Background: ppGalNAc transferases, which initiate O-glycosylation, possess a poorly understood lectin domain.
Results: The lectin domain directs glycosylation in an N- or C- terminal direction in an isoform-specific manner.
Conclusion: Unanticipated isoform-specific directionality was revealed for modification of glycopeptide substrates.
Significance: A novel mechanism of controlling of mucin type O-glycosylation has been discovered based on tethered lectin domains specifying N- or C-terminal modification of glycopeptide substrates.
Keywords: Convertases, Glycobiology, Glycoprotein Biosynthesis, Glycosyltransferases, Lectin, Mucins, Edman Sequencing, O-Glycosylation, Control of Glycosylation, Random Glycopeptide
Abstract
Mucin type O-glycosylation is initiated by a large family of polypeptide GalNAc transferases (ppGalNAc Ts) that add α-GalNAc to the Ser and Thr residues of peptides. Of the 20 human isoforms, all but one are composed of two globular domains linked by a short flexible linker: a catalytic domain and a ricin-like lectin carbohydrate binding domain. Presently, the roles of the catalytic and lectin domains in peptide and glycopeptide recognition and specificity remain unclear. To systematically study the role of the lectin domain in ppGalNAc T glycopeptide substrate utilization, we have developed a series of novel random glycopeptide substrates containing a single GalNAc-O-Thr residue placed near either the N or C terminus of the glycopeptide substrate. Our results reveal that the presence and N- or C-terminal placement of the GalNAc-O-Thr can be important determinants of overall catalytic activity and specificity that differ between transferase isoforms. For example, ppGalNAc T1, T2, and T14 prefer C-terminally placed GalNAc-O-Thr, whereas ppGalNAc T3 and T6 prefer N-terminally placed GalNAc-O-Thr. Several transferase isoforms, ppGalNAc T5, T13, and T16, display equally enhanced N- or C-terminal activities relative to the nonglycosylated control peptides. This N- and/or C-terminal selectivity is presumably due to weak glycopeptide binding to the lectin domain, whose orientation relative to the catalytic domain is dynamic and isoform-dependent. Such N- or C-terminal glycopeptide selectivity provides an additional level of control or fidelity for the O-glycosylation of biologically significant sites and suggests that O-glycosylation may in some instances be exquisitely controlled.
Introduction
Mucin type protein O-glycosylation, as defined by the α-GalNAc-O-Ser/Thr linkage, is one of the most common types of protein glycosylation found in higher organisms. This modification is initiated in the Golgi by a large family (∼20 members in mammals and about half the number in the fly and Caenorhabditis elegans (see Ref. 1 for a review) of polypeptide GalNAc transferases (ppGalNAc Ts)3 that transfer GalNAc from UDP-GalNAc to Ser and Thr. Subsequent chain elongation of the GalNAc residue by the stepwise action of additional specific transferases results in a vast array of heterogeneous O-linked glycan structures. In contrast to the initiating ppGalNAc Ts, nearly all of these O-glycan elongating transferases occur in small families of 1–4 members. Despite its prevalence, the biological roles and control of mucin type O-glycosylation remain incompletely understood.
Mucin type O-glycans have typically been associated with so-called mucin domains, where high numbers of Ser and Thr residues are glycosylated, producing a chemical- and protease-resistant extended peptide structural motif. Glycoproteins containing O-glycosylated mucin domains commonly function in the protection of the cell surface and the modulation of cell-cell interactions and hence play important roles, for example in inflammation, the immune response, metastasis, and tumorigenesis (2–6). Because the role of the mucin domain in these glycoproteins has been thought to produce a stable extended hydrophilic motif, the actual glycosylation pattern has usually been thought to be of relatively low importance. This is supported by the very low sequence conservation across mammalian and insect species of the glycosylated domains of the secreted mucins (7, 8). Nevertheless, it cannot be ruled out that in some cases, O-glycosylated domains may present a molecular code for the specific recognition of additional binding partners, thus playing potential roles in protein sorting, targeting, and turnover (9–15).
Recent proteome-wide analysis demonstrates that mucin type O-glycosylation is widely distributed on most proteins passing through the secretory pathway and that most have only one or few isolated O-linked glycans (16–18). Thus, it is likely that O-linked glycans at specific sites may play other important biological roles. One emerging role is the co-regulation of the proprotein processing process (19, 20), where O-glycans modulate cleavage of nearby proprotein convertase cleavage sites (i.e. phosphaturic factor, FGF23, and angiopoietin-like protein 3 (ANGPTL3) (21, 22)). Other studies have shown that specific O-glycosylation can modulate receptor activities (i.e. TGF-βR II and the death receptor DR5) by presently unknown mechanisms (23, 24). Several of the above O-glycosylation events are targeted by specific ppGalNAc T isoforms (i.e. ppGalNAc T2,T3, T14, and T16); therefore, some occurrences of O-glycosylation may be actively regulated to serve as modulators of complex biological processes and perhaps even signaling. Unsurprisingly, a number of distinct ppGalNAc T isoforms have been shown to cause or be associated with human disease. The loss of ppGalNAc T3 causes familial tumoral calcinosis due to abnormal processing of FGF23 (21, 25). ppGalNAc T2, T5, and T19 (also known as WBSCR17)4 are linked to Williams-Beuren syndrome (15), hereditary multiple exostoses (26), and levels of HDL cholesterol and coronary artery disease (22, 27, 28), respectively, whereas ppGalNAc T3, T6, T7, T9, and T12 are associated with various cancers (29–34). Except for the known role of ppGalNAc T3 in modulating the cleavage and inactivation of FGF23, the mechanisms for causing disease by the other transferase isoforms are still uncertain.
Recent studies have further demonstrated the critical role of O-glycosylation in vertebrate and invertebrate development (35–39). In the fly, several ppGalNAc T isoforms are required to complete development (38), whereas in the mouse, ppGalNAc T1 modulates salivary gland organogenesis (40). Interestingly, the loss of the elongating transferase, T-synthase (which adds a β-Gal to the 3-position of the peptide GalNAc) leads to embryonic lethality in the mouse (35). Although a few target proteins have been implicated in the above studies, little is known about the actual site(s) of glycosylation and their specificity, and even less is known of the actual molecular mechanism(s) leading to their biological function.
Structurally, members of the ppGalNAc T family (except h-ppGalNAc T20 (41)) possess a unique two-domain architecture consisting of a transmembrane tethered N-terminal catalytic domain linked via a short flexible segment to a C-terminal ricin-like lectin domain containing three potential carbohydrate binding sites (see Fig. 1 for the crystal structure of ppGalNAc T2 with bound EA2 peptide substrate (42)). Presently, the roles of the catalytic and lectin domains in peptide and glycopeptide recognition and specificity remain unclear. Our recent studies utilizing short random (glyco)peptide substrates have shown that the ppGalNAc Ts possess specific binding preferences that vary among isoforms for peptide and even GalNAc-O-Ser/Thr-containing glycopeptide substrates (43–45). Interestingly, we and others have shown that the placement of a neighboring GalNAc-O-Ser/Thr residue near a glycosylation site produces a range of transferase-specific effects (i.e. a relative inhibition of glycosylation (i.e. ppGalNAc T1 and T2 (46, 47)), an alteration or shift in glycosylation site (ppGalNAc T2 and T4 (48–51)), or even a large apparent rate enhancement (i.e. requirement for glycosylated substrate) (ppGalNAc T7 and T10 (45, 51–53)). Interestingly, the alteration in site preference observed for ppGalNAc T2 and T4 resides in their lectin domain (48, 50, 51), whereas the nearly absolute glycopeptide requirement of ppGalNAc T10 resides completely in its catalytic domain (45, 51). Recent studies characterizing the properties of the lectin domains of ppGalNAc T2 and T3 further suggest that they may recognize glycopeptide sequence context (54) and only glycopeptides having O-linked α-GalNAc residues (55), respectively. Because, to date, there have been no truly systematic studies of ppGalNAc T glycopeptide substrate utilization, we have now extended our studies to the function of the lectin domain, utilizing a series of random glycopeptides containing N- or C- terminally placed GalNAc-O-Thr residues. Our results show that the presence and the N- or C-terminal location of a GalNAc-O-Thr site in these glycopeptides are indeed important determinants of the overall catalytic activity and specificity of these enzymes, which can significantly differ between transferase isoforms. Because of the large differences in glycosylation rates observed for some isoforms between an N- or C-terminally placed GalNAc-O-Thr, we believe we have now uncovered another level of control of mucin type O-glycosylation that will further advance our understanding of the regulation of this important modification.
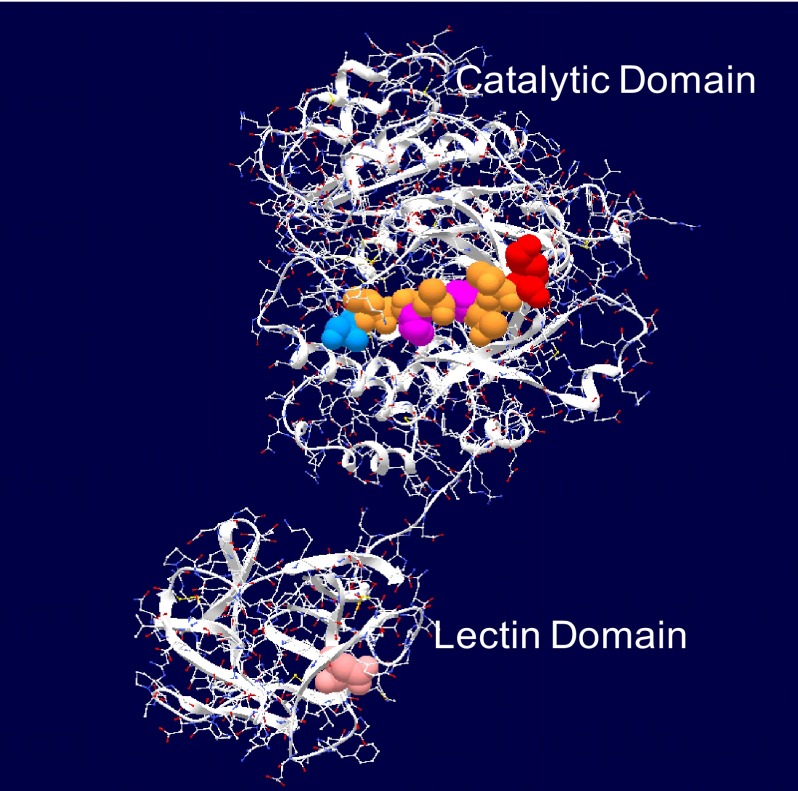
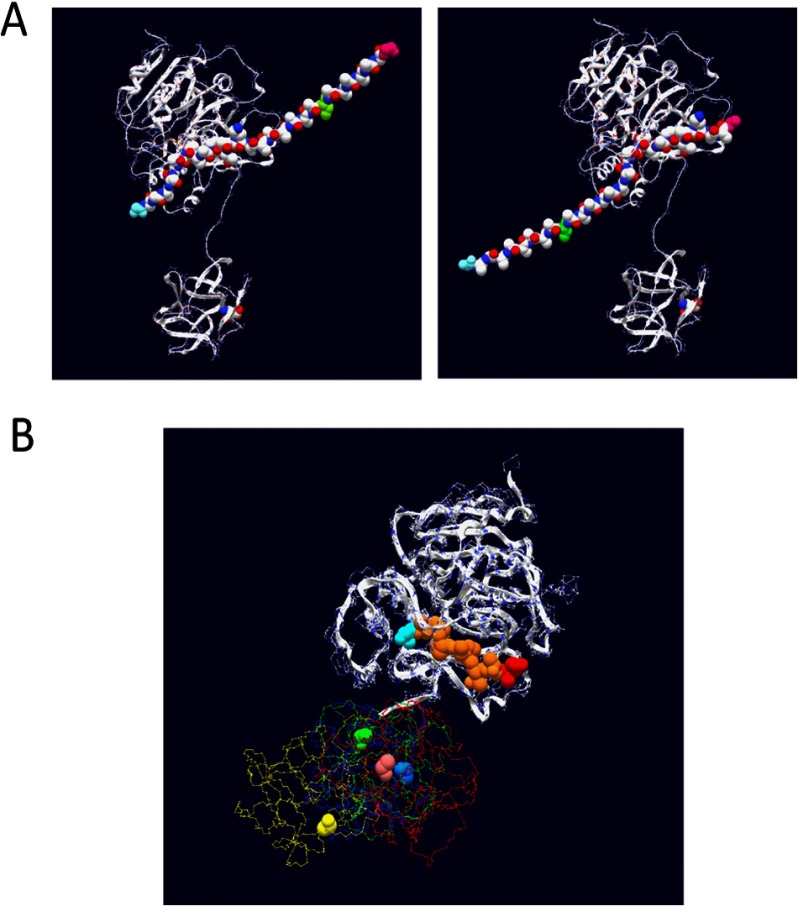
FIGURE 1.
Crystal structure of ppGalNAc T2 with bound peptide substrate EA2 (42) showing the tethered catalytic and lectin domains. Observed EA2 residues (5STTPAPTTK13) bound to the catalytic domain are space-filled (blue, N-terminal Ser; red, C-terminal Lys; purple, Pro; brown, Ser or Thr). The α-lectin subdomain Asp458 residue that is required for glycopeptide binding activity is space-filled in pink (50, 54) (see Table 2).
EXPERIMENTAL PROCEDURES
Reagents and Random Peptide Substrates
Fully N-acetylated UDP-[3H]GalNAc (C6-[3H]CH2-labeled) and UDP-[14C]GalNAc (NAc-[14C]CH3-labeled) were purchased from American Radiolabeled Chemicals Inc. Dowex 1X8 anion exchange resin (100–200-mesh) was purchased from Acros Organics. Sephadex G-10 was obtained from Amersham Biosciences and GE Healthcare. Random (glyco)peptide substrates GPIV, GPIV-C, GPV, and GPV-C (see “Results” and Table 1 for sequences) were custom synthesized by Sussex Research (Ottawa, CA). Random (glyco)peptide substrates were lyophilized from water several times, adjusted to pH 7.5 with dilute NaOH and/or HCl, and stored frozen at a concentration of 50 mg/ml (15–17 mm). Similar random (glyco)peptide X and Z residue compositions were confirmed by Edman amino acid sequencing (data not shown). Edman amino acid sequencing was performed on an Applied Biosystems Procise 494 peptide sequencer as described previously (43–45, 56). Liquid scintillation counting was performed in a Beckman model LS6500 scintillation counter.
TABLE 1.
ppGalNAc T lectin domain probing random (glyco)peptide substrates
X, GARPNEYVT; Z, GARPNEYV; T*, GalNAc-O-Thr.
| (Glyco)peptide | Sequence |
|---|---|
| GPIV | GAGAXXXXXXXXXXXXZZZZZT*ZZZZZAG |
| GPIV-C | GAGAXXXXXXXXXXXXZZZZZAZZZZZAG |
| GPV | GAGAZZZZZT*ZZZZZXXXXXXXXXXXXAG |
| GPV-C | GAGAZZZZZAZZZZZXXXXXXXXXXXXAG |
Transferases
N-terminally truncated and affinity-tagged ppGalNAc Ts used in this work were obtained from multiple sources and multiple expression systems and used as one or more of the following forms: medium supernatant, affinity-purified soluble transferase, or transferase bound to affinity beads. Experiments completed using transferases from different sources yielded consistent results. Soluble affinity-purified bovine ppGalNAc T1 (from Sf9 insect cells) and human ppGalNAc T2 and T3 (from Pichia pastoris) have been described previously (43–45, 56). ppGalNAc T3 was also used as medium supernatant from P. pastoris as described (43). Poly-His-tagged ppGalNAc T3, T6, and T13, expressed from Xpress SF+ insect cells (Protein Sciences Corp., Meriden, CT), were used directly bound to Ni-NTA affinity beads (Thermo Fisher) after extensive washing.5 Soluble N-terminal poly-His-tagged ppGalNAc T5 and T16 were expressed from HEK293f cells and purified by Ni-NTA superflow (Qiagen) nickel affinity chromatography by procedures analogous to those for the production of rat ST6GAL1 (57).5 Soluble N-terminal poly-His-tagged ppGalNAc T14 and T16 were expressed from High Five insect cells and purified on Ni-NTA-agarose (Invitrogen) and MonoQ 5/50 GL ion exchange chromatography (GE Healthcare) essentially as described previously (58).
Glycosylation of Random Lectin Glycopeptide Substrates
Typical reaction conditions utilizing random lectin glycopeptides substrates GPIV, GPIV-C, GPV, and GPV-C were as follows: 100 mm HEPES, pH 7.5 (ppGalNAc T1, T2, and T3) or 68 mm sodium cacodylate, pH 6.5 (ppGalNAc T3, T5, T6, T13, T14, and T16), 1.8 mm 2-mercaptoethanol, 10 mm MnCl2, 50 μm 3H-radiolabeled UDP-GalNAc (∼6 × 108 dpm/μmol), and 1.5–1.7 mm (5 mg/ml) of glycopeptide substrates and up to 50 μl of soluble transferase or transferase-bound affinity beads. UDP-GalNAc concentrations were kept low (0.5–50 μm) to achieve high specific activity for subsequent Edman sequence analysis (described below). After the addition of transferase, reaction mixtures (75 μl in 500 μl of capped Eppendorf tubes) were incubated at 37 °C in a TAITEC shaking Microincubator M-36. Aliquots of 15 μl were taken out at time points of 15, 45, 120, 240, and ∼1200 min (overnight) after initiating the reaction and quenched by the addition of 1 volume of 250 mm EDTA. UDP and non-hydrolyzed UDP-GalNAc were removed by passing the sample through a column of ∼3 ml of Dowex 1 × 8 anion exchange resin. [3H]GalNAc incorporation was determined by scintillation counting or of the sample before and after passing through the Dowex column. Data are typically reported as plots of the mole fraction of total UDP-GalNAc transferred (including hydrolysis) versus time. Confirmation of [3H]GalNAc transfer to peptide and the extent of UDP-[3H]GalNAc hydrolysis was determined by Sephadex G10 gel filtration analysis of selected reaction time points as described previously (44). For all transferases reported in this study, the extent of hydrolysis was negligible (see “Results”). Incubations were performed using a range of transferase concentrations (determined by trial and error) that would transfer between 10 and 50% of the total [3H]GalNAc to the optimal substrate after an overnight incubation (giving a range of ∼0.003–0.016 mol of GalNAc/mol of glycopeptide). In total, 2–4 independent experiments were performed at 2–3 different transferase concentrations (except for ppGalNAc T14).
Determination of Optimal Reaction Conditions and Initial Observations
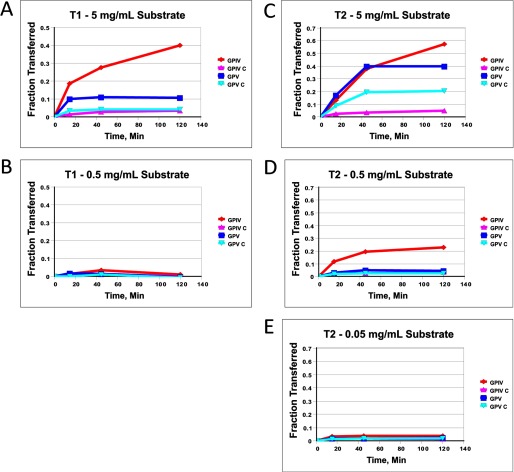
Initial use of the GPIV and GPV series of substrates (see Table 1) with ppGalNAc T1 and T2 gave results that seemed to vary with (glyco)peptide concentration. For example, at the lowest substrate concentrations (∼0.16 or ∼0.016 mm (glyco)peptide with T1 and T2, respectively), all four substrates were essentially inactive, whereas at the highest substrate concentration (1.5–1.7 mm), differential activities were observed, with the glycopeptide substrates displaying significantly greater activity compared with the control peptide substrates (see Fig. 2). We attribute these differential effects to peptide concentrations initially below the minimum Km value of the substrate and therefore have chosen to perform our studies utilizing substrate concentrations of 1.5–1.7 mm (5 mg/ml). Because of the random and multiple acceptor site nature of these substrates, a continuum of Km and Vmax values is expected to be observed; therefore, we have not pursued a detailed kinetic analysis of these substrates. It was further observed that the relative rates of glycosylation of the four substrates varied somewhat with the amount of transferase used in the assay; nevertheless, overall transferase preferences were preserved (data not shown).
FIGURE 2.
(Glyco)peptide GPIV, GPIV-C, GPV, and GPV-C substrate concentration dependence for ppGalNAc T1 and T2. A and B, ppGalNAc T1; 5 mg/ml (1.5–1.7 mm) (A) and 0.5 mg/ml (0.15–0.17 mm) (B). C–E, ppGalNAc T2; 5 mg/ml (1.5–1.7 mm) (C), 0.5 mg/ml (0.15–0.17 mm) (D), and 0.05 mg/ml (0.015–0.017 mm) (E). UDP-GalNAc concentration was 0.5 μm.
A commonly observed feature of the transferase activity plots shown below is the plateau in transfer of GalNAc that is often observed with high transferase activities, even for the least active substrates. We attribute this plateau to a combination of several likely factors. For the most active substrates (approaching 50% GalNAc utilization), the plateau in incorporation is most likely dominated by the depletion of UDP-GalNAc donor. For the less active substrates, we attribute the slowing of the rate to the sequential loss of the optimal or best acceptor sites as glycosylation of the random (glyco)peptide progresses. Because the half-lives of ppGalNAc T1 and T2 are about 5 and 2.5 days (59), it is unlikely that transferase inactivation is the source of the plateau with these transferases. However, transferase inactivation cannot be ruled out for the remaining transferases.
Determination of GalNAc Acceptor Sites of Incorporation by Edman Sequencing
Lyophylized post-Dowex [3H]GalNAc-glycosylated peptide products were isolated by Sephadex G10 chromatography, and the glycopeptide fraction was pooled according to its 3H radioactivity. After multiple lyophylizations from water, glycopeptides were subject to Edman degradation using a custom PTH analyzer program that diverted the PTH-derivatives from each cycle directly into 7-ml HDPE scintillation vials (Fisher) loaded in a 100-tube rack on an ISCO Foxy 200 fraction collector. It was observed that collecting directly into scintillation vials significantly reduced nonspecific losses of the radiolabeled PTH-derivatives compared with collecting in glass or plastic test tubes. To improve the signal/noise ratio, counting was performed for 5 min/vial and typically repeated 2–3 times, with the resulting dpm values averaged before plotting. To reduce nonspecific binding of the [3H]GalNAc-O-Thr-PTH-derivative to the glass fiber sample loading disk (determined by counting the disk after sequencing), ∼0.25 mg of nonreacted “cold” (glyco)peptide substrate was commonly added to the sample to be analyzed. Nevertheless, significant variability in recovery of radiolabel was observed. A preview in radioactivity observed in the N-terminal residues, particularly noticeable in the GPV and GPV-C (glyco)peptides, is thought to be due to nonspecific release of (glyco)peptide from the glass filter during the Edman sequencing.
RESULTS
Design of Lectin Glycopeptide Substrates
The goal of these studies was to develop a series of “universal” substrates that could be used with any ppGalNAc T isoform to access the role of each isoform's lectin domain. Peptides were designed with a single C- or N-terminal GalNAc-O-Thr residue (T*), designed to interact with the lectin domain, flanked by five positions of randomized residues (Z) containing no acceptor (Ser or Thr) residues (Table 1). The Z regions flanking the T* contained no acceptor Thr sites specifically to eliminate glycopeptide activities arising from the direct binding of GalNAc-O-Thr in the catalytic domain as is observed for ppGalNAc T10, which optimally glycosylates a Ser/Thr directly preceding a GalNAc-O-Ser/Thr (45, 51). Beyond these residues, in an N- or C-terminal direction, respectively, are 12 positions of randomized X residues also containing Thr that would serve as the acceptor region by its interaction with the catalytic domain. As a control for lectin domain specificity, peptides (GPIV-C and GPV-C) were also synthesized with the T* replaced with an Ala residue. The rationale behind utilizing randomized residues, X and Z, rather than a specific peptide sequence was based on our goal to eliminate (or at least reduce) transferase-specific bias due to a particular peptide sequence. With this series of (glyco)peptides the role of the lectin domain of the ppGalNAc Ts against glycopeptide substrates can now be systematically accessed, specifically addressing 1) the utilization of the lectin domain on overall transferase activity; 2) the preferred N- or C-terminal orientation, if any, of the T* relative to the site of glycosylation; and 3) the optimal number of residues between the lectin-bound T* and those residues glycosylated by the catalytic domain.
Glycosylation of the GPV-GPIV Series of (Glyco)peptides by ppGalNAc Ts
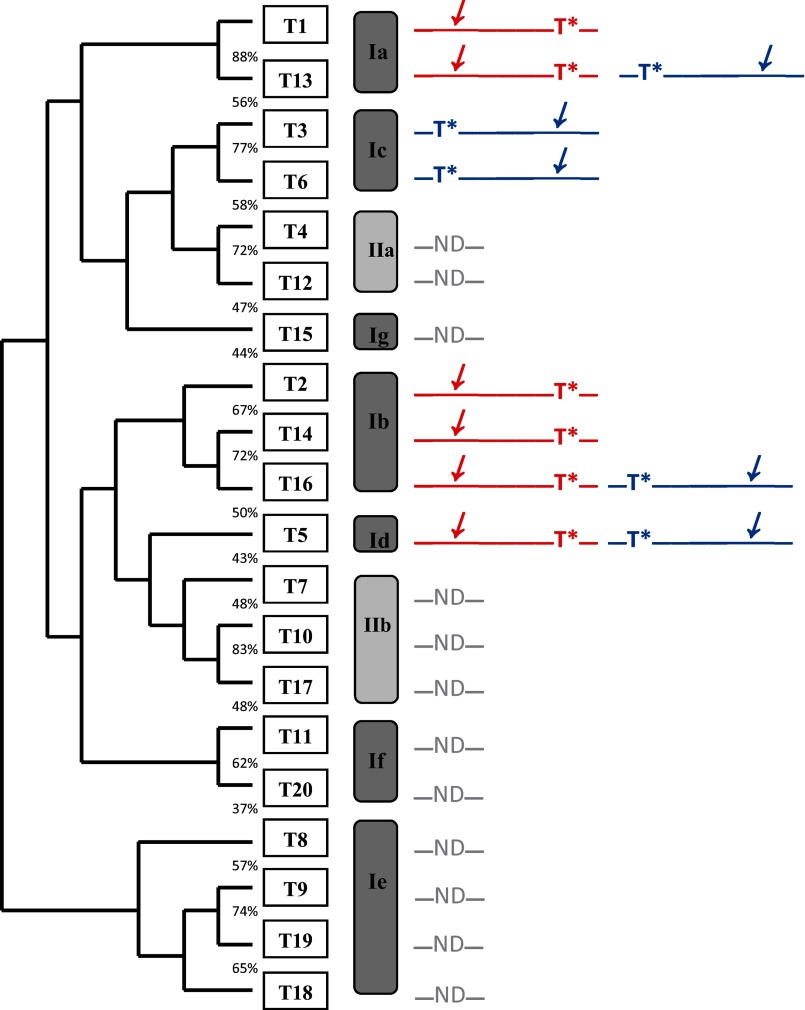
Representative time course plots and gel filtration profiles demonstrating the differential transfer of [3H]GalNAc into peptides GPIV, GPIV-C, GPV, and GPV-C by the eight ppGalNAc Ts under study are given in Fig. 3. As shown by the right-hand panels of Fig. 3, the G10 gel filtration analysis reveals relatively little UDP-GalNAc hydrolysis activity for all of the transferases characterized (i.e. high 3H in peptide fractions ∼27–33 and little 3H where free GalNAc would appear, fractions ∼37–43). Therefore, the time plots of the ion exchange column pass-through (Fig. 3, left-hand panels) can be taken as a direct measure of transfer of GalNAc to (glyco)peptide. To aid in the discussion, a phylogenetic tree and a subfamily classification of the human transferases (1) are shown in Fig. 4, where a graphical summary of glycopeptide utilization deduced from the data in Fig. 3 is also given. We will discuss transferase isoforms with common behaviors below.
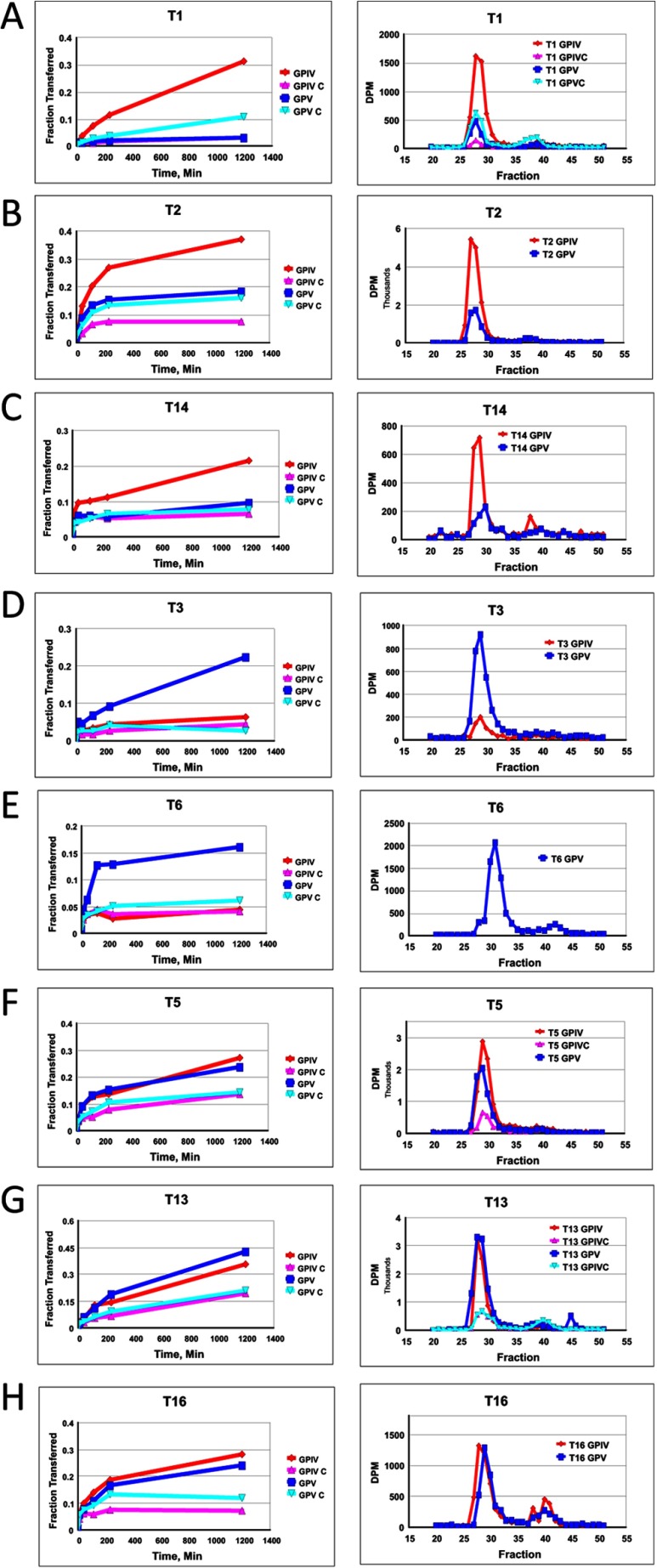
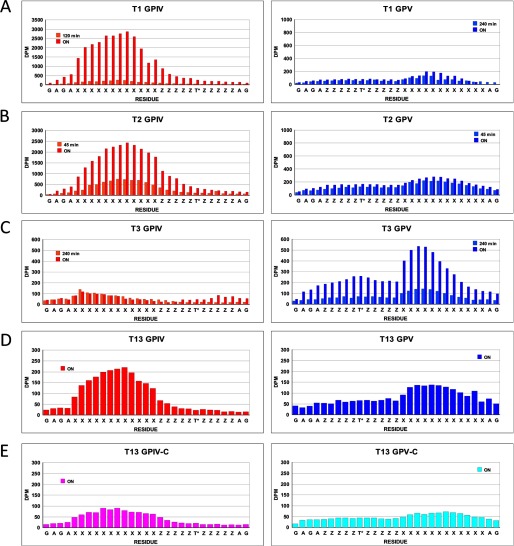
FIGURE 3.
Glycopeptide substrate specificity of ppGalNAc T isoforms. Shown is random (glyco)peptide substrate, GPIV (red, diamond), GPIV-C (purple, upward triangle), GPV (blue, square), and GPV-C (light blue, inverted triangle), glycosylated by ppGalNAc T1 (A panels), T2 (B panels), T14 (C panels), T3 (D panels), T6 (E panels), T5 (F panels), T13 (G panels), and T16 (H panels). Left panels represent mole fraction of [3H]GalNAc (50 μm) transferred onto (glyco)peptide substrate (1.5–1.7 mm) as a function of time as described under “Experimental Procedures.” Right-hand panels represent G10 gel filtration chromatograms of overnight incubations, demonstrating incorporation of [3H]GalNAc into (glyco)peptide substrate (fractions 27–33) with minimal hydrolysis (i.e. free GalNAc, fractions 37–43).
FIGURE 4.
Human ppGalNAc T phylogenetic tree and subfamilies based on Bennett et al. (1). Schematics to the right represent the glycopeptide preferences obtained from this work (GPIV in red and GPV in blue), where T* represents the position of the GalNAc-O-Thr and the slanted arrow indicates the N- or C-terminal region glycosylated by the transferase. ND, transferases whose glycopeptide substrate preferences have not been determined.
GPIV-preferring Transferases; ppGalNAc T1, T2, and T14
As shown in Fig. 3, A–C (and data not shown), these subfamily Ia and Ib transferases are significantly more active against the C-terminal glycosylated glycopeptide, GPIV, compared with its nonglycosylated control and the N-terminal glycosylated analog (glyco)peptide substrates, GPV and GPV-C. This observation suggests that the binding of the C-terminal T* of GPIV to the transferase lectin domain enhances its activity compared with the other substrates. This may be due to both an effective increase in substrate concentration due to repeated lectin binding and release or to an optimal alignment of the acceptor in the catalytic domain due to specific lectin binding.
To confirm that lectin binding is involved with the enhanced GPIV activity, the effects of adding free GalNAc competitor were examined on ppGalNAc T2. As shown in Fig. 5, A and B, the addition of free GalNAc significantly reduced the activity of GPIV while not affecting the activity of the nonglycosylated control peptide. It should be further noted that under the higher (glyco)peptide and transferase and lower UDP-GalNAc concentrations utilized in this and many of our initial experiments, GPV also shows an elevated activity relative to GPV-C (see Fig. 2C) that is completely inhibited by added free GalNAc (Fig. 5, C and D). This suggests that the GPV glycopeptide substrate can interact with the lectin domain of ppGalNAc T2 under certain conditions, also leading to a rate enhancement.
FIGURE 5.
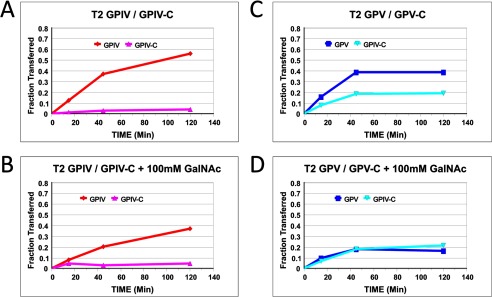
Effects of added GalNAc on ppGalNAc T2 (glyco)peptide utilization. A–D, GPIV and GPIV-C (A), GPIV and GPIV-C in the presence of 100 mm GalNAc (B), GPV and GPV-C (C), and GPV and GPV-C in the presence of 100 mm GalNAc (D). Note the decrease in GPIV and GPV 3H pass-through (fraction transferred) in the presence of 100 mm GalNAc. (Glyco)peptide substrate concentration was 5 mg/ml (1.5–1.7 mm). UDP-GalNAc concentration was 0.5 μm.
The x-ray crystal structure of ppGalNAc T2 (see Fig. 1) shows nine residues of the EA2 peptide substrate bound to the catalytic domain in a specific N to C terminus orientation (42). By model building, we have extended the EA2 peptide to mimic the bound GPIV and GPV glycopeptides, as shown in Fig. 6A (left- and right-hand panels, respectively). In the models, we observe that the glycosylated C terminus (∼XZZZZZT*ZZZZZAG) of GPIV is extended to the right of the catalytic domain, whereas the glycosylated N-terminal (GAGAZZZZZT*ZZZZZX∼) of GPV is extended to the left of the catalytic domain. Note that for simplicity, the extended regions were modeled as Ala residues in an extended conformation; however, in reality, an ensemble of dynamic structures would be expected. The observed elevated activity of GPIV over GPV for ppGalNAc T1, T2, and T14, therefore, would suggest that the lectin domain of these transferases would be positioned toward the right of the catalytic domain binding the T* of GPIV such that its Xaa residues would be aligned optimally in the catalytic domain. Such domain flexibility seems plausible in solution, because the relative positions of the catalytic and lectin domains are highly variable among the several crystal structures of ppGalNAc T1, T2, and T10 shown in Fig. 6B (42, 60, 61). This orientational flexibility is undoubtedly the result of the ∼20-residue extended linker that connects the domains (see Table 2).
FIGURE 6.
A, model building of (glyco)peptide substrates GPIV and GPV onto the EA2-bound ppGalNAc T2 crystal structure (42). Left, GPIV modeled with its Xaa residues placed in the catalytic domain in the same N- to C-terminal orientation as the 9 residues of the bound EA2 peptide. Right, GPV modeled in the catalytic domain. For simplicity, the 9 EA2 residues were maintained as in the original structure (see Fig. 1), whereas 3 additional Ala residues were added to complete the representation of the 12-Xaa acceptor region. Likewise, Ala residues were used to represent the Zaa residues, whereas a Ser residue (green) was used to represent the location of the GalNAc-O-Thr. Note that for simplicity, both regions were modeled as static extended structures but in reality would be relatively flexible, comprising an ensemble of structures. B, superimposition of the catalytic domains of ppGalNAc T2-bound EA2 (yellow trace), ppGalNAc T2 bound UDP (blue trace), ppGalNAc T10-bound UDP and GalNAc (red trace), and ppGalNAc T1 (green trace), showing the positional variability of the lectin domain. The critical α-lectin subdomain Asp residue (see Table 2) is space-filled. Protein Data Bank accession numbers are as follows: 2FFU (42), 2FFV (42), 2D7I (61), and 1XHB (60), respectively.
TABLE 2.
ppGalNAc T linker domain and lectin domain motif alignments
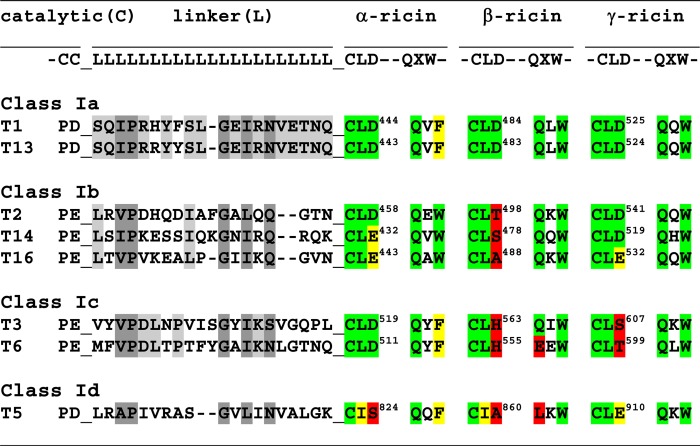
a Linker domain (L) residues conserved (or similar) in all eight transferases are shaded dark grey, whereas residues conserved only within a transferase class are shaded light grey.
b α-, β-, and γ-ricin lectin subdomain canonical motif residues, CLD and QXW, are shaded in green, those residues that are similar to the canonical motif are shaded in yellow, and dissimilar, non-canonical residues are shaded in red (66–69).
c Note that the last residues of the catalytic domain (C) and the first residues of the lectin domain, CLD, are contiguous with the linker domain residues (L).
Glycopeptide GPV-preferring Transferases; ppGalNAc T3 and T6
These subfamily Ic transferases (Fig. 3, D and E) (and data not shown) display the reversed preferences of ppGalNAc T1, T2, and T14, showing GPV as the most active substrate with the remaining substrates being relatively inactive. This suggests that the lectin domains of ppGalNAc T3 and T6 preferentially interact with T* residues N-terminal of the site of subsequent glycosylation, suggesting that the lectin domain of these transferases must be positioned toward the left of the catalytic domain, as shown in the right panel of Fig. 6A.
Transferases with Preferences for both Glycopeptide Substrates GPIV and GPV; ppGalNAc T5, T13, and T16
As shown by the plots in Fig. 3, F–H (and data not shown), ppGalNAc T5, T13, and T16 possess nearly equally enhanced preferences for both glycopeptide substrates GPIV and GPV compared with their control nonglycosylated peptides. ppGalNAc T5 is the only member of subfamily Id, whereas ppGalNAc T13 and T16 are members of subfamilies Ia and Ib, respectively, whose other members, ppGalNAc T1 and ppGalNAc T2 and T14, were shown above to prefer the GPIV glycopeptide substrate (see Fig. 3, A–C). This indicates that even within a subfamily, the N- and C-glycopeptide preferences can vary. The lack of directional glycopeptide specificity for ppGalNAc T5, T13, and T16 may be due to extensive lectin domain conformational dynamics, suggesting that the domain may be equally populated at the right and left of the catalytic domain and/or due to an elevated local concentration of glycopeptide substrate due to lectin binding and release.
Edman Sequencing of GPIV and GPV Series of (Glyco)peptides
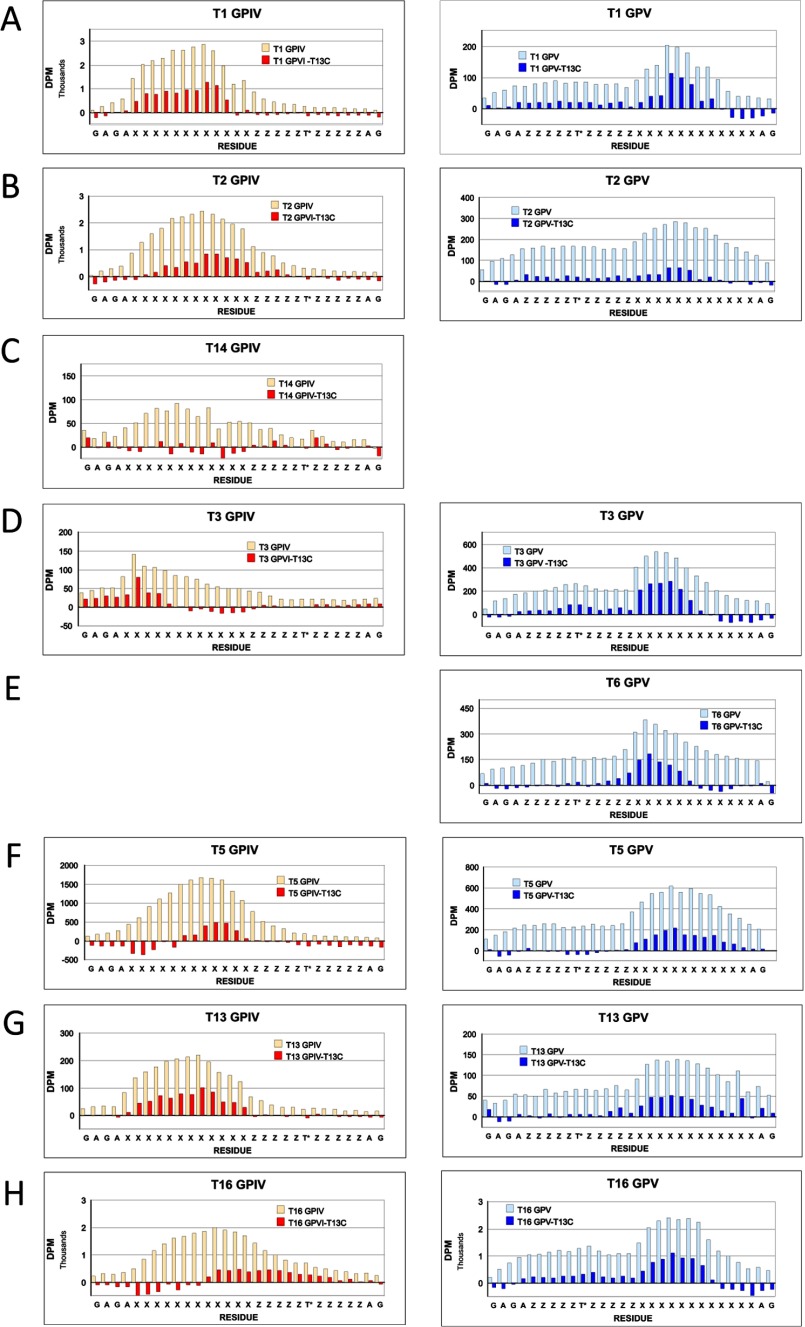
In an attempt to determine whether there is an optimal distance between the lectin-bound T* residue and the Xaa residues glycosylated by the catalytic domain, selected (glyco)peptide products were Edman sequenced, and each residue's [3H]GalNAc content was determined. Fig. 7, A–D, shows representative plots for ppGalNAc T1, T2, T3, and T13 with glycopeptides GPIV and GPV (left- and right-hand panels, respectively, roughly scaled to approximate their relative levels of incorporation).6 Fig. 7E shows incorporation for the control peptides, GPIV-C and GPV-C, glycosylated by ppGalNAc T13. Rather than observing a sharp peak of maximal [3H]GalNAc incorporation, a relatively broad distribution of incorporation is observed for all substrates. As might be expected, the largest differences are found between ppGalNAc T3 and ppGalNAc T1, T2, and T13 for both GPIV and GPV, whereas only subtle differences are observed in the patterns among ppGalNAc T1, T2, and T13. The distance from the T* to the peak in incorporation in GPIV is ∼10 residues for ppGalNAc T1, T2, and T13 and 16 residues for T3. For GPV, the distance from the T* and peak in incorporation is 10–11 residues for ppGalNAc T1, T2, and T13 and 8–9 residues for ppGalNAc T3.
FIGURE 7.
[3H]GalNAc incorporation into the Xaa residues of random (glyco)peptides. A–D, incorporation of [3H]GalNAc into GPIV (left-hand panels) and GPV (right-hand panels) for ppGalNAc T1, T2, T3, and T13, respectively. E, incorporation of [3H]GalNAc into GPIV-C (left) and GPV-C (right) for ppGalNAc T13.6 Note that for A and B, substrates were glycosylated using 0.5 μm, 100-fold specific activity UDP-[3H]GalNAc, whereas in C–E, substrates were glycosylated using 50 μm standard specific activity UDP-[3H]GalNAc.
In an attempt to reveal the extent that the lectin domain may alter the pattern of [3H]GalNAc incorporation, the distributions of the nonglycosylated control peptides, GPIV-C and GPV-C (glycosylated by ppGalNAc T13, Fig. 7E), were subtracted from the distributions obtained for the glycosylated peptides GPIV and GPV, respectively.7 These difference plots for all eight transferase isoforms, grouped by glycopeptide utilization, are shown in Fig. 8. It was anticipated that these plots would reveal the effects of lectin domain binding on the pattern of glycosylation and perhaps further reveal differences between transferase class and N- or C-terminal preference. Except for ppGalNAc T3 on GPIV (Figs. 7C and 8D), the difference plots were not appreciably different from the original distribution, although in a few cases, the distributions may have shifted slightly (i.e. for ppGalNAc T2, T5, and T16 on GPIV). From these results, we conclude that the binding of glycopeptide substrate for the most part (i.e. except for ppGalNAc T3 on GPIV) does not greatly alter the pattern of glycosylation compared with the nonglycopeptide substrate. Furthermore, no consistent differences were observed between isoform classes or between transferases within a class that had different N- or C-terminal glycopeptide preferences (i.e. ppGalNAc T1, T2, and T14 versus ppGalNAc T13, T16, and T5). These findings are consistent with a highly mobile lectin domain where the lectin-bound glycopeptide substrate would have a broad range of acceptor interacting distances with the catalytic domain. Alternatively, these results could be consistent with just a regiospecific increase in substrate concentration simply due to rapid binding and release from a conformationally fixed lectin domain.
FIGURE 8.
Plots of [3H]GalNAc incorporation into the Xaa residues of random glycopeptides GPIV and GPV (open bars) and after normalization by subtraction of control peptides GPIV-C or GPV-C that were glycosylated by ppGalNAc T13 (filled bars). A–H, plots of GPIV (left) and GPV (right) for ppGalNAc T1, T2, T14, T3, T6, T5, T13, and T16, respectively. Note that for A and B, substrates were glycosylated using 0.5 μm, 100-fold specific activity UDP-[3H]GalNAc, whereas in C–H, substrates were glycosylated using 50 μm standard specific activity UDP-[3H]GalNAc.
DISCUSSION
Among eukaryote glycosyltransferases, the ppGalNAc T family is unique by possessing a separate carbohydrate binding lectin domain. Interestingly, such carbohydrate binding modules are common to bacterial glycoside hydrolases, serving to both increase local enzyme-substrate concentrations and to impart substrate specificity to nominally nonspecific catalytic domains by targeting the enzyme to specific substrates or substrate features (62–65). Numerous studies on the ppGalNAc Ts have indeed demonstrated that their lectin domains serve similar functions to modulate glycopeptide substrate recognition and specificity (48–53), but to date there has been no systematic study of the family against a common set of (glyco)peptide substrates. In this work, we have characterized eight nonglycopeptide-requiring ppGalNAc T isoforms against the series of random (glyco)peptide substrates listed in Table 1. Our findings have unambiguously revealed that prior GalNAc-O-Thr(Ser) substrate glycosylation can be recognized by these transferases in a specific N- or C-terminal direction that varies with ppGalNAc T isoform (see Figs. 3 and 4). Thus, for all eight transferases, at least one of the glycopeptide substrates, GPIV and/or GPV, shows significantly elevated activity over its nonglycosylated control. We attribute this N- or C-terminal specificity (i.e. elevated activity) to the binding of the glycosylated Thr residue of the substrate to the lectin domain in such a manner that the acceptor region of the substrate is oriented at the catalytic domain for optimal glycosylation. These findings strongly suggest that the tethered lectin domain on the ppGalNAc Ts may be mobile and that its location relative to the catalytic domain varies among isoforms. Such domain mobility is supported by the superimposition of the x-ray crystal structures of ppGalNAc T1, T2, and T10, as shown in Fig. 6B (42, 60, 61). For those transferases with similar N- and C-terminal glycopeptide enhancements (Fig. 4), it is possible that the lectin domain may be sufficiently mobile that it can enhance glycosylation from both an N- and C-terminal direction. Alternatively, the lectin domain may serve to equally increase the N- and C-terminal glycopeptide substrate concentrations by a simple bind and release mechanism, as shown for the glycoside hydrolases (62–65). Further evidence for a highly flexible lectin domain may be found in the broad distribution of glycosylation that is observed in the Xaa region of both the glycosylated and nonglycosylated substrates (see Figs. 7 and 8). The only significant alteration in distribution is observed for ppGalNAc T3 glycosylating its nonpreferred glycopeptide substrate GPIV (Fig. 7). This suggests that the lectin domain of this transferase is involved to some extent in directing the glycosylation of this substrate. Obviously, further studies are necessary to fully understand the dynamics of the lectin domain of these transferases.
In an attempt to correlate the different N- and C-terminal glycopeptide specificities to transferase peptide sequence we have compared the sequences of their linker domain and selected lectin domain motifs, as shown in Table 2. From the alignment of the linker domains, no obvious differences in length or sequence can readily account for the different behavior observed within a class (i.e. ppGalNAc T2 and T14, which prefer glycopeptide GPIV, are only 1 residue shorter than T16, which shows preferences for both glycopeptide substrates, whereas the linker domains of ppGalNAc T1 and T13 are identical except for two conservative substitutions, although they show different glycopeptide specificities). By contrast, the linker domains of ppGalNAc T3 and T6 are only 50% identical but have the same elevated GPV preferences.
Numerous studies of the ricin lectin α-, β-, and γ-subdomain repeats across multiple organisms have revealed several binding motifs, including the CLD and QXW sequences (66–69). Mutagenesis studies of the ppGalNAc T lectin domains have shown that the Asp residue of the α-subdomain CLD motif is typically required for lectin binding activities (i.e. ppGalNAc T1-D444A (54, 70, 71), ppGalNAc T2-D458H (50, 54), and ppGalNAc T3-D519H (54, 55) are lectin-inactivating mutants). A similar mutation in the ppGalNAc T1 β-ricin subdomain (D484A) only modestly decreases lectin-modulated activity (71, 72), whereas mutation of the γ-ricin subdomains of ppGalNAc T1 and T2 (D525A and D541A, respectively) shows no effects on their lectin domain activities (50, 71, 72). Mutation of the α-subdomain of ppGalNAc T4 (D459H) also eliminates its lectin activity (48). As shown in Table 2, the β-subdomains of ppGalNAc T2, T14, T16, T3, T6, and T5 and the γ-subdomains of T3 and T6 lack this Asp. Several transferases possess Glu instead of Asp (i.e. ppGalNAc T14 and T16 in the α-subdomain and T16 and T5 in the γ-domain); whether these domains can bind GalNAc is presently unknown. The β-subdomains of ppGalNAc T6 and T5 lack the QXW motif having EEW and LKW and would be expected to be inactive. Interestingly, the co-crystal structure of ppGalNAc T10 shows GalNAc-O-Ser bound to its β-subdomain (having canonical motifs CFD and QLW) (61). The ricin subdomain motifs of ppGalNAc T5 would be expected to have the weakest lectin binding activities because all three subdomains lack the critical Asp residue, although, in its γ-subdomain, the Asp is replaced by a Glu. Nevertheless, because ppGalNAc T5 displays clear glycopeptide specificities, one or more of its lectin subdomains (perhaps its γ-subdomain) must possess significant binding activity to provide glycopeptide enhancements. Generally, glycopeptide binding to of any of the ppGalNAc Ts lectin domains has been difficult to directly detect, further suggesting its weak nature (50, 54, 55). As with the analysis of the linker domains, we conclude that we cannot discern any obvious correlation between the likely activities of specific lectin subdomain motif and a given transferase's observed glycopeptide specificity.
From the above discussion of the lectin domain motifs listed in Table 2, it is likely, for all but ppGalNAc T5, that glycopeptide binding would occur predominantly at the α-subdomain. As shown in the superimposed crystal structures of the ppGalNAc Ts in Fig. 6B, the critical Asp residue of the CLD motif in the α-subdomain (highlighted in the structures) is found in a wide range of positions relative to the catalytic domain. One could easily assume that the conformational flexibility of the lectin domain relative to the catalytic domain in solution would be greater than that observed in Fig. 6B. Thus, both the structural and experimental results are consistent with a highly mobile lectin domain, whose dynamics and relative position with respect to the catalytic domain varies widely with transferase isoform. Whether additional domain-domain interactions may modulate the relative positions of the lectin domain to the catalytic domain is presently unknown, but it seems highly plausible that such interactions could assist in the positioning of the lectin domain. Future studies will address these questions.
Prior Evidence for the Lectin Domain Targeting N- or C-terminal Glycopeptide Glycosylation
The glycosylation of the Muc 7 peptide (PTPSATT7PAPPSS13S14APPET19T20AAK) by the subclass Ia transferases ppGalNAc T1 and T13 has been examined by Zhang et al. (73). Interestingly, they observed that although Thr7 was initially glycosylated by both transferases, only ppGalNAc T13 was capable of further glycosylating the boldface C-terminal Ser and Thr residues of the peptide. These findings are entirely consistent with the transferase glycopeptide preferences summarized in Fig. 4, where ppGalNAc T1 prefers to glycosylate sites N-terminal of a prior site of glycosylation, whereas ppGalNAc T13 is capable of glycosylating sites both N- and C-terminal of a prior site of glycosylation. Raman et al. (51) have extensively characterized the glycosylation of a series of MUC5AC glycopeptides by ppGalNAc T2 and its catalytic domain. Based on comparative binding studies, it was concluded that ppGalNAc T2 bound glycopeptide when the acceptor site was 10 residues N-terminal of an existing site of GalNAc glycosylation but not a glycopeptide with the reverse orientation and that this was due to the presence of the lectin domain, an observation consistent with our studies of ppGalNAc T2 (see Fig. 4).8 Likewise, studies on the glycan binding requirements of the lectin domain of ppGalNAc T3 suggest that this transferase utilizes its lectin domain to glycosylate residues C-terminal of the site of prior glycosylation (55), again in keeping with our findings for ppGalNAc T3. ppGalNAc T3 was previously shown to be the only isoform capable of glycosylating the proprotein processing region of FGF23 (IHFNT171PIPR RHT178R↓SAEDD) (21, 25, 74), and in unpublished work,9 Kato et al. have found that ppGalNAc T3 glycosylates the Thr178 site in a lectin-dependent manner by first glycosylating Thr171 in a lectin-independent manner, both in in vitro enzyme assays and ex vivo in CHO cells stably transfected with ppGalNAc T3 variants. The Thr171 site is also glycosylated by several other ppGalNAc-T isoforms (21). Our previous work (43) utilizing transferase specific preferences, obtained from random peptide studies, also suggests that Thr171 would be a moderate to good substrate for ppGalNAc T1, T2, and T3, whereas Thr178 would not be a substrate for ppGalNAc T1 or T2 and would be only a modest substrate for ppGalNAc T3 (see Table 2 of Ref 43). This suggests that the prior glycosylation of Thr171 may serve to target ppGalNAc T3 glycosylation of Thr178, 7 residues C-terminal of Thr171. The glycosylation distribution plots for ppGalNAc T3 for glycopeptide GPV (Figs. 7C and 8D) are entirely consistent with this, showing a peak of glycosylation 8–9 residues C-terminal of the original site of glycosylation. By contrast, the prior glycosylation of Thr171 would fail to enhance the glycosylation of Thr178 by ppGalNAc T1 and T2 because these transferases possess the reverse glycopeptide specificities. Thus, glycosylation of Thr171 enhances the glycosylation of Thr178 by ppGalNAc T3 while effectively reducing the activities of those transferases with the reverse glycopeptide preferences. Thus, prior glycosylation of Thr171 serves as a targeting switch or enhancer for the glycosylation of Thr178, whose glycosylation is required to inhibit the proprotein convertase cleavage of FGF23 in vivo and in vitro (20, 21, 74).
The control of site-specific mucin type O-glycosylation and the need for such a large family of initiating ppGalNAc Ts is not well understood.10 In this work, we have identified a previously unappreciated level of control, whereby prior O-glycosylation is used to target and enhance the glycosylation of specific N- or C-terminal sites in an isoform-specific manner. Our studies and those of others strongly suggest that this N- or C- terminal selectivity is due to weak glycopeptide binding to the lectin domain, whose orientation relative to the catalytic domain is highly mobile and isoform-dependent. This glycopeptide selectivity can provide an additional level of control or fidelity for the glycosylation of biologically significant sites and suggests that O-glycosylation in some instances may be exquisitely controlled. Furthermore, our observations that homologous ppGalNAc T isoforms within a given subfamily (that presumably have similar peptide substrate specificities; see Ref. 56) may possess different N- or C-terminal glycopeptide preferences may help explain the large number of ppGalNAc T family members thereby maintaining peptide specificity while altering glycopeptide specificity. These studies clearly demonstrate that the biological control mucin type O-glycosylation is highly complex and that further structural, biochemical, and biological studies are necessary to fully understand this important modification.
Acknowledgments
We thank Oliver Jamison for initial assistance with designing the lectin peptide substrates and Dr. Vivian Yee for assistance with the molecular modeling.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA78834 (to T. A. G.) and P41-GM103390 and P41-RR005351 (to James Prestegard (Complex Carbohydrate Center, University of Georgia) and K. M. and D. L. J.) and a National Institutes of Health, NIDCR, intramural program grant (to L. A. T.). This work was also supported by Danish National Research Foundation Grant DNRF107 (to H. C.).
We will be using the transferase isoform numbering nomenclature of Bennett et al. (1).
For detailed methods, see the University of Georgia's, Resource for Integrated Glycotechnology National Institutes of Health-funded program “Development of a Repository for Glycan-related Enzymes” (K. Moremen, Josh LaBaer (Arizona State University), and D. L. Jarvis, available on the World Wide Web.
Note that two different incorporation time points are plotted in Fig. 7, A–C, for ppGalNAc T1, T2, and T3 and that the distributions obtained at the two different time points are identical when plotted at the same relative scale (data not shown). This observation suggests that comparisons of the patterns of incorporation among the different transferase isoforms can be made even between data obtained with different extents of glycosylation. This is reasonable because under the typical reaction conditions, the maximum possible incorporation of the UDP-GalNAc would give a (glyco)peptide GalNAc content of about 3 mol %.
GPIV-C and GPV-C control sequencing data were not successfully obtained from most transferases due to very low incorporation; therefore, the successfully obtained data from both control peptides for ppGalNAc T13 were utilized. The only other successfully obtained control data, from ppGalNAc T2, were nearly identical to ppGalNAc T13.
Note that Raman et al. (51) have also proposed in their kinetic analysis that the ppGalNAc T2 lectin domain may also direct glycosylation to sites C-terminal of a GalNAc-O-Thr residue by assisting in product release rather than assisting in substrate binding.
K. Kato, C. Jeanneau, I. Dar, E. P. Bennett, K. T. Schjoldager, A. Benet-Pagès, T. M. Strom, and H. Clausen, unpublished results.
ppGalNAc T1 and T2 are ubiquitously expressed in nearly all mammalian tissues and cell lines, whereas the remaining ppGalNAc T isoforms studied in this work are selectively expressed (see Refs. 1, 16, and 75). For example, ppGalNAc T3 is the dominant transferase expressed in the testis and is also highly expressed in the kidney (21, 75, 76), and ppGalNAc T16 is more highly expressed than ppGalNAc T1 in the heart and brain (41), whereas ppGalNAc T13 is specifically expressed in neurons (73). How the multiple transferases work in concert in glycosylating their target proteins is largely unknown, as is their regulation at both the protein and transcriptional levels, although, judging from observations in our laboratory and other laboratories, large differences in protein stabilities exist.
- ppGalNAc T
- UDP-α-GalNAc:polypeptide N-α-acetylgalactosaminyltransferase
- PTH
- phenylthiohydantoin amino acid derivative of Edman sequencing
- Ni-NTA
- nickel-nitrilotriacetic acid
- GPIV, GPIV-C, GPV, and GPV-C
- random (glyco)peptides as defined in Table 1
- HDPE
- high density polyethylene.
REFERENCES
- 1. Bennett E. P., Mandel U., Clausen H., Gerken T. A., Fritz T. A., Tabak L. A. (2012) Control of mucin-type O-glycosylation. A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22, 736–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hollingsworth M. A., Swanson B. J. (2004) Mucins in cancer. Protection and control of the cell surface. Nat. Rev. Cancer 4, 45–60 [DOI] [PubMed] [Google Scholar]
- 3. Brockhausen I. (2006) Mucin-type O-glycans in human colon and breast cancer. Glycodynamics and functions. Endocrinology 7, 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gill D. J., Clausen H., Bard F. (2011) Location, location, location. New insights into O-GalNAc protein glycosylation. Trends Cell Biol. 21, 149–158 [DOI] [PubMed] [Google Scholar]
- 5. Wopereis S., Lefeber D. J., Morava E., Wevers R. A. (2006) Mechanisms in protein O-glycan biosynthesis and clinical and molecular aspects of protein O-glycan biosynthesis defects. A Review. Clin. Chem. 52, 574–600 [DOI] [PubMed] [Google Scholar]
- 6. Tian E., Ten Hagen K. (2009) Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lang T., Hansson G. C., Samuelsson T. (2007) Gel-forming mucins appeared early in metazoan evolution. Proc. Natl. Acad. Sci. U.S.A. 104, 16209–16214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carmon A., Wilkin M., Hassan J., Baron M., MacIntyre R. (2007) Concerted evolution within the Drosophila dumpy gene. Genetics 176, 309–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alfalah M., Jacob R., Preuss U., Zimmer K. P., Naim H., Naim H. Y. (1999) O-Linked glycans mediate apical sorting of human intestinal sucrase-isomaltase through association with lipid rafts. Curr. Biol. 9, 593–596 [DOI] [PubMed] [Google Scholar]
- 10. Tani M., Iida H., Ito M. (2003) O-Glycosylation of mucin-like domain retains the neutral ceramidase on the plasma membranes as a type II integral membrane protein. J. Biol. Chem. 278, 10523–10530 [DOI] [PubMed] [Google Scholar]
- 11. Li F., Wilkins P. P., Crawley S., Weinstein J., Cummings R. D., McEver R. P. (1996) Post-translational modifications of recombinant P-selectin glycoprotein ligand-1 required for binding to P- and E-selectin. J. Biol. Chem. 271, 3255–3264 [PubMed] [Google Scholar]
- 12. Lee S. H., Yu S. Y., Nakayama J., Khoo K. H., Stone E. L., Fukuda M. N., Marth J. D., Fukuda M. (2010) Core2 O-glycan structure is essential for the cell surface expression of sucrase isomaltase and dipeptidyl peptidase-IV during intestinal cell differentiation. J. Biol. Chem. 285, 37683–37692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Potter B. A., Hughey R. P., Weisz O. A. (2006) Role of N- and O-glycans in polarized biosynthetic sorting. Am. J. Physiol. Cell Physiol. 290, C1–C10 [DOI] [PubMed] [Google Scholar]
- 14. Kinlough C. L., Poland P. A., Gendler S. J., Mattila P. E., Mo D., Weisz O. A., Hughey R. P. (2011) Core-glycosylated mucin-like repeats from MUC1 are an apical targeting signal. J. Biol. Chem. 286, 39072–39081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakayama Y., Nakamura N., Oki S., Wakabayashi M., Ishihama Y., Miyake A., Itoh N., Kurosaka A. (2012) A putative polypeptide N-acetylgalactosaminyltransferase/Williams-Beuren syndrome chromosome region 17 (WBSCR17) regulates lamellipodium formation and macropinocytosis. J. Biol. Chem. 287, 32222–32235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T. B., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Paul Bennett E., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H. (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schjoldager K. T., Vakhrushev S. Y., Kong Y., Steentoft C., Nudelman A. S., Pedersen N. B., Wandall H. H., Mandel U., Bennett E. P., Levery S. B., Clausen H. (2012) Probing isoform-specific functions of polypeptide GalNAc-transferases using zinc finger nuclease glycoengineered SimpleCells, Proc. Natl. Acad. Sci. U.S.A. 109, 9893–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vakhrushev S. Y., Steentoft C., Vester-Christensen M. B., Bennett E. P., Clausen H., Levery S. B. (2013) Enhanced mass spectrometric mapping of the human GalNAc-type O-glycoproteome with SimpleCells. Mol. Cell. Proteomics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schjoldager K. T., Clausen H. (2012) Site-specific protein O-glycosylation modulates proprotein processing. Deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim. Biophys. Acta 1820, 2079–2094 [DOI] [PubMed] [Google Scholar]
- 20. Schjoldager K. T., Vester-Christensen M. B., Goth C. K., Petersen T. N., Brunak S., Bennett E. P., Levery S. B., Clausen H. (2011) A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J. Biol. Chem. 286, 40122–40132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato K., Jeanneau C., Tarp M. A., Benet-Pagès A., Lorenz-Depiereux B., Bennett E. P., Mandel U., Strom T. M., Clausen H. (2006) Polypeptide GaINAc-transferase T3 and familial tumoral calcinosis. Secretion of FGF23 requires O-glycosylation. J. Biol. Chem. 281, 18370–18377 [DOI] [PubMed] [Google Scholar]
- 22. Schjoldager K. T., Vester-Christensen M. B., Bennett E. P., Levery S. B., Schwientek T., Yin W., Blixt O., Clausen H. (2010) O-Glycosylation modulates proprotein convertase activation of angiopoietin-like protein 3. J. Biol. Chem. 285, 36293–36303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herr P., Korniychuk G., Yamamoto Y., Grubisic K., Oelgeschläger M. (2008) Regulation of TGF-β signalling by N-acetylgalactosaminyltransferase-like 1, Development 135, 1813–1822 [DOI] [PubMed] [Google Scholar]
- 24. Wagner K. W., Punnoose E. A., Januario T., Lawrence D. A., Pitti R. M., Lancaster K., Lee D., von Goetz M., Yee S. F., Totpal K., Huw L., Katta V., Cavet G., Hymowitz S. G., Amler L., Ashkenazi A. (2007) Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat. Med. 13, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 25. Ichikawa S., Sorenson A. H., Austin A. M., Mackenzie D. S., Fritz T. A., Moh A., Hui S. L., Econs M. J. (2009) Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression. Endocrinology 150, 2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simmons A. D., Musy M. M., Lopes C. S., Hwang L. Y., Yang Y. P., Lovett M. (1999) A direct interaction between EXT proteins and glycosyltransferases is defective in hereditary multiple exostoses. Hum. Mol. Genet. 8, 2155–2164 [DOI] [PubMed] [Google Scholar]
- 27. Holleboom A. G., Karlsson H., Lin R. S., Beres T. M., Sierts J. A., Herman D. S., Stroes E. S., Aerts J. M., Kastelein J. J., Motazacker M. M., Dallinga-Thie G. M., Levels J. H., Zwinderman A. H., Seidman J. G., Seidman C. E., Ljunggren S., Lefeber D. J., Morava E., Wevers R. A., Fritz T. A., Tabak L. A., Lindahl M., Hovingh G. K., Kuivenhoven J. A. (2011) Heterozygosity for a loss-of-function mutation in GALNT2 improves plasma triglyceride clearance in man. Cell Metab. 14, 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J. Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Hua Zhao J., Yuan X., Luan J., Lamina C., Ziegler A., Zhang W., Zee R. Y., Wright A. F., Witteman J. C., Wilson J. F., Willemsen G., Wichmann H. E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J. G., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W. J. H., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., 'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K. E., Lucas G., Luben R., Loos R. J. F., Lokki M. L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., Konig I. R., Khaw K. T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M. R., Cecile J. W. J., Ingelsson E., Igl W., Kees Hovingh G., Hottenga J. J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Doring A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J. C., de Faire U., Crawford G., Collins F. S., Chen Y. d., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E.-S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor H. A., Jr., Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J. P., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., Kathiresan S. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guda K., Moinova H., He J., Jamison O., Ravi L., Natale L., Lutterbaugh J., Lawrence E., Lewis S., Willson J. K., Lowe J. B., Wiesner G. L., Parmigiani G., Barnholtz-Sloan J., Dawson D. W., Velculescu V. E., Kinzler K. W., Papadopoulos N., Vogelstein B., Willis J., Gerken T. A., Markowitz S. D. (2009) Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc. Natl. Acad. Sci. U.S.A. 106, 12921–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gaziel-Sovran A., Segura M. F., Di Micco R., Collins M. K., Hanniford D., Vega-Saenz de Miera E., Rakus J. F., Dankert J. F., Shang S., Kerbel R. S., Bhardwaj N., Shao Y., Darvishian F., Zavadil J., Erlebacher A., Mahal L. K., Osman I., Hernando E. (2011) miR-30b/30d Regulation of GalNAc Transferases Enhances Invasion and Immunosuppression during Metastasis, Cancer Cell 20, 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Park J. H., Katagiri T., Chung S., Kijima K., Nakamura Y. (2011) Polypeptide N-acetylgalactosaminyltransferase 6 disrupts mammary acinar morphogenesis through O-glycosylation of fibronectin. Neoplasia 13, 320–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taniuchi K., Cerny R. L., Tanouchi A., Kohno K., Kotani N., Honke K., Saibara T., Hollingsworth M. A. (2011) Overexpression of GalNAc-transferase GalNAc-T3 promotes pancreatic cancer cell growth. Oncogene 30, 4843–4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berois N., Mazal D., Ubillos L., Trajtenberg F., Nicolas A., Sastre-Garau X., Magdelenat H., Osinaga E. (2006) UDP-N-acetyl-d-galactosamine. Polypeptide N-acetylgalactosaminyltransferase-6 as a new immunohistochemical breast cancer marker. J. Histochem. Cytochem. 54, 317–328 [DOI] [PubMed] [Google Scholar]
- 34. Berois N., Gattolliat C. H., Barrios E., Capandeguy L., Douc-Rasy S., Valteau-Couanet D., Bénard J., Osinaga E. (2013) GALNT9 gene expression is a prognostic marker in neuroblastoma patients. Clin. Chem. 59, 225–233 [DOI] [PubMed] [Google Scholar]
- 35. Xia L., Ju T., Westmuckett A., An G., Ivanciu L., McDaniel J. M., Lupu F., Cummings R. D., McEver R. P. (2004) Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J. Cell Biol. 164, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. ten Hagen K. G., Zhang L., Tian E., Zhang Y. (2009) Glycobiology on the fly. Developmental and mechanistic insights from Drosophila. Glycobiology 19, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tabak L. A. (2010) The role of mucin-type O-glycans in eukaryotic development. Semin. Cell Dev. Biol. 21, 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tran D. T., Zhang L., Zhang Y., Tian E., Earl L. A., Ten Hagen K. G. (2012) Multiple members of the UDP-GalNAc. Polypeptide N-acetylgalactosaminyltransferase family are essential for viability in Drosophila. J. Biol. Chem. 287, 5243–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tran D. T., Ten Hagen K. G. (2013) Mucin-type O-glycosylation during development. J. Biol. Chem. 288, 6921–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tian E., Hoffman M. P., Ten Hagen K. G. (2012) O-Glycosylation modulates integrin and FGF signalling by influencing the secretion of basement membrane components. Nat. Commun. 3, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raman J., Guan Y., Perrine C. L., Gerken T. A., Tabak L. A. (2012) UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferases. Completion of the family tree. Glycobiology 22, 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fritz T. A., Raman J., Tabak L. A. (2006) Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-2. J. Biol. Chem. 281, 8613–8619 [DOI] [PubMed] [Google Scholar]
- 43. Gerken T. A., Jamison O., Perrine C. L., Collette J. C., Moinova H., Ravi L., Markowitz S. D., Shen W., Patel H., Tabak L. A. (2011) Emerging paradigms for the initiation of mucin type protein O-glycosylation by the polypeptide GalNAc transferase (ppGalNAc T) family of glycosyltransferases. J. Biol. Chem. 286, 14493–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gerken T. A., Raman J., Fritz T. A., Jamison O. (2006) Identification of common and unique peptide substrate preferences for the UDP-GalNAc. Polypeptide α-N-acetylgalactosaminyltransferases T1 and T2 (ppGalNAc T1 and T2) derived from oriented random peptide substrates. J. Biol. Chem. 281, 32403–32416 [DOI] [PubMed] [Google Scholar]
- 45. Perrine C. L., Ganguli A., Wu P., Bertozzi C. R., Fritz T. A., Raman J., Tabak L. A., Gerken T. A. (2009) The glycopeptide preferring polypeptide-GalNAc transferase-10 (ppGalNAc T10), involved in mucin type-O-glycosylation, has a unique GalNAc-O-Ser/Thr binding site in its catalytic domain not found in ppGalNAc T1 or T2. J. Biol. Chem. 284, 20387–20397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gerken T. A., Gilmore M., Zhang J. (2002) Determination of the site-specific oligosaccharide distribution of the O-glycans attached to the porcine submaxillary mucin tandem repeat. Further evidence for the modulation of O-glycan side chain structures by peptide sequence. J. Biol. Chem. 277, 7736–7751 [DOI] [PubMed] [Google Scholar]
- 47. Gerken T. A., Tep C., Rarick J. (2004) Role of peptide sequence and neighboring residue glycosylation on the substrate specificity of the uridine 5′-diphosphate-α-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyl transferases T1 and T2. Kinetic modeling of the porcine and canine submaxillary gland mucin tandem repeats. Biochemistry 43, 9888–9900 [DOI] [PubMed] [Google Scholar]
- 48. Hassan H., Reis C. A., Bennett E. P., Mirgorodskaya E., Roepstorff P., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., Clausen H. (2000) The lectin domain of UDP-N-acetyl-d-galactosamine. Polypeptide N-acetylgalactosaminyltransferase-T4 directs its glycopeptide specificities, J. Biol. Chem. 275, 38197–38205 [DOI] [PubMed] [Google Scholar]
- 49. Hanisch F. G., Reis C. A., Clausen H., Paulsen H. (2001) Evidence for glycosylation-dependent activities of polypeptide N-acetylgalactosaminyltransferases rGalNAc-T2 and -T4 on mucin glycopeptides, Glycobiology 11, 731–740 [DOI] [PubMed] [Google Scholar]
- 50. Wandall H. H., Irazoqui F., Tarp M. A., Bennett E. P., Mandel U., Takeuchi H., Kato K., Irimura T., Suryanarayanan G., Hollingsworth M. A., Clausen H. (2007) The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc. Lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology 17, 374–387 [DOI] [PubMed] [Google Scholar]
- 51. Raman J., Fritz T. A., Gerken T. A., Jamison O., Live D., Liu M., Tabak L. A. (2008) The catalytic and lectin domains of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J. Biol. Chem. 283, 22942–22951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bennett E. P., Hassan H., Hollingsworth M. A., Clausen H. (1999) A novel human UDP-N-acetyl-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T7, with specificity for partial GalNAc-glycosylated acceptor substrates. FEBS Lett. 460, 226–230 [DOI] [PubMed] [Google Scholar]
- 53. Cheng L., Tachibana K., Zhang Y., Guo J., Kahori Tachibana K., Kameyama A., Wang H., Hiruma T., Iwasaki H., Togayachi A., Kudo T., Narimatsu H. (2002) Characterization of a novel human UDP-GalNAc transferase, pp-GalNAc-T10. FEBS Lett. 531, 115–121 [DOI] [PubMed] [Google Scholar]
- 54. Pedersen J. W., Bennett E. P., Schjoldager K. T., Meldal M., Holmér A. P., Blixt O., Cló E., Levery S. B., Clausen H., Wandall H. H. (2011) Lectin domains of polypeptide GalNAc-transferases exhibit glycopeptide binding specificity. J. Biol. Chem. 286, 32684–32696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoshimura Y., Nudelman A. S., Levery S. B., Wandall H. H., Bennett E. P., Hindsgaul O., Clausen H., Nishimura S. (2012) Elucidation of the sugar recognition ability of the lectin domain of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 3 by using unnatural glycopeptide substrates. Glycobiology 22, 429–438 [DOI] [PubMed] [Google Scholar]
- 56. Gerken T. A., Ten Hagen K. G., Jamison O. (2008) Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc transferase orthologue pairs. Glycobiology 18, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barb A. W., Meng L., Gao Z., Johnson R. W., Moremen K. W., Prestegard J. H. (2012) NMR characterization of immunoglobulin G Fc glycan motion on enzymatic sialylation. Biochemistry 51, 4618–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vester-Christensen M. B., Bennett E. P., Clausen H., Mandel U. (2013) in Glycosyltransferases: Methods and Protocols (Brockhausen I., ed), Methods in Molecular Biology, Humana Press, in press [Google Scholar]
- 59. Gerken T. A., Zhang J., Levine J., Elhammer A. (2002) Mucin core O-glycosylation is modulated by neighboring residue glycosylation status. Kinetic modeling of the site-specific glycosylation of the apo-porcine sumbaxillary mucin tandem repeat by UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases T1 and T2, J. Biol. Chem. 277, 49850–49862 [DOI] [PubMed] [Google Scholar]
- 60. Fritz T. A., Hurley J. H., Trinh L. B., Shiloach J., Tabak L. A. (2004) The beginnings of mucin biosynthesis. The crystal structure of UDP-GalNAc:polypeptide α-N-acetylgalactosaminyltransferase-T1. Proc. Natl. Acad. Sci. U.S.A. 101, 15307–15312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kubota T., Shiba T., Sugioka S., Furukawa S., Sawaki H., Kato R., Wakatsuki S., Narimatsu H. (2006) Structural basis of carbohydrate transfer activity by human UDP-GalNAc. Polypeptide α-N-acetylgalactosaminyltransferase (pp-GalNAc-T10). J. Mol. Biol. 359, 708–727 [DOI] [PubMed] [Google Scholar]
- 62. Wakinaka T., Kiyohara M., Kurihara S., Hirata A., Chaiwangsri T., Ohnuma T., Fukamizo T., Katayama T., Ashida H., Yamamoto K. (2013) Bifidobacterial α-galactosidase with unique carbohydrate-binding module specifically acts on blood group B antigen. Glycobiology 23, 232–240 [DOI] [PubMed] [Google Scholar]
- 63. Cuskin F., Flint J. E., Gloster T. M., Morland C., Baslé A., Henrissat B., Coutinho P. M., Strazzulli A., Solovyova A. S., Davies G. J., Gilbert H. J. (2012) How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules. Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hervé C., Rogowski A., Blake A. W., Marcus S. E., Gilbert H. J., Knox J. P. (2010) Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. U.S.A. 107, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fujimoto Z., Kuno A., Kaneko S., Yoshida S., Kobayashi H., Kusakabe I., Mizuno H. (2000) Crystal structure of Streptomyces olivaceoviridis E-86 β-xylanase containing xylan-binding domain. J. Mol. Biol. 300, 575–585 [DOI] [PubMed] [Google Scholar]
- 67. Imberty A., Piller V., Piller F., Breton C. (1997) Fold recognition and molecular modeling of a lectin-like domain in UDP-GalNac:polypeptide N-acetylgalactosaminyltransferases. Protein Eng. 10, 1353–1356 [DOI] [PubMed] [Google Scholar]
- 68. Hazes B. (1996) The (QxW)3 domain. A flexible lectin scaffold. Protein Sci. 5, 1490–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maveyraud L., Niwa H., Guillet V., Svergun D. I., Konarev P. V., Palmer R. A., Peumans W. J., Rougé P., Van Damme E. J., Reynolds C. D., Mourey L. (2009) Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra. Proteins 75, 89–103 [DOI] [PubMed] [Google Scholar]
- 70. Raju T. S., Davidson E. A. (1994) Role of sialic acid on the viscosity of canine tracheal mucin glycoprotein. Biochem. Biophys. Res. Commun. 205, 402–409 [DOI] [PubMed] [Google Scholar]
- 71. Tenno M., Saeki A., Kézdy F. J., Elhammer A. P., Kurosaka A. (2002) The lectin domain of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase 1 is involved in O-glycosylation of a polypeptide with multiple acceptor sites. J. Biol. Chem. 277, 47088–47096 [DOI] [PubMed] [Google Scholar]
- 72. Tenno M., Kézdy F. J., Elhammer A. P., Kurosaka A. (2002) Function of the lectin domain of polypeptide N-acetylgalactosaminyltransferase 1. Biochem. Biophys. Res. Commun. 298, 755–759 [DOI] [PubMed] [Google Scholar]
- 73. Zhang Y., Iwasaki H., Wang H., Kudo T., Kalka T. B., Hennet T., Kubota T., Cheng L., Inaba N., Gotoh M., Togayachi A., Guo J., Hisatomi H., Nakajima K., Nishihara S., Nakamura M., Marth J. D., Narimatsu H. (2003) Cloning and characterization of a new human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase, designated pp-GalNAc-T13, that is specifically expressed in neurons and synthesizes Tn antigen. J. Biol. Chem. 278, 573–584 [DOI] [PubMed] [Google Scholar]
- 74. Garringer H. J., Fisher C., Larsson T. E., Davis S. I., Koller D. L., Cullen M. J., Draman M. S., Conlon N., Jain A., Fedarko N. S., Dasgupta B., White K. E. (2006) The role of mutant UDP-N-acetyl-α-d-galactosamine-polypeptide N-acetylgalactosaminyltransferase 3 in regulating serum intact fibroblast growth factor 23 and matrix extracellular phosphoglycoprotein in heritable tumoral calcinosis. J. Clin. Endocrinol. Metab. 91, 4037–4042 [DOI] [PubMed] [Google Scholar]
- 75. Young W. W., Jr., Holcomb D. R., Ten Hagen K. G., Tabak L. A. (2003) Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isoforms in murine tissues determined by real-time PCR. A new view of a large family. Glycobiology 13, 549–557 [DOI] [PubMed] [Google Scholar]
- 76. Bennett E. P., Hassan H., Mandel U., Hollingsworth M. A., Akisawa N., Ikematsu Y., Merkx G., van Kessel A. G., Olofsson S., Clausen H. (1999) Cloning and characterization of a close homologue of human UDP-N-acetyl- α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase-T3, designated GalNAc-T6. Evidence for genetic but not functional redundancy. J. Biol. Chem. 274, 25362–25370 [DOI] [PubMed] [Google Scholar]