Background: The JIL-1 histone H3 serine 10 (H3S10) tandem kinase functions to maintain active gene expression.
Results: The NH2-terminal domain and both kinase domains are required for H3S10 kinase activity.
Conclusion: The NH2-terminal domain is required for chromatin complex interactions, and H3S10 phosphorylation functions as a causative regulator of chromatin structure.
Significance: Determining how the domain structure of JIL-1 contributes to its function is crucial for understanding regulation of gene expression.
Keywords: Chromatin Histone Modification, Chromatin Remodeling, Drosophila, Phosphorylation, Protein Kinases, H3S10 Phosphorylation, JIL-1 Kinase
Abstract
The JIL-1 kinase localizes to Drosophila polytene chromosome interbands and phosphorylates histone H3 at interphase, counteracting histone H3 lysine 9 dimethylation and gene silencing. JIL-1 can be divided into four main domains, including an NH2-terminal domain, two separate kinase domains, and a COOH-terminal domain. In this study, we characterize the domain requirements of the JIL-1 kinase for histone H3 serine 10 (H3S10) phosphorylation and chromatin remodeling in vivo. We show that a JIL-1 construct without the NH2-terminal domain is without H3S10 phosphorylation activity despite the fact that it localizes properly to polytene interband regions and that it contains both kinase domains. JIL-1 is a double kinase, and we demonstrate that both kinase domains of JIL-1 are required to be catalytically active for H3S10 phosphorylation to occur. Furthermore, we provide evidence that JIL-1 is phosphorylated at serine 424 and that this phosphorylation is necessary for JIL-1 H3S10 phosphorylation activity. Thus, these data are compatible with a model where the NH2-terminal domain of JIL-1 is required for chromatin complex interactions that position the kinase domain(s) for catalytic activity in the context of the state of higher order nucleosome packaging and chromatin structure and where catalytic H3S10 phosphorylation activity mediated by the first kinase domain is dependent on autophosphorylation of serine 424 by the second kinase domain. Furthermore, using a lacO repeat tethering system to target mutated JIL-1 constructs with or without catalytic activity, we show that the epigenetic H3S10 phosphorylation mark itself functions as a causative regulator of chromatin structure independently of any structural contributions from the JIL-1 protein.
Introduction
In Drosophila, histone H3 serine 10 (H3S10)4 phosphorylation by the JIL-1 kinase at interphase functions to maintain active gene expression by serving as a protective epigenetic mark counteracting spreading of H3K9 dimethylation and gene silencing (1–5). Furthermore, JIL-1 is enriched about 2-fold on the male X chromosome and implicated in dosage compensation of transcription due to its association with the male-specific lethal complex (6–9). JIL-1 is a Ser/Thr family tandem kinase that localizes specifically to euchromatic interband regions of polytene chromosomes (6, 10). JIL-1 can be divided into four main domains, including an NH2-terminal domain (NTD), two kinase domains (KDI and KDII), and a COOH-terminal domain (CTD) (6). It has previously been demonstrated that the CTD domain of JIL-1 is sufficient for proper chromatin localization and for rescue of the grossly perturbed polytene chromosome morphology in JIL-1 null mutants (3, 11, 12) and that a JIL-1 construct without the CTD domain has kinase activity for H3S10 despite the fact that it does not localize properly (3, 12). However, in these studies, the properties of the NTD or the relative contributions of the two kinase domains were not determined. Thus, in order to further characterize the domain requirements of the JIL-1 kinase for H3S10 phosphorylation and regulation of chromatin structure, we generated a series of mutated JIL-1 constructs and expressed them in a JIL-1 null mutant background. Our results showed that a JIL-1 construct without the NTD localizes properly to chromatin and rescues mutant polytene chromosome morphology. However, in immunocytochemistry and immunoblot analyses with H3S10ph antibody, no H3S10 phosphorylation could be detected, strongly suggesting that the NTD domain is required for H3S10 kinase activity. Furthermore, we mutated either or both kinase domains to render them “kinase-dead” and assessed their ability to phosphorylate H3S10. The results indicated that both kinase domains are required for kinase activity. In addition, using a LacI/lacO repeat-tethering system (13), we provide evidence that only JIL-1 constructs with H3S10 phosphorylation activity have the capacity to induce a change in higher order chromatin structure from a condensed heterochromatin-like band state to a more open euchromatic interband state.
EXPERIMENTAL PROCEDURES
JIL-1 Fusion Constructs
The full-length LacI-JIL-1 construct (FL) was previously described by Deng et al. (13). LacI-tagged constructs from residues 1–926 lacking the COOH-terminal domain of JIL-1 (ΔCTD), a construct from residues 261–1207 lacking the NH2-terminal domain (ΔNTD), and a COOH-terminal domain construct from residues 927–1207 were cloned into the pUAST vector with the in-frame DNA-binding domain of the LacI repressor from Escherichia coli at the NH2 terminus using standard methods (14). In addition, pUAST constructs were generated using the TransformerTM site-directed mutagenesis kit (Clontech) to introduce K293A (KDI*) and K652A (KDII*) substitutions in the ATP-binding loops for each kinase domain as well as a double mutant combination (KDI*/KDII*). A QuikChange® multisite-directed mutagenesis kit (Agilent) was used to introduce a S425A substitution into the first kinase domain of a full-length JIL-1 cDNA. Subsequently, the mutated JIL-1 cDNA was cloned into the pUASP-HA-FLAG gateway vector according to the manufacturer's instructions (Invitrogen). For protein purification, a TAP-tagged JIL-1 construct from residues 261–1207 lacking the COOH-terminal domain (TAP-JIL-1) was cloned into the pUAST-NTAP vector (15). The TAP tag consists of calmodulin binding peptide and Protein A. The fidelity of all constructs was verified by sequencing at the Iowa State University sequencing facility.
Drosophila melanogaster Stocks
Fly stocks were maintained at 25 °C according to standard protocols (16). The JIL-1z2 null allele is described by Wang et al. (10) and Zhang et al. (17), whereas the Lac operator insertion line P11.3 and the GFP-LacI fusion line are described by Li et al. (18) and Danzer and Wallrath (19). All transgenic lines were generated by standard P-element transformation (BestGene, Inc.), and expression of the transgenes was driven using Sgs3-GAL4 or hsp70-GAL4 drivers introduced by standard genetic crosses. Recombinant JIL-1z2, hsp70-GAL4 chromosomes were generated as described by Ji et al. (20). The hsp70 promoter is leaky and was used without heat shock treatment as described previously (12). Expression levels of each of the JIL-1 constructs were monitored by immunoblot analysis as described below. All driver lines were obtained from the Bloomington Stock Center. Balancer chromosomes and markers are described by Lindsley and Zimm (21). Viability assays were performed by crossing JIL-transgene/JIL-transgene; JIL-1z2/TM6 flies with JIL-1z2 Hsp70-GAL4/TM6 flies. In these crosses, the TM6 chromosome was identified by the Stubble marker. Consequently, the experimental genotypes could be distinguished from balanced heterozygotic flies by the absence of the Stubble marker. The expected Mendelian ratio of non-Stubble to Stubble flies assuming full rescue by the JIL-1 transgenes in the JIL-1 null background was 1:2 because TM6/TM6 is embryonic lethal. Thus, the percentage of rescue of adult viability for each transgene was calculated as observed non-Stubble flies × 200/observed Stubble flies. In each experiment, the degree of rescue was determined on the basis of the analysis of the genotypes from at least 300 surviving progeny.
Immunohistochemistry
Standard polytene chromosome squash preparations were performed as described by Cai et al. (22) using either 1- or 5-min fixation protocols and labeled with antibody as described by Johansen et al. (23). Primary antibodies used for immunocytochemistry included rabbit anti-H3S10ph (Cell Signaling), rabbit anti-H3K9me2 (Upstate Biotechnology), rabbit anti-JIL-1 (6), chicken anti-JIL-1 (7), and anti-HA antibody (Roche Applied Science). DNA was visualized by staining with Hoechst 33258 (Molecular Probes) in PBS. The appropriate species- and isotype-specific Texas Red-, TRITC-, and FITC-conjugated secondary antibodies (Cappel/ICN, Southern Biotech) were used (1:200 dilution) to visualize primary antibody labeling. The final preparations were mounted in 90% glycerol containing 0.5% n-propyl gallate. The preparations were examined using epifluorescence optics on a Zeiss Axioskop microscope, and images were captured and digitized using a cooled Spot CCD camera. Images were imported into Photoshop, where they were pseudocolored, image-processed, and merged. In some images, non-linear adjustments were made to the channel with Hoechst labeling for optimal visualization of chromosomes.
Immunoblot Analysis
Protein extracts were prepared from adult flies or from dissected third instar larval salivary glands homogenized in a buffer containing 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm EDTA, 1 mm EGTA, 0.2% Triton X-100, 0.2% Nonidet P-40, 2 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 1.5 μg/ml aprotinin. Protein separation by SDS-PAGE and electroblot transfer were performed according to standard procedures (14). For these experiments, we used the Bio-Rad Mini PROTEAN III system, electroblotting to 0.2-μm nitrocellulose with transfer buffer containing 20% methanol and in most cases including 0.04% SDS. Primary antibodies for immunoblot analysis included rabbit anti-H3S10ph (Cell Signaling), rabbit anti-histone H3 (Cell Signaling), rabbit anti-JIL-1 (6), chicken anti-JIL-1 (7), and mouse anti-lamin Dm0 (Developmental Studies Hybridoma Bank). The appropriate anti-mouse or anti-rabbit HRP-conjugated secondary antibody (Bio-Rad) (1:3000) was used for visualization of primary antibody. Antibody labeling was visualized using chemiluminescent detection methods (SuperSignal West Pico Chemiluminescent Substrate, Pierce). The immunoblots were digitized using a flatbed scanner (Epson Expression 1680).
Protein Purification and Mass Spectrometry
For TAP-JIL-1 fusion protein purification, the expressed protein was purified from protein extracts of 200 third instar larval salivary glands homogenized in a lysis buffer containing 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm EDTA, 1 mm EGTA, 0.2% Triton X-100, 0.2% Nonidet P-40, 2 mm Na3VO4, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and 1.5 μg/ml aprotinin. Protein lysate was incubated with IgG-agarose beads (Sigma) for 3 h at 4 °C on a rotation wheel. Bound protein beads were washed with lysis buffer five times. Subsequently, the bound proteins were separated on a 4–12% Tris-glycine gradient gel (Bio-Rad) and stained with Coomassie Blue R250. The TAP-JIL-1 gel band was identified based on size and Coomassie staining intensity and cut out of the gel. The cut band was reduced, alkylated with iodoacetamide, and sent to the University of California, Davis, Proteomics Core Facility for analysis with tandem mass spectrometry using standard methods. Tandem mass spectra were extracted, and charge states were deconvoluted and deisotoped by Scaffold version 3.5.1. All MS/MS samples were analyzed using Sequest version 3.3 (Thermo Fisher Scientific). Sequest was set up to search the Uniprot D. melanogaster database (November 2011; 55,391 entries) along with 101 common laboratory contaminant proteins, assuming the digestion enzyme trypsin. Sequest was searched with a fragment ion mass tolerance of 1.00 kDa and a parent ion tolerance of 20 ppm. Oxidation of methionine, acetylation of lysine, trimethylation of arginine, iodoacetamide derivative of cysteine, and phosphorylation of serine, threonine, and tyrosine were specified in Sequest as variable modifications. Scaffold version 3.5.1 (Proteome Software Inc.) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 80.0% probability as specified by the Peptide Prophet algorithm (24). Protein identifications were accepted if they could be established at greater than 80.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (25). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Using these parameters, a false discovery rate was calculated as 1.7% on the peptide level and 0% on the protein level.
RESULTS
JIL-1 Transgene Expression
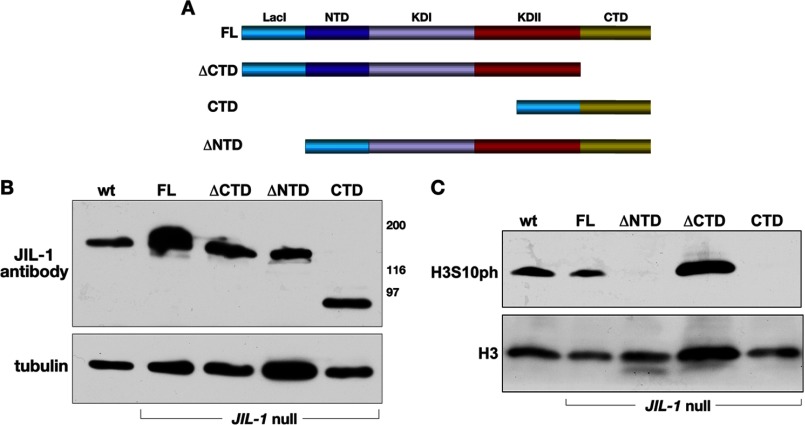
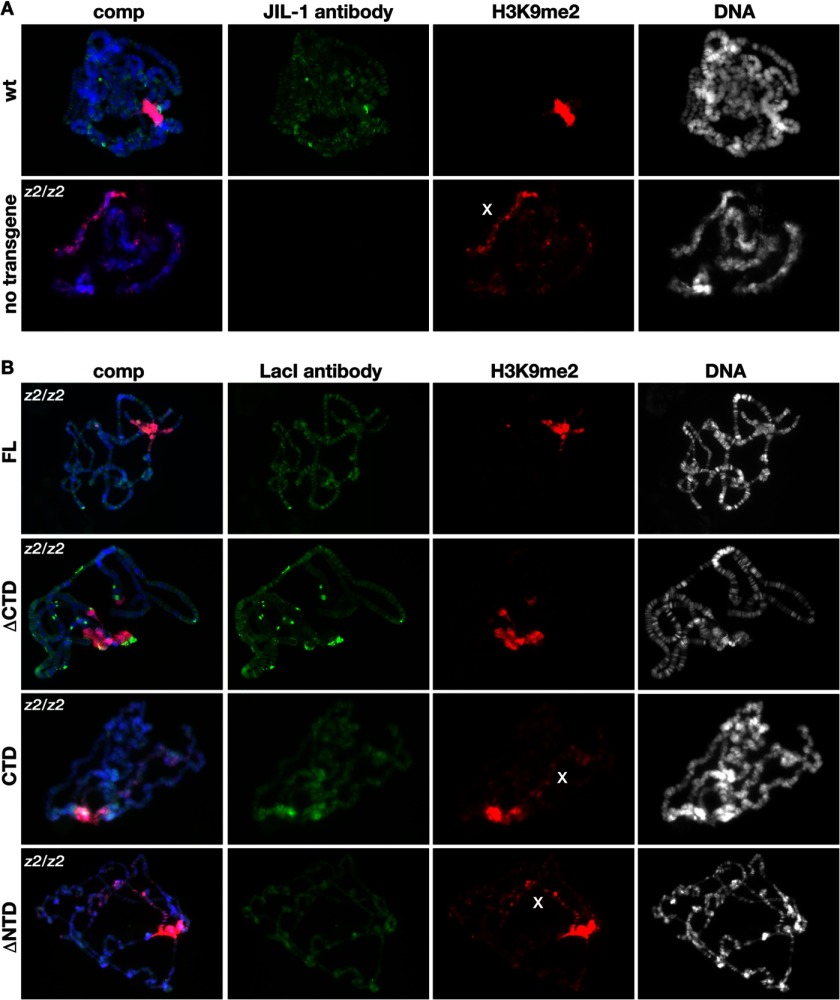
In order to further explore the relative contributions of the different JIL-1 domains to H3S10 phosphorylation and chromatin remodeling, we expressed four LacI-tagged JIL-1 UAS P-element insertion constructs transgenically in wild type and JIL-1 null mutant animals. FL, a construct without the COOH-terminal domain (ΔCTD), a construct without the NH2-terminal domain (ΔNTD), and a construct containing only the COOH-terminal domain (CTD) were made (Fig. 1A). A ΔNTD construct has not been previously characterized; however, the LacI-tagged FL, ΔCTD, and CTD constructs used in this study had properties identical to those reported for similar GFP- or CFP-tagged JIL-1 constructs (3, 12) (Figs. 1 and 2). A transgenic line was selected for each construct that expressed at levels comparable with those of endogenous JIL-1 using a hsp-70-GAL4 driver line as illustrated in Fig. 1B. FL rescued all aspects of the JIL-1 null mutant phenotype, including polytene chromosome morphology (Fig. 2B) and viability, and like endogenous JIL-1, FL was up-regulated on the male X chromosome (data not shown). The ΔCTD lacks the COOH-terminal sequences required for proper chromatin localization, leading to mislocalization of the protein (13). However, it does retain its in vivo kinase activity (Fig. 1C), resulting in ectopic H3S10 phosphorylation (12). The ΔCTD rescues autosome polytene chromosome morphology (Fig. 2B) but only partially rescues that of the male X chromosome in JIL-1 null mutants (12). In contrast, the CTD largely restores JIL-1 null mutant chromosome morphology (Fig. 2B), including that of the male X chromosome (12). Furthermore, when the CTD is expressed in a wild type background, it has a dominant negative effect and displaces endogenous JIL-1 (12).
FIGURE 1.
Expression of transgenic JIL-1 constructs in a JIL-1 null background. A, diagrams of the JIL-1 LacI-tagged constructs analyzed. B, immunoblot labeled with JIL-1 antibody of protein extracts from wild type (wt) and from JIL-1 null salivary glands expressing the FL, the ΔCTD, the ΔNTD, and the CTD, respectively. Labeling with tubulin antibody was used as a loading control. The relative migration of molecular size markers is indicated to the right of the immunoblot in kDa. C, immunoblot labeled with H3S10ph antibody of protein extracts from salivary glands from wild type third instar larvae (wt) and from JIL-1 null salivary glands expressing the FL, the ΔCTD, the ΔNTD, and the CTD, respectively. Labeling with histone H3 antibody was used as a loading control.
FIGURE 2.
Immunocytochemical analysis of polytene chromosome morphology and H3K9 dimethylation in JIL-1 null larvae expressing JIL-1 transgenes. A, comparison of polytene chromosome squash preparations from wild type (wt) and JIL-1 null (z2/z2) third instar larval salivary glands labeled with JIL-1 antibody. B, polytene chromosome squash preparations from JIL-1 null (z2/z2) third instar larval salivary glands expressing the FL, the ΔCTD, the ΔNTD, and the CTD, respectively. Transgenic protein localization (in green) was identified using LacI antibody, H3K9 dimethylation (in red) was identified using H3K9me2 antibody, and DNA (in blue or gray) was labeled by Hoechst.
The NTD Is Required for H3S10 Phosphorylation in Vivo
In order to determine the properties of a JIL-1 construct without the NH2-terminal domain, we expressed the ΔNTD in a JIL-1 null mutant background. As illustrated in Fig. 2B, the ΔNTD restored polytene chromosome morphology to or near the level in wild type preparations (Fig. 2A) and localized correctly to interband regions. Interestingly, in contrast to the ΔCTD, the ΔNTD was not able to phosphorylate H3S10 as documented by immunoblotting of salivary gland protein extracts in Fig. 1C. Furthermore, the polytene squash preparations from larvae expressing the ΔNTD showed that the heterochromatic H3K9me2 mark spread to the chromosome arms (Fig. 2B). Spreading on the X chromosome was especially pronounced in both males and females, as was also the case in the JIL-1 null background (Fig. 2A) and as when the CTD that also is without H3S10 kinase activity (Fig. 1C) was expressed (Fig. 2B). These findings are consistent with the hypothesis that the epigenetic H3S10ph mark itself is necessary for counteracting the spreading of H3K9 dimethylation. Moreover, the results indicate that the NTD of JIL-1 is required for H3S10 phosphorylation to occur in vivo.
H3S10 Phosphorylation Is Sufficient for Chromatin Remodeling
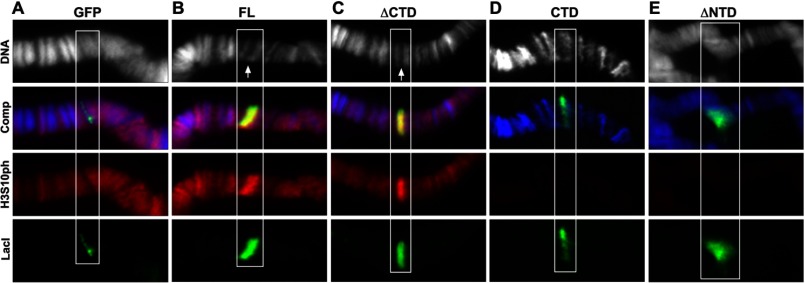
We have previously shown that ectopic targeting of full-length JIL-1 using a LacI-tethering system induces robust H3S10 phosphorylation and a change in higher order chromatin structure from a condensed heterochromatin-like state to a more open euchromatic state (13). However, the surprising finding that the CTD of JIL-1 alone is sufficient to rescue JIL-1 null mutant chromosome defects, including those of the male X chromosome restoring euchromatic interband regions (12) (Fig. 2B), raised the question of whether the CTD also would be sufficient to cause ectopic chromatin remodeling independently of H3S10 phosphorylation. Therefore, we further explored the LacI-tethering paradigm by targeting the various LacI-JIL-1 deletion constructs with and without kinase activity, including the CTD, to chromatin in a condensed heterochromatin-like state. As illustrated in Fig. 3, tethering of the FL or the ΔCTD construct to lacO repeats inserted into the middle of a polytene chromosome band in region 96C1-2 resulted in ectopic H3S10 phosphorylation and “opening” of the band. In contrast, targeting of GFP-LacI or the CTD resulted in neither H3S10 phosphorylation nor chromatin structure changes. In addition, as illustrated in Fig. 3, the CTD displaces endogenous JIL-1, as reported previously (12), leading to a striking decrease in the levels of H3S10 phosphorylation in the interband polytene chromosome regions. Thus, these findings strongly indicate that the CTD of JIL-1 does not contribute to “opening” of the band when full-length JIL-1 is targeted but rather that these chromatin structure changes are caused solely by H3S10 phosphorylation. Supporting this conclusion, when the ΔNTD was targeted, it accumulated at the target site without any detectable H3S10 phosphorylation or chromatin decondensation and, similarly to the CTD, resulted in decreased overall levels of H3S10 phosphorylation. However, expression of the ΔNTD additionally induced chromatin structure perturbations, especially at the targeting site, as well as ectopic contacts between non-homologous chromatin regions. These results suggest that expression of the ΔNTD, like expression of the CTD, reduced global H3S10 phosphorylation levels by displacing native JIL-1 from the normal binding sites of JIL-1 but that the ΔNTD had an additional dominant negative effect due to the presence of the two kinase domains.
FIGURE 3.
Tethering of LacI-tagged JIL-1 constructs to a polytene chromosome band LacO insertion site. The panels show triple labelings of polytene squash preparations from third instar larvae homozygous for the lacO repeat line P11.3, which is inserted into the middle of a polytene band in region 96C1-2. GFP was tethered to the lacO repeats (A), FL (B), ΔCTD (C), CTD (D), and ΔNTD (E). LacI antibody labeling is shown in green, H3S10ph antibody labeling in red, and Hoechst labeling of DNA in blue or gray. The white boxes indicate the location of the polytene band with the lacO repeat insertion site. The white arrows indicate the “split” in the polytene bands, reflecting decondensation of the chromatin when FL or ΔCTD is tethered to the band, in contrast to its wild type morphology when GFP, CTD, or ΔNTD is tethered and where there is no ectopic up-regulation of H3S10 phosphorylation. Note that expression of the CTD and ΔNTD has a dominant negative effect and displaces endogenous JIL-1, resulting in a chromosome-wide reduction in the levels of H3S10ph at interband regions (D and E).
JIL-1-mediated H3S10 Phosphorylation Requires That Both Kinase Domains Are Catalytically Active
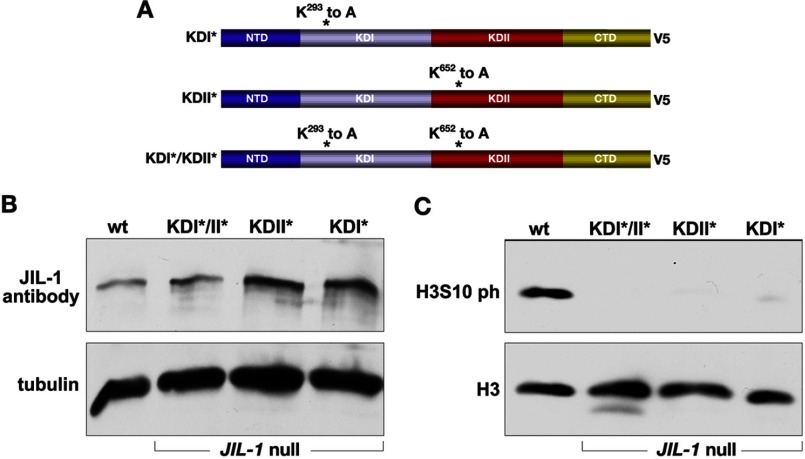
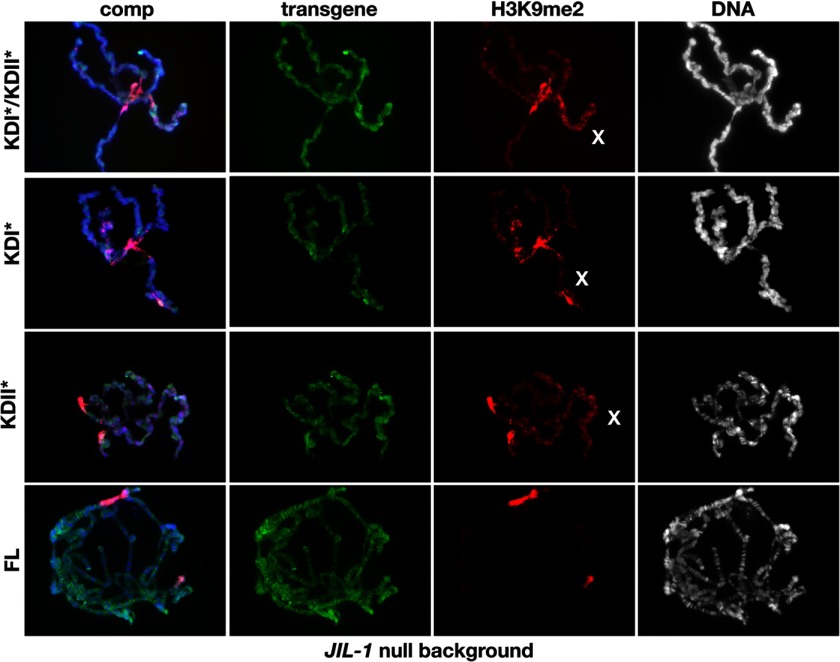
It has recently been demonstrated that a kinase-dead version of full-length JIL-1 (KDI*/KDII*) in which the crucial lysine for catalytic activity in each of the two kinase domains (Lys293 and Lys652) was changed to alanine is without kinase activity (13) in a JIL-1 null mutant background. However, these experiments did not address whether one or both kinase domains are functional and required for H3S10 phosphorylation activity. Therefore, we made two additional constructs (KDI* and KDII*), in each of which only one of the lysines was changed to an alanine, as diagrammed in Fig. 4A, and expressed them in a JIL-1 mutant background. A transgenic line was selected for each construct that expressed at levels comparable with those of endogenous JIL-1 using an hsp70-GAL4 driver (Fig. 4B). The results showed that the properties of both the single kinase-dead versions, KDI* and KDII*, were indistinguishable from those obtained by expressing a construct where both kinase domains (KDI*/KDII*) were mutated. As illustrated in Fig. 4C, none of the three constructs were able to phosphorylate H3S10, as documented by immunoblotting of salivary gland protein extracts. Furthermore, in polytene squash preparations from JIL-1 null larvae expressing KDI*, KDII*, or KDI*/KDII* (Fig. 5), there was no rescue of chromosome morphology, and spreading of the H3K9me2 mark to the chromosome arms was similar to that observed in JIL-1 null preparations (Fig. 2A). These results strongly indicate that both kinase domains of JIL-1 are required to be catalytically active for H3S10 phosphorylation to occur.
FIGURE 4.
Expression of transgenic JIL-1 kinase-dead constructs in a JIL-1 null background. A, diagrams of the JIL-1 V5-tagged constructs analyzed. B, immunoblot labeled with JIL-1 antibody of protein extracts from wild type (wt) and from JIL-1 null salivary glands expressing the KDI*/KDII*, KDII*, and KDI*, respectively. Labeling with tubulin antibody was used as a loading control. C, immunoblot labeled with H3S10ph antibody of protein extracts from salivary glands from wild type third instar larvae (wt) and from JIL-1 null salivary glands expressing KDI*/KDII*, KDII*, and KDI*, respectively. Labeling with histone H3 antibody was used as a loading control.
FIGURE 5.
Immunocytochemical analysis of polytene chromosome morphology and H3K9 dimethylation in JIL-1 null larvae expressing JIL-1 kinase-dead transgenes. The panels show polytene chromosome squash preparations from JIL-1 null (z2/z2) third instar larval salivary glands expressing the KDI*/KDII*, KDII*, KDI*, and FL, respectively. Protein localization (in green) was identified using JIL-1 antibody, H3K9 dimethylation (in red) was identified using H3K9me2 antibody, and DNA (in blue or gray) was labeled by Hoechst.
Phosphorylation of Ser424 Is Required for JIL-1 H3S10 Kinase Activity
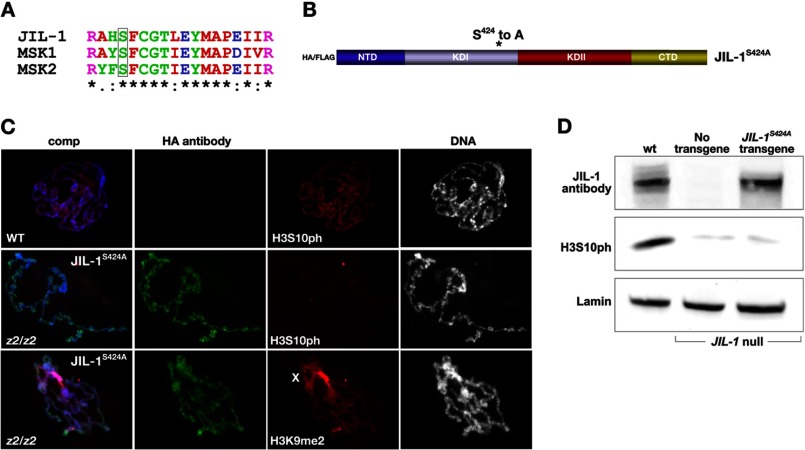
Phylogenetic analysis suggests that JIL-1 is most closely related to the mammalian tandem mitogen and stress-activated kinases 1 and 2 (MSK1/2) in a clade with 95% bootstrap support that is distinct from the RSK (ribosomal S6 p90 kinase) family of tandem kinases (26). The NH2-terminal kinase domain of JIL-1 shares 63% amino acid identity with the corresponding domain in human MSK1 but only 47% identity with that of Drosophila RSK (6). MSKs regulate transcription by phosphorylation of transcription factors, including CREB and ATF1 (27), and chromatin-associated proteins, including HMG-14 and histone H3 (28, 29). Similarly to JIL-1, MSK1/2 phosphorylates H3 at Ser10 (29); however, unlike JIL-1 (30, 31), MSK1/2 additionally has histone H3 Ser28 phosphorylation activity (29, 32). The molecular mechanism of MSK activation is complex and requires multisite phosphorylation by upstream kinases and subsequent autophosphorylation of Ser212 and Ser376 by its COOH-terminal kinase domain (33, 34). The JIL-1 kinase is constitutively active and is autophosphorylated (6, 30). Moreover, the region in human MSK1 containing Ser212 in the NH2-terminal kinase domain is highly conserved in JIL-1 (MSK1 Ser212 corresponds to JIL-1 position 424, Fig. 6A), whereas the residue corresponding to MSK1 Ser376 in the linker region has been substituted with a threonine in JIL-1 (JIL-1 Thr589). Thus, to investigate whether these residues also play a role in JIL-1 activity in vivo, we expressed a tandem affinity-tagged JIL-1 construct that included the NH2-terminal domain, the two kinase domains, and the linker region (TAP-JIL-1) transgenically in flies. The expressed protein was purified from protein extracts of third instar larval salivary glands and separated using SDS-PAGE, and the enriched TAP-JIL-1 band identified by size and Coomassie Blue staining was cut out and analyzed by tandem mass spectrometry for phosphorylated residues. Of the five peptides recovered containing JIL-1 Thr589, none were phosphorylated; however, in six peptides of 10 containing JIL-1 Ser424, this residue was phosphorylated. Thus, these results indicate that in salivary glands, JIL-1 is phosphorylated at Ser424 but not at detectable levels at Thr589. No other phosphorylated sites were identified in the samples.
FIGURE 6.
Immunocytochemical and immunoblot analysis of a transgenic JIL-1S424A point mutation construct expressed in a JIL-1 null background. A, alignment of the amino acid sequences flanking a conserved serine (box) in the first kinase domain of JIL-1 and human MSK1/2. B, diagram of the JIL-1 HA/FLAG-tagged JIL-1S424A point mutation construct analyzed. C, polytene chromosome squash preparations from wild type (WT) as well as JIL-1 null (z2/z2) third instar larval salivary glands expressing the JIL-1S424A construct. JIL-1S424A localization (in green) was identified using HA antibody, H3K9 dimethylation (in red) was identified using H3K9me2 antibody, H3S10 phosphorylation (in red) was identified using H3S10ph antibody, and DNA (in blue or gray) was labeled by Hoechst. D, immunoblot labeled with H3S10ph antibody of protein extracts from salivary glands from wild type third instar larvae (wt), from JIL-1 null salivary glands, and from JIL-1 null salivary glands expressing the JIL-1S424A construct. Endogenous JIL-1 and JIL-1S424A construct expression were detected by JIL-1 antibody labeling. Labeling with lamin antibody was used as a loading control.
In order to determine whether phosphorylation of Ser424 is required for H3S10 kinase activity, we made a JIL-1 construct where Ser424 was mutated to an alanine (JIL-1S424A; Fig. 6B) and expressed it transgenically in a JIL-1 mutant background. A transgenic line was selected that expressed at levels comparable with those of endogenous JIL-1 using a hsp70-GAL4 driver (Fig. 6D). As illustrated in Fig. 6D, JIL-1S424A had little or no H3S10 phosphorylation activity, as documented by immunoblotting of salivary gland protein extracts. Furthermore, in polytene squash preparations from JIL-1 null larvae expressing JIL-1S424A (Fig. 6C), there was no detectable H3S10 phosphorylation, and spreading of the H3K9me2 mark to the chromosome arms was similar to that observed in JIL-1 null preparations (Fig. 2A). These results strongly indicate that phosphorylation of Ser424 is required for JIL-1 to be catalytically active and for H3S10 phosphorylation to occur. However, it should be noted that although chromosome morphology was still perturbed in JIL-1 null preparations expressing the JIL-1S424A construct, the morphology was discernibly improved compared with preparations expressing KDI*, KDII*, or KDI*/KDII* (Fig. 5).
The properties of the JIL-1 constructs analyzed in the present study, including viability, are summarized in Table 1. The homozygous JIL-1z2 genotype is a late larval lethal with no escapers (17). Interestingly, all of the deletion and mutated constructs with or without H3S10 phosphorylation activity restored some degree of viability, indicating, as suggested previously (12), that multiple and independent mechanisms may be contributing to the lethality of the JIL-1 null mutant.
TABLE 1.
Properties of JIL-1 constructs expressed in a JIL-1z2/JIL-1z2 null background
| Construct | Localization to chromatin | Rescue of autosome morphology | H3S10 phosphorylation | H3K9me2 spreading | Rescue of adult viabilitya (n) |
|---|---|---|---|---|---|
| % | |||||
| JIL-1-FL | Yes | Yes | Yes | No | 58.6 (644) |
| CTD | Yes | Yes | No | Yes | 13.6 (515) |
| ΔCTD | Yesb | Yes | Yes | No | 4.4 (591) |
| ΔNTD | Yes | Yes | No | Yes | 18.9 (428) |
| KDI*/KDII* | Yes | No | No | Yes | 22.9 (447) |
| KDI*/KDII | Yes | No | No | Yes | 14.3 (329) |
| KDI/KDII* | Yes | No | No | Yes | 12.3 (568) |
| JIL-1S424A | Yes | Partial | No | Yes | 14.7 (439) |
a The rescue of adult viability in the JIL-1 null mutant background by each construct was calculated as described under “Experimental Procedures.” n indicates the total number of eclosed progeny in each experiment.
b Ectopic chromatin localization.
DISCUSSION
In this study, we show that a JIL-1 construct without the NH2-terminal domain is without H3S10 phosphorylation activity despite the fact that it localizes properly to polytene interband regions and that it contains both kinase domains. This is in contrast to a JIL-1 construct without the COOH-terminal domain that, although mislocalized to ectopic chromatin regions, retains its H3S10 phosphorylation capability (12) (this study). Interestingly, although the CTD of JIL-1 binds to the NH2-terminal tail of histone H3, modeling of the three-dimensional structure of JIL-1 relative to nucleosome structure suggested that JIL-1 is likely to phosphorylate H3 of one or more nucleosomes some distance away from this binding site.5 Taken together, these findings suggest a model where the NTD of JIL-1 is required for chromatin complex interactions that position the kinase domain(s) for catalytic activity in the context of the state of higher order nucleosome packaging and chromatin structure, whereas sequences in the CTD mediate binding to specific chromatin sites. Furthermore, it has been demonstrated that sequences within both the NTD and CTD are required for enrichment of JIL-1 on the male X chromosome (12).
JIL-1 is a double kinase, raising the question of the function and relative contributions of the two kinase domains. In this study, we show that the properties of both of the single kinase-dead versions of JIL-1, KDI* and KDII*, were indistinguishable from those obtained by expressing a construct where both kinase domains (KDI*/KDII*) were mutated. Neither construct was able to phosphorylate H3S10, rescue JIL-1 null polytene chromosome morphology, or prevent spreading of the H3K9me2 silencing mark, strongly indicating that both kinase domains of JIL-1 are required to be catalytically active for H3S10 phosphorylation to occur. Furthermore, we provide evidence that JIL-1 is phosphorylated at Ser424 in vivo and that this phosphorylation is required for JIL-1 H3S10 phosphorylation activity. This phosphorylation site is conserved in the mammalian MSK family of tandem kinases that are the closest homologs in the databases to the JIL-1 kinase (26). Upon activation of MSKs by upstream kinases, this site has been demonstrated to be autophosphorylated by the COOH-terminal MSK kinase domain and to be required for catalytic activity (33, 34). Thus, the data presented here are compatible with a model where autophosphorylation of Ser424 by the second kinase domain of JIL-1 is similarly required for catalytic activity and H3S10 phosphorylation mediated by the first kinase domain. The molecular mechanism of MSK activation involves multisite phosphorylation that is context- and pathway-dependent (33, 34). In contrast, the JIL-1 kinase is constitutively active (6, 30), and therefore the requirements for its activation are likely to be less complex. Supporting this hypothesis, we did not find evidence for phosphorylation of sites other than Ser424, including Thr589, whose corresponding residue is required to be autophosphorylated for activation of the MSKs.
An important issue of epigenetic chromatin structure regulation by histone phosphorylation is whether the H3S10ph mark itself has the capacity to induce chromatin changes or whether it only plays a reinforcing or maintenance role. Using a LacI tethering system, Deng et al. (13) provided evidence that ectopic H3S10 phosphorylation by the JIL-1 kinase is sufficient to cause striking changes in chromatin packaging from a condensed to an open state. This effect was absent when a kinase-dead LacI-JIL-1 construct that is without H3S10 phosphorylation activity was expressed (13). This indicates that the observed chromatin structure changes depended on JIL-1 kinase-mediated H3S10 phosphorylation and not on the tethering of the LacI-JIL-1 construct itself. However, the surprising finding that the CTD of JIL-1 alone is sufficient to rescue JIL-1 null mutant chromosome defects, including those of the male X chromosome, restoring euchromatic interband regions, suggested that alternative or redundant mechanisms of direct chromatin structure modification may be mediated by the CTD (12). One possibility is that the CTD may serve as a binding platform for an unidentified protein or protein complex that has the ability to induce changes in chromosome morphology but that can function only after proper localization through interactions with the CTD of JIL-1. Another possibility is that binding of the CTD to chromatin by itself is sufficient to alter chromatin structure through bridging interactions with surrounding molecules. However, in this study, we demonstrate using the same experimental paradigm as Deng et al. (13) that LacI-mediated tethering of neither the CTD nor the ΔNTD of JIL-1 resulted in H3S10 phosphorylation or transformation of a polytene chromatin band into an interband and that such chromatin remodeling only is effectuated by JIL-1 constructs with H3S10 kinase activity. Thus, these findings provide further evidence that the epigenetic H3S10 phosphorylation mark itself functions as a causative regulator of chromatin structure independently of any structural contributions from the JIL-1 protein.
Acknowledgments
We thank members of the laboratory for discussion, advice, and critical reading of the manuscript. We especially thank Dr. L. Wallrath for providing fly stocks.
This work was supported, in whole or in part, by National Institutes of Health Grant GM62916 (to K. M. J. and J. J.).
W. Cai, unpublished results.
- H3S10
- histone H3 serine 10
- H3K9
- histone H3 lysine 9
- NTD
- NH2-terminal domain
- CTD
- COOH-terminal domain
- KDI
- kinase domain I
- KDII
- kinase domain II
- FL
- full-length LacI-JIL-1
- MSK
- mitogen- and stress-activated kinase
- RSK
- ribosomal S6 p90 kinase
- H3S10ph
- phosphorylated H3S10
- H3K9me2
- H3K9 dimethylation
- CFP
- cyan fluorescent protein
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Zhang W., Deng H., Bao X., Lerach S., Girton J., Johansen J., Johansen K. M. (2006) The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 133, 229–235 [DOI] [PubMed] [Google Scholar]
- 2. Deng H., Cai W., Wang C., Lerach S., Delattre M., Girton J., Johansen J., Johansen K. M. (2010) JIL-1 and Su(var)3–7 interact genetically and counterbalance each others' effect on position effect variegation in Drosophila. Genetics 185, 1183–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang C., Cai W., Li Y., Deng H., Bao X., Girton J., Johansen J., Johansen K. M. (2011) The epigenetic H3S10 phosphorylation mark is required for counteracting heterochromatic spreading and gene silencing in Drosophila melanogaster. J. Cell Sci. 124, 4309–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang C., Girton J., Johansen J., Johansen K. M. (2011) A balance between euchromatic (JIL-1) and heterochromatic (SU(VAR)2–5 and SU(VAR)3–9) factors regulates position-effect variegation in Drosophila. Genetics 188, 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang C., Cai W., Li Y., Girton J., Johansen J., Johansen K. M. (2012) H3S10 phosphorylation by the JIL-1 kinase regulates H3K9 dimethylation and gene expression at the white locus in Drosophila. Fly 6, 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jin Y., Wang Y., Walker D. L., Dong H., Conley C., Johansen J., Johansen K. M. (1999) JIL-1. A novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell 4, 129–135 [DOI] [PubMed] [Google Scholar]
- 7. Jin Y., Wang Y., Johansen J., Johansen K. M. (2000) JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the MSL dosage compensation complex. J. Cell Biol. 149, 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lerach S., Zhang W., Deng H., Bao X., Girton J., Johansen J., Johansen K. M. (2005) JIL-1 kinase, a member of the male-specific lethal (MSL) complex, is necessary for proper dosage compensation of eye pigmentation in Drosophila. Genesis 43, 213–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C. I., Alekseyenko A. A., LeRoy G., Elia A. E., Gorchakov A. A., Britton L. M., Elledge S. J., Kharchenko P. V., Garcia B. A., Kuroda M. I. (2013) Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat. Struct. Mol. Biol. 20, 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y., Zhang W., Jin Y., Johansen J., Johansen K. M. (2001) The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105, 433–443 [DOI] [PubMed] [Google Scholar]
- 11. Deng H., Zhang W., Bao X., Martin J. N., Girton J., Johansen J., Johansen K. M. (2005) The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma 114, 173–182 [DOI] [PubMed] [Google Scholar]
- 12. Bao X., Cai W., Deng H., Zhang W., Krencik R., Girton J., Johansen J., Johansen K. M. (2008) The COOH-terminal domain of the JIL-1 histone H3S10 kinase interacts with histone H3 and is required for correct targeting to chromatin. J. Biol. Chem. 283, 32741–32750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng H., Bao X., Cai W., Blacketer M. J., Belmont A. S., Girton J., Johansen J., Johansen K. M. (2008) Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development 135, 699–705 [DOI] [PubMed] [Google Scholar]
- 14. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 15. Veraksa A., Bauer A., Artavanis-Tsakonas S. (2005) Analyzing protein complexes in Drosophila with tandem affinity purification mass spectrometry. Dev. Dyn. 232, 827–834 [DOI] [PubMed] [Google Scholar]
- 16. Roberts D. B. (1998) Drosophila: A Practical Approach, 2nd Ed., IRL Press, Oxford, UK [Google Scholar]
- 17. Zhang W., Jin Y., Ji Y., Girton J., Johansen J., Johansen K. M. (2003) Genetic and phenotypic analysis of alleles of the Drosophila chromosomal JIL-1 kinase reveals a functional requirement at multiple developmental stages. Genetics 165, 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., Danzer J. R., Alvarez P., Belmont A. S., Wallrath L. L. (2003) Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130, 1817–1824 [DOI] [PubMed] [Google Scholar]
- 19. Danzer J. R., Wallrath L. L. (2004) Mechanisms of HP1-mediated gene silencing in Drosophila. Development 131, 3571–3580 [DOI] [PubMed] [Google Scholar]
- 20. Ji Y., Rath U., Girton J., Johansen K. M., Johansen J. (2005) D-Hillarin, a novel W180-domain protein, affects cytokinesis through interaction with the septin family member Pnut. J. Neurobiol. 64, 157–169 [DOI] [PubMed] [Google Scholar]
- 21. Lindsley D. L., Zimm G. G. (1992) The Genome of Drosophila melanogaster. Academic Press, Inc., New York [Google Scholar]
- 22. Cai W., Jin Y., Girton J., Johansen J., Johansen K. M. (2010) Preparation of polytene chromosome squashes for antibody labeling. J. Vis. Exp., 10.3791/1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johansen K. M., Cai W., Deng H., Bao X., Zhang W., Girton J., Johansen J. (2009) Methods for studying transcription and epigenetic chromatin modification in Drosophila polytene chromosome squash preparations using antibodies. Methods 48, 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 25. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 26. Johansen K. M., Johansen J., Jin Y., Walker D. L., Wang D., Wang Y. (1999) Chromatin structure and nuclear remodeling. Crit. Rev. Eukaryotic Gene Expr. 9, 267–277 [DOI] [PubMed] [Google Scholar]
- 27. Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., Arthur J. S. (2002) MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell Biol. 22, 2871–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomson S., Clayton A. L., Hazzalin C. A., Rose S., Barratt M., Mahadevan L. C. (1999) The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential H3/HMG-14 kinase. EMBO J. 18, 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., Arthur J. S. (2003) MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22, 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Regnard C., Straub T., Mitterweger A., Dahlsveen I. K., Fabian V., Becker P. B. (2011) Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet. 7, e1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang C., Yao C., Li Y., Cai W., Bao X., Girton J., Johansen J., Johansen K. M. (2013) Evidence against a role for the JIL-1 kinase in H3S28 phosphorylation and 14-3-3 recruitment to active genes in Drosophila. PLoS One 8, e62484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dyson M. H., Thomson S., Inagaki M., Goto H., Arthur S. J., Nightingale K., Iborra F. J., Mahadevan L. C. (2005) MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J. Cell Sci. 118, 2247–2259 [DOI] [PubMed] [Google Scholar]
- 33. McCoy C. E., Campbell D. G., Deak M., Bloomberg G. B., Arthur J. S. (2005) MSK1 activity is controlled by multiple phosphorylation sites. Biochem. J. 387, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy C. E., MacDonald A., Morrice N. A., Campbell D. G., Deak M., Toth R., McIlrath J., Arthur J. S. (2007) Identification of novel phosphorylation sites in MSK1 by precursor ion scanning MS. Biochem. J. 402, 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]