Background: Cytokinesis requires formation of the centralspindlin complex.
Results: The neck linker regions in ZEN-4 are highly mobile when free, but their mobility is greatly restricted by CYK-4 binding.
Conclusion: Central spindle assembly requires a binding event that modulates the structure of the kinesin ZEN-4.
Significance: Structural changes in the centralspindlin complex contribute to cell division.
Keywords: Cytokinesis, Electron Paramagnetic Resonance (EPR), Kinesin, Microtubules, Molecular Motors
Abstract
Centralspindlin is a critical regulator of cytokinesis in animal cells. It is a tetramer consisting of ZEN-4/MKLP1, a kinesin-6 motor, and CYK-4/MgcRacGAP, a Rho GTPase-activating protein. At anaphase, centralspindlin localizes to a narrow region of antiparallel microtubule overlap and initiates central spindle assembly. Central spindle assembly requires complex formation between ZEN-4 and CYK-4. However, the structural consequences of CYK-4 binding to ZEN-4 are unclear as are the mechanisms of microtubule bundling. Here we investigate whether CYK-4 binding induces a conformational change in ZEN-4. Characterization of the structure and conformational dynamics of the minimal interacting regions between ZEN-4 and CYK-4 by continuous wave EPR and double electron-electron resonance (DEER) spectroscopy reveals that CYK-4 binding dramatically stabilizes the relative positions of the neck linker regions of ZEN-4. Additionally, our data indicate that each neck linker is similarly structured in the bound and unbound states. CYK-4 binding decreases the rate of ZEN-4-mediated microtubule gliding. These results constrain models for the molecular organization of centralspindlin.
Introduction
Cytokinesis, the final event in the cell cycle, results in the generation of two identical daughter cells from a single parental cell. In animal cells, cytokinesis requires a structure called the central spindle (1). Assembly of the central spindle occurs at anaphase onset, when a set of antiparallel, nonkinetochore microtubules become bundled at their overlapping plus ends by microtubule-associated proteins and kinesin family motor proteins (1). Bundling of the antiparallel microtubules that comprise the central spindle requires centralspindlin, a heterotetrameric complex comprised of two proteins: a kinesin-6 family motor protein, ZEN-4/MKLP1, and a Rho GTPase-activating protein, CYK-4/MgcRacGAP (2). Centralspindlin specifically localizes to the narrow region of antiparallel microtubule overlap immediately following chromosome segregation at anaphase, where it initiates central spindle assembly (3). The central spindle can also dictate the site of cleavage furrow formation (4–9) and is required for proper completion of cytokinesis (10–13). Intriguingly, bundling of microtubules in vitro requires the presence of both ZEN-4/MKLP1 and CYK-4/MgcRacGAP and their ability to bind with high affinity (14). Similarly, the centralspindlin complex, but not the individual subunits alone, can support microtubule bundling and central spindle assembly in vivo (11–13).
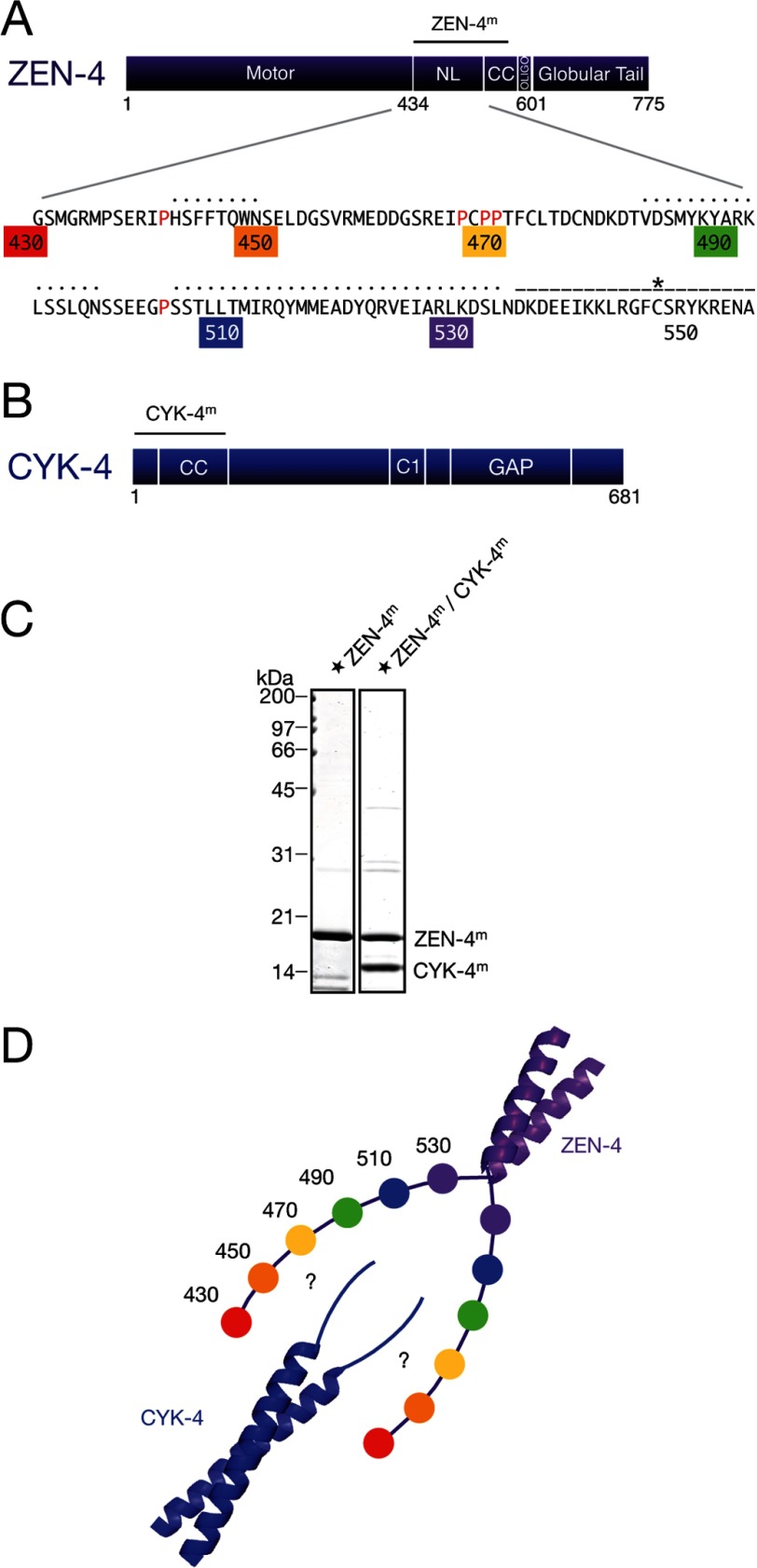
Caenorhabditis elegans ZEN-4 is comprised of an N-terminal motor domain, an atypically long neck linker region, a parallel coiled coil, and a C-terminal globular tail domain (Fig. 1A). C. elegans CYK-4 has a short, N-terminal region followed by a parallel coiled coil, a central C1 domain, and a C-terminal RhoGAP domain (Fig. 1B) (11). ZEN-4 and CYK-4 directly interact both in vitro and in vivo, forming a stoichiometric 2:2 tetramer (2). The binding interface between the two subunits of centralspindlin is formed by the N-terminal region of CYK-4 and a region of ∼100 amino acids in ZEN-4 positioned between the motor domain and the coiled coil (2) (Fig. 1, A and B). The coiled coils of CYK-4 and ZEN-4 contribute to the high affinity interaction, although they do not interact directly as they can be substituted by alternate parallel coiled coils (14).
FIGURE 1.
Design of ZEN-4m and CYK-4m. A, domain schematic of ZEN-4 highlighting the sequence of the neck linker and first ∼20 residues of the predicted coiled coil domain. Proline residues within the neck linker are highlighted in red. Residue numbers are colored for reference and indicate the regions of the neck linker where cysteine residues were introduced for spin labeling. Black dotted lines indicate the regions of the neck linker predicted to be α-helical by the PHYRE2 and PRALINE secondary structure prediction servers. Dashed line marks the residues predicted to form the coiled coil domain in ZEN-4m (NL, neck linker; CC, coiled coil; oligo, oligomerization). Asterisk marks cysteine 547, a native cysteine predicted to lie in the internal register (a or d) of the coiled coil. B, domain schematic of CYK-4. CC, coiled coil. C, purified ZEN-4m and centralspindlinm derivatives analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. Tandem affinity purification of ZEN-4m and CYK-4m reveals that they form a stoichiometric complex. Star indicates spin-labeled ZEN-4m. D, schematic of ZEN-4m and CYK-4m indicating possible interacting regions. Residue numbers are colored for reference and indicate the approximate positions of spin labels within the neck linker. This labeling scheme is used throughout to indicate the approximate position of the spin labels.
ZEN-4 is a member of the kinesin family of motor proteins. Kinesin motors use the energy derived from ATP hydrolysis to translocate along microtubules and execute a variety of intracellular functions. Kinesin-1, the most well characterized, prototypical kinesin, is a dimeric molecular motor that consists of an N-terminal catalytic domain, a short, unstructured neck linker region, a region that forms a parallel coiled coil, and a C-terminal globular tail domain (15). Kinesin-1 motility involves a hand-over-hand mechanism (16, 17), and kinesin-1 is highly processive, taking hundreds of precise, 8-nm steps toward the plus-end of a single microtubule before dissociating (18–20).
The neck linker region is a critical structural element for kinesin motility. The neck linker of kinesin-1 consists of ∼15 amino acids. This region undergoes nucleotide-dependent conformational changes that are critical for microtubule plus-end-directed motility (21, 22). This region is highly conserved in most N-terminal kinesins, and it makes intramolecular interactions with highly conserved residues in the motor domain (23).
The analogous region in ZEN-4 between the motor domain and the coiled coil is atypical compared with all other kinesins with N-terminal motor domains (2). In contrast to the neck linker of kinesin-1, the corresponding region in ZEN-4 is ∼100 residues long, and its structure and function are unclear. The linker region appears to lack the highly conserved residues that make direct contacts with the motor core, yet the corresponding residues in the motor core appear conserved (2, 23). Although the primary sequence of this region is not well conserved, it has a conserved role in binding to CYK-4 (2, 24). This region is predicted to be partially α-helical in diverse kinesin-6 family members, although it contains several proline-rich regions, and therefore it is unlikely to be entirely α-helical. For comparison, artificial extension of the neck linker of kinesin-1 alters the step-size and processivity of the motor and impairs its ability to couple the energy from ATP hydrolysis into forward steps (25). Importantly, CYK-4 binds ZEN-4 in the linker region, raising the possibility that CYK-4 binding may allosterically modify the structural and/or the biochemical properties of ZEN-4. Indeed, one attractive model is that the ZEN-4 linker is disordered unless bound by CYK-4.
Although the myriad functions of centralspindlin have been defined genetically and the molecular composition of centralspindlin has been characterized biochemically (2), the mechanistic details of how centralspindlin bundles antiparallel microtubules are not adequately understood. Using continuous wave electron paramagnetic resonance (CW-EPR)3 and double electron-electron resonance (DEER) spectroscopy methods, we demonstrate that CYK-4 binding results in a conformational change in the neck linker region of ZEN-4. These changes reduce the rate of microtubule gliding by ZEN-4, and they are likely important for antiparallel microtubule bundling in vivo. These structural changes place strong constraints on potential models for centralspindlin function in central spindle assembly.
EXPERIMENTAL PROCEDURES
Secondary Structure Prediction
Secondary structure predictions were performed with PHYRE2 (26) and PRALINE (27).
Protein Preparation
For EPR spectroscopy, C. elegans ZEN-4m (residues 430–555, molecular mass 17 kDa) variants were expressed in Escherichia coli BL-21 (DE3) RIL cells as N-terminal chitin binding domain (CBD) fusions. C. elegans CYK-4m (residues 1–120, molecular mass 14 kDa) was expressed in E. coli BL-21 (DE3) RIL cells as an N-terminal glutathione S-transferase (GST) fusion protein. Protein expression was induced by addition of 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. Both the ZEN-4m variants and CYK-4m were expressed at 25 °C for 4 h shaking at 275 rpm. Bacteria were lysed in 10 mm HEPES (pH 7.7), 1 mm EGTA (pH 8.0), 1 mm MgCl2, 0.1% Triton X-100, 250 mm NaCl, 0.1 mm ATP, 1 mm DTT, 10 μg/ml leupeptin/pepstatin, and 1 mm phenylmethylsulfonyl fluoride (PMSF), and containing 0.5 mg/ml lysozyme 30 min prior to sonication. Lysates were clarified by centrifugation at 20,000 rpm at 4 °C for 25 min in a JA-20 Beckman rotor. For all experiments, with the exception of Fig. 1C, CYK-4 and ZEN-4 were prepared as follows. Chitin beads (ZEN-4) (New England BioLabs) or glutathione-Sepharose beads (CYK-4) (BioWorld) were preequilibrated in lysis buffer without PMSF and lysozyme, added to the cleared lysates, and incubated for 4 h at 4 °C with mixing for binding. Following washes, ZEN-4 derivatives were incubated overnight at 4 °C with tobacco etch virus protease for removal of the N-terminal CBD tag, and the resulting proteins were spin-labeled for EPR or stored in aliquots at −80 °C. CYK-4m was eluted with buffer containing 10 mm reduced glutathione, cleaved with PreScission protease to remove the N-terminal GST tag, and further purified by Mono S cation exchange chromatography (GE Healthcare). The resulting protein was used immediately in experiments or stored in aliquots at −80 °C. In Fig. 1C, lysates expressing CBD-ZEN-4 and GST-CYK-4 were mixed and bound to chitin affinity resin, eluted with tobacco etch virus protease, bound to GST affinity resin, and eluted with PreScission protease.
For microtubule gliding assays, ZEN-4 (1–585, molecular mass 68 kDa) was expressed as an N-terminal CBD fusion, with a C-terminal flexible linker and biotin-acceptor sequence (28). Extraction and chromatography were performed as described for the ZEN-4m variants. Purified protein was stored in 10 mm HEPES (pH 7.7), 1 mm EGTA (pH 8.0), 1 mm MgCl2, 0.1% Triton X-100, 250 mm NaCl, 25 μm ATP, 1 mm DTT, and 10 μg/ml leupeptin/pepstatin in aliquots at −80 °C.
Spin Labeling of ZEN-4m Derivatives
ZEN-4m derivatives were exchanged into EPR buffer containing 10 mm HEPES (pH 7.7), 1 mm EGTA (pH 8.0), 1 mm MgCl2, 250 mm NaCl, and 10% glycerol using a PD-10 desalting column (GE Healthcare) or FPLC Fast Desalting column (GE Healthcare). ZEN-4m variants were spin-labeled with (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate spin label (MTSL, Toronto Research Chemicals) for ∼2 h at 4 °C. Immediately following labeling, excess spin label was removed using PD-10 desalting columns or a Superdex-200 gel filtration column (GE Healthcare) and further purified by Mono Q anion exchange chromatography (GE Healthcare).
Assembly of the Centralspindlinm Complex
To assemble centralspindlinm complexes, a 1.5–2-fold excess of CYK-4m was mixed with ZEN-4m and incubated for at least 4 h at 4 °C with mixing. The successful formation of stoichiometric complexes was verified by gel filtration using a Bio-Sil SEC 125 column (Bio-Rad), in which all the ZEN-4m was observed to shift to a larger molecular mass complex in the presence of CYK-4m. Dynamic light scattering confirmed that both ZEN-4m and purified centralspindlinm complex were homogeneous and that centralspindlinm was larger than ZEN-4m alone.
Disulfide Cross-linking
Recombinant ZEN-4m variants with single, unique cysteine residues at sites close to the predicted coiled coil were incubated in EPR buffer, in the presence or absence of CYK-4 and 20 mm oxidized glutathione. Reactions were incubated at room temperature for 40 min with mixing and analyzed by nonreducing SDS-PAGE followed by Coomassie Brilliant Blue staining.
EPR Spectroscopy
ZEN-4m and centralspindlinm derivatives were concentrated to >300 μm for EPR experiments using centrifugal concentrators (Millipore). Experiments were performed in EPR buffer: 10 mm HEPES (pH 7.7), 1 mm EGTA (pH 8.0), 1 mm MgCl2, 250 mm NaCl, and 10% glycerol.
CW-EPR
X-band CW-EPR spectroscopic measurements were performed at room temperature using a Bruker ELEXYS spectrometer (Bruker Biospin, Billerica, MA) equipped with a loop-gap resonator and a gas-permeable TPX capillary with the following parameters: 9.8-GHz frequency, 2-mW incident microwave power, 100-kHz modulation frequency, 1-G modulation amplitude, 41-ms conversion time, and 41-ms time constant.
DEER Spectroscopy
DEER measurements were carried out using a Bruker ELEXYS E580 X-band pulsed spectrometer operating near 9.7 GHz equipped with a split-ring MS2 resonator at a temperature of 55 K. Spin-labeled ZEN-4m and centralspindlinm derivatives were transferred to quartz 1.1 × 1.6 × 100-mm capillaries and flash-frozen to 55 K in the resonator at an estimated concentration of >300 μm in EPR buffer. All measurements were performed using a constant-time version of the four-pulse DEER sequence π/2(vobs) - τ1 - π(vobs) - t′ - π(vpump) - (τ1+ τ2 - t′) - π(vobs) - τ2 - echo (29), where time t′ is varied, using pulseSPEL program. The resonator was overcoupled to Q < 100–200, the pump frequency (vpump) was set to the center of the resonator dip and coincided with the maximum of the nitroxide EPR spectrum, while the observer frequency (vobs) was set to 65–70 MHz higher and coincided with the low field local maximum of the spectrum. The pulse lengths for π/2 and π were 16 ns and 32 ns, respectively, and the pump pulse length was 28–36 ns. In each case, the pump pulse and inversion pump lengths were optimized using nutation experiments. In all experiments, a τ1 of 200 ns was used. Data were typically recorded at steps of 12 ns. Accumulation times for the different datasets varied between 4 and 12 h. Data were processed and analyzed using the program DeerAnalysis2008 (30). A homogeneous three-dimensional background model was used to subtract intermolecular background from the raw data. The criterion for choosing background subtraction was the frequency-domain spectrum, where neither a positive spike nor an obvious hole is present in the center of the Pake pattern. Computation of the L-curve was performed to choose the optimal value for the Tikhonov regularization parameter, which corresponded to the corner of the L-curve.
Motility Assays
For microtubule gliding assays, recombinant ZEN-4(1–585) motors, biotinylated at the C terminus, alone or in complex with GST-CYK-4(1–120), were immobilized on a coverslip sequentially coated with biotin-BSA and NeutrAvidin (Molecular Probes). Microtubules were diluted into motility buffer containing 80 mm PIPES (pH 6.8), 1 mm EGTA (pH 8.0), 5 mm MgCl2, 6.25 mm glucose, 50 mm KCl, 2 mm ATP, 0.1 mg/ml BSA, 0.5 mg/ml casein, 1% β-mercaptoethanol, 5 μm paclitaxel (Sigma-Aldrich), 1× gloxy (31), and 0.5% glycerol, and flowed into the chamber. Images were acquired every 10 s for ∼10 min. To measure velocities as a function of motor density, flow chambers containing ZEN-4 alone were assayed. Subsequently, a 10-fold excess of GST-CYK-4m (or GST alone) in motility buffer was flowed into the chamber, and the sample was reassayed. Microtubule gliding velocities were quantified using the MTrackJ (32) plugin for ImageJ (33).
Circular Dichroism (CD) Spectroscopy
CD experiments were performed using an Aviv 202 CD spectrometer (Aviv Biomedical, Lakewood NJ) with a 0.1-cm path length. Spectra were collected from 190 to 260 nm at 25 °C at a protein concentration of 1–60 μm in EPR buffer. Data were fit using CONTINLL (34).
RESULTS
Biochemical Characterization of Centralspindlinm
We created a cysteine-less ZEN-4 construct, ZEN-4m, consisting of the neck linker, the minimal CYK-4 binding region, and the first 20 amino acids of the predicted coiled coil domain (amino acids 430–555, 17 kDa) (Fig. 1A). This mutant retained the ability to form a stoichiometric complex with cysteine-light CYK-4m (1–120, 14 kDa) comprising a coiled coil homodimer containing an N-terminal extension required for ZEN-4 binding (Fig. 1, B and C). Circular dichroism measurements indicate that ZEN-4m is ∼70% helical (data not shown). For clarity, we will refer to these truncation derivatives of ZEN-4 and CYK-4 as ZEN-4m and CYK-4m, and the complex as centralspindlinm (Fig. 1D) hereafter. ZEN-4m and centralspindlinm each migrates as a homogeneous species on a size exclusion column (data not shown).
Disulfide Cross-linking of ZEN-4m Variants in the Presence and Absence of CYK-4m
We first used a cross-linking approach to define more precisely the limits of the coiled coil region and to probe for potential conformational changes. For these experiments, we introduced single, unique cysteine residues at sites close to the predicted coiled coil in ZEN-4m (Fig. 1D). ZEN-4m derivatives containing single cysteine residues were assayed for disulfide bridge formation under oxidizing conditions followed by nonreducing gel electrophoresis to measure dimer formation in the presence and absence of CYK-4m. As a positive control, we assayed the cross-linking efficiency of cysteine 547, which is predicted to lie in the internal region of the parallel coiled coil region in ZEN-4 (position a or d) (14). As seen previously with ZEN-4(1–555), a cysteine at this position allows efficient formation of cross-linked dimers, both in the presence and absence of CYK-4m (Fig. 2). If the coiled coil continued with a heptad repeat, 533 would be predicted to lie in the same register as cysteine 547 (a or d) and cross-link with high efficiency. However, only a small fraction of the molecules form dimers when a cysteine residue is placed at position 533. Neighboring cysteines at residues 530, 531, 532, and 534 did not show any cross-linking in either the bound or unbound states (Fig. 2 and data not shown).
FIGURE 2.
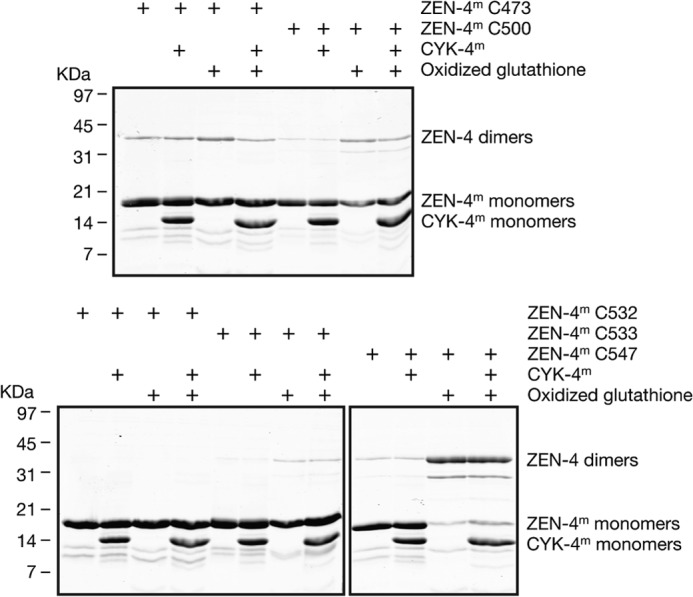
Disulfide cross-linking of ZEN-4m derivatives in the presence and absence of CYK-4m. Recombinant ZEN-4m fragments containing single cysteines engineered at the indicated sites were incubated under oxidizing conditions in the presence and absence of CYK-4m and assayed for disulfide bridge formation by nonreducing gel electrophoresis. As a positive control, we assayed the ability of ZEN-4m containing a cysteine at position 547 that is predicted to be in the a or d register of the coiled coil to form disulfide bridges under oxidizing conditions.
In addition to analyzing cross-linking efficiency at positions predicted to lie near the coiled coil, we also determined whether selected cysteines engineered at other residues throughout the neck linker region of ZEN-4m were sufficiently close in proximity (≤0.7 nm) (35) to cross-link efficiently in the presence and absence of CYK-4m. A fraction of ZEN-4m containing cysteines at 473 or 500 formed dimers with low efficiency in presence and absence of CYK-4m. We conclude from these data that the coiled coil domain in ZEN-4m likely begins at about residue 535 and that the coiled coil domain in ZEN-4m comprises ∼20 residues, which is consistent with the minimum number of residues required to form a stable coiled coil, reported to be ∼21–28 residues (3–4 heptad repeats) (36).
CYK-4m Binding Results in a Conformational Change That Stabilizes the Relative Positions of the Two Neck Linker Regions in ZEN-4m
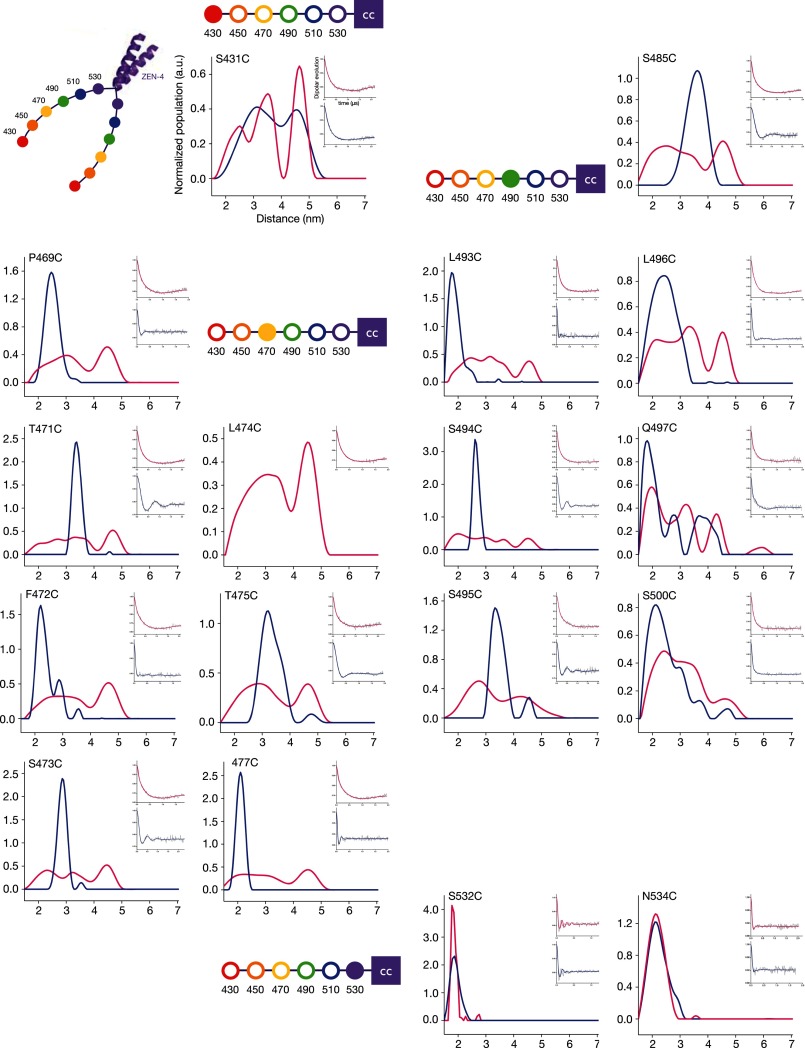
To determine more precisely whether CYK-4 binding induces conformational changes in the neck linker region of ZEN-4m, we measured distances between pairs of sites in the ZEN-4m dimer in the presence and absence of CYK-4m by DEER spectroscopy. DEER is a pulsed-EPR approach, a robust technique to measure distances and distance distributions of spin pairs to monitor the conformational dynamics of macromolecules (37–39). We introduced single, unique cysteine residues for spin labeling and performed DEER spectroscopy with specimens labeled at 17 sites throughout the linker region in ZEN-4m (Fig. 1D). The spectra are grouped according to the approximate position of the attached spin labels as indicated by the filled circles in the schematics adjacent to each set of spectra.
Spin labeling of positions 532 and 534 of ZEN-4m, near the beginning of the predicted coiled coil domain, resulted in a very sharp distance distribution in both the unbound and bound states, with single populations centered around 2 nm (Fig. 3). The separation between spin labels was slightly less at cysteine 532 compared with 534, with mean distances of 1.87 ± 0.17 nm and 2.19 ± 0.37 nm, respectively. The spectra from these two positions are quite similar and indicate that the structure of this region of ZEN-4m is rather rigid and not altered significantly upon CYK-4m binding.
FIGURE 3.
Distance measurements in the ZEN-4m dimer in the presence and absence of CYK-4m measured by DEER spectroscopy. DEER distance distribution profiles for ZEN-4m spin-labeled at the indicated positions in the bound (blue) and unbound (pink) states are shown. ZEN-4m schematics highlight the position in the neck linker nearest to the labeled cysteine for reference. Insets show the dipolar evolution curves for the bound and unbound states.
In contrast, labeling of the N terminus of the neck linker/minimal CYK-4m-interacting region, position 431, resulted in a very broad distance distribution (with three predominant populations centered at 2.46 ± 0.8 nm, 3.46 ± 0.55 nm, and 4.66 ± 0.44 nm) in the unbound state. This distribution transitioned to two predominant populations (at 3.17 ± 1.26 nm and 4.59 ± 0.79 nm) in the presence of CYK-4m (Fig. 3).
Most interestingly, we found that CYK-4m induces significant structural changes at 14 sites distributed throughout the linker region. In all cases, the unbound state exhibited broad distributions with multiple populations that transition into short and defined distances of ∼2–4.5 nm upon CYK-4m binding (Figs. 3 and 4). The most dramatic changes in the distance distributions upon CYK-4m binding were observed at positions 469, 471–473, 475, 477, 485, and 493–497 (Fig. 3). Here we observed two to three maxima in the distance distributions, possibly corresponding to two to three conformers in the unbound state and a single, predominant population (likely corresponding to a single conformer) in the bound state (Figs. 3 and 4). ZEN-4m labeled at L474C was unable to bind to CYK-4m, suggesting that this residue may lie in the binding interface between ZEN-4m and CYK-4m. Residues 481–497 and 506–532 are predicted to lie in regions with a conserved potential α-helical propensity (within the kinesin-6 family of motor proteins) (Fig. 1A). Analysis of residues in this region of ZEN-4m reveals some apparent periodicity in the distances measured at consecutive, or nearly consecutive, residues which is consistent with α-helical secondary structure (Fig. 1A).
FIGURE 4.
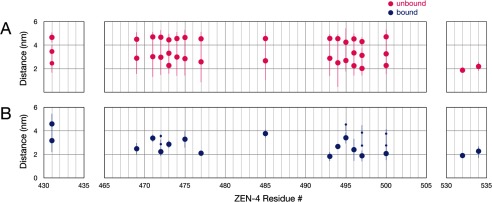
CYK-4m binding results in a conformational change that stabilizes the relative positions of the two neck linker regions in ZEN-4m. Mean distance measurements of individual populations of spin labels in the unbound (A) and bound (B) states are shown. The sizes of the dots reflect differences in the sizes of the individual populations plotted (refer to the distance distributions in Fig. 3).
In summary, the distance measurements determined by pulsed-EPR provide detailed structural insight into the conformational states of the neck linker regions in ZEN-4m in the presence and absence of CYK-4m. Except at the extreme termini of ZEN-4m, we observe very broad distance distributions in the unbound state and a very sharp distance distribution in the bound state, strongly suggesting that CYK-4m functions to stabilize the positions of the neck linker regions of ZEN-4m relative to each other, resulting in distances of 2–4.5 nm at all positions examined. Consistent with the cross-linking results, the distance measurements between spin labels positioned near the predicted coiled coil domain suggest that these residues are not in a coiled coil configuration.
CYK-4m Binding Alters the Local Environment of the Spin Probe at Specific Residues in the Neck Linker Region of ZEN-4m
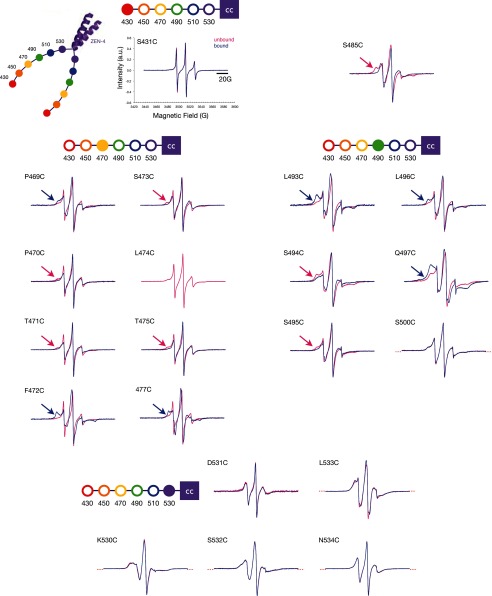
To gain insight into the local environment of the spin label and by extension, into the secondary structure of the neck linker region of ZEN-4m, we used CW-EPR. We found that the spectra from 16 of 21 sites distributed throughout the neck linker region of ZEN-4m are largely similar in the bound and unbound states (Fig. 5). CYK-4m binding does induce significant structural changes at a subset of residues. In particular, prominent decreases in the mobility of the spin label (measured by the inverse width of the central line resonance, ΔH0−1), were observed at 5/21 positions: 469, 472, 477, 493, and 497 (Fig. 6). Subtle changes are detected at 10 positions (Figs. 5 and 6). The latter changes largely indicate gain or loss of a small additional immobile fraction. In addition, spectra from positions 469, 472, 477, 493, 496, and 497 in ZEN-4m indicate significant immobile fractions of spins (at low magnetic field) in the bound state that are absent or significantly smaller in the spectra of ZEN-4m alone (Fig. 5, blue arrows). In contrast, the spectra from positions 470, 471, 473, 475, 485, 494, and 495 contain immobile fractions of spins in the unbound state that are reduced upon CYK-4m binding (Fig. 5, pink arrows). These differences in mobility are indicative of differences in the local secondary structural environment of the spin label involving a subpopulation of the spins in the bound and unbound states. Finally, two sites, 475 and 500, show significant increases in mobility, as measured by the mobility parameter (ΔH0−1), in the centralspindlinm complex compared with free ZEN-4m.
FIGURE 5.
CW-EPR spectra of ZEN-4m variants in the presence and absence of CYK-4m. CW-EPR spectra of ZEN-4m derivatives in the unbound (pink) and CYK-4m-bound (blue) states are shown. Flanking red dotted lines indicate spectra that overlay perfectly in the bound and unbound states. Blue arrows indicate immobile populations of spins present at low magnetic field in the bound state. Red arrows indicate immobile components present in the unbound state.
FIGURE 6.
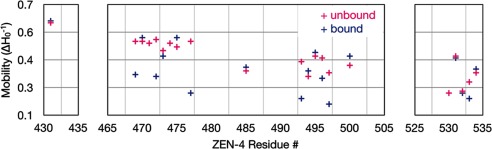
CYK-4m binding alters the mobility of the spin label at several positions in the ZEN-4m dimer. Mobility values (ΔH0−1) in the bound and unbound states were determined from the inverse width of the central line resonance.
The CW-EPR data reveal that the mobility of the spin label is largely similar in the bound and unbound states at numerous positions spread out over ∼100 residues in the linker region. These results indicate that although there are some local changes that occur upon complex formation, the linker is probably similarly structured in both the bound and unbound conformations. Comparison of the mobility changes (ΔH0−1) with the changes in distance between sites in the ZEN-4m dimer reveals that the sites that show the greatest reduction in mobility exhibit the greatest reduction in interlabel distances.
CYK-4 Alters the Motility Behavior of ZEN-4 in Vitro
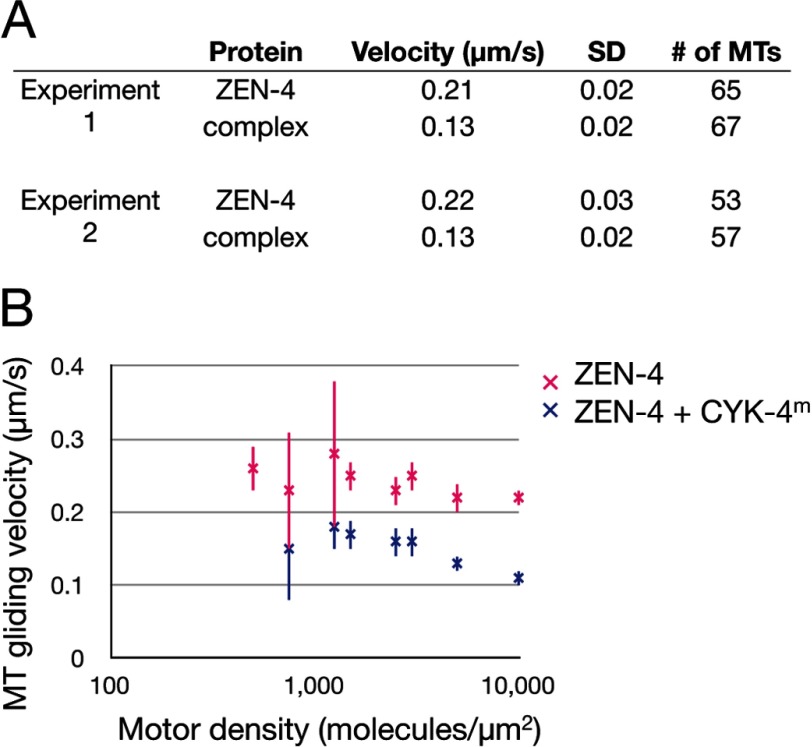
Because the minimal interacting region between CYK-4 and ZEN-4 lies within the neck linker region of ZEN-4 (2) and because conformational changes in the neck linker are important for the mechanism of motility of other N-kinesins, we hypothesized that CYK-4 binding may alter the microtubule gliding activity and/or processivity of ZEN-4. Therefore, we performed microtubule motility assays with dimeric ZEN-4(1–585) in the presence and absence of dimeric CYK-4(1–120). Analysis of the rates of microtubule gliding by ZEN-4 indicates that CYK-4 binding decreases the rate of motility of ZEN-4 by ∼40% (Fig. 7A). ZEN-4-mediated gliding requires a threshold of motor densities (>500 motors/μm2). The CYK-4-induced reduction in velocity was observed over a large range of motor concentrations, and no significant change in the threshold density for motility was observed (Fig. 7B). Additional analysis at the single-molecule level will be required to determine whether CYK-4 alters the run-length, step-size, dwell time, or processivity of dimeric ZEN-4.
FIGURE 7.
CYK-4m binding alters the motility behavior of dimeric ZEN-4. A, in vitro microtubule gliding assays indicating that CYK-4m binding alters the motility behavior of ZEN-4. The difference in gliding velocities is significant (t test, p ≪ 0.001). B, microtubule gliding assays at the indicated motor densities. ZEN-4 is a nonprocessive motor, and gliding is not observed below 500 motors/μm2. Error bars indicate S.D. MT, microtubule.
DISCUSSION
We investigated the molecular consequences of CYK-4 binding to ZEN-4, to gain insight into how centralspindlin executes its first of many functions in central spindle assembly: bundling of antiparallel microtubules. We sought to address whether CYK-4m binding induces a conformational change in the neck linker region of ZEN-4m which could result in some degree of ordering in the bound state (i.e. folding, positioning). These CYK-4-dependent structural changes could, by extension, alter the spatial arrangement of the two dimeric motor domains of ZEN-4 which could greatly impact the microtubule bundling activity and/or motility behavior of ZEN-4 in vivo. Using a structural and biochemical approach, we have more precisely defined the interaction between ZEN-4 and CYK-4, providing insight into the molecular mechanism by which centralspindlin bundles microtubules. The ability of EPR spectroscopy to detect multiple populations rather than bulk average was invaluable to observe the conformational variability of ZEN-4 and the CYK-4-ZEN-4 complex.
The N-terminal region of CYK-4 binds the neck linker region in ZEN-4 that connects the motor domain to the coiled coil (2). The interaction between ZEN-4 and CYK-4 is conserved; however, the sequences that mediate their interaction are not highly conserved. In addition, the ∼100-residue neck linker in ZEN-4 is structurally distinct compared with the corresponding 15-residue region in all other plus-end-directed kinesins, the functional consequences of which are unclear.
Models for the Molecular Organization of Centralspindlin
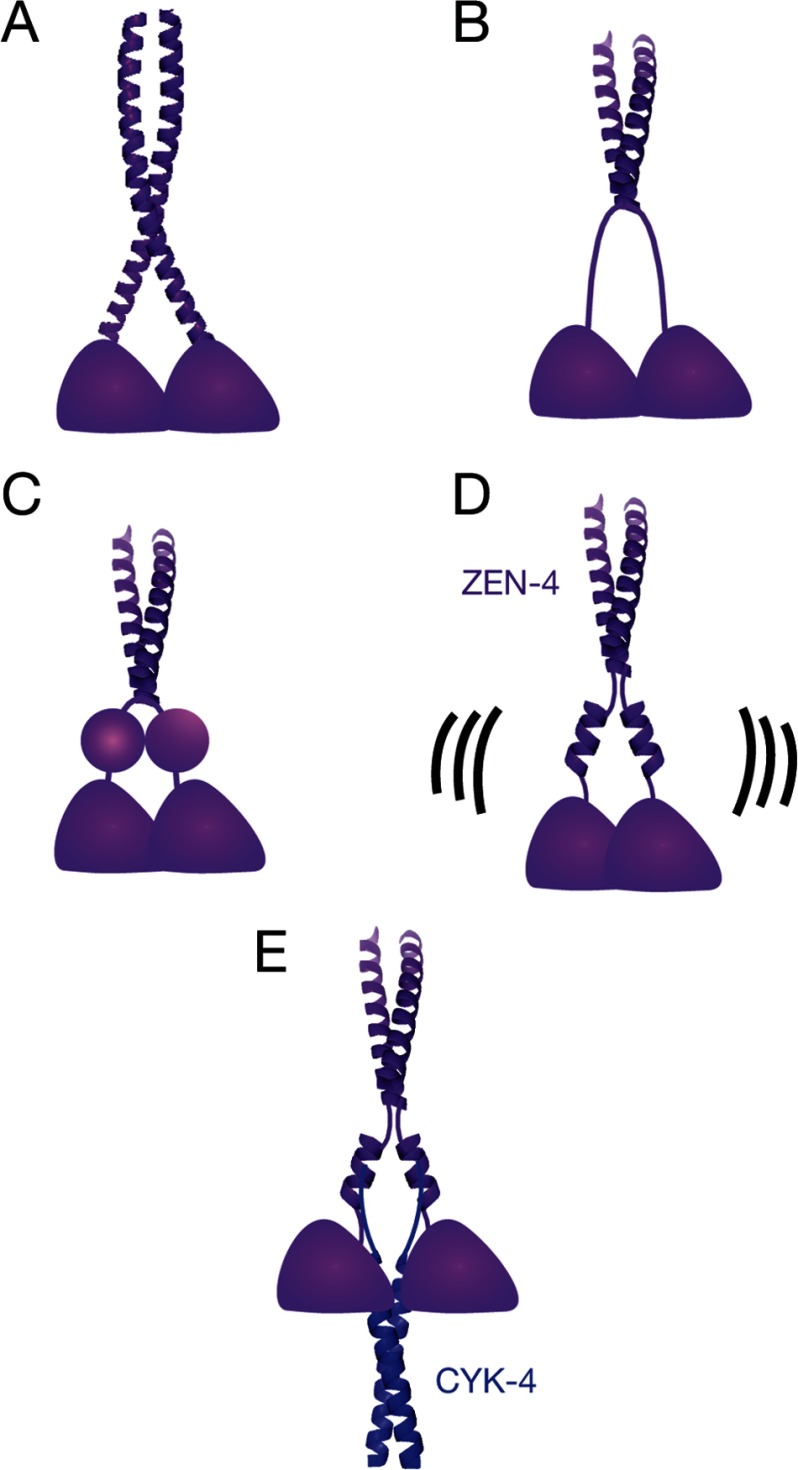
There are a number of possible arrangements of the ZEN-4 linker region: (i) random coil; (ii) extended coiled coil or continuous α-helices in a non-coiled coil configuration; (iii) small, folded domain; and (iv) nonassociated α-helices interrupted by several intervening unstructured/loop regions. Primary and secondary structure analyses suggest that the extended coiled coil or extended α-helices models are unlikely (Fig. 8A). The presence of multiple proline residues in the neck linker is inconsistent with either model. The distance measurements provide further evidence against an extended coiled coil because that model would predict distances that lie in the range from 0.5 to 2 nm. We do not observe any residues that are less than 1 nm, and we observe many that are significantly greater than 2 nm. Finally, the high degree of mobility of the spin label, analyzed in detail by CW-EPR at some of the 21 positions distributed throughout the neck linker, are inconsistent with a stable structural element like a coiled coil. In contrast, positions 530–534, near the coiled coil, provide clear examples of reduced mobility.
FIGURE 8.
Models for the molecular organization of centralspindlin. A, extended coiled coil or continuous α-helices in a non-coiled coil configuration. B, random coil. C, globular domain. D and E, interrupted α-helices in the unbound (D) and bound (E) states. The helices may be interrupted at several places. Structures are modeled from the crystal structures of GCN4 (Protein Data Bank ID code 1ZIK) (42) and AmiC-AmiR (Protein Data Bank ID code 1QO0) (43).
As an alternative, we considered the possibility that the neck linkers of ZEN-4m form extended, random coils (Fig. 8B). In this model, the mean-squared intradimer distances in the ZEN-4m linker should increase as a function of (2n) (40), where n is the number of residues N-terminal to the beginning of the coiled coil. However, our DEER results do not reveal an increase in distance as the spin label is moved away from the coiled coil toward the N terminus of the neck linker region, suggesting that the neck linker regions of ZEN-4m are not unfolded, random coils. There is also little evidence from the CW-EPR spectra that CYK-4m binding induces a random coil-to-folded transition in ZEN-4m.
The neck linkers could also form small, folded domains (Fig. 8C). For example, the 50-residue SH3 domain is ∼2.3 nm in diameter (41). In ZEN-4m, the positions of these domains could be variable, yet fixed upon CYK-4m binding. There are a limited number of possibilities in which these domains could be arranged such that pairs of corresponding residues distributed throughout the domain would all be between 2 and 5 nm. If they were rotationally symmetric, intradimer distances (d) would range from x < d < x + 4.6 nm, where x is the distance between the domains. We did not find distances that differ by this magnitude; however, if the neck linkers are translationally symmetric, distances between all positions would be ∼x, which could be consistent with the distance measurements.
A fourth model, which we favor, postulates that the neck linker regions may form interrupted α-helices, likely discontinuous at multiple sites, in the presence and absence of CYK-4m and that CYK-4m binding dramatically alters the positioning of the neck linkers, stabilizing a particular conformation (Fig. 8, D and E). Indeed, the intradimer distances at each of 13 residues distributed over a stretch of 31 amino acids changes from a wide distribution of distances, suggesting intradimer mobility, to a single, narrow distance distribution, suggesting a much more constrained conformation. This model is consistent with the high degree of helicity, the distance measurements within the ZEN-4m dimer, the mobility of the spin label at positions throughout the entire neck linker region, and the presence of multiple proline residues within the neck linker.
Structural Insights into the Binding Interface between ZEN-4 and CYK-4
The linker domains of ZEN-4m appear to be similarly structured in the bound and unbound states. In the bound state, the fixed intradimer distances at nearly all positions suggest that intact centralspindlinm has a well defined structure. The CW-EPR spectra of at individual residues distributed in most of the 100 residues throughout the linker region of ZEN-4m are similar in the bound and unbound states, suggesting that the local environment of most residues in the unbound state is similar to that of the bound state. Thus, if the linker region of ZEN-4m is structured when bound to CYK-4m, it must also be similarly structured when free, albeit with variable distances between the dimers.
A subset of residues does exhibit significant changes in their local environment. Residues 469, 472, 477, 493, and 497 undergo significant decreases in probe mobility (ΔH0−1) upon CYK-4m binding. These large reductions in mobility suggest that these residues may lie near the binding interface between ZEN-4m and CYK-4m. Consistent with this interpretation, we were not able to measure distances in the bound state at the nearby residue 474 because the spin-labeled variant was not able to form a complex, suggesting that this residue interacts directly with CYK-4m. The fact that we were able to form complexes with species labeled at many other positions indicates either a high degree of plasticity within the binding interface between ZEN-4m and CYK-4m such that the substitution of the native amino acids to MTSL-cysteine are tolerated. Alternatively, these residues may not form critical contacts with CYK-4m.
Importantly, the residues in ZEN-4m that exhibit the greatest attenuation of mobility upon CYK-4m binding, likely due to direct binding, correlate well with those that undergo the greatest changes in distance. In the bound state, these sites exhibit the smallest intradimer distances of all the sites we measured. In light of this correlation, we speculate that CYK-4m binds to the inner face of the ZEN-4m dimer.
Two sites, 475 and 500, show significant increases in mobility (ΔH0−1) in the centralspindlinm complex compared with free ZEN-4m. Positions 470, 471, 475, 485, 494, and 495 all have smaller, albeit reproducible, immobile spectral components in the unbound state that are not present upon CYK-4m binding, which are also indicative of increased mobility in the centralspindlinm complex compared with free ZEN-4m. Residues 493–497 lie in a predicted α-helix that extends from ∼481 to 497. The sites that show a clear increase in mobility in the bound state are ± 1–3 residues from the sites where the mobility (ΔH0−1) decreases the most upon binding. The residues where the spin label exhibits greater mobility in the bound state may be positioned on a different face of an α-helix from the residues that are stabilized (less mobile) upon binding. The residues that show increased mobility upon CYK-4m binding could participate in an intramolecular interaction that is mutually exclusive with CYK-4 binding. Consistent with this idea, genome-wide searches for suppressors of temperature-sensitive mutations in CYK-4 and ZEN-4 have suggested the possibility of autoinhibition (2, 14), due to the identification of single ZEN-4 mutations that rescue mutations in both CYK-4 and ZEN-4.
In summary, we have shown that CYK-4m binding induces substantial conformational changes in ZEN-4m. Additionally, we demonstrate that the consequence of these conformational changes is a dramatic stabilization of the relative positions of the dimeric neck linker regions of ZEN-4m. Finally, we show that the neck linker regions of ZEN-4m are partially structured and dynamic (exist in multiple conformations) in the unbound state, and CYK-4m binding stabilizes one particular conformation that may be important for antiparallel microtubule bundling in vivo. We hypothesize that aspects of neck linker function are retained in this motor protein. In particular, we note that the residues of the kinesin core that interact with the conserved neck linker region (23) are also conserved in ZEN-4 and its orthologs, despite its divergent neck linker. We speculate that part of the extended ZEN-4 neck linker region will dock with the motor core as in conventional kinesins. CYK-4 binding to the neck linker could impact the spatial arrangement of the motor domains of ZEN-4 and therefore the mechanism of microtubule binding by centralspindlin, which could underlie the observed changes in motility rates.
Footnotes
- CW-EPR
- continuous wave EPR
- CBD
- chitin binding domain
- DEER
- double electron-electron resonance.
REFERENCES
- 1. Glotzer M. (2009) The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mishima M., Kaitna S., Glotzer M. (2002) Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell 2, 41–54 [DOI] [PubMed] [Google Scholar]
- 3. Mishima M., Pavicic V., Grüneberg U., Nigg E. A., Glotzer M. (2004) Cell cycle regulation of central spindle assembly. Nature 430, 908–913 [DOI] [PubMed] [Google Scholar]
- 4. Cao L. G., Wang Y. L. (1996) Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol. Biol. Cell 7, 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eckley D. M., Ainsztein A. M., Mackay A. M., Goldberg I. G., Earnshaw W. C. (1997) Chromosomal proteins and cytokinesis: patterns of cleavage furrow formation and inner centromere protein positioning in mitotic heterokaryons and mid-anaphase Cells. J. Cell Biol. 136, 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adams R. R., Wheatley S. P., Gouldsworthy A. M., Kandels-Lewis S. E., Carmena M., Smythe C., Gerloff D. L., Earnshaw W. C. (2000) INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 10, 1075–1078 [DOI] [PubMed] [Google Scholar]
- 7. Bonaccorsi S., Giansanti M. G., Gatti M. (1998) Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J. Cell Biol. 142, 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Giansanti M. G., Bonaccorsi S., Williams B., Williams E. V., Santolamazza C., Goldberg M. L., Gatti M. (1998) Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 12, 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alsop G. B., Zhang D. (2003) Microtubules are the only structural constituent of the spindle apparatus required for induction of cell cleavage. J. Cell Biol. 162, 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wheatley S. P., Wang Y. (1996) Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 135, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jantsch-Plunger V., Gönczy P., Romano A., Schnabel H., Hamill D., Schnabel R., Hyman A. A., Glotzer M. (2000) CYK-4: a Rho family GTPase-activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149, 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Powers J., Bossinger O., Rose D., Strome S., Saxton W. (1998) A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 8, 1133–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raich W. B., Moran A. N., Rothman J. H., Hardin J. (1998) Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell 9, 2037–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pavicic-Kaltenbrunner V., Mishima M., Glotzer M. (2007) Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol. Biol. Cell 18, 4992–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozielski F., Sack S., Marx A., Thormählen M., Schönbrunn E., Biou V., Thompson A., Mandelkow E. M., Mandelkow E. (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell 91, 985–994 [DOI] [PubMed] [Google Scholar]
- 16. Asbury C. L., Fehr A. N., Block S. M. (2003) Kinesin moves by an asymmetric hand-over-hand mechanism. Science 302, 2130–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yildiz A., Tomishige M., Vale R. D., Selvin P. R. (2004) Kinesin walks hand-over-hand. Science 303, 676–678 [DOI] [PubMed] [Google Scholar]
- 18. Vale R. D., Funatsu T., Pierce D. W., Romberg L., Harada Y., Yanagida T. (1996) Direct observation of single kinesin molecules moving along microtubules. Nature 380, 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mori T., Vale R. D., Tomishige M. (2007) How kinesin waits between steps. Nature 450, 750–754 [DOI] [PubMed] [Google Scholar]
- 20. Toprak E., Yildiz A., Hoffman M. T., Rosenfeld S. S., Selvin P. R. (2009) Why kinesin is so processive. Proc. Natl. Acad. Sci. U.S.A. 106, 12717–12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rice S., Lin A. W., Safer D., Hart C. L., Naber N., Carragher B. O., Cain S. M., Pechatnikova E., Wilson-Kubalek E. M., Whittaker M., Pate E., Cooke R., Taylor E. W., Milligan R. A., Vale R. D. (1999) A structural change in the kinesin motor protein that drives motility. Nature 402, 778–784 [DOI] [PubMed] [Google Scholar]
- 22. Case R. B., Rice S., Hart C. L., Ly B., Vale R. D. (2000) Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol. 10, 157–160 [DOI] [PubMed] [Google Scholar]
- 23. Wade R. H., Kozielski F. (2000) Structural links to kinesin directionality and movement. Nat. Struct. Biol. 7, 456–460 [DOI] [PubMed] [Google Scholar]
- 24. Somers W. G., Saint R. (2003) A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell 4, 29–39 [DOI] [PubMed] [Google Scholar]
- 25. Yildiz A., Tomishige M., Gennerich A., Vale R. D. (2008) Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell 134, 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 27. Simossis V. A., Heringa J. (2003) The PRALINE online server: optimising progressive multiple alignment on the web. Comput. Biol. Chem. 27, 511–519 [DOI] [PubMed] [Google Scholar]
- 28. Beckett D., Kovaleva E., Schatz P. J. (1999) A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pannier M., Veit S., Godt A., Jeschke G., Spiess H. W. (2000) Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 142, 331–340 [DOI] [PubMed] [Google Scholar]
- 30. Jeschke G., Zimmermann H., Godt A. (2006) Isotope selection in distance measurements between nitroxides. J. Magn. Reson. 180, 137–146 [DOI] [PubMed] [Google Scholar]
- 31. Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 [DOI] [PubMed] [Google Scholar]
- 32. Meijering E., Dzyubachyk O., Smal I. (2012) Methods for cell and particle tracking. Methods Enzymol. 504, 183–200 [DOI] [PubMed] [Google Scholar]
- 33. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Provencher S. W., Glöckner J. (1981) Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 35. Falke J. J., Dernburg A. F., Sternberg D. A., Zalkin N., Milligan D. L., Koshland D. E. (1988) Structure of a bacterial sensory receptor: a site-directed sulfhydryl study. J. Biol. Chem. 263, 14850–14858 [PubMed] [Google Scholar]
- 36. Fairman R., Chao H. G., Mueller L., Lavoie T. B., Shen L., Novotny J., Matsueda G. R. (1995) Characterization of a new four-chain coiled-coil: influence of chain length on stability. Protein Sci. 4, 1457–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., Jung H. (2006) DeerAnalysis2006: a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30, 473–498 [Google Scholar]
- 38. Torbeev V. Y., Raghuraman H., Hamelberg D., Tonelli M., Westler W. M., Perozo E., Kent S. B. (2011) Protein conformational dynamics in the mechanism of HIV-1 protease catalysis. Proc. Natl. Acad. Sci. U.S.A. 108, 20982–20987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jeschke G. (2012) DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 63, 419–446 [DOI] [PubMed] [Google Scholar]
- 40. Fitzkee N. C., Rose G. D. (2004) Reassessing random-coil statistics in unfolded proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12497–12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Musacchio A., Noble M., Pauptit R., Wierenga R., Saraste M. (1992) Crystal structure of a Src-homology 3 (SH3) domain. Nature 359, 851–855 [DOI] [PubMed] [Google Scholar]
- 42. Gonzalez L., Jr., Woolfson D. N., Alber T. (1996) Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat. Struct. Biol. 3, 1011–1018 [DOI] [PubMed] [Google Scholar]
- 43. O'Hara B. P., Norman R. A., Wan P. T., Roe S. M., Barrett T. E., Drew R. E., Pearl L. H. (1999) Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 18, 5175–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]