Background: DNA/RNA stimulates the ATPase function of a helicase by an unclear mechanism.
Results: A primuline derivative specifically stimulates the ATPase function of only the HCV genotype 1b NS3 helicase.
Conclusion: Small molecules can mimic RNA to stimulate the NS3 ATPase by binding near NS3 residues Arg-393, Glu-493, and Ser-231.
Significance: Compounds reveal key residues needed for ATP hydrolysis to fuel helicase movements.
Keywords: ATPases, Chemical Biology, Drug Discovery, High Throughput Screening (HTS), Molecular Motors, RNA Helicase
Abstract
ATP hydrolysis fuels the ability of helicases and related proteins to translocate on nucleic acids and separate base pairs. As a consequence, nucleic acid binding stimulates the rate at which a helicase catalyzes ATP hydrolysis. In this study, we searched a library of small molecule helicase inhibitors for compounds that stimulate ATP hydrolysis catalyzed by the hepatitis C virus (HCV) NS3 helicase, which is an important antiviral drug target. Two compounds were found that stimulate HCV helicase-catalyzed ATP hydrolysis, both of which are amide derivatives synthesized from the main component of the yellow dye primuline. Both compounds possess a terminal pyridine moiety, which was critical for stimulation. Analogs lacking a terminal pyridine inhibited HCV helicase catalyzed ATP hydrolysis. Unlike other HCV helicase inhibitors, the stimulatory compounds differentiate between helicases isolated from various HCV genotypes and related viruses. The compounds only stimulated ATP hydrolysis catalyzed by NS3 purified from HCV genotype 1b. They inhibited helicases from other HCV genotypes (e.g. 1a and 2a) or related flaviviruses (e.g. Dengue virus). The stimulatory compounds interacted with HCV helicase in the absence of ATP with dissociation constants of about 2 μm. Molecular modeling and site-directed mutagenesis studies suggest that the stimulatory compounds bind in the HCV helicase RNA-binding cleft near key residues Arg-393, Glu-493, and Ser-231.
Introduction
Helicases are motor proteins fueled by ATP hydrolysis that translocate on nucleic acids to separate strands, rearrange base pairs, or dislodge nucleic acid-binding proteins. Because nucleic acids in turn stimulate helicase-catalyzed ATP hydrolysis, it is conceivable that a small molecule might bind the protein to elicit the same effect. Such compounds might be useful as tools to design drugs that target helicases or as probes of helicase function, but until now none have been reported. Here we report the discovery that a small molecule synthesized from the main component of the yellow dye primuline (1) binds the helicase encoded by the hepatitis C virus (HCV)2 in such a manner that it mimics RNA to stimulate helicase-catalyzed ATP hydrolysis.
HCV is a positive-sense, single-stranded virus that infects ∼170 million people worldwide. The HCV genome encodes a ∼3,000 amino acid long polypeptide, which is processed by cellular and viral proteases into three structural proteins (core, E1 and E2) and seven non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). The HCV helicase is part of NS3, which is multifunctional, and has an N-terminal serine protease function in addition to its C-terminal helicase function. The recently approved antiviral drugs boceprevir (2) and telaprevir (3) are potent NS3 protease inhibitors. Less progress has been made toward developing NS3 helicase inhibitors as antivirals.
The C-terminal portion of NS3 (called NS3h) can be expressed in Escherichia coli as a NS3 fragment that retains helicase function but lacks protease function. NS3h has two well-defined ligand binding sites. ATP binds NS3h between its two conserved helicase motor domains (4), and one strand of RNA (or DNA) binds in a cleft that separates the two N-terminal helicase motor domains from a third C-terminal helical domain (5). X-ray crystallography has revealed that helicase inhibitors such as nucleotide analogs (4) and the triphenylmethane dye Soluble Blue HT (6) bind NS3h in its ATP binding site. The interaction of small molecules with the NS3 RNA binding cleft has not yet been directly observed, but numerous compounds are known to competitively inhibit the ability of NS3 to interact with DNA or RNA, suggesting that they bind NS3 reversibly in place of nucleic acids. Such compounds include peptide nucleic acids (7), symmetrical benzimidazoles (8), and nucleotide mimics (9). Unlike the natural substrates of NS3, these compounds either inhibit helicase-catalyzed ATP hydrolysis or they have no apparent effect on helicase-catalyzed ATP hydrolysis. Until now, no small molecules, other than DNA and RNA oligonucleotides, have been reported to stimulate NS3 helicase-catalyzed ATP hydrolysis.
Primuline is a fluorescent dye composed of a mixture of benzothiazole oligomers terminating with p-aminobenzene groups, and it prevents proteins from binding ssDNA or RNA (1, 10). Li et al. synthesized a series of chemical derivatives from a benzothiazole dimer purified from primuline, and they showed that some of these compounds potently and specifically inhibit HCV helicase (1). Later, Mukherjee et al. (10) showed that these primuline derivatives inhibit NS3h by displacing the protein after it is bound to its nucleic acid substrate. Unlike other compounds that simply compete with the DNA, the primuline derivatives, and related benzothiazole polymers like the dye titan yellow, cause NS3 to fall from DNA even in the absence of ATP (8, 10), suggesting they can actively induce a protein conformational change needed to release the helicase protein from the nucleic acid tract on which it translocates (11, 12).
Ndjomou et al. characterized a small subset of primuline derivatives with regard to their effects on the ability of NS3 to cleave ATP and peptides, and their antiviral effects in cells. All 13 primuline derivatives that Ndjomou et al. tested inhibited NS3h-catalyzed ATP hydrolysis, but much more of the primuline derivatives were needed to inhibit NS3h-catalyzed ATP hydrolysis than that were needed to inhibit helicase catalyzed DNA or RNA unwinding. These results suggest that primuline derivatives do not act nonspecifically (e.g. through protein aggregation), and that they do not inhibit helicase action simply by binding in place of ATP (13).
Here, we have analyzed the rest of the primuline derivatives in the Li et al. (1) collection with regard to their effect on NS3 helicase-catalyzed ATP hydrolysis. We report the surprising discovery that two primuline derivatives stimulate helicase-catalyzed ATP hydrolysis. Since such an activity has not yet been reported for any small molecule (other than DNA or RNA oligonucleotides), we also report a biochemical characterization of the interaction of stimulatory primuline derivatives with NS3h. Data reveal that the compounds appear to bind near residues in the NS3h RNA-binding cleft. The compounds should therefore be useful to design more specific NS3 helicase inhibitors that could be used as molecular probes or as the basis to design antiviral drugs to be used alone, in combination with NS3 protease inhibitors, or with other antiviral drugs.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
All oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). The truncated C-terminal His-tagged NS3 proteins lacking the protease domain (i.e. NS3h) were purified as described before: NS3h_1a(JFH1), NS3h_1b(J4), NS3h_2a(J6) (14), NS3h_1b(con1) (15), NS3h_2a(JFH1) (8), NS3h_R393A (16), NS3h_E493Q (17). An open reading frame expressing NS3h_S231A was generated for this study from a plasmid expressing NS3h_1b(con1) by Mutagenex Inc. (Hillsborough, NJ). The plasmid was used to express the NS3h_S231A protein, which was purified using the same procedure used to purify wild type NS3h.
Primuline derivatives were prepared as described previously (1). Compound 1 (CID 50930756) and compound 2 (CID 50930734), were formerly compounds 31 & 32 in Li et al. (1) respectively. Compound 3 (CID 49849280) was formerly compound 5 in Li et al. (1).
ATP Hydrolysis (ATPase) Assays
NS3h-catalyzed ATP hydrolysis was monitored using a colorimetric assay that measures inorganic phosphate released from ATP (assay 1), and two coupled assays that monitor helicase-catalyzed ADP release from ATP (assays 2 & 3).
Assay 1 (Malachite Green Assay)
This assay was used to generate data shown in Figs. 1, A, C, D and 2, 3 and 5. Reactions (30 μl) were initiated by adding 3 μl of ATP of 10 mm ATP to 27 μl of assembled reagents, such that final concentrations were 25 mm MOPS pH 6.5, 1.25 mm MgCl2, 1 mm ATP, 33 μg/ml BSA, 0.07% (v/v) Tween 20, 0.3 mm DTT, 10% DMSO, and 50 nm HCV helicase with indicated compound concentrations. After 15 min at 37 °C, reactions were terminated by adding 200 μl of the malachite green reagent (3 volumes 0.045% (w/v) malachite green: 1 volume 4.2% ammonium molybdate in 4N HCl: 0.05 volume 20% Tween 20), followed immediately by adding 30 μl of 35% sodium citrate. After 30 min, A630 was read, and net phosphate produced was calculated from a phosphate standard curve after subtracting values obtained in control reaction lacking helicase. In Figs. 1, 3 & 5, data are expressed relative to control reaction performed in the absence of compound.
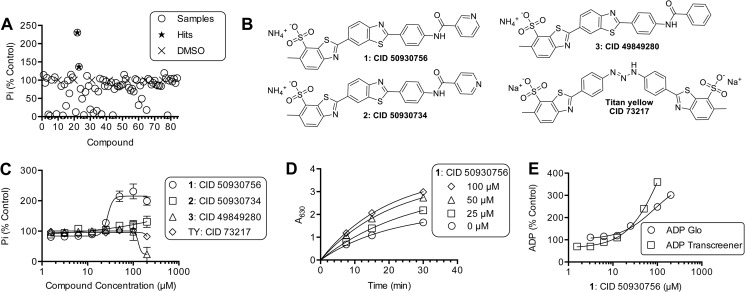
FIGURE 1.
Primuline derivatives that stimulate HCV helicase-catalyzed ATP hydrolysis. A, ATP hydrolysis catalyzed by 100 nm NS3h_1b(con1) was monitored using assay 1 (“Experimental Procedures”) in the presence of 88 different primuline derivatives. All reactions contained 5% DMSO and 100 μm compound (circles). Percent activity remaining (y axis) was calculated by normalizing observed phosphate concentrations to average phosphate observed in reactions containing DMSO only (x). The reactions were incubated for 30 min at 23 °C. B, structures of two hits (1 and 2), a compound with the terminal pyridine replaced with benzene (compound 3) and the related soluble dye titan yellow. C, NS3h-catalyzed phosphate released from ATP in the presence of indicated concentrations of compounds 1–3 and titan yellow. D, kinetics of NS3h catalyzed ATP hydrolysis in the presence of indicated concentrations of compound 1. Data are fit to a first order rate equation. E, NS3h-catalyzed ADP release from ATP in the presence of various concentrations of compound 1, as measured by either the ADP-Glo assay (Promega, Assay 2 methods) or the ADP transcreener assay (BellBrook labs, Assay 3 methods).
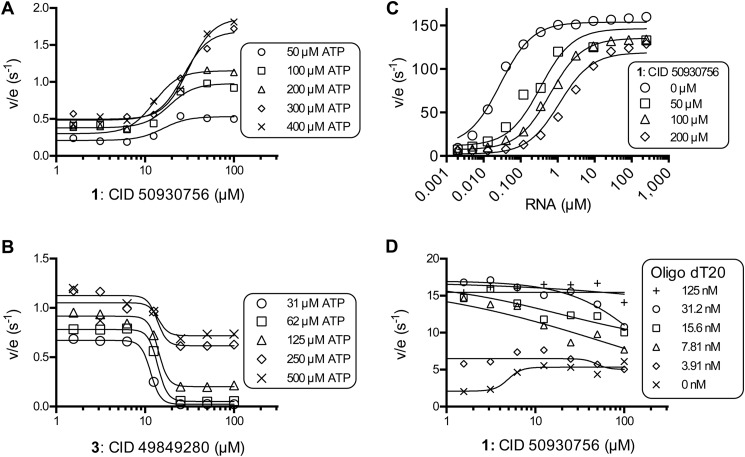
FIGURE 2.
Effect of primuline derivatives on NS3h-catalyzed ATP hydrolysis in the presence of various concentrations of ATP and RNA. A, specific activities (nmol PO4/s/nmol NS3h) observed in the presence of various concentrations of compound 1 at the indicated initial concentrations of ATP. B, specific activities observed in the presence of various concentrations of compound 3 at the indicated initial concentrations of ATP. C, specific activities observed in the presence of compound 1 and various concentrations of PolyU RNA. Data are fitted to Equation 1 (“Experimental Procedures”), with kslow of 3 ± 2 s−1, kfast of 142 ± 7 s−1, KRNA of 26 ± 7 nm, Ki of 4 ± 1.2 μm, and Kii of 920 ± 400 μm. Uncertainties represent 95% confidence intervals for the global non-linear regression. D, specific activities observed in the presence of compound 1 and various concentrations of the oligonucleotide dT20 (5′-TTT TTT TTT TTT TTT TTT TT). In panels A, B, and D, data are fitted to a standard 4-parameter concentration-response equation using GraphPad Prism (v. 6).
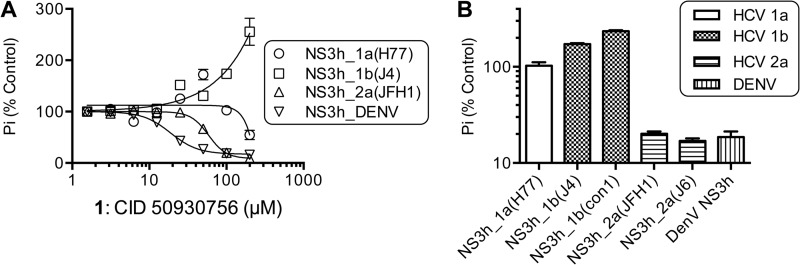
FIGURE 3.
Compound 1 only stimulates NS3h encoded by HCV genotype 1b strains. A, relative rates of ATP hydrolysis catalyzed in the presence of various concentrations of compound 1 by NS3h proteins derived from various HCV genotypes and dengue virus (DENV). B, relative rates of ATP hydrolysis catalyzed by various NS3h enzymes in the presence of 100 μm of compound 1. Data are expressed relative to control reactions containing the same proteins that were performed in the absence of compound 1. All reactions were performed in triplicate, points are average values, and error bars depict standard deviations.
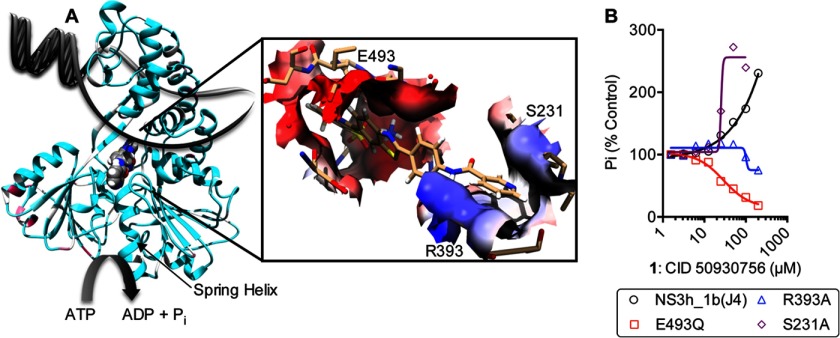
FIGURE 5.
Possible molecular interactions between compound 1 and HCV helicase. A, molecular model of compound 1 docked in PDB file 3KQN (genotype 1b(con1)). NS3 helicase is depicted as ribbons colored by conservation with a gradient from cyan (identical) to maroon (most variable). Compound 1 is shown as a space-filled model, which is bound between the motor domains perpendicular to the nucleic acid binding cleft. The inset shows the surface of NS3 that contacts compound 1 (sticks) in a model of the binding surface colored based on electrostatic potential using UCSF Chimera. Residues targeted for site directed mutagenesis are shown as sticks and labeled. B, response of NS3 harboring amino acid substitution in the putative compound 1 binding cleft to various concentrations of compound 1.
In Fig. 2, concentration of ATP cleaved in 15 min (v) and total enzyme concentration (Et) were used to calculate specific activities (v/Et). Data in Figs. 2, A, B, and D were fitted to a standard dose response equation (GraphPad Prism). In Fig. 2C, data were globally fitted to PolyU RNA concentration using GraphPad Prism (v. 6.0) and Equation 1.
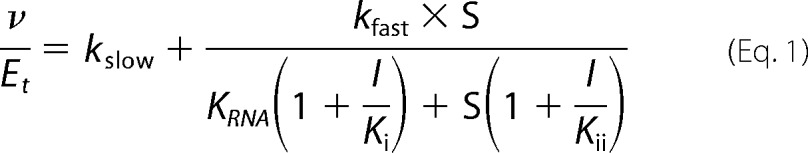 |
In Equation 1, v is the initial rate of ATP hydrolysis, Et is NS3h concentration, kslow is a first order rate constant describing ATP hydrolysis in the absence of RNA, kfast is a first order rate constant describing ATP hydrolysis in the presence of RNA, KRNA is the concentration of RNA leading to a half maximal rate of ATP hydrolysis, I is compound concentration, Ki is the apparent dissociation constant for NS3h and compound when RNA is absent, Kii is the apparent dissociation constant for NS3h and compound when RNA is bound to NS3h.
Assay 2 (ADP GloTM)
Reactions were carried out in a total volume of 5 μl in Greiner Bio-One 384-well small volume microtiter plates and included 100 nm NS3h_1b(con1), 25 mm MOPS pH 6.5, 1 mm MgCl2, 1 mm ATP, 10% DMSO, 40 μg/ml BSA, and 0.08% Tween 20. After incubation for 15 min at 37 °C, reactions were terminated by adding 5 μl of ADP-GloTM reagent (Promega). Further incubation for 40 min at room temperature was completed before the addition of 10 μl of Kinase Detection ReagentTM (Promega). After 60 min at room temperature, luminescence was read using a BMG Labtech OMEGA plate reader (positioning delay, 0.2 s; gain, 2500). The amount of ADP produced was determined from an ATP/ADP standard curve.
Assay 3 (Transcreener® ADP2)
Helicase-catalyzed ATP hydrolysis was monitored using the Transcreener ADP2 Fluorescent Intensity (FI) kit (BellBrook Labs), per the manufacturer's instructions. Briefly, reactions were carried out a 10 μl total volume in Greiner 384-well flat bottom black polystyrol small volume microtiter plates and included 150 nm NS3h_1b(con1), 25 mm MOPS pH 6.5, 1.25 mm MgCl2, 1 mm ATP, 5% DMSO, 5 μg/ml BSA, and 0.01% Tween 20. After incubation for 15 min at 37 °C, reactions were terminated by adding 10 μl of Transcreener ADP Detection Mixture (BellBrook Labs). Further incubation for 60 min at room temperature was performed before fluorescence intensity was read using a Tecan Infinite M1000 plate reader (excitation wavelength 580 nm; emission wavelength of 620 nm, bandwidth 20 nm). The amount of ADP produced was determined from an ATP/ADP standard curve.
Intrinsic Protein Fluorescence
Intrinsic Protein Fluorescence (IPF) was monitored in a 1 cm cuvette in a Varian Cary Eclipse fluorescence spectrophotometer (Agilent Technologies). NS3h (200, 100, or 50 nm) was present in solution containing 25 mm MOPS, pH 7.0, 1.25 mm MgCl2, 5% DMSO, and 0.01% Tween 20. Fluorescence (Excitation 280 nm, Emission 340 nm with 5 nm slit widths) was recorded after each (1 μl) addition of compound dissolved in the same buffer, after stirring 23 °C for 2 min. Fluorescence values were corrected for dilution and inner filter effects using Equation 2.
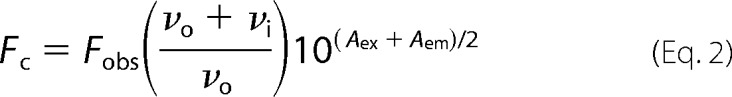 |
In Equation 2, Fc is corrected fluorescence, Fobs is observed fluorescence, vo is initial sample volume, vi is total volume of titrant added, Aex is the absorbance of the solution at the excitation wavelength (280 nm), and Aem is the absorbance of the solution at the observed emission wavelength (340 nm).
The resulting corrected fluorescence values were fitted to the Hill equation to calculate the ligand concentration needed for half occupation (Kd), using Equation 3.
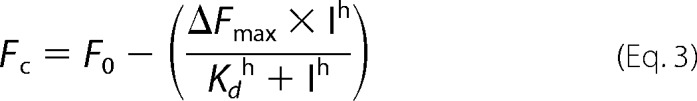 |
In Equation 3, I is compound concentration, Fc is corrected fluorescence, F0 is fluorescence in the absence of compound, ΔFmax is the maximum change in fluorescence, h is the Hill coefficient.
Molecular Modeling
Compound 1 was docked into PDB files 3KQN and 3KQL that were stripped of their ligands (4). Briefly, water molecules and counterions were removed from each PDB file, and incomplete side chains were filled using UCSF Chimera 1.6.2 (18). Using UCSF Chimera's Dock Prep module, histidine protonation states were calculated, and incomplete side chains were automatically filled. A three-dimensional conformation of the compound 1 was generated using Open Babel GUI, was saved as a PDB file, and positioned using a rigid body orienting code in UCSF DOCK 6.5 (19). During modeling, the ligand-binding site of the protein was constrained to be the largest cluster of spheres surrounding the molecular surface of the protein as generated by UCSF DOCKS's sphgen module. No additional constraints were made on the resolution of the binding site.
RESULTS
Identification of HCV Helicase Inhibitors That Stimulate Helicase-catalyzed ATP Hydrolysis
This project was initiated to understand how NS3h-catalyzed ATP hydrolysis is influenced by primuline derivatives. Primuline derivatives were previously shown to inhibit the ability of NS3 helicase to unwind its DNA substrates (1). To understand how primuline derivatives affect the ability of ATP to fuel unwinding, all compounds in the primuline derivative collection described by Li et al. (1) were analyzed for their effect on NS3h-catalyzed ATP hydrolysis using a colorimetric assay (Fig. 1A). All ATPase assays in this screen were performed in the absence of RNA (or DNA) because primuline derivatives are known to prevent NS3 from binding nucleic acids (10). RNA stimulates helicase-catalyzed ATP hydrolysis, and any compound that prevents RNA from binding NS3h would inhibit ATP hydrolysis in the presence of RNA. About two-thirds of the compounds in the collection inhibited NS3h-catalyzed ATP hydrolysis, about one-third had no apparent effect at the highest concentration tested (200 μm). Surprisingly, two compounds stimulated helicase-catalyzed ATP hydrolysis: Compound 1, i.e. 6-methyl-2-[2-[4-(pyridine-3-carbonylamino)phenyl]-1,3-benzothiazol-6-yl]-1,3-benzothiazole-7-sulfonate (PubChem (20) CID 50930756) and compound 2, i.e. 6-methyl-2-[2-[4-(pyridine-4-carbonylamino)phenyl]-1,3-benzothiazol-6-yl]-1,3-benzothiazole-7-sulfonate (PubChem (20) CID 50930734) (Fig. 1A).
Both stimulatory compounds were similar because they were the only two compounds in the collection with a terminal pyridine moiety (Fig. 1B). The position of the nitrogen of the pyridine ring was critical for stimulation, as evidenced by the observation that compound 2 stimulates helicase-catalyzed ATP hydrolysis about 30%, whereas compound 1 stimulates ATP hydrolysis 2.5-fold. All compounds in the collection in which the pyridine was replaced with benzene (e.g. compound 3 (CID 49849280)) inhibited NS3h-catalyzed ATP hydrolysis, typically with IC50 values that were greater than the concentration of compound 1 needed to stimulate ATP hydrolysis by 50%. For example, compound 3 inhibited ATP hydrolysis with an IC50 value of 130 ± 70 μm, and compound 1 stimulated ATP hydrolysis with an EC50 value of 27 ± 6 μm (Fig. 1C).
Li et al. previously showed that the solubility of compound 3 (formerly compound 5 in Ref. 1), and other primuline derivatives terminated with substituted benzenes, is low (i.e. < 50 μm) in helicase assay buffer. However, they also showed that compound 1 is more than 5 times more soluble; compound 1 is fully soluble at the highest concentration tested by Li et al. (125 μm) and unlike related compounds, compound 1 is soluble in buffer lacking any non-ionic detergents (1). It is therefore unlikely that the solubility of compound 1 (or 2) limits its activity in the buffers used here, but it is possible that the inhibition seen with compound 3 is due to compound-induced aggregation at high concentration. Therefore, we chose a freely soluble benzothiazole dye, titan yellow (CID 73217), which also inhibits the ability of NS3h to load onto DNA substrates, to compare with compounds 1 and 2. Titan yellow inhibits NS3h catalyzed DNA unwinding with an IC50 of 12 μm (10), but titan yellow has little effect on NS3h-catalyzed ATP hydrolysis, inhibiting the reaction by only 18% at 200 μm (Fig. 1C).
To determine if the apparent stimulation of ATP hydrolysis by 1 and 2 was simply due to the fact that the compounds enhanced the absorbance of the colored product in the phosphate assay, each was subjected to a simple counter screen where 100 μm of each was mixed with 70 μm inorganic phosphate, followed by the addition of the colorimetric reagents. Neither compound enhanced (or diminished) absorbance of the phosphomolybdate: malachite green complex. Further evidence that stimulation is not due to a chemical effect was obtained from the observations that absorbance in the phosphate assay increased both with time and with compound concentration (Fig. 1D), and that compound 1 stimulated two ATPase assays that monitor ADP rather than the phosphate product of ATP hydrolysis. Compound 1 stimulated both ADP assays to similar extents, and in a concentration-dependent fashion similar to that seen with the phosphate assay (Fig. 1E).
PolyU RNA Prevents Compound 1 from Stimulating NS3h-catalyzed ATP Hydrolysis
Compound 1 stimulated NS3h-catalyzed ATP hydrolysis in a concentration-dependent manner regardless of the concentration of ATP in the reaction (Fig. 2A). However, at higher ATP concentrations, more compound 1 was needed to stimulate the reaction by 50%. At 50 μm ATP, the EC50 value for compound 1 was 15 μm, but in the presence of 400 μm ATP, compound 1 stimulated NS3h with an EC50 of 31 μm. A similar phenomenon has been seen before with RNA and DNA. Both RNA and DNA increase the apparent Km for ATP in the helicase reaction, and both bind NS3h weaker in the presence of ATP (21). In contrast, compound 3 (i.e. the benzene analog) inhibited steady-state rates of NS3h-catalyzed ATP hydrolysis at each ATP concentration tested, and more compound 3 was needed to inhibit reactions performed at higher ATP concentrations to a similar extent (Fig. 2B). When data with compound 3 were plotted as velocity versus ATP concentration (not shown) and fitted to the Michaelis-Menten equation, compound 2 lowered both apparent Vmax and raised apparent Km, in a concentration-dependent manner. Apparent Km and apparent Vmax (or Vmax/Km) were not linearly dependent on compound 3 concentration, however, suggesting that this scaffold does not bind the same site as ATP, nor does it act as a classic non-competitive inhibitor of ATP.
To examine the possibility that compound 1 binds to the NS3h RNA binding site, as has been observed with other HCV helicase inhibitors (8), the ability of RNA to compete with compound 1 to stimulate helicase-catalyzed ATP hydrolysis was examined. In the absence of compound 1, RNA stimulated NS3h-catalyzed ATP hydrolysis by 165-fold and the concentration of RNA needed to stimulate a half-maximal rate of ATP hydrolysis (KRNA) was 20 nm (expressed in terms of nucleotide concentration). Compound 1 increased KRNA over 100-fold to 2,300 nm, and it also decreased the maximum to which RNA stimulated ATP hydrolysis (Fig. 2C). Unlike other compounds that act by binding in place of RNA, such as symmetrical benzimidazoles (8), data did not fit a steady-state rate equation that assumes compound 1 competes with RNA for the same site that activates helicase-catalyzed ATP hydrolysis. Reasonable fits were obtained from models that assume that compound 1 acts as a mixed inhibitor of RNA stimulation (Equation 1, see “Experimental Procedures”). The model assumes compound 1 binds the enzyme with a Ki of 4 ± 1.2 μm and that compound 1 binds the enzyme RNA complex with an Kii of = 920 ± 400 μm (Fig. 2C). Similar experiments with DNA oligonucleotides (not shown) yielded similar results.
In experiments designed to observe RNA stimulation of NS3h-catalyzed ATP hydrolysis, the 2–4-fold stimulation of NS3-catalyzed ATP hydrolysis by compound 1 is difficult to observe because RNA stimulates hydrolysis by 50–100-fold. We therefore also examined the ability of compound 1 to compete with a DNA oligonucleotide that stimulates ATP hydrolysis by only about 15-fold (Fig. 2D). In assays performed with the oligonucleotide, compound 1 stimulated helicase-catalyzed ATP hydrolysis in the absence of DNA, but it inhibited ATP hydrolysis in the presence of saturating DNA concentrations (Fig. 2D).
Compound 1 Only Stimulates ATP Hydrolysis Catalyzed by NS3h Encoded by HCV Genotype 1b
All the above assays were performed with truncated NS3 lacking its protease function (i.e. NS3h) that was isolated from the con1 strain of HCV genotype 1b (GenBankTM AB114136). The con1 HCV strain is of interest because the strain is the basis for HCV subgenomic replicons that are frequently used to test HCV antiviral agents (22). To examine compound specificity, the same assays above were repeated using NS3h isolated from another genotype 1b strain (J4, GenBankTM AF054254 (23)), HCV genotype 1a strain H77 (GenBankTM AF011751 (24)), HCV genotype 2a strain J6 (GenBankTM AF177036 (25)), and the HCV genotype 2a JFH1 strain (GenBankTM AJ238799 (26)), and Dengue Virus strain 2 (DENV) (GenBankTM 2BMF (8)). Remarkably, compound 1 stimulated ATP hydrolysis catalyzed by NS3h from the second HCV genotype 1b strain tested, but it inhibited all other NS3h proteins to some extent (Fig. 3). Compound 1 stimulated NS3h_1b(J4) to a similar extent that it stimulated NS3h_1b(con1). The potency and extent to which compound 1 inhibited the other enzymes varied. Compound 1 was the most effective inhibitor of ATP hydrolysis catalyzed by DENV NS3h, with an IC50 value of 18 ± 2 μm (Fig. 3A).
Interaction of Compound 1 with NS3 as Monitored Using Intrinsic Protein Fluorescence
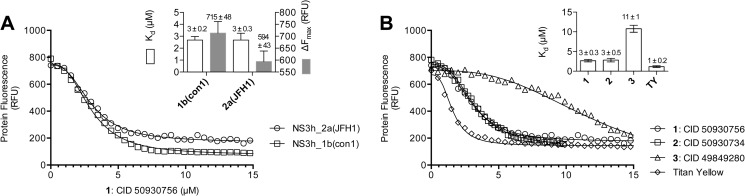
We next sought to understand if the stimulatory primuline derivatives bound NS3 in the absence of the ATP substrate. To this end, we examined the effect of each compound on NS3h intrinsic protein fluorescence. Ndjomou et al. recently showed that primuline derivatives quench intrinsic protein fluorescence with apparent dissociation constants in the low micromolar range similar to the IC50 values seen in helicase assays (13). Similar results were obtained here for compounds 1, 2, 3, and titan yellow, with helicases isolated from either genotype 1b or 2a (Fig. 4A). Compounds 1 and 2 appear to bind NS3h about 3-times weaker than titan yellow, and both ∼4-times more tightly than compound 3 (Fig. 4B).
FIGURE 4.
Interaction of primuline derivatives and titan yellow with NS3 helicase in the absence of RNA and ATP. Intrinsic protein fluorescence was monitored in the presence of various concentrations of compounds 1–3 and titan yellow. A, shown are two representative titrations of either 200 nm NS3h_1b(con1) or 200 nm NS3h_2a(JFH1) with compound 1. The inset shows average (± S.D.) Kd and ΔFmax values (Equation 3, methods) obtained from 3 separate titrations. B, shown are three representative titrations of 200 nm NS3h_2a(JFH1) with each compound. Three data sets were obtained with each compound and either 50 nm, 100 nm or 200 nm NS3h. Titrations were fitted to Equation 3 (“Experimental Procedures”), and the resulting dissociation constants were averaged to yield the dissociation constants shown on the inset. Error bars are standard deviations from the three repeats.
Effect of Compound 1 on NS3h with Altered Residues Needed for RNA Binding
To better localize where compound 1 might bind NS3, we next examined possible binding sites using molecular modeling. For compound 1 to stimulate helicase catalyzed ATP hydrolysis, it would need to interact when ATP is also bound. To examine possible interactions with NS3h in the presence of ATP, we docked compound 1 into two x-ray crystal structures that show ATP analogs bound to NS3h. The first, Protein Data Bank (PDB) file 3KQN, shows the enzyme bound to ADP-BeF3, which mimics the NS3h:ATP ground state. The second, PDB file 3KQL, shows NS3h bound to ADP-AlF4, which mimics the transition state that occurs during ATP hydrolysis (4). Ligands, water molecules, and counterions were removed from each PDB file using UCSF Chimera 1.6.2, and UCSF DOCK 6.5 (19) was used to position compound 1 in the cleft surrounding the RNA binding site.
Modeling with either PDB file 3KQN or 3KQL predicted that compound 1 binds NS3h such that it makes intimate contacts with several residues known to be involved in the helicase-catalyzed unwinding reaction (Fig. 5A). These residues include Arg-393, which helps clamp RNA in the RNA-binding cleft (16), Glu-493, whose ionization state affects the ability of NS3h to unwind DNA and RNA (17), and Ser-231, which forms a hydrogen bond with the DNA backbone (27). Since changing these residues by site directed mutagenesis affects DNA binding but not the ability of NS3h to hydrolyze ATP (16, 17), we tested if NS3h harboring these mutations responded differently to compound 1. Remarkably, compound 1 inhibited both R393A and E493Q. In contrast, the S231A protein was stimulated, but less compound 1 was needed to stimulate ATP hydrolysis to the same extent, suggesting that the compound bound more tightly to S231A than it did to the wild type protein (Fig. 5B).
DISCUSSION
This study reports the discovery of the first non-nucleosides that stimulate HCV helicase-catalyzed ATP hydrolysis. Besides RNA and DNA, the only other compounds previously reported to stimulate HCV helicase catalyzed ATP hydrolysis are ribavirin (28), and other nucleoside analogs, similar to DNA or RNA building blocks (29, 30). The compounds here might therefore provide a more novel starting point to design specific antiviral drugs targeting the viral helicase.
HCV helicase is a notoriously difficult drug target. HCV helicase is needed for HCV replication (31), the protein was first crystallized over 15 years ago, and numerous high throughput assays are available that can be used to identify inhibitors (32–34). However, relatively few HCV helicase inhibitors have been reported and the mechanism of action of most known HCV helicase inhibitors is still not clear. The primuline scaffold was recently optimized to find other compounds that are either specific HCV helicase inhibitors (e.g. CID 50930730), or compounds that inhibit HCV replication in cells (e.g. CID 50930749). The compounds reported here, which possess the unique ability to stimulate helicase-catalyzed ATP hydrolysis, were originally designed to enhance the solubility of the primuline derivative scaffold, by substituting the terminal benzene with a pyridine (1). The pyridine moiety in these compounds is critical for stimulation, as evidenced by the fact that replacing it with benzene leads to inhibition rather than stimulation. The results here shed new light on how the primuline scaffold interacts with NS3h, suggesting that compounds in this series mimic nucleic acid to the extent that some can elicit the same biological response as DNA, or RNA, i.e. they stimulate helicase-catalyzed ATP hydrolysis. Prior studies have shown that both compounds 1 and 2 inhibit the ability of NS3h to unwind DNA with IC50 values of 22 μm and 50 μm, respectively. When administered to cells harboring HCV subgenomic replicons at 10 μm, the compounds decrease replicon content by about 50%, but do not reduce cell viability (1).
Unlike other compounds that have been previously reported to stimulate ATP hydrolysis catalyzed by other helicases, compound 1 does not stimulate NS3h-catalyzed ATP hydrolysis in the presence of RNA (Fig. 2). Other than nucleosides, there is one noteworthy example of a compound that stimulates helicase-catalyzed ATP hydrolysis in the presence of nucleic acids. Bordeleau et al. (35) found that the eukaryotic translation inhibitor pateamine (CID 10053416) stimulates ATP hydrolysis catalyzed by eukaryotic initiation factor 4A (eIF4A) but only in the presence of nucleic acids. Unlike the compounds described here, pateamine enhances the affinity of the helicase for RNA, leading to both enhanced helicase unwinding activity and enhanced helicase-catalyzed ATP hydrolysis. Compound 1 inhibits NS3h-catalyzed ATP hydrolysis in the presence of RNA and causes the enzyme to bind RNA weaker (Fig. 2C). As a consequence, unlike pateamine (35), compound 1 inhibits the ability of a helicase to bind and unwind its nucleic acid substrates (1).
The magnitude by which compound 1 stimulates HCV helicase-catalyzed ATP hydrolysis varies with the concentration of ATP present in the reaction. Stimulation also depends on ATP concentrations such that less stimulation is observed at lower ATP concentrations. Compound 1 also appears to modulate HCV helicase-catalyzed ATP hydrolysis similar to RNA (or DNA), in that both cause the apparent Km of ATP in the reaction to increase. Increasing concentrations of ATP similarly cause a reciprocal increase in the amount of RNA (21, 36, 37) or compound 1 (Fig. 2A) needed to stimulate NS3h-catalyzed ATP hydrolysis by 50%. Compound 1 also causes the apparent affinity of NS3 for RNA to decrease (Fig. 2C), but not as a linear competitive inhibitor of RNA binding, as has been observed with other compounds (8).
Another remarkable property of the compounds described here is their unique ability to differentiate between NS3 helicases derived from different HCV genotypes. Whereas most NS3 protease inhibitors show a clear preference for certain HCV genotypes (38), other helicase inhibitors tested to date on various HCV genotypes act similarly against NS3 purified from different genotypes (8, 39). While it is important for a potential HCV antiviral to act on a wide variety of strains, the lack of specificity seen with helicase inhibitors might be indicative of the fact that they act by an indirect or nonspecific mechanism. It is therefore important to note here that the primuline derivatives only stimulated ATP hydrolysis catalyzed by helicase isolated from HCV genotype 1b. In contrast, compound 1 inhibited ATP hydrolysis catalyzed by NS3 purified from other HCV genotypes (e.g. 1a and 2a) or related flaviviruses (e.g. Dengue virus) (Fig. 3).
Neither ATP nor RNA is needed for compound 1 to interact with NS3h, as shown by the ability of compound 1 to quench intrinsic protein fluorescence in a dose dependent manner with a dissociation constant of 3 ± 0.3 μm. This observation suggests that the compound will be bound to NS3h under all conditions where stimulation is observed (Fig. 4). NS3h has four tryptophan residues, all of which are in the C-terminal helical domain (Trp-501, Trp-532, Trp-578, and Trp-582). The key Trp-501 is present in the NS3h RNA binding cleft and it acts like a bookend to trap nucleic acid in the enzyme (40, 41). Again, the presence of the pyridine moiety influences the interaction observed with protein fluorescence, and when it is replaced with benzene, the compound binds about four times more weakly (Fig. 4B). We also observed a slight difference in the fluorescence change that compounds induce in the different NS3h proteins, which might be correlated with the fact that compound 1 stimulates genotype 1b NS3h but it inhibits genotype 2a NS3h catalyzed ATP hydrolysis (Fig. 4A). This subtle difference, however, might simply be due to the fact that genotype 2a helicase has three fewer phenylalanines (Phe-197, Phe-418, and Phe-473). All are tyrosines in genotype 2a strains. Of these, Phe-418 is the most likely to play a role in ATPase stimulation because it lies in the ATP binding cleft (42).
We attempted to use molecular modeling to clarify the molecular basis for this intriguing compound specificity. The first structure used one recently reported by Gu & Rice (4) that shows HCV genotype 1b(con1) NS3h bound to ADP-AlF4, a non-hydrolysable ATP analog that mimics the pentavalent transition state that presumably occurs during NS3h-catalyzed ATP hydrolysis (PDB 3KQL). Compound 1 docked perpendicular to the nucleic acid binding site in the cleft between the helicase motor domains. We speculate that in this conformation, compound 1 might stabilize the motor domains, so they catalyze ATP hydrolysis. When we repeated the DOCK procedure with PDB 3KQN, which displays NS3h bound to the ground state analog ADP-BeF3 (4), compound 1 docked in the same cleft but was rotated by about 180°, with the sulfonate moiety contacting the positively charged Arg-393 highlighted in Fig. 5.
Our models predict that compound 1 re-aligns the NS3h ATP-binding site in the same manner as DNA and RNA. Gu & Rice (4) observed that in order to stabilize the transition state, a helix lining the ATP-binding cleft stretches, which they call the “spring helix.” The spring helix runs from Val-232 to His-246, and movements coupled through the spring helix reorient other key residues in the ATP binding cleft such as the catalytic base Glu-291 (21). The tip of this “spring helix” (Fig. 5A) also contacts bound nucleic acid, and Gu & Rice propose that the spring helix helps couple ATP hydrolysis to helicase movement on nucleic acids (4). In our model of compound 1 docked with NS3 (Fig. 5A), the spring helix contacts the pyridine moiety of compound 1, and we speculate that this contact helps re-organize the ATP binding site so that the compound stimulates ATP hydrolysis. Analogs lacking the pyridine (e.g. compound 3) might fail to stimulate due to less favorable interactions with the spring helix. We are presently testing this hypothesis by crystalizing NS3h in the presence of compound 1.
Sequence alignments of various HCV NS3 helicases, which have been published before (14), did not reveal any variable residues that might clearly explain why the compounds stimulated NS3h purified from genotypes 1b strains but inhibited NS3h purified from other HCV genotypes (Fig. 5A). Variable residues within 5 Å of docked compound 1 include Ser-294, Thr-295, Ser-297, and Pro-482. Pro-482, Ser-294, and Thr-295 are all identical in both stimulated 1b helicases and the inhibited genotype 1a helicase. Ser-297 is identical in 2a(J6) and the genotype 1b helicases, but Ser-297 is an Ala in genotype 1a and 2a(JFH1). Variable residues in the ATP binding cleft include spring helix residue Met-242, and domain 2 residue Met-470, but again, neither is identical only in the genotype 1b helicases. We have located seven residues that are the same in the genotype 1b (i.e. the stimulated) helicases and different in the inhibited (genotype 1a and 2a) helicases: Met-175, Ile-356, Thr-477, Ser-510, Leu-517, Ser-534, and Asp-555. All are far from known ligand binding sites except for Asp-555, which is near the known RNA binding cleft and is a Glu in the genotype 1a and 2a strains.
Finding residues explaining compound 1 specificity will likely require a long process of analyzing many site-directed mutants. However, key residues known to be involved in the helicase reaction were observed to contact docked compound 1 (Fig. 5A). Glu-493 is known to help propel the helicase along its nucleic acid tract (17). Arg-393 clamps the NS3h on nucleic acids (16). A third residue, Ser-231 was also targeted for mutagenesis because in the molecular model, it formed a hydrogen bond with the nitrogen of the terminating pyridine in compound 1. Previous studies have shown that Ser-231 contacts nucleic acids but is not critical for the NS3 ATPase or unwinding reactions (27). Importantly, NS3h harboring mutation in any of these amino acids responds differently to compound 1 than the wild type (Fig. 5), lending support to the assumption that the modeled compound 1-NS3h complex might be biologically relevant.
In conclusion, compounds 1 and 2 bind NS3 in a manner unlike other compounds that target the enzyme. They make specific contacts with HCV helicase such that they can mimic the nucleic acid substrate to stimulate helicase-catalyzed ATP hydrolysis. The compounds could serve as templates to design antivirals that target HCV helicase, or they could be used as probes to better understand how nucleic acid binding modulates ATP hydrolysis or how the protein ATPase function fuels helicase movements. Other members of this class of compounds might also be useful to probe related enzymes that play important roles in DNA replication, DNA repair, transcription, translation, or recombination.
Acknowledgments
We thank Jean Ndjomou, Rajesh Kolli, and Alicia Hanson for helpful advice and technical assistance with this study.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI088001 and U54 HG005031, and a Research Growth Initiative Award from the UWM Research Foundation.
- HCV
- hepatitis C virus
- NS3
- nonstructural protein 3
- NS3h
- NS3 helicase fragment lacking the NS3 protease domain
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Li K., Frankowski K. J., Belon C. A., Neuenswander B., Ndjomou J., Hanson A. M., Shanahan M. A., Schoenen F. J., Blagg B. S., Aubé J., Frick D. N. (2012) Optimization of potent hepatitis C virus NS3 helicase inhibitors isolated from the yellow dyes thioflavine S and primuline. J. Med. Chem. 55, 3319–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bacon B. R., Gordon S. C., Lawitz E., Marcellin P., Vierling J. M., Zeuzem S., Poordad F., Goodman Z. D., Sings H. L., Boparai N., Burroughs M., Brass C. A., Albrecht J. K., Esteban R. (2011) Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 364, 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zeuzem S., Andreone P., Pol S., Lawitz E., Diago M., Roberts S., Focaccia R., Younossi Z., Foster G. R., Horban A., Ferenci P., Nevens F., Müllhaupt B., Pockros P., Terg R., Shouval D., van Hoek B., Weiland O., Van Heeswijk R., De Meyer S., Luo D., Boogaerts G., Polo R., Picchio G., Beumont M. (2011) Telaprevir for retreatment of HCV infection. N. Engl. J. Med. 364, 2417–2428 [DOI] [PubMed] [Google Scholar]
- 4. Gu M., Rice C. M. (2010) Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc. Natl. Acad. Sci. U.S.A. 107, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim J. L., Morgenstern K. A., Griffith J. P., Dwyer M. D., Thomson J. A., Murcko M. A., Lin C., Caron P. R. (1998) Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure 6, 89–100 [DOI] [PubMed] [Google Scholar]
- 6. Chen C. S., Chiou C. T., Chen G. S., Chen S. C., Hu C. Y., Chi W. K., Chu Y. D., Hwang L. H., Chen P. J., Chen D. S., Liaw S. H., Chern J. W. (2009) Structure-based discovery of triphenylmethane derivatives as inhibitors of hepatitis C virus helicase. J. Med. Chem. 52, 2716–2723 [DOI] [PubMed] [Google Scholar]
- 7. Tackett A. J., Wei L., Cameron C. E., Raney K. D. (2001) Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 29, 565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belon C. A., High Y. D., Lin T. I., Pauwels F., Frick D. N. (2010) Mechanism and specificity of a symmetrical benzimidazolephenylcarboxamide helicase inhibitor. Biochemistry 49, 1822–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gemma S., Butini S., Campiani G., Brindisi M., Zanoli S., Romano M. P., Tripaldi P., Savini L., Fiorini I., Borrelli G., Novellino E., Maga G. (2011) Discovery of potent nucleotide-mimicking competitive inhibitors of hepatitis C virus NS3 helicase. Bioorg. Med. Chem. Lett. 21, 2776–2779 [DOI] [PubMed] [Google Scholar]
- 10. Mukherjee S., Hanson A. M., Shadrick W. R., Ndjomou J., Sweeney N. L., Hernandez J. J., Bartczak D., Li K., Frankowski K. J., Heck J. A., Arnold L. A., Schoenen F. J., Frick D. N. (2012) Identification and analysis of hepatitis C virus NS3 helicase inhibitors using nucleic acid binding assays. Nucleic Acids Res. 40, 8607–8621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levin M. K., Gurjar M., Patel S. S. (2005) A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nat. Struct. Mol. Biol. 12, 429–435 [DOI] [PubMed] [Google Scholar]
- 12. Beran R. K., Bruno M. M., Bowers H. A., Jankowsky E., Pyle A. M. (2006) Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 358, 974–982 [DOI] [PubMed] [Google Scholar]
- 13. Ndjomou J., Kolli R., Mukherjee S., Shadrick W. R., Hanson A. M., Sweeney N. L., Bartczak D., Li K., Frankowski K. J., Schoenen F. J., Frick D. N. (2012) Fluorescent primuline derivatives inhibit hepatitis C virus NS3-catalyzed RNA unwinding, peptide hydrolysis and viral replicase formation. Antiviral Res. 96, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lam A. M., Keeney D., Eckert P. Q., Frick D. N. (2003) Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 77, 3950–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heck J. A., Lam A. M., Narayanan N., Frick D. N. (2008) Effects of mutagenic and chain-terminating nucleotide analogs on enzymes isolated from hepatitis C virus strains of various genotypes. Antimicrob. Agents Chemother. 52, 1901–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam A. M., Keeney D., Frick D. N. (2003) Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J. Biol. Chem. 278, 44514–44524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frick D. N., Rypma R. S., Lam A. M., Frenz C. M. (2004) Electrostatic analysis of the hepatitis C virus NS3 helicase reveals both active and allosteric site locations. Nucleic Acids Res. 32, 5519–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 19. Moustakas D. T., Lang P. T., Pegg S., Pettersen E., Kuntz I. D., Brooijmans N., Rizzo R. C. (2006) Development and validation of a modular, extensible docking program: DOCK 5. J. Comput. Aided Mol. Des. 20, 601–619 [DOI] [PubMed] [Google Scholar]
- 20. Wang Y., Xiao J., Suzek T. O., Zhang J., Wang J., Bryant S. H. (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 37, W623–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frick D. N., Banik S., Rypma R. S. (2007) Role of divalent metal cations in ATP hydrolysis catalyzed by the hepatitis C virus NS3 helicase: magnesium provides a bridge for ATP to fuel unwinding. J. Mol. Biol. 365, 1017–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lohmann V., Körner F., Koch J., Herian U., Theilmann L., Bartenschlager R. (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 [DOI] [PubMed] [Google Scholar]
- 23. Yanagi M., St Claire M., Shapiro M., Emerson S. U., Purcell R. H., Bukh J. (1998) Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244, 161–172 [DOI] [PubMed] [Google Scholar]
- 24. Yanagi M., Purcell R. H., Emerson S. U., Bukh J. (1997) Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. U.S.A. 94, 8738–8743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yanagi M., Purcell R. H., Emerson S. U., Bukh J. (1999) Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262, 250–263 [DOI] [PubMed] [Google Scholar]
- 26. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin C., Kim J. L. (1999) Structure-based mutagenesis study of hepatitis C virus NS3 helicase. J. Virol. 73, 8798–8807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borowski P., Niebuhr A., Mueller O., Bretner M., Felczak K., Kulikowski T., Schmitz H. (2001) Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol. 75, 3220–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang N., Chen H. M., Koch V., Schmitz H., Minczuk M., Stepien P., Fattom A. I., Naso R. B., Kalicharran K., Borowski P., Hosmane R. S. (2003) Potent inhibition of NTPase/helicase of the West Nile Virus by ring-expanded (“fat”) nucleoside analogues. J. Med. Chem. 46, 4776–4789 [DOI] [PubMed] [Google Scholar]
- 30. Zhang N., Chen H. M., Koch V., Schmitz H., Liao C. L., Bretner M., Bhadti V. S., Fattom A. I., Naso R. B., Hosmane R. S., Borowski P. (2003) Ring-expanded (“fat”) nucleoside and nucleotide analogues exhibit potent in vitro activity against flaviviridae NTPases/helicases, including those of the West Nile virus, hepatitis C virus, and Japanese encephalitis virus. J. Med. Chem. 46, 4149–4164 [DOI] [PubMed] [Google Scholar]
- 31. Lam A. M., Frick D. N. (2006) Hepatitis C virus subgenomic replicon requires an active NS3 RNA helicase. J. Virol. 80, 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Belon C. A., Frick D. N. (2008) Monitoring helicase activity with molecular beacons. BioTechniques 45, 433.- 40, 442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frick D. N., Ginzburg O., Lam A. M. (2010) A method to simultaneously monitor hepatitis C virus NS3 helicase and protease activities. Methods Mol. Biol. 587, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanson A. M., Hernandez J. J., Shadrick W. R., Frick D. N. (2012) Identification and analysis of inhibitors targeting the hepatitis C virus NS3 helicase. Methods Enzymol. 511, 463–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bordeleau M. E., Matthews J., Wojnar J. M., Lindqvist L., Novac O., Jankowsky E., Sonenberg N., Northcote P., Teesdale-Spittle P., Pelletier J. (2005) Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. U.S.A. 102, 10460–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Preugschat F., Averett D. R., Clarke B. E., Porter D. J. (1996) A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 271, 24449–24457 [DOI] [PubMed] [Google Scholar]
- 37. Levin M. K., Gurjar M. M., Patel S. S. (2003) ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J. Biol. Chem. 278, 23311–23316 [DOI] [PubMed] [Google Scholar]
- 38. Thibeault D., Bousquet C., Gingras R., Lagacé L., Maurice R., White P. W., Lamarre D. (2004) Sensitivity of NS3 serine proteases from hepatitis C virus genotypes 2 and 3 to the inhibitor BILN 2061. J. Virol. 78, 7352–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Najda-Bernatowicz A., Krawczyk M., Stankiewicz-Drogoń A., Bretner M., Boguszewska-Chachulska A. M. (2010) Studies on the anti-hepatitis C virus activity of newly synthesized tropolone derivatives: identification of NS3 helicase inhibitors that specifically inhibit subgenomic HCV replication. Bioorg. Med. Chem. 18, 5129–5136 [DOI] [PubMed] [Google Scholar]
- 40. Preugschat F., Danger D. P., Carter L. H., 3rd., Davis R. G., Porter D. J. (2000) Kinetic analysis of the effects of mutagenesis of W501 and V432 of the hepatitis C virus NS3 helicase domain on ATPase and strand-separating activity. Biochemistry 39, 5174–5183 [DOI] [PubMed] [Google Scholar]
- 41. Kim J. W., Seo M. Y., Shelat A., Kim C. S., Kwon T. W., Lu H. H., Moustakas D. T., Sun J., Han J. H. (2003) Structurally conserved amino acid w501 is required for RNA helicase activity but is not essential for DNA helicase activity of hepatitis C virus NS3 protein. J. Virol. 77, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Neumann-Haefelin C., Frick D. N., Wang J. J., Pybus O. G., Salloum S., Narula G. S., Eckart A., Biezynski A., Eiermann T., Klenerman P., Viazov S., Roggendorf M., Thimme R., Reiser M., Timm J. (2008) Analysis of the evolutionary forces in an immunodominant CD8 epitope in hepatitis C virus at a population level. J. Virol. 82, 3438–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]