Background: The C5-hydroxylation reaction of coenzyme Q biosynthesis in Escherichia coli is catalyzed by an unknown enzyme.
Results: The UbiI protein is responsible for the C5-hydroxylation reaction.
Conclusion: The three monooxygenases involved in aerobic Q biosynthesis are now identified.
Significance: We report the characterization of a gene of unknown function and the first crystal structure of a monooxygenase involved in Q biosynthesis.
Keywords: Coenzyme Q, Crystal Structure, Escherichia coli, Flavin, Hydroxylase, Monooxygenase, ubi Genes
Abstract
Coenzyme Q (ubiquinone or Q) is a redox-active lipid found in organisms ranging from bacteria to mammals in which it plays a crucial role in energy-generating processes. Q biosynthesis is a complex pathway that involves multiple proteins. In this work, we show that the uncharacterized conserved visC gene is involved in Q biosynthesis in Escherichia coli, and we have renamed it ubiI. Based on genetic and biochemical experiments, we establish that the UbiI protein functions in the C5-hydroxylation reaction. A strain deficient in ubiI has a low level of Q and accumulates a compound derived from the Q biosynthetic pathway, which we purified and characterized. We also demonstrate that UbiI is only implicated in aerobic Q biosynthesis and that an alternative enzyme catalyzes the C5-hydroxylation reaction in the absence of oxygen. We have solved the crystal structure of a truncated form of UbiI. This structure shares many features with the canonical FAD-dependent para-hydroxybenzoate hydroxylase and represents the first structural characterization of a monooxygenase involved in Q biosynthesis. Site-directed mutagenesis confirms that residues of the flavin binding pocket of UbiI are important for activity. With our identification of UbiI, the three monooxygenases necessary for aerobic Q biosynthesis in E. coli are known.
Introduction
Coenzyme Q (ubiquinone or Q)3 is a widespread redox-active lipid essential for electron and proton transport in the respiratory chains from bacteria to mammals (1, 2). Q is composed of a fully substituted benzoquinone ring to which is attached a polyisoprenyl tail with a number of isoprenyl units varying from one species to another (6 in Saccharomyces cerevisiae, 10 in humans, and 8 in Escherichia coli). Biosynthesis of Q is a highly conserved pathway that involves a large number of genes that have been identified mostly from seminal genetic studies using S. cerevisiae and E. coli (3, 4). However, our genetic understanding of Q biosynthesis is still incomplete, and very few Q biosynthetic proteins have been isolated and characterized biochemically.
Biosynthesis of Q8 in E. coli requires 9 Ubi proteins (UbiA–H and UbiX); most of them are involved in reactions that modify the aromatic ring of the 4-hydroxybenzoic acid (4-HB) universal precursor (Fig. 1). After prenylation of 4-HB catalyzed by UbiA, UbiX and UbiD are proposed to be involved in the decarboxylation (5). Three methylation (reactions d, f, and h, Fig. 1) and three hydroxylation reactions (on C5, C1, and C6, reactions c, e, and g, respectively) follow to yield Q8. UbiG catalyzes both O5- and O6-methylation (6) whereas UbiE is the C2-methyltransferase (7). The genes encoding the enzymes responsible for the aerobic C1- and C6-hydroxylation reactions have been identified as ubiH and ubiF, respectively (8, 9) (Fig. 1). UbiF and UbiH share 32% of sequence identity and are defined as members of class A flavoprotein monooxygenases from sequence analysis (10), even though they have not been isolated in pure form and characterized in vitro. In contrast, the gene responsible for the C5-hydroxylation reaction in E. coli (reaction c on Fig. 1) remains unknown. UbiB has been proposed to be involved in this reaction based on the accumulation of OPP (Fig. 1) in a ubiB− strain (11, 12). However, accumulation of OPP does not necessarily reflect a defect in C5-hydroxylation because a strain deficient in the methyltransferase UbiG also accumulates OPP (13). Moreover, the primary sequence of UbiB does not contain any motif characteristic for hydroxylases but instead displays conserved motifs found in a superfamily of ancestral kinases (14).
FIGURE 1.
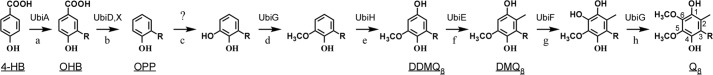
Biosynthetic pathway of coenzyme Q8 in E. coli. The numbering of the aromatic carbon atoms used throughout this study is shown on Q8, and the octaprenyl tail is represented by R on carbon 3 of the different biosynthetic intermediates. The name of the enzymes catalyzing the reactions (each labeled with a lowercase letter) is indicated. The enzyme catalyzing the C5-hydroxylation (reaction c) is unknown (see full text for further details). Abbreviations for coenzyme Q8 (Q8), C6-demethoxy-Q8 (DMQ8), C1-demethyl-C6-demethoxy-Q8 (DDMQ8), 3-octaprenyl-4-hydroxybenzoic acid (OHB), and 3-octaprenylphenol (OPP) are underlined.
Searching the gene responsible for the C5-hydroxylation, we reasoned that visC, a gene of unknown function, could be a good candidate to investigate because it locates immediately downstream of ubiH within the same operon (9). Furthermore, visC is predicted to encode a protein with the typical domains shared by flavoprotein monooxygenases and with 30 and 39% sequence identity to UbiH and UbiF, respectively (supplemental Fig. S1) (10). In the present study, we report genetic and biochemical experiments that unambiguously establish that VisC participates in coenzyme Q8 biosynthesis by catalyzing the aerobic C5-hydroxylation reaction. We therefore propose to change the name of VisC into UbiI. We succeeded in purifying the protein and crystallizing a truncated form of UbiI. The crystal structure shows a high degree of similarity with that of para-hydroxybenzoate hydroxylase (PHBH) a well characterized FAD-dependent monooxygenase (15). Albeit incomplete, our structure of UbiI is the first crystal structure of a monooxygenase associated with ubiquinone biosynthesis.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
E. coli strains were grown in Luria-Bertani (LB)-rich medium at 37 °C. Anerobic cultures were performed in sealed 15-ml Falcon tubes filled to the top with culture medium. Strains used in this study are listed in supplemental Table S1. Ampicillin (50 μg/ml) and kanamycin (25 μg/ml) were added when necessary. All of the strains are E. coli K-12 derivatives. Strains JW2874, JW2875, JW0659, and JW5581 from the Keio collection (16) were kindly provided by P. Moreau (LCB, Marseille) and were the donor of the ΔubiI::kan, ΔubiH::kan, ΔubiF::kan, and ΔubiE::kan mutations. The ΔubiI strain was cured to yield ΔubiIc which was then used to construct the ΔubiFΔubiI and ΔubiEΔubiI strains. Each mutation was transduced via P1 phage into MG1655. Mutations were confirmed using colony PCR with primers flanking the mutation.
S. cerevisiae was transformed using lithium acetate and was grown in YNB without pABA and folate (MP Biomedicals) with the indicated carbon sources (17).
Plasmid Construction
Plasmids are listed in supplemental Table S1, and their construction is detailed in the supplemental Materials and Methods.
Quinone Extraction and Quantification by HPLC-Electrochemical Detection (ECD) Analysis
5 ml of cells in exponential phase were centrifuged, and the pellet mass was determined. Glass beads (100 μl), 50 μl of 0.15 m KCl, and a volume of a Q solution (used as an internal standard) proportional to the wet weight were added to cell pellet. Q10 was used for E. coli samples and Q4 for S. cerevisiae samples. Lipids extraction was performed by adding 0.6 ml of methanol, vortexing for 10 min, then adding 0.4 ml of petroleum ether (boiling range 40–60 °C) and vortexing for 3 min. The phases were separated by centrifugation 1 min, 5000 rpm. The upper petroleum ether layer was transferred to a fresh tube. Petroleum ether (0.4 ml) was added to the glass beads and methanol-containing tube, and the extraction was repeated. The petroleum ether layers were combined and dried under nitrogen. The dried samples were stored at −20 °C and were resuspended in 100 μl of 98% ethanol, 20 mm lithium perchlorate. Aliquots were analyzed by reversed-phase HPLC with a C18 column (Betabasic-18, 5 mm, 4.6 × 150 mm; Thermo Scientific) at a flow rate of 1 ml/min using 40% ethanol, 40% acetonitrile, and 20% of a mix of 90% isopropyl alcohol and 10% lithium perchlorate (1 m) as a mobile phase. The mobile phase used to analyze yeast extracts was 5% isopropyl alcohol, 20% acetonitrile, and 75% of a mix of 98% methanol and 2% lithium perchlorate (1 m). Quinones were quantified with an ESA Coulochem III electrochemical detector and a 5011A analytical cell (E1, −650 mV; E2, 650 mV). Hydroquinones present in samples were oxidized with a precolumn 5020 guard cell set in oxidizing mode (E, +650 mV). Different volumes of the standard Q solutions were injected in the same conditions to correct for sample loss during the organic extraction on the basis of the recovery of the Q internal standard. Standard curves for Q4 and Q10 were used for Q6 and Q8 quantification, respectively.
4-HP8 Purification and Characterization
12 liters of LB culture of ΔubiI cells in stationary phase were centrifuged (4000 rpm, 15 min), and cell pellet mass was determined (30 g). Lipid extraction was performed for at least ten cycles at 50 °C with 150 ml of acetone using a Soxhlet extractor. The extract was concentrated with a rotary evaporator, resuspended in chloroform and applied on a silica column (M60; Macherey Nagel). Elution of molecules was realized with 100% of chloroform. Compounds of interest were collected in glass tubes, and 4-HP8 was further purified by HPLC with 40% acetonitrile, 40% ethanol, and 20% of a mix of 90% isopropyl alcohol and 10% of water. Oxidized and reduced forms of 4-HP8 were purified based on their characteristic UV spectra (see supplemental Fig. S3D). NMR spectra were recorded on a Bruker EMX 300 MHz spectrometer with D6-ethanol as solvent. Mass spectrometry measurements were carried out as described in the supplemental Materials and Methods.
Expression and Purification
Overexpression of UbiI was achieved by using BL21 (DE3) E. coli strain (Novagen) transformed with pET-ubiI-His6. The cultures were grown at 37 °C until they reached A600 = 0.4 and then shifted to 20 °C for induction. Isopropyl 1-thio-β-d-galactopyranoside was added to 0.1 mm, and growth continued overnight. All subsequent operations were carried out at 4 °C. Cells were harvested by centrifugation. The cell pellet was resuspended in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT, 30% glycerol (v/v), and 1 mm Pefabloc (VWR). After sonication, the lysate was centrifuged at 180,000 × g for 90 min. The resulting supernatant was loaded onto nickel-nitrilotriacetic acid Superflow resin (Qiagen) equilibrated with buffer 50 mm Tris-HCl, pH 7.5, 25 mm imidazole, 150 mm NaCl, 5 mm DTT, and 30% glycerol (v/v). UbiI was eluted with 250 mm imidazole, and after imidazole removal by ultrafiltration with Amicon Ultra 30K columns (Millipore), the purified protein was found to be >95% pure as judged by SDS-PAGE. The purified protein was aliquoted and stored at −80 °C in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm DTT, and 30% glycerol (v/v). Selenomethionylated UbiI was overexpressed according to the indicated protocol (18) and purified with the same protocol as the native UbiI.
Limited Trypsin Proteolysis
To optimize the conditions of limited proteolysis, experiments were first conducted on an analytical scale (500 μg of UbiI) where ratios of enzyme (trypsin) over substrate (UbiI) ranged from 1:15 to 1:1000 (w/w) in a final volume of 200 μl. Digestion time was then optimized for a fixed enzyme over substrate ratio. Tryptic digestion analysis was performed by SDS-PAGE. Large scale preparation of cleaved UbiI was then carried out in 50 mm Tris-HCl, pH 7.5, 30% glycerol (v/v) at a protein concentration of 3 mg/ml. After 2 h of incubation of UbiI (21 mg) with trypsin (42 μg) at 37 °C, the reaction was stopped by the addition of Pefabloc (1 mm), followed by analytical SDS-PAGE. The N-terminal 40-kDa domain, which lacks the C-terminal His6 tag, was purified on a nickel-nitrilotriacetic acid Superflow resin by collecting the flow-through and concentrated to 6–8 mg/ml in 25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% (v/v) glycerol.
Protein Analysis
Mass spectrometry analysis (MALDI-TOF) was performed to delineate the C-terminal boundary of the N-terminal 40-kDa domain of UbiI, with the MASCOT software and Findpept tool (SiCaPS platform of IMAGIF (Gif-sur-Yvette, France). SEC-MALLS experiments were performed to determine the oligomeric state of the protein, in 25 mm Tris-HCl, pH 7.3, 150 mm NaCl, 10% (v/v) glycerol (IMAGIF Structural and Proteomic Biology Unit, Gif-sur-Yvette, France).
Crystallization
Crystals were grown at 19 °C by the hanging drop vapor diffusion method. 1 μl of protein (6–8 mg/ml) was mixed with 1 μl of precipitant solution consisting of 0.1 m Tris, pH 8.5, 12% (w/v) polyethylene glycol 4000, 0.1 m NaCl, 0.2 m MgCl2, which led to plate crystals within 2–3 days. For the selenomethionylated protein, the precipitant solution was 0.1 m Bistris propane, pH 9.0, 14% (w/v) polyethylene glycol 4000, 0.1 m NaCl, 0.15 m MgCl2.
Data collection and structure determination were carried out as described in the supplemental Materials and Methods.
Docking Methods
A potential binding site of FAD in UbiItr was explored using docking calculations. Comparing the binding site residues of FAD in PHBH (1PBE) with those in the crystal structure of UbiItr, we noted that a number of residues are not favorably oriented to allow the docking of FAD. Preliminary to docking calculations, a UbiItr monomer was thus prepared as follows: (i) Rotamers of Arg-49, Arg-35, and Asp-164 were selected to allow the one-step insertion of a rigid model of FAD in its extended conformation as observed in 1PBE. (ii) The binding site residues were further energy minimized using CHARMm in Discovery Studio 3.1. The resulting protein structure was then used as the receptor, and FAD was docked as a flexible ligand using Autodock software (19). The docking results place FAD in a range of plausible positions spanning the length of the binding pocket of UbiItr. Among the best docked structures, we detail here a potential location of the FAD cofactor in UbiI selected for its highest similarity with the conformation observed in 1PBE. There is a rather good correspondence for ligand-receptor interactions between PHBH-FAD (1PBE) and the modeled UbiItr-FAD complex (supplemental Table S4).
RESULTS
E. coli ΔubiI Synthesizes Low Q8 Level in Aerobic Conditions
We constructed a ΔubiI strain in which the ORF of ubiI has been replaced with a kanamycin resistance cassette. ΔubiI cells display a growth comparable with WT cells both on glucose and on succinate media (supplemental Fig. S2A) unlike ΔubiH cells, as observed previously (9). Evaluation of the cellular content in electroactive isoprenoid quinones by using ECD of lipid extracts after separation by HPLC (17, 20) revealed that Q8 content in ΔubiI cells accounted for only 7% of that in WT cells (Fig. 2A). Furthermore, an unknown electroactive compound X could be detected (Fig. 2B). The levels of the isoprenoid naphthoquinones, demethylmenaquinone and menaquinone (MK8), were not significantly different in WT and ΔubiI cells (Fig. 2B and supplemental Fig. S2B). Transformation of ΔubiI cells with a plasmid containing the ubiI ORF restored Q8 content and abolished the accumulation of compound X (Fig. 2, A and B). ΔubiI cells grown anaerobically had a content of Q8 comparable with that in WT cells (Fig. 2, C and D), and accumulation of compound X was abolished. As expected, a strong increase in MK8 content was observed in cells grown anaerobically (21). Reversion of the Q8 biosynthetic defect by anaerobic growth of ΔubiI cells is reminiscent of the phenotype of a ΔubiH strain (22) (Fig. 2, C and D). Collectively, these data establish that UbiI is involved in aerobic Q8 biosynthesis but not in MK8 biosynthesis.
FIGURE 2.
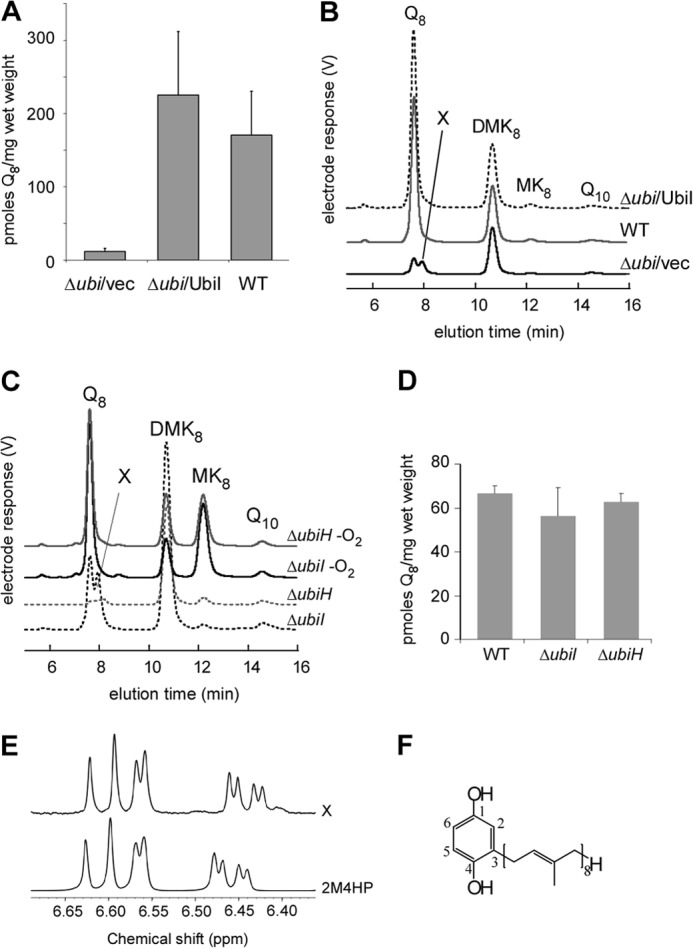
ΔubiI cells have a decreased Q8 content in aerobic conditions and accumulate 4-HP8. A, quantification of cellular Q8 content (n = 6–8) in lipid extracts from WT and ΔubiI cells containing an empty vector pBAD (vec) or pBAD-ubiI. Error bars are S.D. B, HPLC-ECD of lipid extracts from 2 mg of the same cells as in A. The peaks corresponding to Q8, compound X, demethylmenaquinone (DMK8), MK8, and to the Q10 standard are marked. C, HPLC-ECD of lipid extracts from 2 mg of ΔubiI cells (in black) or ΔubiH cells (in gray) grown in aerobic (dashed lines) or anaerobic conditions (solid lines). D, quantification of cellular Q8 content of cells described in A (n = 3). Error bars are S.D. E, superimposition of the aromatic region of the 1H NMR spectra of compound X and of commercial 2-methyl-4-hydroxyphenol (2M4HP) in D6-ethanol. F, Chemical structure of 4-HP8.
E. coli ΔubiI Accumulates 4-HP8
Compound X was purified from ΔubiI cells and found to be present in two redox states. Analysis by high resolution mass spectrometry in positive mode gave a m/z ratio (M+H+) of 655.5445 for reduced compound X and 653.5297 for the oxidized form corresponding respectively to chemical formulas of C46H70O2 +H+ (M = C46H70O2: 654.53755; ppm, 1.3) and C46H68O2 +H+ (M = C46H68O2: 652.5219; ppm, 0). Fragmentation of the 653.5 molecular ion yielded a characteristic tropylium-like ion (C7H7O2+), often found for aromatic compounds containing a benzyl unit, in addition to fragments corresponding to loss of methine, methylene, or methyl groups of an octaprenyl tail in the molecule (supplemental Fig. S3A). These data are consistent with compound X being a phenyl ring functionalized with an octaprenyl tail and either two OH groups in the reduced (quinol) C46H70O2 molecule or two keto groups in the oxidized (quinone) C46H68O2 molecule (supplemental Fig. S3B). The 1H NMR spectrum of the purified C46H70O2 compound shows well defined peaks both in the aromatic and aliphatic regions (supplemental Fig. S3C). Most of the signals in the 3.2–5.3 ppm region could be attributed to the vinylic and allylic protons of the octaprenyl substituent. The aromatic region of the spectrum shows more relevant information. Three distinguishable massifs, each corresponding to a unique proton, can be observed at 6.44 ppm (doublet of doublet with 4J and 3J coupling constants), 6.57 ppm (doublet with a 4J coupling constant), and 6.61 ppm (doublet with a 3J coupling constant) (Fig. 2E). These results confirm a 1,2,4-trisubstitution of the phenyl ring. The NMR spectrum of the commercially available 2-methyl-4-hydroxyphenol shows a highly similar pattern with close chemical shifts and coupling constants (Fig. 2E). The NMR data thus establish that the two hydroxyl groups of the C46H70O2 compound are in para relative position. Accordingly, the UV spectra of oxidized and reduced forms of compound X and of 2-methyl-4-hydroxyphenol are comparable (supplemental Fig. S3, D and E). Collectively, all of these data identify 4-HP8 (Fig. 2F) as the compound that accumulates in ΔubiI cells.
UbiI Complements a C5-hydroxylation Defect in S. cerevisiae
Drastic decrease of Q8 and accumulation of 4-HP8, lacking a OH or a OCH3 group at C5 are strong indications that the C5-hydroxylation reaction is deficient in E. coli ΔubiI cells and that UbiI functions as a C5-hydroxylase in Q8 biosynthesis. To verify this hypothesis, we tested whether heterologous expression of UbiI was able to complement a S. cerevisiae Δcoq6 strain which lacks the C5-hydroxylase Coq6 (17). In combination with Coq8 overexpression, required for maintaining the integrity of the Q biosynthetic complex in S. cerevisiae (23), the mitochondria-targeted E. coli UbiI protein was found to partially restore respiratory growth of Δcoq6 cells on lactate-glycerol medium, a process that requires coenzyme Q (Fig. 3A). HPLC-ECD analysis of cellular lipid extracts of Δcoq6/Coq8+UbiI cells demonstrated the presence of Q6 together with 4-HP6 (Fig. 3B) whereas Δcoq6/Coq8 cells accumulated exclusively 4-HP6 (17). Complementation by UbiI is not optimal but is nevertheless significant because Δcoq6/Coq8+UbiI cells contain approximately 12% of the amount of Q6 present in Δcoq6/Coq6 cells (Fig. 3C). These results establish that UbiI can function as a C5-hydroxylase in S. cerevisiae.
FIGURE 3.
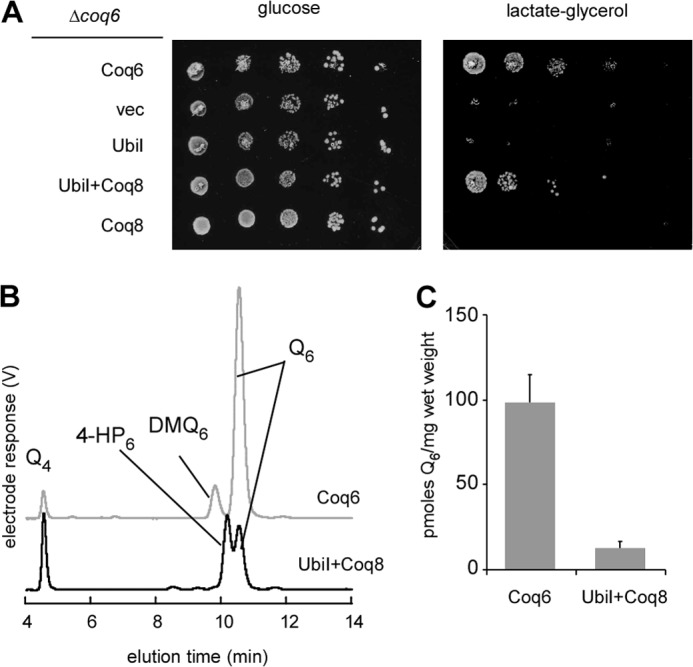
UbiI displays C5-hydroxylase activity in S. cerevisiae. A, S. cerevisiae Δcoq6 cells transformed with pRS416-COQ6 plasmid encoding Coq6, an empty vector (vec), or with pRS426 TDH3-ubiI, an episomal vector encoding UbiI in combination or not with pRS423-COQ8 grown in YNB-pABA-folate 2% galactose 0.4% glucose for 24 h. Serial dilutions were spotted onto agar plates. The plates contained synthetic medium-pABA-folate + 10 μm 4-HB with either 2% glucose or 2% lactate-2% glycerol. The plates were incubated for 2 days (glucose) or 4 days (lactate-glycerol) at 30 °C. B, HPLC-ECD of lipid extracts from 2 mg of Δcoq6 cells expressing Coq6 or from 6 mg of Δcoq6 cells expressing UbiI in combination with Coq8. Cells were grown in YNB-pABA-folate 2% lactate-2% glycerol supplemented with 10 μm 4-HB. Peaks corresponding to demethoxy-Q6 (DMQ6), Q6, 4-HP6, and to the Q4 standard are marked. C, Q6 content of Δcoq6 cells containing either pRS416-COQ6 or pRS426 TDH3-ubiI in combination with pRS423-COQ8 grown in the same medium as in B. Error bars are S.D. (n = 4).
The residual C5-hydroxylase Activity in ΔubiI Cells Is Provided by UbiF
ΔubiI cells synthesize a small amount of Q8 (Fig. 2A), reflecting that the C5-hydroxylation activity, although limited, is still taking place. To test the involvement of genes of the Q pathway in this biosynthetic activity, we constructed ΔubiF ΔubiI and ΔubiE ΔubiI mutants. ΔubiE cells accumulate the biosynthetic intermediate C2-demethyl-C6-demethoxy-Q8 (DDMQ8, peak at 6.87 min in Fig. 4A), as published before (7). Deletion of ubiI in a ΔubiE strain led to the accumulation of 4-HP8 and of a diminished amount of DDMQ8 (Fig. 4A). In contrast, deletion of ubiI in the ΔubiF strain caused the expected accumulation of 4-HP8 but also the complete disappearance of DMQ8 (Fig. 4B), showing that the C5-hydroxylation reaction is totally abolished in the ΔubiF ΔubiI strain. Collectively, these results demonstrate that the residual C5-hydroxylase activity observed in ΔubiI cells is provided by UbiF, the C6-monooxygenase.
FIGURE 4.
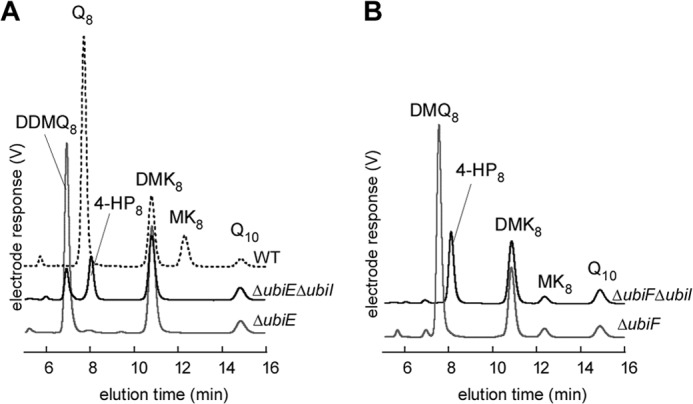
UbiF is responsible for the residual C5-hydroxylation in the absence of UbiI. A, HPLC-ECD of lipid extracts from 2 mg of WT cells (dashed black line), ΔubiE cells (in gray), or ΔubiEΔubiI cells (in black). B, HPLC-ECD of lipid extracts from 2 mg of ΔubiF cells (in gray) or ΔubiFΔubiI cells (in black).
Crystal Structure of a C-terminal Truncated Form of UbiI
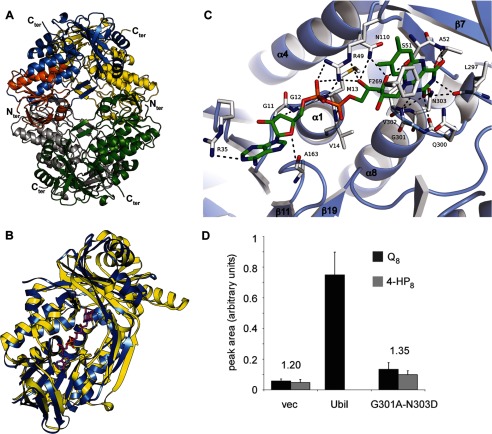
We overexpressed and purified the UbiI-His6 protein which contains a C-terminal His6 tag. However, the protein was cofactor-free, as shown by UV-visible spectroscopy, and highly unstable, with a significant propensity to aggregate and precipitate. Limited trypsin proteolysis of UbiI-His6 generated a soluble fragment (molecular mass ∼40 kDa, supplemental Fig. S4A). MALDI-TOF mass spectrometry of the UbiItr (supplemental Fig. S4B) together with the result that UbiItr does not bind to nickel-nitrilotriacetic acid, suggested that ∼35 residues at the C terminus had been removed (two cleavage sites after residue 364 and 365, respectively), as confirmed below by the crystal structure of UbiItr. The truncated UbiItr protein expressed from a plasmid was not able to complement the ΔubiI strain because the quinone profile was similar to that of ΔubiI cells containing an empty vector (supplemental Fig. S4C). UbiItr behaved as a tetramer in solution, as shown by SEC-MALLS experiments (supplemental Fig. S4D), and crystallized in space group P21212.
The structure of the native UbiItr protein was solved at 2.8 Å resolution by the single anomalous dispersion method using the selenomethionylated protein and refined to 2.0 Å resolution (supplemental Table S2 and supplemental Fig. S5). The overall tertiary structure of the tetramer reveals that each monomer contains a typical FAD-binding domain with a Rossman-like β/α/β-fold (Fig. 5A). A search for proteins with similar fold with DALI (24) indicates that the structure of UbiItr is similar to that of several flavin-dependent monooxygenases, including PHBH) from Pseudomonas fluorescens (supplemental Table S3). Superimposition of the structure of UbiItr with that of PHBH in complex with FAD (Fig. 5B) supports the presence of a FAD binding pocket in UbiItr.
FIGURE 5.
Crystal structure of a truncated form of UbiI and its FAD binding site. A, ribbon diagram of the crystal structure of tetrameric UbiItr. Each subunit is represented with a different color with the Rossman-like β/α/β-fold of one subunit highlighted in orange. B, DALI superimposition of UbiItr (blue) with PHBH (yellow; Protein Data Bank ID code 1PBE) and the FAD of PHBH shown as ball and sticks. The root mean square deviation is 3.0 Å for 327 Cα. C, Docking of FAD in UbiItr. Residues involved in hydrogen bonding and π-type interactions are represented. See details in supplemental Table S4. D, corrected integration of the electrochemical signal for the peaks corresponding to Q8 (in black) and 4-HP8 (in gray) after HPLC analysis of lipid extracts from ΔubiI cells containing an empty vector, pBAD-ubiI, or pBAD-ubiI(G301A/N303D) (n = 4). The ratio of the area of the Q8 and 4-HP8 peaks is indicated.
The FAD Binding Site in UbiItr
The existence of a binding site for a FAD cofactor in UbiI was confirmed with docking experiments and minimization simulations using a UbiItr monomer and the extended conformation of FAD from PHBH (1PBE) as the starting position (25). The results of these calculations are shown in Fig. 5C. The docked FAD in UbiItr exhibits a number of canonical interactions reminiscent of those involving FAD in PHBH, especially at the isoalloxazine ring and the ribityl chain (Fig. 5C and supplemental Table S4).
Fig. 5C shows that the isoalloxazine ring is located within hydrogen bonding distances of the main-chain nitrogens of Leu-297, Val-302, Asn-303, and Ala-52, and the side-chain amide nitrogen atom of Asn-303. Also, it may be stabilized by π-stacking with Phe-269, an interaction shared in a number of other flavoproteins (26–28). The ribityl chain is hydrogen-bonded to side chains of Arg-49 and Asn-110, as it is in PHBH using Arg-44 and Gln-102. Finally, the negatively charged pyrophosphate group is electrostatically complemented by: (i) the α1 helix dipole of the β/α/β-fold at the N terminus as in PHBH; (ii) hydrogen bonding with the side chain of Arg-49 residue (Arg-44 in PHBH) and the main chain of Met-13, a residue within the conserved GXGXXG motif of flavoproteins (Ser-13 in PHBH).
Our model of the docked FAD in UbiItr was validated by site-directed mutagenesis. We indeed mutated both residues Gly-301 and Asn-303, which are conserved in flavoprotein monooxygenases (29) (supplemental Fig. S1). These residues form the bottom of the isoalloxazine binding pocket in UbiItr. Expression of UbiI-G301A-N303D in the ΔubiI strain led to a Q8/4-HP8 ratio comparable with that found in ΔubiI cells containing an empty vector (Fig. 5D). In contrast, expression of UbiI led to the complete disappearance of 4-HP8 (Fig. 2B). These results show that the G301A/N303D double mutation strongly impairs UbiI activity probably as the consequence of a steric effect from the G301A mutation and the substitution of a hydrogen-bond stabilization by a repulsive electrostatic interaction resulting from the N303D mutation, both predicted to substantially affect the binding of FAD.
DISCUSSION
By using a combination of genetic, biochemical, and structural approaches, we have established that visC, a gene of unknown function in E. coli, encodes a protein that plays an important role in coenzyme Q biosynthesis under aerobic conditions. Consequently, we have renamed this gene ubiI. We show here that the UbiI protein functions in the C5-hydroxylation reaction of aerobic Q biosynthesis in E. coli.
More precisely, the function of the UbiI protein is to introduce the OH group on the C5 carbon of the phenyl ring of ubiquinone as supported by several lines of evidence. First, 4-HP8 that accumulates in ΔubiI cells is diagnostic of a deficiency in the C5-hydroxylation reaction. Indeed, 4-HP8 results from decarboxylation of OHB (reaction b, Fig. 1) to form OPP, which is then hydroxylated at position C1 (reaction e) without C5-hydroxylation (reaction c) and O5-methylation (reaction d) taking place. Likewise, we have shown that S. cerevisiae strains deficient for the C5-hydroxylation accumulate 4-HP6 (17, 20). Second, UbiI complements the C5-hydroxylation defect of Δcoq6 S. cerevisiae cells (Fig. 3), which shows that UbiI is a functional ortholog of the C5-hydroxylase Coq6. Given the modest sequence identity between UbiI and Coq6 (25%) and the fact that Coq proteins are organized in a multiprotein complex (3), it is not surprising that UbiI does not fully complement the deletion of coq6 (Fig. 3C). Third, we have established that UbiI displays a number of characteristics of FAD-dependent monooxygenases: (i) it shows high sequence homology with members of this class of enzymes, including the well studied PHBH and also UbiH, UbiF, and Coq6 (supplemental Fig. S1); (ii) the crystal structure of UbiItr shows great structural homology with PHBH, including at the level of a binding pocket for a FAD cofactor, as confirmed by docking experiments; (iii) Gly-301 and Asn-303, which contribute to the FAD binding pocket, are important for the C5-hydroxylase activity of UbiI. On the basis of these results, we propose that UbiI is a FAD-dependent monooxygenase that catalyzes the O2-dependent C5-hydroxylation reaction of coenzyme Q8 biosynthesis in aerobic conditions.
It should be noted that very few Ubi proteins involved in Q8 ring modification have been characterized biochemically, maybe as a result of limited stability and solubility when these proteins are expressed individually, as encountered with UbiI. In fact, only the purified O-methyltransferase UbiG has been shown to display in vitro activity so far (6). In the case of UbiI, we had to work with a truncated form lacking 34 amino acids in the C terminus to obtain a homogeneous protein in solution that we could crystallize. Unfortunately, this protein was inactive when assayed in vivo and could not bind FAD in vitro. Further studies are required to obtain a pure active UbiI enzyme. Nevertheless, our structure of UbiI is the first x-ray crystal structure of a monooxygenase implicated in Q biosynthesis.
A fascinating question raised by our work concerns the role of UbiB in Q biosynthesis. We clearly show that UbiI is responsible for the C5-hydroxylation reaction, and therefore UbiB is not catalyzing this step as this has been thought for years (4). An attractive possibility is that UbiB functions in regulating Q8 biosynthesis via its putative kinase activity as originally proposed by Clarke and co-workers (12). If so, the function of UbiB would be consistent with the emerging function of its eukaryotic homologs from S. cerevisiae (Coq8) (23) and humans (ACDK3/CABC1) (30).
Our characterization of ubiI yielded intriguing observations regarding the hydroxylation reactions. First, inactivation of ubiI does not completely abrogate Q8 biosynthesis. We have shown that in the absence of UbiI, the C6 monooxygenase UbiF is able to hydroxylate the neighboring C5, which points to a relatively poor regioselectivity of UbiF. However, there is no doubt that the main C5-hydroxylase is UbiI and not UbiF because UbiF only promotes the biosynthesis of 7% of WT Q8 levels in ΔubiI cells and because the C5-hydroxylation is not deficient in ΔubiF cells because only DMQ8 but no 4-HP8 is detected in these cells. Second, we have shown that UbiI is dispensable for anaerobic Q8 biosynthesis similarly to what was reported for UbiH and UbiF (22). As dioxygen is the source of the three oxygen atoms that are incorporated during the aerobic hydroxylation reactions (reactions c, e, and g, Fig. 1) of Q8 biosynthesis (31), the flavin-dependent monooxygenases UbiF, UbiH, and UbiI do not function in anaerobic conditions. Therefore, an alternative unidentified enzymatic system must substitute for UbiI, UbiF, and UbiH to catalyze the hydroxylation reactions during anaerobic Q8 biosynthesis. The next challenge will be to identify the anaerobic hydroxylation system.
Supplementary Material
Acknowledgments
We thank Christophe Velours and Laila Sago for performing the SEC-MALLS and mass spectrometry analysis on the IMAGIF platform, Dimitri Chérier for technical assistance, and Djemel Hamdane for fruitful discussions. We thank SOLEIL for provision of synchrotron radiation facilities and Beatriz Guimaraez for assistance in using beamline PROXIMA 1. We thank Dr. Nassos Typas (EMBL, Heidelberg) and Prof. Carol Gross (University of California, San Francisco) for sharing unpublished results at the beginning of this study.
This work was supported by Agence Nationale de la Recherche Grant JCJC SVSE 8-2011, Project pABACoQ (to F. P.) and Sup'Biotech (to A. I.).

This article contains supplemental Figs. S1–S5, Tables S1–S4, Materials and Methods, and additional references.
The atomic coordinates and structure factors (code 4K22) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Q
- coenzyme Q
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- DDMQ8
- C1-demethyl-C6-demethoxy-Q8
- ECD
- electrochemical detection
- 4-HB
- 4-hydroxybenzoic acid
- 4-HP6
- 3-hexaprenyl-4-hydroxyphenol
- 4-HP8
- 3-octaprenyl-4-hydroxyphenol
- MK8
- menaquinone
- OPP
- 3-octaprenylphenol
- pABA
- para-aminobenzoic acid
- PHBH
- para-hydroxybenzoate hydroxylase
- Q4
- coenzyme Q4
- Q8
- coenzyme Q8
- Q10
- coenzyme Q10
- SEC-MALLS
- size exclusion chromatography-multiangle laser light scattering
- UbiItr
- truncated UbiI.
REFERENCES
- 1. Nowicka B., Kruk J. (2010) Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797, 1587–1605 [DOI] [PubMed] [Google Scholar]
- 2. Bentinger M., Tekle M., Dallner G. (2010) Coenzyme Q: biosynthesis and functions. Biochem. Biophys. Res. Commun. 396, 74–79 [DOI] [PubMed] [Google Scholar]
- 3. Tran U. C., Clarke C. F. (2007) Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 7, S62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meganathan R. (2001) Ubiquinone biosynthesis in microorganisms. FEMS Microbiol. Lett. 203, 131–139 [DOI] [PubMed] [Google Scholar]
- 5. Gulmezian M., Hyman K. R., Marbois B. N., Clarke C. F., Javor G. T. (2007) The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch. Biochem. Biophys. 467, 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poon W. W., Barkovich R. J., Hsu A. Y., Frankel A., Lee P. T., Shepherd J. N., Myles D. C., Clarke C. F. (1999) Yeast and rat Coq3 and Escherichia coli UbiG polypeptides catalyze both O-methyltransferase steps in coenzyme Q biosynthesis. J. Biol. Chem. 274, 21665–21672 [DOI] [PubMed] [Google Scholar]
- 7. Lee P. T., Hsu A. Y., Ha H. T., Clarke C. F. (1997) A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J. Bacteriol. 179, 1748–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwon O., Kotsakis A., Meganathan R. (2000) Ubiquinone (coenzyme Q) biosynthesis in Escherichia coli: identification of the ubiF gene. FEMS Microbiol. Lett. 186, 157–161 [DOI] [PubMed] [Google Scholar]
- 9. Nakahigashi K., Miyamoto K., Nishimura K., Inokuchi H. (1992) Isolation and characterization of a light-sensitive mutant of Escherichia coli K-12 with a mutation in a gene that is required for the biosynthesis of ubiquinone. J. Bacteriol. 174, 7352–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Berkel W. J., Kamerbeek N. M., Fraaije M. W. (2006) Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689 [DOI] [PubMed] [Google Scholar]
- 11. Cox G. B., Young I. G., McCann L. M., Gibson F. (1969) Biosynthesis of ubiquinone in Escherichia coli K-12: location of genes affecting metabolism of 3-octaprenyl-4-hydroxybenzoic acid and 2-octaprenylphenol. J. Bacteriol. 99, 450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poon W. W., Davis D. E., Ha H. T., Jonassen T., Rather P. N., Clarke C. F. (2000) Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182, 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsu A. Y., Poon W. W., Shepherd J. A., Myles D. C., Clarke C. F. (1996) Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry 35, 9797–9806 [DOI] [PubMed] [Google Scholar]
- 14. Lagier-Tourenne C., Tazir M., López L. C., Quinzii C. M., Assoum M., Drouot N., Busso C., Makri S., Ali-Pacha L., Benhassine T., Anheim M., Lynch D. R., Thibault C., Plewniak F., Bianchetti L., Tranchant C., Poch O., DiMauro S., Mandel J. L., Barros M. H., Hirano M., Koenig M. (2008) ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am. J. Hum. Genet. 82, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Entsch B., Cole L. J., Ballou D. P. (2005) Protein dynamics and electrostatics in the function of p-hydroxybenzoate hydroxylase. Arch. Biochem. Biophys. 433, 297–311 [DOI] [PubMed] [Google Scholar]
- 16. Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ozeir M., Mühlenhoff U., Webert H., Lill R., Fontecave M., Pierrel F. (2011) Coenzyme Q biosynthesis: Coq6 Is required for the C5-hydroxylation reaction and substrate analogs rescue Coq6 deficiency. Chem. Biol. 18, 1134–1142 [DOI] [PubMed] [Google Scholar]
- 18. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 19. Trott O., Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Mühlenhoff U., Ozeir M., Lill R., Fontecave M. (2010) Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17, 449–459 [DOI] [PubMed] [Google Scholar]
- 21. Shestopalov A. I., Bogachev A. V., Murtazina R. A., Viryasov M. B., Skulachev V. P. (1997) Aeration-dependent changes in composition of the quinone pool in Escherichia coli: evidence of post-transcriptional regulation of the quinone biosynthesis. FEBS Lett. 404, 272–274 [DOI] [PubMed] [Google Scholar]
- 22. Alexander K., Young I. G. (1978) Alternative hydroxylases for the aerobic and anaerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry 17, 4750–4755 [DOI] [PubMed] [Google Scholar]
- 23. Xie L. X., Ozeir M., Tang J. Y., Chen J. Y., Jaquinod S. K., Fontecave M., Clarke C. F., Pierrel F. (2012) Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq-null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287, 23571–23581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schreuder H. A., Prick P. A., Wierenga R. K., Vriend G., Wilson K. S., Hol W. G., Drenth J. (1989) Crystal structure of the p-hydroxybenzoate hydroxylase-substrate complex refined at 1.9 Å resolution: analysis of the enzyme-substrate and enzyme-product complexes. J. Mol. Biol. 208, 679–696 [DOI] [PubMed] [Google Scholar]
- 26. Xia C., Hamdane D., Shen A. L., Choi V., Kasper C. B., Pearl N. M., Zhang H., Im S. C., Waskell L., Kim J. J. (2011) Conformational changes of NADPH-cytochrome P450 oxidoreductase are essential for catalysis and cofactor binding. J. Biol. Chem. 286, 16246–16260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Correll C. C., Batie C. J., Ballou D. P., Ludwig M. L. (1992) Phthalate dioxygenase reductase: a modular structure for electron transfer from pyridine nucleotides to [2Fe-2S]. Science 258, 1604–1610 [DOI] [PubMed] [Google Scholar]
- 28. Boyd J. M., Endrizzi J. A., Hamilton T. L., Christopherson M. R., Mulder D. W., Downs D. M., Peters J. W. (2011) FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins. J. Bacteriol. 193, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palfey B. A., Entsch B., Ballou D. P., Massey V. (1994) Changes in the catalytic properties of p-hydroxybenzoate hydroxylase caused by the mutation Asn300Asp. Biochemistry 33, 1545–1554 [DOI] [PubMed] [Google Scholar]
- 30. Xie L. X., Hsieh E. J., Watanabe S., Allan C. M., Chen J. Y., Tran U. C., Clarke C. F. (2011) Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta 1811, 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Alexander K., Young I. G. (1978) Three hydroxylations incorporating molecular oxygen in the aerobic biosynthesis of ubiquinone in Escherichia coli. Biochemistry 17, 4745–4750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.