Background: The peripheral light-harvesting complexes of photosystem II (LHCII) switch between light energy capturing and dissipative states.
Results: An NMR analysis is presented of the arginines in LHCII prepared in its two states.
Conclusion: The protein core of arginine-glutamate interlocked helices is preserved, whereas moderate changes occur in the stromal loop region.
Significance: This work reveals consequences for the molecular switching of LHCII.
Keywords: Bioenergetics, Photosynthesis, Photosystem II, Protein Conformation, Solid-state NMR, LHC Family, Non-photochemical Quenching, Photosynthetic Light Harvesting
Abstract
Light-harvesting antennae of the LHC family form transmembrane three-helix bundles of which two helices are interlocked by conserved arginine-glutamate (Arg-Glu) ion pairs that form ligation sites for chlorophylls. The antenna proteins of photosystem II have an intriguing dual function. In excess light, they can switch their conformation from a light-harvesting into a photoprotective state, in which the excess and harmful excitation energies are safely dissipated as heat. Here we applied magic angle spinning NMR and selective Arg isotope enrichment as a noninvasive method to analyze the Arg structures of the major light-harvesting complex II (LHCII). The conformations of the Arg residues that interlock helix A and B appear to be preserved in the light-harvesting and photoprotective state. Several Arg residues have very downfield-shifted proton NMR responses, indicating that they stabilize the complex by strong hydrogen bonds. For the Arg Cα chemical shifts, differences are observed between LHCII in the active, light-harvesting and in the photoprotective, quenched state. These differences are attributed to a conformational change of the Arg residue in the stromal loop region. We conclude that the interlocked helices of LHCII form a rigid core. Consequently, the LHCII conformational switch does not involve changes in A/B helix tilting but likely involves rearrangements of the loops and helical segments close to the stromal and lumenal ends.
Introduction
The elementary step in photosynthesis is the capture of solar energy by the light-harvesting antenna. In eukaryotic organisms, this step is performed by a family of pigment-binding proteins called light-harvesting complexes (LHCs)2 that absorb sunlight and transfer the excitation energy toward the reaction centers, where charge separation takes place (1). The Lhcb antenna proteins of higher plants and moss and Lhcbm antenna proteins of LHCs in green algae contain three membrane-spanning helices A, B, and C, and their tertiary structure is typified by two crossing A/B helices interconnected by Arg-Glu ion pairs that form ligation sites for Chls.
The most abundant LHC in plants and green algae is the major light-harvesting complex II (LHCII), which captures about 50% of all land-bound chlorophylls. The LHCII complex is trimeric, and each monomer forms a scaffold for eight Chla and six Chlb molecules, two luteins (Lut), one neoxanthin, and one violaxanthin, which is reversibly replaced by zeaxanthin upon de-epoxidation during the xanthophyll cycle (2–4).
In addition to their light-harvesting function, LHCII complexes are involved in several regulatory mechanisms that balance the incoming excitation energies and prevent photodamage (1, 4–6). Under high sunlight conditions, the photosynthetic antenna can rapidly switch from light-harvesting into a photoprotective state in which excess light energy is safely dissipated as heat (7). This photoprotective mechanism is called non-photochemical quenching (NPQ) and protects oxygenic organisms against photooxidative damage. The major component of NPQ, qE, depends on the transmembrane proton gradient ΔpH. In plants, a decrease in lumenal pH triggers protonation of PsbS (8, 9) and of LHCs in photosystem II (10) and activates the conversion of the LHC-bound xanthophyll cycle carotenoid from violaxanthin to zeaxanthin (11). In green algae, PsbS is absent, and the NPQ state is triggered by LhcSR, a pigment-binding complex with a short fluorescence lifetime that senses the decrease in lumenal pH (12–14). In algae, LHCII also participates in quenching, in particular the Lhcbm1 component (15, 16). On a supramolecular scale, a reorganization of the thylakoid membrane takes place in which the LHCII complexes dissociate from photosystem II and self-associate or associate with PsbS (17–20). The supramolecular rearrangements, xanthophyll exchange, and decrease in lumenal pH were proposed to promote subtle conformational changes inside the photosystem II light-harvesting proteins, by which altered chromophore configurations create a dissipation channel for the incoming light energy (5).
To date, the molecular basis for the photophysical process of NPQ is under much debate (22–28), and it is unclear how the LHCs respond to environmental changes and energy dissipative channels are formed. The LHCII complexes of plants have been studied extensively and were shown to reversibly switch their conformation between active, unquenched and photoprotective quenched conformational states (24, 29, 30). In the photoprotective state, Chl excitations in LHCII are quenched by Chl-Lut energy transfer (24) or by low lying Chl-Lut excitonic states (23), whereas Chl-Chl charge transfer states have also been proposed (25).
There is a controversy whether the LHCII x-ray structures represent the active or quenched form of the protein (22, 31, 32). LHCII complexes reconstituted in lipid nanodiscs retained their fluorescent state but showed small spectral changes when compared with LHCII in detergent micelles (33), suggesting that the protein can adopt slightly different conformations within its active state. The quenched state is associated with a twist in the configuration of the LHCII-bound neoxanthin that may promote conformational changes at the Lut L1 site (24). Protonation of specific acidic residues under low lumenal pH conditions could trigger conformational changes of the short helix D and the BC loop (2), and these residues are important for stabilizing the complex at different pH values (34). Under NPQ conditions, changes in the thickness of the thylakoid membrane have been observed, and it was proposed that LHCII adopts a more condensed structure (35).
In this work, we applied MAS-NMR techniques in combination with selective Arg isotope labeling as a noninvasive method to obtain high-resolution structural information of LHCII in its active and photoprotective states. Photosynthetic light-harvesting proteins are accessible for NMR via uniform or selective isotope enrichment of the photosynthetic organisms (36). This way, pigment-protein interactions could be detected in atomic detail inside intact purple bacterial light-harvesting oligomers (37–39). More recently, we performed a MAS-NMR analysis of LHCII from uniformly 13C-enriched Chlamydomonas reinhardtii green algae cells (40, 41). The C. reinhardtii Lhcbm sequences have a high similarity with Lhcb sequences of higher plants (40), making C. reinhardtii LHCII a suitable in vitro model system for the structural flexibility of plant Lhcb and algae Lhcbm proteins.
Here we analyze LHCII complexes from an Arg-auxotrophic strain of C. reinhardtii that was supplied with 13C6-15N4 Arg. One-dimensional 13C and 15N CP-MAS and two-dimensional 1H-13C dipolar heteronuclear correlation (HetCor) Arg NMR spectra are presented of LHCII in quenched and unquenched states. The Arg residues that stabilize the interlocked helix pair in LHCII are identified because of Chl ring current-induced shifts of their NMR responses. We show that these protein sites are preserved in the photoprotective state, confirming a structural view in which the flexible loop regions of the LHCII polypeptides are complemented by a relatively rigid scaffolding of the protein interior (42).
EXPERIMENTAL PROCEDURES
Isotope Enrichment and Purification of LHCII Trimers
C. reinhardtii strain cc424, an Arg auxotrophic strain obtained commercially from the Chlamydomonas Connection, was grown in liquid TAP (Tris-acetate-phosphate) medium (43) with 50 mg/liter Arg used (44). Cells were cultured at room temperature under continuous illumination at 60 microeinsteins of flux. Arginine was added separately to the cell cultures, and for selective Arg enrichment, it was substituted by [13C6,15N4]Arg purchased from Silantes GmbH. The cells were harvested in mid-log phase by centrifugation (4000 rpm, 6 min, 4 °C), and thylakoids were prepared as described in Ref. 45 with a few modifications described in Ref. 46. Thylakoids were separated from other materials on a discontinuous gradient (24,000 rpm, 1 h, 4 °C) in a TST-41.14 swing-out rotor. Thylakoid membranes were washed two times, first with 10 mm HEPES, pH 7.5, and 5 mm EDTA and finally with 10 mm HEPES, pH 7.5. Thylakoids were resuspended in solubilization buffer (10 mm HEPES, pH 7.5) to a Chl concentration of 1 mg/ml, and equal amounts of β-dodecylmaltoside (β-DM) were added to get a final concentration of 0.6% β-DM. The suspension was vortexed a few seconds and centrifuged (15,000 rpm, 10 min, 4 °C) to remove unsolubilized material, and the supernatant was loaded on a sucrose density gradient, prepared by 0.65 m sucrose, 10 mm Tricine, pH 7.8, 0.03% β-DM and ultracentrifuged for 17 h at 37,000 rpm. The top band of the sucrose gradient contained LHCII trimer complexes with more than 90% purity, verified by FPLC.
For obtaining LHCII in its quenched state, isolated LHCII trimers in 0.03% β-DM were dialyzed for 72 h against detergent-free buffer. The quenched state of the detergent-depleted LHCII aggregates was verified by low temperature fluorescence spectroscopy, whereas the unquenched, light-harvesting state of the LHCII trimers in β-DM was verified by time-resolved fluorescence spectroscopy.
Fluorescence Experiments
Steady-state fluorescence excitation and emission spectra were measured with a commercial spectrophotometer (HORIBA Scientific, FluoroLog). For 77 K fluorescence emission measurements, the samples were diluted in 40% HEPES/β-DM buffer and 60% glycerol (v/v) and cooled in a nitrogen-bath cryostat to 77 K.
Time-resolved fluorescence emission measurements were performed at room temperature with a Streak camera setup. The sample was measured front-face using a 1-mm quartz cuvette. To minimize the effects of photodamage, about 10 spectra of maximum 1 min per scan were acquired. It was verified that within this time period of illumination, no degradation of sample occurred. Excitation pulses of 400 nm (∼100 fs) with vertical polarization were generated using a titanium:sapphire laser (Coherent Vitesse) with a regenerative amplifier (Coherent, MIRA seed, and RegA) that was used to pump an optical parametric amplifier (Coherent, OPA). The repetition rate was 50 kHz with pulse energies of ∼0.2 nJ. A 10-fold increase or 2-fold decrease of the excitation pulse energy did not affect the fluorescence lifetimes, confirming that experiments were performed in the annihilation-free regime. The obtained streak data were analyzed with Glotaran3 (47).
Solid-state NMR Experiments
One-dimensional 13C and 15N and two-dimensional 1H-13C frequency-switched Lee-Goldburg heteronuclear correlation experiments were performed with a Bruker AV-750 spectrometer equipped with a 4-mm triple resonance MAS probe head, using a 13C radio frequency of 188.6 MHz. The temperature was lowered to 220–240 K under slow spinning of the sample. For the NMR experiments, spinning frequencies of 13 kHz were used. The chemical shift scale was calibrated from an FSLG spectrum of solid tyrosine HCl salt.
Density Functional Theory Calculations
Density functional theory (DFT) calculations were performed in vacuum within the DFT framework and using the Gaussian 03 package (48). The BLYP exchange correlation function (49–51) was applied to produce the NMR chemical shifts (48, 52). The geometric arrangements of Arg-70, Glu-180, and Chl610 were extracted from the 2BHW crystallographic structure of pea LHCII (3). The phytyl group of the Chl610 was truncated at the ester group and replaced by a hydrogen atom. The truncation had no effect on the electronic structure of porphyrin ring. The geometries were partially optimized, preserving the planar structure of the Chl macrocycle and of the backbone of Arg-70 and Glu-180. The 1H, 13C, and 15N NMR chemical shieldings were calculated by using the gauge-independent atomic orbital (54–57) on the whole complex, and subsequently, NMR calculations were performed. For both partial optimization and NMR chemical shift calculations, the BLYP/6-311G** basis set was used.
RESULTS
Fluorescence Conditions of LHCII in Detergent-solubilized and Aggregated State
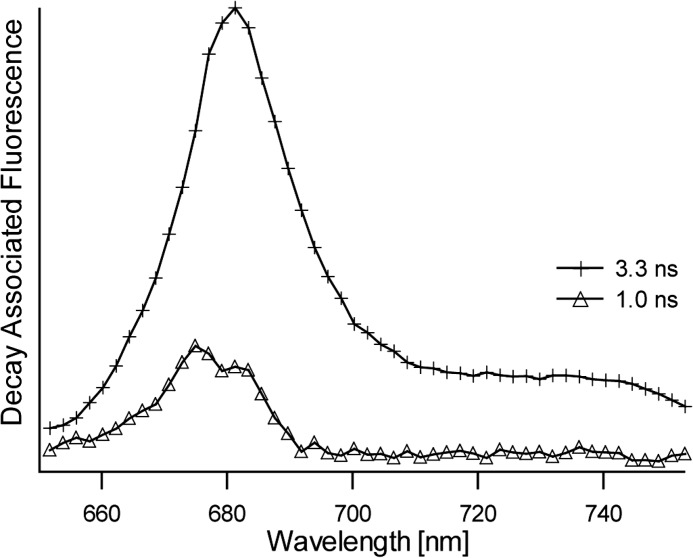
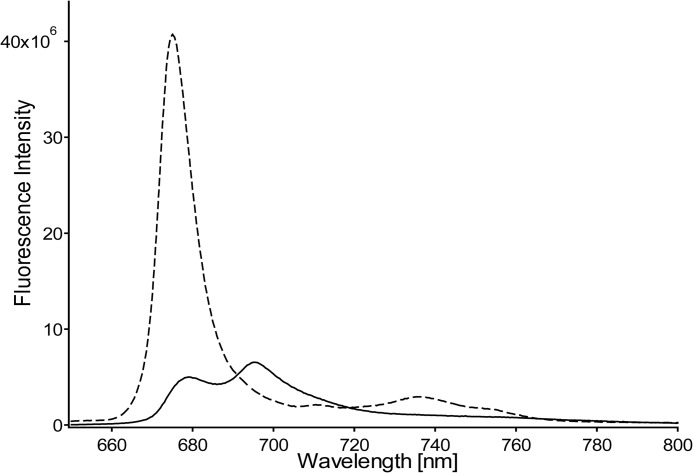
LHCII was prepared in its active, light-harvesting state by solubilizing the LHCII complexes in β-DM buffer. Time-resolved fluorescence experiments on LHCII complexes concentrated in β-DM buffer solution verified that the light-harvesting state of LHCII was retained in the highly concentrated form required for the NMR experiments. Fig. 1 shows the decay-associated spectra obtained by a global analysis fitting of the streak camera images of LHCII concentrated in β-DM solution. The average fluorescence lifetime 〈τf〉 of this sample is 3.0 ns and verifies that the concentrated sample of LHCII in β-DM buffer retained its unquenched state. LHCII aggregates that reflect the photoprotective state of LHCII were produced by dialysis against detergent-free buffer. For the aggregate sample, the fluorescence signal was below the detection limit of the Streak camera setup, and its quenched state was verified by a 77 K steady-state fluorescence emission spectrum. Fig. 2 presents a 77 K fluorescence spectrum of LHCII in β-DM buffer (dashed spectrum) and after dialysis against detergent-free buffer (solid spectrum). Significant fluorescence quenching of LHCII aggregates formed upon dialysis is confirmed by the 10-fold decrease and 4-nm red shift of the fluorescence peak at 675 nm and the appearance of an additional fluorescence band at 695 nm, which has been associated with quenching and aggregation of LHCII (30).
FIGURE 1.
Decay-associated fluorescence spectra and associated lifetimes of LHCII concentrated in β-DM buffer solution.
FIGURE 2.
77 K fluorescence spectra of LHCII. Dashed spectrum, LHCII in β-DM buffer. Solid spectrum, LHCII in detergent-free buffer after extensive dialysis for removal of the β-DM.
Arg NMR Responses for LHCII in Its Active and Dissipative State
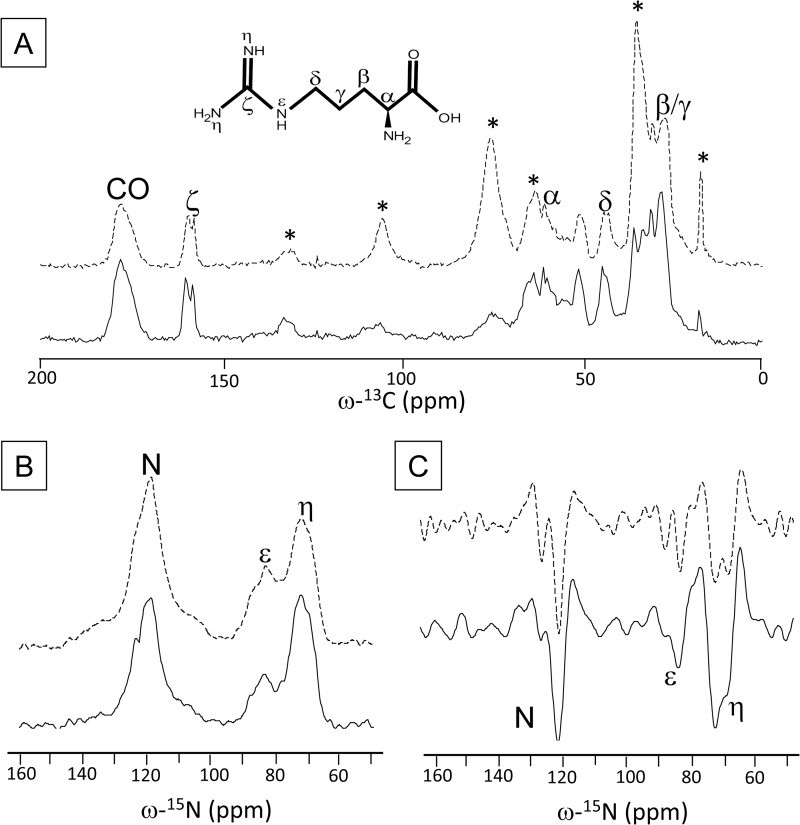
Fig. 3 shows the one-dimensional 13C (panel A) and 15N (panel B) CP-MAS spectra of quenched (solid) and unquenched (dashed) LHCII. In panel C, the second derivative of the 15N spectra is drawn. The Arg chemical structure is drawn with the spectra in panel A. The natural abundance 13C NMR responses of the β-DM detergent molecules is denoted with asterisks in the 13C spectrum of unquenched LHCII. Selective [13C6,15N4]Arg enrichment of the LHCII samples is confirmed by the characteristic Cζ Arg peaks of ∼158 ppm and Nη and Nϵ responses of ∼72 and ∼82 ppm, respectively. Both samples show splitting of NMR responses for the Arg Cζ, Cδ, and Cα and for the Nη, Nϵ, and nitrogen atoms.
FIGURE 3.
13C CP-MAS spectra of unquenched (dashed line) and quenched (solid line) LHCII. A, detergent peaks in unquenched LHCII are denoted with asterisks. The chemical structure of Arg is also shown. B, 15N CP-MAS spectra of unquenched (dashed line) and quenched (solid line) LHCII. C, second derivatives of the 15N spectra in B.
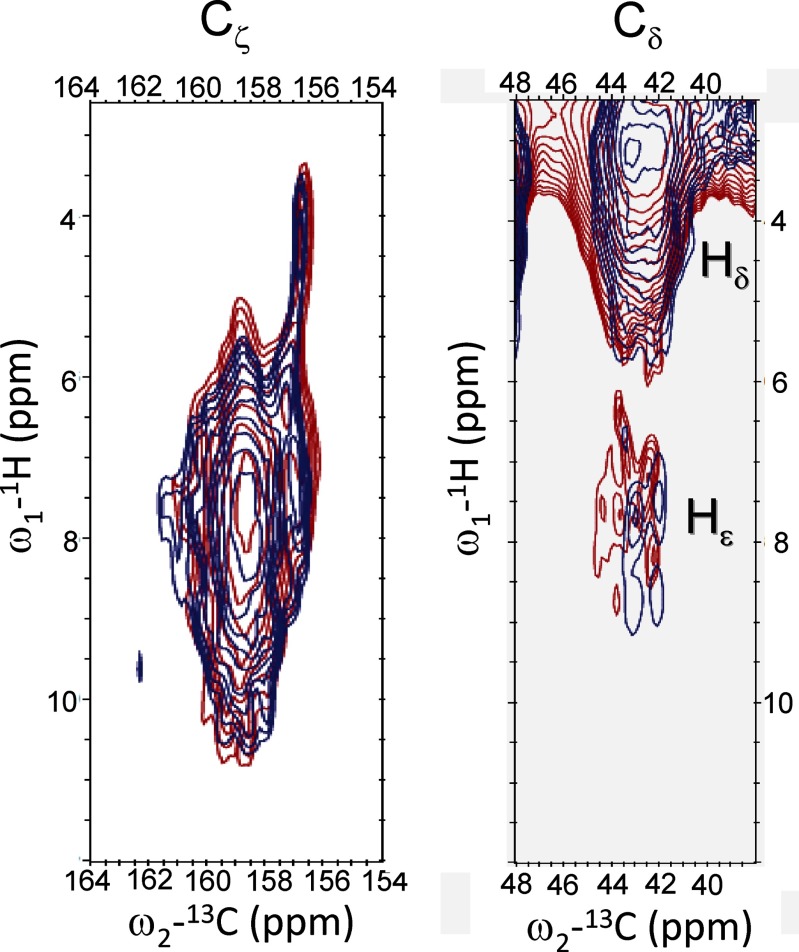
Fig. 4 shows an overlay of 1H-13C HetCor spectra of quenched (red) and unquenched (blue) LHCII in the Arg 13Cζ region (left panel) and 13Cδ region (right panel). The left panel presents the correlations between Cζ and Hηi/Hϵ, whereas the right panel presents the correlations between Cδ and Hδ (proton range 3–5 ppm) and between Cδ and Hϵ (proton range 6–8.5 ppm). Because the Hϵ proton signals (right panel) are in the range of 6.5–8.5 ppm, the proton responses in the left panel between 8.5 and 10.5 ppm are attributed to the Hηi protons. The left panel also shows very peculiar well resolved correlation signals of narrow Cζ responses at 156.8 ppm 13C associated with very upfield shifted proton responses centered around 4.2 ppm 1H.
FIGURE 4.
Heteronuclear 1H-13C correlation spectra of unquenched (blue) and quenched (red) LHCII in the Arg 13Cζ region (left) and in the 13Cδ region (right).
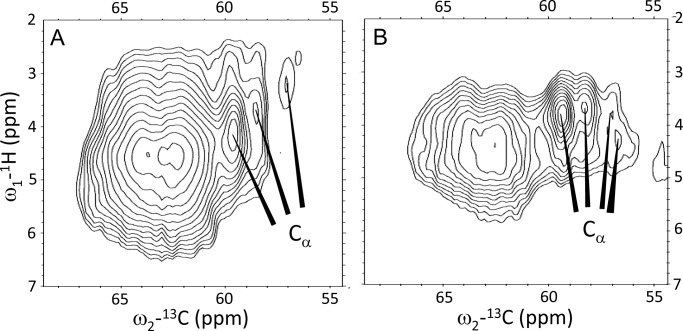
Fig. 5 shows the 1H-13C HetCor spectra in the region of the Arg 13Cα responses (panel A, unquenched LHCII, and panel B, quenched LHCII). The correlation signals in the range 62–64 ppm are attributed to 13C natural abundance chemical shift signals of the detergent molecules. The Arg Cβ and Cγ responses are strongly obscured by overlap with the detergent signals in the range 15–35 ppm and could not be resolved (data not shown). A comparison of Fig. 5, A and B, shows that the Cα correlation peak signal at 57 ppm 13C and 3.2 ppm 1H in the spectrum of unquenched LHCII (Fig. 5A) apparently shifts considerably downfield in the 1H dimension and produces a doubled response for quenched LHCII (Fig. 5B).
FIGURE 5.
Heteronuclear 1H-13C correlation spectra of unquenched (A) and quenched (B) LHCII in the Arg 13Cα region.
DISCUSSION
The LHCII trimers of C. reinhardtii are isomers composed of different Lhcbm polypeptides. These Lhcbm polypeptides contain 6 Arg residues that are conserved in the Lhcb sequences of Arabidopsis thaliana (40). The Lhcbm1, Lhcbm5, and Lhcbm10 polypeptides contain additional Arg residues (2 residues for Lhcbm1 and 10 and 1 residues for Lhcbm5) very close to the N terminus, in a flexible part of LHCII that was not resolved in the LHCII x-ray structures. Most likely, the NMR responses of these additional Arg are also unresolved in our NMR datasets because they are only present in a subpopulation of the LHCII isomers. Moreover, their NMR responses are likely to be weakened due to intrinsic disorder of the N-terminal part, which causes dynamic broadening of the NMR lines and poor cross-polarization. For example, in MAS NMR datasets of purple bacterial antenna proteins, the terminal ends of the α- and β-polypeptides were not resolved (58).
Fig. 6A shows the homology structure of the C. reinhardtii Lhcbm1 monomer, based on the pea LHCII x-ray structure (2BHW) (3), with the 6 conserved Arg residues highlighted. Five Arg residues reside in the α-helical part of the protein, whereas the 6th Arg is located in the stromal loop region. The arginines that form ion pairs with Glu and ligate Chls, Arg-70, Arg-185, and Arg-160, are indicated in the figure, as well as Arg-25 in the loop region. Fig. 6B shows the arrangement of one of the stabilizing Arg-Glu pairs in detail: Arg-70 and Glu-180 that link helix A and B and ligate Chl610.
FIGURE 6.
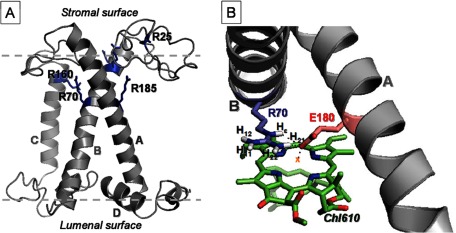
A, homology structure of C. reinhardtii LHCII highlighting the Arg residues. B, the geometric arrangements of Arg-70, Glu-180, and Chl610 taken from the 2BHW (3) LHCII x-ray structure.
According to the plant LHCII x-ray structures, the 6 Arg residues are capable of forming hydrogen bonds via their side chain Hϵ or Hη amides. The strong downfield NMR shifts of the Arg proton responses in the HetCor spectrum in Fig. 4, left panel, suggest that in C. reinhardtii LHCII, strong hydrogen bonds are also formed to the Arg residues that involve the NHηi amide protons.
Upfield Shifted NMR Responses Are Explained by Chl Ring Current Shifts for Arg-70 and Arg-185
In the HetCor spectrum in Fig. 4, left panel, the 13Cζ response at 156.8 ppm correlates with unusual upfield shifted proton responses at 4.2 ppm that are ∼3 ppm shifted relative to the average values (7.4 and 6.8–6.9 ppm, respectively) found for Arg Hϵ and Hηi in the Biological Magnetic Resonance Bank (59). To find an explanation for these unusual shifts, we estimated the ring current effects that are produced by the Chl macrocycle rings. Chl ring currents can induce large shifts for the NMR responses of atoms in close vicinity to the ring center distances (60). Close inspection of the LHCII x-ray structures shows that both Arg-70 and Arg-185 have their side chains hanging over the macrocycle planes of the ligating Chls (Chl602 and Chl610), which positions the Arg side chain atoms close to the Chl rings. The third Arg residue involved in Chl ligation, Arg-160, forms an ion pair with a Glu residue of the same helix (helix C) and has its side chain oriented perpendicular to the macrocycle plane of its ligating Chl (Chl609). For Arg-160, only one proton is positioned close enough to the Chl ring center that it could experience significant ring current effects.
The geometric arrangements of Arg-70, Glu-180, and Chl610 as shown in Fig. 6B were taken from the 2BHW crystal structure and partially optimized as described under “Experimental Procedures.” Quantum-mechanical DFT calculations were performed for three structure models: 1) Arg-70, 2) Arg-70···Glu-180, and 3) Arg-70···Glu-180···Chl610 to estimate the magnitude of Chl ring current and H-bonding-induced shifts of the Arg-70 NMR responses. Table 1 shows how the chemical shifts of Arg-70 (model 1) are affected by interaction with Glu-180 (model 2) and by combined interaction with Glu-180 and Chl610 (model 3). The effects of a partial geometry optimization were most critically obvious for the calculated chemical shifts of the Arg-70 Hϵ and H21 atoms. For these protons also, non-optimized calculated shifts values are presented in which the geometries were taken directly from the x-ray structure.
TABLE 1.
DFT-predicted NMR chemical shift displacements for Arg-70
The NMR chemical shifts of R70 were calculated using model 1 (Arg-70), model 2 (Arg-70···Glu-180), and model 3: (Arg-70···Glu-180···Chl610).
| Atom | Arg-70···Glu-180 (model 2) (Δσ)a | Arg-70···Arg-180···Chl610 (model 3) (Δσ)b |
|---|---|---|
| ppm | ppm | |
| Hϵ | +6.0 | +1.0 |
| Hϵ | +3.5c | −1.1c |
| H11 | −1.1 | −1.5 |
| H12 | −0.9 | −1.7 |
| H21 | +7.0 | +3.7 |
| H21 | +3.0c | −0.6c |
| H22 | −0.8 | −2.1 |
| Cζ | −1.2 | −2.7 |
| Cσ | −1.2 | −2.4 |
| Nζ | −3.3 | +1.3 |
| N1 | +11.6 | +9.5 |
| N2 | −8.1 | −6.8 |
a Chemical shift differences between model 2 and model 1.
b Chemical shift differences between model 3 and model 1.
c Non-optimized.
The results in Table 1 show that for both the unmodified and the partially optimized structures, the presence of the Chl causes strong ring current shifts for the Arg-70 Hϵ and H21 responses of ∼−5 and ∼−3.5 ppm (i.e. the differences between the NMR responses calculated for models 2 and 3), which we attribute primarily to the ring currents in the Chl610 that counteract the large downfield shifts due to H-bonding of these protons to the Glu-180 carboxyl. For Hϵ, the H-bond interaction with the Glu-180 carboxyl is weakened when a ligand to magnesium is formed via the same oxygen atom, which may also affect the Hϵ chemical shift.
The net effect of H-bonding and ring current shifts in our calculations are that the Hϵ and H21 NMR responses are shifted between +3.5 and −1 ppm, depending on the optimization procedure that was applied. The net effects for the other amide protons H11, H12, and H22 are shifts between −1.5 and 2 ppm, whereas the side chain carbon responses of Cζ and Cδ are shifted −2.7 and −2.4 ppm, respectively.
Although the exact geometries of the Arg-Glu-Chl structures may differ in C. reinhardtii LHCII from the plant LHCII x-ray structures, the trends of the calculated chemical shift changes match with the experimental observations of (i) Cζ carbon responses that are shifted 2–3 ppm upfield from the bulk of Cζ signals and that correlate with (ii) proton responses that are shifted 0–2 ppm downfield in addition to (iii) proton responses with significant upfield shifts because for the H11, H12, and H22, the interaction with Glu-180 and Chl610 produces shifts of −1.5 to −2 ppm. Hence, according to the DFT chemical shift calculations, the upfield shifted 1H-13Cζ correlations that are well resolved in Fig. 4, left panel, could originate from Arg-Glu-Chl interactions involving the two Arg residues that form an important structural motif by locking helix A and B.
Our limited models do not take into account hydrogen bonds to other parts of the protein. In the LHCII x-ray structures, one amide proton of Arg-70 is very close to the C13 keto carbonyl of Chl608. This proton is too far from the Chl ring centers to experience any ring current shift. A strong H-bond of this proton to the Chl608 carbonyl should induce a strong downfield shift of its NMR response. Instead, no strong downfield shifts are observed in the 1H dimension for the Cζ signal at 156.8 ppm. This suggests that the H-bonding patterns to the Chl side chains in C. reinhardtii LHCII differ from plant LHCII. In fact, in an earlier study on uniform 13C-labeled C. reinhardtii LHCII, we estimated that the number of Chls with H-bonded keto carbonyls is lower in C. reinhardtii LHCII than in plant LHCII (40).
When comparing the Cζ regions in the HetCor spectra for quenched and unquenched LHCII (Fig. 4), no significant changes are observed, and the spectra are almost identical. In particular, the unusual correlation signal with very upfield proton shifts that we attribute to the helix-connecting Arg residues is preserved. The data imply that the Arg-Glu-Chl geometric arrangements are preserved in the quenched state, relative to the unquenched state. Because the Arg-Glu pairs interlock helix A and B, the conserved chemical shift patterns suggest that the orientations of these two transmembrane helices, which define the tertiary structure, are also preserved in the two states. In addition, the Arg hydrogen-bonding patterns appear to be preserved in the two forms of LHCII.
The DFT-calculated nitrogen chemical shifts in Table 1 predict displacements of the Arg amide 15N NMR responses when a hydrogen bond is formed to Glu (model 2), which breaks the symmetry of the Nηi responses and induces an upfield shift of the Nϵ response. The splitting of the Arg 15Nη signal (Fig. 3, B and C) therefore can be explained by the induced asymmetry when a hydrogen bond is formed to one of the two Nη atoms, whereas splitting of the Arg Nϵ signal is explained by heterogeneity of the Arg structures with respect to H-bonding of their side chain Nϵ atoms.
In Quenched LHCII, Arg-25 Changes Its Backbone Conformation
The Arg 13Cα chemical shift responses reflect moderate conformational changes between the quenched and unquenched forms of LHCII. In the spectrum of unquenched LHCII, two Cα correlation signals appear at 59.5 and 58.5 ppm, and a smaller peak appears at 57 ppm. In the spectrum of quenched LHCII, the latter peak is split into two weaker signals.
The LHCII x-ray structures 2BHW (3) and 1RWT (2) were used to predict the Arg backbone chemical shifts using the SHIFTX2 server. The structure-predicted Arg Cα shifts range between 58 and 60 ppm for the residues in the α-helical stretches, whereas the Arg residue in the stromal loop region, Arg-25, has a predicted Cα shift of 56 ppm. In the NMR spectrum of unquenched LHCII (Fig. 5A), the relative signal intensities of the Cα peaks at 59.5, 58.5, and 57 ppm are roughly in the order of 3:2:1, matching with 5 Arg residues located in the α-helical stretches that have Cα shifts in the range 58–60 ppm and one Arg residue in the loop region with a more upfield shifted Cα response. Therefore we tentatively assign the response at 57 ppm to the Cα of Arg-25 in the stromal loop region. We cannot exclude that the response at 57 ppm 13Cα includes the additional Arg residues at the N-terminal site, appearing in a subpopulation of LHCII. The backbone NMR chemical shifts of the additional Arg residues will likely fall in the chemical shift range for random coil structures, i.e. 54–58 ppm.
Apparently, the backbone conformation of Arg-25 (and/or of the additional Arg at the N-terminal ends) is changed in the photoprotective state (Fig. 5B). Arg-25 is in the water-exposed stromal region of the protein and this part of the protein is probably affected by a change in hydrophobicity or by protein-protein interactions inside the LHCII aggregates.
Stability of the Interlocked A and B Helices in the LHCII Core and Implications for Possible Conformational Changes
Early studies of Murakami and Packer (61) showed a thinning of the thylakoid membrane bilayer upon illumination together with an increase in membrane hydrophobicity, and similar effects were observed in electron microscopy micrographs of granal thylakoid membranes (17, 19). Johnson et al. (35) performed an extended analysis and demonstrated that apparent changes in the appressed membrane zone upon illumination correlated with structural changes in membrane thickness. The authors suggested that the observed decrease in membrane thickness reflects conformational changes of LHCII, forming a more condensed state (35). A similar condensed state is proposed to form upon aggregation in vitro due to protein-protein interactions and changes in hydrophobicity (5). Here we elaborate on this theory and reason that a hypothetical compressed conformation of LHCII can be established in two ways.
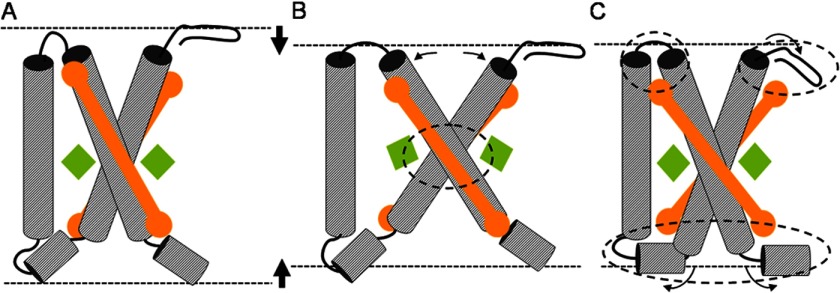
First, it can be established by increased helix tilting, an effect that is a common response of membrane helices to hydrophobic mismatch (53, 62). The LHCII monomers can be compressed by increasing the angles of the membrane-spanning helices A and B with respect to the membrane normal. This is illustrated in Fig. 7B. The LHC structure in Fig. 7A is compressed by increased tilting of the interlocked helices, resulting in the structure drawn in Fig. 7B. However, such a mechanical movement would modify the geometric arrangements of the Glu and Arg residues linking the two helices and of the Arg-Glu ligated Chls (Fig. 7B, encircled area). According to our results in Fig. 4, left panel, this scenario is unlikely. The NMR data show that the responses of the involved Arg residues are almost identical for the quenched and unquenched forms of LHCII, whereas our modeling results predict that these responses are very sensitive to the specific orientations of the interacting Glu, Arg, and Chls. Our findings match with the results of Dockter et al. (42), which show that the LHCII polypeptides have low flexibility in the core region.
FIGURE 7.
Graphic illustrating the possible effects of membrane thinning on LHCII. Compression of the LHC structure in A is achieved by (i) increased tilting of the transmembrane helices, causing structural changes in the Arg-Glu interlocked core (B; the affected core region is encircled) or (ii) reorientation of protein segments close to the water interface, causing structural changes in the regions near the lumenal and stromal sites (C; the affected end regions are encircled). A structural change as depicted in B will affect the orientations of Chl602 and Chl610 (green diamonds) that are ligated to the Arg-Glu interlocked core. However, such a structural rearrangement is unlikely according to the preserved NMR chemical shifts of the Chl-ligating Arg. The structural change depicted in C could reorient the luteins (orange rods) that have their head groups bound to the affected protein end regions.
Second, alternatively, a condensed state of LHCII can be created by rearrangement of protein segments located near the water interface. This is illustrated in Fig. 7C, where segments at the stromal and lumenal ends of the structure in Fig. 7A are refolded (encircled areas) to create a more compact conformation. Such conformational changes are in line with the NMR data and can explain the variability of the 13Cα Arg response, which we tentatively attribute to rearrangement of Arg-25 in the stromal region (Fig. 5). Although under our labeling conditions there are no residues reporting at the LHCII lumenal site, it has been proposed that changes in lumenal pH under NPQ conditions could enhance local refolding of the lumenal loop region by protonation of acidic residues (2, 34).
Photophysical quenching models have been proposed based on altered carotenoid-Chl interactions, in specific between lutein Lut620 and Chla molecules in the terminal emitter domain (Chl610, Chl611, and Chl612) (23, 24). According to our data results here on [13C-15N]Arg LHCII, the ring current effects of Chl610 and Chl602 acting on Arg-70 and Arg-185 do not change in the quenched state. This is a strong indication that the orientations of Chl610 and of Chl602, which are in close distance with either of the two luteins in LHCII, are preserved. The position of Chl612 is also likely to be conserved because its ligand Asn-183 is close to the Arg-Glu interlocked helical core. Thus, photophysical quenching models based on altered lutein-Chla interactions must involve a change in the orientation of a lutein chromophore with respect to the fixed positions of the adjacent Chls. Indeed, the quenched state of LHCII has been associated with a conformational change of the Lut620 carotenoid (21). A change in the orientation of a lutein could affect the NMR responses of closely spaced Chl carbons, explaining the quenching-related changes of specific LHCII Chla NMR responses that were observed in Ref. 41. Lut620 is stabilized by protein interactions via helix D and the loop segment connecting helix A and C. In a hypothetical, mechanistic model, changes in pH and hydrophobicity may reorient these protein segments while maintaining the position of the interlocked helices A and B, resulting in a more compact protein structure as illustrated in Fig. 7C and moving Lut620 with respect to the fixed positions of Chl610 and Chl612, creating a photophysical quencher state.
Conclusion
We used a noninvasive method to selectively probe the structure and environment of the Arg residues in C. reinhardtii LHCII without the need for recombinant approaches. Our approach shows that solid-state NMR is a powerful method to determine the molecular structure of light-harvesting proteins while controlling their functional states. The conformations of the Arg residues in the α-helical regions near the stromal site and in the interlocked core are preserved in the light-harvesting and photoprotective states. In contrast, moderate changes are observed for the Arg in the stromal loop region. The results fit into a mechanistic picture where conformational changes of the LHCII end segments under NPQ conditions may reorient a carotenoid with respect to the fixed positions of the Chls in the terminal emitting domain, rendering a photophysical response for dissipation of harmful excitation energies.
Acknowledgments
Drs. Karthick Babu Sai Sankar Gupta, Fons Lefeber, and Kees Erkelens (Leiden, The Netherlands) are gratefully acknowledged for help with the solid-state NMR spectroscopy, and Dr. Francesco Buda is gratefully acknowledged for advice and discussion on the DFT calculations.
This work was supported by the HARVEST Marie Curie Research Training Network (Grant PITN-GA-2009-238017).
- LHC
- light-harvesting complex
- LHCII
- light-harvesting complex II
- Chl
- chlorophyll
- Lut
- lutein
- NPQ
- non-photochemical quenching
- MAS
- magic angle spinning
- CP-MAS
- cross-polarization-MAS
- HetCor
- heteronuclear correlation
- β-DM
- β-dodecylmaltoside
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- DFT
- density functional theory.
REFERENCES
- 1. Ballottari M., Girardon J., Dall'osto L., Bassi R. (2012) Evolution and functional properties of photosystem II light harvesting complexes in eukaryotes. Biochim. Biophys. Acta 1817, 143–157 [DOI] [PubMed] [Google Scholar]
- 2. Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Ä resolution. Nature 428, 287–292 [DOI] [PubMed] [Google Scholar]
- 3. Standfuss J., Terwisscha van Scheltinga A. C., Lamborghini M., Kühlbrandt W. (2005) Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 Ä resolution. EMBO J. 24, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barros T., Kühlbrandt W. (2009) Crystallisation, structure, and function of plant light-harvesting Complex II. Biochim. Biophys. Acta 1787, 753–772 [DOI] [PubMed] [Google Scholar]
- 5. Ruban A. V., Johnson M. P., Duffy C. D. P. (2012) The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta 1817, 167–181 [DOI] [PubMed] [Google Scholar]
- 6. Horton P. (2012) Optimization of light harvesting and photoprotection: molecular mechanisms and physiological consequences. Philos. Trans. R Soc. Lond. B Biol. Sci. 367, 3455–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szabó I., Bergantino E., Giacometti G. M. (2005) Light and oxygenic photosynthesis: energy dissipation as a protection mechanism against photo-oxidation. EMBO Rep. 6, 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li X. P., Björkman O., Shih C., Grossman A. R., Rosenquist M., Jansson S., Niyogi K. K. (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403, 391–395 [DOI] [PubMed] [Google Scholar]
- 9. Li X. P., Gilmore A. M., Caffarri S., Bassi R., Golan T., Kramer D., Niyogi K. K. (2004) Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J. Biol. Chem. 279, 22866–22874 [DOI] [PubMed] [Google Scholar]
- 10. Walters R. G., Ruban A. V., Horton P. (1996) Identification of proton-active residues in a higher plant light-harvesting complex. Proc. Natl. Acad. Sci. U.S.A. 93, 14204–14209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Müller P., Li X. P., Niyogi K. K. (2001) Non-photochemical quenching. A response to excess light energy. Plant Physiol. 125, 1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonente G., Ballottari M., Truong T. B., Morosinotto T., Ahn T. K., Fleming G. R., Niyogi K. K., Bassi R. (2011) Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 9, e1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonente G., Passarini F., Cazzaniga S., Mancone C., Buia M. C., Tripodi M., Bassi R., Caffarri S. (2008) The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem. Photobiol. 84, 1359–1370 [DOI] [PubMed] [Google Scholar]
- 14. Peers G., Truong T. B., Ostendorf E., Busch A., Elrad D., Grossman A. R., Hippler M., Niyogi K. K. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521 [DOI] [PubMed] [Google Scholar]
- 15. Elrad D., Niyogi K. K., Grossman A. R. (2002) A major light-harvesting polypeptide of photosystem II functions in thermal dissipation. Plant Cell 14, 1801–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferrante P., Ballottari M., Bonente G., Giuliano G., Bassi R. (2012) LHCBM1 and LHCBM2/7 polypeptides, components of major LHCII complex, have distinct functional roles in photosynthetic antenna system of Chlamydomonas reinhardtii. J. Biol. Chem. 287, 16276–16288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson M. P., Goral T. K., Duffy C. D. P., Brain A. P. R., Mullineaux C. W., Ruban A. V. (2011) Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell 23, 1468–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goral T. K., Johnson M. P., Brain A. P. R., Kirchhoff H., Ruban A. V., Mullineaux C. W. (2010) Visualizing the mobility and distribution of chlorophyll proteins in higher plant thylakoid membranes: effects of photoinhibition and protein phosphorylation. Plant J. 62, 948–959 [DOI] [PubMed] [Google Scholar]
- 19. Kirchhoff H., Hall C., Wood M., Herbstová M., Tsabari O., Nevo R., Charuvi D., Shimoni E., Reich Z. (2011) Dynamic control of protein diffusion within the granal thylakoid lumen. Proc. Natl. Acad. Sci. U.S.A. 108, 20248–20253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Betterle N., Ballottari M., Zorzan S., de Bianchi S., Cazzaniga S., Dall'osto L., Morosinotto T., Bassi R. (2009) Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J. Biol. Chem. 284, 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ilioaia C., Johnson M. P., Liao P. N., Pascal A. A., van Grondelle R., Walla P. J., Ruban A. V., Robert B. (2011) Photoprotection in plants involves a change in lutein 1 binding domain in the major light-harvesting complex of photosystem II. J. Biol. Chem. 286, 27247–27254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barros T., Royant A., Standfuss J., Dreuw A., Kühlbrandt W. (2009) Crystal structure of plant light-harvesting complex shows the active, energy-transmitting state. EMBO J. 28, 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bode S., Quentmeier C. C., Liao P. N., Hafi N., Barros T., Wilk L., Bittner F., Walla P. J. (2009) On the regulation of photosynthesis by excitonic interactions between carotenoids and chlorophylls. Proc. Natl. Acad. Sci. U.S.A. 106, 12311–12316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruban A. V., Berera R., Ilioaia C., van Stokkum I. H. M., Kennis J. T. M., Pascal A. A., van Amerongen H., Robert B., Horton P., van Grondelle R. (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450, 575–578 [DOI] [PubMed] [Google Scholar]
- 25. Miloslavina Y., Wehner A., Lambrev P. H., Wientjes E., Reus M., Garab G., Croce R., Holzwarth A. R. (2008) Far-red fluorescence: a direct spectroscopic marker for LHCII oligomer formation in non-photochemical quenching. FEBS Lett. 582, 3625–3631 [DOI] [PubMed] [Google Scholar]
- 26. Liao P. N., Pillai S., Kloz M., Gust D., Moore A. L., Moore T. A., Kennis J. T. M., van Grondelle R., Walla P. J. (2012) On the role of excitonic interactions in carotenoid-phthalocyanine dyads and implications for photosynthetic regulation. Photosynth. Res. 111, 237–243 [DOI] [PubMed] [Google Scholar]
- 27. Berera R., Herrero C., van Stokkum I. H. M., Vengris M., Kodis G., Palacios R. E., van Amerongen H., van Grondelle R., Gust D., Moore T. A., Moore A. L., Kennis J. T. M. (2006) A simple artificial light-harvesting dyad as a model for excess energy dissipation in oxygenic photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 5343–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holt N. E., Zigmantas D., Valkunas L., Li X. P., Niyogi K. K., Fleming G. R. (2005) Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307, 433–436 [DOI] [PubMed] [Google Scholar]
- 29. Krüger T. P. J., Novoderezhkin V. I., Ilioaia C., van Grondelle R. (2010) Fluorescence spectral dynamics of single LHCII trimers. Biophys. J. 98, 3093–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruban A. V., Horton P. (1992) Mechanism of ΔpH-dependent dissipation of absorbed excitation-energy by photosynthetic membranes. 1. Spectroscopic analysis of isolated light-harvesting complexes. Biochim. Biophys. Acta 1102, 30–38 [Google Scholar]
- 31. Pascal A. A., Liu Z., Broess K., van Oort B., van Amerongen H., Wang C., Horton P., Robert B., Chang W., Ruban A. (2005) Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature 436, 134–137 [DOI] [PubMed] [Google Scholar]
- 32. van Oort B., Maréchal A., Ruban A. V., Robert B., Pascal A. A., de Ruijter N. C. A., van Grondelle R., van Amerongen H. (2011) Different crystal morphologies lead to slightly different conformations of light-harvesting complex II as monitored by variations of the intrinsic fluorescence lifetime. Phys. Chem. Chem. Phys. 13, 12614–12622 [DOI] [PubMed] [Google Scholar]
- 33. Pandit A., Shirzad-Wasei N., Wlodarczyk L. M., van Roon H., Boekema E. J., Dekker J. P., de Grip W. J. (2011) Assembly of the major light-harvesting complex II in lipid nanodiscs. Biophys. J. 101, 2507–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang C., Lambrev P., Chen Z., Jávorfi T., Kiss A. Z., Paulsen H., Garab G. (2008) The negatively charged amino acids in the lumenal loop influence the pigment binding and conformation of the major light-harvesting chlorophyll a/b complex of photosystem II. Biochim. Biophys. Acta 1777, 1463–1470 [DOI] [PubMed] [Google Scholar]
- 35. Johnson M. P., Brain A. P. R., Ruban A. V. (2011) Changes in thylakoid membrane thickness associated with the reorganization of photosystem II light harvesting complexes during photoprotective energy dissipation. Plant Signal. Behav. 6, 1386–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pandit A., de Groot H. J. M. (2012) Solid-state NMR applied to photosynthetic light-harvesting complexes. Photosynth. Res. 111, 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pandit A., Buda F., van Gammeren A. J., Ganapathy S., de Groot H. J. M. (2010) Selective chemical shift assignment of bacteriochlorophyll a in uniformly [13C-15N]-labeled light-harvesting 1 complexes by solid-state NMR in ultrahigh magnetic field. J. Phys. Chem. B 114, 6207–6215 [DOI] [PubMed] [Google Scholar]
- 38. Pandit A., Wawrzyniak P. K., van Gammeren A. J., Buda F., Ganapathy S., de Groot H. J. M. (2010) Nuclear magnetic resonance secondary shifts of a light-harvesting 2 complex reveal local backbone perturbations induced by its higher-order interactions. Biochemistry 49, 478–486 [DOI] [PubMed] [Google Scholar]
- 39. van Gammeren A. J., Buda F., Hulsbergen F. B., Kiihne S., Hollander J. G., Egorova-Zachernyuk T. A., Fraser N. J., Cogdell R. J., de Groot H. J. M. (2005) Selective chemical shift assignment of B800 and B850 bacteriochlorophylls in uniformly [13C,15N]-labeled light-harvesting complexes by solid-state NMR spectroscopy at ultra-high magnetic field. J. Am. Chem. Soc. 127, 3213–3219 [DOI] [PubMed] [Google Scholar]
- 40. Pandit A., Morosinotto T., Reus M., Holzwarth A. R., Bassi R., de Groot H. J. M. (2011) First solid-state NMR analysis of uniformly 13C-enriched major light-harvesting complexes from Chlamydomonas reinhardtii and identification of protein and cofactor spin clusters. Biochim. Biophys. Acta 1807, 437–443 [DOI] [PubMed] [Google Scholar]
- 41. Pandit A., Reus M., Morosinotto T., Bassi R., Holzwarth A. R., De Groot H. J. M. (2013) An NMR comparison of the light-harvesting complex II (LHCII) in active and photoprotective states reveals subtle changes in the chlorophyll a ground-state electronic structures. Biochim. Biophys. Acta 1827, 738–744 [DOI] [PubMed] [Google Scholar]
- 42. Dockter C., Müller A. H., Dietz C., Volkov A., Polyhach Y., Jeschke G., Paulsen H. (2012) Rigid core and flexible terminus: structure of solubilized light-harvesting chlorophyll a/b complex (LHCII) measured by EPR. J. Biol. Chem. 287, 2915–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gorman D. S., Levine R. P. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Naumann B., Stauber E. J., Busch A., Sommer F., Hippler M. (2005) N-terminal processing of Lhca3 is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280, 20431–20441 [DOI] [PubMed] [Google Scholar]
- 45. Drop B., Webber-Birungi M., Fusetti F., Kouřil R., Redding K. E., Boekema E. J., Croce R. (2011) Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J. Biol. Chem. 286, 44878–44887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Germano M., Yakushevska A. E., Keegstra W., van Gorkom H. J., Dekker J. P., Boekema E. J. (2002) Supramolecular organization of photosystem I and light-harvesting complex I in Chlamydomonas reinhardtii. FEBS Lett. 525, 121–125 [DOI] [PubMed] [Google Scholar]
- 47. Snellenburg J. J., Laptenok S. P., Seger R., Mullen K. M., van Stokkum I. H. M. (2012) Glotaran: A Java-based graphical user interface for the R package TIMP. J. Stat. Softw. 49, 1–22 [Google Scholar]
- 48. Wawrzyniak P. K., Alia A., Schaap R. G., Heemskerk M. M., de Groot H. J. M., Buda F. (2008) Protein-induced geometric constraints and charge transfer in bacteriochlorophyll-histidine complexes in LH2. Phys. Chem. Chem. Phys. 10, 6971–6978 [DOI] [PubMed] [Google Scholar]
- 49. Becke A. D. (1988) Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A 38, 3098–3100 [DOI] [PubMed] [Google Scholar]
- 50. Miehlich B., Savin A., Stoll H., Preuss H. (1989) Results obtained with the correlation energy density functionals of Becke and Lee, Yang and Parr. Chem. Phys. Lett. 157, 200–206 [Google Scholar]
- 51. Lee C., Yang W., Parr R. G. (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 37, 785–789 [DOI] [PubMed] [Google Scholar]
- 52. Facelli J. C. (1998) Density functional theory calculations of the structure and the 15N and 13C chemical shifts of methyl bacteriopheophorbide a and bacteriochlorophyll a. J. Phys. Chem. B 102, 2111–2116 [Google Scholar]
- 53. Kim T., Im W. (2010) Revisiting hydrophobic mismatch with free energy simulation studies of transmembrane helix tilt and rotation. Biophys. J. 99, 175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ditchfield R. (1981) Theoretical studies of the temperature dependence of magnetic shielding tensors: H2, HF, and LiH. Chem. Phys. 63, 185–202 [Google Scholar]
- 55. Wolinski K., Hinton J. F., Pulay P. (1990) Efficient implementation of the gauge-independent atomic orbital method for NMR chemical shift calculations. J. Am. Chem. Soc. 112, 8251–8260 [Google Scholar]
- 56. McWeeny R. (1962) Perturbation theory for the Fock-Dirac density matrix. Phys. Rev. 126, 1028–1034 [Google Scholar]
- 57. Woliński K., Sadlej A. J. (1980) Self-consistent perturbation theory: Open-shell states in perturbation-dependent non-orthogonal basis sets. Mol. Phys. 41, 1419–1430 [Google Scholar]
- 58. van Gammeren A. J., Hulsbergen F. B., Hollander J. G., de Groot H. J. M. (2005) Residual backbone and side-chain 13C and 15N resonance assignments of the intrinsic transmembrane light-harvesting 2 protein complex by solid-state Magic Angle Spinning NMR spectroscopy. J. Biomol. NMR 31, 279–293 [DOI] [PubMed] [Google Scholar]
- 59. Ulrich E. L., Akutsu H., Doreleijers J. F., Harano Y., Ioannidis Y. E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., Nakatani E., Schulte C. F., Tolmie D. E., Kent Wenger R., Yao H., Markley J. L. (2008) BioMagResBank. Nucleic Acids Res. 36, D402–D408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Abraham R. J., Smith K. M. (1983) NMR spectra of porphyrins. 21. Applications of the ring-current model to porphyrin and chlorophyll aggregation. J. Am. Chem. Soc. 105, 5734–5741 [Google Scholar]
- 61. Murakami S., Packer L. (1970) Protonation and chloroplast membrane structure. J. Cell Biol. 47, 332–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park S. H., Opella S. J. (2005) Tilt angle of a trans-membrane helix is determined by hydrophobic mismatch. J. Mol. Biol. 350, 310–318 [DOI] [PubMed] [Google Scholar]