Background: Type IV P-type ATPases (P4-ATPases) Drs2 and Dnf1 are known to recognize different phospholipid headgroups.
Results: Dnf1 preferentially recognizes lysophospholipid, but mutations in transmembrane (TM) segments 1, 2, and 3 allow recognition of diacyl phosphatidylserine.
Conclusion: Residues within TM1–3 cooperate with the proline + 4 residue in TM4 to define P4-ATPase specificity.
Significance: We provide insight into how a P-type ATPase subgroup evolved a new substrate.
Keywords: Lipid Transport, Membrane Function, Membrane Proteins, Membrane Transport, Phosphatidylserine, Yeast, P4-ATPase, Flippase, Membrane Asymmetry
Abstract
Type IV P-type ATPases (P4-ATPases) use the energy from ATP to “flip” phospholipid across a lipid bilayer, facilitating membrane trafficking events and maintaining the characteristic plasma membrane phospholipid asymmetry. Preferred translocation substrates for the budding yeast P4-ATPases Dnf1 and Dnf2 include lysophosphatidylcholine, lysophosphatidylethanolamine, derivatives of phosphatidylcholine and phosphatidylethanolamine containing a 7-nitro-2-1,3-benzoxadiazol-4-yl (NBD) group on the sn-2 C6 position, and were presumed to include phosphatidylcholine and phosphatidylethanolamine species with two intact acyl chains. We previously identified several mutations in Dnf1 transmembrane (TM) segments 1 through 4 that greatly enhance recognition and transport of NBD phosphatidylserine (NBD-PS). Here we show that most of these Dnf1 mutants cannot flip diacylated PS to the cytosolic leaflet to establish PS asymmetry. However, mutation of a highly conserved asparagine (Asn-550) in TM3 allowed Dnf1 to restore plasma membrane PS asymmetry in a strain deficient for the P4-ATPase Drs2, the primary PS flippase. Moreover, Dnf1 N550 mutants could replace the Drs2 requirement for growth at low temperature. A screen for additional Dnf1 mutants capable of replacing Drs2 function identified substitutions of TM1 and 2 residues, within a region called the exit gate, that permit recognition of dually acylated PS. These TM1, 2, and 3 residues coordinate with the “proline + 4” residue within TM4 to determine substrate preference at the exit gate. Moreover, residues from Atp8a1, a mammalian ortholog of Drs2, in these positions allow PS recognition by Dnf1. These studies indicate that Dnf1 poorly recognizes diacylated phospholipid and define key substitutions enabling recognition of endogenous PS.
Introduction
Active transport of substrates across lipid bilayers is an essential process for cell survival. Both prokaryotes and eukaryotes spend considerable energy to actively transport ions, nutrients, waste products, and lipids across the cellular membranes. P-type ATPases include a large family of ion transporters involved in establishing gradients across membranes at the expense of ATP. These ion gradients are required for establishing the resting membrane potential in neurons, signaling at the neural synapse, muscle contraction, and acidification of the stomach lumen.
P-type ATPases cycle through conformational states described by the Post-Albers cycle and characteristically form a phosphorylated intermediate when the γ-phosphate from ATP is transferred temporarily to a conserved aspartate residue on the protein. As the cycle proceeds forward, the phosphate is hydrolyzed from the protein, ensuring a single direction in the reaction cycle. P-type ATPases are grouped into five subclasses on the basis of substrate specificity (1). Type I, II, and III transport heavy metals, cations, and protons, respectively. Interestingly, the type IV (or P4) ATPases are only present in eukaryotic cells and flip phospholipid molecules across the membrane bilayer (flippases). Type V has unknown substrate specificity and is also only present in eukaryotes.
The P4-ATPase3 family has biological functions in establishing and maintaining phospholipid asymmetry and facilitating vesicle-mediated protein trafficking (2). In mammals, P4-ATPases have been implicated in spermatogenesis (3), B cell development (4, 5), neuronal growth (6), hippocampal-dependent learning (7), and cell migration (8). They also play roles in pathological conditions such as progressive hearing loss (9), neurodegenerative diseases (10), intellectual disability disorder (11), obesity and type II diabetes (12), intrahepatic cholestasis (13), and male infertility (14). Mammals have 14 (sometimes 15) P4-ATPases, whereas Saccharomyces cerevisiae has five (Drs2, Dnf1, Dnf2, Dnf3, and Neo1) (1, 3, 15).
A central conundrum involving P-type ATPases is how they have evolved the ability to transport such a wide range of substrates, from ions to phospholipids. Ion transporters use a highly conserved ion binding site within the center of the transmembrane domain highlighted by the crystal structures of the Ca2+ ATPase (16). We have recently described a series of residues involved in defining the substrate specificity of P4-ATPases that implicated a novel transport mechanism for a P-type ATPase using two gates to select phospholipid on opposing membrane faces (17, 18). Substrate-selecting residues near the extracellular or lumenal leaflet surface are proposed to form an “entry gate” where phospholipid is initially selected for transport, and substrate-selecting residues near the cytosolic leaflet surface are proposed to form the “exit gate.”
Drs2 and its mammalian homologs Atp8a1/Atp8a2 are flippases that catalyze “flip” of phosphatidylserine (PS) and phosphatidylethanolamine (PE) within the Golgi-endosomal system (although PS seems to be the preferred substrate) (19–23). Even though Drs2 is closely related to Dnf1, they differ in their substrate specificity and localization. Dnf1 and Dnf2 recognize and transport lyso-PC (24), lysophosphatidylethanolamine (25), and C6-NBD derivatives of these lipids (26), but it is unknown whether these pumps recognize intact, dually acylated PC and PE. Our first description of residues involved in phospholipid selection highlighted a residue of particular interest: the “proline + 4” (P + 4) position in TM4. This residue is four amino acids away from a proline that is completely conserved in all P-type ATPases. The presence of a tyrosine at the P + 4 position restricted PS flip, whereas a phenylalanine allowed PS flip. Surprisingly, Atp8a1 and Atp8a2 have a leucine at this position. When we introduced a leucine into either Dnf1 or Drs2, the overall activity of these proteins was reduced, but we did not observe a change in the specificity.
Here we describe new Dnf1 variants that are capable of flipping endogenous, unlabeled PS. Surprisingly, we observed a striking difference among Dnf1 variants capable of flipping NBD-labeled PS in their ability to restore the endogenous PS asymmetry to a drs2Δ mutant. Identification of new Dnf1 alleles capable of supporting endogenous PS flip revealed substitutions corresponding to the analogous residues in the mammalian PS flippases Atp8a1/Atp8a2. Coordination between the TM4 P + 4 and residues on TMs 1, 2, and 3 define a portion of a phospholipid substrate binding site at the exit gate used by P4-ATPases.
EXPERIMENTAL PROCEDURES
Reagents
All lipids were purchased from Avanti Polar Lipids, Inc. 5-fluoroorotic acid was purchased from Zymo Research. Duramycin was purchased from Sigma-Aldrich. Papuamide B was a gift from Raymond Andersen (University of British Columbia).
Cold Resistance Screen
To generate random mutations, we used procedures reported previously (27). Briefly, we cotransformed a dnf1,2,3Δdrs2Δ strain (ZHY704) with mutagenized PCR products and pRS313-DNF1 gapped with restriction enzymes AflII and MluI to allow for gap repair through homologous recombination. The transformants were plated onto synthetic dropout plates with 5-fluoroorotic acid, grown for 4 days at 20 °C, and then colonies were picked into 96-well plates. We compared growth of the strains expressing dnf1 mutants strains to those expressing WT DNF1 or WT DRS2 and sequenced dnf1 alleles from the strains that grew strongly at 20 °C.
Strains and Culture
Escherichia coli strain DH5α was used for molecular cloning. All strains and plasmids used in this study are listed in supplemental Tables S1 and S2. Yeast strains were cultured with glucose in standard rich medium (yeast extract, peptone, dextrose (YPD)) or synthetic minimal medium (SD) at the indicated temperatures. Yeast transformation used the lithium acetate method (28).
Growth Assay
To test for drug sensitivity, mid-log phase yeast was distributed into 96-well plates at 0.1 A600/ml. Plates were incubated at 30 °C for 20 h, and the A600/ml was measured using a multimode plate reader Synergy HT (Bio-Tek, Winooski, VT). Relative growth was determined by dividing A600 values from drug-treated cells by mock-treated cells in the assay.
Serial Dilutions
50,000 cells were spotted with 10-fold serial dilutions onto YPD plates, YPD + 100 μg/ml calcofluor white, SD, or SD + 5-fluoroorotic acid. Plates were grown at 20 °C or 30 °C for 3–5 days before imaging.
Lipid Uptake
Lipid uptake was performed essentially as described previously (29). Briefly, overnight cultures were subcultured to 0.15 A600/ml and allowed to grow to early mid-log phase. 500 μl of cells was harvested, resuspended in ice-cold SD medium containing 2 μg/ml NBD lipid (∼2.5 μm), and incubated on ice for 30 min. Cells were washed twice with SA medium (SD medium, 2% sorbitol (w/v), 20 mm NaN3) and 4% BSA (w/v), washed once with SA medium, and then resuspended in SA medium prior to analysis by flow cytometry.
To assay lyso-PS (similar to Ref. 30) and dilauryl-PS uptake, overnight cultures of cho1Δ yeast were subcultured to 0.15 A600/ml and allowed to grow to mid-log phase (0.5–0.8 A600/ml). 1500 μl of cells were pelleted, resuspended in SD medium containing the indicated final concentration of lyso-PS (1-oleoyl-2-hydroxy-sn-glycero-3-phospho-l-serine) or DLPS (1,2-dilauroyl-sn-glycero-3-phospho-l-serine) (added from a 10 mg/ml stock in 0.1% Nonidet P-40), and incubated at 30 °C with shaking (or on ice) for 1 h. To end the phospholipid uptake, we added SA medium + 4% BSA (w/v) at a 1:1 ratio with the reaction. The cells were pelleted and resuspended once in SA medium + 4% BSA (w/v), followed by SA medium (no BSA), and, finally, pelleted and resuspended in imaging buffer.
Flow Cytometry
We used a BD LSRII-3 laser (BD Biosciences) using BD FACSDiva v6.1.3. NBD lipid uptake was measured with the FITC filter set (530/30 bandpass with a 525 longpass). Immediately prior to analysis, propidium iodide was added at a final concentration of 5 μm. At least 10,000 events were analyzed using forward/side scatter to identify single cells and propidium iodide fluorescence to exclude dead cells.
Fluorescence Microscopy
Images were collected with an Axioplan microscope (Carl Zeiss, Thornwood, NY) using a Hamamatsu charge coupled device camera and MetaMorph software. To observe GFP-tagged proteins, cells were grown to mid-log phase, pelleted, and resuspended in imaging buffer (10 mm Tris-HCl (pH 7.4), 2% glucose (w/v)). An aliquot of cells was spread onto slides and visualized using a GFP filter set.
Data Analysis
Within each NBD-phospholipid (NBD-PL) uptake experiment, at least three independently isolated transformants containing the same construct were assayed. The data from at least three separate experiments were reported as mean ± S.E. Uptake of NBD-PL for a dnf1,2Δ strain with an empty vector was subtracted, and NBD-PC uptake by WT Dnf1 in the same strain was used to normalize the data. Each value is reported relative to NDB-PC uptake by WT Dnf1 at 30 min. Substrate preference ratios were determined by the dividing NBD-PE or NBD-PS uptake values by the NBD-PC uptake value for each independent replicate (n ≥ 9). The data from each substrate preference ratio is reported as mean ± S.E. For drug sensitivity assays, at least two independently isolated transformants were assayed in at least two separate experiments, and data are reported as mean ± S.E.
RESULTS
Dnf1 N550S Flips Endogenous PS
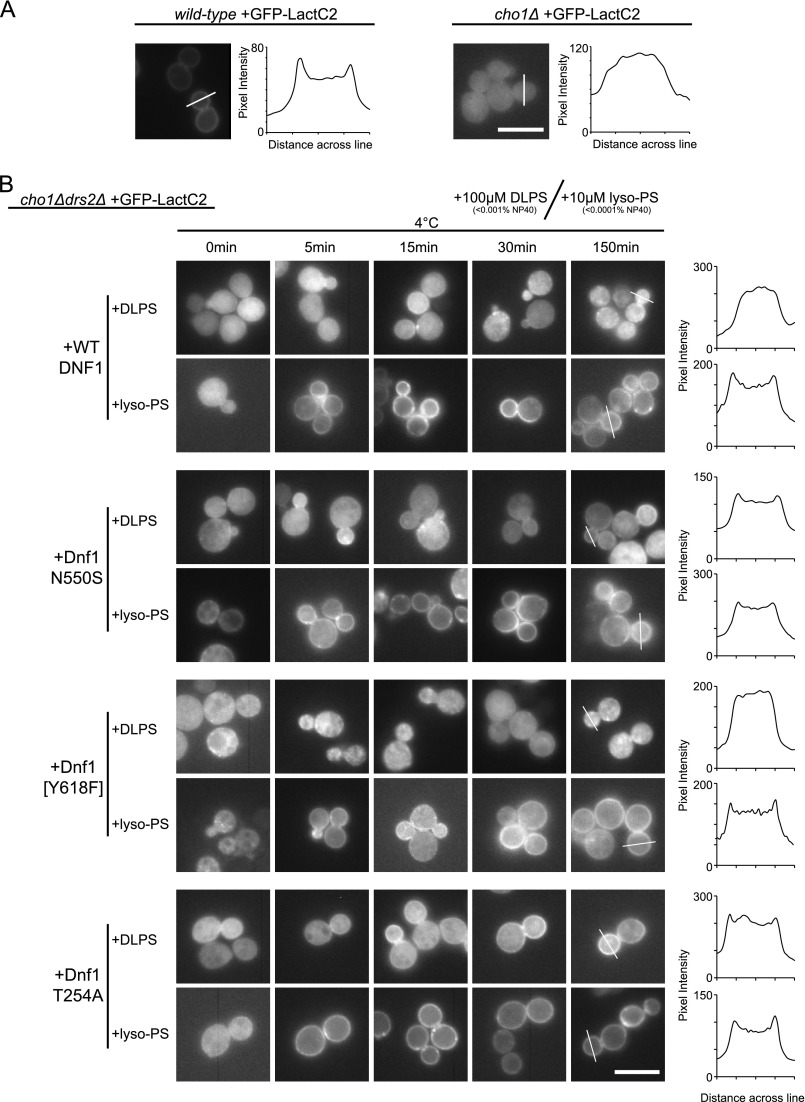
Deletion of DRS2 disrupts plasma membrane asymmetry for PS and PE. Phospholipid asymmetry can be monitored by sensitivity to pore-forming toxins that specifically interact with either PE (duramycin) or PS (papuamide B, PapB) exposed on the outer leaflet of the plasma membrane. We reported previously that WT Dnf1 and Dnf1[Y618F] (a P + 4 mutation; the brackets are used to indicate the replacement of a Dnf1 residue with a Drs2 residue) were unable to restore the phospholipid asymmetry of a drs2Δ strain even though Dnf1[Y618F] was capable of NBD-PS transport (17). Importantly, the reciprocal change in Drs2 specifically reduced the ability of this pump to flip endogenous PS, supporting a role for the P + 4 residue in recognizing the phospholipid headgroup (17). We then tested other Dnf1 mutants reported to transport NBD-PS (18), and like Dnf1[Y618F], neither Dnf1[GA→QQ] (a TM1 entry gate mutation) nor Dnf1[GA→QQ, Y618F] were able to restore PapB resistance and, therefore, PS asymmetry, to a drs2Δ strain (Fig. 1A). Incredibly, Dnf1 N550S (a TM3 exit gate mutation) was unique among the Dnf1 mutants that recognize NBD-PS in its ability to fully restore the PS asymmetry of a drs2Δ strain to near WT levels (Fig. 1A).
FIGURE 1.
Dnf1 variants differ in their suppression of drs2Δ PS asymmetry defects and cold-sensitive growth. A–C, a drs2Δ strain harboring either an empty vector or a vector expressing the indicated Drs2 or Dnf1 variant, were grown for 20 h at 30 °C with increasing concentrations of papuamide B (A and C) or duramycin (B). Among Dnf1 variants competent for NBD-PS transport, Dnf1 N550S is uniquely capable of restoring plasma membrane asymmetry in drs2Δ better than WT Dnf1 for PS asymmetry (A) but not PE asymmetry (B). C, overexpressing WT Dnf1 or Dnf1[Y618F] provides minor suppression of the PS asymmetry defect. D, growth of drs2Δ harboring the indicated Drs2 or Dnf1 variants at 30 °C or 20 °C. Dnf1 N550S suppresses drs2Δ cold-sensitive growth better than WT Dnf1 or other Dnf1 NBD-PS-transporting alleles.
The N550S influence on membrane asymmetry was rather specific to PS because Dnf1 N550S conferred only a slight increase in duramycin resistance relative to the other Dnf1 mutants (Fig. 1B). Thus, PE asymmetry was not fully restored in drs2Δ cells expressing Dnf1 N550S. Even when overexpressed from a 2-μm plasmid, neither WT Dnf1 nor Dnf1[Y618F] were able to correct the PS asymmetry defect of drs2Δ as well as Dnf1 N550S (compare Fig. 1A to C). This suggested that Dnf1[Y618F] was unable to transport endogenous PS, although it was capable of NBD-PS transport.
Because Drs2 is required for growth at 20 °C, we also tested the ability of the Dnf1 NBD-PS-transporting alleles to suppress the cold-sensitive growth defect. Again, only Dnf1 N550S was capable of suppressing the drs2Δ cold sensitivity (Fig. 1D). The localization patterns and expression levels of these Dnf1 mutants were similar and, therefore, unlikely to account for the observed phenotypic differences (18). Therefore, the ability to flip endogenous PS and restore membrane asymmetry in vivo correlated with the suppression of drs2Δ cold sensitivity.
Dnf1 Suppressors of Drs2 Function
We previously used an extensive mutagenesis screen targeted to TM3–4 in Dnf1 and identified several residues within this region involved in NBD-PS selection (18). The observation that Dnf1 N550S was uniquely capable of recognizing endogenous PS and suppressing drs2Δ cold-sensitive growth suggested that a screen for dnf1 alleles that suppress drs2Δ cold-sensitive growth would identify additional changes, allowing Dnf1 to flip endogenous PS. Random mutagenesis targeted to TM1–2 produced about 500 dnf1 mutants (of 5000 transformants) that supported growth of a dnf1,2,3Δdrs2Δ strain at 20 °C. We selected 20% of the colonies that were largest, picked these into liquid culture, and grew the strains at 20 °C. We recovered 25 strains that grew similarly to dnf1,2,3Δdrs2Δ harboring WT DRS2 at 20 °C. From these strains we sequenced the DNF1 constructs, recovering 17 alleles coding for single amino acid substitutions. There were four positions at which we recovered multiple alleles encoding the same amino acid substitution: F213S, A244V, T254A, and D258E. Random mutagenesis was also targeted to TM3–4 in Dnf1. Screening a similar number of dnf1 mutants yielded only a single new Dnf1 substitution that strongly suppressed drs2Δ (Dnf1 V553E). Targeting random mutagenesis to TM5–6 failed to produce dnf1 mutants capable of strongly suppressing drs2Δ cold sensitivity. Except for A244V, these suppressor alleles each arose independently because the codons recovered were different.
We next transformed these DNF1 alleles into a drs2Δ strain to determine whether the suppression was plasmid-linked or extragenic (Fig. 2A). In fact, DNF1 A244V failed to suppress drs2Δ cold sensitivity any better than WT DNF1, and so the original mutation was not plasmid-linked. Each of the other alleles did suppress the cold sensitivity of a drs2Δ strain, although to varying degrees (Fig. 2A). We then assayed the NBD-PL uptake activity for each of the new dnf1 alleles in a dnf1,2Δ background. With the exception of Dnf1 A244V, we observed an increase in the NBD-PS uptake for each Dnf1 mutant. Most of the Dnf1 mutants were able to translocate NBD-PC and NBD-PE at near WT levels, although Dnf1 A244V had a reduced activity for all substrates and Dnf1 V553E displayed a reduction in NBD-PC uptake activity (Fig. 2B). A measure of substrate preference independent of overall activity can be obtained by plotting the uptake ratios of NBD-PE to NBD-PC and NBD-PS to NBD-PC (Fig. 2C) (18). Ratios outside of the shaded confidence interval are considered significant changes in substrate preference. By these criteria, the N550S, F213S, T254A, D258E, and V553E mutants have a significantly increased ability to recognized NBD-PS relative to WT Dnf1, and V553E also showed an increased preference for NBD-PE, caused by reduced uptake of NBD-PC.
FIGURE 2.

Dnf1 suppressors of drs2Δ cold-sensitive growth transport NBD-PS across the plasma membrane and restore endogenous PS asymmetry. A, growth of drs2Δ expressing the indicated Drs2 and Dnf1 variants at 30 °C or 20 °C. B, dnf1Δdnf2Δ cells expressing the indicated Dnf1 variants were incubated with NBD-phospholipid for 30 min at 4 °C as described under “Experimental Procedures.” The amount of lipid transported was normalized to NBD-PC uptake by WT Dnf1. C, the data shown in B was graphed as the ratio of NBD-PE to NBD-PC uptake (open bars) or NBD-PS to NBD-PC uptake (gray bars) to assess relative changes in substrate preference. The dotted lines are the ratios for WT Dnf1, and the shaded region above and below these lines are confidence intervals. Values extending outside of the confidence intervals represent a significant change in substrate specificity. D, the Dnf1 variants indicated were tested for their ability to restore PapB resistance (PS asymmetry) to a drs2Δ strain. The Dnf1 suppressors of drs2Δ cold-sensitive growth restore PS asymmetry better than WT Dnf1.
Because Dnf1 N550S was capable of flipping NBD-PS (18), suppressing drs2Δ cold sensitivity (Fig. 1D), and flipping endogenous PS (Fig. 1A), whereas Dnf1[Y618F] could only flip NBD-PS, we suspected that the new Dnf1 cold-sensitive suppressor alleles would also flip endogenous PS. In fact, these mutants displayed a range of PapB resistance with the strongest being the original N550S, closely followed by T254A (Fig. 2D). Dnf1 F213S, Dnf1 D258E, and Dnf1 V553E provided greater PapB resistance than WT Dnf1 but not as strong as T254A.
Recognition of Mono- versus Diacylated Phospholipid
Each of these dnf1 mutants were competent for NBD-PS uptake but displayed varying resistance to PapB. We speculated that Dnf1 N550S and Dnf1 T254A were capable or recognizing endogenous, unlabeled PS, whereas Dnf1[Y618F] and Dnf1[GA→QQ] were unable to do so. To test this possibility, we made use of GFP-LactC2, a probe that binds specifically to PS, and a cho1Δ strain (30, 31). CHO1 (PSS1) encodes the phosphatidylserine synthase in S. cerevisiae, and cho1Δ mutants completely lack PS (32, 33). GFP-LactC2 localizes primarily to the inner leaflet of the plasma membrane of wild-type cells but is diffusely localized to the cytosol in cho1Δ cells (Fig. 3A) (30). A line scan of fluorescence intensity through the cells clearly shows peaks for the plasma membrane of the wild-type cell that is lacking in the cho1Δ cells. When cho1Δ cells are supplemented with lyso-PS applied extracellularly at 4 °C, GFP-LactC2 is recruited to the plasma membrane because this substrate is flipped to the cytosolic leaflet (30). The low temperature is sufficient to block endocytosis and reduce the activity of flippases that could pump the lyso-PS back to the outer leaflet (34).
FIGURE 3.
Dnf1 suppressors of drs2Δ flip diacyl PS. A, localization of the PS-specific probe GFP-LactC2 in the WT strain or PS-deficient strain (cho1Δ). In WT yeast, PS recruits GFP-LactC2 to the plasma membrane, whereas in the cho1Δ strain, GFP-LactC2 is present primarily in the cytosol. The plot of pixel intensity across the lines in the images indicates that GFP-Lac2 is present at the plasma membrane in WT cells but not cho1Δ. B, a cho1Δdrs2Δ PS-deficient strain expressing GFP-LactC2 and the indicated Dnf1 variants were incubated with 10 μm monoacyl lyso-PS or 100 μm DLPS at 4 °C for the times indicated. The ability of each strain to translocate unlabeled PS substrate across the membrane is indicated by recruitment of GFP-LactC2 from the cytosol to the inner leaflet of the plasma membrane. Dnf1 N550S and Dnf1 T254A can flip dually acylated PS (DLPS) to the cytosolic leaflet, but WT Dnf1 and Dnf1[Y618F] cannot. The right column is a plot of pixel intensity along the lines drawn across cell buds at 150 min. Scale bars = 10 μm.
Using a cho1Δdrs2Δ strain carrying an extra copy of WT DNF1, lyso-PS was rapidly flipped across the plasma membrane at 4 °C allowing GFP-LactC2 recruitment to the cytosolic leaflet of the plasma membrane within 5 min of substrate addition (Fig. 3B). In contrast, DLPS (C12 fatty acyl chains in the sn-1 and sn-2 positions) was not appreciably flipped across the membrane, and GFP-LactC2 remained mostly cytosolic up to 150 min of incubation. We then tested whether Dnf1 N550S, Dnf1[Y618F], or Dnf1 T254A were capable of transporting dually acylated DLPS. Consistent with the ability to restore plasma membrane PS asymmetry, DLPS was flipped into the cell by Dnf1 N550S and Dnf1 T254A but not by Dnf1[Y618F]. GFP-LactC2 plasma membrane fluorescence was weak for Dnf1 N550S cells incubated with DLPS, but the Dnf1 T254A cells treated with DLPS displayed a more robust LactC2 plasma membrane fluorescence. Line scans for cells expressing Dnf1 N550S and Dnf1 T254A and incubated 150 min with DLPS showed peaks of enrichment at the plasma membrane that was similar to the same cells treated with lyso-PS. Lyso-PS was quickly taken up across the plasma membrane of cells expressing each Dnf1 variant (Fig. 3B).
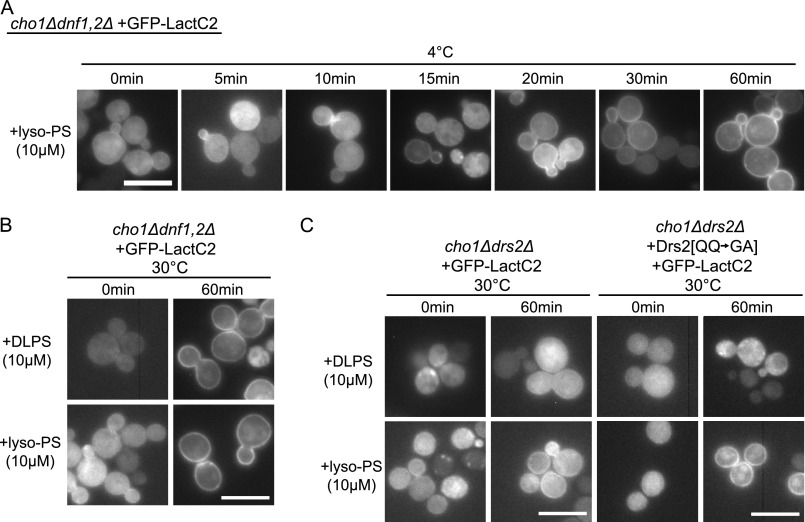
Lyso-PS flip was partially mediated by Dnf1 and Dnf2 even though Dnf1 recognizes NBD-PS poorly. After 30 min of incubation with a cho1Δ dnf1,2Δ strain at 4 °C, lyso-PS crossed the membrane sufficiently to recruit the GFP-LactC2, but it was at least 4-fold slower than with Dnf1 and Dnf2 present (Fig. 4A compared with Fig. 3). This is consistent with a previous report suggesting that yeast cells have another undefined transporter at the plasma membrane capable of taking up NBD-PS or lyso-PS that is independent of Dnf1,2,3 and Drs2 (35). For reasons that are unclear, we could not transform the cho1Δ dnf1,2Δ strain with plasmids carrying functional alleles of DNF1, whereas empty plasmid transformants were readily obtained. Because Dnf1 appears to be toxic to the cho1Δ dnf1,2Δ strain, we could not assay the Dnf1 variants for lipid uptake in this background.
FIGURE 4.
Requirement for Dnf1, Dnf2, and Drs2 in the translocation of lyso-PS and DLPS. A, a cho1Δdnf1Δdnf2Δ strain expressing GFP-LactC2 was incubated with 10 μm lyso-PS at 4 °C for the times indicated prior to imaging. Transport of lyso-PS was significantly slower than with the strains used for Fig. 3 that expressed Dnf1 and Dnf2. B, at 30 °C, a condition permitting endocytosis, both lyso-PS and DLPS are flipped to the cytosolic leaflet of a cho1Δdnf1,2Δ strain. C, a cho1Δdrs2Δ strain carrying an empty plasmid or expressing Drs2[QQ→GA] failed to transport DLPS to the cytosolic leaflet at 30 °C. Drs2[QQ→GA] is specifically deficient for PS recognition. Scale bars = 10 μm.
We next performed similar experiments at 30 °C, a condition that permits endocytosis, allowing the lyso-PS or DLPS to enter the lumenal leaflet of the Golgi-endosomal system. In cho1Δdnf1,2Δ, after 1 h at 30 °C, both the lyso-PS- and DLPS-treated cells displayed GFP-LactC2 localized to the plasma membrane (Fig. 4B). We suspected that this may be due to the lipid being flipped when it reached Drs2 in the Golgi or early endosome. To test this possibility, we examined GFP-LactC2 localization in cho1Δdrs2Δ cells supplemented with either lyso-PS or DLPS at 30 °C. In the absence of Drs2 or in the presence of Drs2[QQ→GA] (strongly deficient for PS recognition (18)), we observed no recruitment of GFP-LactC2 to the plasma membrane of cells incubated with DLPS (Fig. 4C). GFP-LactC2 was recruited to the plasma membrane of cells incubated with lyso-PS.
In total, these data indicate that Dnf1 and Dnf2 are unable to flip dually acylated PS across the plasma membrane but have a weak capacity to flip lyso-PS or NBD-PS. One class of mutations in Dnf1 (e.g. Y618F) enhances recognition of NBD-PS but does not allow PS recognition. A second class of mutations in Dnf1 (e.g. N550S and T254A) strongly enhances recognition of NBD-PS and endogenous PS. Drs2, in contrast, flips endogenous PS and DLPS efficiently.
P4-ATPase Phospholipid Recognition with P + 4 Leucine
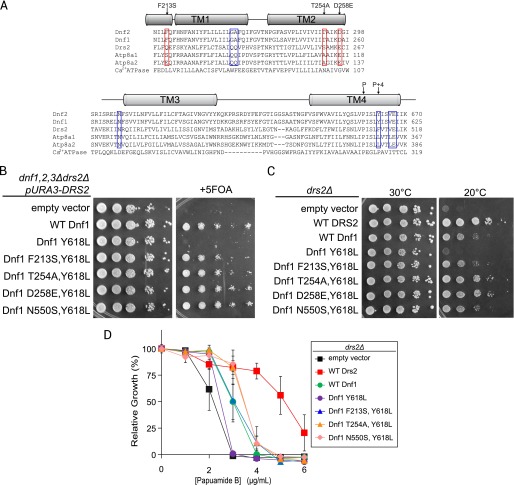
An unresolved question regarding the mechanism of phospholipid recognition involves the “P + 4 position” in TM4. The Drs2 homologs Atp8a1 and Atp8a2 also preferentially flip PS, but have a leucine at the P + 4 position rather than a phenylalanine (19, 21). Substitution of leucine for Tyr-618 or Phe-511 (P + 4 residues) in Dnf1 or Drs2, respectively, causes a reduction in overall activity with no apparent influence on PS recognition (17). Several of the drs2Δ suppressor mutations we identified in Dnf1 through unbiased, random mutagenesis (Fig. 2) were substitutions for the identical residues in Atp8a1 and/or Atp8a2 (Fig. 5A, red boxes). These mutations allowed recognition of endogenous PS and we wondered whether these substitutions combined with a P + 4 leucine could generate a functional Dnf1 capable of PS flip.
FIGURE 5.
Substitutions of specific TM1–3 residues restore function to a Dnf1 P + 4 leucine variant. A, sequence alignment of TM1–2 and TM3–4 from lyso-PC flippases (Dnf1 and Dnf2), PS flippases (Drs2, Atp8a1, Atp8a2), and Ca2+ ATPase. The blue boxes include residues reported previously as contributing to PS specificity. The red boxes indicate newly identified residues contributing to PS specificity in Dnf1 mutants that correspond to equivalent Atp8a1 residues. B, double mutants combining a p + 4 leucine (Y618L) with Atp8a1 residues (F213S, T254A, or D258E) allows Dnf1 complementation of a dnf1,2,3Δdrs2Δ strain. The 5-fluoroorotic acid kills cells that were unable to lose the URA3-marked plasmid carrying DRS2 in the parental strain. Strains that grow express a Dnf1 variant that can support the viability of dnf1,2,3Δdrs2Δ in the absence of Drs2. C, the same set of Dnf1 variants were tested for the ability to support growth of drs2Δ at 20 °C. D, similarly, each Y618L double mutant tested partially restored membrane PS asymmetry to drs2Δ relative to Y618L alone.
To test the potential cooperation of residues in TM1–3 with a P + 4 leucine, we generated combinations of Dnf1 Y618L with substitutions corresponding to Atp8a1 (F213S), Atp8a1/2 (T254A, D258E), or the PS transporting variant identified earlier (N550S). As reported previously, Dnf1 Y618L was unable to support growth of a dnf1,2,3Δdrs2Δ strain (Fig. 5B and Ref. 17). Remarkably, these substitutions combined with Y618L complemented the growth defect of a dnf1,2,3Δdrs2Δ strain, indicating that function had been restored (Fig. 5B). All of the double mutants suppressed the cold sensitivity of drs2Δ much more effectively than the Dnf1 Y618L single mutant and at least as well as WT Dnf1. In addition, Dnf1 T254A, Y618L and Dnf1 D258E, Y618L supported growth at 20 °C nearly as well as WT Drs2 (Fig. 5C).
To test whether these combinations of Y618L and TM1–3 substitutions were functional as flippases, we tested for the ability to restore PS asymmetry in drs2Δ cells. In each case, the double mutants restored PS asymmetry better than Dnf1 Y618L (Fig. 5D). Thus, the endogenous TM1,2 residues Phe-213, Thr-254, and Asp-258 couple with Tyr-618 in Dnf1 to prevent PS translocation and cannot function with a Leu at position 618 (P + 4). However, substituting the mammalian residues of Ser-213, Ala-254, or Glu-258 allow PS flip with either a Tyr or Leu in the P + 4 position.
We next tested whether the function of the Drs2 P + 4 leucine variant (Drs2 F511L) could be restored with these Atp8a1/a2 substitutions. In contrast to Drs2 F511L, each of the Drs2 double mutants tested (F511L combined with Q220S, S261A, or N445S) supported growth of a dnf1,2,3Δdrs2Δ strain (Fig. 6A). In each case, these double mutants also allowed increased cold-resistant growth of drs2Δ relative to Drs2 F511L (Fig. 6B). Each of the Drs2 TM1,2 single mutants tested supported growth of the dnf1,2,3Δdrs2Δ or drs2Δ strains (Fig. 6, C and D). Thus, the mammalian TM1,2 residues can function well with the endogenous P + 4 Phe in Drs2. When tested for the ability to restore PS membrane asymmetry, the double mutants all conferred greater resistance to PapB than Drs2 F511L (Fig. 6E). Although the double mutants tested did not complement the mutant phenotypes as well as WT Drs2, they were significantly better that Drs2 F511L. These results indicate that the P + 4 leucine is compatible with PS recognition when it is combined with the appropriate residues on TM1–3.
FIGURE 6.
Drs2 functions are restored with a P + 4 leucine (F511L) when combined with substitutions corresponding to Atp8a1 residues. A, in Drs2, substitutions of Atp8a1 residues combined with F511L partially restores the ability to complement a dnf1,2,3Δdrs2Δ strain. The same Drs2 variants restore cold resistance to drs2Δ (B) and support membrane PS asymmetry (E) better than Drs2 F511L alone. Substituting the individual residues also allows Drs2 to support growth of dnf1,2,3Δdrs2Δ (C) and cold-resistant growth of drs2Δ (D). 5-FOA, 5-fluoroorotic acid.
DISCUSSION
One interesting question preceding this work was why Dnf1 mutants capable of flipping NDB-PS were not able to restore PS asymmetry in drs2Δ cells (17). The fact that some Dnf1 mutants are fully capable of NBD-PS flip but seemingly unable to flip endogenous PS suggests that these pumps have either the improper localization, abundance, or substrate specificity to restore PS asymmetry in drs2Δ cells. However, the localization and abundance of Dnf1 N550S was indistinguishable from Dnf1 [Y618F] or Dnf1 [GA→QQ] (18). Here we show that Dnf1 N550S can replace Drs2 function in establishing endogenous PS asymmetry, whereas Dnf1 Y618F or Dnf1 [GA→QQ] cannot, even though all three mutant forms of Dnf1 can flip NBD-PS across the plasma membrane. The use of GFP-LactC2 localization in PS-deficient cho1Δ cells to monitor translocation of unmodified lyso-PS and DLPS provided an explanation as to why Dnf1[Y618F] was unable to establish PS asymmetry; it does not recognize dually acylated PS very well. In fact, WT Dnf1 also appears to flip lyso-PS much better than diacyl PS. Although WT Dnf1 does not flip endogenous PS very well, it seems to have a minor activity to NBD-PS, which can be increased in certain cases (17, 26). Dnf1[Y618F] and Dnf1[GA→QQ] certainly identified residues important in headgroup recognition because reciprocal changes in Drs2 abrogated its ability to recognize endogenous PS, not just NBD-PS. NBD-PL may be capable of mimicking a lyso-PL, a natural substrate for Dnf1, as well as the dually acylated PL substrate of Drs2. However, like drs2Δ, a dnf1,2Δ strain is also sensitive to PapB (36). This could be because of an increased exposure of lyso-PS on the extracellular leaflet of the dnf1,2Δ bilayer. Lyso-PS accounts for about 10% of PS in S. cerevisiae, which would likely be enough for PapB to permeabilize the cell if exposed on the outer leaflet of the plasma membrane (30).
A second question is why does yeast maintain two P4-ATPases that appear to function only as lyso-phospholipid transporters? Is this an apparent role for scavenging phospholipid from the environment? Could they be functioning to flip phospholipids that have been damaged by phospholipases in the extracellular environment? Yeast produces lyso-PL when using phospholipids as an acyl donor in triglyceride synthesis (37), and this species can be flipped by Pdr5 and Yor1 to the extracellular leaflet (38). Dnf1,2 may be required to retain the lyso-PL species on the cytosolic leaflet, where they are available to repair enzymes for acylation by Ale1 (39, 40). The ecological positioning of true wild-type yeast is in and around plants and animals, scavenging nutrients from the environment. The ability to scavenge nearly whole lipids (lyso-PL) would be a strong selective advantage, saving considerable energy from synthesis in the Kennedy pathway (41). Additionally, it is becoming increasingly apparent that lyso-PL species are involved in signaling events. Recent work has implicated the accumulation of long-chain lyso-PS species in a mammalian neurodegenerative condition (42). Lyso-PC is an early metabolic serum marker for diabetes (43) and also serves as a signaling lipid in plants, enhancing responsiveness for a Na+ activated H+ channel (44) and activating the plasma membrane proton pump AHA2 (45).
Drs2 may not flip lyso-PL as well as diacyl PL. The fact that Dnf1 N550S rescued drs2Δ growth defects much better than Dnf1 or Dnf1 [Y618F] implies that dually acylated PS is a crucial substrate for Drs2. In addition, Atp8a1 was found to have a much weaker transport activity toward lyso-PS relative to dioleoyl-PS or DLPS (20). Even with the non-preferred lipid PC, Atp8a1 transported the diacyl variants more quickly than lyso-PC (20). This may reflect a difference between the yeast and mammalian proteins, but it seems more likely that Drs2 also prefers intact phospholipid as its substrate. Therefore, we suggest different cellular niches for Drs2 and Dnf1/Dnf2, with Drs2 pumping PS and PE to the cytosolic leaflet and Dnf1/Dnf2 pumping lyso-PC, lysophosphatidylethanolamine, and lyso-PS (in this order of preference).
This work highlights additional residues involved in substrate specificity within TM1–4 in yeast P4-ATPases. The observation that Dnf1 N550S could effectively suppress drs2Δ mutant phenotypes suggested that a screen for additional DNF1 mutations that suppress drs2Δ would identify residues important for recognizing dually acylated PS. This prediction was met with the identification of F213S in TM1 and T254A and D258E in TM2 that allowed Dnf1 to flip PS and replace Drs2 function. These residues all cluster near the cytosolic side of the membrane, and we propose they form part of the exit gate where phospholipid is selected prior to release to the cytosolic leaflet. No residues in the entry gate, where phospholipid is initially selected from the exofacial leaflet, were recovered in our screen, implying that distinguishing monoacylated from diacylated substrate is a function of the exit gate.
The newly identified residues provide potential insight to how mammalian Atp8a1/a2, with a P + 4 leucine, recognizes PS. Strikingly, the randomly induced TM1–2 mutations allowing PS flip were substitutions of Dnf1 residues for analogous residues in Atp8a1. These changes could support PS translocation in the presence of the naturally occurring P + 4 Y618. But, perhaps more interestingly, these TM1–2 substitutions rescued the activity of Dnf1 with a P + 4 leucine (Y618L). The same TM1–2 Atp8a1 residues also rescued function of Drs2 with the F511L P + 4 mutation. These data strongly imply cooperation between the P + 4 position in TM4 and TM1–2 residues for phospholipid selection at the exit gate. For Dnf1, a Tyr in TM4 is coupled to an Asn (TM3), Asp and Thr (TM2), and Phe (TM1) to prevent transport of PS (Fig. 7A). To allow PS transport, these residues are Phe (TM4), Asn (TM3), Glu, Ser (TM2), and Gln (TM1) in Drs2 and, presumably, Leu (TM4), Asn (TM3), Glu, Ala (TM2), and Ser (TM1) in Atp8a1 (Fig. 4A).
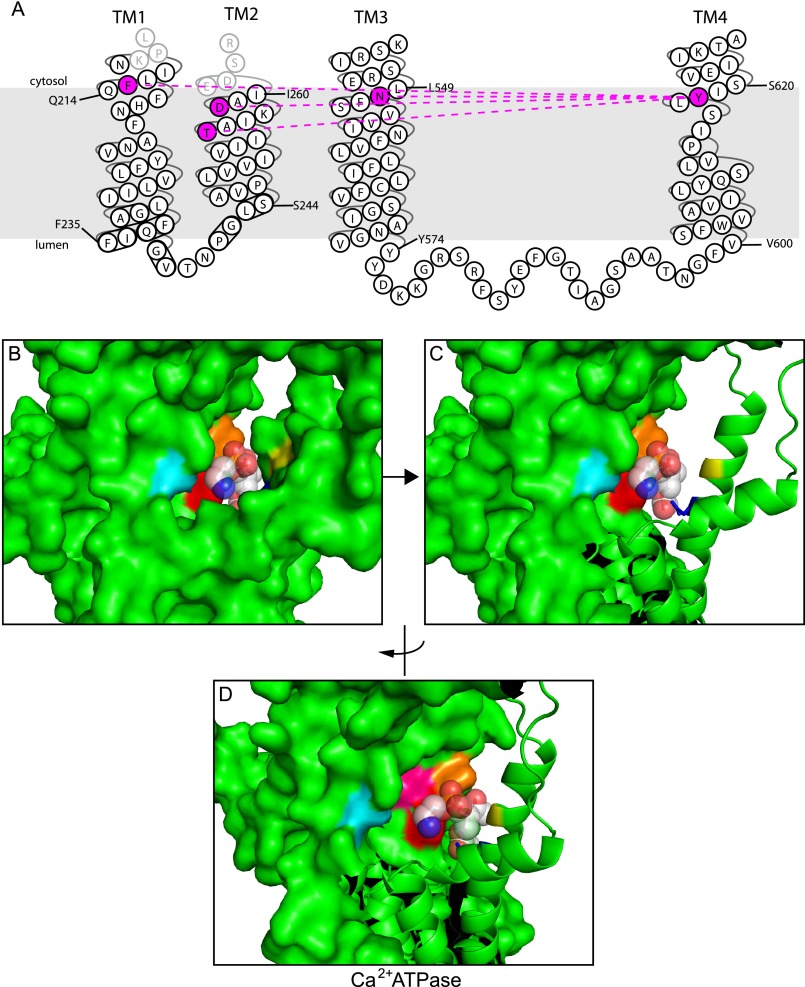
FIGURE 7.
Cooperation between the P + 4 position in TM4 and residues in TM1–3 for PS selection. A, the Dnf1 residues highlighted in purple can be mutated to residues that allow PS transport. Mutations in the highlighted TM1 and 2 positions to Atp8a1 residues can rescue function of a Drs2 or Dnf1 P + 4 leucine mutant. B–D, Ca2+ ATPase crystal structure in the E2 conformation with a PE molecule cocrystallized. The equivalent residues contributing to phospholipid specificity in Dnf1 and Drs2 have been color-coded for reference: cyan, Asp-254 (equivalent to Dnf1 Asn-550, Drs2 Asn-445); red, Pro-312 (Dnf1 Tyr-618, Drs2 Phe-511); pink, Ile-315 (Dnf1 Val-621, Drs2 Val-514); orange, Thr-316 (Dnf1 Glu-622, Drs2 Glu-515); blue, Asn-101 (Dnf1 Thr-254, Drs2 Ser-261); and yellow, Gly-105 (Dnf1 Asp-258, Drs2 Glu-265) (PDB code 2AGV (46).
Our results further support the “boundary lipid to substrate” hypothesis for the evolution of P4-ATPases from a primordial cation pump. The TM1–4 residues in the Ca2+ ATPase analogous to the P4-ATPase exit gate residues form a binding pocket for a phospholipid molecule when the pump is in the E2 conformation (Fig. 7B and Ref. 46). This binding pocket is inaccessible to phospholipid when the pump is in the E1 conformation, so phospholipid presumably enters and exits this site from the cytosolic leaflet with every ATP consumed. We suggest that this boundary lipid binding site evolved into a substrate binding site in the P4-ATPases. The P4-ATPases also evolved an entry gate to load the phospholipid into this binding site from the exofacial leaflet, thus flipping the phospholipid substrate.
Supplementary Material
Acknowledgments
We thank Scott Emr, Greg Fairn, and Sergio Grinstein for plasmids used in this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM62367 (to T. R. G.). This work was also supported by Cellular and Molecular Microbiology Training Program T32AI007611-11 (to R. D. B.), by Vanderbilt Ingram Cancer Center Grant P30 CA68485, and by Vanderbilt Digestive Disease Research Center Grant DK058404.

This article contains supplemental Tables S1 and S2.
- P4-ATPase
- type IV P-type ATPase
- PL
- phospholipid
- PS
- phosphatidylserine
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- P + 4
- proline + 4
- TM
- transmembrane
- SD
- synthetic minimal medium
- NBD
- 7-nitro-2-1,3-benzoxadiazol-4-yl
- DLPS
- dilauryl-phosphatidylserine
- PapB
- papuamide B.
REFERENCES
- 1. Axelsen K. B., Palmgren M. G. (1998) Evolution of substrate specificities in the P-Type ATPase superfamily. J. Mol. Evol. 46, 84–101 [DOI] [PubMed] [Google Scholar]
- 2. Sebastian T. T., Baldridge R. D., Xu P., Graham T. R. (2012) Phospholipid flippases. Building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu P., Okkeri J., Hanisch S., Hu R. Y., Xu Q., Pomorski T. G., Ding X. Y. (2009) Identification of a novel mouse P4-ATPase family member highly expressed during spermatogenesis. J. Cell Sci. 122, 2866–2876 [DOI] [PubMed] [Google Scholar]
- 4. Yabas M., Teh C. E., Frankenreiter S., Lal D., Roots C. M., Whittle B., Andrews D. T., Zhang Y., Teoh N. C., Sprent J., Tze L. E., Kucharska E. M., Kofler J., Farell G. C., Bröer S., Goodnow C. C., Enders A. (2011) ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat. Immunol. 12, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Siggs O. M., Arnold C. N., Huber C., Pirie E., Xia Y., Lin P., Nemazee D., Beutler B. (2011) The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat. Immunol. 12, 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu Q., Yang G.-Y., Liu N., Xu P., Chen Y.-L., Zhou Z., Luo Z.-G., Ding X. (2012) P4-ATPase ATP8A2 acts in synergy with CDC50A to enhance neurite outgrowth. FEBS Lett. 586, 1803–1812 [DOI] [PubMed] [Google Scholar]
- 7. Levano K., Punia V., Raghunath M., Debata P. R., Curcio G. M., Mogha A., Purkayastha S., McCloskey D., Fata J., Banerjee P. (2012) Atp8a1 Deficiency is associated with phosphatidylserine externalization in hippocampus and delayed hippocampus-dependent learning. J. Neurochem. 120, 302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kato U., Inadome H., Yamamoto M., Emoto K., Kobayashi T., Umeda M. (2013) Role for phospholipid flippase complex of ATP8A1 and CDC50A in cell migration. J. Biol. Chem. 288, 4922–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stapelbroek J. M., Peters T. A., van Beurden D. H., Curfs J. H., Joosten A., Beynon A. J., van Leeuwen B. M., van der Velden L. M., Bull L., Oude Elferink R. P., van Zanten B. A., Klomp L. W., Houwen R. H. (2009) ATP8B1 is essential for maintaining normal hearing. Proc. Natl. Acad. Sci. U.S.A. 106, 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu X., Libby R. T., de Vries W. N., Smith R. S., Wright D. L., Bronson R. T., Seburn K. L., John S. W. (2012) Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8, e1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Onat O. E., Gulsuner S., Bilguvar K., Nazli Basak A., Topaloglu H., Tan M., Tan U., Gunel M., Ozcelik T. (2013) Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur. J. Hum. Genet. 21, 281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dhar M. S., Sommardahl C. S., Kirkland T., Nelson S., Donnell R., Johnson D. K., Castellani L. W. (2004) Mice heterozygous for Atp10c, a putative amphipath, represent a novel model of obesity and type 2 diabetes. J. Nutr. 134, 799–805 [DOI] [PubMed] [Google Scholar]
- 13. Bull L. N., van Eijk M. J., Pawlikowska L., DeYoung J. A., Juijn J. A., Liao M., Klomp L. W., Lomri N., Berger R., Scharschmidt B. F., Knisely A. S., Houwen R. H., Freimer N. B. (1998) A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 18, 219–224 [DOI] [PubMed] [Google Scholar]
- 14. Wang L., Beserra C., Garbers D. L. (2004) A novel aminophospholipid transporter exclusively expressed in spermatozoa is required for membrane lipid asymmetry and normal fertilization. Dev. Biol. 267, 203–215 [DOI] [PubMed] [Google Scholar]
- 15. Hua Z., Fatheddin P., Graham T. R. (2002) An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 13, 3162–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 [angst] resolution. Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 17. Baldridge R. D., Graham T. R. (2012) Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 109, E290-E298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baldridge R. D., Graham T. R. (2013) Two gate mechanism for phospholipid selection and transport by type-IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 110, E358-E367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ding J., Wu Z., Crider B. P., Ma Y., Li X., Slaughter C., Gong L., Xie X. S. (2000) Identification and functional expression of four isoforms of ATPase II, the putative aminophospholipid translocase. J. Biol. Chem. 275, 23378–23386 [DOI] [PubMed] [Google Scholar]
- 20. Smriti, Nemergut E. C., Daleke D. L. (2007) ATP-dependent transport of phosphatidylserine analogues in human erythrocytes. Biochemistry 46, 2249–2259 [DOI] [PubMed] [Google Scholar]
- 21. Coleman J. A., Kwok M. C., Molday R. S. (2009) Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 284, 32670–32679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Natarajan P., Wang J., Hua Z., Graham T. R. (2004) Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. U.S.A. 101, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou X., Graham T. R. (2009) Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl. Acad. Sci. U.S.A. 106, 16586–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riekhof W. R., Wu J., Gijón M. A., Zarini S., Murphy R. C., Voelker D. R. (2007) Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae. The role of P-type ATPases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 282, 36853–36861 [DOI] [PubMed] [Google Scholar]
- 25. Riekhof W. R., Voelker D. R. (2006) Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J. Biol. Chem. 281, 36588–36596 [DOI] [PubMed] [Google Scholar]
- 26. Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., Holthuis J. C. (2003) Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 14, 1240–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wilson D. S., Keefe A. D. (2001) Random mutagenesis by PCR. Curr. Protoc. Mol. Biol. 51, 8.3.1–8.3.9 [DOI] [PubMed] [Google Scholar]
- 28. Gietz R. D., Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 29. Hanson P. K., Nichols J. W. (2001) Energy-dependent flip of fluorescence-labeled phospholipids is regulated by nutrient starvation and transcription factors, PDR1 and PDR3. J. Biol. Chem. 276, 9861–9867 [DOI] [PubMed] [Google Scholar]
- 30. Fairn G. D., Hermansson M., Somerharju P., Grinstein S. (2011) Phosphatidylserine is polarized and required for proper Cdc42 localization and for development of cell polarity. Nat. Cell Biol. 13, 1424–1430 [DOI] [PubMed] [Google Scholar]
- 31. Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., Grinstein S. (2008) Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 [DOI] [PubMed] [Google Scholar]
- 32. Atkinson K. D., Jensen B., Kolat A. I., Storm E. M., Henry S. A., Fogel S. (1980) Yeast mutants auxotrophic for choline or ethanolamine. J. Bacteriol. 141, 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Atkinson K., Fogel S., Henry S. A. (1980) Yeast mutant defective in phosphatidylserine synthesis. J. Biol. Chem. 255, 6653–6661 [PubMed] [Google Scholar]
- 34. Kean L. S., Fuller R. S., Nichols J. W. (1993) Retrograde lipid traffic in yeast. Identification of two distinct pathways for internalization of fluorescent-labeled phosphatidylcholine from the plasma membrane. J. Cell Biol. 123, 1403–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stevens H. C., Malone L., Nichols J. W. (2008) The putative aminophospholipid translocases, DNF1 and DNF2, are not required for 7-nitrobenz-2-oxa-1,3-diazol-4-yl-phosphatidylserine flip across the plasma membrane of Saccharomyces cerevisiae. J. Biol. Chem. 283, 35060–35069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen S., Wang J., Muthusamy B.-P., Liu K., Zare S., Andersen R. J., Graham T. R. (2006) Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7, 1503–1517 [DOI] [PubMed] [Google Scholar]
- 37. Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J. T., Sturley S. L. (2000) A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275, 15609–15612 [DOI] [PubMed] [Google Scholar]
- 38. Decottignies A., Grant A. M., Nichols J. W., de Wet H., McIntosh D. B., Goffeau A. (1998) ATPase and Multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 273, 12612–12622 [DOI] [PubMed] [Google Scholar]
- 39. Riekhof W. R., Wu J., Jones J. L., Voelker D. R. (2007) Identification and Characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282, 28344–28352 [DOI] [PubMed] [Google Scholar]
- 40. Jain S., Stanford N., Bhagwat N., Seiler B., Costanzo M., Boone C., Oelkers P. (2007) Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 282, 30562–30569 [DOI] [PubMed] [Google Scholar]
- 41. Voelker D. R. (2000) Interorganelle transport of aminoglycerophospholipids. Biochim. Biophys. Acta 1486, 97–107 [DOI] [PubMed] [Google Scholar]
- 42. Blankman J. L., Long J. Z., Trauger S. A., Siuzdak G., Cravatt B. F. (2013) ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. Proc. Natl. Acad. Sci. U.S.A. 110, 1500–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ferrannini E., Natali A., Camastra S., Nannipieri M., Mari A., Adam K.-P., Milburn M. V., Kastenmüller G., Adamski J., Tuomi T., Lyssenko V., Groop L., Gall W. E. (2013) Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes 62, 1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Viehweger K., Dordschbal B., Roos W. (2002) Elicitor-activated phospholipase A2 generates lysophosphatidylcholines that mobilize the vacuolar H+ pool for pH signaling via the activation of Na+-dependent proton fluxes. Plant Cell 14, 1509–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palmgren M. G., Christensen G. (1994) Functional comparisons between plant plasma membrane H+-ATPase isoforms expressed in yeast. J. Biol. Chem. 269, 3027–3033 [PubMed] [Google Scholar]
- 46. Obara K., Miyashita N., Xu C., Toyoshima I., Sugita Y., Inesi G., Toyoshima C. (2005) Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc. Natl. Acad. Sci. U.S.A. 102, 14489–14496 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.