Abstract
Background
Meat quality is an important economic trait in chickens. To identify loci and genes associated with meat quality traits, we conducted a genome-wide association study (GWAS) of F2 populations derived from a local Chinese breed (Beijing-You chickens) and a commercial fast-growing broiler line (Cobb-Vantress).
Results
In the present study, 33 association signals were detected from the compressed mixed linear model (MLM) for 10 meat quality traits: dry matter in breast muscle (DMBr), dry matter in thigh muscle (DMTh), intramuscular fat content in breast muscle (IMFBr), meat color lightness (L*) and yellowness (b*) values, skin color L*, a* (redness) and b* values, abdominal fat weight (AbFW) and AbFW as a percentage of eviscerated weight (AbFP). Relative expressions of candidate genes identified near significant signals were compared using samples of chickens with High and Low phenotypic values. A total of 14 genes associated with IMFBr, meat color L*, AbFW, and AbFP, were differentially expressed between the High and Low phenotypic groups. These genes are, therefore, prospective candidate genes for meat quality traits: protein tyrosine kinase (TYRO3) and microsomal glutathione S-transferase 1 (MGST1) for IMFBr; collagen, type I, alpha 2 (COL1A2) for meat color L*; and RET proto-oncogene (RET), natriuretic peptide B (NPPB) and sterol regulatory element binding transcription factor 1 (SREBF1) for the abdominal fat (AbF) traits.
Conclusions
Based on the association signals and differential expression of nearby genes, 14 candidate loci and genes for IMFBr, meat L* and b* values, and AbF are identified. The results provide new insight into the molecular mechanisms underlying meat quality traits in chickens.
Keywords: Chicken, GWAS, Meat quality traits, Abdominal fat, Candidate genes
Background
Meat quality in chickens is an important trait and includes pH, meat color, drip loss, tenderness, intramuscular fat (IMF) content, and other fat traits such as the contents and proportions of abdominal and subcutaneous fat. The selection of broiler chickens, initially focused on increasing growth performance and improving body composition [1], also led to indirect and often deleterious effects on meat quality traits, particularly excessive deposition of abdominal fat (AbF), the formation of which represents inefficient use of feed [2,3]. The elucidation of the molecular mechanisms underlying meat quality traits in chickens will have both biological and economic consequences.
Quantitative trait loci (QTLs) for many traits in chicken have been studied for over 20 years; 52 QTLs for meat-quality traits and 272 for abdominal fat traits have been detected in a variety of chicken chromosomal regions [4]. These QTLs were detected by linkage analysis and by candidate gene analysis. Both of these methods have limitations: the identified QTL regions are generally large and require subsequent fine mapping to identify closely linked markers or causative variants. Candidate genes, based on putative physiological roles, may exclude the identification of novel genes or pathways that influence the target traits [5].
The currently available chicken 60 K SNP chip covers the entire genome [5,6]. Genome-wide association studies (GWAS) can aid in more precisely identifying the genes and variants underlying important traits. In chicken, GWAS have already been performed for growth [7,8], egg production and quality [9] and disease resistance [10]. In the present study, we have performed a GWAS of several meat-quality traits in an F2 resource population derived from a cross between a Chinese local breed (Beijing-You, highly regarded for its meat quality) and a commercial rapidly-growing broiler line (Cobb-Vantress) to identify candidate genes.
Results
Phenotype statistics
The descriptive statistics for 16 meat quality traits in the F2 resource population used for the present GWAS are shown in Table 1. All non-normal phenotypic data, intramuscular fat content in thigh muscle (IMFTh), drip loss (DL), meat redness value (a*) and yellowness value (b*) of breast muscle, shear force (SF) of the pectoral major muscle, skin a* and b*, were normalised by Box-Cox or Johnson transformation except those for abdominal fat weight (AbFW), percentage of AbFW to eviscerated weight (AbFP) and the ultimate pH (24 h) of breast muscle (pHu).
Table 1.
Descriptive statistics for the meat quality traits
| Traits (Unit)1 | N | Mean | SD | Min | Max | CV |
|---|---|---|---|---|---|---|
| DMBr (%) |
316 |
27.87 |
1.30 |
20.97 |
33.29 |
4.67 |
| DMTh (%) |
309 |
24.39 |
1.14 |
20.47 |
29.62 |
4.69 |
| IMFBr (%) |
316 |
2.73 |
0.97 |
0.48 |
6.04 |
35.67 |
| IMFTh (%) |
310 |
7.17 |
2.23 |
1.39 |
15.24 |
31.10 |
| AbFW (g) |
324 |
37.47 |
29.60 |
0.00 |
128.30 |
88.43 |
| AbFP (%) |
324 |
1.43 |
1.21 |
0.00 |
5.64 |
84.61 |
| SFT (mm) |
323 |
5.22 |
1.82 |
1.16 |
11.36 |
34.75 |
| pHu |
304 |
5.55 |
0.25 |
5.00 |
6.30 |
4.43 |
| DL (%) |
315 |
7.03 |
2.94 |
1.66 |
18.67 |
41.78 |
| SF (kg/cm2) |
311 |
3.77 |
0.95 |
2.04 |
6.45 |
0.03 |
| Meat color |
|
|
|
|
|
|
| L* |
293 |
57.62 |
3.47 |
48.61 |
66.48 |
6.02 |
| a* |
292 |
12.18 |
2.12 |
7.37 |
19.45 |
17.44 |
| b* |
294 |
15.85 |
3.39 |
8.18 |
25.31 |
21.42 |
| Skin color |
|
|
|
|
|
|
| L* |
270 |
65.33 |
4.79 |
50.72 |
75.90 |
7.34 |
| a* |
266 |
8.61 |
3.40 |
1.80 |
17.24 |
39.55 |
| b* | 267 | 11.03 | 4.53 | 1.12 | 24.98 | 41.04 |
1DMBr dry matter content in breast muscle, DMTh dry matter content in thigh muscle, IMFBr intramuscular fat content in breast muscle, IMFTh intramuscular fat content in thigh muscle, AbFW abdominal fat weight, AbFP percentage of AbFW to eviscerated weight, SFT subcutaneous fat thickness, pHu the ultimate pH (24 h) of breast muscle, DL drip loss, SF shear force of the pectoral major muscle, L* lightness value; a* redness value and b* yellowness value. The same abbreviations are used in the following table.
GWAS analysis
A total of 6,695 independent SNP markers, distributed on all autosomes, were obtained with r2 = 0.2 Multidimensional scaling (MDS) analysis of these SNPs using the first two principal components (Figure 1) indicated that chickens within each full-sib family were clustered together. To correct for population stratification, the first MDS component was used as a covariate in a general linear model (GLM) and a compressed mixed linear model (MLM), as suggested in previous studies [8,11]. The relative kinship matrix was constructed from these independent SNP markers as a random effect in the compressed MLM.
Figure 1.
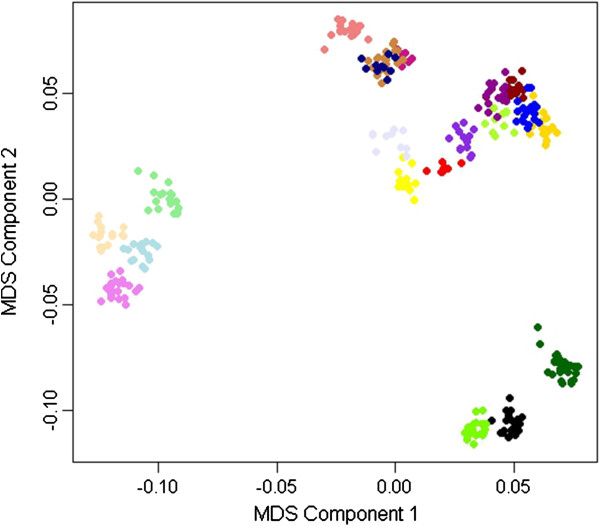
Population structure identification with multidimensional scaling analysis. Full-sib families are shown in the same color.
As observed in Figure 2, the compressed MLM is more effective than the GLM for controlling population structure, as described in previous studies [12-17]. The compressed MLM was, therefore, the preferred model to identify association signals. In addition, the compressed MLM also increases false negatives while false positives are reduced [13-16]. The suggestive significance threshold for p-value was set at 1.0 × 10-4 in the MLM analyses and those reaching genome-wide significance (p < 2.98 × 10-6) from the GLM analysis are also indicated. The 33 SNPs with association signals (p < 1.0 × 10-4) are listed in Tables 2 and 3 and Manhattan plots are shown in Figures 3 and 4 and Additional file 1: Figure S1. Seven of the 33 reached genome-wide significance based on the GLM analysis.
Figure 2.
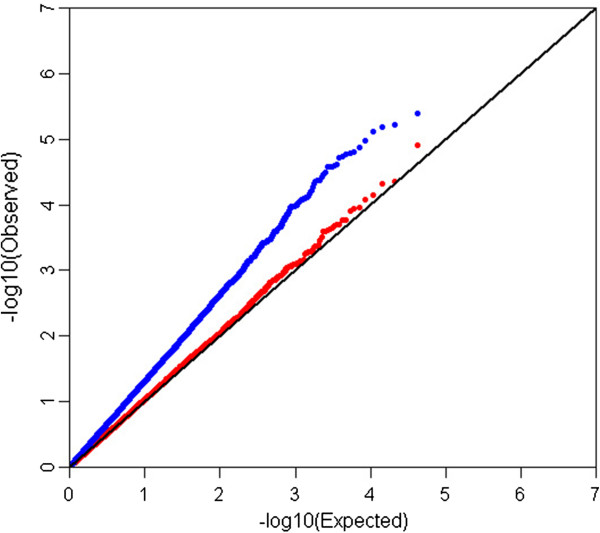
Quantile-quantile plot of the general linear model (GLM) (blue) and compressed mixed linear model (MLM) (red) for abdominal fat weight (AbFW).
Table 2.
Location of and gene information for SNPs associated (P < 1 × 10-4) with meat quality traits
| Trait | Chromosome | SNPs | Position | Alleles | MAF1 | P_value2 | P_adjust3 | R2 4 | Nearest Gene | Distance (Kb)5 |
|---|---|---|---|---|---|---|---|---|---|---|
| DMBr |
2 |
Gga_rs14206277 |
78152161 |
G/A |
0.338 |
2.01E-05 |
0.34 |
0.071 |
FAM105A |
within |
| DMTh |
14 |
Gga_rs14086206 |
14720651 |
A/G |
0.340 |
8.38E-05 |
1.40 |
0.077 |
TBC1D24 |
D 20.6 |
| DMBr |
Z |
GGaluGA346234 |
1626487 |
G/A |
0.016 |
1.16E-05 |
0.19 |
0.075 |
ST8SIA5 |
within |
| IMFBr |
1 |
Gga_rs13878108 |
65617257 |
G/A |
0.240 |
6.49E-05 |
1.09 |
0.063 |
MGST1 |
D 183.1 |
| IMFBr |
3 |
GGaluGA217414 |
40654853 |
A/G |
0.116 |
9.30E-05/2.59E-06 |
1.56 |
0.061 |
NTPCR |
U 109.9 |
| IMFBr |
4 |
GGaluGA255658 |
43311293 |
G/A |
0.100 |
3.46E-05/5.61E-09 |
0.58 |
0.057 |
AGA |
U 330.8 |
| IMFBr |
5 |
Gga_rs14524377 |
27009717 |
G/A |
0.193 |
5.41E-05 |
0.91 |
0.066 |
TYRO3 |
U 73.6 |
| IMFBr |
Z |
GGaluGA348872 |
18701354 |
A/G |
0.025 |
2.14E-05/4.28E-07 |
0.36 |
0.059 |
KIF2A |
U 122.3 |
| Meat color |
|
|
|
|
|
|
|
|
|
|
| L* |
2 |
GGaluGA137635 |
23472767 |
A/G |
0.302 |
7.04E-05 |
1.18 |
0.068 |
COL1A2 |
U 58.0 |
| L* |
18 |
GGaluGA121429 |
6990031 |
C/A |
0.412 |
8.96E-05 |
1.50 |
0.065 |
KPNA2 |
U 0.2 |
| L* |
18 |
GGaluGA121456 |
7112093 |
G/A |
0.383 |
7.19E-05 |
1.21 |
0.068 |
PSMD12 |
U 34.3 |
| b* |
27 |
Gga_rs15237721 |
1597620 |
G/A |
0.364 |
4.85E-05 |
0.81 |
0.077 |
FTSJ3 |
U 2.4 |
| Skin color |
|
|
|
|
|
|
|
|
|
|
| L* |
2 |
GGaluGA159190 |
99182734 |
G/A |
0.122 |
7.62E-05 |
1.28 |
0.073 |
PTPN2 |
U 130.6 |
| L* |
13 |
Gga_rs14058862 |
10586210 |
A/G |
0.114 |
9.39E-05 |
1.57 |
0.071 |
SPINK5 |
U 28.1 |
| L* |
13 |
Gga_rs15697794 |
11284741 |
G/A |
0.049 |
9.61E-05 |
1.61 |
0.058 |
CLINT1 |
U 15.6 |
| L* |
21 |
Gga_rs16180462 |
3645146 |
G/A |
0.162 |
8.76E-05 |
1.47 |
0.072 |
PGD |
U 62.7 |
| a* |
19 |
Gga_rs15044922 |
2083895 |
G/A |
0.213 |
5.10E-05 |
0.85 |
0.077 |
MIR1587 |
D 301.0 |
| b* | 6 | GGaluGA305921 | 33306370 | A/G | 0.412 | 3.24E-05 | 0.54 | 0.089 | BUB3 | D 195.5 |
1MAF minor allele frequency; 2the bold p-values of SNPs are the genome-wide significance by the general linear model (GLM); 3p-adjust indicates the p-value adjusted by “LD adjusted” Bonferroni; 4R2, SNP indicates the ratio of phenotypic variation; 5U indicates that the SNP is upstream of the gene, while D indicates that the SNP is downstream of the gene. These abbreviations are also used in Table 3.
Table 3.
Location of and gene information for SNPs associated (P < 1 × 10-4) with abdominal fat traits
| Trait | Chromosome | SNPs | Position | Alleles | MAF | P_value | P_adjust | R2 | Nearest Gene | Distance (Kb) |
|---|---|---|---|---|---|---|---|---|---|---|
| AbFW |
3 |
Gga_rs14385159 |
83714232 |
A/G |
0.248 |
4.79E-05 |
0.80 |
0.052 |
COL12A1 |
D 65.5 |
| AbFW |
4 |
Gga_rs15634423 |
85973459 |
A/G |
0.203 |
1.22E-05 |
0.20 |
0.059 |
MXD4 |
D 82.4 |
| AbFW |
4 |
Gga_rs16445604 |
86019649 |
A/G |
0.239 |
8.28E-05 |
1.39 |
0.049 |
MXD4 |
D 128.6 |
| AbFW |
6 |
Gga_rs14564481 |
5917270 |
A/G |
0.121 |
7.03E-05/4.11E-07 |
1.18 |
0.050 |
RET |
within |
| AbFW |
22 |
Gga_rs14708241 |
2145071 |
G/A |
0.104 |
4.46E-05/2.34E-07 |
0.75 |
0.052 |
LOC431251 |
U 124.3 |
| AbFP |
2 |
Gga_rs15112628 |
68073875 |
A/G |
0.223 |
9.62E-05 |
1.61 |
0.059 |
FOXC1 |
U 124.7 |
| AbFP |
2 |
Gga_rs13710186 |
68939093 |
G/A |
0.166 |
3.53E-05 |
0.59 |
0.066 |
VPS4B |
U 63.6 |
| AbFP |
2 |
Gga_rs14201030 |
69308190 |
G/A |
0.290 |
8.49E-05 |
1.42 |
0.060 |
BCL2 |
D 160.4 |
| AbFP |
4 |
Gga_rs15634423 |
85973459 |
A/G |
0.203 |
6.37E-05 |
1.07 |
0.062 |
MXD4 |
D 82.4 |
| AbFP |
5 |
GGaluGA276197 |
15378139 |
G/A |
0.132 |
6.59E-05 |
1.10 |
0.062 |
BRSK2 |
U 54.0 |
| AbFP |
6 |
Gga_rs14564481 |
5917270 |
A/G |
0.121 |
4.88E-05/2.62E-08 |
0.82 |
0.064 |
RET |
within |
| AbFP |
21 |
Gga_rs14285437 |
5764093 |
A/G |
0.247 |
6.40E-05 |
1.07 |
0.062 |
NPPB |
within |
| AbFP |
21 |
Gga_rs14285449 |
5774061 |
G/A |
0.258 |
7.30E-05 |
1.22 |
0.061 |
NPPB |
D 8.3 |
| AbFP |
22 |
Gga_rs14708241 |
2145071 |
G/A |
0.104 |
9.23E-06/6.58E-08 |
0.15 |
0.075 |
LOC431251 |
U 124.3 |
| AbFP | 14 | GGaluGA101040 | 4941190 | G/A | 0.245 | 9.94E-05 | 1.67 | 0.059 | SREBF1 | D 6.3 |
Figure 3.
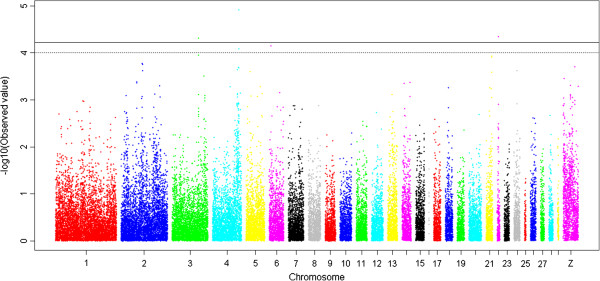
A Manhattan plot showing the association of all SNPs with abdominal fat weight (AbFW) from the compressed mixed linear model (MLM). SNPs are plotted on the x-axis according to their position on each chromosome against their association with AbFW on the y-axis (as -log10 p-value). The dashed line indicates genome-wide association (p = 1.00 × 10-4), and the solid line indicates significance with a p-value threshold of 5.96 × 10-5.
Figure 4.
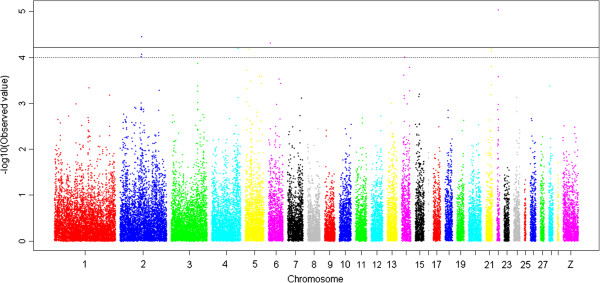
A Manhattan plot showing the association of all SNPs with percentage of abdominal fat (AbFP). The data and form of presentation are exactly as described for Figure 3 except the trait is AbFP, the proportion of eviscerated weight represented by abdominal fat.
Dry matter content in breast (DMBr) and (DMTh)
Two SNPs were associated with DMBr (p = 1.16 × 10-5 and 2.01 × 10-5). They were located at 0.16 Mb on chicken (Gallus gallus) chromosome Z (GGZ) and 7.81 Mb on GGA2, within the genes for ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 (ST8SIA5) and family with sequence similarity 105, member A (FAM105A), respectively. One SNP (Gga_rs14086206), located at 1.47 Mb on GGA14, was associated with DMTh (p = 8.38 × 10-5) and was located 20.6 Kb downstream from the TBC1 domain family member 24 gene (TBC1D24).
Intramuscular fat content in breast muscle (IMFBr)
Genes related to lipid metabolism would be predicted to influence IMFBr. Five SNPs associated with this trait were identified by the compressed MLM (p < 1.0 × 10-4), of which three were of genome-wide significance by GLM analysis (p < 2.98 × 10-6). One SNP (GGaluGA348872), located at 1.87 Mb on GGZ, had highly significant association with IMFBr (p = 2.14 × 10-5) and is located 122.3 Kb upstream of the kinesin heavy chain member 2A (KIF2A) gene. Another SNP (GGaluGA255658), located at 4.33 Mb on GGA4, was also associated with IMFBr (p = 3.46 × 10-5) and is located 330.8 Kb upstream of the aspartylglucosaminidase (AGA) gene. Three more SNPs (Gga_rs14524377, Gga_rs13878108 and GGaluGA217414) having association with IMFBr were in the proximity of protein tyrosine kinase (TYRO3), microsomal glutathione S-transferase 1 (MGST1) and nucleoside-triphosphatase, cancer-related (NTPCR) genes, on GGA5, GGA1 and GGA3, respectively.
Meat color traits
Three SNPs were associated with meat color lightness (L*) (p < 1.0 × 10-4). One SNP, located at 2.35 Mb on GGA2, is 58.0 Kb away from the collagen, type I, alpha 2 (COL1A2) gene. The other two SNPs are located within the 0.12 Mb segment (between 7.11 Mb to 6.99 Mb) on GGA18. These SNPs are approximately 34.3 and 0.20 Kb away from the proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (PSMD12) and karyopherin alpha 2 (RAG cohort 1, importin alpha 1) (KPNA2) genes, respectively. The SNP (Gga_rs15237721), located at 0.16 Mb on GGA27, was associated (p = 4.85 × 10-5) with meat color b*; the nearest gene, FtsJ homolog 3 (E. coli) (FTSJ3) gene, is located 2.4 Kb upstream.
Skin color traits
Four SNPs were found to be associated with skin color L* (p < 1.0 × 10-4). One, located at 9.92 Mb on GGA2, is 130.6 Kb upstream of the tyrosine-protein phosphatase non-receptor type 2 (PTPN2) gene. The SNP identified on GGA21 is 62.7 Kb head of the phosphogluconate dehydrogenase (PGD) gene. Two SNPs, located within a 0.70 Mb segment (10.59 Mb to 11.28 Mb) on GGA13, are approximately 28.1 and 15.6 Kb ahead of the serine peptidase inhibitor, Kazal type 5 (SPINK5) and clathrin interactor 1 (CLINT1) genes, respectively.
The SNP Gga_rs15044922 on GGA19 was associated with skin a* (p = 5.10 × 10-5), and is located distal (301 Kb) to a microRNA cluster that includes three microRNAs (MIR1587, MIR1354 and MIR1567). Another SNP, GGaluGA305921, located at 3.33 Mb on GGA6, was associated with skin b* (p = 3.24 × 10-5). The known gene nearest to this SNP (195 Kb) is budding uninhibited by benzimidazoles 3 homolog (yeast) (BUB3).
Abdominal fat (AbF) traits
Variants at 15 loci were identified as SNPs associated with AbF traits by compressed MLM (p < 1.0 × 10-4), and four were of genome-wide significance by GLM (p < 2.98 × 10-6) (Table 3, and Figures 3, 4). Two SNPs (Gga_rs15634423 and Gga_rs16445604) located within a 46.19 Kb region on GGA4 were associated with AbFW (p < 1.0 × 10-4); these SNPs are both downstream (82.4 Kb and 128.6 Kb) of the max-interacting transcriptional repressor MAD4 (MXD4) gene. The SNP Gga_rs15634423 was also associated with AbFP (p = 6.37 × 10-5). Another SNP (Gga_rs14708241), located at 0.21 Mb on GGA22, was associated with both AbFW and AbFP (p = 4.46 × 10-5 and p = 9.23 × 10-6, respectively). This SNP is 124.3 Kb away from the nearest known gene, LOC431251, which encodes prolactin-releasing peptide receptor-like protein (PrRPR), also known as G-protein coupled receptor 10 (GPR10). SNP Gga_rs14385159, located at 8.37 Mb on GGA3, is 65.5 Kb downstream of Collagen, type XII, alpha 1 (COL12A1) and was associated with AbFW (p = 4.79 × 10-5). Notably, a SNP (Gga_rs14564481) within the RET proto-oncogene (RET) gene on GGA6, was associated with both AbFW and AbFP (p < 1.0 × 10-4).
Three SNPs located within a 1.23 Mb region between 68.07 Mb and 69.31 Mb on GGA1 were associated with AbFP (p <1.0 × 10-4). One of these SNPS is 63.6 Kb upstream of the vacuolar protein sorting-associated protein 4B (VPS4B) gene, and another is approximately 160.4 Kb away from the known B-cell lymphoma 2 (BCL2) gene. The third SNP is located 124.7 Kb upstream of the forkhead box C1 (FOXC1) gene. On GGA21, two SNPs were associated with AbFP (p < 1.0 × 10-4). These SNPs are within 8.3 Kb of the natriuretic peptide B (NPPB) gene. Another SNP, located at 54.0 Kb on GGA5, was associated with AbFP and is 54.0 Kb away from the nearest known gene, BR serine/threonine kinase 2 (BRSK2). Interestingly, one SNP located at 0.49 Mb on GGA14 was associated with AbFP and is 6.3 Kb downstream from the gene for adipocyte determination- and differentiation-dependent factor 1, the sterol regulatory element binding transcription factor 1 (SREBF1).
No SNPs were found to be associated with IMFTh, SFT, pHu, DL, SF and meat color a* of breast muscle.
The mRNA expression study of candidate genes identified by the GWAS
Based on the GWAS analysis, 17 candidate genes containing or in proximity of SNPs with trait association were further evaluated by real-time quantitative PCR (Q-PCR) in subsets of six chickens with lowest or highest trait phenotypic values (Low, High). Significant differential expression (p < 0.05) between the Low and High birds was demonstrated for 14 of the 17 (Table 4).
Table 4.
Relative mRNA abundance in the low and high groups
| Traits | Genes |
Groups1 |
|
|---|---|---|---|
| Low | High | ||
| IMFBr |
KIF2A |
1.00 ± 0.05** |
0.55 ± 0.23 |
| |
TYRO3 |
1.00 ± 0.10** |
0.25 ± 0.11 |
| |
MGST1 |
1.01 ± 0.13** |
0.24 ± 0.04 |
| |
NTPCR |
1.00 ± 0.04** |
0.35 ± 0.22 |
| AbFW |
COL12A1 |
1.02 ± 0.24* |
0.57 ± 0.17 |
| |
RET |
1.02 ± 0.23 |
31.16 ± 0.68** |
| AbFP |
VPS4B |
1.03 ± 0.29** |
0.48 ± 0.17 |
| |
NPPB |
1.12 ± 0.14 |
72.08 ± 9.21** |
| |
BRSK2 |
1.01 ± 0.17** |
0.10 ± 0.04 |
| |
FOXC1 |
1.04 ± 0.33** |
0.30 ± 0.12 |
| |
SREBF1 |
1.02 ± 0.24** |
0.07 ± 0.04 |
| Meat color L* |
COL1A2 |
1.01 ± 0.19** |
0.24 ± 0.19 |
| |
PSMD12 |
1.01 ± 0.16* |
0.67 ± 0.04 |
| KPNA2 | 1.00 ± 0.09 | 1.76 ± 0.37* | |
1High group (n = 6) consisting of samples from chicken with the highest trait values and Low group (n = 6) with the lowest trait values; * p < 0.05, **p < 0.01.
For IMFBr, transcript abundance of four (of the five) genes identified near significant SNPs was significantly down-regulated in the High group compared to the Low group (p < 0.01); the differentially expressed genes were KIF2A, TYRO3, MGST1 and NTPCR.
For meat color L*, three of four chosen genes were differentially expressed between Low and High phenotypic groups (p < 0.05 or 0.01). Transcript abundance of COL1A2 and PSMD12 was significantly down-regulated in the High group (p < 0.01, 0.05), and that of KPNA2 was significantly higher (p < 0.01).
Of the eight genes identified in connection with AbF traits, seven were differentially expressed between the Low and High group (p < 0.01). Compared to the Low group, the expression of RET and NPPB was significantly increased (p < 0.01), and that of COL12A1, VPS4B, BRSK2, FOXC1 and SREBF was decreased (p < 0.05 or 0.01) in the High group.
Discussion
Genome-wide association analysis
There has been effective use of GWAS in meat quality and carcass traits in other species and narrow regions or SNPs associated with pork and beef quality have been revealed [18,19]. Here, we present a GWAS of meat quality traits in an F2 chicken population derived from a cross of Beijing-You chickens and commercial fast-growing broilers.
Meat quality traits
Dry matter content (DM) is a primary muscle characteristic. Nones et al. (2012) mapped a QTL for the water content (100-DM) of a chicken carcass at 23 cM on GGA27 in a half-sib linkage analysis [20]. In the present study, two loci for DM content in breast muscle (DMBr) and one for DMTh have been identified. Two SNPs within ST8SIA5 and FAM105A genes were associated with DMBr. ST8SIA5 encodes a type II membrane protein, a member of glycosyltransferase family 29 playing a role in the synthesis of some gangliosides and having a function in cellular recognition and cell-to-cell communication [21]. The pro-apoptotic gene FAM105A, when overexpressed, leads to cell apoptosis [22]. A SNP near TBC1D2 was identified to be associated with DMTh. This gene encodes a protein with a conserved domain, the TBC domain, common in proteins interacting with GTPases and has been related to endocytic trafficking [23]. Function characterization of these genes, near SNPs associated with muscle DM in chickens, is not yet clear.
Intramuscular fat (IMF) is an important determinant of meat quality influencing the tenderness, juiciness and flavour of meat [24]. In the present study, four genes (TYRO3, MGST1, KIF2A, and NTPCR) are shown to be potentially related to IMFBr. These genes were all differentially expressed in chickens with low and high IMFBr (Table 4), and some are known to play roles in lipid metabolism. TYRO3 plays an important role in cell proliferation and differentiation and has been associated with adipocyte size in moderately obese individuals in a clinical study [25]. The protein encoded by MGST1 catalyses the conjugation of glutathione to electrophiles and the reduction of lipid hydroperoxides. This gene was differentially expressed in the longissimus dorsi of Northeastern Indigenous and Large White pigs [26] and was previously identified in a QTL for chicken IMFBr[27]. KIF2A encodes the kinesin-like protein KIF2A, a microtubule-associated motor protein. It can regulate microtubule dynamics at the growth cone edge by depolymerizing microtubules and plays a role in the suppression of collateral branch extension [28]. NTPCR is a cancer-related gene with a presumed role in human tumorigenesis [29]. No previous studies have linked KIF2A or NTPCR with IMFBr and, as the associations were strong (Table 2) and the differential expression was quite large, further study of these genes seems to be warranted.
Meat color is an important quality that influences consumer acceptance of poultry meat and has significant positive correlations with pH and water-holding capacity [30]. Le Bihan-Duval et al. (2011) identified an influence of beta-carotene dioxygenase 1 (BCDO1) on chicken breast meat color using classical QTL analysis and gene expression QTL (eQTL) [31]. In the present study, we did not observe an influence of BCDO2 on meat color, probably because a different chicken population was tested. Three new genes for breast meat color were expressed differentially between the High and Low phenotypic groups. COL1A2, which encodes one of the chains of type I collagen, was differentially expressed here (4-fold) and also between the red and white skeletal muscles of Chinese Meishan pigs [32]; this gene could well be a candidate gene for meat color in chickens. PSMD12, encoding a proteasomal regulatory subunit, has been associated with liver function in humans [33]. KPNA2 encodes an importin, functioning in retrograde transport of signaling molecules from the axonal growth cone to the nucleus [34]. How the PSMD12 and KPNA2 genes might function in influencing meat color in chickens is not known.
The color of chicken skin influences consumer appeal and hence is also an important phenotype. A previous study found that yellow skin was caused by one or more cis-acting and tissue-specific regulatory mutation(s) that inhibit expression of BCDO2 in the skin [35]. Six SNPs were identified here as being associated with skin color but the only one associated with yellowness was near the BUB3 gene, with unknown function in chickens. One of genes with known effects on skin color, close to a SNP of significance, is SPINK5. The skin of SPINK5-deficient mice has large intensely blue areas close to colorless regions [36].
Abdominal fat traits
In addition to IMF, other fat traits, and especially abdominal fat, are important selection criteria in chicken breeding. Abdominal fat (AbF) is under complex genetic control and has medium heritability (h2 = 0.62 for AbFW, and 0.24 for AbFP) in one of the founding breeds used here [37]. As the trait is economically important but requires post-slaughter measurement, marker-assisted selection could be a more efficient method for genetic selection. Abdominal fat traits have been a focus of QTL mapping studies of chickens and several chromosomes are involved [4].
Seven genes (RET, NPPB, SREBF1, COL12A1, VPS4B, BRSK2, FOXC1) containing or near SNPs associated with abdominal fat traits, identified here, had significant different expression in AbF from cohorts of birds with highest and lowest AbF content. Three of the genes, RET, NPPB and SREBF1, are known to be related to lipid metabolism in other species. RET increases lipid accumulation in humans, based on a high-throughput siRNA screen with primary (pre)adipocytes [38]. In the present study, RET transcripts were profoundly increased in chickens with high AbFW compared to those in the low group. Even more striking was the 72-fold increased abundance of NPPB transcripts in birds with high AbFP. Natriuretic peptide B is implicated in a variety of actions [39,40], including an important role in obesity and insulin resistance [41]. SREBF1 encodes a transcription factor with roles in adipocyte differentiation and regulation of lipogenesis [42]. Expression of this gene was greatly diminished in the chickens with high AbFP and it is expressed at lower levels in adipose tissue from obese human subjects [43].
Expression of the other genes examined (COL12A1, VPS4B, BRSK2, and FOXC1) was significantly lower in chickens with high AbFW or AbFP, though the magnitude of the differential expression was less.
The authors are aware that these traits, measured here at d 93 for their relevance to chicken meat production, reflect cumulative cellular, developmental and metabolic processes, some of which are set in place at much earlier stages. The leads provided by the present GWAS, and general verification by the expression analyses comparing phenotypic extremes, now require a systematic ontogenic analysis from before hatching, where feasible. In the case of adipose tissue, which is non-discernible pre-hatch, attempts to functionally characterize the relevant genes may be possible with preadipocytes, differentiated in vitro.
The present approach has used GWAS and mRNA expression analysis to identify loci and genes influencing meat quality traits in chicken. Fine mapping of causal variants in these associated regions and more thorough functional characterization of these genes will be required, using systematic post-GWAS strategies [44,45].
Conclusions
In summary, the present GWAS has exposed a total of 33 SNPs having significant association with ten meat quality traits (DMBr, DMTh, IMFBr, AbFW, AbFP, meat color L* and b* values, and skin color L*, a* and b* values). Of the 17 genes near the SNPs associated with IMFBr, meat L*, b* values and AbF, 14 were differentially expressed in breast muscle or abdominal fat among subsets of chickens with lowest and highest phenotypic values. These results provide new insight into the molecular mechanisms underlying meat quality traits in chickens.
Methods
Ethics Statement
The study was conducted in accordance with the Guidelines for Experimental Animals established by the Ministry of Science and Technology (Beijing, China).
Experimental animals
The Chinese Academy of Agricultural Science (CAAS) chicken F2 resource population was used. The chickens were raised in stair-step cages under the same recommended environmental and nutritional conditions at the conservation farm of the Institute of Animal Sciences (IAS), CAAS. The population was derived from a cross between Beijing-You (BJY) chickens and Cobb broilers (CB, Cobb-Vantress, Inc.). BJY is a slow-growing Chinese indigenous breed, and CB is a commercial fast-growing broiler strain. Six BJY males were each mated to 12 CB females to generate the F1 generation from which six males and 20 females produced the F2 progeny. F1 males were mated to non-related females using artificial insemination. A total of 367 (184 male and 183 female) F2 chickens in five batches, hatched at two-week intervals, were used.
Phenotypic traits
At 56 days of age, blood was collected from the brachial vein of chickens by venipuncture using citrated syringes during a routine health inspection. At 93 days, chickens were weighed and killed by stunning and exsanguination, 12 h after feed was withheld. After the carcass composition traits were determined, meat quality traits were measured using methods previously described in detail [24,46,47]. The meat quality traits included subcutaneous fat thickness (SFT), AbFW, AbFP, DMBr, DMTh, IMFBr, IMFTh, pHu, DL, SF, and the color of muscle and skin, L*, a*, and b*. Samples from the breast muscle and abdominal fat tissues were snap-frozen in liquid nitrogen then held at −80°C until analysis of relative mRNA expression.
Genotyping and quality control
Genomic DNA (gDNA) was extracted from blood samples using the phenol-chloroform method. Genotyping was performed by DNALandMarks Inc., Saint-Jean-sur-Richelieur, PQ, Canada using Illumina 60 K Chicken SNP Beadchips. Thirty-nine samples were excluded due to sample call rate < 90%. A total of 15,051 SNPs were removed for failing to meet one or more of the following conditions: SNP call rate < 90%, minor allele frequency (MAF) < 3%, Hardy-Weinberg equilibrium (HWE) test p of < 10-6 and SNPs with no assigned chromosome or linkage group. After these quality control steps, 42,585 SNPs remained and were distributed among 28 chromosomes and one linkage group (LGE22). The average physical distance between two neighbouring SNPs was approximately 20.4 Kb (Additional file 2: Table S1).
Statistical analysis
The population structure was assessed by MDS analysis using PLINK 1.07 software [7,48]. Independent SNP markers were obtained on all autosomes using the indep-pairwise option, with a window size of 25 SNPs, a step of five SNPs, and an r2 threshold of 0.2. Pairwise identity-by-state (IBS) distances were calculated between all individuals using these independent SNP markers, and MDS components were acquired using the mds-plot option based on the IBS matrix. The relative kinship matrix was also constructed from these independent SNP markers.
The descriptive statistics of the traits were analysed using the MEANS procedure in SAS 8.0 software (SAS Institute Inc., Cary, NC, USA). Some traits deviated from normality, and Box-Cox or Johnson transformations were implemented with Minitab 15 (Minitab Inc., Quality Plaza, PA, USA).
The GWAS analysis for meat quality traits used the GLM and compressed MLM procedures [17] and was performed by Tassel 3.0 software [49] with 42,585 SNPs passing quality control. Both models were performed with the first MDS component as covariates, with batch and sex as fixed effects. In the compressed MLM, relative kinship matrix was a random effect. For AbFW, eviscerated weight (EW) was used as a covariate in both models. The statistical models were,
| (I) |
| (II) |
where, Yijklmn are phenotypic values, μi is the common mean, C1j is the effect of the first principal component, Sk is the effect of sex, Bl is the effect of batch of hatching (l = 1 - 5), Gm is the effect of the SNP, Kn is the random effect of the relative kinship matrix, which was constructed by matrix simple matching coefficients based on the independent SNPs, and this step was followed by compression [17], and eijklmn is the random residual.
The p-value thresholds of “LD adjusted” Bonferroni genome-wide significance were calculated based on the estimated number of effective markers and LD blocks [8,50]. The F2 population was estimated to have 16,760 effective SNPs (Additional file 3: Table S2), based on the “solid spine of LD” algorithm with a minimum D′ value of 0.8 calculated by Haploview [51]. The two significant threshold p-values were 5.96×10-5 (1/16,760) for suggestive significance and 2.98 × 10-6 (0.05/16,760) for genome-wide significance. A Manhattan plot of the p-value results from the GWAS was produced using R 2.13.2 software [52] with the “gap” package [53].
Quantitative measurements of the expression of candidate genes by Q-PCR
Candidate genes for IMFBr, meat color L* and b* in breast muscle, and AbFW and AbFP, exposed by the GWAS, were assessed in the relevant tissues using Q-PCR. Tissue samples from chickens at the extremes of the phenotypic rankings were assembled as High (n = 6) and Low (n = 6) groups (Table 5). Total RNA was isolated from breast muscle and abdominal fat tissue with the RNAsimple Total RNA kit (TIANGEN BIOTECH, Beijing, China). First-strand cDNA was synthesised from 2 μg total RNA using the Reverse Transcription Kit (Promega, Beijing, China). Power SYBR®Green PCR Master Mix (Applied Biosystems, USA) was used to analyse mRNA expression of the selected genes. Quantitative Real-Time PCR was performed with an ABI 7500 Real-time Detection System (Applied Biosystems, USA). The primers (Additional file 4: Table S3) were designed using Primer Premier 5.0 based on chicken sequences. The amplification was performed in a total volume of 20 μl containing 10 μl of 2 × PCR Master Mix, 100 ng cDNA, 0.5 μl of each primer (10 μmol), and 8.0 μl ddH2O. To ensure similar PCR efficiencies (close to 100 %) between the target genes and the reference gene (β-actin), the concentrations of primers and cDNA were optimized, if needed. The following PCR conditions were used: 95°C for 10 min, followed by 40 amplification cycles of 95°C for 15s, 60°C for 20 s and 72°C for 32s. To determine fold-changes in gene expression, the comparative CT method was used [54], calculated as 2-ΔΔCT. The results are expressed as the mean fold-change in gene expression from triplicate analyses, using the L group samples as the calibrator (assigned an expression level of 1 for each gene).
Table 5.
Phenotypic values of the traits in the Low and High groups
| Traits1 |
Groups2 |
|
|---|---|---|
| Low (n = 6) | High (n = 6) | |
| IMFBr (%) |
1.22 ± 0.31 |
3.95 ± 0.49** |
| AbFW (g) |
3.11 ± 1.32 |
70.48 ± 12.96** |
| AbFP (%) |
0.21 ± 0.09 |
2.95 ± 0.63** |
| Meat color L* |
51.53 ± 1.74 |
62.50 ± 1.14** |
| Meat color b* | 11.07 ± 0.98 | 21.23 ± 1.34** |
1IMFBr intramuscular fat content in breast muscle, AbFW abdominal fat weight, AbFP percentage of AbFW to eviscerated weight, L* lightness value, b* yellowness value, 2High group (n = 6) consisting of samples from chicken with the highest trait values and Low group (n = 6) with the lowest trait values; **p < 0.01.
Abbreviations
a*: Redness value; AbFP: AbFW as a percentage of eviscerated weight; AbFW: Abdominal fat weight; AGA: aspartylglucosaminidase; b*: Yellowness value; BCL2: B-cell lymphoma 2; BRSK2: BR serine/threonine kinase 2; BUB3: Benzimidazoles 3 homolog (yeast); CAAS: Chinese Academy of Agricultural Science; CLINT1: Clathrin interactor 1; COL1A2: Collagen type I alpha 2; COL12A1: Collagen type XII alpha 1; CV: Coefficient of variation; DL: Drip loss; DM: Dry matter content; DMBr: Dry matter content in breast muscle (% by weight); DMTh: Dry matter content in thigh muscle (% by weight); EW: Eviscerated weight; FAM105A: Family with sequence similarity 105 member A; FOXC1: Forkhead box C1; FTSJ3: FtsJ homolog 3 (E. coli); GGA: Chicken (Gallus gallus) chromosome; GDNA: Genomic DNA; GLM: General linear model; GPR10: G-protein coupled receptor 10; GWAS: Genome-wide association studies; HWE: Hardy-Weinberg equilibrium; IAS: Institute of Animal Sciences; IBS: Pairwise identity-by-state; IMFBr: Intramuscular fat content in breast muscle (% by weight); IMFTh: Intramuscular fat content in thigh muscle (% by weight); KIF2A: Kinesin heavy chain member 2A; Kb: Kilobase; KPNA2: Karyopherin alpha 2 (RAG cohort 1 importin alpha 1); L*: Lightness value; LD: Linkage disequilibrium; MAF: Minor allele frequency; MDS: Multidimensional Scaling; MGST1: Microsomal glutathione S-transferase 1; MLM: Mixed linear model; MXD4: Max-interacting transcriptional repressor MAD4; NPPB: Natriuretic peptide B; NTPCR: Nucleoside-triphosphatase cancer-related; PHu: The ultimate pH (at 24h) of breast muscle; PGD: Phosphogluconate dehydrogenase; PrRPR: Prolactin-releasing peptide receptor-like protein; PSMD12: Proteasome (prosome macropain) 26S subunit non-ATPase, 12; PTPN: Tyrosine-protein phosphatase non-receptor type 2; Q-PCR: Real-time quantitative PCR; QTLs: Quantitative trait loci; RET: RET proto-oncogene; SF: Shear force of the pectoral major muscle; SFT: Subcutaneous fat thickness; SNP: Single-nucleotide polymorphism s; SPINK5: Serine peptidase inhibitor Kazal type 5; SREBF1: Sterol regulatory element binding transcription factor 1; ST8SIA5: Alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5; TBC1D24: TBC1 domain family member 24; TYRO3: TYRO3 protein tyrosine kinase; VPS4B: Vacuolar protein sorting-associated protein 4B.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YS contributed to the slaughter experiment, genotyped SNPs, performed the Q-PCR experiment and wrote the manuscript. GZ participated in the design of the study, the slaughter experiment and data analysis. RL contributed to the design of the study, the slaughter experiment, data analysis and drafting of the manuscript. MZ participated in the design of the study and the slaughter experiment. YH, DW and LZ participated in the slaughter experiment and genotyped SNPs. PL participated in the feeding and management of chickens and slaughter experiment. JW supervised the study and participated in its design and coordination. All authors read and approved the final manuscript.
Supplementary Material
A Manhattan plot showing the association of all SNPs with meat quality traits from the compressed mixed linear model (MLM). SNPs are plotted on the x-axis according to their position on each chromosome against their association with these traits on the y-axis (shown as -log10 p-value). The dashed line indicates genome-wide association (p-value = 1.00 × 10-4), and the solid line indicates significance with a p-value threshold of 5.96 × 10-5.
Basic information for SNP markers on a physical map.
LD blocks in the F2 population.
Q-PCR primers used in this study.
Contributor Information
Yanfa Sun, Email: boysun2010@163.com.
Guiping Zhao, Email: zhgp0402@iascaas.net.cn.
Ranran Liu, Email: ranran112@gmail.com.
Maiqing Zheng, Email: wheatzheng@163.com.
Yaodong Hu, Email: yaodong.hu@yahoo.com.
Dan Wu, Email: wdcaas@yahoo.cn.
Lei Zhang, Email: aleiyishi@126.com.
Peng Li, Email: lipengnd@yahoo.com.cn.
Jie Wen, Email: jiewen@iascaas.net.cn.
Acknowledgements
The authors would like to acknowledge Dr. W. Bruce Currie (Emeritus Professor, Cornell University, USA) and Md. Shahjahan (Institute of Animal Sciences, Chinese Academy of Agricultural Sciences) for their assistance in preparing the manuscript. The research was supported by grants: the National High-tech R&D Program (Grant Number: 2013AA102501); the National Key Technology R&D Program (2011BAD28B03); the earmarked fund for modern agro-industry technology research system (CARS-42).
References
- Berri C, Wacrenier N, Millet N, Le Bihan-Duval E. Effect of selection for improved body composition on muscle and meat characteristics of broilers from experimental and commercial lines. Poult Sci. 2001;80(7):833–838. doi: 10.1093/ps/80.7.833. [DOI] [PubMed] [Google Scholar]
- Nadaf J, Gilbert H, Pitel F, Berri CM, Feve K, Beaumont C, Duclos MJ, Vignal A, Porter TE, Simon J. et al. Identification of QTL controlling meat quality traits in an F2 cross between two chicken lines selected for either low or high growth rate. BMC Genomics. 2007;8:155. doi: 10.1186/1471-2164-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abasht B, Pitel F, Lagarrigue S, Le Bihan-Duval E, Le Roy P, Demeure O, Vignoles F, Simon J, Cogburn L, Aggrey S. et al. Fatness QTL on chicken chromosome 5 and interaction with sex. Genet Sel Evol. 2006;38(3):297–311. doi: 10.1186/1297-9686-38-3-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicken QTLdb. http://www.animalgenome.org/cgi-bin/QTLdb/GG/index.
- Fan B, Du ZQ, Gorbach DM, Rothschild MF. Development and application of high-density SNP arrays in genomic studies of domestic animals. Asian Austral J Anim. 2010;23(7):833–847. doi: 10.5713/ajas.2010.r.03. [DOI] [Google Scholar]
- Groenen MAM, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RPMA, Vereijken A, Okimoto R, Muir WM, Cheng HH. The development and characterization of a 60 K SNP chip for chicken. BMC Genomics. 2011;12:274. doi: 10.1186/1471-2164-12-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu XR, Feng CG, Ma L, Song C, Wang YQ, Da Y, Li HF, Chen KW, Ye SH, Ge CR. et al. Genome-wide association study of body weight in chicken F2 resource population. PLoS One. 2011;6(7):e21872. doi: 10.1371/journal.pone.0021872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Luo CL, Zhang CG, Zhang R, Tang J, Nie QH, Ma L, Hu XX, Li N, Da Y. et al. Genome-wide association study identified a narrow chromosome 1 region associated with chicken growth traits. PLoS One. 2012;7(2):e30910. doi: 10.1371/journal.pone.0030910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WB, Li DF, Liu JF, Chen SR, Qu LJ, Zheng JX, Xu GY, Yang N. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. PLoS One. 2011;6(12):e28600. doi: 10.1371/journal.pone.0028600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fife MS, Howell JS, Salmon N, Hocking PM, van Diemen PM, Jones MA, Stevens MP, Kaiser P. Genome-wide SNP analysis identifies major QTL for Salmonella colonization in the chicken. Anim Genet. 2011;42(2):134–140. doi: 10.1111/j.1365-2052.2010.02090.x. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovski CS, Stram DO. A kinship-based modification of the armitage trend test to address hidden population structure and small differential genotyping errors. PLoS One. 2009;4(6):e5825. doi: 10.1371/journal.pone.0005825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen WW, Araus JL, Shah T, Cairns J, Mahuku G, Banziger M, Torres JL, Sanchez C, Yan JB. Molecular characterization of a diverse maize inbred line collection and its potential utilization for stress tolerance improvement. Crop Sci. 2011;51(6):2569–2581. doi: 10.2135/cropsci2010.08.0465. [DOI] [Google Scholar]
- Cappa EP, Martínez MC, Garcia MN, Villalba PV, Poltri SNM. Effect of population structure and kinship relationships on the results of association mapping tests of growth and wood quality traits in four Eucalyptus populations. BMC Proc. 2011;5(Suppl 7):23. [Google Scholar]
- Huang XH, Wei XH, Sang T, Zhao QA, Feng Q, Zhao Y, Li CY, Zhu CR, Lu TT, Zhang ZW. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM. et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42(4):355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Cheng D, Chen S, Wang L, Li Y, Ma X, Song X, Liu X, Li W, Liang J. Genome-wide association analysis of meat quality traits in a porcine large white × minzhu intercross population. Int J Biol Sci. 2012;8(4):580. doi: 10.7150/ijbs.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolormaa S, Neto LP, Zhang Y, Bunch R, Harrison B, Goddard M, Barendse W. A genome-wide association study of meat and carcass traits in Australian cattle. J Anim Sci. 2011;89(8):2297–2309. doi: 10.2527/jas.2010-3138. [DOI] [PubMed] [Google Scholar]
- Nones K, Ledur MC, Zanella EL, Klein C, Pinto LF, Moura AS, Ruy DC, Baron EE, Ambo M, Campos RL. et al. Quantitative trait loci associated with chemical composition of the chicken carcass. Anim Genet. 2012;43(5):570–576. doi: 10.1111/j.1365-2052.2012.02321.x. [DOI] [PubMed] [Google Scholar]
- Fishman PH, Brady RO. Biosynthesis and function of gangliosides. Science. 1976;194(4268):906–915. doi: 10.1126/science.185697. [DOI] [PubMed] [Google Scholar]
- Mannherz O, Mertens D, Hahn M, Lichter P. Functional screening for proapoptotic genes by reverse transfection cell array technology. Genomics. 2006;87(5):665–672. doi: 10.1016/j.ygeno.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Serva A, Knapp B, Tsai Y-T, Claas C, Lisauskas T, Matula P, Harder N, Kaderali L, Rohr K, Erfle H. miR-17-5p Regulates Endocytic Trafficking through Targeting TBC1D2/Armus. PLoS One. 2012;7(12):e52555. doi: 10.1371/journal.pone.0052555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WJ, Zhao GP, Chen JL, Zheng MQ, Wen J. Influence of dietary vitamin E supplementation on meat quality traits and gene expression related to lipid metabolism in the Beijing-you chicken. Brit Poul Sci. 2009;50(2):188–198. doi: 10.1080/00071660902755409. [DOI] [PubMed] [Google Scholar]
- Rizkalla SW, Prifti E, Cotillard A, Pelloux V, Rouault C, Allouche R, Laromiguiere M, Kong L, Darakhshan F, Massiera F. et al. Differential effects of macronutrient content in 2 energy-restricted diets on cardiovascular risk factors and adipose tissue cell size in moderately obese individuals: a randomized controlled trial. Am J Clin Nutr. 2012;95(1):49–63. doi: 10.3945/ajcn.111.017277. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang YH, Jiang H, Xiao SQ, Wang S, Ma Q, Sun GJ, Li FJ, Deng Q, Dai LS. et al. Detection of differentially expressed genes in the longissimus dorsi of Northeastern Indigenous and Large White pigs. Genet Mol Res. 2011;10(2):779–791. doi: 10.4238/vol10-2gmr1170. [DOI] [PubMed] [Google Scholar]
- Jennen DG, Vereijken AL, Bovenhuis H, Crooijmans RM, van der Poel JJ, Groenen MA. Confirmation of quantitative trait loci affecting fatness in chickens. Genet Sel Evol. 2005;37(2):215–228. doi: 10.1186/1297-9686-37-3-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma N, Takei Y, Tanaka Y, Nakata T, Terada S, Kikkawa M, Noda Y, Hirokawa N. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 2003;114(2):229–239. doi: 10.1016/S0092-8674(03)00522-1. [DOI] [PubMed] [Google Scholar]
- Placzek WJ, Almeida MS, Wüthrich K. NMR structure and functional characterization of a human cancer-related nucleoside triphosphatase. J Mol Biol. 2007;367(3):788–801. doi: 10.1016/j.jmb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Qiao M, Fletcher DL, Smith DP, Northcutt JK. The effect of broiler breast meat color on pH, moisture, water-holding capacity, and emulsification capacity. Poult Sci. 2001;80(5):676–680. doi: 10.1093/ps/80.5.676. [DOI] [PubMed] [Google Scholar]
- Le Bihan-Duval E, Nadaf J, Berri C, Pitel F, Graulet B, Godet E, Leroux SY, Demeure O, Lagarrigue S, Duby C. et al. Detection of a Cis eQTL controlling BMCO1 gene expression leads to the identification of a QTG for chicken breast meat color. PLoS One. 2011;6(7):e14825. doi: 10.1371/journal.pone.0014825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu ZY, Li HY, Xiong YZ, Zuo B. Differential transcriptional analysis between red and white skeletal muscle of Chinese Meishan pigs. Int J Biol Sci. 2010;6(4):350–360. doi: 10.7150/ijbs.6.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-Y, Byun M-J, Kim H. A replication study of genome-wide CNV association for hepatic biomarkers identifies nine genes associated with liver function. Biochem & Mol Biol Reports. 2011;44(9):578–583. doi: 10.5483/bmbrep.2011.44.9.578. [DOI] [PubMed] [Google Scholar]
- Estrada-Bernal A, Sanford SD, Sosa LJ, Simon GC, Hansen KC, Pfenninger KH. Functional complexity of the axonal growth cone: a proteomic analysis. PLoS One. 2012;7(2):e31858. doi: 10.1371/journal.pone.0031858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson J, Larson G, Gunnarsson U, Bed'hom B, Tixier-Boichard M, Stromstedt L, Wright D, Jungerius A, Vereijken A, Randi E. et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4(2):e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descargues P, Deraison C, Bonnart C, Kreft M, Kishibe M, Ishida-Yamamoto A, Elias P, Barrandon Y, Zambruno G, Sonnenberg A. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat Genet. 2004;37(1):56–65. doi: 10.1038/ng1493. [DOI] [PubMed] [Google Scholar]
- Chen JL, Zhao GP, Zheng MQ, Wen J, Yang N. Estimation of genetic parameters for contents of intramuscular fat and inosine-5 '-monophosphate and carcass traits in Chinese Beijing-You chickens. Poult Sci. 2008;87(6):1098–1104. doi: 10.3382/ps.2007-00504. [DOI] [PubMed] [Google Scholar]
- Sohle J, Machuy N, Smailbegovic E, Holtzmann U, Gronniger E, Wenck H, Stab F, Winnefeld M. Identification of new genes involved in human adipogenesis and fat storage. PLoS One. 2012;7(2):e31193. doi: 10.1371/journal.pone.0031193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell NH, Tang G, Yu C, Mercurio F, DiDonato JA, Lin A. Activation of NF-kappa B is required for hypertrophic growth of primary rat neonatal ventricular cardiomyocytes. Proc Natl Acad Sci USA. 2001;98(12):6668–6673. doi: 10.1073/pnas.111155798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Cheh C, Larson MG, Vasan RS, Levy D, Bloch KD, Surti A, Guiducci C, Kathiresan S, Benjamin EJ, Struck J. et al. Association of common variants in NPPA and NPPB with circulating natriuretic peptides and blood pressure. Nat Genet. 2009;41(3):348–353. doi: 10.1038/ng.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AM, Cheng S, Magnusson M, Larson MG, Newton-Cheh C, McCabe EL, Coviello AD, Florez JC, Fox CS, Levy D. et al. Cardiac natriuretic peptides, obesity, and insulin resistance: evidence from two community-based studies. J Clin Endocrinol Metab. 2011;96(10):3242–3249. doi: 10.1210/jc.2011-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Xu Z, Han X, Wang F, Wang L. The pattern of development for gene expression of sterol regulatory element binding transcription factor 1 in pigs. Czech J Anim Sci. 2006;51(6):248–252. [Google Scholar]
- Oberkofler H, Fukushima N, Esterbauer H, Krempler F, Patsch W. Sterol regulatory element binding proteins: relationship of adipose tissue gene expression with obesity in humans. Biochim Biophys Acta. 2002;1575(1–3):75–81. doi: 10.1016/s0167-4781(02)00279-8. [DOI] [PubMed] [Google Scholar]
- Freedman ML, Monteiro AN, Gayther SA, Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C, Duggan D. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43(6):513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV, Friesen ML, McIntyre LM. Genotype–phenotype mapping in a post-GWAS world. Trends Genet. 2012;28(9):421–426. doi: 10.1016/j.tig.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GP, Chen JL, Zheng MQ, Wen J, Zhang Y. Correlated responses to selection for increased intramuscular fat in a Chinese quality chicken Line. Poult Sci. 2007;86(11):2309–2314. doi: 10.1093/ps/86.11.2309. [DOI] [PubMed] [Google Scholar]
- Cui HX, Zheng MQ, Liu RR, Zhao GP, Chen JL, Wen J. Liver dominant expression of fatty acid synthase (FAS) gene in two chicken breeds during intramuscular-fat development. Mol Biol Rep. 2012;39(4):3479–3484. doi: 10.1007/s11033-011-1120-8. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics. 2008;9:516. doi: 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- R: A Language and Environment for Statistical Computing. http://www.r-project.org/
- Zhao JH. gap: Genetic analysis package. J Stat Softw. 2007;23(8):1–18. [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A Manhattan plot showing the association of all SNPs with meat quality traits from the compressed mixed linear model (MLM). SNPs are plotted on the x-axis according to their position on each chromosome against their association with these traits on the y-axis (shown as -log10 p-value). The dashed line indicates genome-wide association (p-value = 1.00 × 10-4), and the solid line indicates significance with a p-value threshold of 5.96 × 10-5.
Basic information for SNP markers on a physical map.
LD blocks in the F2 population.
Q-PCR primers used in this study.


