Background: How SET8 is regulated is not fully understood.
Results: MicroRNA-7 down-regulates SET8 and inhibits H4K20 monomethylation, suppresses metastasis of breast cancer cells, and sensitizes cells to DNA damages.
Conclusion: MicroRNA-7 is a negative regulator of SET8.
Significance: This work aids our understanding of the biological function of microRNA-7, supporting the pursuit of microRNA-7 as a potential target for breast cancer intervention.
Keywords: Breast Cancer, DNA Damage, DNA Damage Response, Epithelial Mesenchymal Transition, MicroRNA, MicroRNA-7, SET8
Abstract
SET8 (SET domain containing 8) is a histone H4 lysine 20 (H4K20)-specific monomethyltransferase in higher eukaryotes that exerts diverse functions in transcription regulation, DNA repair, tumor metastasis, and genome integrity. The activity of SET8 is tightly controlled during cell cycle through post-translational modifications, including ubiquitination, phosphorylation, and sumoylation. However, how the expression of SET8 is regulated is not fully understood. Here, we report that microRNA-7 is a negative regulator of SET8. We demonstrated that microRNA-7 inhibits H4K20 monomethylation and suppresses epithelial-mesenchymal transition and the invasive potential of breast cancer cells. We showed that microRNA-7 promotes spontaneous DNA damages and sensitizes cells to induced DNA damages. Our experiments provide a molecular mechanism for the regulation of SET8 and extend the biological function of microRNA-7 to DNA damage response, supporting the pursuit of microRNA-7 as a potential target for breast cancer intervention.
Introduction
MicroRNAs (miRNAs)2 are a cluster of evolutionarily conserved, non-encoding single-stranded RNA molecules of 20–24 nucleotides. These molecules function to silence gene expression by binding to partially complementary recognition sequences of the 3′-UTRs of target mRNAs, leading to either mRNA degradation or translation inhibition (1, 2). More than 30% of human genes appear to have been under selective pressure to maintain their pairing to miRNA seeds, suggesting a role of miRNAs in posttranscriptional repression of these genes. With a growing number of oncogenes and tumor suppressor genes that have been found to be under the control of miRNAs, the importance of miRNAs in cell cycle progression, cell proliferation, genome stability, and malignant transformation is becoming increasingly recognized (3).
MicroRNA-7 (miR-7) is an intronic miRNA that resides in three different gene loci in the human genome and is conserved among all species (4). Aberrant expression of miR-7 was observed in various malignancies, including breast cancer (5, 6), glioblastomas (7), and gastric and colorectal cancers (8). Functionally, miR-7 has been characterized as a putative tumor suppressor through its suppression of the expression of multiple EGF receptor signaling-related genes, including EGF receptor as well as its downstream kinase RAF1 (9), p21-activated kinase 1, and insulin receptor substrate-2 (10). Moreover, a range of functionally important genes, including Kruppel-like factor 4 (5), Yin Yang 1 (YY1) (11), focal adhesion kinase (6), and insulin-like growth factor 1 receptor (12), have also been reported to be miR-7 targets, suggesting its profound role in malignant transformation and metastasis.
SET8 (also known as PR-Set7/9, SETD8, KMT5A), a member of the SET domain-containing methyltransferase family that specifically targets H4K20 for monomethylation, has been implicated in a diverse array of biological processes, including controlling gene transcription (13, 14), maintaining genome integrity (15, 16), regulating cell cycle progression (17, 18), and mediating DNA damage and repair (19, 20). In addition to H4K20, SET8 was also reported to methylate p53 at lysine residue 382 (p53K382me1) and to repress p53-mediated transcriptional activation of target genes (21). Previously, we reported that SET8 is functionally linked to TWIST and acts, via its H4K20 monomethylation activity, as a dual epigenetic modifier on the promoters of the TWIST target genes E-cadherin and N-cadherin (22).
Although the importance of SET8 in cell proliferation and homeostasis is well recognized and its activity is believed to be tightly controlled during cell cycle through post-translational modifications, including ubiquitination, phosphorylation, and sumoylation, how the expression of SET8 is regulated is poorly understood. In this report, we identified miR-7 as a negative regulator of SET8. We showed that miR-7 inhibits H4K20 monomethylation through targeting the 3′-UTR of SET8 mRNA. We demonstrated that miR-7 suppresses epithelial-mesenchymal transition (EMT) and invasion of breast cancer cells. We found that miR-7 promotes spontaneous DNA damages and sensitizes cells to induced DNA damages.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
The source of antibodies against following proteins was as follows. Tubulin and p53 were from B & M BIOTECH CO., Ltd.; SET8, RAD51, and 53BP1 were from Cell Signaling Technology; E-cadherin, α-catenin, β-catenin, N-cadherin, and γ-catenin were from BD Biosciences; fibronectin, H4, H3K4me3, H3K9me3, H3K27me3, H4K36me3, and H4K20me1 were from Abcam; γH2AX was from Millipore. Control siRNA, SET8 siRNA, and miR-7 mimics were synthesized by Shanghai GeneChem, Inc. (Shanghai, China). MiR-7 inhibitors were obtained from Dharmacon.
Cell Culture and Cell Transfection
MCF-7 and MDA-MB-231 cells were from the American Type Culture Collection (Manassas, VA). MCF-7 cells were maintained in DMEM (Hyclone) supplemented with 10% FBS. MDA-MB-231 cells were cultured in Leibovitz's L-15 medium with 10% FBS at 37 °C without CO2. Plasmids transfection was carried out using Megatran (version 1.0, Origene) according to the manufacturer's recommendations.
Plasmid Construction and Luciferase Assays
The wild-type and mutant SET8 3′-UTR were amplified by PCR and cloned in pMIR-REPORT (Ambion) with firefly luciferase. These PCR products were digested and ligated into the SpeI-Hind III sites of pMIR-REPORT vectors to generate a series of reporter constructs. The luciferase assays were performed in MCF-7 cells as described previously (23). MCF-7 cells treated with control, miR-7 mimics, or miR-7 inhibitors were transfected with wild-type or mutants of SET8 3′-UTR luciferase reporters together with Renilla plasmid. 48 h after transfection, the firefly and Renilla luciferases were assayed according to the manufacturer's instructions (Promega), and the firefly luciferase activity was normalized to that of Renilla luciferase. Each experiment was repeated in triplicate. SET8 3′-UTR wt-1 (forward primer), 5′-CTTCTTCAAAGGACAAAGTGCC-3′; SET8 3′-UTR wt-1 (reverse primer), 5′-TACAAAGCTAAACCACAAACAGG-3′; SET8 3′-UTR wt-2 (forward primer), 5′-GGACTAGTCTTCAAAGGACAAAGTGC-3′, SET8 3′-UTR wt-2 (reverse primer), 5′-GGGCCGGCACGTTAGGGGAACAAGAG-3′; SET8 3′-UTR wt-3 (forward primer), 5′-GGACTAGTACTCAGCACAGGTTTTAGA-3′; SET8 3′-UTR wt-3 (reverse primer), 5′-GGGCCGGCATTTCTGGTCCCACTACA-3′; SET8 3′-UTR wt-4 (forward primer), 5′-ATGCAGTCAAAGACTCAGCACAG-3′; SET8 3′-UTR wt-4 (reverse primer), 5′-GACAGCAGGTCTGGAACTTTCAA-3′; SET8 3′-UTR mut-1 (forward primer), 5′-GCCGAACGTTTGTGCCCTCCGTGTGCATGCAGTCAAAGAC-3′; SET8 3′-UTR mut-1 (reverse primer), 5′-GTCTTTGACTGCATGCACACGGAGGGCACAAACGTTCGGC-3′; SET8 3′-UTR mut-2 (forward primer), 5′-GTTTTTGCAGTAGCTAGACCTTCCCTCTGCTTTCTCGAA-3′, SET8 3′-UTR mut-2 (reverse primer), 5′-TTCGAGAAAGCAGAGGGAAGGTCTAGCTTACTGCAAAAAC-3′; SET8 3′-UTR mut-3 (forward primer), 5′-CCGGGCATAGATTTCCACGTACACAAGCTGCCGCTTTTCT-3′, SET8 3′-UTR mut-3 (reverse primer), 5′-AGAAAAGCGGCAGCTTGTGTACGTGGAAATCTATGCCCGG-3′; and SET8 3′-UTR mut-4 (forward primer), 5′-ATGCAGTCAAAGACTCAGCACAG-3′; SET8 3′-UTR mut-4 (reverse primer), 5′-GACAGCAGGTCTGGAACTTTCAA-3′.
RNA Isolation and Real-time Quantitative PCR
Total cellular RNAs were isolated with the TRIzol (Invitrogen) and the first strand cDNA synthesis was performed with the reverse transcription system (Promega). Quantization of all gene transcripts was done by real-time quantitative PCR using Power SYBR Green PCR Master Mix and an ABIPRISM 7300 sequence detection system (Applied Biosystems, Foster City, CA) with the expression of GAPDH or U6 as the internal control. The primer pairs used were as follows: GAPDH (forward primer), 5′-CCCACTCCTCCACCTTTGAC-3′ and GAPDH (reverse primer), 5′-CATACCAGGAAATGAGCTTGACAA-3′; SET8 (forward primer), 5′-ACTTACGGATTTCTACCCTGTC-3′ and SET8 (reverse primer), 5′-CGATGAGGTCAATCTTCATTCC-3′; miR-7 (reverse transcription primer), 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAACAAAAT-3′; miR-7 (forward primer), 5′-ACACTCCAGCTGGGTGGAAGACTAGTGAT-3′ and miR-7 (reverse primer), 5′-TGGTGTCGTGGAGTCG-3′; U6 (reverse transcription primer), 5′-AAAATATGGAACGCTTCACGAATTTGC-3′; U6 (forward primer), 5′-CCTGCTTCGGCAGCACA-3′ and U6 (reverse primer), 5′-TGGAACGCTTCACGAA-3′.
Fluorescence Confocal Microscopy
MCF-7 cells were plated into six-well chamber slides and were transfected with control, miR-7 mimics, or miR-7 inhibitors with/without SET8 cDNA lacking 3′-UTR for 36 h. Cells were washed with PBS, fixed in 4% (w/v) paraformaldehyde, permeabilized with 0.1% (v/v) Triton X-100 in PBS, blocked with 0.8% BSA, and incubated with appropriate primary antibodies followed by staining with FITC or RITC-conjugated secondary antibodies. Cells were washed four times and a final concentration of 0.1 μg/ml DAPI (Sigma) was included in the last washing to stain nuclei. Images were visualized and recorded with an Olympus FV1000S confocal microscope.
Transwell Invasion Assay
The Transwell invasion assays were performed using the Transwell chamber (Chemicon, Inc.) with a Matrigel-coated filter. MDA-MB-231 cells were transfected with control, miR-7 mimics, or miR-7 inhibitors with/without SET8 cDNA lacking 3′-UTR. 48 h later, cells were deprived in serum-free Leibovitz's L-15 medium. After 18 h of deprivation, cells were harvested, washed three times in PBS, and resuspended in serum-free culture medium. Afterward, 1 × 106 of these cells in 300 μl of serum-free medium were plated onto the upper chamber of the transwell. The upper chamber was then transferred to a well containing 500 μl of medium supplemented with 10% FBS and incubated for 24 h. Cells may actively migrate from the upper to the lower side of the filter due to FBS as attractant. Cells on the upside were removed using cotton swabs, and the invasive cells on the lower side were fixed, stained with 0.1% crystal violet solution, and counted using light microscope. The experiments were repeated three times.
Homologous Recombination (HR) and Nonhomologous End-Joining (NHEJ) Assays
DR-GFP-U2OS and EJ5-GFP-HEK293 stable cell lines (kindly provided by Dr. Xingzhi Xu, The Capital Normal University) were transfected with control or miR-7 mimics with/without SET8 cDNA lacking 3′-UTR for 6 h prior to transfection with PCBASce or pCAGGS-Ds-red (transfection indicator). After 48 h of transfection, HR or NHEJ repair efficiency was measured with the percentage of GFP-positive by FACScan.
Clonogenic Assay
MCF-7 cells were transfected with control or miR-7 mimics with/without SET8 cDNA lacking 3′-UTR. After 48 h of transfection, cells were challenged with different concentrations of VP16 or camptothecin, and then were plated in triple wells at 1000 cells/60-mm2 dish. After 14 days, the colonies were stained with 0.1% crystal violet solution, and counted using light microscope. Colonies were defined as groups of 30 or more cells and survival potential was calculated as a percentage of colonies relative to untreated controls. The experiment was repeated three times.
RESULTS
Histone Methyltransferase SET8 Is a Downstream Target of miR-7
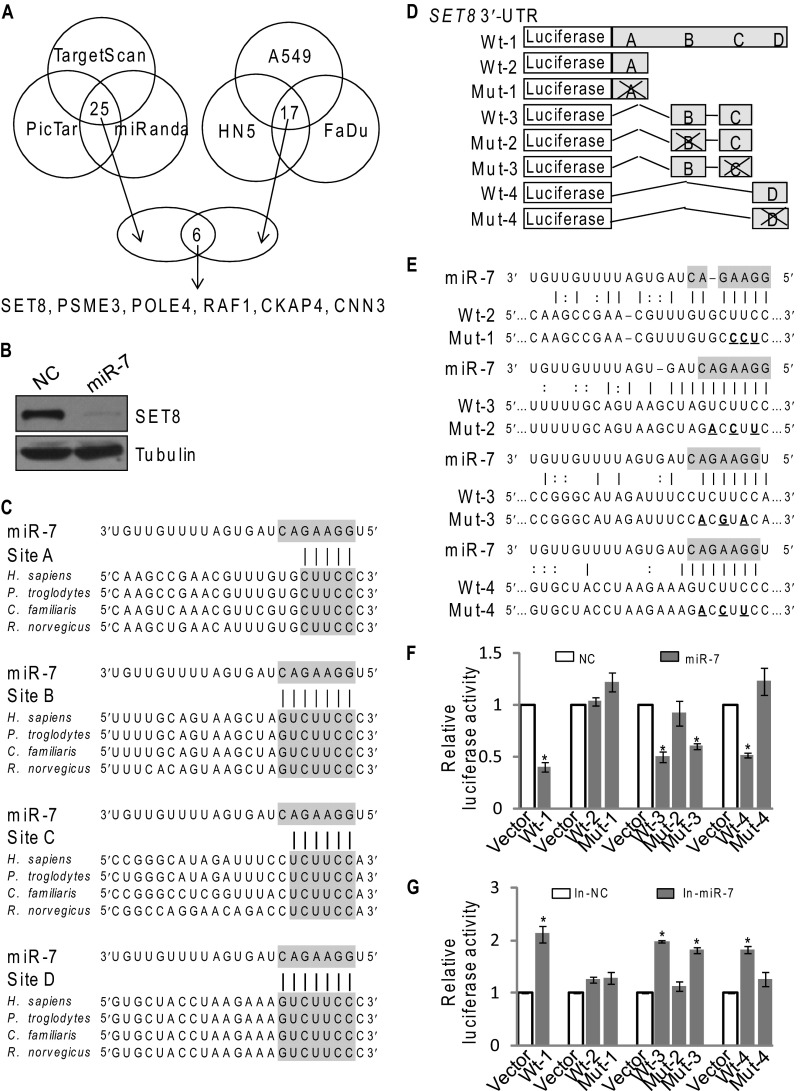
MiR-7 is implicated in cell proliferation and apoptosis, and its expression is dysregulated in many types of tumors, including breast cancer (10). To further explore the functional role of miR-7 in pathophysiological settings, we sought to identify additional targets for miR-7. Analyses based on three public algorithms (TargetScan, PicTar, and miRanda) and of the data about miR-7-downregulated genes in three cell lines (HN5, A549, and FaDu) from microarrays (9, 24) yielded several potential targets for miR-7 with high confident scores (Fig. 1A), including SET8, PSME3, POLE4, CKAP4, and CNN3, as well as RAF1, a known target of miR-7. In light of our previous observation that SET8 is implicated in the development and progression of breast cancer (22), we focused our study on SET8. Western blotting analysis showed a marked reduced level of SET8 protein in miR-7 mimic-transfected MCF-7 cells (Fig. 1B), and further bioinformatics analysis of miRanda indicated that the SET8 3′-UTR harbors four potential miR-7 target sites (A, B, C, and D) (Fig. 1C). These sites are either completely (B and D) or partially (A and C) complementary to a heptamer motif 5′-GGAAGAC-3′ that is found in the seed region of human miR-7 (Fig. 1C).
FIGURE 1.
Histone methyltransferase SET8 is a downstream target of miR-7. A, the intersection of the left Venn diagram displays 25 potential targets for miR-7 predicted through PicTar, TargetScan, and miRanda algorithms with high confident scores. The intersection of the right Venn diagram displays 17 down-regulated genes (>2.0-fold) based on microarray data in miR-7-overexpressed A549, HN5, and FaDu cells. The intersection of the lower Venn diagram is an overlap between algorithm prediction and microarray data analysis, representing six candidate targets for miR-7. B, MCF-7 cells were transfected with control or miR-7 mimics for 48 h. The endogenous SET8 protein expression was measured by Western blotting. C, sequence alignment of SET8 3′-UTR between human (Homo sapiens), chimpanzee (Pan troglodytes), dog (Canis familiaris), and rat (Rattus norvegicus). The miR-7 seed sequence (5′-GGAAGAC-3′) and the corresponding complementary sequences of SET8 3′-UTR are shaded. D, the diagram of luciferase reporter constructs for full-length wild-type (wt-1), truncated wild-type (wt-2, 3, 4), and mutants (mut-1, 2, 3, 4) of SET8 3′-UTR. E, sequences of wild-type and mutant target sites for miR-7 in SET8 3′-UTR are shown. miR-7 seed sequence (5′-GGAAGAC-3′) is shaded. Three point mutations (underlined) predicted to abolish miRNA-mRNA binding were made in miR-7 recognition region. F, luciferase reporter assays were performed to identify the binding of miR-7 to SET8 3′-UTR. MCF-7 cells were co-transfected with negative control (NC) or miR-7 mimics (miR-7) together with the luciferase gene driven by either wild-type or mutated SET8 3′-UTR sequences, including the full-length and three deletions of the wild-type SET8 3′-UTR (wt-1, 2, 3, 4) as well as four mutants of SET8 3′-UTR (mut-1, 2, 3, 4). The normalized luciferase activity in the control group was set as relative luciferase activity. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. G, luciferase reporter assays were performed in MCF-7 cells that co-transfected with negative control (In-NC) or miR-7 inhibitors (In-miR-7) together with the luciferase gene driven by either wild-type or mutated SET8 3′-UTR sequences, including the full-length and three deletions of the wild-type SET8 3′-UTR (wt-1, 2, 3, 4) as well as four mutants of SET8 3′-UTR (mut-1, 2, 3, 4). The normalized luciferase activity in the control group was set as relative luciferase activity. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05.
To verify the proposition that SET8 is a downstream target of miR-7, reporter assays were performed in MCF-7 cells with the luciferase gene driven by either wild-type or mutated SET8 3′-UTR sequences (Fig. 1D), including the full-length and three deletions of the wild-type SET8 3′-UTR (wt-1, 2, 3, 4) as well as 4 mutants of SET8 3′-UTR (mut-1, 2, 3, 4), with each containing three point mutations (Fig. 1E). The results showed a 60% reduction in luciferase activity with wt-1 (the full-length wild-type SET8 3′-UTR) in MCF-7 cells transfected with miR-7 mimics. Consistent with our prediction, the sites B and D play a more important role in the binding of miR-7 to SET8 3′-UTR, as there were more evident decreases in luciferase activity with wt-3, wt-4 and mut-3, compared with that with wt-2 (Fig. 1F). Moreover, the binding of miR-7 to sites B and D of SET8 3′-UTR was specific, as the point mutation of corresponding sites (mut-2 and mut-4) disrupted miR-7 repressive activity on reporter gene expression, whereas mutation of sites A (mut-1) did not change luciferase activity (Fig. 1F). Consistently, treatment with miR-7 inhibitors led to a reverse trend of luciferase activity with wt-1, 3, 4, and mut-3 (Fig. 1G). Taken together, these results suggest that miR-7 binds to SET8 3′-UTR and that SET8 is a downstream target of miR-7.
miR-7 Promotes SET8 mRNA Degradation and Inhibits H4K20 Monomethylation
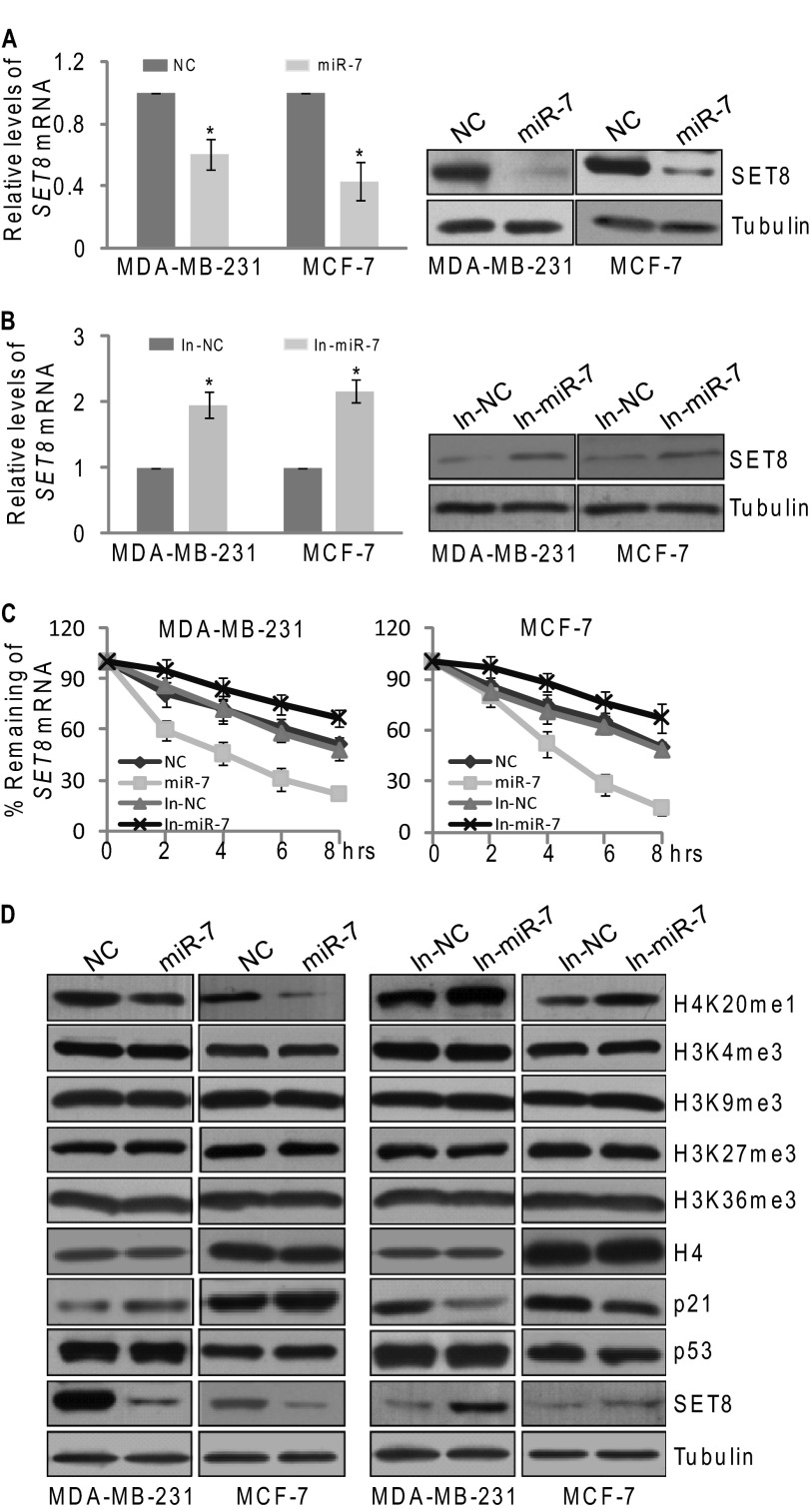
In view of the fact that miRNAs silence genes by translational inhibition and mRNA destabilization (25), we next investigated the effect of miR-7 on the expression of SET8 mRNA and protein. For this purpose, MDA-MB-231 and MCF-7 cells were transfected with control or miR-7 mimics, followed by the measurement of SET8 mRNA and protein expression by real-time quantitative PCR (qPCR) and Western blotting, respectively. The results showed that miR-7 overexpression led to ∼40 and 60% decrease in SET8 mRNA in MDA-MB-231 and MCF-7 cells, respectively (Fig. 2A), and Western blotting analysis also showed a marked reduction of SET8 protein expression in cells transfected with miR-7 mimics (Fig. 2A). Consistently, treatment of the cells with miR-7 inhibitors resulted in an ∼2-fold increase in SET8 mRNA (Fig. 2B) and a pronounced elevation in SET8 protein expression (Fig. 2B). To determine the impact of miR-7 on SET8 mRNA stability, MCF-7 cells and MDA-MB-231 cells were transfected with miR-7 mimics or miR-7 inhibitors followed by treatment with transcription inhibitor actinomycin D. Real-time qPCR measurement showed that miR-7 depletion was associated with an increased SET8 mRNA half-time, whereas miR-7 overexpression led to a reduced SET8 mRNA half-time (from 8 to 4 h) (Fig. 2C), further supporting the notion that miR-7 represses SET8 expression through destabilization of SET8 mRNA.
FIGURE 2.
miR-7 promotes SET8 mRNA degradation and inhibits H4K20 monomethylation. A and B, MiR-7 represses SET8 mRNA and protein expression. The endogenous SET8 mRNA (left panel) and protein expression (right panel) were measured by real-time qPCR and Western blotting, respectively. MDA-MB-231 and MCF-7 cells were transfected with control, miR-7 mimics, or miR-7 inhibitors for 48 h. The relative level of SET8 mRNA was normalized to GAPDH. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. C, miR-7 destabilizes SET8 mRNA. MDA-MB-231 and MCF-7 cells were transfected with control, miR-7 mimics, or miR-7 inhibitors for 36 h and then treated with actinomycin D (5 μg/ml) for the indicated times. The endogenous SET8 mRNA was measured by qPCR, and the relative level of SET8 mRNA was normalized to GAPDH. Each bar represents the mean ± S.D. for triplicate experiments. D, miR-7 inhibits H4K20 mono-methylation and promotes p53 transcriptional activity. MDA-MB-231 and MCF-7 cells were transfected with control, miR-7 mimics, or miR-7 inhibitors. Cellular lysates were prepared for Western blotting using antibodies against the indicated proteins. The loading controls were α-H4 and α-tubulin.
As stated above, SET8 is a member of SET domain-containing methyltransferase family that specifically catalyzes mono-methylation of histone H4 Lys-20 (H4K20me1). The observation that miR-7 influences the mRNA level of SET8 prompted us to investigate whether miR-7 could impact on monomethylation of H4K20. Consistent with decreased expression of SET8 protein, reduction of H4K20me1 was observed in miR-7 mimic-transfected MCF-7 cells and MDA-MB-231 cells, whereas no significant difference in H3K4me3, H3K9me3, H3K27me3, H3K36me3, and total histone H4 were detected in these cells (Fig. 2D). In contrast, cells that were treated with miR-7 inhibitors exhibited increased levels of H4K20me1 (Fig. 2D), supporting the argument that miR-7 inhibits H4K20 monomethylation by promoting SET8 mRNA degradation.
It was reported that SET8 could also methylate p53 at lysine 382 (p53K382) and suppress p53-mediated transcription activation of target genes such as p21 protein (21). Therefore, we tested whether miR-7 could regulate the activity of p53 and the expression of its downstream genes in breast cancer cells. Western blotting analysis showed an elevation of p21 expression in miR-7 mimic-treated cells, whereas miR-7 inhibitor treatment led to a reverse trend of p21 protein but not p53 itself (Fig. 2D). Collectively, these data indicate that miR-7 inhibits H4K20 monomethylation and promotes p53 transcriptional activity by targeting SET8.
miR-7 Suppresses EMT and Invasion of Breast Cancer Cells by Down-regulating SET8
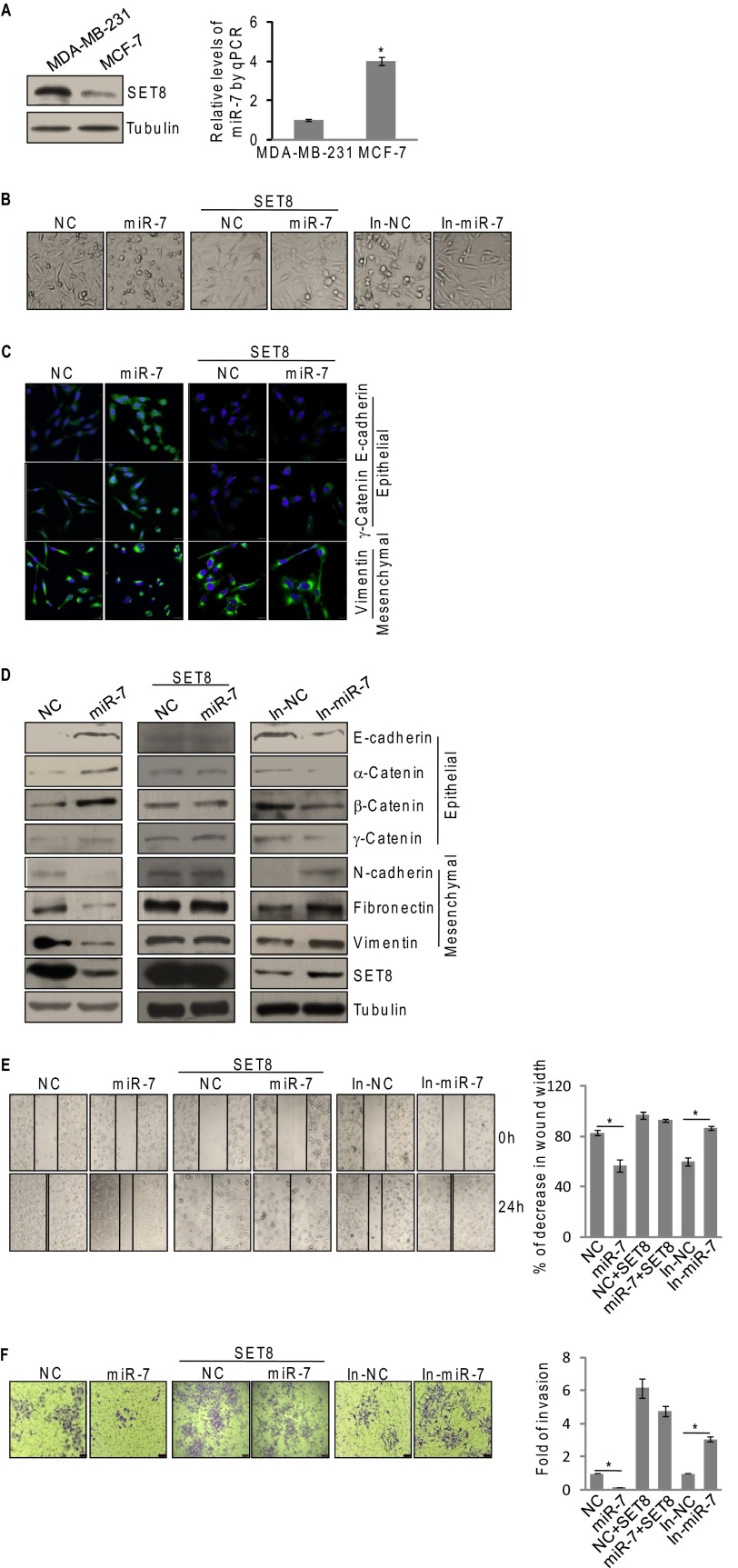
We next investigated the biological significance of miR-7-downregulated SET8 expression. To this end, we first examined the expression of endogenous SET8 protein and miR-7 expression in breast cancer cells. As shown in Fig. 3A, higher expression of SET8 and lower miR-7 levels were observed in metastatic MDA-MB-231 cells compared with that in non-metastatic MCF-7 cells. Based on our previous report that SET8 promotes EMT and metastasis of breast cancer (22), it is reasonable to postulate that miR-7 also play a role in EMT and metastasis of breast cancer cells. To test this hypothesis, we examined cell morphology and expression of epithelial/mesenchymal markers in miR-7 mimic- or miR-7 inhibitor-transfected MDA-MB-231 cells by microscopy and Western blotting, respectively. Although control MDA-MB-231 cells maintained a spindle-like, fibroblastic morphology representing mesenchymal cell morphology, miR-7 mimic-transfected cells displayed a scattering and cobble stone-like appearance (Fig. 3B). Meanwhile, miR-7 inhibitor-treated cells still kept mesenchymal cell morphology and even became longer and thinner (Fig. 3B). Consistent with this, immunofluorescent microscopy showed an elevation of E-cadherin and γ-catenin staining in miR-7-overexpressing MDA-MB-231 cells compared with that in control cells, whereas vimentin, a mesenchymal marker, exhibited a reverse trend (Fig. 3C). In agreement with the results, we found that overexpression of miR-7 resulted in increases in the expression levels of epithelial markers (E-cadherin, α-catenin, β-catenin, and γ-catenin) and decreases in that of mesenchymal markers (N-cadherin, fibronectin, and vimentin), as shown by immunoblotting (Fig. 3D). Conversely, repression of miR-7 with specific inhibitors in MDA-MB-231 cells was associated with an evident derepression of the mesenchymal markers and a significant deactivation of the epithelial markers, as shown by immunoblotting (Fig. 3D). Moreover, miR-7 expressing MDA-MB-231 cells that were co-transfected with the SET8 expression construct containing SET8 coding sequence but lacking the SET8 3′-UTR, which is resistant to miR-7, attenuated the morphological alterations and epithelial/mesenchymal marker changes induced by miR-7 (Fig. 3, B–D). The restoration of SET8 expression was confirmed by immunoblotting (Fig. 3D), supporting a notion that the impact of miR-7 on EMT and expression of epithelial/mesenchymal markers in breast cancer cells is mediated by SET8.
FIGURE 3.
miR-7 suppresses EMT and invasive potential of breast cancer cells by down-regulating SET8. A, the endogenous expressions of SET8 and miR-7 were measured by Western blotting (left panel) and qPCR (right panel) in MDA-MB-231 and MCF-7 cells. Transcript level of miR-7 was normalized to that of U6. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. B, the effect of miR-7 on cell morphology in MDA-MB-231 cells. Cells were transfected with miR-7 mimics or miR-7 inhibitors with/without SET8 cDNA lacking the 3′-UTR. The morphological alterations were observed by phase-contrast microscopy. C, the effect of miR-7 on expression of epithelial/mesenchymal markers in MDA-MB-231 cells. Cells were transfected with control or miR-7 mimics and/or SET8 cDNA lacking the 3′-UTR. The immunofluorescence staining of epithelial (E-cadherin and γ-catenin) and mesenchymal (vimentin) markers (green) were visualized by confocal microscopy, and DAPI staining (blue) was included to visualize the cell nucleus. D, cells were transfected with miR-7 mimics or miR-7 inhibitors with/without SET8 cDNA lacking the 3′-UTR. Immunoblotting examination of the epithelial/mesenchymal markers in MDA-MB-231 cells using antibodies against the indicated proteins. E, MDA-MB-231 cells were transfected with miR-7 mimics or miR-7 inhibitors with/without SET8 cDNA lacking the 3′-UTR. Wound healing assays were determined by phase-contrast microscopy at the indicated time points. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. F, MDA-MB-231 cells were transfected with miR-7 mimics or miR-7 inhibitors with/without SET8 cDNA lacking the 3′-UTR for 48 h. Cells were starved for 18 h before transwell invasion assay were performed using Matrigel transwell filters. The invaded cells were stained and counted. The images represent one field under microscopy in each group. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. NC, negative control; In-miR-7, miR-7 inhibitors.
To substantiate the role of miR-7 in tumor migration and invasion, the effect of gain-of-function or loss-of-function of miR-7 on the invasive potential in MDA-MB-231 cells was investigated by wound-healing assays. The results showed that miR-7 inhibitor-treated cells were more efficient in wound healing, whereas miR-7 mimic-treated cells were somewhat resistant to wound healing (Fig. 3E). In addition, transwell invasion assays showed that miR-7 overexpression inhibited the invasion of MDA-MB-231 cells by 4.5-fold, whereas miR-7 knockdown promoted the invasion by 3-fold (Fig. 3F). Moreover, miR-7-suppressed EMT of breast cancer cells was probably mediated by SET8, as ectopic expression of SET8 was able to rescue the effect of miR-7 on tumor migration and invasion (Fig. 3, E and F). Together, these results support an argument that miR-7 suppresses EMT and invasion of breast cancer cells and that this activity is, at least partially, mediated by targeting SET8.
miR-7 Promotes Spontaneous DNA Damages and Sensitizes Cells to Induced DNA Damages
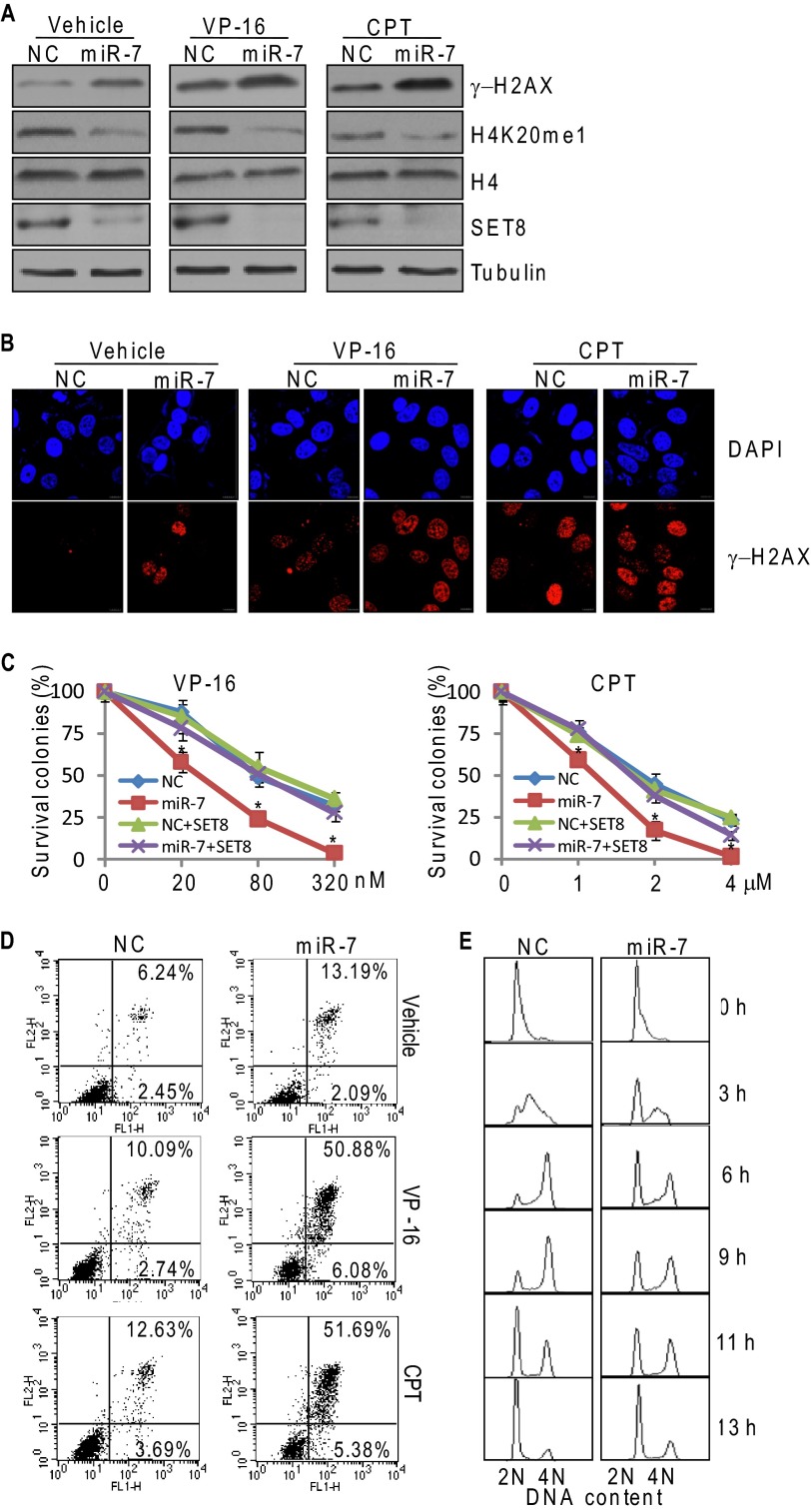
It was reported that loss of function of SET8 was associated with decreases in H4K20me1 and increases in DNA double-strand breaks (15). In light of our observation that SET8 is subjected to negative regulation by miR-7, we asked the question what role, if any, miR-7 plays in DNA damage response. For this purpose, MCF-7 cells were treated with control or miR-7 mimics for 36 h and challenged with DNA-damaging agents, etoposide (VP-16), or camptothecin for 4 h. Western blotting analysis indicated the levels of phosphorylated H2A.X (γH2AX) increased in miR-7 mimic-treated cells both in the presence and absence of VP-16 or camptothecin (Fig. 4A). Consistently, immunofluorescent microscopy showed that, compared with control, miR-7 mimic-treated cells were enriched with nuclear foci of γH2AX, suggesting that miR-7 promotes the accumulation of spontaneous DNA damages (Fig. 4B). Treatment with VP-16 or camptothecin led to intensified enrichments of γH2AX foci, suggesting that miR-7 sensitizes cells to induced DNA damages (Fig. 4B).
FIGURE 4.
miR-7 promotes spontaneous DNA damages and sensitizes cells to induced DNA damages. A, MCF-7 cells were treated with control or miR-7 mimics for 36 h and challenged with 80 nm etoposide (VP-16) or 2 μm camptothecin (CPT) for 4 h. Cellular lysates were analyzed by Western blotting using antibodies against the indicated proteins. B, MCF-7 cells were treated with control or miR-7 mimics for 36 h and challenged with 80 nm VP-16 or 2 μm camptothecin for 4 h. Immunofluorescence staining of γ-H2AX foci (red) was visualized by confocal microscopy and DAPI staining (blue) was included to visualize the cell nucleus. C, the effect of miR-7 on cell survival under the exposure of genotoxic agents. MCF-7 cells were transfected with control or miR-7 mimics, and/or SET8 cDNA lacking the 3′-UTR, challenged with the indicated concentrations of VP-16 or camptothecin, and subjected to cologenic survival assays. Cells were stained with crystal violet and air-dried after 14 days. Numbers of colonies (≥30 per colony) were scored by light microscope. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. D, the effect of miR-7 on cell apoptosis under the exposure of genotoxic agents. MCF-7 cells were treated with control or miR-7 mimics for 24 h, challenged with 40 nm etoposide (VP-16) or 1 μm camptothecin for 36 h, and stained with annexin V/propidium iodide before flow cytometry. E, the effect of miR-7 on cell cycle progression. MCF-7 cells treated with control or miR-7 mimics were synchronized by double thymidine block and released into the cell cycle for the indicated times. Cells were collected for cell cycle analysis by propidium iodide staining and flow cytometry. Experiments were repeated by three times, and the data from a representative experiment are shown. NC, negative control.
To further consolidate miR-7-induced cellular sensitivity to DNA damage, we tested the effect of miR-7 on cell survival and apoptosis under the exposure of DNA damaging agents. For this purpose, MCF-7 cells treated with control or miR-7 mimics were challenged with VP-16 or camptothecin. Clonogenic assays showed that a more pronounced decrease in the cell viability was detected upon exposure of miR-7-overexpressing MCF-7 cells to DNA damaging agents of increased concentrations (Fig. 4C), which could be partially reversed by reconstitution of SET8. Consistently, transfection of miR-7 mimics in MCF-7 cells resulted in an increased number of cells that underwent apoptosis, which was further exacerbated when cells were exposed to VP-16 or camptothecin (Fig. 4D). Moreover, flow cytometry showed miR-7 overexpression caused a delay of cell cycle progression, revealing that miR-7-induced DNA damage could potentially cause the activation of checkpoint and cell cycle arrest (Fig. 4E). Collectively, these observations suggest that miR-7 promotes cellular susceptibility to genotoxic agents, and the effect is, at least partially, through its targeting of SET8.
miR-7 Impedes both HR and NHEJ of DNA Repair
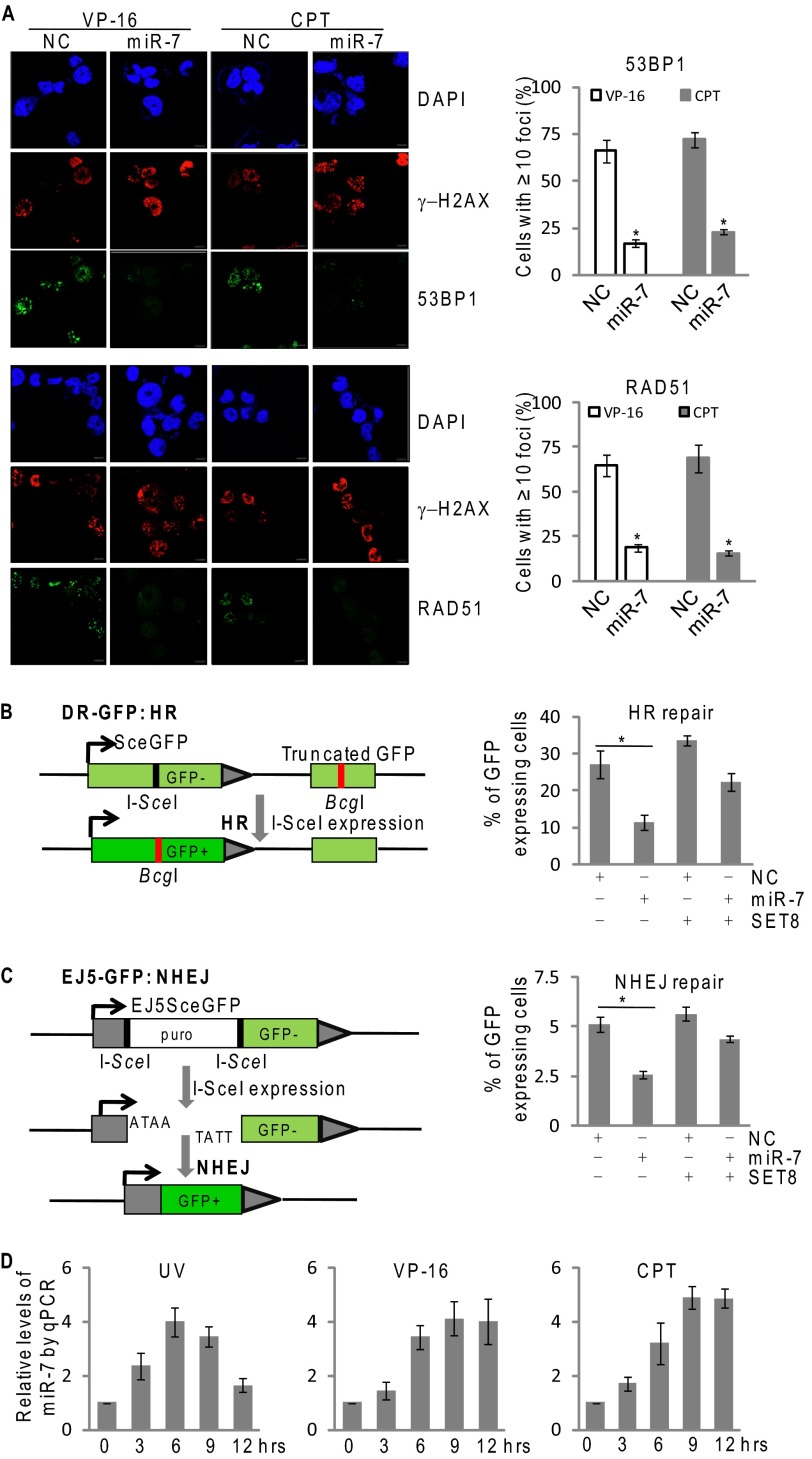
Given the importance of H4K20 methylation in DNA damage response (26, 27), we next explored how miR-7 influences DNA repair. Immunofluorescent staining showed that miR-7 suppressed the formation of nuclear foci of 53BP1 and RAD51, two key proteins that are recruited at damage sites of chromosomes during DNA repair (28–30), in MCF-7 cells under the exposure of VP-16 or camptothecin (Fig. 5A).
FIGURE 5.
miR-7 impedes both HR and NHEJ of DNA repair. A, the immunofluorescence staining of RAD51 (green), 53BP1 (green), and γ-H2AX nuclear foci (red) were visualized by confocal microscopy, and DAPI staining (blue) was included to visualize the cell nucleus in MCF-7 cells. Cells were transfected with control or miR-7 mimics for 36 h and challenged with 80 nm VP-16 or 2 μm camptothecin for 4 h. 53BP1 or RAD51 foci (≥10) were scored and quantified in at least 200 cells. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. B, left panel: schematic illustration of I-SceI mediated double-strand break induction and repair using DR-GFP transgenes. Right panel: FACS analysis of relative repair rate in the treated DR-GFP transgenic U2OS cells. Cells were transfected with control or miR-7 mimics with/without SET8 cDNA lacking 3′-UTR, and were subjected to HR assays as described in methods. Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *p < 0.05. (C) Left panel: schematic illustration of I-SceI mediated double-strand break induction and repair using EJ5-GFP transgenes. Right panel: FACS analysis of relative repair rate in the treated EJ5-GFP transgenic 293 cells. Cells were treated with control or miR-7 mimics with/without SET8 cDNA lacking 3′-UTR, and were subjected to NHEJ assays as described under “Experimental Procedures.” Each bar represents the mean ± S.D. for triplicate experiments. p values were determined by Student's t test. *, p < 0.05. (D) MCF-7 cells were challenged with 10 mJ/cm2 UV radiation, 40 nm VP-16 or 1 μm camptothecin. The endogenous expression of miR-7 was measured by qPCR at the indicated times after appropriate treatment. The transcript level of miR-7 was normalized to that of U6. Each bar represents the mean ± S.D. for triplicate experiments. NC, negative control; CPT, camptothecin.
To understand further the role of miR-7 in DNA repair, we investigated the pathway where miR-7 impacts DNA repair, HR, or NHEJ. For this purpose, we utilized two stable cell lines, DR-GFP-U2OS and EJ5-GFP-HEK293, which were designed to measure HR and NHEJ, respectively (26, 27). The systems utilize GFP as a recombination reporter and I-SceI endonuclease for introduction of double-strand breaks. The repair by HR or NHEJ restores functional GFP expression, and a fluorescent signal can be detected by flow cytometry (31). As shown in Fig. 5, B and C, the efficiency of both HR and NHEJ decreased by ∼2-fold in miR-7 mimic-treated cells. Ectopic expression of SET8 partially restored repair potential of the cells. It appears that miR-7 impedes both homologous recombination and non-homologous end joining of DNA repair, and the effect is, at least partially, through its targeting of SET8. Significantly, increased miR-7 levels were also detected in MCF-7 cells exposed to VP-16, camptothecin, or UV (Fig. 5D), as measured by qPCR, suggesting that miR-7 could be induced in DNA damage responses.
DISCUSSION
Epigenetic regulation, by means of DNA methylation, histone modification, chromatin remodeling, and miRNAs, play important roles in a wide range of biological processes, such as X-chromosome inactivation (32), imprinting (33), reprogramming (34), and gene silencing (35). Each of the epigenetic mechanisms does not work alone but forms an essential and intricate network with each other. For example, it has been reported that the promoters in about half of miRNAs are associated with CpG islands that may be repressed by DNA methylation (36). Moreover, a series of histone-modifying enzymes are subject to miRNA modulation. For example, the expression of enhancer of Zeste 2 (EZH2) is inhibited by miR-101 (37), miR-26a (38) and miR-214 (39) in cancer cell lines; miR-137 targets LSD1 and forms a feedback regulatory loop to participate in neural stem cell proliferation and differentiation during neural development (40, 41). We report here that miR-7 is a negative regulator of histone methyltransferase SET8 that inhibits H4K20 monomethylation, aiding the understanding of the complexity of the epigenetic regulatory network.
Thus far, more than 1500 human miRNAs are annotated, which regulate ∼30% of the total genomic mRNAs, many of which are associated with a carcinogenic potential (42). It has been reported that miR-7 targets, in addition to BCL-2 (43) and YY1 (11), gene transcripts whose protein products are linked to EGF receptor signaling, including EGF receptor itself (9), RAF-1 (9), p21-activated kinase 1 (44), IRS-1 (45), and insulin receptor substrate-2 (10), to regulate cell proliferation and apoptosis. A recent report showed that miR-7 functions as an anti-metastatic microRNA in gastric cancer by targeting IGFR1 (12). Our current study demonstrated that SET8 is an additional target of miR-7. Significantly, we showed that miR-7 suppressed EMT and invasion of MDA-MB-231 cells, consistent with our previous observation that SET8 and TWIST are functionally interdependent in promoting EMT and the invasive potential of breast cancer cells (22).
We demonstrated that miR-7 promoted spontaneous DNA damages and impedes DNA repair, extending its role in DNA damage response. Recent studies indicate that miR-421 and miR-182 are also implicated in DNA damage response and affect cellular sensitivity to DNA damaging agents by targeting ATM (46) or BRCA1 (47), respectively. It is reasonable to believe that miRNAs, through regulation of the enrichment of various DNA repair factors at the post-transcriptional level, add another layer to the sophisticated regulatory network of DNA repair system, ensuring the efficiency and accuracy of DNA repair.
H4K20 monomethylation has been associated with nucleosome decondensation and DNA damage foci accessible for recruitment of DNA repair proteins following DNA damage (15). Although other as-yet-unidentified substrates cannot be excluded, H4K20 monomethylation appears to be the major effector of SET8 and a crucial mediator of its impact on DNA repair. In support of this argument, it is noteworthy that embryonic stem cells lacking SET8 display massive DNA damage and γH2AX foci (16). In addition, SET8 is somewhat unique among histone methyltransferases in that its protein level oscillates during cell cycle progression. It is highly expressed during G2/M and early G1 and is absent during S phase (48, 49). Thus, tight regulation of SET8 appears to be essential for proper cell cycle progression. Recent studies showed that SET8 is also regulated at the transcriptional level (50, 51). Our observation that SET8 is targeted by miR-7 revealed a mechanism by which SET8 is regulated post-transcriptionally, underscoring the importance of SET8 regulation.
Finally, despite the progresses made in miRNA research, a comprehensive understanding of the interplay/cross-talk among their multiple targets is urgently needed. With the identification of a broad spectrum of target genes for miRNAs, it is difficult and improper to divide the miRNAs into oncogenes and tumor suppressors due to the diverse activities of target genes and various cellular contexts for a particular miRNA. This is probably also true for miR-7. Nevertheless, our experiments provide a molecular mechanism underlying the regulation of SET8 and extend the biological function of miR-7 to DNA damage response, supporting the pursuit of miR-7 as a potential target for breast cancer intervention.
This work was supported by Grants 81071673 (to L. S.) and 91219201 and 81130048 (to Y. S.) from the National Natural Science Foundation of China, Grants 5112017 and 5132020 (to L. S.) from Beijing Natural Science Foundation, and Grant 973 Program 2011CB504204 (to Y. S.) from the Ministry of Science and Technology of China.
- miRNA
- microRNA
- EMT
- epithelial-mesenchymal transition
- HR
- homologous recombination
- NHEJ
- nonhomologous end-joining
- qPCR
- quantitative PCR.
REFERENCES
- 1. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Behm-Ansmant I., Rehwinkel J., Izaurralde E. (2006) MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 71, 523–530 [DOI] [PubMed] [Google Scholar]
- 3. Iorio M. V., Croce C. M. (2009) MicroRNAs in cancer: small molecules with a huge impact. J. Clin. Oncol. 27, 5848–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kredo-Russo S., Ness A., Mandelbaum A. D., Walker M. D., Hornstein E. (2012) Regulation of pancreatic microRNA-7 expression. Exp. Diabetes Res. 2012, 695214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okuda H., Xing F., Pandey P. R., Sharma S., Watabe M., Pai S. K., Mo Y. Y., Iiizumi-Gairani M., Hirota S., Liu Y., Wu K., Pochampally R., Watabe K. (2013) miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 73, 1434–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kong X., Li G., Yuan Y., He Y., Wu X., Zhang W., Wu Z., Chen T., Wu W., Lobie P. E., Zhu T. (2012) MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PloS One 7, e41523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Skalsky R. L., Cullen B. R. (2011) Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PloS One 6, e24248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kong D., Piao Y. S., Yamashita S., Oshima H., Oguma K., Fushida S., Fujimura T., Minamoto T., Seno H., Yamada Y., Satou K., Ushijima T., Ishikawa T. O., Oshima M. (2012) Inflammation-induced repression of tumor suppressor miR-7 in gastric tumor cells. Oncogene 31, 3949–3960 [DOI] [PubMed] [Google Scholar]
- 9. Webster R. J., Giles K. M., Price K. J., Zhang P. M., Mattick J. S., Leedman P. J. (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J. Biol. Chem. 284, 5731–5741 [DOI] [PubMed] [Google Scholar]
- 10. Cochrane D. R., Cittelly D. M., Howe E. N., Spoelstra N. S., McKinsey E. L., LaPara K., Elias A., Yee D., Richer J. K. (2010) MicroRNAs link estrogen receptor α status and Dicer levels in breast cancer. Horm. Cancer 1, 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang N., Li X., Wu C. W., Dong Y., Cai M., Mok M. T., Wang H., Chen J., Ng S. S., Chen M., Sung J. J., Yu J. (2012) Oncogene, DOI 10.1038/onc.2012.526 [DOI] [PubMed] [Google Scholar]
- 12. Zhao X., Dou W., He L., Liang S., Tie J., Liu C., Li T., Lu Y., Mo P., Shi Y., Wu K., Nie Y., Fan D. (2013) MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 32, 1363–1372 [DOI] [PubMed] [Google Scholar]
- 13. Congdon L. M., Houston S. I., Veerappan C. S., Spektor T. M., Rice J. C. (2010) PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J. Cell. Biochem. 110, 609–619 [DOI] [PubMed] [Google Scholar]
- 14. Li Z., Nie F., Wang S., Li L. (2011) Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc. Natl. Acad. Sci. U.S.A. 108, 3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Houston S. I., McManus K. J., Adams M. M., Sims J. K., Carpenter P. B., Hendzel M. J., Rice J. C. (2008) Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J. Biol. Chem. 283, 19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oda H., Okamoto I., Murphy N., Chu J., Price S. M., Shen M. M., Torres-Padilla M. E., Heard E., Reinberg D. (2009) Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol. Cell. Biol. 29, 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jørgensen S., Elvers I., Trelle M. B., Menzel T., Eskildsen M., Jensen O. N., Helleday T., Helin K., Sørensen C. S. (2007) The histone methyltransferase SET8 is required for S-phase progression. J. Cell Biol. 179, 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Centore R. C., Havens C. G., Manning A. L., Li J. M., Flynn R. L., Tse A., Jin J., Dyson N. J., Walter J. C., Zou L. (2010) CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol. Cell 40, 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanders S. L., Portoso M., Mata J., Bähler J., Allshire R. C., Kouzarides T. (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119, 603–614 [DOI] [PubMed] [Google Scholar]
- 20. Oda H., Hübner M. R., Beck D. B., Vermeulen M., Hurwitz J., Spector D. L., Reinberg D. (2010) Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol. Cell 40, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi X., Kachirskaia I., Yamaguchi H., West L. E., Wen H., Wang E. W., Dutta S., Appella E., Gozani O. (2007) Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell 27, 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang F., Sun L., Li Q., Han X., Lei L., Zhang H., Shang Y. (2012) SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 31, 110–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lal A., Pan Y., Navarro F., Dykxhoorn D. M., Moreau L., Meire E., Bentwich Z., Lieberman J., Chowdhury D. (2009) miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 16, 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalinowski F. C., Giles K. M., Candy P. A., Ali A., Ganda C., Epis M. R., Webster R. J., Leedman P. J. (2012) Regulation of epidermal growth factor receptor signaling and erlotinib sensitivity in head and neck cancer cells by miR-7. PLoS One 7, e47067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 26. Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Solomon D. A., Cardoso M. C., Knudsen E. S. (2004) Dynamic targeting of the replication machinery to sites of DNA damage. J. Cell Biol. 166, 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schultz L. B., Chehab N. H., Malikzay A., Halazonetis T. D. (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hays S. L., Firmenich A. A., Berg P. (1995) Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc. Natl. Acad. Sci. U.S.A. 92, 6925–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muñoz M. C., Laulier C., Gunn A., Cheng A., Robbiani D. F., Nussenzweig A., Stark J. M. (2012) RING finger nuclear factor RNF168 is important for defects in homologous recombination caused by loss of the breast cancer susceptibility factor BRCA1. J. Biol. Chem. 287, 40618–40628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pierce A. J., Johnson R. D., Thompson L. H., Jasin M. (1999) XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Panning B., Jaenisch R. (1998) RNA and the epigenetic regulation of X chromosome inactivation. Cell 93, 305–308 [DOI] [PubMed] [Google Scholar]
- 33. Fedoriw A., Mugford J., Magnuson T. (2012) Genomic imprinting and epigenetic control of development. Cold Spring Harb. Perspect. Biol. 4, a008136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cantone I., Fisher A. G. (2013) Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 20, 282–289 [DOI] [PubMed] [Google Scholar]
- 35. Youn J. I., Kumar V., Collazo M., Nefedova Y., Condamine T., Cheng P., Villagra A., Antonia S., McCaffrey J. C., Fishman M., Sarnaik A., Horna P., Sotomayor E., Gabrilovich D. I. (2013) Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat. Immunol. 14, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park J. H., Park J., Choi J. K., Lyu J., Bae M. G., Lee Y. G., Bae J. B., Park D. Y., Yang H. K., Kim T. Y., Kim Y. J. (2011) Identification of DNA methylation changes associated with human gastric cancer. BMC Med. Genomics 4, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Varambally S., Cao Q., Mani R. S., Shankar S., Wang X., Ateeq B., Laxman B., Cao X., Jing X., Ramnarayanan K., Brenner J. C., Yu J., Kim J. H., Han B., Tan P., Kumar-Sinha C., Lonigro R. J., Palanisamy N., Maher C. A., Chinnaiyan A. M. (2008) Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322, 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu J., He M. L., Wang L., Chen Y., Liu X., Dong Q., Chen Y. C., Peng Y., Yao K. T., Kung H. F., Li X. P. (2011) MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 71, 225–233 [DOI] [PubMed] [Google Scholar]
- 39. Juan A. H., Kumar R. M., Marx J. G., Young R. A., Sartorelli V. (2009) Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell 36, 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun G., Ye P., Murai K., Lang M. F., Li S., Zhang H., Li W., Fu C., Yin J., Wang A., Ma X., Shi Y. (2011) miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Asangani I. A., Ateeq B., Cao Q., Dodson L., Pandhi M., Kunju L. P., Mehra R., Lonigro R. J., Siddiqui J., Palanisamy N., Wu Y. M., Cao X., Kim J. H., Zhao M., Qin Z. S., Iyer M. K., Maher C. A., Kumar-Sinha C., Varambally S., Chinnaiyan A. M. (2013) Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol. Cell 49, 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giovannetti E., van der Velde A., Funel N., Vasile E., Perrone V., Leon L. G., De Lio N., Avan A., Caponi S., Pollina L. E., Gallá V., Sudo H., Falcone A., Campani D., Boggi U., Peters G. J. (2012) High-throughput microRNA (miRNAs) arrays unravel the prognostic role of MiR-211 in pancreatic cancer. PLoS One 7, e49145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiong S., Zheng Y., Jiang P., Liu R., Liu X., Chu Y. (2011) MicroRNA-7 inhibits the growth of human non-small cell lung cancer A549 cells through targeting BCL-2. Int. J. Biol. Sci. 7, 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Reddy S. D., Ohshiro K., Rayala S. K., Kumar R. (2008) MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 68, 8195–8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rai K., Takigawa N., Ito S., Kashihara H., Ichihara E., Yasuda T., Shimizu K., Tanimoto M., Kiura K. (2011) Liposomal delivery of MicroRNA-7-expressing plasmid overcomes epidermal growth factor receptor tyrosine kinase inhibitor-resistance in lung cancer cells. Mol. Cancer Ther. 10, 1720–1727 [DOI] [PubMed] [Google Scholar]
- 46. Mansour W. Y., Bogdanova N. V., Kasten-Pisula U., Rieckmann T., Köcher S., Borgmann K., Baumann M., Krause M., Petersen C., Hu H., Gatti R. A., Dikomey E., Dörk T., Dahm-Daphi J. (2013) Aberrant overexpression of miR-421 downregulates ATM and leads to a pronounced DSB repair defect and clinical hypersensitivity in SKX squamous cell carcinoma. Radiother. Oncol. 106, 147–154 [DOI] [PubMed] [Google Scholar]
- 47. Moskwa P., Buffa F. M., Pan Y., Panchakshari R., Gottipati P., Muschel R. J., Beech J., Kulshrestha R., Abdelmohsen K., Weinstock D. M., Gorospe M., Harris A. L., Helleday T., Chowdhury D. (2011) miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 41, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Havens C. G., Shobnam N., Guarino E., Centore R. C., Zou L., Kearsey S. E., Walter J. C. (2012) Direct role for proliferating cell nuclear antigen in substrate recognition by the E3 ubiquitin ligase CRL4Cdt2. J. Biol. Chem. 287, 11410–11421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu S., Rice J. C. (2011) A new regulator of the cell cycle: the PR-Set7 histone methyltransferase. Cell Cycle 10, 68–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Driskell I., Oda H., Blanco S., Nascimento E., Humphreys P., Frye M. (2012) The histone methyltransferase Setd8 acts in concert with c-Myc and is required to maintain skin. EMBO J. 31, 616–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wakabayashi K., Okamura M., Tsutsumi S., Nishikawa N. S., Tanaka T., Sakakibara I., Kitakami J., Ihara S., Hashimoto Y., Hamakubo T., Kodama T., Aburatani H., Sakai J. (2009) The peroxisome proliferator-activated receptor γ/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell. Biol. 29, 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]