the Hannon laboratory FastX toolkit. rRNA sequences were removed using bowtie (27) alignment against mouse rRNA sequences, and all unaligned sequences were retained for further processing.
Alignment to RefSeq mRNA Sequences
RefSeq fasta sequences were obtained from the UCSC genome browser (mm9 assembly, RefSeq Genes were downloaded in November of 2012). These fasta files were reduced to a single entry for each mRNA corresponding to the longest transcript isoform. For CDS alignments, the first 15 and last 3 codons were excluded to avoid bias caused by altered footprint density at the initiation and termination codons. Sequences that aligned to only a single site within the database, allowing for two mismatches, were identified using the bowtie short sequence aligner (27). Technical variability was addressed by linear regression analysis of all RefSeq mRNAs with greater than 0.01 RPFKM or RPKM values (11,918 mRNAs) for WT samples obtained from mice fed diets supplemented with 0.1 or 2.0 ppm selenium. In each comparison R2 values were >0.98, and >92% of mRNAs revealed less than a 2-fold difference in RPFKM or RPKM values (data not shown).
Ribosome profiling and total RNA sequences were aligned separately to selenoprotein and Gapdh mRNAs and were determined using the bowtie alignment parameters described above against the following RefSeq entries: NM_027652.2 Seli, NM_027905 Selo, NM_007860.3 Dio1, NM_013759.2 Sepx1, NM_024439.3 Sels, NM_015762.2 Txnrd1, NM_008160.6 Gpx1, NM_008162.2 Gpx4, NM_009155.3 Sepp1, NM_053102.2 Sep15, NM_013711.3 Txnrd2, NM_019979.2 Selk, NM_001040396.2 Selt, NM_009156.2 Sepw1, NM_009266.3 Sephs2, NM_030677.2 Gpx2, NM_001178058.1 Txnrd3, NM_029100.2 Sepn1, NM_053267.2 Selm, NM_008161.3 Gpx3, NM_001033166.2 Selh, NM_172119.2 Dio3, NM_010050.2 Dio2, NM_175033.3 Selv, NM_008084.2 Gapdh.
Quantitative Analysis and A-site Assignment
The 5′ end of ribosome profiling and RNA reads were offset to the predicted first nucleotide of the A-site using the following rule: for sequence lengths ≤34 nt (+17 nt from the 5′ end of the sequence) and for sequence lengths ≥35 nt (+18 nt from the 5′ end of the sequence). These values were determined as the optimum to position initiating ribosome P-sites at the AUG codon for all RefSeq mRNAs (see supplemental Figs. S1 and S3). Total mapped reads used to derive RPKM calculation were determined by aligning sequences to the CDSs of the RefSeq database described above. Total number of aligned reads were: WT 0 ppm selenium 23,945,053 (ribosome profiling) and 16,709,701 (RNA); WT 0.1 ppm selenium 30,296,837 (ribosome profiling) and 14,220,047 (RNA); WT 2.0 ppm selenium 17,763,406 (ribosome profiling) and 12,399,318 (RNA); TrspA37G 0 ppm selenium 28,864,210 (ribosome profiling) and 12,403,067 (RNA); TrspA37G 0.1 ppm selenium 29,816,461 (ribosome profiling) and 10,669,213 (RNA); TrspA37G 2.0 ppm 22,139,115 (ribosome profiling) and 13,073,779 (RNA). Analysis of ribosome footprinting upstream of the UGA-Sec codon excluded the first 15 codons after the initiation codon and the 5 codons preceding the UGA-Sec codon. Ribosome footprinting RPKMs downstream of the UGA-Sec were calculated from the second codon after UGA-Sec to the third codon preceding the stop codon. For reads assigned to A-site codons (Fig. 6 and supplemental Fig. S3), only ribosome footprints of 32 to 36 nt were utilized. The reads that mapped to the first nucleotide of a codon were then summed with those mapping to the adjacent −1 and +1 nucleotide positions to determine the number of footprints with A-sites assigned to each codon.
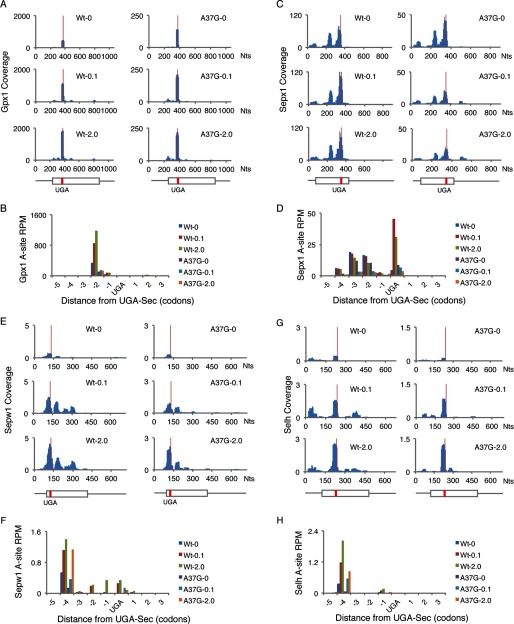
FIGURE 6.
Ribosome footprints near UGA-Sec codons. The number of ribosome footprints mapping to each nucleotide across the mRNAs of Gpx1 (A), Sepx1 (C), Sepw1 (E), and Selh (G) normalized to total mapped reads (Coverage) is shown for each mRNA in livers of WT or TrspA37G (A37G) mice fed diets supplemented with 0, 0.1, or 2.0 ppm selenium. Below the histograms in A, C, E, and G is a schematic of each mRNA. Thin lines represent UTRs, boxes represent open reading frames, and red lines indicate the position of the UGA-Sec codons. The number of ribosome footprints with A-site codons assigned to the −5 to +3 codons relative to UGA-Sec, normalize to total mapped reads (reads per million mapped reads, rpm, is shown for Gpx1 (B), Sepx1 (D), Sepw1 (F), and Selh (H).
Western Blot Analysis
Protein extracts were prepared from WT and TrspA37G mouse liver (n = 2) by homogenizing the liver in ice-cold lysis buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 1 mm EDTA, 0.1% Igepal, and Complete Mini Protease Inhibitor (Roche Applied Science)). Total protein was electrophoresed on 4–12% NuPage polyacrylamide gels (Invitrogen), transferred onto PVDF membranes (Bio-Rad), and blotted with antibodies against Txnrd1, Gpx4, Sep15 (Epitomics), Gpx1 (Abcam), Sepw1 (Rockland), Sepx1 or Selt (26). After incubation of the primary antibody, membranes were washed with TBS-T (Tris-buffered saline; 20 mm Tris/HCl (pH 7.5), 150 mm NaCl containing 0.1% Tween 20) and incubated in anti-rabbit HRP-conjugated secondary antibody (Thermo Scientific). Membranes were washed with TBS-T, incubated in Supersignal West Dura Extended Duration Substrate (Thermo Scientific), and exposed to x-ray film.
Quantitative PCR
Total RNA was isolated from WT and TrspA37G mouse liver (n = 3) using TRIzol (Invitrogen), and two-step quantitative real-time-PCR was performed to determine the relative expression of genes using the primer sequences as given (28). For each sample, total RNA (1 μg) was reverse-transcribed using an iScript cDNA Synthesis kit (Bio-Rad) according to the vendor's instructions and used for quantitative real-time-PCR using DyNAmo SYBR Green (Thermo Scientific). Reactions were carried out in triplicate, and RNA levels were normalized to Gapdh.
RESULTS
Selenium-deficient Availability and Sec-tRNA[Ser]Sec Um34 Modification Affect Global Translation of the Selenoproteome
Ribosome profiling involves the isolation and deep sequencing of ribosome-protected fragments (ribosome footprints or RPFs) after digestion of unprotected mRNA with RNase1 (29–31). Ribosome density and position on thousands of mRNAs can be determined by quantification and localization of ribosome footprints. An alternative method involving sucrose gradient fractionation of intact polysomes and microarray hybridization to determine mRNA position in the gradient was developed previously and applied to estimate overall ribosome density on mRNAs (32, 33). However, we selected ribosome profiling for analyzing selenoprotein expression due to its ability to measure both mRNA ribosome density and ribosome position at codon resolution. As previous in vitro studies have shown that UGA redefinition to encode Sec is in competition with termination (34, 35), we postulated that the ability to quantify ribosomes located 5′ and 3′ of UGA-Sec codons would provide a means of estimating changes in Sec incorporation efficiency and a more accurate method for measuring changes in full-length selenoprotein synthesis. Herein, we have applied this technique to examine the effects of altered dietary selenium levels and Sec-tRNA[Ser]Sec modification status on synthesis of the liver selenoproteome as outlined in Fig. 1A.
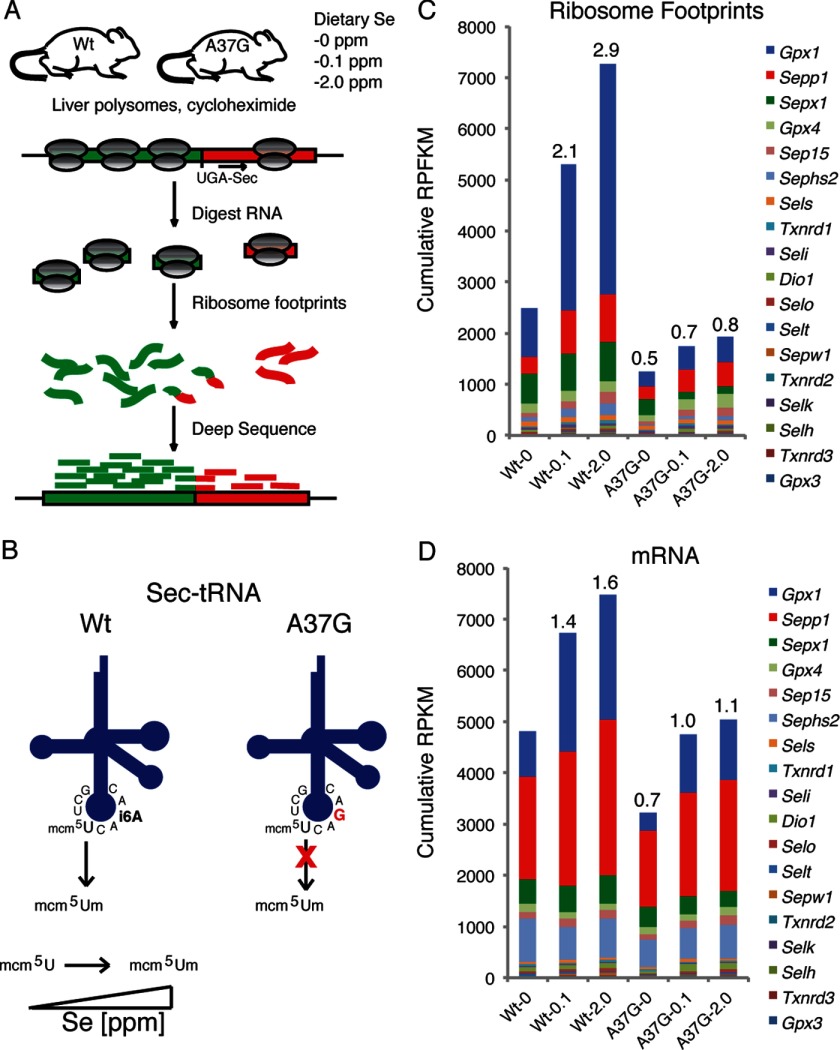
FIGURE 1.
Overview of experimental design and changes in global selenoproteome synthesis as a function of dietary selenium and Um34 methylation of Sec-tRNA[Ser]Sec. A, livers were harvested from WT mice and TrspA37G transgenic mice (A37G) fed diets supplemented with 0, 0.1, or 2.0 ppm selenium. Liver tissue was subjected to ribosome profiling, deep sequencing, and alignment to selenoprotein mRNAs. B, a schematic shows WT tRNA[Ser]Sec and A37G tRNA[Ser]Sec in a cloverleaf model with the base sequence in the anticodon loop and locations of mcm5U, mcm5Um, isopentenyladenosine, and A37G indicated. Increasing dietary selenium levels enhance Um34 synthesis, whereas the A37G change prevents Um34 synthesis. C, shown is the cumulative fraction of ribosome footprints (RPFKM, ribosome footprint/kilobase/million mapped reads) mapping to each selenoprotein mRNA coding sequence. D, shown is the cumulative fraction of RNA sequence reads (RPKM, RNA reads/kilobase/million mapped reads) mapping to each selenoprotein mRNA. The fold change relative to WT mice fed selenium-deficient diets is shown above each column in C and D.
Um34 synthesis is the last step in maturation of tRNA[Ser]Sec and is dependent on an intact tertiary structure (36), prior synthesis of all base modifications, including isopentenyladenosine at position 37 (24, 26), and aminoacylation of the tRNA (37). When mammalian cells or tissues are selenium-deficient, the mcm5U isoform is enriched relative to the mcm5Um isoform, whereas the opposite is true under selenium sufficiency (Fig. 1B). A37G prevents Um34 synthesis (24, 26). The TrspA37G transgenic mouse, which encodes 20 copies of the A37G transgene generates a Sec-tRNA[Ser]Sec population consisting of a much higher proportion of mutant than wild type tRNA[Ser]Sec (24).
We fed WT and TrspA37G mice defined diets supplemented with 0, 0.1, or 2.0 ppm selenium. Livers were harvested, and ribosome profiling experiments were performed as described under “Experimental Procedures.” Undigested total RNA was processed for RNA-Seq in parallel to estimate changes in mRNA abundance. Ribosome-protected fragments and total RNA fragments were aligned to an mRNA database derived from RefSeq. Analysis of tissue-derived ribosome footprint mRNA alignments to all CDSs longer than 1000 nt (supplemental Fig. S1A) revealed, as expected, that ribosome footprints were highly enriched in CDSs relative to the UTRs, and strong triplet phasing was observed corresponding to the step size of the ribosome. Importantly, no global bias was observed in the average ribosome protection toward the 3′ end of CDSs, indicating ribosomes were rapidly captured during tissue lysis. As has been previously reported (29, 31), a number of RPFs mapped to 5′-UTRs, likely reflecting ribosome occupancy of upstream open reading frames, and there was an elevation in RPFs found near the initiation codon. Thus, we show here that ribosome profiling in intact mammalian tissue captures the expected features of actively translating ribosomes.
The effects of altered dietary selenium and Sec-tRNA[Ser]Sec modification on translation and mRNA abundance of the selenoproteome were examined by aligning ribosome footprint or total RNA sequence reads to the mRNA reference sequences for all 24 selenoproteins in mice. Ribosome density and mRNA abundance were assessed by counting the number of reads mapping to each mRNA corrected for the mRNA length and normalized to the total mapped reads for each sample (reads per kilobase per million mapped reads: RPFKM for ribosome footprints and RPKM for total RNA fragments) (38). The global effect of dietary selenium levels on ribosome activity and mRNA abundance across the selenoproteome is shown as the cumulative ribosome footprint RPFKM (Fig. 1C) and mRNA RPKM (Fig. 1D) for all selenoproteins with greater than 25 mapped ribosome-protected fragments or mRNA sequences in the sample derived from WT mice fed diets supplemented with 0.1 ppm selenium. The addition of 0.1 or 2.0 ppm selenium to the diets of WT mice increased global selenoprotein translation and mRNA abundance compared with the levels seen in mice fed selenium-deficient diets. Selenoprotein translational activity, mRNA abundance, and magnitude of response to increased dietary selenium were reduced in TrspA37G mice. Although both mRNA abundance and translational activity of the selenoproteome were affected, the greatest changes in response to increased dietary selenium levels in WT mice (∼2–3×) was observed for ribosome footprint RPFKMs, demonstrating that translational control plays a central role in the selenium-dependent regulation of the liver selenoproteome.
Selenium Availability and A37G Sec-tRNA[Ser]Sec Have Gene-specific Effects on Selenoprotein mRNA Abundance and Ribosome Density
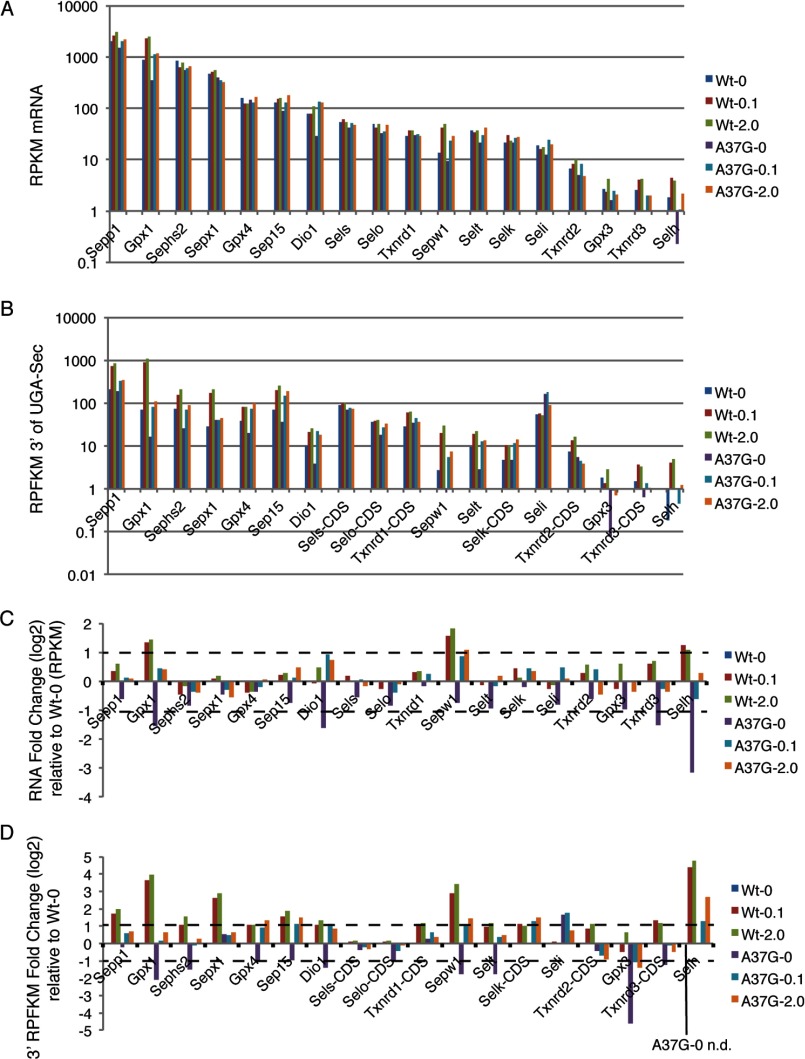
We applied RNA-Seq to quantify selenoprotein mRNA abundance as a function of dietary selenium and Um34 methylation. The RNA RPKM measurements were first validated by comparison to quantitative real-time PCR (see supplemental Fig. S2). RPKM and quantitative real-time PCR measurements for the selenoprotein mRNAs in each sample were highly correlated (r > 0.95). Selenoprotein mRNA levels (Fig. 2A) span nearly 3 orders of magnitude with selenoprotein P (Sepp1), Gpx1, selenophosphate synthetase 2 (Sephs2), and Sepx1 being highly abundant in mouse liver. In WT mice, Gpx1, Sepw1, and selenoprotein H (Selh) mRNA levels were most sensitive to dietary selenium availability with the addition of 0.1 or 2.0 ppm selenium to diets, resulting in a >2-fold increase in mRNA abundance (Fig. 2C and supplemental Fig. S2). In selenium-deficient TrspA37G mice, the abundance of these three mRNAs was reduced relative to that seen in selenium-deficient WT mice, and RNA levels were restored to varying degrees by the addition of selenium to the diet. Deiodinase 1 (Dio1) showed a marked decrease in mRNA abundance in selenium-deficient TrspA37G mice that was increased by ∼5-fold in the TrspA37G mutants fed selenium- supplemented diets. An effect of this magnitude on Dio1 mRNA was not observed in samples from WT mice.
FIGURE 2.
Quantitative analysis of selenoprotein mRNA levels and ribosome footprints. A, selenoprotein mRNA levels were determined by RNA abundance analysis (RPKM, RNA reads/kilobase/million mapped reads) of deep sequencing data for WT and TrspA37G mice (A37G) fed diets supplemented with 0, 0.1, or 2.0 ppm selenium. B, analysis was the same as A except for RPFKMs (ribosome footprint reads/kilobase/million mapped reads). RPFKMs were determined for all selenoproteins 3′ of the UGA-Sec codon with the exception of Sels, Txnrd1, Selk, Selo, Txnrd2, and Txnrd3 that have UGA-Sec codons located near the C terminus. For these selenoproteins, CDS ribosome footprinting RPFKMs are reported (Sels-CDS, Txnrd1-CDS, Selk-CDS, Selo-CDS, Txnrd2-CDS, and Txnrd3-CDS). C, analysis was the same as A and was expressed as the log2 changes in selenoprotein mRNA levels measured by RPKM relative to the corresponding values observed in WT selenium-deficient mice. D, analysis was the same as B, expressed as the log2 change in selenoprotein ribosome footprint RPFKM relative to the corresponding RPFKM values observed in selenium-deficient WT mice. n.d., not determined.
Next we examined translational activity on selenoprotein mRNAs. As Sec incorporation efficiency is not 100%, we surmised that ribosome density downstream of the UGA-Sec codon would provide the most accurate measure of translational activity leading to the production of full-length protein. CDS RPFKM measurements downstream of the UGA-Sec codon are shown in Fig. 2B and supplemental Fig. S2. For some selenoprotein mRNAs, the UGA-Sec codon is near the 3′ end of the CDS (selenoprotein S (Sels), selenoprotein O (Selo), Txnrd1, Txnrd2, Txnrd3, and selenoprotein K (Selk)). In these cases, ribosome footprint RPFKMs were calculated for the entire CDS and, consequently, may not reflect the production of functional full-length Sec-containing protein due to the fraction of ribosomes that may terminate at the UGA-Sec codon. For all selenoproteins with non-C-terminal UGA-Sec codons with the exception of Seli (discussed below) and Gpx3, there was an increase in RPFKMs downstream of the UGA-Sec codon in WT mice that ranged from ∼2 to nearly 30-fold in mice fed selenium supplemented diets relative to those on selenium-deficient diets (Fig. 2D). The largest increases were seen for Gpx1 (13× and 16×), Sepx1 (6× and 8×), Sepw1 (7× and 11×), and Selh (21× and 27×). Similarly, selenium addition to the diets of TrspA37G mice resulted in an increase in 3′ ribosome footprint RPFKMs, although in many cases to a lesser extent than seen in WT mice. Notably, there was a substantial reduction in the total RPFKMs in TrspA37G mice fed selenium-supplemented diets relative to their WT counterparts for Gpx1, Sepx1, Sepw1, Selh (and to a lesser extent for Sepp1, Sephs2, Selenoprotein T (Selt), and Gpx3), suggesting that Um34 methylation was required to varying degrees for efficient synthesis of these selenoproteins. In contrast, 3′ RPFKMs in TrspA37G mice were restored to WT levels by selenium supplementation for Gpx4, Sep15, and Dio1.
The relative selenium-dependent change in abundance of the three proteins in which 3′ RPFKMs were significantly reduced in the TrspA37G mutant (Gpx1, Sepw1, Sepx1) and four where RPFKMs were unaffected or affected to a lesser degree (Gpx4, Sep15, Selt, Txnrd1) was determined by Western blot analysis (Fig. 3). Comparisons to the changes in RPFKMs confirmed qualitatively similar results supporting the validity of utilizing ribosome profiling to approximate changes in selenoprotein levels.
FIGURE 3.
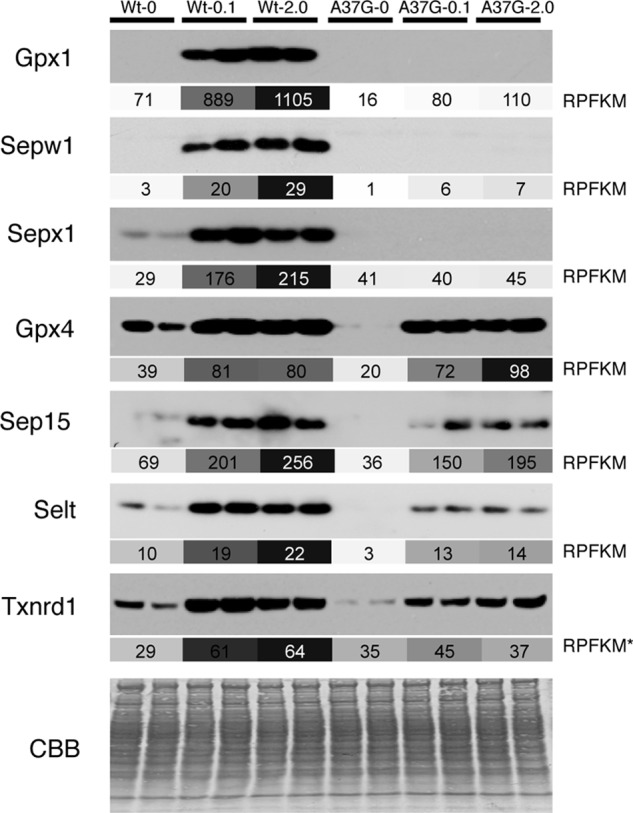
Comparison of Western blots and ribosome profiling measurements of selenoprotein synthesis. Western blots of Gpx1, Sepw1, Sepx1, Gpx4, Sep15, Selt, and Txnrd1 from two biological samples are shown above the corresponding RPFKM (ribosome footprint reads/kilobase/million mapped reads) values in shaded boxes. For each selenoprotein, RPFKM values are shaded according to a continuous scale from 0 RPFKM (white) to the highest RPFKM value for that gene (dark gray). RPFKM values are for the portion of the mRNA downstream of the UGA-Sec codon except for Txnrd1, where the RPFKM value is for the CDS (RPFKM*). CBB, Coomassie Blue staining.
Sec Incorporation Efficiency Accounts for the Selenium-dependent Translational Control of Selenoprotein Synthesis
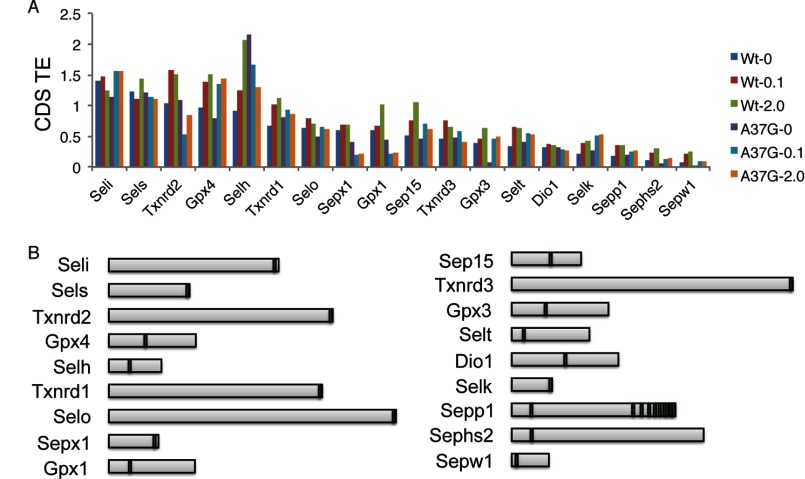
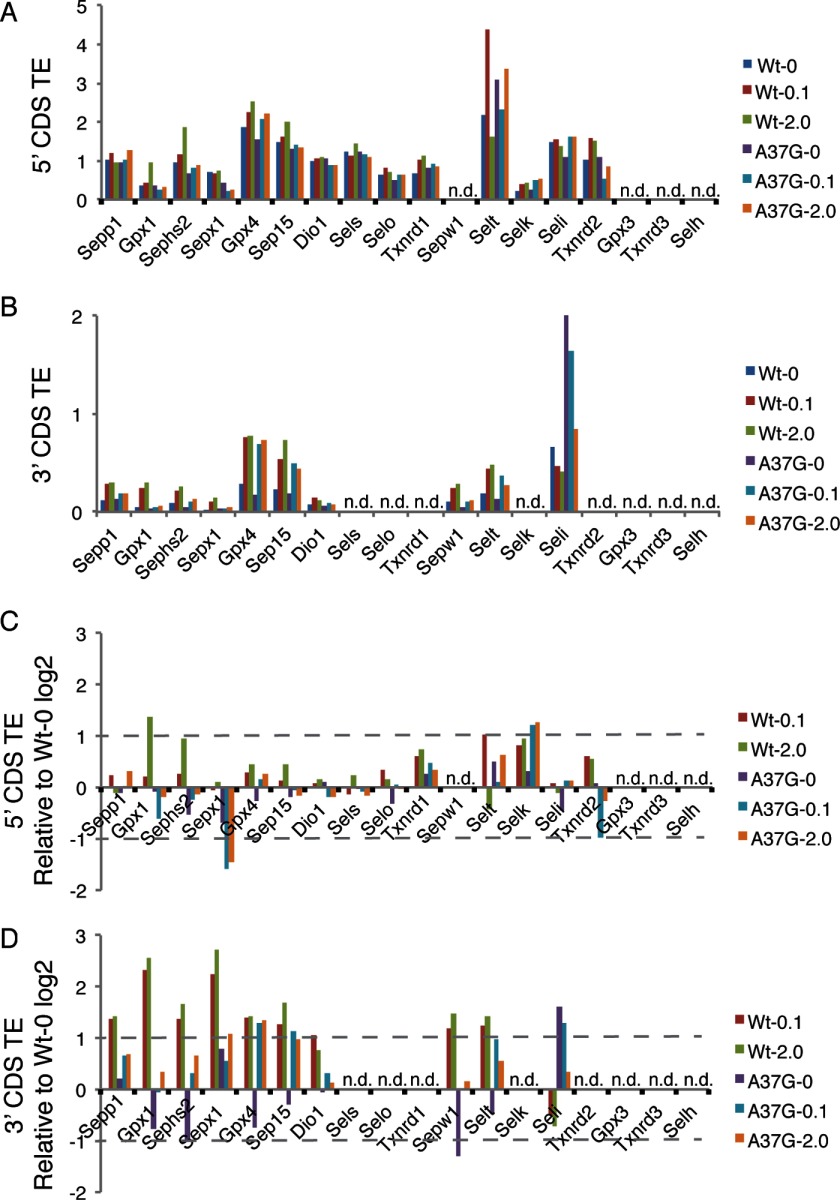
Ribosome footprint RPFKMs reflect the number of ribosomes actively synthesizing protein for each mRNA, whereas translational efficiency (TE) (the amount of ribosome footprints normalized to mRNA abundance, RPFKM/RPKM) provides a means of estimating translational regulation. In Fig. 4A, the TE across the CDS is shown for abundant selenoproteins in liver. A broad range of TEs is observed with Seli, Sels, and Txnrd2, examples of selenoprotein mRNAs with apparent high translational efficiency (see below), whereas Sepp1, Sephs2, and Sepw1 mRNAs are mRNAs that appear to have low translational efficiency. Assuming that the average translation elongation rate preceding and following the UGA-Sec codon is constant, CDS TE is primarily determined by both the efficiency of translation initiation and the efficiency of Sec insertion. As Sec incorporation efficiency is not 100% and it is conceivable that slow decoding of the UGA-Sec codon could slow the elongation rates of upstream ribosomes, the overall CDS TE may be strongly influenced by both the efficiency of Sec incorporation and the position of the UGA-Sec codon within the CDS (Fig. 4B), thus making interpretation of whole CDS RPFKM values challenging. To circumvent these limitations, we determined the TE for regions upstream (5′ TE) and downstream (3′ TE) of the UGA-Sec codon, independently. However, for some selenoproteins it was not possible to obtain TE measurements either because the UGA-Sec codon was too near the 5′ or 3′ ends or the interrogated region had RPKM or RPFKM values below a threshold value of 10 (“Experimental Procedures” and Ref. 26).
FIGURE 4.
Translational efficiency of selenoprotein coding sequences. A, translational efficiency was calculated as the ribosome footprint RPFKMs/RNA RPKMs across each CDS for WT and TrspA37G mice fed diets supplemented with 0, 0.1, or 2.0 ppm selenium diets. B, relative length and position of the UGA-Sec codon (vertical bars) are shown for each selenoprotein.
The selenoproteins revealed a range of 5′ TEs, with Selt and Gpx4 having the highest TE across all dietary selenium levels in both WT and TrspA37G mice, whereas Gpx1 and Selk are examples of selenoproteins with low 5′ TEs (Fig. 5A). The addition of 0.1 or 2.0 ppm selenium resulted in less than a 2-fold increase in 5′ TE relative to selenium-deficient WT mice with the exception of Gpx1 in WT mice fed 2.0 ppm selenium diets, where the 5′ TE increased 2.6-fold, and for Selk in TrspA37G mice fed 0.1 and 2.0 ppm selenium diets, where the 5′ TE increased 2.3- and 2.4-fold, respectively (Fig. 5C). Furthermore, the 5′ TE of Sepx1 in TrspA37G mice was reduced by 0.63-, 0.33-, and 0.37-fold compared with selenium-deficient WT mice, suggesting that the mcm5U isoform of Sec-tRNA[Ser]Sec may have an effect on either translation initiation or elongation rates upstream of the UGA-Sec codon of Sepx1.
FIGURE 5.
Translational efficiency 5′ and 3′ of UGA-Sec codons. Translational efficiency (RPFKM/RPKM) of selenoprotein-coding sequences located either 5′ (A) or 3′ (B) of UGA-Sec codon (5′ or 3′ of the first UGA-Sec codon for Sepp1) in samples from WT and TrspA37G (A37G) mice fed diets supplemented with 0, 0.1, or 2.0 ppm selenium. C, same as A, expressed as the log 2 change in 5′ TE relative to the corresponding values measured in WT mice fed selenium-deficient diets. D, same as B expressed as the log 2 change in 3′ TE relative to the corresponding values measured in WT mice fed selenium-deficient diets. Selenoprotein mRNAs were excluded from this analysis (n.d.) if 5′ or 3′ RPFKM values were <10 in Wt-0.1 sample or if the UGA-Sec was located near the start (5′ TE) or stop codon (3′ TE).
In contrast to the relatively small changes in 5′ TE, a complex pattern of selenium-dependent changes in 3′ TE was observed reflecting changes in Sec incorporation efficiency depending on both dietary selenium levels and Um34 methylation (Fig. 5, B and D). For all mRNAs examined with the exception of Seli in TrspA37G mice, the 3′ TE was reduced compared with the 5′ TE, indicating that Sec incorporation is not 100% efficient (compare Fig. 5, A to B). In WT mice, the addition of selenium to the diet increased 3′ TEs for Gpx1, Sepx1, Sepw1, Sepp1, Sephs2, Dio1 Gpx4, Sep15, and Selt from ∼2-fold to nearly 8-fold (Fig. 5D). In contrast, we found the opposite effect for Seli, where the 3′ TE for Seli was slightly decreased in WT mice fed selenium- supplemented diets and increased under all selenium dietary levels in TrspA37G mice (3.1-fold in diets with 0 ppm selenium, 2.4-fold with 0.1 ppm selenium, and 1.3-fold with 2.0 ppm selenium, compared with WT selenium-deficient mice). In the TrspA37G mouse samples, three selenoprotein mRNAs, Gpx4, Sep15, and Selt, showed a selenium-dependent increase in 3′ TE to approximately the same levels as was observed in WT mice. In contrast, Gpx1, Sepx1, Sepw1, Dio1, Sepp1, and Sephs2 showed a reduced response to dietary selenium supplementation in the TrspA37G mouse samples, suggesting that translation of these four mRNAs requires Um34 methylation (to varying degrees) for efficient Sec incorporation and translational regulation by dietary selenium.
Ribosome Density Near the UGA-Sec Codon and in the 5′-UTR
Slow decoding of UGA-Sec codons resulting in ribosome pausing has been postulated to be a kinetic feature of Sec incorporation (34, 39). Here we find little evidence of excess ribosome footprints near UGA-Sec codons with the notable exceptions of four selenoprotein mRNAs, Gpx1, Sepx1, Sepw1, and Selh, where the number of ribosome footprints either at the UGA codon or in the 5 preceding codons exceeded 20% of the total footprints mapping to the respective mRNA (Table 1). Fig. 6 shows the ribosome footprint coverage across the mRNA (Fig. 6, A, C, E, and G) and the A-site codon location (Fig. 6. B, D, F, and H) of ribosome footprints mapping near the UGA-Sec codon for these mRNAs. Only for Sepx1 were there a large number of footprints with A-sites mapping to the UGA-Sec codon (Fig. 6D). The majority of footprints mapped to the −2 codon relative to the UGA-Sec codon for Gpx1 (Fig. 6B) and to the −4 codon for Sepw1 and Selh (Fig. 6, F and H).
TABLE 1.
Fraction of ribosome footprints near UGA-Sec codons
The number of ribosome footprints with A-sites mapping near the UGA-Sec codon (−5 to +1 codons) is shown as a percentage of the total ribosome footprints mapping to each respective selenoprotein gene. Data are shown for WT or TrspA37G (A37G) mice fed diets supplemented with 0, 0.1, or 2.0 ppm selenium.
| Wt-0 | Wt-0.1 | Wt-2.0 | A37G-0 | A37G-0.1 | A37G-2.0 | |
|---|---|---|---|---|---|---|
| Gpx1 | 80 | 64 | 68 | 86 | 71 | 64 |
| Selh | 38 | 36 | 49 | 50 | 54 | 58 |
| Sepw1 | 34 | 26 | 27 | 35 | 23 | 36 |
| Sepx1 | 31 | 42 | 32 | 37 | 30 | 29 |
| Dio1 | 10 | 7 | 11 | 9 | 8 | 5 |
| Gpx3 | 7 | 3 | 8 | 0 | 4 | 11 |
| Selt | 4 | 3 | 2 | 8 | 2 | 2 |
| Sep15 | 3 | 3 | 4 | 7 | 4 | 2 |
| Selk | 3 | 2 | 1 | 6 | 4 | 4 |
| Sepp1 | 2 | 1 | 2 | 3 | 2 | 1 |
| Gpx4 | 2 | 2 | 2 | 1 | 2 | 3 |
| Seli | 2 | 2 | 1 | 3 | 1 | 2 |
| Sephs2 | 1 | 1 | 2 | 2 | 1 | 1 |
| Selo | 1 | 1 | 1 | 1 | 1 | 1 |
| Sels | 1 | 2 | 2 | 1 | 1 | 2 |
| Txnrd1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Txnrd2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Txnrd3 | 0 | 0 | 1 | 0 | 0 | 2 |
To confirm accurate assignment of A-site codons, ribosome footprints were mapped to the Xbp1 mRNA, which is known to have a translational pause site at Asn-256 (31, 40), and this site is found to be a predominant site of ribosome pausing in our experiment (supplemental Fig. S3). The A-site relative to AUG initiation codons and UGA termination codons of RefSeq mRNAs are also shown in supplemental Fig. S3. The A-site codons start abruptly at the +1 codon (the AUG is in the P-site during initiation) and stop at the codon immediately preceding UGA termination codons. Ribosome footprint size was also examined at UGA-Sec codons to determine if the size was altered in such a way that it might affect A-site assignment at UGA-Sec codons. The results show that altered footprint size was not skewed relative to the size of ribosome footprints at other CDS locations.
Finally, we observed elevated mRNA protection in the 5′-UTR of several selenoprotein mRNAs. The highest levels of 5′-UTR protection were observed for Selh, Sephs2, Txnrd1, and Selt, where the fraction approached or exceeded 10% that of the total ribosome-protected fragments mapping to each respective mRNA (Table 2). Ribosome footprint coverage of 5′-UTRs is shown for Selh (Fig. 6G) as well as Sephs2 and Selt (Fig. 7). 5′-UTR ribosome protection were also observed, albeit at low levels, for Sepx1 (Fig. 6C), Sep15, Selk, and Sepp1.
TABLE 2.
Fraction of ribosome footprints in 5′-UTRs
The number of ribosome footprints with A-sites mapping within the 5′-UTR (excluding the two codons preceding the AUG codon) are shown as a percentage of the total ribosome footprints mapping to each respective selenoprotein gene. Data are shown for WT or TrspA37G (A37G) mice fed diets supplemented with 0, 0.1, or 2.0 ppm selenium.
| Wt-0 | Wt-0.1 | Wt-2.0 | A37G-0 | A37G-0.1 | A37G-2.0 | |
|---|---|---|---|---|---|---|
| Gpx1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Selh | 42 | 33 | 17 | 38 | 16 | 11 |
| Sepw1 | 13 | 2 | 4 | 0 | 6 | 5 |
| Sepx1 | 5 | 4 | 5 | 4 | 4 | 5 |
| Dio1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gpx3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Selt | 9 | 8 | 13 | 8 | 8 | 10 |
| Sep15 | 4 | 2 | 1 | 4 | 2 | 1 |
| Selk | 6 | 8 | 7 | 1 | 8 | 7 |
| Sepp1 | 7 | 4 | 5 | 5 | 4 | 4 |
| Gpx4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Seli | 1 | 1 | 2 | 1 | 1 | 1 |
| Sephs2 | 33 | 19 | 18 | 31 | 25 | 23 |
| Selo | 0 | 0 | 0 | 0 | 0 | 0 |
| Sels | 0 | 0 | 0 | 0 | 0 | 0 |
| Txnrd1 | 10 | 10 | 14 | 3 | 6 | 10 |
| Txnrd2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Txnrd3 | 3 | 0 | 0 | 0 | 0 | 0 |
FIGURE 7.
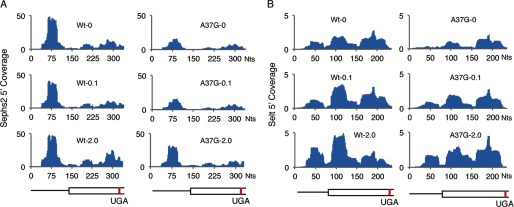
Ribosome protection in the 5′-UTR. The number of ribosome footprints mapping to each nucleotide across the 5′-UTR and coding sequence extending 10 nt beyond the UGA-Sec codon of Sephs2 (A) and Selt (B), normalized to total mapped reads (Coverage) is shown for each mRNA in livers of WT or TrspA37G (A37G) mice fed diets supplemented with 0, 0.1, or 2.0 ppm selenium. Below the histograms in A and B is a schematic of each mRNA. Thin lines represent the 5′-UTR, and boxed regions are the open reading frame up to, and extending 10 nt beyond the UGA-Sec codon. Red lines indicate the position of the UGA-Sec codon.
DISCUSSION
Regulation of the selenoproteome by dietary selenium intake is a multifaceted process involving gene-specific changes in both mRNA abundance and ribosome activity (Fig. 1). We find that selenoprotein mRNA levels in WT mouse liver are relatively resistant to changing levels of dietary selenium with the notable exception of Gpx1, Sepw1, and Selh in both WT and TrspA37G mice as well as Dio1 in selenium-deficient TrspA37G mice (Fig. 2 and supplemental Fig. S2). Although changes in transcriptional regulation by dietary selenium cannot be excluded as a contributing factor, we suggest that altered rates of mRNA turnover and, more specifically, susceptibility to NMD based on the efficiency of Sec insertion relative to translational termination at the UGA-Sec codon is the more likely explanation. In support of this notion are the observations that Gpx1 is a known target of NMD (18, 41) with the degree of susceptibility to NMD depending upon selenium availability. Also, Sepw1 mRNA abundance in rat L8 cells is affected by mRNA turnover, not transcription, when selenium levels are altered (42). Dio1 may be an exceptional case, as a reduction in mRNA abundance was only observed in selenium-deficient TrspA37G mice. The three deiodinases in mammals (the other two deiodinases, Dio2 and Dio3, were below detection limits in liver) are involved in the biosynthesis and interconversion of thyroid hormones between the inactive and active form, T4 and T3, respectively (43). As the Dio1 gene contains thyroid hormone response promoter elements (44), it is a distinct possibility that altered levels of the active T3 hormone, due to changing deiodinase activity in liver or other tissues, may create a feedback loop affecting transcription of Dio1 that may in part contribute to the large decrease in Dio1 mRNA observed in selenium-deficient TrspA37G mice.
We find that the selenoprotein mRNAs in liver that reveal the largest changes in abundance due to altered dietary selenium levels (Gpx1, Sepx1, Sepw1, and Selh) are the same as those that reveal the greatest changes in active translation and Sec incorporation efficiency. The data presented in Fig. 5 further clarify the mechanism of translational control. Although selenium availability appears to have modest effects on translation initiation estimated by changing translational efficiency upstream of the UGA-Sec codon, we find dynamic selenium-dependent changes in ribosome density downstream of the UGA-Sec codon, supporting a model in which Sec incorporation is a limiting step during biosynthesis of selenoproteins and a primary target for selenium regulation of selenoprotein expression.
It has been previously reported that the selenium- induced methylation of Um34 is required for expression of a subset of stress-related selenoproteins (see Fig. 3 and Refs. 24 and 25). Here we extend these findings by showing that either selenium deficiency or expression of an A37G mutant of Sec-tRNA[Ser]Sec severely reduced ribosome density downstream of the UGA-Sec codon for Gpx1, Sepx1, Sepw1, and Selh, reflecting diminished Sec incorporation efficiency. In addition, under selenium-adequate conditions, Sec insertion efficiency remains relatively unaffected by expression of the A37G mutant Sec-tRNA[Ser]Sec for the housekeeping selenoprotein mRNAs, including Gpx4 and Sep15, with many selenoproteins showing intermediate changes. Seli is notable among the selenoproteins in that selenium-adequate or -supplemented diets do not appear to increase Sec insertion efficiency in WT mice, whereas in TrspA37G mice Sec insertion efficiency is increased, suggesting the unexpected possibility that the unmethylated mcm5U Sec-tRNA[Ser]Sec isoform may be preferentially utilized for UGA-Sec decoding of Seli, thus increasing its expression under selenium- limiting conditions. It should be noted that the UGA-Sec codon is near the termination codon of Seli, and thus ribosome density 3′ of the UGA-Sec codon may also be influenced by the efficiency of termination. The Seli protein is a putative CDP-alcohol phosphatidyltransferase involved in the production of phospholipids (45), an activity that may be important for replacing damaged phospholipids due to oxidative stress induced by selenium deficiency.
Furthermore, we find evidence for a high level of ribosome protection immediately upstream of the UGA-Sec codons of Gpx1, Sepx1, Sepw1, and Selh, the same genes that are most affected by dietary selenium levels. Assignment of ribosome A-sites to the 3′ end of RefSeq mRNAs (supplemental Fig. S3) demonstrates a sharp drop-off at the codon preceding standard UGA termination codons. Consequently, the accumulation of ribosomes immediately upstream of UGA-Sec codons must be interpreted with caution. In one scenario, ribosomes may pause at the UGA-Sec codon due to slow decoding, resulting in tightly stacked ribosomes (34, 46) behind the UGA-Sec codon. If ribosomes decoding UGA-Sec codons are not effectively captured by cycloheximide treatment and are released during tissue processing, this could allow for limited movement of the preceding paused ribosomes to positions immediately upstream of the UGA-Sec codon. Alternatively, ribosome pausing may occur before the UGA-Sec codon enters the A-site of the ribosome. In this case, slow decoding of upstream codons could allow time for assembly of the Sec incorporation machinery and reprogramming of ribosomes before decoding of the UGA-Sec codon, thus minimizing competition with termination. Several previous studies have demonstrated that secondary structures downstream of UGA-Sec codons, which may slow local ribosome elongation rates, enhance Sec incorporation efficiency (47, 48). In addition, nascent peptides in the exit channel of the ribosome may provide an alternative mechanism to slow translation in the region preceding the UGA-Sec codon. Furthermore, additional caution is warranted due to library construction biases that may influence quantification of ribosome-protected fragments at any given position. Nevertheless, experimental analysis of the UGA-Sec sequence context of these mRNAs is warranted to illuminate the mechanisms contributing to increased ribosome protection and its contribution to selenium-dependent regulation of Sec incorporation during synthesis of these selenoproteins.
Increased ribosome protection was observed in the 5′-UTRs of several selenoproteins. For Selh, Sephs2, and Selt, a contiguous open reading frame exists between the 5′-UTR footprints and the annotated AUG codons, suggesting the possibility of N-terminal extensions to the selenoprotein or, alternatively, translation of peptides derived from small out-of-frame upstream upstream open reading frames. Extensive genome-wide translation of upstream open reading frames, initiated by both AUG and non-AUG codons, has recently been described in studies utilizing ribosome profiling in mammalian cells and in yeast (29–31). Although the fraction of total ribosome-protected fragments mapping to the 5′-UTR did have an inverse correlation with dietary selenium levels for Selh and Sephs2 in our experiments (Table 2), the relationship between 5′-UTR translation and selenium-dependent regulation of these selenoproteins remains unclear, and the effect is likely to be small based on our observation that 5′ TE, which should in part reflect initiation efficiency, is relatively unaffected by increasing selenium availability. Given the complex relationship between 5′-UTR translation and gene translation, it is possible that 5′-UTR translation is not related to dietary selenium levels but is utilized under other circumstances to regulate selenoprotein translation or to produce selenoproteins with N-terminal extensions.
The data presented here demonstrate that dietary selenium levels can alter the efficiency by which the standard rules of genetic decoding are redefined to allow UGA codons to encode Sec. Importantly, we show that UGA-Sec redefinition is inefficient and a primary determinant of selenoprotein synthesis rates in vivo. Our study provides direct support for a model in which the mcm5U and mcm5Um isoforms of Sec-tRNA[Ser]Sec are differentially utilized to determine Sec incorporation efficiencies in a manner that depends upon the selenoprotein mRNA. Likely candidates for factors involved in discrimination between the two Sec-tRNA[Ser]Sec isoforms include differences between SECIS elements, the local sequence context of the UGA-Sec codon, and their influence on either recruitment of Sec-tRNA[Ser]Sec isoform-specific eEFSec ternary complexes or isoform-specific recognition of the UGA-Sec codon. The efficiency of Sec insertion relative to termination for each selenoprotein mRNA further impacts mRNA turnover rate and abundance. As dietary selenium levels determine the ratio of mcm5U and mcm5Um Sec-tRNA[Ser]Sec isoforms (23), the preferential utilization of one isoform over the other and the resulting effect of altered Sec insertion efficiency on mRNA abundance provides a unifying mechanism to explain how dietary selenium regulates the readout of the genetic code and gene-specific selenoprotein expression.
Supplementary Material
Acknowledgments
We thank Drs. Ray Gesteland and John Atkins (University of Utah) for helpful discussions and Dr. Brian Dalley (University of Utah) for assistance with library construction and deep sequence.
This work was supported, in whole or in part, by the National Institutes of Health (NIH Grant R21ES022716; to M. T. H.) and the Intramural Research Program of the NIH, NCI, Center for Cancer Research (to D. L. H.). This work was also supported by a University of Utah Seed grant (to M. T. H.).

This article contains supplemental Figs. S1–S3.
- Sec
- selenocysteine
- RPF
- ribosome-protected fragment
- UGA-Sec
- UGA selenocysteine codon
- SECIS
- Sec insertion sequence element
- NMD
- nonsense-mediated decay
- mcm5U
- 5-methoxycarbonylmethyluridine
- mcm5Um
- 5-methoxycarbonylmethyl-2′-O-methyluridine
- RPFKM
- reads per kilobase per million mapped reads for ribosome footprints
- RPKM
- reads per kilobase per million mapped reads for total RNA
- CDS
- coding sequence
- Txnrd1
- thioredoxin reductase 1
- Gpx4
- glutathione peroxidase 4
- TE
- translational efficiency
- nt
- nucleotides.
REFERENCES
- 1. Keshan, Disease Research Group (1979) Epidemiologic studies on the etiologic relationship of selenium and Keshan disease. Chin. Med. J. 92, 477–482 [PubMed] [Google Scholar]
- 2. Yang G. Q., Wang S. Z., Zhou R. H., Sun S. Z. (1983) Endemic selenium intoxication of humans in China. Am. J. Clin. Nutr. 37, 872–881 [DOI] [PubMed] [Google Scholar]
- 3. Nuttall K. L. (2006) Evaluating selenium poisoning. Ann. Clin. Lab. Sci. 36, 409–420 [PubMed] [Google Scholar]
- 4. Hatfield D., Berry MJ, Gladyshev VN. (ed) (2012) Selenium: Its Molecular Biology and Role in Human Health, Springer-Verlag New York Inc., New York [Google Scholar]
- 5. Kryukov G. V., Castellano S., Novoselov S. V., Lobanov A. V., Zehtab O., Guigó R., Gladyshev V. N. (2003) Characterization of mammalian selenoproteomes. Science 300, 1439–1443 [DOI] [PubMed] [Google Scholar]
- 6. Mariotti M., Ridge P. G., Zhang Y., Lobanov A. V., Pringle T. H., Guigo R., Hatfield D. L., Gladyshev V. N. (2012) Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS ONE 7, e33066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berry M. J., Banu L., Chen Y. Y., Mandel S. J., Kieffer J. D., Harney J. W., Larsen P. R. (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353, 273–276 [DOI] [PubMed] [Google Scholar]
- 8. Berry M. J., Banu L., Harney J. W., Larsen P. R. (1993) Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 12, 3315–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hill K. E., Lloyd R. S., Burk R. F. (1993) Conserved nucleotide sequences in the open reading frame and 3′ untranslated region of selenoprotein P mRNA. Proc. Natl. Acad. Sci. U.S.A. 90, 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen Q., Chu F. F., Newburger P. E. (1993) Sequences in the 3′-untranslated region of the human cellular glutathione peroxidase gene are necessary and sufficient for selenocysteine incorporation at the UGA codon. J. Biol. Chem. 268, 11463–11469 [PubMed] [Google Scholar]
- 11. Copeland P. R., Driscoll D. M. (1999) Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 274, 25447–25454 [DOI] [PubMed] [Google Scholar]
- 12. Berry M. J., Tujebajeva R. M., Copeland P. R., Xu X. M., Carlson B. A., Martin G. W., 3rd, Low S. C., Mansell J. B., Grundner-Culemann E., Harney J. W., Driscoll D. M., Hatfield D. L. (2001) Selenocysteine incorporation directed from the 3′UTR. Characterization of eukaryotic EFsec and mechanistic implications. Biofactors 14, 17–24 [DOI] [PubMed] [Google Scholar]
- 13. Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A. (2000) Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19, 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hadley K. B., Sunde R. A. (2001) Selenium regulation of thioredoxin reductase activity and mRNA levels in rat liver. J. Nutr. Biochem. 12, 693–702 [DOI] [PubMed] [Google Scholar]
- 15. Hill K. E., Lyons P. R., Burk R. F. (1992) Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem. Biophys. Res. Commun. 185, 260–263 [DOI] [PubMed] [Google Scholar]
- 16. Lei X. G., Evenson J. K., Thompson K. M., Sunde R. A. (1995) Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J. Nutr. 125, 1438–1446 [DOI] [PubMed] [Google Scholar]
- 17. Christensen M. J., Burgener K. W. (1992) Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J. Nutr. 122, 1620–1626 [DOI] [PubMed] [Google Scholar]
- 18. Moriarty P. M., Reddy C. C., Maquat L. E. (1998) Selenium deficiency reduces the abundance of mRNA for selenium-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol. Cell. Biol. 18, 2932–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weiss S. L., Sunde R. A. (1998) Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA 4, 816–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Budiman M. E., Bubenik J. L., Miniard A. C., Middleton L. M., Gerber C. A., Cash A., Driscoll D. M. (2009) Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol. Cell 35, 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Low S. C., Grundner-Culemann E., Harney J. W., Berry M. J. (2000) SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 19, 6882–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Squires J. E., Stoytchev I., Forry E. P., Berry M. J. (2007) SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol. Cell. Biol. 27, 7848–7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diamond A. M., Choi I. S., Crain P. F., Hashizume T., Pomerantz S. C., Cruz R., Steer C. J., Hill K. E., Burk R. F., McCloskey J. A. (1993) Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA[Ser]Sec. J. Biol. Chem. 268, 14215–14223 [PubMed] [Google Scholar]
- 24. Moustafa M. E., Carlson B. A., El-Saadani M. A., Kryukov G. V., Sun Q. A., Harney J. W., Hill K. E., Combs G. F., Feigenbaum L., Mansur D. B., Burk R. F., Berry M. J., Diamond A. M., Lee B. J., Gladyshev V. N., Hatfield D. L. (2001) Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol. Cell. Biol. 21, 3840–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carlson B. A., Xu X. M., Gladyshev V. N., Hatfield D. L. (2005) Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J. Biol. Chem. 280, 5542–5548 [DOI] [PubMed] [Google Scholar]
- 26. Carlson B. A., Moustafa M. E., Sengupta A., Schweizer U., Shrimali R., Rao M., Zhong N., Wang S., Feigenbaum L., Lee B. J., Gladyshev V. N., Hatfield D. L. (2007) Selective restoration of the selenoprotein population in a mouse hepatocyte selenoproteinless background with different mutant selenocysteine tRNAs lacking Um34. J. Biol. Chem. 282, 32591–32602 [DOI] [PubMed] [Google Scholar]
- 27. Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shrimali R. K., Irons R. D., Carlson B. A., Sano Y., Gladyshev V. N., Park J. M., Hatfield D. L. (2008) Selenoproteins mediate T cell immunity through an antioxidant mechanism. J. Biol. Chem. 283, 20181–20185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerashchenko M. V., Lobanov A. V., Gladyshev V. N. (2012) Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 109, 17394–17399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ingolia N. T., Lareau L. F., Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arava Y., Boas F. E., Brown P. O., Herschlag D. (2005) Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 33, 2421–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hendrickson D. G., Hogan D. J., McCullough H. L., Myers J. W., Herschlag D., Ferrell J. E., Brown P. O. (2009) Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7, e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fletcher J. E., Copeland P. R., Driscoll D. M. (2000) Polysome distribution of phospholipid hydroperoxide glutathione peroxidase mRNA. Evidence for a block in elongation at the UGA/selenocysteine codon. RNA 6, 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nasim M. T., Jaenecke S., Belduz A., Kollmus H., Flohé L., McCarthy J. E. (2000) Eukaryotic selenocysteine incorporation follows a nonprocessive mechanism that competes with translational termination. J. Biol. Chem. 275, 14846–14852 [DOI] [PubMed] [Google Scholar]
- 36. Kim L. K., Matsufuji T., Matsufuji S., Carlson B. A., Kim S. S., Hatfield D. L., Lee B. J. (2000) Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. Rna 6, 1306–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim J. Y., Carlson B. A., Xu X. M., Zeng Y., Chen S., Gladyshev V. N., Lee B. J., Hatfield D. L. (2011) Inhibition of selenocysteine tRNA[Ser]Sec aminoacylation provides evidence that aminoacylation is required for regulatory methylation of this tRNA. Biochem. Biophys. Res. Commun. 409, 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 39. Stoytcheva Z., Tujebajeva R. M., Harney J. W., Berry M. J. (2006) Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol. Cell. Biol. 26, 9177–9184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yanagitani K., Imagawa Y., Iwawaki T., Hosoda A., Saito M., Kimata Y., Kohno K. (2009) Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell 34, 191–200 [DOI] [PubMed] [Google Scholar]
- 41. Sunde R. A., Raines A. M. (2011) Selenium regulation of the selenoprotein and nonselenoprotein transcriptomes in rodents. Adv. Nutr. 2, 138–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gu Q. P., Ream W., Whanger P. D. (2002) Selenoprotein W gene regulation by selenium in L8 cells. Biometals 15, 411–420 [DOI] [PubMed] [Google Scholar]
- 43. Bianco A. C., Salvatore D., Gereben B., Berry M. J., Larsen P. R. (2002) Biochemistry, cellular, and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr. Rev. 23, 38–89 [DOI] [PubMed] [Google Scholar]
- 44. Toyoda N., Zavacki A. M., Maia A. L., Harney J. W., Larsen P. R. (1995) A novel retinoid X receptor-independent thyroid hormone response element is present in the human type 1 deiodinase gene. Mol. Cell. Biol. 15, 5100–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Horibata Y., Hirabayashi Y. (2007) Identification and characterization of human ethanolaminephosphotransferase1. J. Lipid Res. 48, 503–508 [DOI] [PubMed] [Google Scholar]
- 46. Wolin S. L., Walter P. (1988) Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7, 3559–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Howard M. T., Aggarwal G., Anderson C. B., Khatri S., Flanigan K. M., Atkins J. F. (2005) Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J. 24, 1596–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Howard M. T., Moyle M. W., Aggarwal G., Carlson B. A., Anderson C. B. (2007) A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA 13, 912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.