Background: The identification of palmitoyl substrate-enzyme pairs is important for elucidating physiological roles of protein palmitoylation.
Results: In silico screening revealed neurochondrin palmitoylation. DHHC1/3/10 were identified as neurochondrin palmitoylating enzymes and were essential for targeting of neurochondrin to Rab5-positive endosomes.
Conclusion: Neurochondrin and the DHHC1/10 and DHHC3/7 subfamilies represent novel substrate-enzyme pairs.
Significance: In silico palmitoyl screening is useful for clarifying functions of palmitoylation.
Keywords: Enzymes, Neurobiology, Neuroscience, Post Translational Modification, Protein Palmitoylation, DHHC Protein, Rab5, Endosomes, Neurochondrin
Abstract
Protein palmitoylation, a common post-translational lipid modification, plays an important role in protein trafficking and functions. Recently developed palmitoyl-proteomic methods identified many novel substrates. However, the whole picture of palmitoyl substrates has not been clarified. Here, we performed global in silico screening using the CSS-Palm 2.0 program, free software for prediction of palmitoylation sites, and selected 17 candidates as novel palmitoyl substrates. Of the 17 candidates, 10 proteins, including 6 synaptic proteins (Syd-1, transmembrane AMPA receptor regulatory protein (TARP) γ-2, TARP γ-8, cornichon-2, Ca2+/calmodulin-dependent protein kinase IIα, and neurochondrin (Ncdn)/norbin), one focal adhesion protein (zyxin), two ion channels (TRPM8 and TRPC1), and one G-protein-coupled receptor (orexin 2 receptor), were palmitoylated. Using the DHHC palmitoylating enzyme library, we found that all tested substrates were palmitoylated by the Golgi-localized DHHC3/7 subfamily. Ncdn, a regulator for neurite outgrowth and synaptic plasticity, was robustly palmitoylated by the DHHC1/10 (zDHHC1/11; z1/11) subfamily, whose substrate has not yet been reported. As predicted by CSS-Palm 2.0, Cys-3 and Cys-4 are the palmitoylation sites for Ncdn. Ncdn was specifically localized in somato-dendritic regions, not in the axon of rat cultured neurons. Stimulated emission depletion microscopy revealed that Ncdn was localized to Rab5-positive early endosomes in a palmitoylation-dependent manner, where DHHC1/10 (z1/11) were also distributed. Knockdown of DHHC1, -3, or -10 (z11) resulted in the loss of Ncdn from Rab5-positive endosomes. Thus, through in silico screening, we demonstrate that Ncdn and the DHHC1/10 (z1/11) and DHHC3/7 subfamilies are novel palmitoyl substrate-enzyme pairs and that Ncdn palmitoylation plays an essential role in its specific endosomal targeting.
Introduction
Protein palmitoylation, a post-translational modification of proteins with lipid palmitate, increases the hydrophobicity of proteins, promotes their association with intracellular and plasma membranes, and regulates protein trafficking and functions (1–5). Unlike other lipid modifications, such as myristoylation or prenylation, palmitoylation is a reversible reaction that may be regulated by extracellular signals. The reversible nature of palmitoylation allows proteins to shuttle between intracellular compartments, relocalize in physiological contexts, and participate in diverse aspects of cellular signaling (1–5).
The dynamic palmitoylation level is finely controlled by palmitoyl acyltransferases/palmitoylating enzymes and palmitoyl protein thioesterases/depalmitoylating enzymes. Palmitoylating enzymes were originally identified by forward genetic screening in yeast and contains the conserved Cys-rich DHHC (Asp-His-His-Cys) domain and four or six transmembrane domains (6–8). A family of the mammalian DHHC proteins was identified (9), and it can be categorized into several subfamilies based on the homology of catalytic DHHC domains (9). The discovery of the mammalian DHHC protein family and the establishment of the simple screening system using the DHHC palmitoylating enzyme library have facilitated identification of palmitoyl substrate-enzyme pairs (4, 5). Importantly, a subfamily of DHHC proteins often shares its substrates. However, the identified number of the substrate-enzyme pairs is still limited. Thus, the enzymatic activities of some DHHC proteins, such as the DHHC1/10 (zDHHC1/11; z1/11) subfamily, remain unknown because none of their physiological substrates have been found (4, 5).
Recently, several groups have developed proteomic methods by which palmitoylated proteins are purified from cultured cells or tissues and identified by mass spectrometry. These purification methods of palmitoylated proteins include the acyl-biotinyl exchange (ABE)2 method (10–13) and click chemistry (14–16). Although these powerful proteomic analyses have identified many new palmitoyl proteins (11, 12, 14–16), the method using mass spectrometry has some limitations due to the biochemical properties of proteins. First, some proteins are expressed at too low levels to be identified by mass spectrometry. Second, some membrane proteins, such as G-protein-coupled receptors or ion channels, are barely extracted from cells by detergent and also not efficiently ionized for mass spectrometry (17, 18). Third, some proteins that are not palmitoylated but are just co-purified with palmitoylated proteins might be contaminated (i.e. false-positive results). Last, identifying palmitoylation sites by mass spectrometry is not easily accessible to every laboratory (19). Our method used in this study, in silico prediction of palmitoylation sites of proteins, is able to complementally overcome those problems and identify novel palmitoyl substrates.
Here, we identified neurochondrin (Ncdn)/norbin as a novel palmitoyl substrate. Ncdn was originally identified as a gene whose expression is up-regulated when long term potentiation is chemically induced in rat hippocampus (20). Ncdn is a 75-kDa neuronal cytoplasmic protein with no obvious domain structures (21). Overexpression of Ncdn in Neuro2a cells promotes neurite outgrowth (20). KO of Ncdn in mice leads to early embryonic lethality (22). Nervous system-specific conditional KO of Ncdn in mice results in epileptic seizure (23). Forebrain-specific Ncdn KO attenuates metabotropic glutamate receptor 5 (mGluR5)-dependent stable change in synaptic transmission in the hippocampus and results in a behavioral phenotype associated with a rodent model of schizophrenia (24). This study also showed that Ncdn regulates the surface expression of mGluR5 (24). Taken together, these findings suggest that Ncdn is a promising central nervous system regulator and functions as a scaffolding protein or an adaptor protein. However, its precise subcellular localization in neurons and regulatory mechanism remains unclear.
In this study we systematically screened the mouse whole protein database for palmitoyl substrates through computational prediction and experimentally verified at least 10 novel palmitoyl substrates including Ncdn. We found that the DHHC1/10 (z1/11) and DHHC3/7 subfamilies quantitatively palmitoylated Ncdn at Cys-3 and Cys-4. We also found that Ncdn palmitoylation by DHHC1/3/10 (z1/3/11) plays an essential role in targeting Ncdn to Rab5-positive early endosomes in dendrites.
EXPERIMENTAL PROCEDURES
Global in Silico Prediction of Palmitoyl Proteins
For the global prediction of palmitoyl proteins with their sites, we used the software, CSS-Palm 2.0 (Palmitoylation Site Prediction Using a Clustering and Scoring Strategy) (25). This software algorithm is based on experimentally verified palmitoylation sites: 263 palmitoylation sites from 109 distinct palmitoyl proteins (25). The mouse protein sequence data consisting of 62,695 redundant sequences was downloaded from the UniProt database (26). Sequence files were saved as separate text files in FASTA format programmed by Perl and used as the input of the locally installed CSS-Palm 2.0 (without setting a threshold). To run CSS-Palm 2.0 automatically for all sequence files, the applications used were Windows XP Professional SP3 32 bit, UWSC Version 4.6c, and Java Access Bridge for Microsoft Windows Operating System Version 2.0.1. The output CSV file included results with positions of predicted palmitoyl cysteines, surrounding sequences, and CSS-Palm scores for each protein. All the datasets from CSS-Palm 2.0 included 59,157 files of proteins that have cysteine residues. Protein files with only score 0 cysteines were then removed. The resultant 59,136 proteins (with score >0) were listed in descending order of scores in the CSV file format (supplemental Table S1), which included ∼19,000 proteins with a score greater than the cut-off score of 1.8. Redundant sequences and sequences with cysteines in the signal sequence were then removed.
To find palmitoyl candidate proteins that are expressed predominantly in the brain, the listed protein sequences were further analyzed by a gene expression database, BodyMap-Xs (27). The entire dataset for mouse genes (38,632 gene data) was automatically downloaded from BodyMap-Xs, and data files were saved as CSV files. Information about accession numbers (Unigene no.) included in each output data file from BodyMap-Xs was collated with the UniProt annotation in the initial output data files from CSS-Palm 2.0. Then the CSS-Palm-predicted candidates included in datasets from the BodyMap-Xs analysis were further extracted. The brain enrichment score was calculated by dividing the score for brain by the sum of scores for all tissues. Because the present expression database is not sufficient for all the proteins, this narrowing-down process may omit 27% of CSS-Palm 2.0 data that is not covered in the BodyMap-Xs database. Proteins with brain enrichment scores ≥0.75 were listed, and proteins containing candidate cysteines only in the extracellular region were manually excluded. The resultant list consisted of 573 promising candidates (supplemental Table S2). In addition, we manually inspected the omitted candidate proteins (with ≥1.8 CSS-Palm scores, but without BodyMap-Xs data), and selected homer 1C, Syd-1, paxillin, zyxin, and Par3 as additional palmitoyl candidates that are enriched in specific membrane domains such as pre- and postsynapses and cell adhesion sites. Instead of cornichon-3 (CSS-Palm score 1.852), cornichon-2 (1.730) was selected because the potential palmitoyl cysteine is conserved in both isoforms and cornichon-2 is predominantly expressed in the hippocampus and has been extensively studied as an AMPA receptor modulator (28).
Cloning and Plasmid Constructions
Rat cDNAs of cornichon-2 (CNIH2, NM_001025132), Ncdn/norbin (AB006461), Syd-1 (NM_001191876), Par3 (amino acids 1–281) (NM_031235), Rab3A (NM_013018), Rab11A (NM_031152), SERCA2b (NM_001110823), and DHHC1 (NM_001039099) were cloned from rat brain total RNA by RT-PCR. Mouse cDNAs of Rab4A (NM_009003), Rab5A (NM_025887), Rab7A (NM_009005), Rab8A (NM_023126), and Rab9A (NM_019773) were cloned from mouse brain total RNA by RT-PCR. The following cDNAs were kindly provided by indicated investigators: human Rab6A, Franck Perez (Institut Curie); rat CaMKIIα, Ulli Bayer (University of Colorado, Denver, CO); human TRPC1, Yasuo Mori (Kyoto University); mouse TRPM8, Ardem Patapoutian (The Scripps Research Institute, La Jolla, CA), and Makoto Tominaga (National Institute for Physiological Sciences, Japan); mouse orexin 2 receptor, Akihiro Yamanaka (Nagoya University); rat TARP γ-2, rat TARP γ-8, and human liprin-α2, David Bredt (Johnson and Johnson Research and Development, San Diego, CA); rat KIF5C (amino acids 1–560), Gary Banker (Oregon Health and Science University, Portland, OR); rat homer 1C, Akihiko Kato (Eli Lilly, Indianapolis, IN); rat kalirin7, Xin-Ming Ma (University of Connecticut Health Science Center, Farmington, CT). Other constructs used in this study were described previously (9, 29, 30).
cDNA of rat Ncdn was subcloned into pCAGGS or pCAGGS-GFP(N) to fuse the GFP tag at the carboxyl terminus. Ncdn C3S, Ncdn C4S, and Ncdn C3S,C4S (NcdnCS) were constructed by site-directed mutagenesis. To construct the knockdown vectors expressing microRNAs (miRNAs or miRs), BLOCK-iT RNAi Designer (Invitrogen) was used to select the targeting sequences, and the following targeting sequences for rat genes were used: miR-Ncdn_803 (5′-TCCTAGGAAGCAAGTTGAGCT-3′), miR-DHHC1_532 (5′-CACAGTGTGGCATCTGCTTTA-3′), miR-DHHC3_735 (5′-TGAGACGGGAATAGAACAATT-3′), miR-DHHC10 (z11)_171 (5′-TGTCACCTTTGGGATCTTCAT-3′), and miR-LacZ (5′-GACTACACAAATCAGCGATTT-3′) (as a negative control). These oligonucleotides were subcloned into pcDNA6.2:EmGFP-miR, and then these miRNA sequences with EmGFP were subcloned into pCAGGS (pCAGGS:EmGFP-miR). To increase the knockdown efficiency, we tandem-chained the miRNA sequences for several experiments (miR-Ncdn_803_803, miR-DHHC1_532_532, miR-DHHC3_735_735, miR-DHHC10 (z11)_171_171, and miR-LacZ_LacZ). For molecular replacement experiments or rescue experiments, miRNA-resistant rat Ncdn (resNcdnWT or resNcdnCS) and mouse DHHC10 (z11) (resDHHC10WT or resDHHC10CS (C158S)) were constructed by site-directed mutagenesis (Ncdn, 5′-TCTTGGGGAGCAAGTTGAGCT-3′, DHHC10 (z11) 5′-TGTCACATTCGGAATCTTCAT-3′; changed nucleotides are underlined). Mouse DHHC3WT or CS (C157S) was resistant to miR-DHHC3_735 and was used for rescue experiments (resDHHC3WT or resDHHC3CS). Then, EmGFP in pCAGGS:EmGFP-miR-LacZ_LacZ, miR-Ncdn803_803, miR-DHHC3_735_735, or miR-DHHC10 (z11)_171_171 was replaced with GST-HA, resNcdnWT/CS-HA, HA-resDHHC3WT/CS, or HA-resDHHC10WT/CS, allowing co-cistronic expression of the miRNA and miRNA-resistant Ncdn or DHHCs. Knockdown of Ncdn by the miRNA vector and knockdown-resistant expression of resNcdn was validated in HEK293 cells (data not shown). All PCR products were analyzed by DNA sequencing.
Antibodies
The following antibodies were used: rabbit polyclonal antibodies to TARPs γ-2/γ-8 (Millipore 07–577), Syd-1 (Abcam ab80402), mGluR5 (Millipore 06-451), and Ncdn (Sigma HPA023676); mouse monoclonal antibodies to HA (clone 16B12; Covance MMS-101P), FLAG (Clone M2; Sigma F3165), V5 (Invitrogen R960-25), CaMKII (Clone 6G9; Millipore MAB3119), PSD-95 (Clone 7E3-1B8; Thermo Fisher Scientific MA1-046), MAP2 (Clone HM-2; Sigma M4403), pan-axonal neurofilament (Clone SMI312; Covance SMI312R), synaptophysin (clone SVP-38; Sigma S5768), GM130 (Clone 35/GM130; BD Transduction Laboratories 610822) and β-catenin (BD Biosciences, 14/β-catenin); rat monoclonal antibody to HA (clone 3F10; Roche Applied Science 11867423001) and chicken polyclonal antibody to GFP (Millipore ab16901). Rabbit polyclonal antibodies to GFP and moesin were described previously (29). Rabbit polyclonal antibody to Ncdn (N-rNcdn) was raised against His6-N-rat Ncdn (amino acids 1–59) and affinity-purified.
Cell Culture
Hippocampal and cortical neuron cultures were prepared from rat embryonic day 18–20 embryos or postnatal day 1 pups. All animal experiments described herein were reviewed and approved by the ethics committee in our institutes and were performed according to the institutional guidelines concerning the care and handling of experimental animals. Neurons were seeded in neurobasal medium (Invitrogen) supplemented with B-27 supplement (Invitrogen) or B-27 plus (MACS) and 2 mm Glutamax (Invitrogen).
Metabolic Palmitate Labeling and Subcellular Fractionation
Both experiments were performed as previously described (9, 29).
ABE Method
The ABE method was performed as previously described (10–12, 29). Briefly, hippocampal neurons were solubilized and incubated with N-ethylmaleimide to mask free cysteines. Free N-ethylmaleimide was removed by chloroform-methanol precipitation. Precipitated proteins were incubated with hydroxylamine (NH2OH) to cleave a thioester bond or Tris/Cl as a control followed by incubation with biotin-HPDP (N-[6-biotinamido]hexyl]-3′-(2′-pyrdyldithio)propionamide) to label new free cysteines. Biotinylated proteins were then purified with NeutrAvidin-agarose (Thermo Fisher Scientific) and analyzed by Western blotting with indicated antibodies. To detect Ncdn palmitoylation, the improved ABE protocol (13) was used to reduce background signals. Here, to extract more proteins, lysis buffer included 4% SDS instead of 2% SDS. Also, to cleave disulfide bonds before alkylation with N-ethylmaleimide, we used tris(2-carboxyethyl)phosphine, a potent reducing agent that does not cleave the thioester bond between palmitate and cysteine residue.
Tandem Affinity Purification
To pull down associated proteins with DHHC palmitoylating enzymes from the brain membrane fraction, HEK293 cells transfected with His6-FLAG-DHHC2, 3 or 10 (z11) were scraped with immunoprecipitation buffer (20 mm Tris/Cl at pH 7.5, 1 mm EDTA, 100 mm NaCl, 2.0% Triton X-100, 50 μg/ml PMSF) and homogenized followed by centrifugation at 100,000 × g at 4 °C for 30 min. P2 membrane fraction was prepared by homogenization of rat whole brain in homogenizing buffer (20 mm Tris/Cl at pH 7.5, 2 mm EDTA, 0.32 m sucrose, 100 μg/ml PMSF) and centrifugation at 20,000 × g at 4 °C for 1 h. The resultant pellet was suspended and mixed with the HEK293 cell supernatant containing tagged DHHC proteins prepared above. After incubation for 1 h at 4 °C, the homogenate was centrifuged at 100,000 × g at 4 °C for 1 h. The supernatant was incubated with FLAG-M2 beads (Sigma) for 2 h at 4 °C. After washing the beads, His6-FLAG-DHHC was eluted with FLAG peptide (0.25 mg/ml) for 1 h at 4 °C. The FLAG eluate was incubated with nickel-nitrilotriacetic acid-agarose for 1 h at 4 °C. After washing the beads, His6-FLAG-DHHC was eluted with the buffer containing 250 mm imidazole.
Transfection and Immunofluorescence Analysis
Cortical or hippocampal neurons (5 × 104 cells) on 12-mm coverslips were transfected by Lipofectamine 2000 (Invitrogen). For knockdown experiments, 6–7 days after transfection, neurons were fixed with 4% paraformaldehyde at room temperature for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with PBS containing 10 mg/ml BSA for 10 min. Neurons were then labeled with rabbit polyclonal anti-Ncdn (N-rNcdn) and Cy3-conjugated donkey anti-rabbit IgG. Neurons transfected with miR-Ncdn expression vector were visualized by co-cistronic expression of GFP. For Fig. 5, D and E, hippocampal neurons were fixed with methanol at −30 °C for 10 min. For confocal imaging, fluorescent images were obtained using an LSM5 Exciter system (Carl Zeiss) with a Plan-Apochromat 63× NA 1.40 oil immersion objective lens. For two-color stimulated emission depletion (STED) imaging, neurons were immunostained by rabbit anti-Ncdn and chicken anti-GFP or anti-V5 antibodies followed by ATTO425 (Rockland) and Alexa488 (Invitrogen)-conjugated secondary antibodies. STED images were obtained by a Leica TCS STED CW with a 100× NA 1.40 oil immersion objective lens. Obtained images were further deconvoluted with the built-in deconvolution algorithms of the Leica LAS-AF software. Also, the same region was acquired by the confocal mode of a Leica TCS SP5 II and directly compared with that of STED imaging. For statistical analysis, one-way analysis of variance with Tukey's HSD or Dunnett post hoc test was performed. Data are presented as the mean ± S.E.
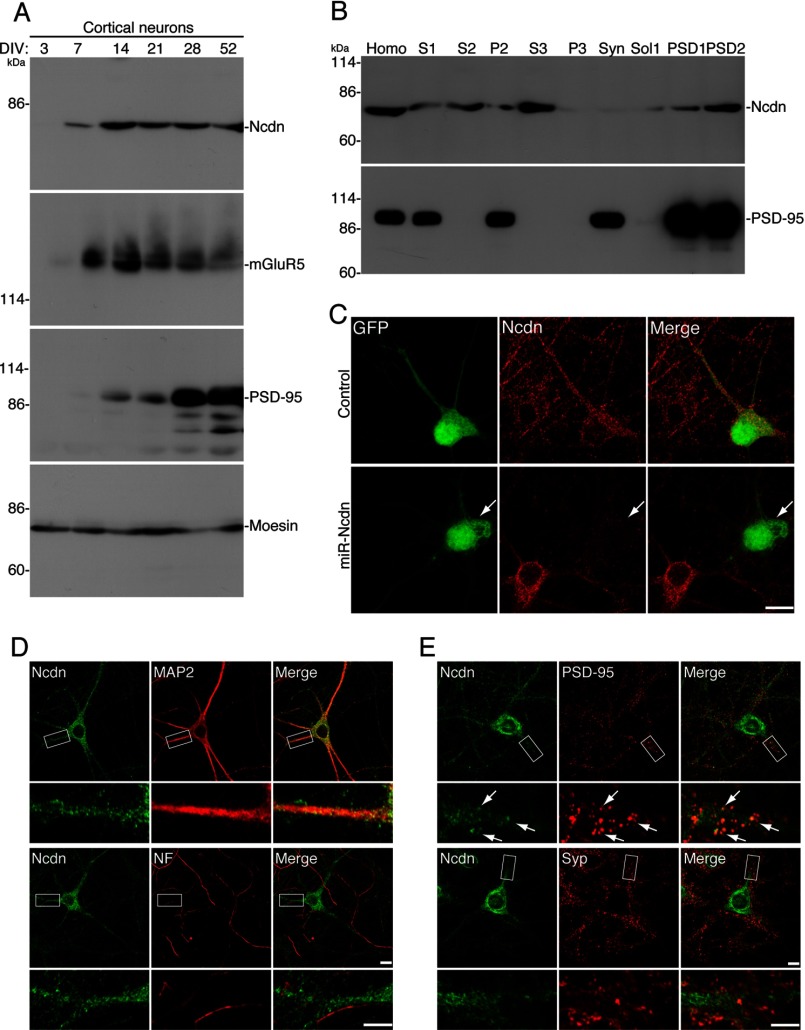
FIGURE 5.
Ncdn expression and localization in neurons. A, expression of Ncdn increases between 7 and 14 DIV. In cultured cortical neurons, Ncdn expression was not detected at 3 DIV, began to be detected at 7 DIV, and was the highest at 14 DIV. This expression pattern of Ncdn was similar to that of mGluR5. PSD-95 expression increased at later DIV than Ncdn. Moesin was an internal control. B, subcellular distribution of Ncdn was examined by biochemically fractionating the brain homogenate (Homo). Ncdn protein was fractionated into both cytoplasmic (S3) and Triton X-100-insoluble (PSD1 and PSD2) fractions. PSD-95 was used as a marker for the PSD fraction. C, Ncdn antibody specifically recognizes endogenous Ncdn in neurons. Ncdn was knocked down in cortical neurons by transfection with miR-Ncdn expression vector and analyzed by immunostaining with anti-Ncdn antibody (red). Neurons transfected with miR-Ncdn expression vector were visualized by co-cistronic expression of GFP (white arrow). Bar, 10 μm. D and E, Ncdn is localized in dendrites, but not in the axon, and partially localized in dendritic spines. Hippocampal neurons were immunostained with anti-Ncdn and either anti-MAP2 (dendritic marker), anti-pan-neurofilament (NF) (axonal marker), anti-PSD-95 (postsynaptic marker) or anti-synaptophysin (Syp, presynaptic marker) antibodies. Upper bars, 10 μm; lower bars, 5 μm. Arrows indicate the co-localization of Ncdn with PSD-95 (upper) (E).
RESULTS
Global in Silico Screening for Novel Palmitoyl Substrates
To identify novel palmitoyl substrates, we took advantage of computational prediction using CSS-Palm 2.0 algorithm, which was recently developed for palmitoylation site prediction (25) and is freely available. A pilot study using protein sequences of known palmitoyl and non-palmitoyl proteins showed that CSS-Palm 2.0 predicted the reported palmitoyl cysteines of representative palmitoyl substrates with high scores (e.g. PSD-95, 5.096; GAP-43, 16.844; H-Ras, 4.024) but gave low scores toward non-palmitoyl proteins (β-actin, 1.296; β-catenin, 0.687). We next applied the comprehensive protein sequences to CSS-Palm 2.0, predicting many novel palmitoyl substrates.
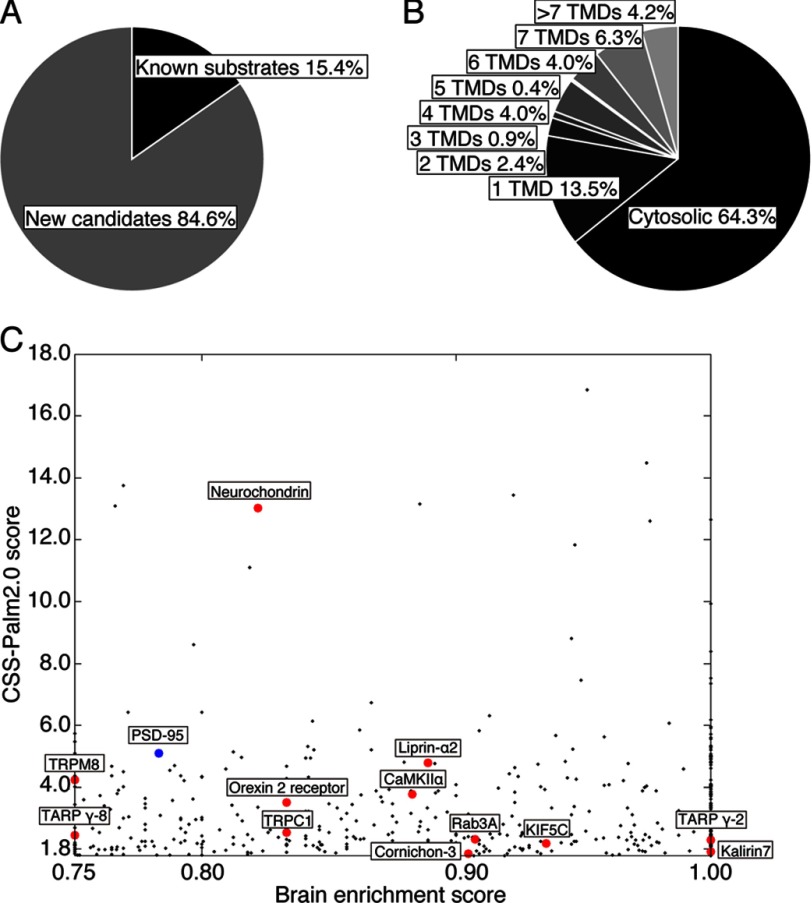
As a comprehensive protein database, we used the UniProt database and selected Mus musculus as an organism (26). To automatically retrieve about 60,000 sequences with redundancies from the UniProt database and apply them to the CSS-Palm 2.0 prediction program, we developed the automatic program (see “Experimental Procedures). Proteins that have cysteines with CSS-Palm score >0 were listed (supplemental Table S1). Because most known palmitoylated substrates showed more than a score 1.8, we set 1.8 as the cut-off value for the CSS-Palm 2.0 score. Brain enrichment score (see “Experimental Procedures”) was used as another criterion for selection (cut-off point is ≥0.75), and 573 proteins were retrieved as candidates for brain-enriched palmitoyl substrates (supplemental Table S2). Of the 573 proteins, 84.6% are novel candidates for palmitoyl proteins (Fig. 1A). The palmitoyl candidates included the substantial number of putative transmembrane proteins (35.7%) (Fig. 1B). Many of the candidate proteins are known to function in signal transduction, for example, as receptors, channels, transporters, GTPases, and kinases (supplemental Table S2). We selected 12 proteins as candidates for novel palmitoyl substrates based on their brain enrichment (Fig. 1C, Table 1, and supplemental Table S2). Given that palmitoyl proteins accumulate at specialized membrane compartments, such as pre- and postsynaptic membranes, focal adhesions, and tight junctions, we selected five proteins whose CSS-Palm 2.0 scores are ≥1.8 but whose brain enrichment scores are <0.75 or not calculated (Table 1).
FIGURE 1.
In silico screening for neuronal palmitoyl candidates. A–C, based on our criteria (≥1.8 CSS-Palm score and ≥0.75 brain enrichment score), we chose 573 proteins as brain-enriched palmitoyl candidates (supplemental Table S2). Note that secreted proteins and proteins with target cysteines only in extracellular regions were excluded. Of the candidate proteins, 84.6% have not yet been reported and are considered as promising new palmitoyl substrates (A). 35.7% of neuronal substrate candidates are membrane proteins (B). TMD, transmembrane domain. Our 12 selected candidates (red) and PSD-95 (blue), a representative neuronal palmitoyl protein, are marked on the scattergram, consisting of CSS-Palm scores (≥1.8) on the y axis and brain-enrichment scores (≥0.75) on the x axis (C).
TABLE 1.
Selected palmitoyl candidates in this study
Highest CSS-Palm score, Position, and Sequence indicate the highest score in the protein given by CSS-Palm 2.0, the position of the corresponding cysteine (underlined in sequence), and the sequence around the cysteine with amino acid positions, respectively. N (bold), N terminus; C (bold), C terminus. ND means no data retrieved from Body-Map database. “Metabolic” indicates the result of metabolic labeling assay with [3H]palmitate: +, palmitoylated; −, not palmitoylated. ABE indicates the result of the ABE method: +, endogenous protein was palmitoylated; NT, not tested.
| Protein name | CSS-Palm prediction |
Experiments |
||||
|---|---|---|---|---|---|---|
| Highest CSS-Palm score | Position | Sequence | Brain enrichment score | Metabolic | ABE | |
| Postsynapse | ||||||
| TARP γ-2 | 1.913 | 121 | -118GGLCIAA124- | 1.000 | + | NT |
| TARP γ-8 | 2.461 | 144 | -141GGVCCVAA147- | 0.750 | + | + |
| Cornichon-2 | 1.730 | 9 | -6AAFCYML12- | 0.901 | + | NT |
| CaMKIIα | 3.765 | 6 | -3TITCTRF9- | 0.883 | + | + |
| Kalirin7 | 2.297 | 1404 | -1401LLTCCEE1407- | 1.000 | − | NT |
| Homer 1C | 2.687 | 365 | -362LLECS-C | ND | − | NT |
| Neurochondrin | 13.016 | 3 | N-MSCCDL6- | 0.822 | + | + |
| Presynapse | ||||||
| Rab3A | 2.313 | 220 | -217DCAC-C | 0.907 | − | NT |
| Syd-1 | 3.078 | 736 | -733INVC l-C | ND | + | + |
| Liprin-α2 | 4.783 | 3 | N-MMCEVM6- | 0.889 | − | NT |
| Motor protein | ||||||
| KIF5C | 2.183 | 7 | -4PAECSIK10- | 0.935 | − | NT |
| Channels | ||||||
| TRPM8 | 4.234 | 1032 | -1029FKCCCKE1035- | 0.750 | + | NT |
| TRPC1 | 2.547 | 736 | -733KVMCCLV739- | 0.833 | + | NT |
| GPCR | ||||||
| Orexin 2 receptor | 3.500 | 381 | -378AFSCCLG384- | 0.833 | + | NT |
| Focal adhesion | ||||||
| Paxillin | 2.270 | 591 | -588KLFC-C | ND | − | NT |
| Zyxin | 2.012 | 404 | -401CFTCHQC407- | ND | + | NT |
| Tight junction | ||||||
| Par3 | 3.061 | 6 | -3VTVCFGR9- | ND | − | NT |
At the postsynapse, many proteins are subjected to palmitoylation (1, 4, 12). Examples include AMPA-type glutamate receptor (AMPAR, subunits GluA1, -2, -3, and -4) (31), NMDA-type receptor (32), and their scaffolding proteins, PSD-95 (33), PSD-93 (34), and GRIP (35). Because AMPAR, which mediates fast excitatory synaptic transmission in the brain (36, 37), and its scaffolding proteins are palmitoylated, we assumed that other AMPAR regulators with high CSS-Palm scores might be palmitoylated as well. We selected TARP γ-2 (CSS-Palm score 1.913), TARP γ-8 (2.461) (37–40), CNIH2 (1.730) (28, 41), and CaMKIIα (3.765) (42) as candidates for palmitoyl substrates (Fig. 1C and Table 1). In addition, kalirin7 (2.297), a Rho guanine nucleotide exchange factor and PSD-95-binding protein (43), and homer 1C (2.687), a regulator of mGluRs (44, 45), were selected. Ncdn/norbin (13.016), a regulator for mGluR5 (24), was also selected.
At the presynapse, numerous SNARE proteins including SNAP-25, synaptotagmin, synaptobrevin, and syntaxin, have been reported as palmitoyl substrates (4, 12). Besides SNARE proteins, we focused on three important presynaptic proteins with high CSS-Palm scores: Rab3A (CSS-Palm score 2.313), Syd-1 (3.078), and liprin-α2 (Syd-2) (4.783) (Fig. 1C and Table 1).
At cell adhesion sites, integrins at focal adhesions (46) and claudins at tight junctions (47) are palmitoylated. We speculated that scaffolding/anchoring proteins at these adhesion sites might be also palmitoylated. We selected focal adhesion proteins, paxillin (CSS-Palm score 2.270) and zyxin (2.012), and a tight junction protein, Par3 (3.061) (Table 1).
In addition, several functional molecules with high CSS-Palm scores, including transient receptor potential (TRP) channels, TRPM8 (4.234) and TRPC1 (2.547), a G-protein-coupled receptor, orexin 2 receptor (3.500), and a kinesin motor protein KIF5C (2.183), were selected (Fig. 1C and Table 1).
Examination of Novel Palmitoyl Substrates by Experimental Approaches
To examine whether those 17 substrate candidates are actually palmitoylated in cells, cDNAs of individual candidate and DHHC3 palmitoylating enzyme were co-transfected into HEK293 cells, and palmitoyl modified proteins were metabolically labeled with [3H]palmitate. Based on our previous studies, DHHC3 functions as a general palmitoylating enzyme (i.e. all palmitoyl substrates we have tested are palmitoylated by DHHC3) (4). Among postsynaptic candidate proteins, TARP γ-2, TARP γ-8, CNIH2, CaMKIIα, and Ncdn were robustly palmitoylated (Fig. 2A and Table 1). Neither kalirin7 nor homer 1C were palmitoylated. The presynaptic candidate Syd-1 was palmitoylated, whereas Rab3A (Fig. 2A) and liprin-α2 (data not shown) were not palmitoylated. Among cell adhesion-related proteins, zyxin incorporated [3H]palmitate, whereas paxillin and Par3 were not palmitoylated. We noted that the expression level of paxillin-GFP was relatively reduced in the presence of DHHC3. Even when the amounts of paxillin-GFP loaded were adjusted between samples, we did not see clear incorporation of [3H]palmitate to paxillin-GFP in the presence of DHHC3 (data not shown). Other candidate TRPM8, TRPC1 and orexin 2 receptor, but not KIF5C, were efficiently palmitoylated by DHHC3. Thus, of the 17 candidates, 10 were verified as palmitoyl substrates (Table 1).
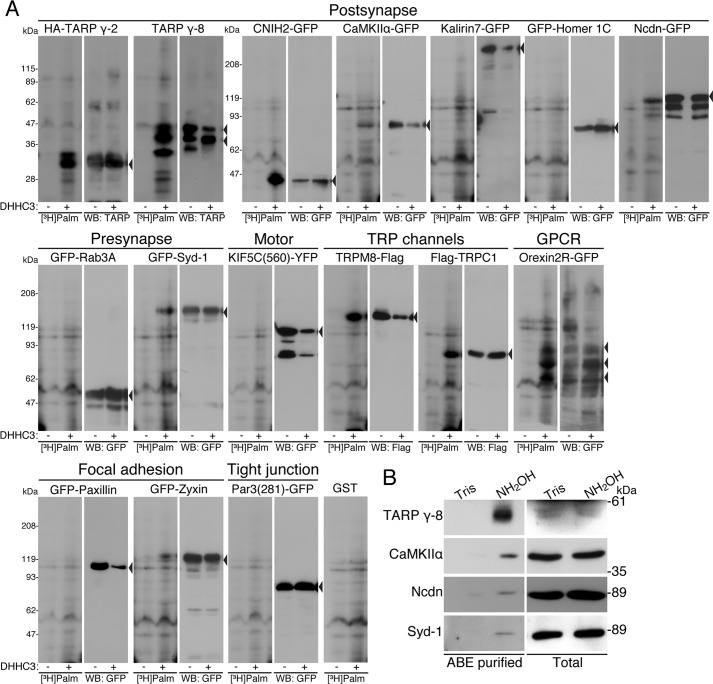
FIGURE 2.
Identification of 10 novel palmitoyl substrates. A, HEK293 cells were co-transfected with expression vectors of indicated candidate cDNAs and DHHC3 palmitoylating enzyme. After metabolic labeling with [3H]palmitate, proteins were separated by SDS-PAGE and analyzed by fluorography ([3H]Palm) and Western blotting (WB). Arrowheads indicate the positions of individual candidate proteins. Actual palmitoylation of TARP γ-2, TARP γ-8, CNIH2, CaMKIIα, Ncdn, Syd-1, TRPM8, TRPC1, orexin 2 receptor, and zyxin was experimentally verified. As a negative control, GST was expressed instead of palmitoyl candidates. B, hydroxylamine (NH2OH)-sensitive palmitoylated proteins were purified from neurons by the ABE method. Then samples were analyzed by Western blotting with indicated antibodies. Endogenous TARP γ-8, CaMKIIα, Ncdn, and Syd-1 were palmitoylated in cultured hippocampal neurons. Tris, Tris treatment as a control of hydroxylamine. GPCR, G-protein coupled receptors
This assay, however, might possibly include false-positive substrates, as both DHHC3 enzyme and a substrate protein are overexpressed in cells. To test whether endogenous proteins are palmitoylated in neurons, we purified palmitoylated proteins in primary hippocampal neurons by the ABE method and analyzed them by Western blotting. Because of the availability of the specific antibodies, only TARP γ-8, CaMKIIα, Ncdn, and Syd-1 were tested. All four endogenous proteins were palmitoylated in the primary hippocampal neurons (Fig. 2B and Table 1). Although we could not examine the other substrates (TARP γ-2, CNIH2, zyxin, TRPM8, TRPC1, and orexin 2 receptor), this result strongly suggests that our screening could identify authentic palmitoyl substrates.
DHHC1, -3, -7, and -10 (z11) Enhance Ncdn Palmitoylation
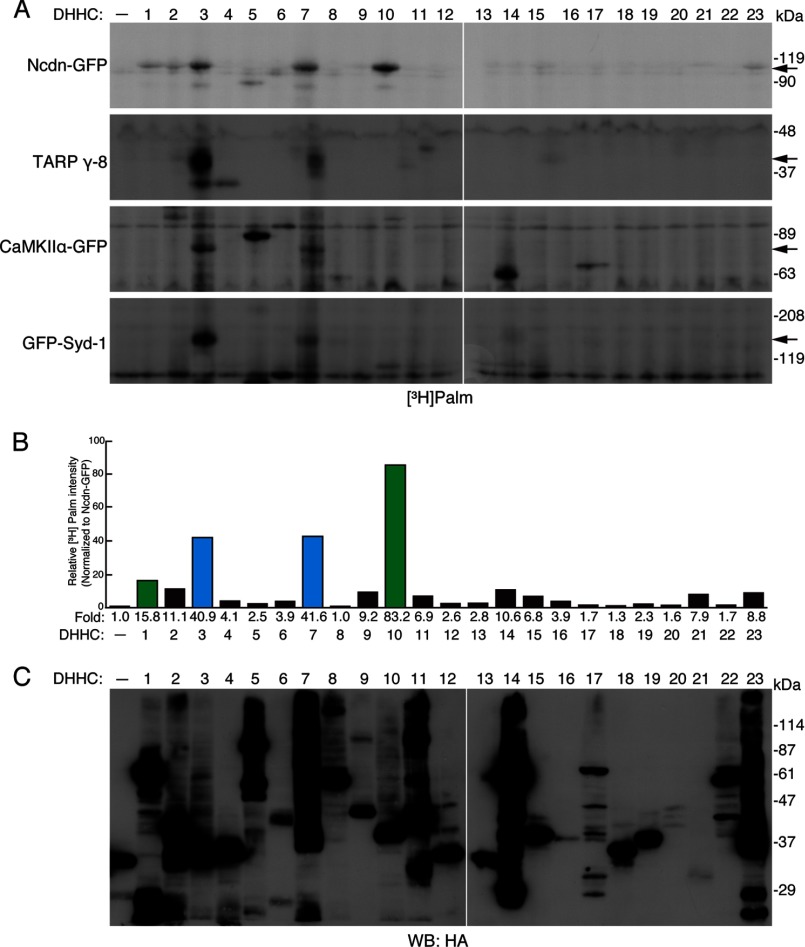
We next investigated which DHHC proteins palmitoylate Ncdn, TARP γ-8, CaMKIIα, and Syd-1. To identify the responsible enzymes for these four substrates, we screened the DHHC palmitoylating enzyme library by metabolic labeling with [3H]palmitate (Fig. 3, A–C) (9, 30). Palmitoylation of all four substrates was enhanced by DHHC3 and -7, which belong to the same subfamily (Fig. 3A) (9). Very uniquely, in addition to DHHC3 and -7, Ncdn was also palmitoylated by DHHC1 and -10 (z11) (Fig. 3, A and B). DHHC1 and -10 (z11) belong to another subfamily (9). Ncdn is the first substrate to be identified for the DHHC1/10 (z1/11) subfamily (4, 5). Moreover, Ncdn is predominantly expressed in the brain, has a very high CSS-Palm score (Fig. 1C, Table 1, and supplemental Table S2), and plays an important role in neurite outgrowth (20, 48) and synaptic plasticity (20, 23, 24). Thus, we subsequently focused on Ncdn palmitoylation. Using the same screening method, we confirmed that seven candidate proteins (kalirin7, homer 1C, paxillin, Par3, liprin-α2, Rab3A, and KIF5C), which had not been palmitoylated by DHHC3 (Fig. 2A), were not palmitoylated by any DHHC proteins (data not shown).
FIGURE 3.
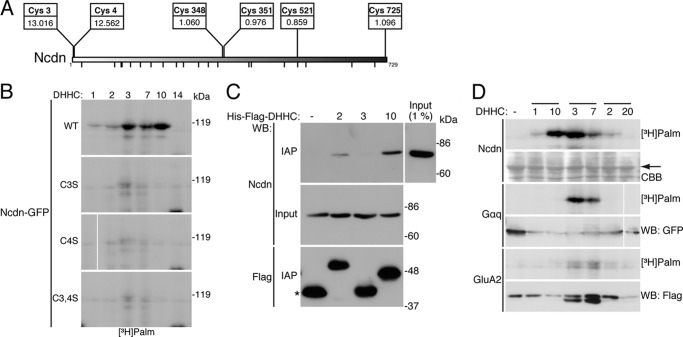
DHHC1, -3, -7, and -10 (z11) palmitoylate Ncdn. A, 23 individual DHHC proteins were co-expressed with Ncdn, TARP γ-8, CaMKIIα, or Syd-1 in HEK293 cells and metabolically labeled with [3H]palmitate, as shown in Fig. 2A. Arrows indicate the positions of palmitoylated substrates. Palmitoylation of Ncdn was enhanced by DHHC1, -3, -7, and -10 (z11), whereas TARP γ-8, CaMKIIα, and Syd-1 were palmitoylated by the DHHC3/7 subfamily. HA-GST was used for mock indicated as –. B, palmitoylation of Ncdn was increased by the DHHC1/10 (z1/11) and DHHC3/7 subfamilies. The band intensity of palmitoylated Ncdn was quantified and normalized to total Ncdn. The -fold increase in relative band intensity (in the presence of DHHC proteins) to the basal intensity (in the absence of DHHC proteins) is indicated below each lane. The average values of three independent experiments are represented. DHHC1 and -10 (z11) belong to the same subfamily in green, and DHHC3 and -7 belong to the other subfamily in blue. C, expression of 23 HA-DHHC proteins was confirmed by Western blotting (WB) with anti-HA antibody (a representative result).
Ncdn Palmitoylation Occurs at Cysteines 3 and 4
Ncdn protein contains 25 cysteine residues, 2 near the amino terminus at positions 3 and 4, and other cysteines scattered throughout the protein. CSS-Palm 2.0 strongly predicted cysteines 3 and 4 as Ncdn palmitoylation sites (Scores are 13.016 and 12.562 at Cys-3 and Cys-4, respectively; Fig. 4A). In contrast, the scores of the other cysteines were less than the 1.8 cut-off value. These scores of Cys-3 and Cys-4 of Ncdn are much higher than those of the other novel substrates, TARP γ-8 (2.461), CaMKIIα (3.765), and Syd-1 (3.078) and even a well known substrate, PSD-95 (5.096) (Fig. 1C and Table 1). In fact, the CSS-Palm score of Ncdn (13.016) was the seventh highest among proteins with high brain enrichment score (≥0.75) (supplemental Table S2). To determine whether Cys-3, Cys-4, or both in Ncdn serves as the palmitoylation sites, those cysteine residues were mutated to serines (Ncdn C3S, Ncdn C4S, and Ncdn C3S,C4S). HEK293 cells were co-transfected with GFP-fused wild-type (WT) or mutant Ncdn together with DHHCs, and palmitoyl proteins were metabolically labeled with [3H]palmitate. Fluorography showed that a mutation on either Cys-3 or Cys-4 and dual mutations completely eliminated palmitoylation of Ncdn (Fig. 4B). These results indicate that the Ncdn palmitoylation sites are Cys-3 and Cys-4 and that DHHC1, -3, -7, and -10 (z11) mediate Ncdn palmitoylation at specific cysteine residues.
FIGURE 4.
Specific palmitoylation of Ncdn by the DHHC1/10 (z1/11) subfamily. A, Cys-3 and Cys-4 of Ncdn are predicted as the most potential cysteines for palmitoylation sites. The positions and CSS-Palm scores of potential cysteines are shown. The small bars at the bottom indicate the positions of cysteines with low scores (<0.8). B, Ncdn palmitoylation sites are Cys-3 and Cys-4. Cys-3, Cys-4, or both were mutated to serine (Ncdn C3S, Ncdn C4S, or Ncdn C3S,C4S). GFP-fused Ncdn WT and mutants were co-expressed with DHHC1, -2, -3, -7, -10 (z11), or -14 in HEK293 cells and analyzed by metabolic labeling assay. Mutations on Cys-3 and/or Cys-4 abolished Ncdn palmitoylation. C, DHHC10 (z11) physically interacted with Ncdn. His6-FLAG- DHHC2, -3 and -10 (z11) were expressed in HEK293 cells, and the lysates were mixed with the membrane fraction from rat brain. Immunoaffinity purified (IAP) His6-FLAG-DHHC proteins were analyzed by Western blotting (WB) with anti-Ncdn or FLAG antibody. An asterisk indicates His6-FLAG-GST expressed instead of DHHCs. D, DHHC1 and DHHC10 (z11) specifically palmitoylate Ncdn. Ncdn-GFP, Gαq-GFP, and GluA2-FLAG were co-expressed with three DHHC subfamily members, DHHC1/10 (z1/11), 3/7, and 2/20 in HEK293 cells and analyzed by metabolic labeling assay. An arrow indicates the position of Ncdn. CBB, Coomassie Brilliant Blue staining.
Because it was reported that some palmitoyl substrates physically interact with their responsible enzymes (49–51), we tested the interaction of Ncdn with DHHC2, -3, and -10 (z11). His6-FLAG-tagged DHHC2, -3, and -10 (z11) were expressed in HEK293 cells, and lysates were mixed with rat brain membrane extracts containing endogenous Ncdn. His6-FLAG-DHHC proteins were tandem affinity-purified with anti-FLAG-antibody-conjugated M2-agarose and then with nickel-nitrilotriacetic acid-agarose. The purified samples were analyzed by Western blotting with anti-Ncdn antibody. DHHC10 (z11), but not DHHC3, preferentially interacted with Ncdn (Fig. 4C). Although DHHC1 was not extracted from the membrane fractions (data not shown), this result suggests that the DHHC1/10 (z1/11) subfamily stably interacts with Ncdn, whereas the DHHC3/7 subfamily palmitoylates Ncdn by transient interaction. We also noted that DHHC2 weakly interacted with Ncdn (Fig. 4C). This is consistent with the data that DHHC2 expression slightly enhanced Ncdn palmitoylation levels (Figs. 3, A and B, and 4, B and D), suggesting that DHHC2 may be also involved in Ncdn palmitoylation. We also found that the DHHC1/10 (z1/11) subfamily specifically palmitoylates Ncdn under the conditions in which the DHHC3/7 subfamily palmitoylates Gαq and GluA2 as well as Ncdn (Fig. 4D). Thus, the DHHC3/7 and DHHC1/10 (z1/11) subfamilies may regulate palmitoylation states of Ncdn differently.
Ncdn Is Localized Specifically in Neuronal Dendrites
Because palmitoylation regulates protein localization, we generated an antibody to Ncdn (N-rNcdn) to examine its subcellular localization in neurons. The antibody to Ncdn specifically recognized a single ∼75-kDa band in rat cultured cortical neurons (Fig. 5A). We first examined the developmental change of Ncdn expression in cultured cortical neurons. At 3 DIV, when an axon just differentiates from the immature neurite, Ncdn was not detected (Fig. 5A). Ncdn expression began to be detected at 7 DIV and was the highest at 14 DIV, when dendrites differentiate from the other immature neurites. Ncdn expression then slightly declined but remained higher than that at 7 DIV. Notably, this expression pattern of Ncdn was very similar to that of mGluR5.
Next, we investigated the subcellular distribution of Ncdn by biochemically fractionating the brain homogenate (Fig. 5B). Ncdn was fractionated into both cytoplasmic (S3) and membrane fractions (P2) and was further fractionated into both TritonX-100-soluble (Sol1) and -insoluble (PSD1 and PSD2) fractions from P2-derived synaptosome fraction. Ncdn was similarly enriched in the S3- and Triton X-100-insoluble fractions, suggesting that there are two major pools, palmitoylated and non-palmitoylated Ncdn.
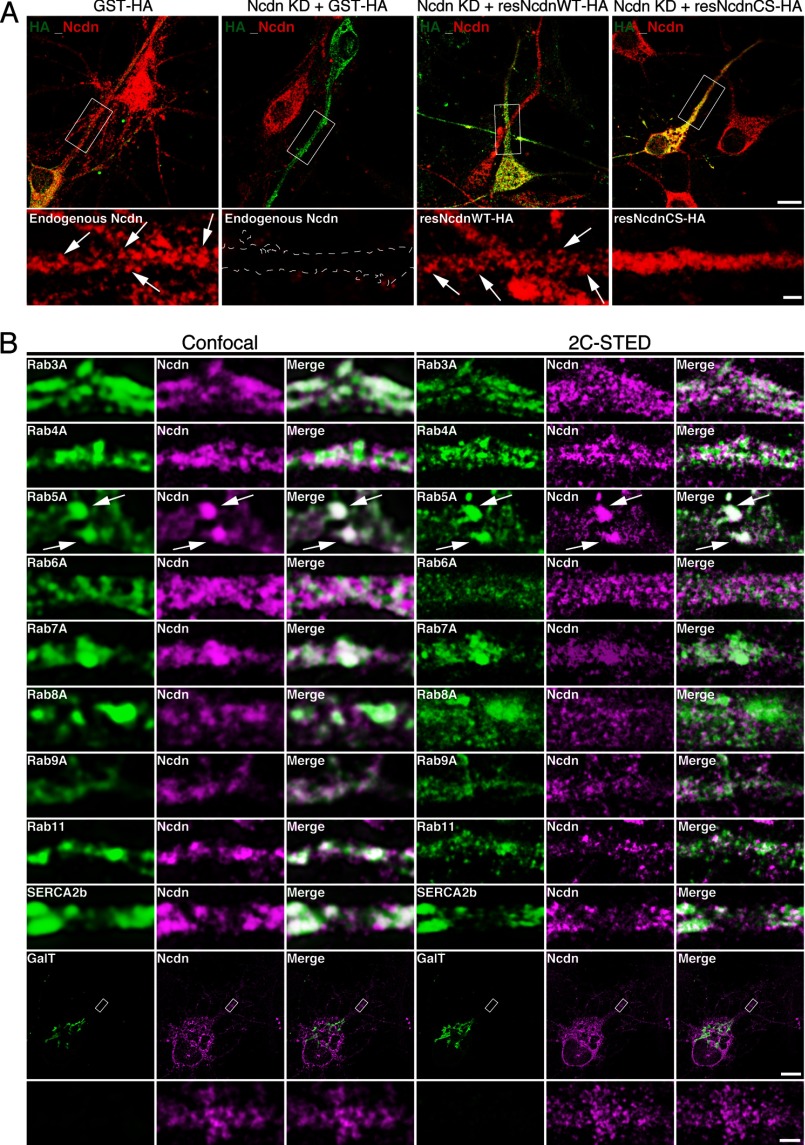
When we immunocytochemically examined the subcellular localization of Ncdn in cultured cortical neurons, strong somato-dendritic signals were observed by the Ncdn antibody (Fig. 5C). This signal is specific as the staining completely disappeared in the validated knockdown vector (miR-Ncdn)-transfected neurons (Fig. 5C). We next tested whether Ncdn is localized in dendrites and/or the axon. Anti-MAP2 and pan-axonal neurofilament antibodies were used as a dendrite marker and an axonal marker, respectively. Ncdn was localized in MAP2-positive dendrites, whereas Ncdn was completely absent in the neurofilament-positive axon (Fig. 5D). Ncdn was distributed in the dendrites and cell body as vesicle-like/punctate structures and was partially co-localized with postsynaptic PSD-95 but not well with presynaptic synaptophysin (Fig. 5E).
Palmitoylation by the DHHC1/10 (z1/11) and DHHC3/7 Subfamilies Targets Ncdn to Rab5-positive Early Endosomes in Dendrites
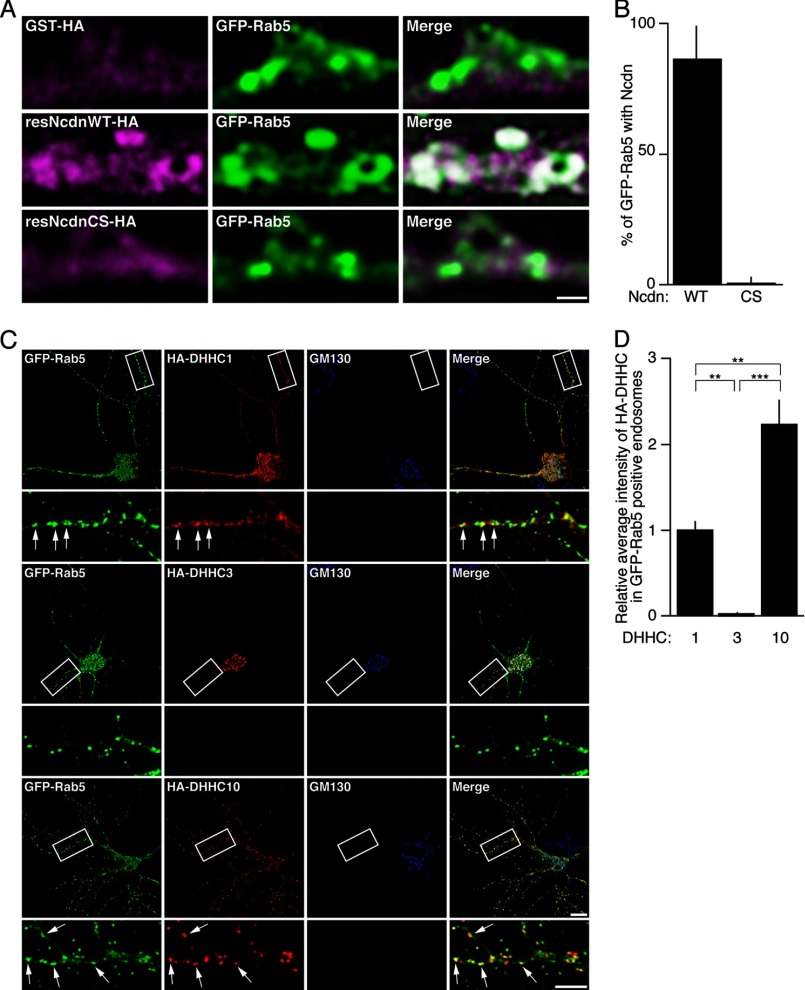
To investigate the role of palmitoylation in Ncdn localization, we took advantage of a molecular replacement strategy that allows simultaneous miRNA-mediated acute knockdown of endogenous Ncdn and expression of miRNA-resistant Ncdn. miRNA-resistant WT (resNcdnWT-HA) or palmitoylation-deficient Ncdn-HA (resNcdnCS-HA) was expressed with miR-Ncdn in neurons. Endogenous Ncdn was specifically detected as vesicle-like/punctate structures in dendrites (Figs. 5C and 6A). This Ncdn staining disappeared after Ncdn knockdown. Under these conditions WT Ncdn (resNcdnWT-HA) showed similar vesicle-like structures as did endogenous Ncdn. In contrast, palmitoylation-deficient resNcdnCS-HA was distributed diffusely in dendrites and the cell body (Fig. 6A), indicating that palmitoylation of Ncdn is essential for its targeting to vesicle-like structures. To identify these Ncdn-positive vesicle-like structures, we next compared the localization of Ncdn with that of various subcellular marker proteins: Rab3A (synaptic vesicle), Rab4A (recycling endosome), Rab5A (early endosome), Rab6A (medial/trans Golgi), Rab7A (late endosome), Rab8A (Trans-Golgi-to-plasma membrane transport), Rab9A (late endosome), Rab11A (recycling endosome), SERCA2b (endoplasmic reticulum), and GalT (Golgi). When we observed their localization with conventional confocal microscopy, Ncdn seemed to be overlapped with various marker proteins including Rab3A, Rab5A, Rab7A, and Rab11A (Fig. 6B). However, super-resolution imaging by STED microscopy revealed that only Rab5A was substantially co-localized with Ncdn (Figs. 6B and 7A). To directly test whether early endosomal localization of Ncdn is palmitoylation-dependent, GFP-Rab5 and either resNcdnWT-HA or resNcdnCS-HA were co-expressed with miR-Ncdn in neurons. About 87% of Rab5-positive vesicles were co-localized with accumulation of resNcdnWT-HA, whereas Rab5-labeled vesicles were hardly co-localized with resNcdnCS-HA (Fig. 7, A and B). Interestingly, the DHHC1/10 (z1/11) subfamily, but not the Golgi-resident DHHC3 (29, 30), was co-localized with Rab5-positive endosomes in dendrites (Fig. 7C). The quantitative data indicates that DHHC10 (z11) was more enriched in Rab5-positive endosomes than DHHC1 in hippocampal neurons (Fig. 7D).
FIGURE 6.
Ncdn is localized in Rab5-positive endosomes. A, miRNA-resistant wild-type Ncdn (resNcdnWT-HA) as well as endogenous Ncdn are localized in vesicle-like structures (arrows), whereas palmitoylation-deficient Ncdn (resNcdnCS-HA) is localized diffusedly in dendrites. Hippocampal neurons were transfected with a molecular replacement plasmid (at 4 DIV) that allows co-cistronic expression of miRNA-resistant Ncdn (WT or CS) and the miRNA. Neurons were then fixed and immunostained with anti-HA (green) and anti-Ncdn (red) at 12 DIV. Dashed lines indicate the dendrite outline of a neuron in which Ncdn was knocked down. Upper bar, 10 μm; lower bar, 2 μm. B, to characterize subcellular localization of Ncdn, hippocampal neurons were transfected with GFP- or V5-tagged subcellular markers and immunostained with anti-Ncdn and either anti-GFP or anti-V5 antibodies. Only Rab5A was overlapped with Ncdn under two color-STED (2C-STED) images (arrows). Confocal and STED images were sequentially acquired in the same regions. Bars, 10 μm (upper) and 1 μm (lower).
FIGURE 7.
Localization of Ncdn to Rab5-positive endosomes is palmitoylation-dependent. A, GFP-Rab5 and either resNcdnWT-HA or resNcdnCS-HA were co-expressed with miR-Ncdn in neurons. Then neurons were immunostained with anti-GFP and anti-HA antibodies. Bar, 1 μm. B, the percentages of GFP-Rab5-positive vesicles overlapped with Ncdn-HA were quantified. n = 6 neurons. C and D, DHHC1 and -10 (z11) are co-localized with Rab5-positive early endosomes. Hippocampal neurons were transfected with HA- DHHC1, -3, or -10 (z11) together with GFP-Rab5 and immunostained with anti-HA, anti-GFP, and anti-GM130 (Golgi marker) antibodies. HA-DHHC1 and -10 (z11) were co-localized with GFP-Rab5 (arrows), whereas DHHC3 was localized in the Golgi apparatus (C). The average fluorescence intensities of HA-DHHC in GFP-Rab5-positive endosomes were quantified blindly and normalized to the average fluorescence intensities of HA-DHHC in the Golgi apparatus (D). The bar graph presents the relative values to the average intensity of HA-DHHC1. Error bars indicate S.E. n = 5 neurons. Analysis of variance p < 0.001; Tukey's HSD, **, p < 0.01; ***, p < 0.001. Bars, 10 μm (upper) and 5 μm (lower).
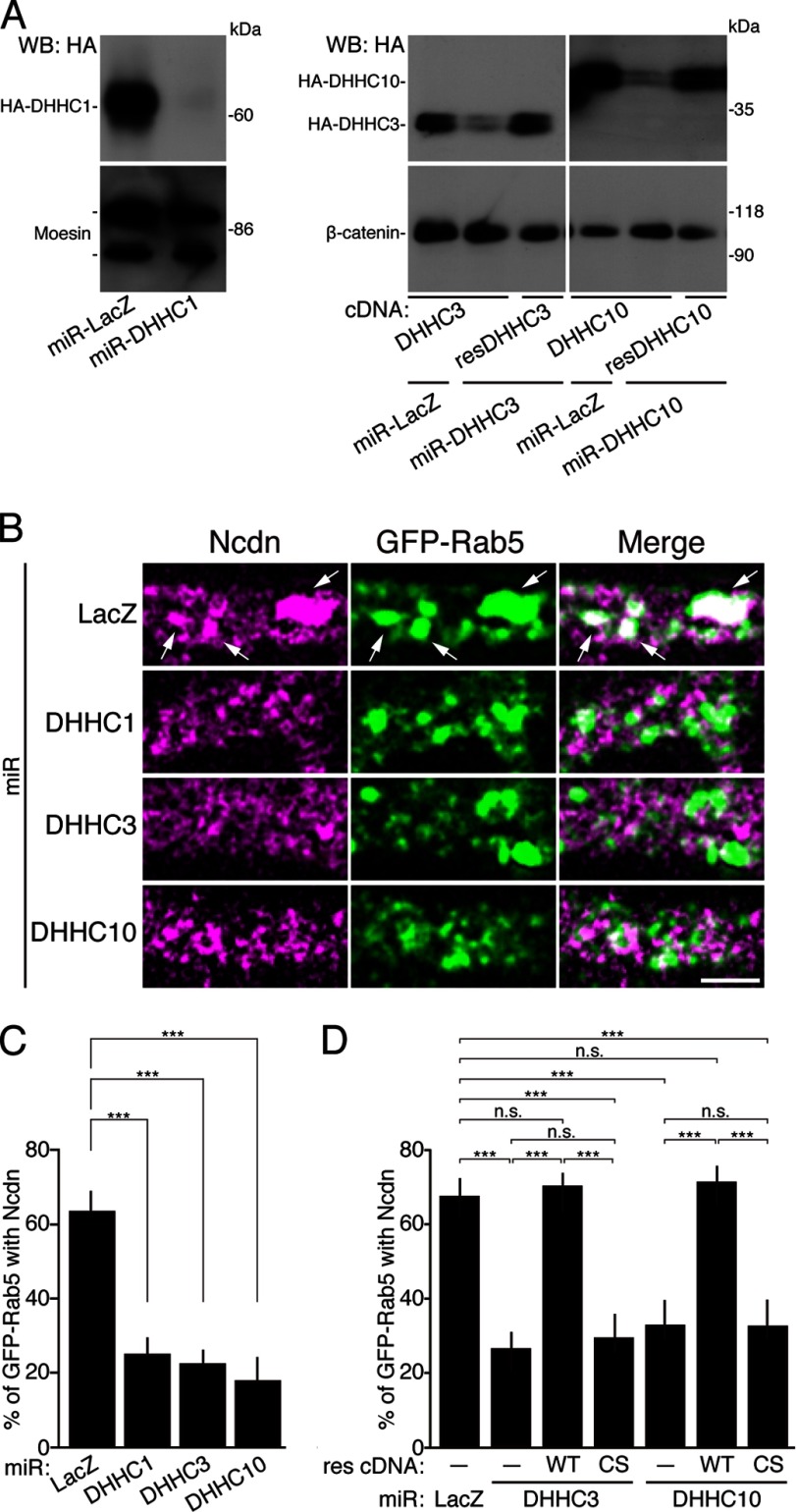
Finally, to investigate whether the DHHC1/10 (z1/11) and/or DHHC3/7 subfamilies mediate the targeting of Ncdn to Rab5-positive endosomes, we knocked-down DHHC1, -3, or -10 (z11). DHHC7, another member of DHHC3/7 subfamily, was excluded from the experiment because of the minimum expression in neurons (29). We found that knockdown of DHHC1, -3, or -10 (z11) by the validated miRNA (miR-DHHCs) resulted in the significant loss of Ncdn from Rab5-positive endosomes (Fig. 8, A–C). This knockdown phenotype was rescued by the miRNA-resistant wild-type DHHC enzyme but not by the catalytically inactive CS mutant (Fig. 8D). These results indicate that palmitoylation of Ncdn by DHHC1/3/10 (z1/3/11) is required for proper localization of Ncdn to Rab5-positive endosomes. Thus, we conclude that Ncdn and the DHHC1/10 (z1/11) and DHHC3/7 subfamilies are novel substrate-enzyme pairs and that the endosome-localized DHHC1/10 (z1/11) subfamily and Golgi-localized DHHC3 non-redundantly regulate the localization of Ncdn to endosomes in neurons.
FIGURE 8.
An essential role for Ncdn palmitoylation by DHHC1/3/10 (z1/3/11) in its localization to early endosomes. A, HEK293 cells were co-transfected with the indicated knockdown (miR) and HA-tagged DHHC expression vectors. The cell lysates were analyzed by Western blotting (WB) with anti-HA and either moesin or β-catenin antibodies. miR-LacZ, control miRNA targeting to LacZ (β-galactosidase); resDHHC, miRNA-resistant DHHC proteins. B, neurons were co-transfected with GFP-Rab5 and indicated miR expression vectors. Then neurons were immunostained with anti-Ncdn, anti-GFP, and anti-HA (not shown) antibodies and analyzed by STED microscopy. Arrows indicate GFP-Rab5 vesicles overlapped with Ncdn. Bar, 0.5 μm. C, knockdown of DHHC1, -3, or -10 (z11) resulted in a significant loss of Ncdn from Rab5-positive endosomes. n = 45 neurons. Analysis of variance, p < 0.001, and Dunnett test, ***, p < 0.001. D, the knockdown phenotype was rescued by the miRNA-resistant (res) WT DHHC enzyme but not by the catalytically inactive CS mutant. Error bars indicate S.E. n = 45 neurons. Analysis of variance p < 0.001; Tukey's HSD, ***, p < 0.001; n.s., not significant.
DISCUSSION
In Silico Prediction of Palmitoyl Proteins
The recent development of purification methods for palmitoylated proteins (the ABE method or click chemistry) has greatly contributed to the identification of novel palmitoyl proteins (11–16). However, these methods have several potential problems that could lead to false-negative or false-positive results. The biochemical properties of proteins, such as detergent insolubility and post-translational modifications may hinder the purification and mass spectrometry detection, leading to false-negative results. Also, non-palmitoylated proteins might be co-purified with palmitoyl substrates, leading to false-positive results. In fact, non-palmitoylated, secreted protein LGI1, which indirectly associates with palmitoylated PSD-95 through ADAM22, was listed as a candidate palmitoyl protein (12). In this study we searched for novel palmitoyl substrates through global in silico screening by the CSS-Palm 2.0 prediction program, which requires only protein sequences and ignores any biochemical properties of target proteins. By automatically applying all mouse protein sequences to CSS-Palm 2.0, we classified the 59,136 obtained protein sequences according to the CSS-Palm scores (supplemental Table S1). In fact, this list included many transmembrane proteins such as G-protein-coupled receptors, which are difficult to be solubilized for purification. Of the 17 candidates we selected, 10 proteins were experimentally verified as novel palmitoyl substrates that were not captured by the proteomic methods. This indicates that the in silico approach is useful for the discovery of novel palmitoyl substrates and complementally functions with the experimental methods. However, of the 17 candidates, palmitoylation of seven proteins were not detected by metabolic labeling assay (i.e. false-positive results). In terms of accuracy, this prediction program may have room for improvement. CSS-Palm 2.0 was developed by referring 263 palmitoylation sites from 109 proteins, which had been identified experimentally (25). Very recently, CSS-Palm has been updated to Version 3.0 based on 439 palmitoylation sites from 194 proteins. We repeated the in silico analysis with the 17 candidate substrates by CSS-Palm 3.0 (supplemental Table S3). However, the scores predicted by CSS-Palm 3.0 did not always reflect our experimental results. For example, the score of palmitoyl TARP γ-2 was 1.9 by CSS-Palm 2.0, whereas the score was 0.7 by CSS-Palm 3.0. Further improvements need to be made for more precise prediction. For example, the increased number of references of known substrates and their palmitoylation sites and the clarification of DHHC subfamily-specific consensus sequences will make the software a much more powerful tool.
Role of Ncdn Palmitoylation in Dendritic Early Endosome Localization
We found that Ncdn, a neuron-specific protein, is localized at the cell body, dendritic shafts, and dendritic spines as vesicle-like structures but not in the axon (Fig. 5, C–E). Moreover, super-resolution imaging with STED microscopy revealed that Ncdn is specifically targeted to Rab5-positive dendritic early endosomes in a palmitoylation-dependent manner. One may wonder where Ncdn is palmitoylated and why Ncdn is specifically associated with early endosomal membranes. We noted that the DHHC1/10 (z1/11) subfamily (although exogenously expressed) was uniquely co-localized with Rab5-labeled early endosomes in neurons among Ncdn-palmitoylating enzymes (DHHC1, -3, -7, and -10 (z11)) (Fig. 7C). In contrast to DHHC1/10 (z1/11), the DHHC3/7 subfamily is exclusively localized at the Golgi apparatus (Fig. 7C) (29, 30). Given that DHHC1 and -10 (z11) represent palmitoylating enzymes associated with early endosomes, DHHC1 and/or 10 (z11) may locally palmitoylate Ncdn and thereby actively recruit Ncdn to early endosomal membranes. Specific antibodies for endogenous DHHC1 and DHHC10 (z11) are needed to address this possibility.
Physiological Role of Ncdn Palmitoylation
Previous studies showed that overexpression of Ncdn promotes neurite outgrowth in Neuro2a cells (20, 48). The first 100 amino acids of Ncdn, including palmitoylation sites, were thought to contain the activity site that induces the differentiation (48). In polarized neurons, which develop two differential processes, an axon and dendrites, we found that Ncdn protein is highly expressed during dendrite development (Fig. 5A) and localized specifically in dendrites (Fig. 5, D and E). These results imply that Ncdn is involved in dendritogenesis. In fact, we found that overexpression of NcdnWT in hippocampal neurons significantly increased the total length of dendrites compared with control neurons (data not shown). However, Ncdn palmitoylation had no effects on dendrite outgrowth because there was no significant difference in effect of overexpression of NcdnWT versus CS mutant. We also examined whether Ncdn palmitoylation affects dendrite morphology using the molecular replacement strategy. However, we could see no significant effects of knockdown or molecular replacement of Ncdn on dendrite morphology (dendrite length and branching) (data not shown). Further studies on physiological roles of Ncdn and its palmitoylation in neurons will be required.
Ncdn palmitoylation may participate in the regulation of synaptic plasticity. Ncdn was originally identified as an inducible gene during the induction of long-term potentiation (20). Very recently, it was reported that Ncdn interacts with mGluR5 and increases its cell surface expression (24). Forebrain-specific Ncdn KO mice show the reduction of dihydroxyphenylglycine-induced long term potentiation and impairment of the induction of long term potentiation in the Schaffer collateral to CA1 synapses (24). Given that Rab5 is required for internalization of AMPAR during long term depression (52), Ncdn might regulate internalization of AMPAR, directly acting on early endosome dynamics or indirectly modulating a signal through mGluR5. Because conditional Ncdn KO mice showed apparent pathological phenotypes (epileptic seizures and schizophrenia-relevant behaviors) (23, 24), genetic experiments using knock-in mice in which Ncdn palmitoylation-deficient mutant is expressed should reveal the roles of Ncdn palmitoylation in these pathophysiological phenotypes.
In this study we attempted the global identification of palmitoyl substrates by using an in silico computational approach and identified several unexpected substrates. Among them, we focused on Ncdn palmitoylation and found that the DHHC1/10 (z1/11) and DHHC3/7 subfamilies enhance Ncdn palmitoylation and regulate its localization to early endosomes in hippocampal neurons. This finding demonstrates for the first time that the DHHC1/10 (z1/11) subfamily proteins are functional palmitoylating enzymes in mammalian cells. In the final stage of our manuscript preparation, another group reported that human DHHC1 and -10 (z11) show the palmitoylating activity in the yeast system (53). Thus, this in silico approach can complement experimental approaches to clarify protein functions regulated by post-translational modifications.
Supplementary Material
Acknowledgments
We greatly thank Drs. Hiroko Iwanari, Yasuhiro Mochizuki, and Takao Hamakubo (University of Tokyo) and Dr. Akiyoshi Fukamizu (Tsukuba University, Japan) for kind support. We gratefully appreciate the generous gifts of plasmids from Drs. Franck Perez (Institut Curie), Ulli Bayer (University of Colorado, Denver, CO), Yasuo Mori (Kyoto University), Ardem Patapoutian (The Scripps Research Institute), Makoto Tominaga (National Institute for Physiological Sciences), Akihiro Yamanaka (Nagoya University), David Bredt (Johnson and Johnson Research and Development), Gary Banker (Oregon Health and Science University), Akihiko Kato (Eli Lilly), and Xin-Ming Ma (University of Connecticut Health and Science Center) (see “Experimental Procedures” for details). We also thank Drs. Jun Noritake, Tsuyoshi Iwanaga, and Norihiko Yokoi for technical assistance and all members of Fukata laboratory for kind help.
This work was supported, in whole or in part, by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 21680029 and 23110520 (to Y. F.) and 20670005 (to M. F.) and by the Funding Program for Next Generation World-Leading Researchers (LS123; to M. F.).

This article contains supplemental Tables 1–3.
- ABE
- acyl-biotinyl exchange
- TARP
- transmembrane AMPAR regulatory protein
- CaMKIIα
- Ca2+/calmodulin-dependent protein kinase IIα
- Ncdn
- neurochondrin
- TRP channel
- transient receptor potential channel
- STED
- stimulated emission depletion
- mGluR5
- metabotropic glutamate receptor 5
- CNIH
- cornichon
- miRNA or miR
- microRNA
- AMPAR
- AMPA receptor
- DIV
- days in vitro.
REFERENCES
- 1. el-Husseini Ael-D, Bredt D. S. (2002) Protein palmitoylation. A regulator of neuronal development and function. Nat. Rev. Neurosci. 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 2. Resh M. D. (2006) Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2, 584–590 [DOI] [PubMed] [Google Scholar]
- 3. Linder M. E., Deschenes R. J. (2007) Palmitoylation. Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 4. Fukata Y., Fukata M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 5. Greaves J., Chamberlain L. H. (2011) DHHC palmitoyl transferases. Substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 6. Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. (2002) Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- 7. Zhao L., Lobo S., Dong X., Ault A. D., Deschenes R. J. (2002) Erf4p and Erf2p form an endoplasmic reticulum-associated complex involved in the plasma membrane localization of yeast Ras proteins. J. Biol. Chem. 277, 49352–49359 [DOI] [PubMed] [Google Scholar]
- 8. Roth A. F., Feng Y., Chen L., Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 10. Drisdel R. C., Green W. N. (2004) Labeling and quantifying sites of protein palmitoylation. Biotechniques 36, 276–285 [DOI] [PubMed] [Google Scholar]
- 11. Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Global analysis of protein palmitoylation in yeast. Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin B. R., Cravatt B. F. (2009) Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yount J. S., Moltedo B., Yang Y. Y., Charron G., Moran T. M., López C. B., Hang H. C. (2010) Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat. Chem. Biol. 6, 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martin B. R., Wang C., Adibekian A., Tully S. E., Cravatt B. F. (2012) Global profiling of dynamic protein palmitoylation. Nat. Methods 9, 84–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mirza S. P., Halligan B. D., Greene A. S., Olivier M. (2007) Improved method for the analysis of membrane proteins by mass spectrometry. Physiol. Genomics 30, 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santoni V., Molloy M., Rabilloud T. (2000) Membrane proteins and proteomics. Un amour impossible? Electrophoresis 21, 1054–1070 [DOI] [PubMed] [Google Scholar]
- 19. Forrester M. T., Hess D. T., Thompson J. W., Hultman R., Moseley M. A., Stamler J. S., Casey P. J. (2011) Site-specific analysis of protein S-acylation by resin-assisted capture. J. Lipid Res. 52, 393–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shinozaki K., Maruyama K., Kume H., Kuzume H., Obata K. (1997) A novel brain gene, norbin, induced by treatment of tetraethylammonium in rat hippocampal slice and accompanied with neurite-outgrowth in neuro 2a cells. Biochem. Biophys. Res. Commun. 240, 766–771 [DOI] [PubMed] [Google Scholar]
- 21. Wang H., Nong Y., Bazan F., Greengard P., Flajolet M. (2010) Norbin. A promising central nervous system regulator. Commun. Integr. Biol. 3, 487–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mochizuki R., Dateki M., Yanai K., Ishizuka Y., Amizuka N., Kawashima H., Koga Y., Ozawa H., Fukamizu A. (2003) Targeted disruption of the neurochondrin/norbin gene results in embryonic lethality. Biochem. Biophys. Res. Commun. 310, 1219–1226 [DOI] [PubMed] [Google Scholar]
- 23. Dateki M., Horii T., Kasuya Y., Mochizuki R., Nagao Y., Ishida J., Sugiyama F., Tanimoto K., Yagami K., Imai H., Fukamizu A. (2005) Neurochondrin negatively regulates CaMKII phosphorylation, and nervous system-specific gene disruption results in epileptic seizure. J. Biol. Chem. 280, 20503–20508 [DOI] [PubMed] [Google Scholar]
- 24. Wang H., Westin L., Nong Y., Birnbaum S., Bendor J., Brismar H., Nestler E., Aperia A., Flajolet M., Greengard P. (2009) Norbin is an endogenous regulator of metabotropic glutamate receptor 5 signaling. Science 326, 1554–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008) CSS-Palm 2.0. An updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Apweiler R., Bairoch A., Wu C. H. (2004) Protein sequence databases. Curr. Opin. Chem. Biol. 8, 76–80 [DOI] [PubMed] [Google Scholar]
- 27. Ogasawara O., Otsuji M., Watanabe K., Iizuka T., Tamura T., Hishiki T., Kawamoto S., Okubo K. (2006) BodyMap-Xs. Anatomical breakdown of 17 million animal ESTs for cross-species comparison of gene expression. Nucleic Acids Res. 34, D628–D631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kato A. S., Gill M. B., Ho M. T., Yu H., Tu Y., Siuda E. R., Wang H., Qian Y. W., Nisenbaum E. S., Tomita S., Bredt D. S. (2010) Hippocampal AMPA receptor gating controlled by both TARP and cornichon proteins. Neuron 68, 1082–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., Matsuura Y., Bredt D. S., Hamakubo T., Fukata M. (2009) Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J. Cell Biol. 186, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsutsumi R., Fukata Y., Noritake J., Iwanaga T., Perez F., Fukata M. (2009) Identification of G protein α subunit-palmitoylating enzyme. Mol. Cell. Biol. 29, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hayashi T., Rumbaugh G., Huganir R. L. (2005) Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron 47, 709–723 [DOI] [PubMed] [Google Scholar]
- 32. Hayashi T., Thomas G. M., Huganir R. L. (2009) Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron 64, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Topinka J. R., Bredt D. S. (1998) N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron 20, 125–134 [DOI] [PubMed] [Google Scholar]
- 34. El-Husseini A. E., Topinka J. R., Lehrer-Graiwer J. E., Firestein B. L., Craven S. E., Aoki C., Bredt D. S. (2000) Ion channel clustering by membrane-associated guanylate kinases. Differential regulation by N-terminal lipid and metal binding motifs. J. Biol. Chem. 275, 23904–23910 [DOI] [PubMed] [Google Scholar]
- 35. DeSouza S., Fu J., States B. A., Ziff E. B. (2002) Differential palmitoylation directs the AMPA receptor-binding protein ABP to spines or to intracellular clusters. J. Neurosci. 22, 3493–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shepherd J. D., Huganir R. L. (2007) The cell biology of synaptic plasticity. AMPA receptor trafficking. Annu. Rev. Cell Dev. Biol. 23, 613–643 [DOI] [PubMed] [Google Scholar]
- 37. Nicoll R. A., Tomita S., Bredt D. S. (2006) Auxiliary subunits assist AMPA-type glutamate receptors. Science 311, 1253–1256 [DOI] [PubMed] [Google Scholar]
- 38. Jackson A. C., Nicoll R. A. (2011) The expanding social network of ionotropic glutamate receptors. TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen L., Chetkovich D. M., Petralia R. S., Sweeney N. T., Kawasaki Y., Wenthold R. J., Bredt D. S., Nicoll R. A. (2000) Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408, 936–943 [DOI] [PubMed] [Google Scholar]
- 40. Hashimoto K., Fukaya M., Qiao X., Sakimura K., Watanabe M., Kano M. (1999) Impairment of AMPA receptor function in cerebellar granule cells of ataxic mutant mouse stargazer. J. Neurosci. 19, 6027–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schwenk J., Harmel N., Zolles G., Bildl W., Kulik A., Heimrich B., Chisaka O., Jonas P., Schulte U., Fakler B., Klöcker N. (2009) Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 42. Wyllie D. J., Nicoll R. A. (1994) A role for protein kinases and phosphatases in the Ca2+-induced enhancement of hippocampal AMPA receptor-mediated synaptic responses. Neuron 13, 635–643 [DOI] [PubMed] [Google Scholar]
- 43. Penzes P., Jones K. A. (2008) Dendritic spine dynamics. A key role for kalirin-7. Trends Neurosci. 31, 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brakeman P. R., Lanahan A. A., O'Brien R., Roche K., Barnes C. A., Huganir R. L., Worley P. F. (1997) Homer. A protein that selectively binds metabotropic glutamate receptors. Nature 386, 284–288 [DOI] [PubMed] [Google Scholar]
- 45. Kato A., Ozawa F., Saitoh Y., Fukazawa Y., Sugiyama H., Inokuchi K. (1998) Novel members of the Vesl/Homer family of PDZ proteins that bind metabotropic glutamate receptors. J. Biol. Chem. 273, 23969–23975 [DOI] [PubMed] [Google Scholar]
- 46. Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004) Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J. Cell Biol. 167, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van Itallie C. M., Gambling T. M., Carson J. L., Anderson J. M. (2005) Palmitoylation of claudins is required for efficient tight-junction localization. J. Cell Sci. 118, 1427–1436 [DOI] [PubMed] [Google Scholar]
- 48. Schwaibold E. M., Brandt D. T. (2008) Identification of neurochondrin as a new interaction partner of the FH3 domain of the diaphanous-related formin Dia1. Biochem. Biophys. Res. Commun. 373, 366–372 [DOI] [PubMed] [Google Scholar]
- 49. Fang C., Deng L., Keller C. A., Fukata M., Fukata Y., Chen G., Lüscher B. (2006) GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses. J. Neurosci. 26, 12758–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fernández-Hernando C., Fukata M., Bernatchez P. N., Fukata Y., Lin M. I., Bredt D. S., Sessa W. C. (2006) Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J. Cell Biol. 174, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M. R., El-Husseini A. (2009) Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 23, 2605–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brown T. C., Tran I. C., Backos D. S., Esteban J. A. (2005) NMDA receptor-dependent activation of the small GTPase Rab5 drives the removal of synaptic AMPA receptors during hippocampal LTD. Neuron 45, 81–94 [DOI] [PubMed] [Google Scholar]
- 53. Ohno Y., Kashio A., Ogata R., Ishitomi A., Yamazaki Y., Kihara A. (2012) Analysis of substrate specificity of human DHHC protein acyltransferases using a yeast expression system. Mol. Biol. Cell 23, 4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.