Background: Exosomes are extracellular vesicles that have been implicated in intercellular communication.
Results: Exosomes that originate from human cells infected with HIV-1 contain virus-derived small noncoding RNA.
Conclusion: Virus-derived small RNA present in exosomes exert functional consequences in naive recipient cells.
Significance: Viral RNA molecules present in exosomes may be critical mediators of intercellular viral spread in infected hosts.
Keywords: AIDS, Apoptosis, Cell Cycle, Exosomes, HIV-1, Gag, Infectivity, Nef, TAR
Abstract
Exosomes are nano-sized vesicles produced by healthy and virus-infected cells. Exosomes derived from infected cells have been shown to contain viral microRNAs (miRNAs). HIV-1 encodes its own miRNAs that regulate viral and host gene expression. The most abundant HIV-1-derived miRNA, first reported by us and later by others using deep sequencing, is the trans-activation response element (TAR) miRNA. In this study, we demonstrate the presence of TAR RNA in exosomes from cell culture supernatants of HIV-1-infected cells and patient sera. TAR miRNA was not in Ago2 complexes outside the exosomes but enclosed within the exosomes. We detected the host miRNA machinery proteins Dicer and Drosha in exosomes from infected cells. We report that transport of TAR RNA from the nucleus into exosomes is a CRM1 (chromosome region maintenance 1)-dependent active process. Prior exposure of naive cells to exosomes from infected cells increased susceptibility of the recipient cells to HIV-1 infection. Exosomal TAR RNA down-regulated apoptosis by lowering Bim and Cdk9 proteins in recipient cells. We found 104–106 copies/ml TAR RNA in exosomes derived from infected culture supernatants and 103 copies/ml TAR RNA in the serum exosomes of highly active antiretroviral therapy-treated patients or long term nonprogressors. Taken together, our experiments demonstrated that HIV-1-infected cells produced exosomes that are uniquely characterized by their proteomic and RNA profiles that may contribute to disease pathology in AIDS.
Introduction
Exosomes are nano-sized vesicles that range from 30 to 100 nm in diameter and are produced by multiple cell types (1–3). Exosomes originate from late endosomal compartments called multivesicular bodies or de novo from the plasma membrane by outward budding (4). Exosomes contain lipids, proteins, and nucleic acids (mRNAs and miRNAs)2 (5, 6). The proteomic composition of exosomes has been well characterized (7–10). Exosomes released into the intercellular space can fuse with multiple target cells and exert regulatory influences on the target cell (11–15). Exosomal components have been explored as potential biomarkers of the cellular disease state, particularly in cancers (10, 16).
Viruses, upon infection, alter the host cell in ways that counter the host's innate immune response and promote their survival and replication. One critical host strategy to combat viral infections is RNA interference (RNAi), which selectively eliminates foreign nucleic acids (17–20). The steps that lead to generation of functional miRNAs have been well studied (21–31). Viruses have co-evolved with the host RNAi machinery by either encoding their own miRNAs or by encoding suppressors of RNAi that can inhibit the host RNAi response (32–37). DNA viruses have been long known to produce their own miRNAs (38–43). The notion that retroviruses such as HIV-1 encode their own miRNAs is a subject of debate. An initial report by Pfeffer et al. in 2005 (44) claimed that there were no HIV-1-encoded viral miRNAs. This claim was later reinstated by Lin and Cullen in 2007 (45) after analysis of roughly 1000 clones of miRNAs obtained from HIV-1-infected cells. It was later reported in 2007 by Klase et al. (46) that the TAR element of HIV-1 was processed to yield a viral miRNA as detected by sensitive RNase protection assays (47). The TAR-derived miRNA was demonstrated to regulate host cell gene expression relevant to suppression of apoptosis in infected cells (48). Within the next 2 years, two independent research groups made confirmatory observations about the existence of HIV-1-derived small noncoding RNAs. Yeung et al. (49) carried out deep sequencing analysis and reported that multiple small viral noncoding RNAs existed in HIV-1-infected cells. The sequencing of a total of 47,773 clones showed that 60% of them represented miRNAs. Within this population, the authors identified 125 noncoding RNAs that were HIV-1-specific. They also reported that the TAR noncoding RNAs were the most abundant followed by the Rev response element and Nef-noncoding RNAs. A similar observation was made by Oullet et al. (50) that the TAR element of HIV-1 was asymmetrically processed to yield a viral miRNA. Viral miRNAs have also been reported to originate from the Nef region of the HIV-1 genome, the RRE-containing element, and miR-H1, also originating from the LTR region (49, 51, 52). Schopman et al. (53) employed the sensitive SOLiD (tm) 3 Plus System to analyze viral interfering RNA accumulation in HIV-1-infected T lymphocytes and reported that HIV-1 may trigger the production of viral siRNAs and viral miRNAs to modulate cellular and/or viral gene expression. A recent study by Klase et al. (54) additionally demonstrated that HIV-1-encoded noncoding RNAs do not negatively influence viral replication.
Many viral miRNAs have been discovered in exosomes. This has been demonstrated in the case of Epstein-Barr virus infections, both in cell culture systems and patient serum samples (55–59). In the case of HIV-1, there is extensive data on viral proteins contained in exosomes derived from infected cells (60–64). The viral Gag protein has been shown to be included in exosomes originating from infected cells, and this inclusion is dependent on the ability of the viral protein to form higher order oligomeric structures with itself, the host ESCRT machinery, and the plasma membrane (4, 65–69).
In this study, we hypothesized that the TAR RNA produced in infected cells may be incorporated into exosomes. We found that there are abundant levels of extracellular TAR RNA in the cell culture supernatants of infected cells and in patient sera. We also found that although HIV-1-infected cell lines produce exosomes with TAR RNA, there were little detectable levels of viral mRNA in these exosomes. Our data demonstrate that exosomes isolated from culture supernatants of primary latently infected cells also contain TAR RNA. We provide evidence that exosomes derived from HIV-1-infected cells contain both 5′ and 3′ TAR miRNAs. Our proteomic analysis of exosomes derived from infected cells showed the presence of the viral proteins, such as Gag and Env. Within exosomes originating from HIV-1-infected cells, we detected Drosha and Dicer proteins, components of the miRNA machinery. Experiments using leptomycin B indicated that CRM1-mediated nuclear export of TAR RNA was required for the inclusion of TAR RNA in exosomes. Purification of exosomes from HIV-1-infected primary cells using density fractionation gradients confirmed inclusion of TAR RNA in exosomes. Treatment of uninfected cells with exosomes purified from HIV-1-infected cells revealed that the exosome-delivered TAR RNA could confer a protective phenotype to recipient cells under conditions of cellular stress. Our data suggest that prior exposure of uninfected target cells to exosomes from infected cells made the target cells more susceptible to HIV-1 infection. Finally, we have also detected TAR RNA and specific components of the host miRNA machinery in serum exosomes of HIV-1-infected patient samples.
EXPERIMENTAL PROCEDURES
Cells
The human embryonic kidney cell line 293T was obtained from the AIDS Reagent program and was maintained in DMEM complete medium. TZM-bl, a modified HeLa cell line expressing CD4 and CCR5, was obtained from the AIDS Reagent program and maintained in DMEM complete medium. Jurkat and CEM cell lines (uninfected T cells), J1.1 cell lines (HIV-1-infected T cells), ACH-2 (HIV-1-infected cell line), 8E5 (LAV-infected T cell line), and U937 (promonocytic) cells were grown in RPMI complete medium. HLM-1 (HIV-1-infected HeLa cell line/Tat−) cells were grown in DMEM complete medium. All cells were maintained at 37 °C and 5% CO2. A detailed description of all the cells used in this study with description of the viruses that were used to infect these cells is provided in supplemental Table 1. Bovine exosomes were excluded from culture media by ultracentrifugation of the FBS at 100,000 × g for 2 h prior to growth of cells for exosome isolation.
Reagents and Antibodies
OptiPrepTM (60% iodixanol w/v in water) was purchased from Sigma. Protein A/G beads were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Complete culture media consisted of RPMI 1640 or DMEM supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% streptomycin/penicillin (Quality Biological). Media used for serum starvation experiments consisted of 0.1% FBS, 1% l-glutamine, and 1% streptomycin/penicillin. Leptomycin B was purchased from Sigma. Antibodies used for Western blots were obtained from Abcam (CD63, ab8219 (1:1000); cytochrome c, ab13575 (1:1000); TRBP, ab42018 (1:1000); Dicer, ab14601 (1:1000); Drosha, ab12286 (1:1000); DGCR8, ab90579 (1:1000); Ago-ab5070 (1:1000); β-actin ab49900 (1:5000)), Santa Cruz Biotechnology (Bim, sc-11425 (1:250); CD45, sc-53666 (1:250); Cdk9, sc-484 (1:1000); Hsp70, sc1060 (1:1000)), and AIDS Reagent program (Nef antibody 3689 (1:250)).
Clinical Samples
The clinical samples used in this study were obtained through the Women's Interagency HIV study at the Washington D. C. site and have been described in detail by Van Duyne et al. (68). LTNPs were defined as being HIV-1-infected but free of disease for a minimum period of at least 5 years, a CD4 count of greater than 500 at all visits, and no prior history of antiretroviral therapy. All samples were obtained after informed consent and were approved by the ethics board of George Washington University.
CD4 T Lymphocytes
Peripheral blood was obtained from healthy donors. CD4+ T cells (CD4TL) were then isolated from fresh PBMCs using negative selection kit according to the manufacturer's instructions (STEMCELL Technologies, Vancouver, British Columbia, Canada). Cells were solicited from anonymous, healthy volunteer donors who had signed an informed consent approved by the Centre Hospitalier de l'Université Laval research ethics review board. These cells were activated with PHA-L (1 μg/ml) and maintained in complete culture medium supplemented with IL-2 (30 units/ml) at a density of 2 × 106 cells/ml for 3 days. Then CD4TL was incubated with NL4–3BalEnv R5 virus (800 ng of p24 for 40 × 106 cells/ml) for 2 h, and after washes, activated CD4TL was added in complete RPMI 1640 medium supplemented with recombinant human IL-2 (30 units/ml) for 5 days. Exosomes were purified after 5 days.
Exosome Purification
Jurkat and J1.1 cells were grown in appropriate media supplemented with 10% exosome-depleted FBS. Exosome preparations were made from 80 to 100 ml of cell culture supernatants (material produced from a culture of 1 × 106 cells per ml for 5 days). Cells were pelleted by centrifugation at 300 × g for 10 min. An additional centrifugation at 2000 × g for 10 min was used to pellet dead cells. The supernatant was collected and ultracentrifuged at 10,000 × g for 30 min to eliminate cell debris. This was followed by ultracentrifugation at 100,000 × g for 70 min two times to pellet the vesicular components. The resulting exosome pellet was then resuspended in an appropriate amount of PBS (15–150 μl). All spins were done at 4 °C. We included a filtration step prior to the ultracentrifugation steps through a 0.22-μm filter to account for contamination of our exosome preparations with other vesicles, and we determined that no significant change to total protein could be detected between filtered and unfiltered samples (data not shown). Protein levels in exosomes were determined by standard Bradford assay. All exosome preparations were quality controlled by electrophoresis in 4–20% Tris-glycine gels followed by Coomassie or silver staining in addition to determination of protein concentration.
Exosomes were purified using ExoQuick reagent as per the manufacturer's instructions. Briefly, for every 250 μl of patient serum, about 65 μl of ExoQuick reagent was added, and the two components were incubated overnight. After 24 h, the exosomes were centrifuged, and the pellets were resuspended in ∼10 times less volume than the starting material with PBS (containing Ca2+ or Mg2+). Although the PBS-resuspended material was directly utilized for TRIzol/RNA extraction, for Western blot analyses, the material was diluted 10 times further in TNE-50 with 0.1% Nonidet P-40 and passed through a Sephadex G-10 spin column (2 min at 2000 rpm) prior to electrophoresis.
OptiPrep Gradient Separation
Iodixanol (OptiPrep) gradients were prepared in PBS in 1.2% increments ranging from 6 to 18%. Ultracentrifuged microvesicle/exosome sample (210 μl) was layered on top of the gradient and centrifuged for 1.5 h at 250,000 × g in an SW41 Ti rotor. Gradient fractions were collected from the top of the gradient in 1-ml increments and were transferred to polycarbonate ultracentrifuge tubes. They were then diluted with 21 ml of PBS and ultracentrifuged at 100,000 × g for 70 min in a Ti-70 rotor. The resulting pellets were then resuspended in 15–100 μl of PBS and concentrated to a volume of 10–15 μl.
Acetylcholinesterase Enzyme (AchE) Activity Assay
AchE activity of exosomes after OptiPrep purification was carried out as described in detail by Cantin et al. (69). Briefly, 50 μl of every fraction was resuspended in 1.25 mm/liter acetylthiocholine and 0.1 mm/liter 5.5-dithiobis(2-nitrobenzoic acid) in a final volume of 1 ml. Changes in absorption were monitored at 412 nm during a 10-min incubation period at 37 °C.
SDS-PAGE and Western Blot Analysis
Cell extracts were resolved by SDS-PAGE on 4–20% Tris-glycine gels (Invitrogen). Gels were stained using Coomassie Blue stain or silver stain by standard procedures. For Western blot analyses, proteins were transferred to Immobilon membranes (Millipore) at 80 mA for 16 h. Membranes were blocked with Dulbecco's phosphate-buffered saline (PBS) + 0.1% Tween 20 + 5% dry milk for 1 h at room temperature. Primary antibodies against specified proteins were incubated with the membranes overnight at 4 °C. Membranes were washed twice with PBS + 0.1% Tween 20 and incubated with HRP-conjugated secondary antibody for 2 h at 4 °C. Membranes were washed two times with PBS + 0.1% Tween 20 and once with PBS prior to imaging. HRP luminescence was elicited with Super Signal West Dura Extended Duration Substrate (Pierce) and visualized by a Molecular Imager ChemiDoc XRS system (Bio-Rad). The band intensities were calculated using Quantity One 4.6.5 software (Bio-Rad).
Transmission Electron Microscopy
Samples were prepared as follows: Jurkat and J1.1 exosomes (5 μg) were adsorbed onto 300 mesh Formvar-coated grids, stabilized with evaporated carbon film (Electron Microscopy Science, FCF300-Ni), and fixed in 4% glutaraldehyde (5 μl) (Electron Microscopy Sciences, 16210) at 4 °C for 5 min. After four rinses with autoclaved deionized water, fixed samples were stained for 2 min with uranium acetate (10 μl), dried for 20 min, and imaged with the transmission electron microscope (JEOL JEM 1200EX).
Ago2 Immunoprecipitation and RT-PCR for TAR miRNA
For immunoprecipitations, up to 3 ml of culture supernatants and 200 μl (∼200 μg) of purified Jurkat and J1.1-derived exosomes were incubated with 10 μg of anti-Ago2 or IgG antibody overnight at 4 °C. The next day, 30 μl of a 30% slurry of protein A/G beads was added to each reaction and incubated at 4 °C for 2 h. The beads were washed twice with TNE buffer, and the Ago2-associated RNA was extracted using TRIzol. Total RNA isolated from the immunoprecipitated complexes was utilized for RT-PCRs using the QuantiMiR small RNA quantification system (SBI) following the manufacturer's instructions. Briefly, 50% of the isolated RNA sample was used as input material for a three-step assay that tags all small RNA species with a poly(A) tail followed by annealing of an oligo(dT) adaptor and cDNA synthesis. One-third of the cDNA was then utilized in RT-PCRs performed using a universal reverse primer and forward primer against miR16, 5′ TAR miRNA, and 3′ TAR miRNA (70, 71). Amplified products were resolved in a 4–20% polyacrylamide gel, stained with ethidium bromide, and visualized using a Molecular Imager ChemiDoc XRS system (Bio-Rad).
Luciferase Assay
For transfections, TZM-bl cells were seeded in a 96-well culture plate (50,000 cells per well). Twenty four hours later, the cells were transfected with 0.2 μg of Tat plasmid (positive control) or incubated with exosomes derived from Jurkat or J1-1 cells. Twenty four hours after transfection/incubation, luciferase activity was assayed using BrightGlo luciferase assay (Promega). Luminescence was measured with Promega GloMax MultiDetection System. Data shown represent the average of three experimental repeats.
MALDI-TOF Mass Spectrometry
Protein bands of interest were excised from silver-stained gels (silver stain kit, Pierce). Gel pieces were vortexed and washed with 100% acetonitrile to dehydrate the gel pieces and were re-swelled with up to 200 ng of trypsin by incubation on ice for 30 min. Residual trypsin was removed; 20 μl of 25 mm NH4HCO3 was added to the gel pieces, and the reactions were incubated overnight at 37 °C. Peptides were extracted with a 1× distilled H2O wash with brief vortexing and sonication, followed by three washes with 60% acetonitrile, 5% TFA. Extracted peptides were pooled together, and a Speed Vacuum was utilized to reduce the volume to ∼10 μl. Twenty microliters of 0.1% TFA was added to each tube, and peptides were desalted using C18 ZipTips (Millipore) according to the manufacturer's instructions. Peptides were spotted on MALDI sample plate 1:1 with α-cyano-4-hydroxycinnaminic acid matrix solution: 10 mg of α-cyano-4-hydroxycinnaminic acid, 500 μl of 100% acetonitrile, 500 μl of 0.1% TFA. Positive control calibration peptide solution of bradykinin, angiotensin II, P14R, and ACTH was spotted along with negative control empty gel slice. Mass peaks obtained were entered into Mascot and ProFound databases for peptide mass fingerprinting analysis.
LC-MS/MS Analysis
Whole exosome preparations were lysed in 8 m urea, and after that, they were reduced using DTT and acetylated using iodoacetamide by standard procedures. The reduced and alkylated proteins were trypsin-digested (trypsin, Promega) overnight at 37 °C. The digested peptides were eluted using ZipTip purification (Millipore), and identification of the peptides was performed by linear trap quadrupole-tandem MS/MS equipped with a reverse phase liquid chromatography nanospray (ThermoFisher). The reverse phase column was slurry-packed in house with 5 μm 200-Å pore size C18 resin (Michrom BioResources) in a 100-μm × 10-cm fused silica capillary (Polymicro Technologies) with a laser-pulled tip. After sample injection, the column was washed for 5 min at 200 nl/min with 0.1% formic acid; peptides were eluted using a 50-min linear gradient from 0 to 40% acetonitrile and an additional step of 80% acetonitrile (all in 0.1% formic acid) for 5 min. The linear trap quadrupole-MS was operated in a data-dependent mode in which each full MS scan was followed by five MS-MS scans where the five most abundant molecular ions were dynamically selected and fragmented by collision-induced dissociation using normalized collision energy of 35%. Tandem mass spectra were matched against the National Center for Biotechnology Information mouse database by Sequest Bioworks software (ThermoFisher) using full tryptic cleavage constraints and static cysteine alkylation by iodoacetamide. For a peptide to be considered accurately identified, it had to be the top number one matched and had to achieve cross-correlation scores of 1.9 for [M + H]1+, 2.2 for [M + 2H]2+, 3.5 for [M + 3H]3+, ΔCin >0.1, and a maximum probability of randomized identification of 0.01.
Fas Antibody Treatment and Cell Cycle Analysis
Jurkat cells (1 × 105 cells/100 μl) were pretreated with exosomes derived from Jurkat or J1.1 cells after which the cells were incubated with Fas antibody. Anti-Fas antibody (clone CH11-05-201) recognizes the human cell surface antigen Fas expressed in various human cells, including myeloid cells, T lymphoblastoid cells, and diploid fibroblasts. Samples at 20 μg/ml were used to induce apoptosis of human Jurkat cells. Samples were treated with exosomes for 2 h followed by incubation with anti-Fas antibody (1, 3, and 5 μl) for 48 h. Cells were washed with 1× PBS and fixed with 70% ice-cold ethanol. Following rehydration in 1× PBS, cells were stained in PBS containing 25 μg/ml propidium iodide (Sigma), 10 μg/ml RNase A (Sigma), and 0.1% Nonidet P-40 (Calbiochem). Cells were analyzed on a Accuri C6 flow cytometer (BD Biosciences). Cell cycle analysis and measurement was performed using Accuri C6-CFlow software (BD Biosciences).
Reverse Transcriptase (RT) Activity Analysis
RT analyses were performed by previously published methods (72). Briefly, 10 μl of cell culture supernatants were incubated in a 96-well plate with RT reaction mixture containing 1× RT buffer (50 mm Tris-HCl, 1 mm DTT, 5 mm MgCl2, 20 mm KCl), 0.1% Triton, poly(A) (1 unit/ml), pd(T) (1 units/ml), and [3H]TTP. The mixture was incubated overnight at 37 °C, and 10 ml of the reaction mix was spotted on a DEAE Filtermat paper, washed four times with 5% Na2HPO4 and three times with water, and dried. RT activity was measured in a Betaplate counter (Wallac, Gaithersburg, MD).
Chromatin Immunoprecipitation Assay (ChIP)
293T cells were transfected (20 μg) with a pol III-driven plasmid expressing scrambled DNA or wild type TAR (generous gift of Dr. John Rossi, City of Hope), collected 48 h post-transfection, and processed for ChIP analysis. For ChIP, ∼5 × 106 cells were used per IP. ChIP assays were performed as described previously (46). Briefly, cells were harvested by trypsinization, washed with PBS, and resuspended in 1% formaldehyde for 10 min at 37 °C. Cells were washed twice with PBS and resuspended in 500 μl of SDS Lysis Buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1) per IP. Cells were sonicated for six 10-s pulses and clarified by centrifugation at 14,000 rpm for 10 min at 4 °C. Supernatants were collected and diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl). Extracts were precleared with ChIP-prepared A/G beads (protein A/G beads with 10 mg/ml salmon sperm DNA and 10 mg/ml BSA) for 1 h at 4 °C. Extracts were then spun at 3000 rpm for 10 min at 4 °C, and lysates were transferred to a new tube. Specific antibodies (5 μg) were incubated overnight, rotating at 4 °C. ChIP-prepared A/G beads were added the next day and allowed to rotate for 2 h at 4 °C. Samples were spun for 5 min at 3000 rpm at 4 °C and washed successively with 1× low salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), 2× high salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 500 mm NaCl), 1× LiCl wash buffer (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.1), and 1× TE. Complexes were eluted off the beads two times with elution buffer (1% SDS, 0.1 m NaHCO3). Eluates were then reverse cross-linked with 5 m NaCl and 50 μg/ml proteinase K for 5 h at 55 °C. DNA was phenol/chloroform extracted, precipitated, and resuspended in 50 μl of TE. PCR was performed against the Cdk9 promoter and the Bim promoter using the following primers: Cdk9, forward 5-′GGAAGAGGCGGGGTCG-3′ and reverse 5′-ACTCCAGGCCCCTCCG-3′; Bim, forward 5′-TAAATATGGGCTCCCACCCC-3′ and reverse 5′-ACACCAGGCGGACAATGTAA-3′.
RNA Isolation and Quantitative RT-PCR
For quantitative analysis of HIV-1 RNA, total RNA was isolated from various samples, including cell culture supernatants, exosome fraction of cell culture supernatants, serum specimens of HIV-1-infected individuals, and lysates of CEM cells treated with exosomes from HIV-1-infected J1.1 cells; RNA was isolated using TRI Reagent-LS (MRC, Cincinnati, OH) according to the manufacturer's protocol. A total of 0.5 μg of RNA from the RNA fraction was treated with 0.25 mg/ml DNase I RNase-free (Roche Applied Science) for 60 min in the presence of 5 mm MgCl2, followed by heat inactivation at 65 °C for 15 min. A 250-ng aliquot of total RNA was used to generate cDNA with the GoScript reverse transcription system (Promega, Madison, WI) using TAR-specific reverse primer TARfll-R (5′-GTGGGTTCCCTAGTTAGC-3′) or oligo(dT) reverse primers. Subsequent quantitative real time PCR analysis was performed with 2 μl of undiluted and 10−1 and 10−2 diluted aliquots of RT reaction mixes using iQ SYBR Green Supermix (Bio-Rad) with the following pairs of primers: 1) TAR-specific primers TARfll-F (5′-GGTCTCTCTGGTTAGACC-3′) and TARfll-R (see above) amplified 60 nt; TAR sequence: 2) LTR-specific primers NFκB1-2-F (5′-TTCCGCTGGGGACTTTCC-3′) and TARfll-R amplified 158 nt; fragment of U3-R sequence of HIV-1 LTR: 3) env-specific primers Env2019F (5′-GGCAAGTCTGTGGAATTGG-3′) and Env2187R (5′-TGGGATAAGGGTCTGAAACG-3′) amplified 168 nt; a fragment of the HIV-1 Env gene. In the RNA samples from culture media of the latently HIV-1-infected cells, the count of Vif, Vpr, Tat, Rev, Vpu, and Nef RNA was measured using quantitative real time PCR analysis with iQ SYBR Green Supermix. The following primer sets were used: 1) Vif-F18 (5′-GGTGATGATTGTGTGGCAAG-3′) and Vif-R245 (5′-CCCAAATGCCAGTCTCTTTC-3′); 2) Vpr-F34 (5′-AGGGAGCCATACAACGAATG-3′) and Vpr-R208 (5′-TAAACGGCAGTTGTTGCAGA-3′); 3) Tat-F33 (5′-GAAGCATCCAGGAAGTCAGC-3′) and Tat-r225 (5′-GGAGGTGGGTTGCTTTGATA-3′); 4) Rev-F107 (AGGCCCGAAGGAATAGAAGA-3′) and Rev-R282 (5′-CGTCCCAGAAGTTCCACAAT-3′); 5) Vpu-F51 (5′-AGCAATAGTTGTGTGGTCCAT-3′) and Vpu-R233 (5′-ATATCCCAAGGAGCATGGTG-3′); and 6) Nef-F31 (5′-ATTGGATGGCCTGCTGTAAG-3′) and Nef-R206 (5′-GGAAAACCCACCTCTTCCTC-3′). Serial dilutions of DNA from 8E5 cells (CEM cell line containing a single copy of HIV-1 LAV provirus per cell) were used as the quantitative standards. A schematic representation of the primers used in the study is included in supplemental Fig. 1. To normalize HIV-1 RNA quantifications in exosome-treated CEM cells, the β-globin gene was also quantified by real time PCR using β-globin-specific primers as follows: forward primer BGF1 (5′-CAACCTCAAACAGACACCATGG-3′) and reverse primer BGR1 (5′-TCCACGTTCACCTTGCCC-3′). Real time PCRs were carried out in triplicate using the PTC-200 Peltier Thermal Cycler with Chromo4 Continuous Fluorescence Detector (both from MJ Research) and Opticon Monitor 2.03 software.
Poly(A) RT-PCR for Small RNA Quantitation
For poly(A) RT-PCR detection of small RNAs, the miRNA fraction was isolated from DNase I-treated total RNA samples using the mirVana miRNA isolation kit (Ambion, Austin, TX). Then the 3′ poly(A) tailing and RT reaction with oligo(dT) primer containing 5′ adaptor sequence were performed in a 500-ng aliquot of miRNA-enriched fractions using the QuantiMiR kit (SBI, Mountain View, CA) according to the manufacturer's protocol. A manufacturer-provided universal reverse primer and specific microRNA forward primers identical in sequence to the microRNA of interest (TAR, miR-16 and U6 snRNA) were used for quantitative real time PCR with iQ SYBR Green Supermix. Serial dilutions of the synthetic Caenorhabditis elegans miRNA cel-mir-238 were used as quantitative standards. Real time PCRs were carried out in triplicate. Comparative Ct (ΔΔCt) method was applied to analyze data and determine copy numbers.
Infectivity Assay Using Exosomes
To determine functional activity of the exosomes, multiple cell types, including CEM, H9, THP1, U937, and U87MG, were grown to early-mid log phase of growth in complete media (10% serum). Cells (0.5 × 106/ml) were seeded in a 12-well plate and incubated with Jurkat- or J1.1-derived exosomes for 24 h. Next, 89.6 virus (0.5 ng/p24/ml; 100 μl) was added to the mixture, and culture supernatants were collected 5 and 7 days post-addition of virus and assayed for reverse transcriptase (RT) activity.
Latent Infection of Primary Cells with HIV-1
Primary cells latently infected with HIV-1 were generated essentially as described in Marini et al. (73), with some modifications. Total CD4+ T cells were enriched by negative selection with the CD4+ T cell isolation kit (Miltenyi Biotec) from the PBMCs of healthy seronegative donors. CD4+ T cells were activated with monocyte-derived dendritic cells (1 monocyte-derived dendritic cell/10 CD4+ T cells), 500 ng/ml Staphylococcus enterotoxin B, and 50 units/ml IL-2. After 4 days, half of the cells were infected with 2 × 103 TCID50/106 cells of HIV-1IIIB (ABI) for 2 h at 37 °C; the other half of the cells was left uninfected. Infected and uninfected cultures were expanded for 10–15 days in medium containing 25 units/ml IL-2. When the infected culture contained 10–15% infected cells as determined by flow cytometry after intracellular staining with anti-p24 antibodies (clone KC57, Beckman Coulter), both infected and uninfected cultures were placed in resting phase for 7 days in medium containing 1 ng/ml IL-7. At the end of the 7-day resting phase, the cultures were restimulated with anti-CD3/CD28 for 3–5 days. At the end of the 3–5 days, cultures were harvested and processed for exosomes.
Statistical Analysis
Quantitative data were analyzed by two-way analysis of variance (OriginPro version 8.0) and Student's t test (Microsoft Excel). Standard deviation was calculated in all quantitative experiments for at least three independent preparations. The difference was considered to be statistically significant when p < 0.05.
RESULTS
TAR RNA Can Be Detected in Sera of HIV-1-infected Patients and in Culture Supernatants of Infected Cells
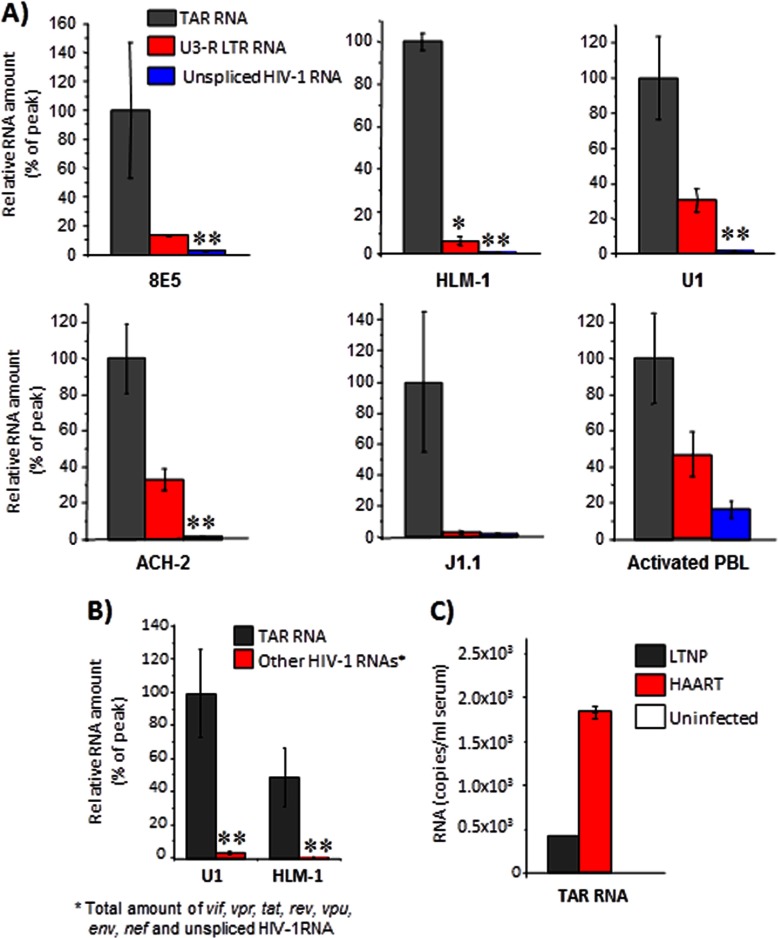
Earlier studies have demonstrated that HIV-1 produces its own viral miRNAs with TAR miRNA being the predominant species (46–53). On average, an infected cell may produce up to 104–105 copies of TAR RNA; however, only a small fraction of that population is utilized by the virus to regulate viral and host transcription. Therefore, we speculated that the excess of TAR RNA exerted its regulatory influence on other bystander cells in an extracellular manner. We tested whether extracellular TAR RNA (unprocessed RNA that contains stem and loop structure, minimally consisting of the first 57 bases or pri-miRNA) could be detected in culture supernatants of early log phase growing HIV-1-infected cells. We carried out qRT-PCR analysis of supernatants from infected 8E5, ACH-2, U1, J1.1, and HLM-1 cells with TAR RNA and control primers. A schematic representation of the primers utilized in the qRT-PCR studies is provided in supplemental Fig. 1. The data revealed the presence of TAR RNA in supernatants of all the infected cell types tested, albeit at different concentrations (Fig. 1A). When compared with the other LTR region (U3 primer, supplemental Fig. 1) and a different HIV-1-derived RNA (Env), we observed that TAR RNA was in vast excess in the culture supernatants of all cell types tested. We performed similar qRT-PCR analysis of TAR RNA in the culture supernatants of HIV-infected primary cells (Fig. 1A, activated PBL) and observed TAR RNA in these supernatants as well. Furthermore, we performed independent qRT-PCRs to determine whether other virus-derived mRNAs were also present in culture supernatants of these cells. To address that question, we performed the same kind of qRT-PCR analysis of RNA isolated (as described above for TAR RNA) from supernatants of U1 and HLM-1 cell cultures with primers that would amplify all other viral mRNAs, including Vif, Vpr, Tat, Rev, Vpu, Env, and Nef, and we found that TAR RNA was in excess over all other viral mRNAs (Fig. 1B). The total copy numbers of these mRNAs did not exceed 5% of the TAR RNA copy number.
FIGURE 1.
TAR RNA is present in infected cell culture supernatants and in sera of HIV-1-infected patients. A, total RNA isolated from culture supernatants of 8E5, ACH-2, U1, HLM-1, J1.1 cells and PHA and IL-2 activated peripheral blood lymphocytes, infected with HIV-1 NL4-3 for 72 h, was quantitated by RT-SYBR Green real time PCR with the primers specific for HIV-1 TAR, U3-R LTR, and Env sequences. Relative RNA count is shown as percentage of the peak count detected for TAR HIV-1 RNA in each sample of the cell culture supernatant. Error bars show the standard deviation from three independent preparations ± S.D. Single asterisk indicates p ≤ 0.05; double asterisk indicates p ≤ 0.01. B, total RNA isolated from culture supernatants of U1 and HLM-1 cells was analyzed by qRT-PCR with primers specific for TAR RNA and viral mRNAs, including vpu, vpr, vif, nef, tat, env, rev, and unspliced HIV-1 RNA as described in A. Results are presented as a mean of three independent measurements ± S.D. Double asterisk indicates p ≤ 0.01. C, total RNA isolated from 1 ml of pooled sera (six individual patients) from control uninfected, HAART-treated, and LTNP patient groups was analyzed by qRT-PCR with TAR-RNA-specific primers as described in A. Error bars show the standard error of two independent measurements.
To check whether the increased amount of extracellular TAR RNA was present in the serum of HIV-1-infected individuals, we quantified TAR RNA in serum specimens from different groups of HIV-1-infected patients. We performed qRT-PCR for TAR RNA using control samples from uninfected serum samples, sera from HIV-1-infected individuals undergoing HAART treatment, and LTNPs (68). Our results demonstrated that the HAART-treated patient serum samples contained ∼1.7 × 103 copies of TAR-pre-miRNA/ml, whereas LTNPs possessed about 4 × 102 copies/ml (Fig. 1C). We had made similar observations of TAR RNA in serum samples of HIV-positive individuals using RNase protection assay as well (data not shown). Collectively, our data demonstrated that HIV-1-derived TAR RNA (pri-miRNA) was detected as an extracellular species in patient samples and in cell culture supernatants.
HIV-1-infected Cells Produced Exosomes with Typical Characteristics
We hypothesized that the extracellular population of TAR RNA is less likely to exist as nascent RNA species and more likely to be enclosed in a protected environment such as membrane-bound vesicles. Our reasoning came from previous experiments where addition of purified, free TAR RNA (T7-based, in vitro synthesized) to culture supernatants or patient serum samples resulted in rapid degradation of TAR in less than 5 min (98% degradation of 105 copies/ml stock at 37 °C; data not shown). Therefore, we focused on exosomes, which have attracted attention in recent years as extracellular membranous vesicles that contain both viral and cellular miRNAs. We chose J1.1 cells, which are chronically infected T cells, produce high-titer viruses without killing the host cell, and are amenable to isolation of virus-free exosomes by centrifugation methods. We isolated exosomes from HIV-1-infected J1.1 and Jurkat cells (uninfected parental cell) using a series of ultracentrifugation steps. First, we determined the kinetics of exosome accumulation in the culture supernatant of Jurkat cells over time. We cultured Jurkat cells in exosome-free media and isolated exosomes after 1, 2, and 5 days. The enriched exosome population and comparable whole cell extracts (WCEs) were separated on 4–20% Tris-glycine gels and stained with Coomassie Blue to visualize total protein levels. Multiple isolates demonstrated that the 5-day-old culture supernatant contained the highest concentration of exosomes from infected or uninfected cells (data not shown).
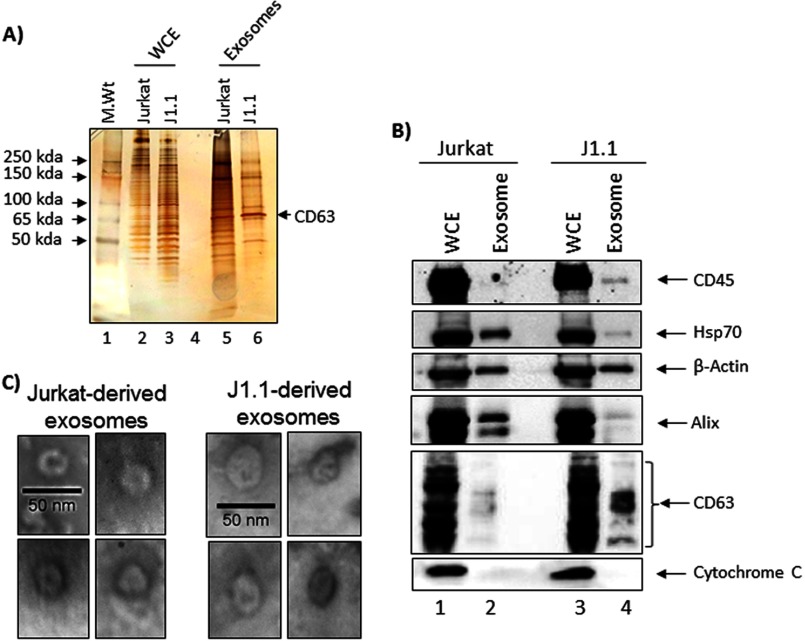
We next asked whether exosomes from these two cell cultures are similar in protein composition. To evaluate the protein profile of Jurkat-derived and J1.1-derived exosomes, we separated equivalent amounts of exosomes (∼20 μg) isolated from these two different cells by SDS-PAGE and silver-stained the gel. WCEs (50 μg) from both cells were included as controls. We observed that exosomes from J1.1-derived cells contained lower overall protein diversity compared with the Jurkat-derived exosomes (Fig. 2A, compare lanes 5 and 6). We observed protein bands that were common and unique to either cell types. We consistently observed an intense band in Fig. 2A, lanes 5 and 6, that migrated close to the 65-kDa mark. The band was excised and subjected to MALDI-TOF mass spectrometry and was determined to be the transmembrane protein CD63, a classical exosomal tetraspanin marker. We then analyzed the Jurkat- and J1.1-derived exosomes for other well defined exosomal marker proteins by Western blots. Equivalent amounts of exosomal proteins and WCEs from both cell types were analyzed by Western blots with antibodies against CD45, Hsp70, β-actin, Alix, and CD63. Although β-actin levels in both sets of exosomes were comparable (compare lanes 2 and 4), there were higher levels of Hsp70 in the Jurkat exosomes (Fig. 2B). J1.1-derived exosomes contained higher amounts of the tetraspanins CD63 and CD45 compared with the Jurkat-derived exosomes (Fig. 2B). Finally, we did not detect cytochrome c (an intracellular protein that is typically absent in exosomes) in our exosome preparations, although cytochrome c could be detected in the corresponding WCEs, thus validating the purity of the exosome preparations.
FIGURE 2.
Exosomes isolated from HIV-1-infected cells possess typical characteristics. A, Jurkat and J1.1-derived exosomes obtained from 5-day-old cell culture supernatants and corresponding WCEs were resolved in a Tris-glycine gel and silver-stained. The band migrating close to 65 kDa was excised from both the Jurkat and J1.1-exosome lanes and analyzed by MALDI-TOF MS. B, Jurkat and J1.1-derived exosomes and corresponding WCEs were analyzed by Western blots with antibodies to CD63, CD45, Hsp70, Alix, cytochrome c, and β-actin. C, Jurkat and J1.1-derived exosomes were observed by transmission electron microscopy and the multiple representative exosomes indicated.
We performed electron microscopy analysis of exosomes isolated from both Jurkat and J1.1 cells and have provided examples of representative exosomes (Fig. 2C). The morphology of Jurkat and J1.1-derived exosomes was similar to published morphological features of exosomes. Cumulatively, we observed that exosomes isolated from Jurkat and J1.1 cells possessed some of the well documented characteristics of exosomal membranous vesicles with differences in some of the other proteins.
J1.1-derived Exosomes Contain TAR RNA
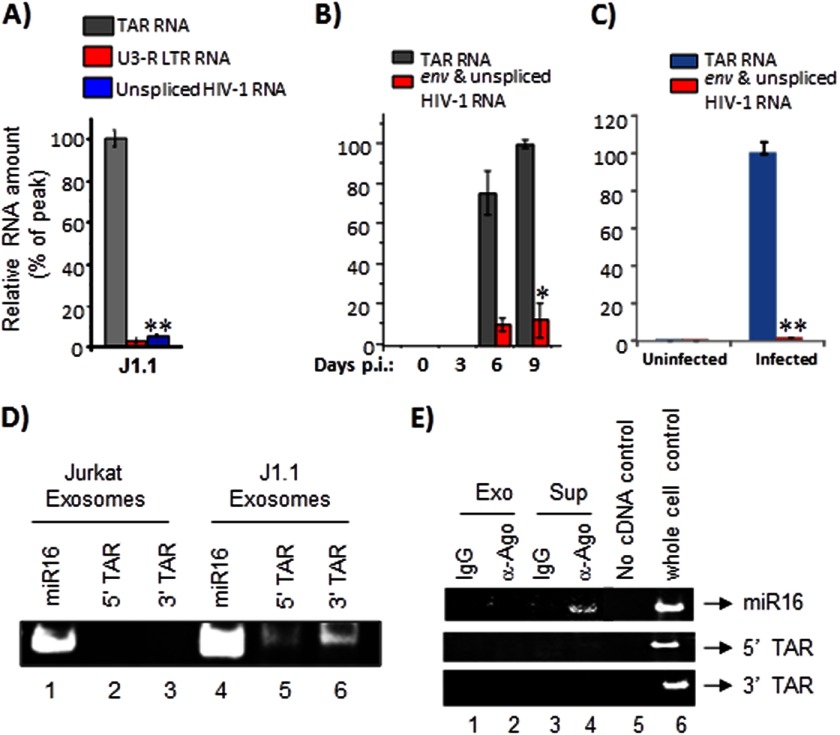
Our experiments above with HIV-1-infected cells demonstrated an excess of TAR RNA in the culture supernatants (Fig. 1A). We next asked whether the exosomes we isolated from J1.1 cells contained TAR RNA. To answer that question, we performed qRT-PCR using the J1.1 exosome material and TAR-specific primers. We determined that there were on average 5 × 105 copies of TAR RNA in the exosomes (Fig. 3A). We also performed similar qRT-PCRs with primers designed to amplify total coding HIV-1 RNA and were not able to detect significant levels of total viral RNA. We performed studies with exosomes isolated from PBMCs infected with dual-tropic 89.6 virus. Quantification of TAR RNA in exosomes by qRT-PCR revealed that high levels of TAR RNA could be detected in this instance as well (Fig. 3B).
FIGURE 3.
Exosomes derived from HIV-1-infected cells contain TAR RNA. A, total RNA was isolated from J1.1-derived exosomes and analyzed by qRT-PCR with primers specific for TAR RNA, U3-R LTR sequence, and unspliced HIV-1 RNA as described in A. Error bars show the standard deviation from three independent RNA preparations; double asterisk indicates p ≤ 0.01. B, primary cells from a healthy donor (PBMCs) were activated with anti-CD3/CD28 and then infected with HIV-1IIIB (ABI). Culture supernatants were harvested in 3, 6, and 9 days after inoculation; exosomal fraction was separated, and then the total RNA was purified and analyzed by quantitative RT-PCR with primers specific for TAR and unspliced HIV-1 RNA. Results are presented as a mean of three independent measurements ±S.D.; the asterisk indicates p ≤ 0.05. C, primary cells were activated as described in B. When 10–15% of the cells were infected, the culture was harvested, washed, and incubated for 7 days in complete medium (exosome-free) supplemented with 1 ng/ml IL-7 (R&D Systems) to allow the cells to revert to quiescence. Uninfected cells were maintained alongside as controls. Total RNA was obtained from exosomes enriched from culture supernatants of uninfected and infected cells and analyzed by qRT-PCR with primers for TAR and env RNA. Error bars show the standard deviation from three independent preparations. Double asterisk indicates p ≤ 0.01. D, total RNA was isolated from Jurkat and J1.1-derived exosomes and analyzed by RT-PCR (QuantiMiR small RNA analysis method) using primers specific to miR16, 5′, and 3′ TAR miRNA. After the RT-PCR, the amplified products were resolved on a 4–20% Tris-glycine polyacrylamide gel and visualized after staining with ethidium bromide. E, Jurkat and J1.1-derived exosomes were immunoprecipitated with anti-Ago2 antibodies. Immunoprecipitations performed with IgG antibodies were used as controls. All RNA associated with the antibodies was extracted and utilized for cDNA synthesis using the QuantiMiR small RNA isolation and analysis protocol. Half of the isolated RNA was utilized in the cDNA amplification procedure, and 30% of the amplified cDNA was used in the RT-PCR with 5′ primers against miR16, 5′ TAR miRNA, and 3′ TAR miRNA. The amplified DNA products were resolved in a polyacrylamide gel, stained with ethidium bromide, and visualized using a Molecular Imager ChemiDoc XRS system (Bio-Rad). Exo, exosomes; Sup, culture supernatant.
We next asked whether exosomes produced by latently infected primary cells also contained TAR RNA. To that effect, exosomes were isolated from latently infected primary cells and analyzed by qRT-PCR with TAR-specific primers. We observed that the exosomes derived from latently infected cells also contained high amounts of TAR RNA (∼5 × 105 copies), although the presence of Env RNA was close to the background (Fig. 3C).
As a next step, we asked whether the TAR RNA present in the exosomes existed as the processed 5′ and 3′ TAR miRNA forms. To address that question, we isolated RNA from exosomes derived from both Jurkat and J1.1 culture supernatants using TRIzol and subjected the isolated RNA to analysis for miRNA content (QuantiMiR miRNA analysis kit). We utilized miR16 as a positive control for the presence of miRNAs in the RNA pool and as a point of reference for migration of miRNAs in an acrylamide gel (Fig. 3D). Accordingly, we could detect miR16 in the Jurkat-derived exosome preparations, although we did not detect any band corresponding to 5′ or 3′ TAR miRNA. In the case of exosomes derived from J1.1 cells, although we found miR16, we were also able to detect bands corresponding to both 5′ and 3′ TAR miRNA (Fig. 3D, lanes 5 and 6). We observed that the levels of 3′ TAR miRNA was roughly three times more than the 5′ TAR miRNA, which is in agreement with our earlier published results (46) and that published by Provost and co-workers (50).
It has recently been reported that the majority of extracellular small noncoding RNAs are present as Ago complexes and not necessarily contained in the exosomes (74, 75). To determine whether extracellular TAR RNA was indeed present in Ago2 complexes, we performed IP reactions utilizing both culture supernatants of Jurkat and J1.1 cells and isolated exosomes from both cell types using anti-Ago2 antibody. The IP reactions were carried out using an excess of culture supernatant (3 ml) equivalent to ∼200 μg of isolated exosomes (as determined by β-actin protein levels). At least 50% of the IP reaction was utilized as input material for RT-PCR-based detection of TAR RNA (QuantiMiR detection assay). miR16, which is a host-derived miRNA and has previously been shown to be present with extracellular Ago2 complexes, was used as a positive control (74). Although we were able to detect miR16 in our Ago2 IPs using RT-PCR, we were unable to detect a signal above the IgG background that corresponded to 3′ or 5′ TAR miRNAs (Fig. 3E). Thus, our results suggested that the majority of extracellular TAR RNA was likely to be contained inside exosomal vesicles and not in extra-exosomal free Ago2 complexes.
Cumulatively, our results demonstrated that exosomes derived from both chronically infected and latently infected cells contain TAR RNA and that a small fraction of this TAR RNA (≥1%) could exist as 5′ and 3′ miRNAs in exosomes. Clear association of TAR miRNA with Ago2 could not be observed.
J1.1-derived Exosomes Contain Viral Proteins
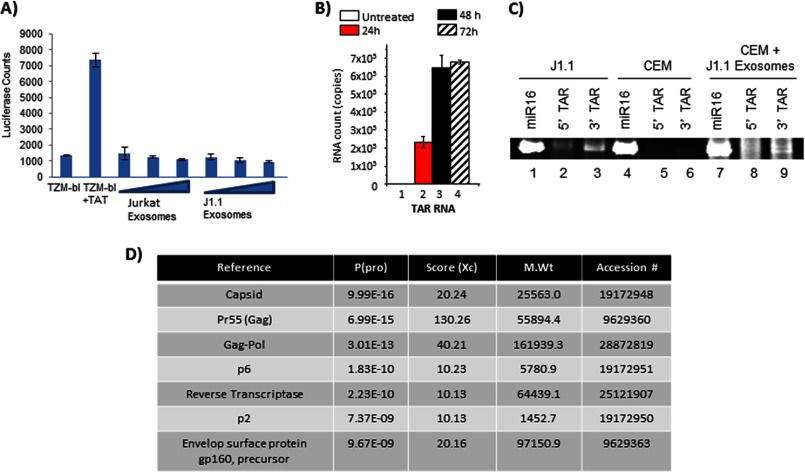
To further verify that the exosomes obtained from J1.1 cells did not contain whole virus, we performed luciferase reporter assays using TZM-bl cells with an integrated HIV-1 LTR. Luciferase expression in the presence of transfected Tat served as a positive control (Fig. 4A). Increasing concentrations of Jurkat-derived exosomes served as a negative control. We observed that incubation of TZM-bl cells with increasing concentrations of J1.1-derived exosomes failed to elicit luciferase expression (Fig. 4A), indicating a lack of intact viral particles or the presence of Tat protein in these exosomes. We observed a similar lack of luciferase induction even when the exosomes were transfected into TZM-bl cells by electroporation (data not shown).
FIGURE 4.
J1.1-derived exosomes contain viral components. A, TZM-bl cells were incubated for 24 h with increasing concentrations of either Jurkat or J1.1-derived exosomes and analyzed by luciferase assay for reporter gene activity. TZM-bl cells transfected with a Tat-expressing construct was included as a positive control for luciferase activity. The error bars show the standard deviation of three independent measurements. B, uninfected CEM cells were incubated with J1.1-derived exosomes for 24, 48, and 72 h. Whole cell pellets were lysed to obtain total RNA. The RNA was then analyzed by qRT-PCR with TAR RNA specific primers. Untreated CEM cells were used as a negative control. Results are presented as a mean for three independent RNA preparations ± S.D. C, uninfected CEM cells were incubated with J1.1-derived exosomes for 72 h after which total RNA was isolated and utilized in the QuantiMiR assay for analysis of small RNA species using primers to miR16, 5′, and 3′ TAR miRNA. Total RNA obtained from CEM cells that were not incubated with exosomes was tested as a negative control, although RNA from J1.1 cells were utilized as the positive control for 5′ and 3′ TAR miRNA. The resultant PCR products were resolved on a polyacrylamide gel, stained with ethidium bromide, and visualized. D, J1.1 exosomes were lysed with urea, trypsinized overnight, and analyzed by LC-MS/MS to identify viral components. The probability of correct peptide identification, Xc score, molecular weight, and accession number of the identified proteins is indicated.
We confirmed that this lack of luciferase induction was not due to the inability of the exosomes to fuse with the target cells and/or release their internal cargo. To address that possibility, we incubated CEM cells with exosomes from J1.1 cells for varying lengths of time (24, 48, and 72 h) and performed qRT-PCR of total RNA from the CEM cell lysates (HIV-1 negative cells) with TAR-specific primers. We reasoned that the only way TAR RNA can be detected in CEM cell lysates would be if the J1.1 exosomes had successfully fused with the target cell and released their cargo inside the target cell. Our qRT-PCR analysis revealed the presence of TAR RNA inside CEM cells 24 h after incubation with J1.1 exosomes, although no TAR RNA was detected in the control untreated cells (Fig. 4B, compare lanes 1 and 2). Although the RNA levels increased at 48 h, the increase was less than 1 log and did not increase by 72 h (Fig. 4B). Thus, our experiment suggested that the majority of the exosomal cargo was released into the target cells by 24 h post-incubation of exosomes with cells.
As a next step, we asked whether the TAR RNA that was introduced into target cells by fusion of exosomes (such as CEM shown in Fig. 4B) could be processed to yield TAR miRNA in the target cells. We incubated CEM cells with J1.1-derived exosomes for 72 h, extracted total RNA using TRIzol, and analyzed for 5′ and 3′ TAR miRNA content using the QuantiMiR microRNA method. We utilized CEM cells, which were not incubated with exosomes, as a negative control and total RNA from J1.1 cells as a positive control for the presence of 5′ and 3′ TAR miRNAs. We utilized miR16 as an internal positive control. The data shown in Fig. 4C demonstrate that J1.1 cells (positive control, lanes 1–3) contain both 5′ and 3′ TAR miRNAs, which are absent in the CEM cells (negative control, lanes 4–6). Both samples contain miR16. In the case of CEM cells that were incubated with J1.1-derived exosomes (Fig. 4C, lanes 7–9), we observed the presence of 3′ TAR miRNA, although there was a comparatively lesser amount of 5′ TAR miRNA. Therefore, our data suggest that at least a part of the exosomal TAR RNA is processed into miRNAs in the recipient cells.
Finally, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of the J1.1-derived exosomes to determine what viral proteins may have been incorporated into the exosomes. The LC-MS/MS study revealed that J1.1-derived exosomes contained multiple molecules of Gag protein (Fig. 4D). This is expected as it has previously been demonstrated that the inherent nature of Gag to form higher order oligomers with itself and interact with the plasma membrane directed its efficient inclusion in extracellular membranous vesicles (65). We detected a precursor form of the HIV-1 Env (gp160), but not the mature gp120 protein. Previous studies have demonstrated the viral protein Nef to be included inside vesicles derived from infected cells (60–64); however, we could not detect reasonable peptide hits corresponding to Nef in J1.1 exosomes by mass spectrometry. Therefore, we carried out Western blot analysis of J1.1-derived exosomes with a Nef-specific antibody. Exosomes obtained from Jurkat culture supernatants were also analyzed in parallel. We were able to identify low levels of Nef in J1.1 exosome preparations only when concentrating the exosome sample by 10-fold (200 μg of total protein utilized) (data not shown). Thus, our experiments demonstrated that TAR RNA introduced into recipient cells, due to exosome fusion, could be processed to miRNAs in the recipient cell. Finally, we observed evidence for the presence of viral proteins in the exosomes derived from HIV-1-infected cells.
J1.1-derived Exosomes Contain Dicer and Drosha Proteins
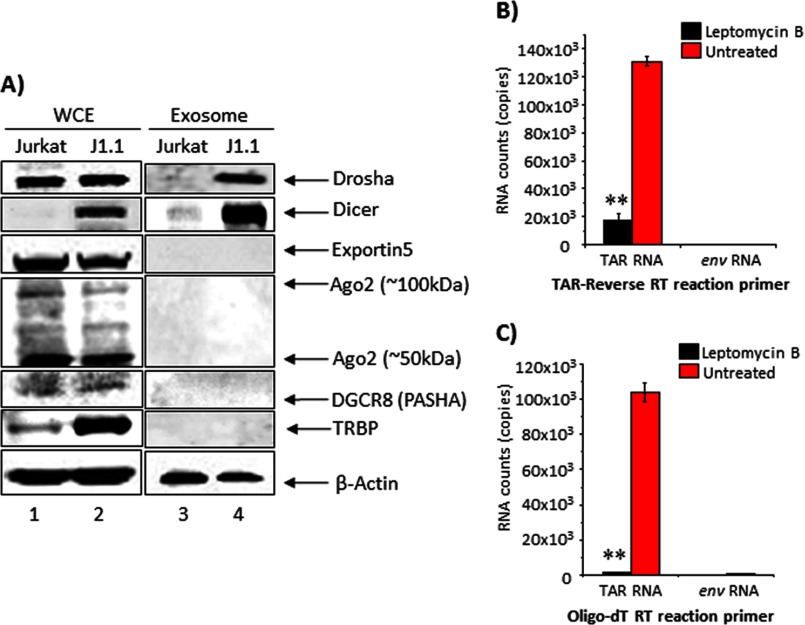
We next asked whether components of the host miRNA machinery, including Dicer, Drosha, Ago2, Exportin, DGCR8 (PASHA), and TRBP, were included in the exosomes along with TAR RNA. We carried out Western blot analyses of J1.1-derived exosomes with antibodies against components of the miRNA machinery (Fig. 5A). We observed that J1.1-derived exosomes contained both Dicer and Drosha proteins. Ago2, Exportin, DGCR8, and TRBP could not be detected in either of the two exosomes. This observation is suggestive of the presence of pri-miRNA (TAR possibly bound to Drosha) in the exosomes.
FIGURE 5.
J1.1-derived exosomes contain components of the RNAi machinery. A, Jurkat- and J1.1-derived whole cell extracts and exosomes were separated on a 4–20% Tris-glycine gel and analyzed by Western blot using antibodies against Dicer, Drosha, exportin, Ago2, DGCR8, TRBP, and β-actin. Total RNA isolated from J1.1-derived exosomes with and without leptomycin B treatment (10 nm) was subjected to qRT-PCR with TAR-reverse primer (B) and oligo(dT) primers (C). cDNA was then quantified by SYBR Green real time PCR with the primer sets specific for HIV-1 TAR and env sequences. Error bars show the standard deviation from three independent RNA preparations. Double asterisk indicates p ≤ 0.01.
The pri-miRNA form (stem loop structure at the 5′ LTR) is generated in the nucleus and is exported to the cytoplasm in a CRM1-dependent manner. To determine whether a CRM1-mediated export of the TAR RNA-Drosha complex was required for TAR RNA to be included in the exosomes, we treated J1.1 cells with an inhibitor of the CRM1 pathway, namely leptomycin B. Leptomycin B treatment was carried out in the nontoxic range (10 nm concentration as determined by prior titrations) for 5 days after which exosomes were isolated. Exosomes isolated from leptomycin B-treated and -untreated cells were subjected to qRT-PCR using TAR RNA-specific primers (Fig. 5B). We observed a decrease in exosomal TAR RNA in the leptomycin B-treated cells supporting the idea that inclusion of TAR RNA in exosomes may require an active CRM1-mediated nuclear export of the TAR-Drosha complex. When we performed a similar qRT-PCR with oligo(dT) primers, we observed that the exosomal TAR RNA contained a polyadenylated 3′ terminus (Fig. 5C). This may be due to the presence of a short stretch of “A” nucleotides in the TAR structure at positions 60, 67, and 70, which also serve as a poly(A) signal. Control qRT-PCRs carried out with Env-specific primers excluded possible contamination of our exosome preparations with virus (Fig. 5, B and C). We evaluated total protein content of exosomes derived from leptomycin B-treated and -untreated cells by PAGE and silver staining. Our analysis revealed comparable total protein in both sets of exosomes and similar levels of β-actin from Western blots (data not shown). Collectively, our results indicated that J1.1-derived exosomes contained components of the host miRNA machinery and that inclusion of TAR RNA in exosomes required CRM1-mediated export from the nucleus.
Exosomes from HIV-1-infected Primary T Cells Contain TAR RNA
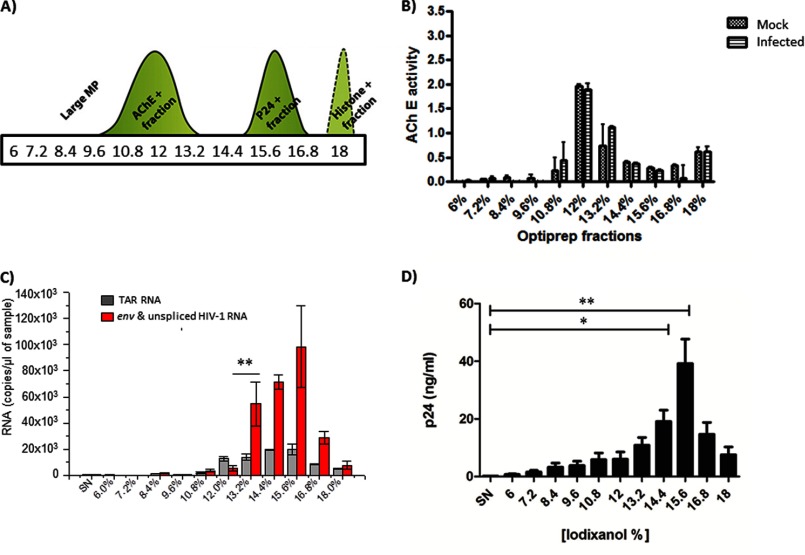
Our studies thus far have been performed utilizing infected cultured cell lines. We next asked whether exosomes originating from primary T cells infected with HIV-1 also contained TAR RNA. To that end primary, activated CD4 cells isolated from healthy donors were infected with HIV-1 (Bal/Env), and exosomes were isolated by a combination of ultracentrifugation and velocity gradient separation (69). The percentages of iodixanol used to generate the gradient and the separation scheme for exosomes (AchE+) and viruses (p24+) are shown in Fig. 6A. Confirmatory AchE activity assays comparing mock-infected and HIV-1-infected primary cells demonstrated the highest AchE activity in the fraction containing 12% iodixanol (Fig. 6B). Therefore, we concluded that the 12% iodixanol fraction was enriched for exosomes. Total RNA was isolated from all the fractions by TRIzol extraction and analyzed by qRT-PCR for TAR RNA. We observed an increase in TAR RNA amounts in 12% iodixanol fraction (Fig. 6D). It was important to note that although the 12% fraction was enriched for TAR RNA, all the other viral RNAs, including Env, were lower in these fractions. This was in contrast to what was observed in the fractions with higher percentages of iodixanol, which also contained TAR RNA, but were highly enriched for all other viral RNAs as well suggesting that those contained viral particles. We also quantified p24 levels in these fractions and observed separation of p24 peaks between exosome and virus fractions particularly in the case of the 12% fraction (Fig. 6D). We were unable to observe Tat protein in these fractions (data not shown). Collectively, these experiments indicated that exosomes derived from primary HIV-1-infected cells also contained TAR RNA.
FIGURE 6.
Exosomes from infected primary cells contain TAR RNA. A, schematic illustration of elution profiles of exosomes and virions after OptiPrep velocity gradient centrifugation of culture supernatants. MP, molecular weight particles. B, acetylcholinesterase activity of fractions was determined to confirm fraction numbers that contained pure exosomes. The x axis shows the % of iodixanol in the appropriate gradients, and the y axis shows relative AchE function. Error bars show the standard error of two independent preparations. C, total RNA isolated from the fractions of infected primary cell culture supernatants were subjected to qRT-PCR with TAR and env-specific primers. D, total protein obtained from the fractions was subjected to Western blot analysis using antibodies to p24 and quantified. C and D, data are presented as a mean for three independent measurements ± S.D. Single asterisk indicates p ≤ 0.05; double asterisk indicates p ≤ 0.01. SN, supernatant.
J1.1-derived Exosomes Down-regulate Apoptosis in Recipient Cells
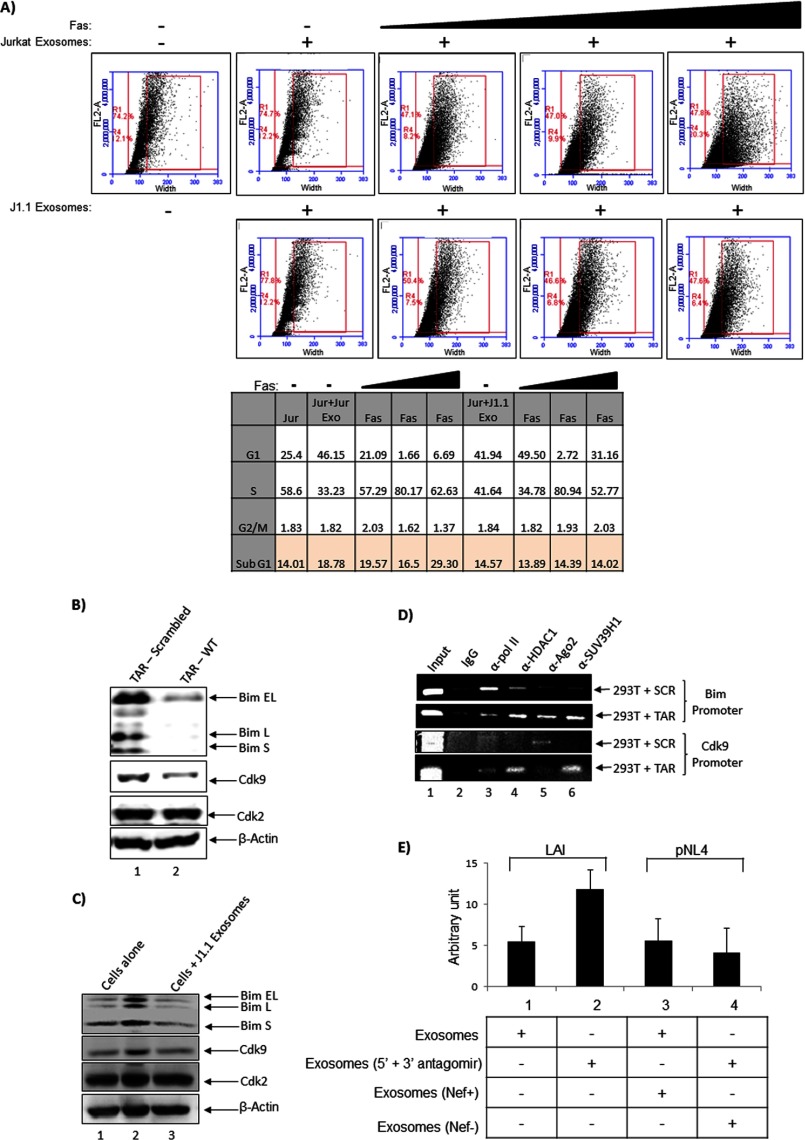
We next performed experiments to decipher the functional significance of exosomes derived from uninfected (Jurkat) and HIV-1-infected (J1.1) cells. Earlier experiments from our laboratory have shown that HIV-1 TAR RNA down-regulated apoptosis in host cells under conditions of cellular stress (48). Therefore, we asked whether TAR RNA present in the exosomes could confer a similar protective phenotype in the recipient uninfected cells when the cells are subjected to cell death-inducing stressors. One such molecule that is documented to induce cell death is the Fas ligand. When cells are treated with Fas ligand, caspase cleavage is activated that results in apoptosis by both the extrinsic and the intrinsic pathway. We therefore incubated recipient Jurkat cells with Jurkat- or J1.1-derived exosomes for 2 h after which increasing concentration of Fas antibody was added. The Fas antibody is expected to function similar to ligand-induced activation of apoptosis. We incubated the cells with Fas antibody for a total of 48 h after which the cells were processed for flow cytometry analysis by propidium iodide staining. The analysis indicated that addition of Jurkat-derived exosomes inherently increased the sub-G1 population when compared with the J1.1-derived exosomes (tabular column, 18.7% in Jurkat + Jurkat exosome lane versus 14.5% in Jurkat + J1.1 exosome lane, Fig. 7A). The increase in the sub-G1 population from Jurkat-derived exosomes was concentration dependent with increasing antibody (tabular column, 19.5, 16.5, and 29.3% at 1, 3, and 5 μl of Fas antibody, Fig. 7A). This was in contrast with what was observed with the J1.1-derived exosomes where addition of Fas antibody did not increase the sub-G1 population (tabular column, 13.8, 14.3, and 14.02% for 1, 3, and 5 μl of Fas antibody, Fig. 7A).
FIGURE 7.
J1.1-derived exosomes confer protection against cell death by down-regulation of Bim. A, Jurkat cells were treated with either Jurkat-derived exosomes or J1.1-derived exosomes for 2 h. Fas antibody was added to the cells at increasing concentrations (1, 3, and 5 μl), and the cells were maintained for another 48 h after which cell cycle distribution was analyzed by flow cytometry. Untreated Jurkat control was maintained alongside for comparison. The panels indicate distribution of recorded events based on size of particles stained with propidium iodide. The outer red box in each panel indicates the percentage of events recorded, and the inner red box indicates the percentage of cells (of the total events) that displayed cell aggregation from IgM aggregation and dying cells (prelude to sub-G1 population). The total percentages of cells determined to be in the various stages of the cell cycle are indicated in a tabular format below. B, 293T cells were transfected with a construct that expressed either wild type TAR or a scrambled TAR sequence under the control of a pol III promoter. Forty eight hours after transfection, whole cell extracts were obtained and analyzed by Western blot for Bim, Cdk9, Cdk2, and β-actin. C, 293T cells were treated with either Jurkat or J1.1-derived exosomes for 48 h. Whole cell lysates were analyzed by Western blot for relative levels of total Bim, Cdk9, Cdk2, and β-actin. D, recruitment of transcription modulators such as HDAC1 and SUV39H1 on Bim and Cdk9 promoters were determined in the presence of wild type or scrambled TAR RNA. ChIP assay was performed using transfected cells and antibodies to RNA pol II, HDAC1, Ago2, and SUV39H1. The precipitated DNA was amplified using primers to Bim and Cdk9, resolved on a 2% agarose gel, and stained with ethidium bromide. E, modulation of Bim levels in 293T cells treated with exosomes obtained from antagomir-treated J1.1 cells (indicated as LAI) or exosomes obtained from Nef+ or Nef− viruses (NL4.3_92BR020.4nef+_IRES_EGFP and NL4.3_92BR020.4nef-_IRES_EGFP respectively, indicated as pNL4) are designated. Error bars show the standard error of two independent measurements.
At the highest concentration of Fas antibody, in the case of Jurkat cells treated with Jurkat-derived exosomes, a pronounced spreading of cells (based on total cell size) was observed (panel 5, top set, Fig. 7A). This spreading effect was mainly due to aggregation of the sub-G1 cells with the IgM pentamer antibody. Such a strong spreading phenomenon was not observed when Jurkat cells were pretreated with J1.1-derived exosomes and Fas antibody (panel 4, bottom set, Fig. 7A). We also quantified the populations of cells in various stages of the cell cycle (G1, S, G2/M) in all samples and have provided the data in a tabular format (Fig. 7A). The numbers indicate that at the highest concentration of Fas antibody, about 29% of the cells exposed to Jurkat-derived exosomes were in the sub-G1 range. In contrast, when cells were exposed to J1.1-derived exosomes, even at the highest concentration of Fas antibody, almost three times less sub-G1 population (14%) was observed. Based on the number of cells in the G1 phase (tabular column), we hypothesize that the increase in sub-G1 population in the case of Jurkat-derived exosomes may have originated from the G1 pool of cells. Cumulatively, these observations underscore our previous observation that TAR RNA exerts a protective phenotype in cells when exposed to cell death-inducing stressors.
As a next step, we attempted to characterize the reason behind the protective phenotype associated with TAR RNA. Previous data obtained from our laboratory had indicated that the proapoptotic protein Bim was modulated in the presence of TAR RNA (data not shown). We performed a sequence analysis of the BIM promoter region (−500 to +500 from the transcriptional start site, NCBI reference sequence NG_029006.1) against 5′ and 3′ TAR miRNA seed sequences (GGGA and AGAGA respectively) and observed complementarity to regions both up- and downstream of the transcriptional start site (nt −29 to −43, −83 to −97, −145 to −159, −178 to −192, −198 to −212, −356 to −370, −447 to −561, +244 to +258, and +312 to +326). Furthermore, TAR miRNA displays complementarity at multiple positions in the BIM mRNA, including one site in the ORF (nt 348–365) and multiple positions in 3′ UTR (nt 895–914, 1008–1029, 1287–1308, 1947–1968, 2303–2337, and 2943–2964). TAR miRNA could therefore potentially exert its protective function by down-regulating Bim expression in a transcriptional and/or post-transcriptional manner. To verify if down-regulation of Bim protein does occur in the presence of TAR in cells, we first transfected a plasmid encoding TAR RNA into 293T cells and 48 h post transfection, cells were lysed and analyzed by Western blot for endogenous levels of Bim protein. We observed that transfection of the TAR plasmid down-regulated levels of Bim protein (Fig. 7B). We also observed that transfected TAR resulted in decreased levels of Cdk9 (complementary target sites at nt 18–42, 285–300, and 560–586), although it did not induce any change in the levels of Cdk2 protein. Therefore, our experiments support the hypothesis that TAR RNA could exert its protective effect by down-regulating the pro-apoptotic protein Bim.
We then asked whether TAR RNA in the exosomes could also down-regulate Bim protein in target cells. We incubated 293T cells with Jurkat- and J1.1-derived exosomes and 24 h later, performed Western blot analysis with Bim and Cdk9 antibodies. The results demonstrated that exosomal TAR RNA from the J1.1 exosomes also down-regulated Bim expression and therefore could contribute to the observed decrease in apoptosis (Fig. 7C). We performed chromatin immunoprecipitation (ChIP) analysis to determine suppression of transcription from the BIM and CDK9 promoter in the presence of TAR RNA. 293T cells were transfected (20 μg) with a pol III-driven plasmid expressing scrambled DNA or wild type TAR and ChIP assays were carried out using antibodies to RNA polymerase II, HDAC1, Ago2, and SUV39H1. Control reactions were performed using IgG antibodies. Our rationale to this approach was that if the promoters of BIM and CDK9 were influenced by TAR, we would observe a change in the recruitment of transcription regulatory factors on the promoters. Accordingly, the ChIP analysis revealed that in the case of the Bim promoter, in the presence of wild type TAR, an increased recruitment of repressive components such as HDAC1 and SUV39H1 was observed. Along these lines, we also observed a decrease in the recruitment of RNA polymerase II on the Bim promoter. Similarly, we observed an increased recruitment of HDAC1 and SUV39H1 on the CDK9 promoter suggestive of transcriptional repression (Fig. 7D).
We then asked whether the presence of TAR in the J1.1-derived exosomes was significant to the down-regulation of Bim in the recipient cells. To address that question, we performed a transfection of antagomirs to both 5′ and 3′ TAR miRNAs into the parental J1.1 cells. Exosomes were purified from both transfected and untransfected J1.1 control cells and were subsequently incubated with the 293T recipient. Total lysates were obtained after 24 h, and Bim Western blots were performed and bands quantified. The data, as shown in Fig. 7E (compare lanes 1 and 2), indicate that in the presence of antagomirs to TAR RNA, the Bim levels increased in the 293T cells. We also performed a second experiment to evaluate the effect of the Nef component in the exosome on the Bim phenotype. To determine whether the presence of Nef in exosomes has any effect on Bim expression in the recipient 293T cells, we purified exosomes from cells infected with Nef+ and Nef− viruses, respectively. The exosomes were then added to 293T cells, and 24 h later, total lysates were analyzed for Bim expression. As shown in Fig. 7E (compare lanes 3 and 4), the total level of Bim in the recipient cell did not change because of the presence or absence of Nef in the exosomes thus indicating that Nef may not play a notable role in modulating Bim expression in the recipient cell. Collectively, our results imply that the TAR RNA contained within J1.1-derived exosomes were functionally competent and down-regulated apoptosis by decreasing Bim and Cdk9 protein levels in recipient cells.
J1.1-derived Exosomes Increase Susceptibility of Naive Cells to HIV-1 Infection
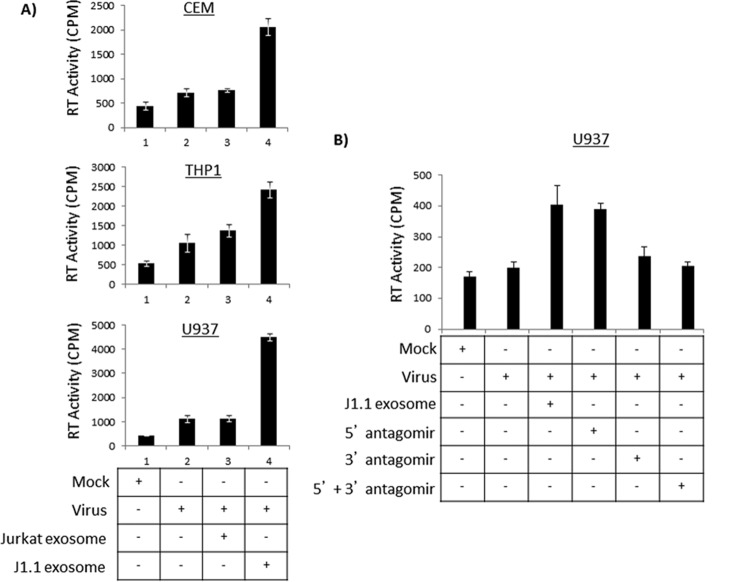
Next, we asked whether J1.1-derived exosomes could exert a bystander effect by altering the pattern of infection in naive cells. Specifically, we asked whether exposure to exosomes derived from infected cells makes a naive target cell more susceptible to infection by low titer HIV-1 infection. To answer that question, we utilized multiple cell lines as recipient cells, including CEM, H9 (T cells), THP1, U937 (pro-monocytic), and U87MG (astroglioma cells). All recipient cells were seeded in a 12-well plate and incubated with Jurkat- or J1.1-derived exosomes for 24 h after which time low concentration of dual-tropic virus (89.6) was added to the cells. Culture supernatants were collected 5 and 7 days post-addition of virus and assayed for the presence of reverse transcriptase (RT) activity in the supernatants. The results were prominent in CEM cells where 5 days post-infection cells pre-exposed to J1.1-derived exosomes showed a 4-fold increase in infectivity when compared with CEM cells infected with virus alone (Fig. 8A, panel CEM, compare lanes 2–4). We observed a similar significant 4-fold increase in infectivity in U937 cells when pre-exposed to J1.1-derived exosomes as compared with cells directly infected with virus (Fig. 8A, panel U937). THP-1 cells also showed a similar increase in susceptibility to HIV-1 infection upon pre-exposure to J1.1-derived exosomes, although the level of infectivity was not as pronounced as observed with CEM and U937 cells. Other cell types such as H9 and U87MGs also demonstrated an increase in infectivity (1.5–3-fold) when the cells were pre-exposed to J1.1-derived exosomes (data not shown). Infectivity analyses performed 7 days post-addition of exosomes followed a similar pattern of viral replication (data not shown).
FIGURE 8.
Prior exposure to J1.1-derived exosomes makes naive recipient cells more susceptible to HIV-1 infection. A, CEM, U937, and THP1 cells were first incubated with Jurkat or J1.1-derived exosomes for 24 h after which they were infected with HIV-1 (89.6 virus strain (0.5 ng/p24/μl; 100 μl)). Untreated, uninfected mock cells were maintained as negative controls, and untreated, virally infected cells were maintained as positive controls. Supernatants obtained from all cells 5 days after viral infection were analyzed by RT assay. Data were obtained from three independent replicates of each sample and are representative of two completely independent experiments. Error bars show ± S.D. B, J1.1 cells were first transfected with 200 nm antagomirs to 5′ and 3′ TAR miRNA using electroporation at 230 V. Both 5′ and 3′ TAR miRNA have previously been described (siTAR1 and siTAR2 (46)). Untransfected J1.1 cells were maintained as controls. Exosomes were isolated from both untransfected and transfected J1.1 cells and used to incubate with naive U937 cells following which the cells were infected with HIV-1 (89.6 virus (0.25 ng/p24/μl; 10 μl)). RT levels were determined using supernatants obtained from all cells at 5 days after viral infection. Data are representative of three replicates of each sample; error bars show the standard deviation.
As a next step, we sought to establish a direct link between the TAR RNA component of the exosome and the increase in susceptibility of naive cells to viral infection. We addressed that by introducing an antagomir to TAR and analyzing for enhanced susceptibility to subsequent viral infection. Accordingly, we transfected in antagomirs to both 5′ and 3′ TAR miRNAs into the parental J1.1 cells. Exosomes were purified from both transfected and untransfected J1.1 control cells and were used in susceptibility studies. Specifically, U937 cells were incubated with either control exosomes obtained from untransfected J1.1 culture supernatants (lane 3, Fig. 8B) or exosomes obtained from J1.1 cells transfected with 5′ TAR antagomir (lane 4), 3′ TAR antagomir (lane 5), or a combination of both (lane 6). After 24 h, the cells were infected with 89.6 virus and monitored for 5 and 7 days. The results shown in Fig. 8B indicate RT activity 5 days post-infection and a similar outcome was observed 7 days post-infection (data not shown). As seen in Fig. 8B, prior exposure of U937 cells to the control J1.1-derived exosomes made the cells more susceptible to a subsequent infection as seen in the increase in RT activity (compare 2nd and 3rd lanes). Upon introduction of an antagomir to 5′ TAR miRNA, we did not observe any significant alteration in susceptibility to subsequent viral infection. However, when we introduced an antagomir to 3′ TAR miRNA, we noticed a 2-fold decrease in the RT activity suggesting that 3′ TAR miRNA played a key role in increasing the susceptibility of target cells to viral infection (Fig. 8B, compare 4th and 5th lanes with 3rd lane). Combining the 5′ and 3′ TAR antagomirs did not display any noticeable synergistic effect. Cumulatively, our data suggest that 3′ TAR miRNA may play a role in priming the recipient cell and increasing its susceptibility to viral infection.
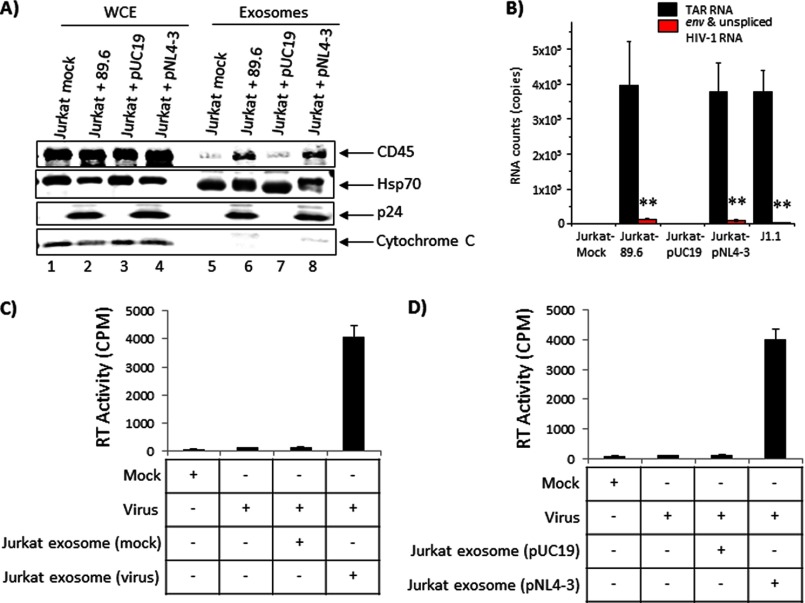
Exosomes Derived from Jurkat Cells Infected with HIV-1 (89.6) or Transfected with pNL4-3 Also Contain TAR RNA
J1.1 is a clonal cell line that displays differences from the parental Jurkat cell line. To directly correlate the presence of TAR RNA in exosomes to HIV-1 infection, we carried out experiments where we transfected Jurkat cells with the pNL4-3 plasmid and isolated exosomes from the culture supernatants 5 days post-transfection. Additionally, we infected Jurkat cells with the dual-tropic virus 89.6 and isolated exosomes from the culture supernatants 5 days post-infection. We validated our exosome preparations by performing standard Western blots for key components such as CD45, Hsp70, and cytochrome c and determined the purity of the preparations (Fig. 9A).
FIGURE 9.
Exosomes derived from transfected (pNL4-3) or infected (89.6) Jurkat cell culture supernatants contain TAR RNA. A, early phase log growing Jurkat cells (2.5 × 107) were transfected with 20 μg of pNL4-3 using electroporation or infected with 89.6 HIV-1 strain (∼200 ng of p24-positive virus) and maintained for 5 days in exosome-free media. Supernatants from these transfected and infected cells (50 ml each) were further processed for the presence of exosomes. pUC19 (20 μg) served as negative control for electroporation. Western blots were carried out using WCE or with exosomes using antibodies against CD45, Hsp70, p24, and cytochrome c. B, exosomes isolated from transfected and infected Jurkats were analyzed by qRT-PCR for the presence of TAR RNA. Error bars show the standard deviation of three independent measurements. Exosomes isolated from infected Jurkat cell culture supernatants (C) and transfected Jurkat culture supernatants (D) were utilized in infectivity experiments using U937 cells. Briefly, U937 cells were incubated for 24 h with each set of exosomes and then infected with 89.6 strain of HIV-1 (0.5 ng/p24/μl; 100 μl). Supernatants were analyzed 5 days post-infection for RT activity. Data were obtained from three independent replicates of each sample. Error bars show ± S.D.
Next, we performed qRT-PCR analysis of the exosomes for the presence of TAR RNA. The results shown in Fig. 9B indicate that exosomes isolated from culture supernatants of Jurkat cells infected with 89.6 virus showed ∼105 copies of TAR RNA, although all other viral RNAs were markedly low. Mock-infected Jurkat cells were maintained as negative controls, and exosomes isolated from mock-infected Jurkat cells did not contain TAR RNA (Fig. 9B). Similarly, exosomes isolated from culture supernatants of Jurkat cells transfected with pNL4-3 also contained comparable levels of TAR RNA as seen with exosomes from infected Jurkat cells. We isolated exosomes from supernatants of pUC19-transfected cells as a negative control and exosomes isolated from J1.1 culture supernatants as a positive control. As expected, although in the exosomes from pUC19-transfected cell culture supernatants there was absence of TAR RNA, ∼105 copies of TAR could be detected in exosomes isolated from J1.1 culture supernatants. Therefore, our data demonstrated that inclusion of TAR RNA in exosomes was a direct consequence of infection and was unlikely to be a cell type (J1.1)-specific phenomenon.
We then asked whether these exosomes could render a naive recipient cell susceptible to subsequent viral infection similar to the phenomenon we observed with exosomes isolated from J1.1 culture supernatants. To that end, we incubated U937 cells with exosomes isolated from culture supernatants of Jurkat cells that were either infected with 89.6 virus (Fig. 9C) or transfected with pNL4-3 (Fig. 9D), 24 h prior to infection with HIV-1. Exosomes isolated from culture supernatants of Jurkat cells that were mock-infected or transfected with pUC19 were utilized in similar experiments alongside as negative controls. Exosomes isolated from J1.1 culture supernatants were utilized as positive control (data not shown). The data demonstrated that prior exposure of U937 cells to exosomes derived from 89.6-infected or pNL4-3-transfected cells made naive U937 cells susceptible to subsequent infection by HIV-1 as determined by RT assays. Taken together, the data obtained from these series of experiments demonstrated that exosomes originating from HIV-1-infected cells contain TAR RNA and exert distinct physiological effects on naive cells in a cell type-independent manner.
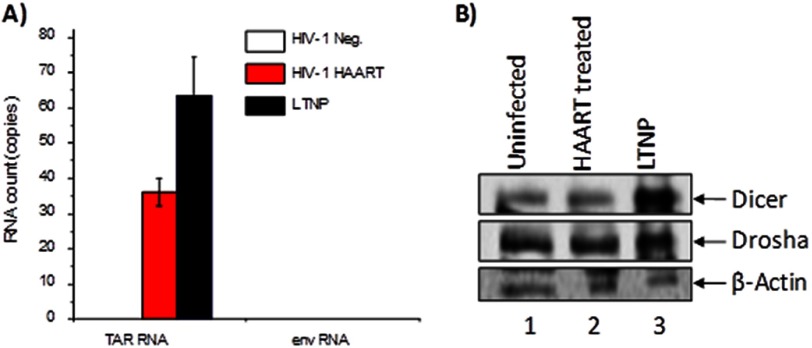
TAR RNA Could Be Detected in Exosomes Isolated from Patient Samples
Our studies performed with cultured cell lines and primary cells have thus far demonstrated the presence of TAR RNA, Dicer, and Drosha inside exosomes. We then asked if samples from infected patients have TAR RNA as well as Dicer and Drosha in their circulating exosomes. To answer that question, we purified exosomes from pooled serum samples that corresponded to the uninfected control group, HAART-treated group, and LTNPs (six samples in each group). Approximately 300 μl of serum samples were utilized for exosome purification using the ExoQuick purification protocol. Pelleted exosomes were resuspended in PBS, and 30% of the material was utilized for the analysis of TAR RNA by qRT-PCR. We observed that every microliter of starting material contained about 17 copies of TAR RNA in HAART-treated samples and about 30 copies in LTNPs (Fig. 10A). Our calculations indicated that there were ∼3.5 × 103 copies per ml of TAR RNA in the serum exosomes of HAART-treated patients. Similarly, there were ∼6 × 103 copies per ml concentration of TAR RNA in the serum exosomes of LTNPs. Env RNA was also analyzed by qRT-PCR and shown to be negative in these pooled samples further indicating that there is little contamination of full-length virus with these crude exosome preparations (use of the ExoQuick purification method).
FIGURE 10.
TAR RNA, Dicer, and Drosha can be detected in serum exosomes. A, exosomes were isolated from pooled sera obtained from uninfected (control), HAART-treated, and LTNP HIV-1-infected patient groups and analyzed by qRT-PCR with TAR- and env-specific primers. Results are presented as a mean of three independent measurements ± S.D. B, serum exosomes were analyzed by Western blot using antibodies against Dicer, Drosha, and β-actin. The ExoQuick-purified material was diluted by a 1:10 ratio (TNE-50 + 0.1% Nonidet P-40), passed through a Sephadex G-10 spin column, and analyzed by Western blot.
We next asked whether we could detect Dicer and Drosha in serum exosomes. We diluted the ExoQuick purified material by a 1:10 ratio, passed the diluted sample through a Sephadex G-10 spin column, and analyzed the diluted sample by Western blot with antibodies against Dicer, Drosha, and β-actin. We were able to detect both Dicer and Drosha in the serum exosomes of HIV-1-infected, HAART-treated patients and LTNPs (Fig. 10B). Our analysis of enriched serum exosomes revealed that although Dicer showed a slight increase in the HAART-treated patient samples over the uninfected control samples, there was an increase in the levels of Dicer in the LTNP samples (Fig. 10B, compare lane 3 with lanes 1 and 2). In the case of Drosha, however, we did not detect the same level of increase in the LTNP samples, although there was a modestly higher amount (∼11%) present than the HAART-treated sample. There was no difference in the levels of β-actin in all three samples. We also found that exosomes isolated from LTNP serum samples contained higher levels of TAR RNA compared with HAART-treated samples. We did not observe any disease-specific association of Ago2 or Exportin with any of the serum exosome preparations (data not shown). Collectively, these studies reveal that exosomes isolated from patient samples also contain TAR RNA, Dicer, and Drosha as we observed in the case of exosomes isolated from infected cells.
DISCUSSION
In recent years, exosomes have attracted much attention as intercellular messengers that mediate multiple functions and decisions involving cell survival or death. In the context of viral infections, multiple recent publications have demonstrated exosome-mediated intercellular transport of small RNA molecules and proteins from infected to naive target cells. In this study, we show that exosomes that originate from HIV-1-infected cells contained TAR RNA (Fig. 3).
Some recent publications have questioned the existence of small RNA molecules in vesicles and have suggested that stable detection of small RNA molecules in an extracellular environment is largely due to the existence of Ago2 ribonucleoprotein complex (74, 75). Turchinovich et al. (75) suggested that the majority of miRNAs in circulation were in fact independent of exosomes. They made this suggestion based on their analysis of three host-derived miRNAs. It is possible that these three miRNAs may be circulating as Ago2 complexes in culture supernatants and/or serum, and this may also partly be a reflection of their synthesis and abundance. This may however not necessarily apply to most or all of the host and virally derived noncoding RNA species. Although our analysis could detect miR16 (one of the miRNAs that was published by Turchinovich et al. (75)) as being present in circulation as an ribonucleoprotein with Ago2, we could not detect any TAR miRNA (either 3′ or 5′ strand) complexed with Ago2 (Fig. 3E). Our data demonstrated that the majority of TAR RNA that we detected in an extracellular environment was in fact TAR pre-miRNA (minimally, the first 57 nucleotides including the stem loop structure) that was transported into exosomes originating from infected cells.
Proteomic analysis of J1.1-derived exosomes for the presence of viral proteins revealed that they contained significant levels of Gag protein, which is in agreement with published data by Fang et al. (65) (Fig. 4D). In addition to Gag, we also detected an unprocessed form of the viral Env protein (Fig. 4D). Inclusion of gp160 in exosomes may be related to modifications/processing of the protein in the endoplasmic reticulum and Golgi regions of the cell. It may be possible that modifications on gp160 may lead to higher order structures that may mediate exosome inclusion. Alternatively, association of gp160 with specific host proteins may drive this phenomenon. These speculations may be amenable to better hypothesis-driven experiments as part of future studies.
It would be intriguing to speculate that the functional relevance of the Env protein may be seen in HIV-associated neurological disease as both Env and Tat proteins have been shown to contribute to the pathogenicity of this form of neurologic damage seen in HIV-1-infected individuals. The viral Env protein has been demonstrated to induce apoptosis of neuronal cells by activating the PTEN signaling cascade (76). Additionally, our data suggested that prior exposure to exosomes from infected cells made U87MGs (astroglioma cells) more susceptible targets to HIV-1 infection (data not shown). Therefore, exosomes from infected cells (i.e. T cells, macrophages, or dendritic cells) may potentially cross the blood-brain barrier and induce neuronal damage in a manner that is independent of an actual infection (77). Along these lines, we are currently trying to determine whether inhibition of exosome production in animals infected with HIV-1 inhibits deleterious by-stander neuronal damage seen in HIV-associated neurological disease.
We were able to observe limited amounts of Nef protein in J1.1-derived exosomes only after concentrating the exosomes 10-fold (i.e. a final concentration of 200 μg) suggesting that the level of Nef protein in these exosomes was low (data not shown). Nef may be incorporated in exosomes by anchoring exosome lipid raft microdomains, i.e. membrane regions rich in cholesterol, through both its N-terminal myristoylation and a stretch of basic amino acids within the α helix 1. Additionally, specific sequence regions in Nef that may interact with host proteins (Mortalin, in the case of Nef) may facilitate inclusion in exosomes (78).
The exact mechanisms that mediate fusion of exosomes with target cells remain a subject of intense investigation. A direct link to fusion of exosomes and target cells are the tetraspanins (79). Both Jurkat- and J1.1-derived exosomes contain CD63 and CD81. These two tetraspanins are among 33 of the total tetraspanins identified to date that are expressed in multiple cell types and have been associated with diverse complex processes that include membrane fusion (80). Tetraspanins have been demonstrated to interact with each other and with multiple integrins that may mediate fusion between the exosome membranes and membranes of target cells (81). These tetraspanin-enriched microdomains are sites of interactions of tetraspanins with integrins and cellular co-receptors such as CXCR4 (82). Exosomal uptake into target cells mediated by tetraspanins may involve clathrin-coated pits, may be AP-2-mediated, may involve caveolae-mediated endocytosis, or may be ceramide-mediated (82, 83). The fact that our proteomic analysis has revealed that Rab GTPases are present in both Jurkat- and J1.1-derived exosomes indicates that multisubunit tethering complexes involving SNAREs may also play a role in exosome (vesicle) fusion with target membranes (84).
We observed that certain cell types displayed increased susceptibility to infection by HIV-1 if they had been exposed to J1.1-derived exosomes (Fig. 8). For example, U937 cells, if exposed to J1.1 exosomes, showed a 4-fold increase in viral replication (Fig. 8A). U937 cells have suboptimal levels of Dicer protein (85). Therefore, the processing of TAR pre-miRNA in these cell types to exert their function may rely on the Dicer that is present in the incoming exosomes. Independent CAT assays suggest that Dicer present in exosomes may be functional in recipient cells (data not shown). Cumulatively, our experiments exploring the functional significance of J1.1-derived exosomes suggested that when the cells were faced with stress conditions that would normally activate apoptotic responses, exosome-mediated regulatory processes might function to down-regulate apoptosis.
We also asked whether our data, which suggest unique phenotypes and compositions associated with J1.1-derived exosomes, might be the result of defective virus contamination in our exosome preparations. A survey of the literature of what was defined as a defective virus included many parameters, including viruses with mutations that would perturb their ability to perform critical functions. Along those lines, mutant viruses that bear small mutations or deletions in the Env region, Integrase domain, and accessory proteins, including Vpr, Vif, and Nef, have been described (86–93). However, qRT-PCRs experiments using exosomes derived from infected cells or patient material did not result in significant amplification of viral mRNA. Our proteomic studies revealed that the unprocessed form of Env (gp160) was present in the exosome preparations, although we could not detect processed gp120 nor gp41 nor other viral proteins, including Tat (Fig. 4D). Thus, many lines of evidence suggest that our exosome preparations contain little contaminating virus.
Finally, we asked the question whether TAR RNA is present in the exosomes of patient sera. Exosomes purified from HAART-treated patients and LTNPs revealed significant levels of TAR RNA (Fig. 10A). The lack of Env RNA attested to the purity of the exosome preparations without viral contamination. We were also able to identify both Dicer and Drosha proteins in the serum exosomes (Fig. 10B). Current experiments using humanized mouse models and human latently infected cells in vitro are in progress to address the amount of TAR associated with the degree of disease and viral progression. Collectively, our data demonstrate that exosomes derived from HIV-1-infected cells are likely to have profound influences on host-virus interaction and disease progression.
Supplementary Material
Acknowledgments
We thank the members of the Kashanchi laboratory for helpful discussions and critical review of the manuscript. Dr. Jonathan Karn (Case Western Reserve University) also helped in the design of the CRM1-mediated export of TAR. We also thank Dr. John Rossi (City of Hope) for the pol III-TAR and control vector and Drs. Jan Münch, Michael Schindler, and Frank Kirchhoff for pNL4.3_92BR020.4nef+_IRES_EGFP and pNL4.3_92BR020.4nef-_IRES_EGFP plasmids obtained via the National Institutes of Health AIDS Research and Reference Reagent Program. Dr. Tim McCaffery (George Washington University Medical Center) generously donated the FAS antibody. Clinical samples utilized in this study were provided by the Washington D. C., Metropolitan Consortium of the Women's Interagency HIV Study Collaborative Study Group (Principal Investigator, Mary Young). The Women's Interagency HIV Study is supported by National Institutes of Health Grants UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590 from NIAID and by the Eunice Kennedy Shriver NICHD Grant UO1-HD-32632.
This work was supported, in whole or in part, by National Institutes of Health Grants AI078859, AI074410, and AI043894 (to F. K.), NCI, NIDA, NIDCD, and NCRR UCSF-CTSI Grant UL1 RR024131, and District of Columbia Developmental Center for AIDS Research Grant 5P30AI087714-02 (to S. I.). This work was also supported by United States Department of Energy Grant DE-SC0001599 (to F. K. and C. B.).

This article contains supplemental Fig. 1 and Table 1.
- miRNA
- microRNA
- TAR
- trans-activation response element
- HAART
- highly active antiretroviral therapy
- LTNP
- long term nonprogressor
- qRT
- quantitative RT
- PBMC
- peripheral blood mononuclear cell
- AchE
- acetylcholinesterase enzyme
- pol
- polymerase
- nt
- nucleotide
- WCE
- whole cell extract
- IP
- immunoprecipitation
- TRBP
- TAR RNA-binding protein.
REFERENCES
- 1. Mathivanan S., Ji H., Simpson R. J. (2010) Exosomes: extracellular organelles important in intercellular communication. J. Proteomics 73, 1907–1920 [DOI] [PubMed] [Google Scholar]
- 2. Denzer K., Kleijmeer M. J., Heijnen H. F., Stoorvogel W., Geuze H. J. (2000) Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113, 3365–3374 [DOI] [PubMed] [Google Scholar]
- 3. Stoorvogel W., Kleijmeer M. J., Geuze H. J., Raposo G. (2002) The biogenesis and functions of exosomes. Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 4. Booth A. M., Fang Y., Fallon J. K., Yang J. M., Hildreth J. E., Gould S. J. (2006) Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 172, 923–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 6. Eldh M., Ekström K., Valadi H., Sjöstrand M., Olsson B., Jernås M., Lötvall J. (2010) Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS One 5, e15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Simpson R. J., Lim J. W., Moritz R. L., Mathivanan S. (2009) Exosomes: proteomic insights and diagnostic potential. Expert Rev. Proteomics 6, 267–283 [DOI] [PubMed] [Google Scholar]
- 8. Rana S., Zöller M. (2011) Exosome target cell selection and the importance of exosomal tetraspanins: a hypothesis. Biochem. Soc. Trans. 39, 559–562 [DOI] [PubMed] [Google Scholar]
- 9. Wang Z., Hill S., Luther J. M., Hachey D. L., Schey K. L. (2012) Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT). Proteomics 12, 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dimov I., Jankovic Velickovic L., Stefanovic V. (2009) Urinary exosomes. Scientific World Journal 9, 1107–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ludwig A. K., Giebel B. (2012) Exosomes: small vesicles participating in intercellular communication. Int. J. Biochem. Cell Biol. 44, 11–15 [DOI] [PubMed] [Google Scholar]
- 12. Anand P. K. (2010) Exosomal membrane molecules are potent immune response modulators. Commun. Integr. Biol. 3, 405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Record M., Subra C., Silvente-Poirot S., Poirot M. (2011) Exosomes as intercellular signalosomes and pharmacological effectors. Biochem. Pharmacol. 81, 1171–1182 [DOI] [PubMed] [Google Scholar]
- 14. Théry C., Ostrowski M., Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 15. Simons M., Raposo G. (2009) Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 [DOI] [PubMed] [Google Scholar]
- 16. Duijvesz D., Luider T., Bangma C. H., Jenster G. (2011) Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 59, 823–831 [DOI] [PubMed] [Google Scholar]
- 17. Agrawal N., Dasaradhi P. V., Mohmmed A., Malhotra P., Bhatnagar R. K., Mukherjee S. K. (2003) RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geanacopoulos M. (2005) An introduction to RNA-mediated gene silencing. Sci. Prog. 88, 49–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grishok A. (2005) RNAi mechanisms in Caenorhabditis elegans. FEBS Lett. 579, 5932–5939 [DOI] [PubMed] [Google Scholar]
- 20. Montgomery M. K. (2004) RNA interference: historical overview and significance. Methods Mol. Biol. 265, 3–21 [DOI] [PubMed] [Google Scholar]
- 21. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 22. Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghildiyal M., Zamore P. D. (2009) Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Winter J., Jung S., Keller S., Gregory R. I., Diederichs S. (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234 [DOI] [PubMed] [Google Scholar]
- 25. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V. N. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 [DOI] [PubMed] [Google Scholar]
- 26. Perron M. P., Provost P. (2008) Protein interactions and complexes in human microRNA biogenesis and function. Front. Biosci. 13, 2537–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perron M. P., Provost P. (2009) Protein components of the microRNA pathway and human diseases. Methods Mol. Biol. 487, 369–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Naqvi A. R., Islam M. N., Choudhury N. R., Haq Q. M. (2009) The fascinating world of RNA interference. Int. J. Biol. Sci. 5, 97–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lejeune E., Allshire R. C. (2011) Common ground: small RNA programming and chromatin modifications. Curr. Opin. Cell Biol. 23, 258–265 [DOI] [PubMed] [Google Scholar]
- 30. Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S. I., Moazed D. (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zofall M., Grewal S. I. (2006) RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harbor Symp. Quant. Biol. 71, 487–496 [DOI] [PubMed] [Google Scholar]
- 32. de Vries W., Berkhout B. (2008) RNAi suppressors encoded by pathogenic human viruses. Int. J. Biochem. Cell Biol. 40, 2007–2012 [DOI] [PubMed] [Google Scholar]
- 33. Haasnoot J., Berkhout B. (2011) RNAi and cellular miRNAs in infections by mammalian viruses. Methods Mol. Biol. 721, 23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schütz S., Sarnow P. (2006) Interaction of viruses with the mammalian RNA interference pathway. Virology 344, 151–157 [DOI] [PubMed] [Google Scholar]
- 35. van Rij R. P., Andino R. (2006) The silent treatment: RNAi as a defense against virus infection in mammals. Trends Biotechnol. 24, 186–193 [DOI] [PubMed] [Google Scholar]
- 36. Bivalkar-Mehla S., Vakharia J., Mehla R., Abreha M., Kanwar J. R., Tikoo A., Chauhan A. (2011) Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res. 155, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scaria V., Hariharan M., Maiti S., Pillai B., Brahmachari S. K. (2006) Host-virus interaction: a new role for microRNAs. Retrovirology 3, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boss I. W., Plaisance K. B., Renne R. (2009) Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol. 17, 544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dölken L., Pfeffer S., Koszinowski U. H. (2009) Cytomegalovirus microRNAs. Virus Genes 38, 355–364 [DOI] [PubMed] [Google Scholar]
- 40. Grundhoff A., Sullivan C. S., Ganem D. (2006) A combined computational and microarray-based approach identifies novel microRNAs encoded by human γ-herpesviruses. RNA 12, 733–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nair V., Zavolan M. (2006) Virus-encoded microRNAs: novel regulators of gene expression. Trends Microbiol. 14, 169–175 [DOI] [PubMed] [Google Scholar]
- 42. Plaisance-Bonstaff K., Renne R. (2011) Viral miRNAs. Methods Mol. Biol. 721, 43–66 [DOI] [PubMed] [Google Scholar]
- 43. Zhu J. Y., Pfuhl T., Motsch N., Barth S., Nicholls J., Grässer F., Meister G. (2009) Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 83, 3333–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grässer F. A., van Dyk L. F., Ho C. K., Shuman S., Chien M., Russo J. J., Ju J., Randall G., Lindenbach B. D., Rice C. M., Simon V., Ho D. D., Zavolan M., Tuschl T. (2005) Identification of microRNAs of the herpesvirus family. Nat. Methods 2, 269–276 [DOI] [PubMed] [Google Scholar]
- 45. Lin J., Cullen B. R. (2007) Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J. Virol. 81, 12218–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Klase Z., Kale P., Winograd R., Gupta M. V., Heydarian M., Berro R., McCaffrey T., Kashanchi F. (2007) HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 8, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Narayanan A., Kehn-Hall K., Bailey C., Kashanchi F. (2011) Analysis of the roles of HIV-derived microRNAs. Expert Opin. Biol. Ther. 11, 17–29 [DOI] [PubMed] [Google Scholar]
- 48. Klase Z., Winograd R., Davis J., Carpio L., Hildreth R., Heydarian M., Fu S., McCaffrey T., Meiri E., Ayash-Rashkovsky M., Gilad S., Bentwich Z., Kashanchi F. (2009) HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yeung M. L., Bennasser Y., Watashi K., Le S. Y., Houzet L., Jeang K. T. (2009) Pyrosequencing of small noncoding RNAs in HIV-1-infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37, 6575–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ouellet D. L., Plante I., Landry P., Barat C., Janelle M. E., Flamand L., Tremblay M. J., Provost P. (2008) Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 36, 2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lamers S. L., Fogel G. B., McGrath M. S. (2010) HIV-miR-H1 evolvability during HIV pathogenesis. Biosystems 101, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Brisibe E. A., Saksena N. K., Fujii Y. R. (2004) HIV-1 Nef suppression by virally encoded microRNA. Retrovirology 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schopman N. C., Willemsen M., Liu Y. P., Bradley T., van Kampen A., Baas F., Berkhout B., Haasnoot J. (2012) Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 40, 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Klase Z., Houzet L., Jeang K. T. (2011) Replication competent HIV-1 viruses that express intragenomic microRNA reveal discrete RNA-interference mechanisms that affect viral replication. Cell Biosci. 1, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., van Eijndhoven M. A., Hopmans E. S., Lindenberg J. L., de Gruijl T. D., Würdinger T., Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pegtel D. M., van de Garde M. D., Middeldorp J. M. (2011) Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim. Biophys. Acta 1809, 715–721 [DOI] [PubMed] [Google Scholar]
- 57. Zomer A., Vendrig T., Hopmans E. S., van Eijndhoven M., Middeldorp J. M., Pegtel D. M. (2010) Exosomes: Fit to deliver small RNA. Commun. Integr. Biol. 3, 447–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barth S., Meister G., Grässer F. A. (2011) EBV-encoded miRNAs. Biochim Biophys Acta 1809, 631–640 [DOI] [PubMed] [Google Scholar]
- 59. Gourzones C., Gelin A., Bombik I., Klibi J., Vérillaud B., Guigay J., Lang P., Témam S., Schneider V., Amiel C., Baconnais S., Jimenez A. S., Busson P. (2010) Extracellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. Virol. J. 7, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Campbell T. D., Khan M., Huang M. B., Bond V. C., Powell M. D. (2008) HIV-1 Nef protein is secreted into vesicles that can fuse with target cells and virions. Ethn. Dis. 18, S2–14-19 [PMC free article] [PubMed] [Google Scholar]
- 61. Lenassi M., Cagney G., Liao M., Vaupotic T., Bartholomeeusen K., Cheng Y., Krogan N. J., Plemenitas A., Peterlin B. M. (2010) HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ali S. A., Huang M. B., Campbell P. E., Roth W. W., Campbell T., Khan M., Newman G., Villinger F., Powell M. D., Bond V. C. (2010) Genetic characterization of HIV type 1 Nef-induced vesicle secretion. AIDS Res. Hum. Retroviruses 26, 173–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Muratori C., Cavallin L. E., Krätzel K., Tinari A., De Milito A., Fais S., D'Aloja P., Federico M., Vullo V., Fomina A., Mesri E. A., Superti F., Baur A. S. (2009) Massive secretion by T cells is caused by HIV Nef in infected cells and by Nef transfer to bystander cells. Cell Host Microbe 6, 218–230 [DOI] [PubMed] [Google Scholar]
- 64. Shelton M. N., Huang M. B., Ali S. A., Powell M. D., Bond V. C. (2012) Secretion modification region-derived peptide disrupts HIV-1 Nef's interaction with mortalin and blocks virus and Nef exosome release. J. Virol. 86, 406–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fang Y., Wu N., Gan X., Yan W., Morrell J. C., Gould S. J. (2007) Higher order oligomerization targets plasma membrane proteins and HIV Gag to exosomes. PLoS Biol. 5, e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gan X., Gould S. J. (2011) Identification of an inhibitory budding signal that blocks the release of HIV particles and exosome/microvesicle proteins. Mol. Biol. Cell 22, 817–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shen B., Wu N., Yang J. M., Gould S. J. (2011) Protein targeting to exosomes/microvesicles by plasma membrane anchors. J. Biol. Chem. 286, 14383–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Van Duyne R., Guendel I., Kehn-Hall K., Easley R., Klase Z., Liu C., Young M., Kashanchi F. (2010) The identification of unique serum proteins of HIV-1 latently infected long-term nonprogressor patients. AIDS Res. Ther. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cantin R., Diou J., Bélanger D., Tremblay A. M., Gilbert C. (2008) Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J. Immunol. Methods 338, 21–30 [DOI] [PubMed] [Google Scholar]
- 70. Carpio L., Klase Z., Coley W., Guendel I., Choi S., Van Duyne R., Narayanan A., Kehn-Hall K., Meijer L., Kashanchi F. (2010) Erratum to: microRNA machinery is an integral component of drug-induced transcription inhibition in HIV-1 infection. J. RNAi Gene Silencing 6, E386. [PMC free article] [PubMed] [Google Scholar]
- 71. Carpio L., Klase Z., Coley W., Guendel I., Choi S., Van Duyne R., Narayanan A., Kehn-Hall K., Meijer L., Kashanchi F. (2010) microRNA machinery is an integral component of drug-induced transcription inhibition in HIV-1 infection. J. RNAi Gene Silencing 6, 386–400 [PMC free article] [PubMed] [Google Scholar]
- 72. Easley R., Carpio L., Dannenberg L., Choi S., Alani D., Van Duyne R., Guendel I., Klase Z., Agbottah E., Kehn-Hall K., Kashanchi F. (2010) Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat activated transcription. Virology 405, 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Marini A., Harper J. M., Romerio F. (2008) An in vitro system to model the establishment and reactivation of HIV-1 latency. J. Immunol. 181, 7713–7720 [DOI] [PubMed] [Google Scholar]
- 74. Arroyo J. D., Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., Mitchell P. S., Bennett C. F., Pogosova-Agadjanyan E. L., Stirewalt D. L., Tait J. F., Tewari M. (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Turchinovich A., Weiz L., Langheinz A., Burwinkel B. (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zou S., El-Hage N., Podhaizer E. M., Knapp P. E., Hauser K. F. (2011) PTEN gene silencing prevents HIV-1 gp120(IIIB)-induced degeneration of striatal neurons. J. Neurovirol. 17, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M. J. (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345 [DOI] [PubMed] [Google Scholar]
- 78. Bentham M., Mazaleyrat S., Harris M. (2006) Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J. Gen. Virol. 87, 563–571 [DOI] [PubMed] [Google Scholar]
- 79. Boucheix C., Rubinstein E. (2001) Tetraspanins. Cell. Mol. Life Sci. 58, 1189–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Charrin S., le Naour F., Silvie O., Milhiet P. E., Boucheix C., Rubinstein E. (2009) Lateral organization of membrane proteins: tetraspanins spin their web. Biochem. J. 420, 133–154 [DOI] [PubMed] [Google Scholar]
- 81. Yunta M., Lazo P. A. (2003) Tetraspanin proteins as organisers of membrane microdomains and signalling complexes. Cell. Signal. 15, 559–564 [DOI] [PubMed] [Google Scholar]
- 82. Pols M. S., Klumperman J. (2009) Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315, 1584–1592 [DOI] [PubMed] [Google Scholar]
- 83. Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 84. Bröcker C., Engelbrecht-Vandré S., Ungermann C. (2010) Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 20, R943–R952 [DOI] [PubMed] [Google Scholar]
- 85. Coley W., Van Duyne R., Carpio L., Guendel I., Kehn-Hall K., Chevalier S., Narayanan A., Luu T., Lee N., Klase Z., Kashanchi F. (2010) Absence of DICER in monocytes and its regulation by HIV-1. J. Biol. Chem. 285, 31930–31943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brandano L., Stevenson M. (2012) A highly conserved residue in the C-terminal helix of HIV-1 matrix is required for envelope incorporation into virus particles. J. Virol. 86, 2347–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Furnes C., Andresen V., Szilvay A. M. (2008) Functional rescue of an oligomerization-defective HIV-1 Rev mutant by fusion with an oligomeric tag. Arch. Virol. 153, 357–362 [DOI] [PubMed] [Google Scholar]
- 88. Mohammed K. D., Topper M. B., Muesing M. A. (2011) Sequential deletion of the integrase (Gag-Pol) carboxyl terminus reveals distinct phenotypic classes of defective HIV-1. J. Virol. 85, 4654–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Paolucci S., Gulminetti R., Maserati R., Dossena L., Baldanti F. (2011) Accumulation of defective HIV-1 variants in a patient with slow disease progression. Curr. HIV Res. 9, 17–22 [DOI] [PubMed] [Google Scholar]
- 90. Poe J. A., Smithgall T. E. (2009) HIV-1 Nef dimerization is required for Nef-mediated receptor down-regulation and viral replication. J. Mol. Biol. 394, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sandonís V., Casado C., Alvaro T., Pernas M., Olivares I., García S., Rodríguez C., del Romero J., López-Galíndez C. (2009) A combination of defective DNA and protective host factors are found in a set of HIV-1 ancestral LTNPs. Virology 391, 73–82 [DOI] [PubMed] [Google Scholar]
- 92. Venkatachari N. J., Walker L. A., Tastan O., Le T., Dempsey T. M., Li Y., Yanamala N., Srinivasan A., Klein-Seetharaman J., Montelaro R. C., Ayyavoo V. (2010) Human immunodeficiency virus type 1 Vpr: oligomerization is an essential feature for its incorporation into virus particles. Virol. J. 7, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang S. H., Xing H., He X., Zhu F. X., Meng Z. F., Ruan Y. H., Shao Y. M. (2008) Nef mutations in long-term nonprogressors from former plasma donors infected with HIV-1 subtype B in China. Biomed. Environ. Sci. 21, 485–491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.