Abstract
The genomes of many cyanobacteria contain the sequence for a small protein with a common “Domain of Unknown Function” grouped into the DUF683 protein family. While the biological function of DUF683 is still not known, their genomic location within nitrogen fixation clusters suggests that DUF683 proteins may play a role in the process. The diurnal cyanobacterium Cyanothece sp. PCC 51142 contains a gene for a protein that falls into the DUF683 family, cce_0567 (78 aa, 9.0 kDa). In an effort to elucidate the biochemical role DUF683 proteins may play in nitrogen fixation, we have determined the first crystal structure for a protein in this family, cce_0567, to 1.84 Å resolution. Cce_0567 crystallized in space group P21 with two protein molecules and one Ni2+ cation per asymmetric unit. The protein is composed of two α-helices, residues P11 to G41 (α1) and L49–E74 (α2), with the second α-helix containing a short 310-helix (Y46–N48). A four-residue linker (L42–D45) between the helices allows them to form an anti-parallel bundle and cross over each other towards their termini. In solution it is likely that two molecules of cce_0567 form a rod-like dimer by the stacking interactions of ~1/2 of the protein. Histidine-36 is highly conserved in all known DUF683 proteins and the N2 nitrogen of the H36 side chain of each molecule in the dimer is coordinated with Ni2+ in the crystal structure. The divalent cation Ni2+ was titrated into 15N-labeled cce_0567 and chemical shift perturbations were observed only in the 1H–15N HSQC spectra for residues at, or near, the site of Ni2+ binding observed in the crystal structure. There was no evidence for an increase in the size of cce_0567 upon binding Ni2+, even in large molar excess of Ni2+, indicating that a metal was not required for dimer formation. Circular dichroism spectroscopy indicated that cce_0567 was extremely robust, with a melting temperature of ~62 °C that was reversible.
Keywords: Metalloprotein, Nitrogen fixation, Cyanobacteria, Circadian rhythm, Nickel-binding
1. Introduction
Cyanobacteria, the progenitors of chloroplasts in plants and algae [1, 2], radically altered the chemistry of our planet starting about 2.3 billion years ago through the release of a photosynthesis byproduct, oxygen [3, 4]. Some cyanobacteria can also still fix nitrogen [5] with nitrogenase, a multimeric enzyme complex [6]. One of the byproducts of the process, hydrogen, is currently of much interest as a clean source of energy [7]. However, nitrogen fixation is biochemically intolerant of the presence of oxygen because nitrogenase is irreversibly inhibited by molecular oxygen [8–10]. To perform the two mutually exclusive functions, photosynthesis and nitrogen fixation, some cyanobacteria have temporally separated them into daytime and nighttime activities, respectively. Indeed, cyanobacteria are the simplest known organisms that display circadian rhythms [11–13]. To better understand the details of how cyanobacteria control the diurnal processes, the genome of the marine diazotrophic cyanobacterium, Cyanothece ATCC 51142, was recently sequenced and deposited into the GenBank database (accession nos. CP000806–CP000811) [14]. Present is a gene, cce_0567, that falls into a family of proteins with a similar “Domain of Unknown Function”, DUF683, that is conserved among various prokaryotic species. Excluding Cyanothece, 54 DUF683 proteins are currently found in GenBank with similarity searches and the conserved domain is also listed 39 times in the Pfam database (PF05082) [15]. Genes that fall into the DUF683 family are typically under 100 amino acid residues in size, are always observed in the nitrogen fixation gene cluster of the genome, and appear to be most prevalent in cyanobacteria. In Cyanothece, the cce_0567 gene is observed between NifW, a gene that codes for a protein associated with nitrogen fixation whose precise function is uncertain, and cce_0566, a gene that falls into the DUF269 family of genes found exclusively in nitrogen fixation operons. The appearance of the cce_0567 gene in nitrogen fixation operons suggests that it may play a role in the process, a role that is still undefined. To obtain clues as to the biochemical function of proteins in the DUF683 family of conserved proteins [16, 17] we have determined the first crystal structure of a protein in the DUF683 family, cce_0567, to a resolution of 1.8 Å. Because the solved protein structure contained a nickel cation, the metal binding properties of cce_0567 towards the divalent cations Ni2+, Zn2+, and Co2+ were assessed by NMR spectroscopy using previously determined backbone NMR assignments [18]. The thermal stability of cce_0567 was assessed by circular dichroism spectroscopy and verified by NMR spectroscopy. On the basis of our crystal structure for cce_0567, a recently determined NMR structure of another member of the DUF683 protein family (Q60C73_METCA from Methylococcus capsulatus (2JS5)), and sequence homology of all proteins in the DUF683 family, the biochemically significant regions and potential biological role of proteins in the DUF683 family are speculated upon.
2. Materials and methods
2.1. Cloning, expression, and purification
The cloning, expression, and purification protocol for 15N-labeled DUF683 has previously been reported [18]. The procedure for preparing selenomethionine-substituted DUF683 was similar using a protocol that inhibited the methionine biosynthesis pathway [19,20]. Basically, after growing the cells at 310 K to mid-log phase (OD600nm~0.8) in M9 minimal medium supplemented with 34 µg/mL kanamycin (RPI Corporation, Prospect, IL),120 µg/mL MgSO4, 11 µg/mL CaCl2, 10 ng/mL Fe2Cl3, 50 µg/mL NaCl, and 4 mg/mL glucose, the temperature was lowered to 298 K. At this point lysine (0.1 mg/mL), phenylalanine (0.1 mg/mL), threonine (0.1 mg/mL), isoleucine (0.05 mg/mL), valine (0.05 mg/mL), and selenomethionine (SeMet) (Acrös Organics, Geel, Belgium) (0.06 mg/mL) were added followed by the induction of protein expression ~15 min later with isopropyl β-d-1-thiogalactopyranoside (0.026 mg/mL). Approximately 6 h later the cells were harvested by mild centrifugation and then frozen at 193 K. From this point forward the protocol was identical to the previously described method with the protein exchanged into the buffer (500 mM NaCl, 20 mM Tris, 1.0 mM dithiothreitol, pH 7.2) used for the crystallization, NMR, and circular dichroism experiments. Although NMR spectroscopy and other evidence indicated that cce_0567 was primarily in the metal-free form at this stage (>90%), protein was treated with 2 mM EDTA and then dialyzed extensively to remove the EDTA for metal titration studies.
2.2. Nuclear magnetic resonance spectroscopy
One microliter aliquots of divalent metal cation were titrated into a 250 µL NMR sample of 15N-labeled cce_0567 (~0.4 mM) using 0.1 M stock solution of NiCl2, CoCl2, and ZnCl2 (Hampton Reseach, Aliso Viejo, CA). Following mild agitation, two-dimensional 1H–15N HSQC spectra were collected on a Varian Inova-750 spectrometer equipped with a triple resonance probe and pulse field gradients. Spectra were recorded after every addition at a metal:protein molar ratio of 0.5, 1.0, 2.0, and 4.0 to 1.0. To test the efficiency of EDTA in removing the metal from the protein, a solution of cce_0567 and Ni2+ (2:1 metal:protein molar ratio) was treated with excess EDTA and an 1H–15N HSQC spectrum collected. To assay the effect of heating, a 0.4 mM sample of 15N-labeled cce_0567 (metal-free) was heated to 80 °C for 30 min and an 1H–15N HSQC spectrum collected after cooling back to room temperature. All NMR data was processed using Felix2007 (Felix NMR, Inc., San Diego, CA) software with 1H and 15N chemical shifts referenced to DSS (DSS = 0 ppm) using indirect methods [21]. The assigned 1H, 13C, and 15N chemical shifts for cce_0567 have been deposited in the BioMagResBank database (www.bmrb.wisc.edu) under the accession number BMRB-15539 [18].
2.3. Circular dichroism spectroscopy
Circular dichroism data were obtained on an Aviv Model 62DS spectropolarimeter calibrated with an aqueous solution of ammonium d-(+)camphorsulfonate. Measurements on cce_0567were made in the same buffer used for the NMR studies (500 mM NaCl, 20 mM TrisHCl, 1 mM DTT, pH 7.2). A thermal denaturation curve for cce_0567 was obtained on a 30 µM solution in a quartz cell of 0.1 cm path length by recorded the ellipticity at 220 nm in 2.5 °C intervals from 10 to 80 °C. Using the same sample, a far-UV wavelength spectrum between 200 and 250 nm was recorded at 25, 80, and again at 25 °C. Spectra were the result of averaging three consecutive scans with a bandwidth of 1.0 nm and a time constant of 1.0 s. The wavelength spectra were processed by subtracting a blank spectrum, correcting the base-line, and then noise reduction.
2.4. Crystallization, structure determination and refinement
Vapor-diffusion crystallization trials using hanging drops were set up on SeMet-labeled cce_0567 at room temperature (~295 K) using screens from Hampton Research (Aliso Viejo, CA). Crystals began appearing 24–48 h later under one condition that was then optimized by adjusting the protein concentration. Crystals were harvested 3–4 days after mixing 2 µL of protein (~2 mg/mL) with 2 µL of reservoir buffer containing 30% (w/v) PEG 1500. Protein crystals of similar morphology were also grown under the same conditions with precipitant containing 1 mM NiCl2 (Hampton Research). Crystals were directly mounted in nylon CryoLoops (Hampton Research), flash-frozen in liquid nitrogen, stored under liquid nitrogen, and shipped to the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory for X-ray data collection.
2.5. X-ray data collection and processing
All X-ray diffraction data were collected at the X29A beamline with an ADSC Q315 CCD detector. Data was initially collected on crystals grown in the absence of 1.0 mM NiCl2 in the precipitant solution which contained 30% (w/v) PEG 1500 that served well as a cryoprotectant when cryocooling the crystals in a nitrogen stream (100 K) during data collection. A three-wavelength SAD data set was collected on these first crystals to a resolution of 2.6 Å, however, it was not possible to generate an interpretable electron density map from the data suggesting that perhaps there was too much disorder in the crystal lattice. In an attempt to remove this disorder, crystallization screens were repeated in the same precipitant in the presence of 12 different metal additives (Hampton Research, Aliso Viejo, CA). In the presence of these additives, crystals were only obtained in the presence of 1 mM NiCl2 and they indexed to the same space group (P21) as the original crystals. Such crystals diffracted to a resolution of 1.84 Å and yielded excellent electron density maps. X-ray fluorescence spectroscopy on the single crystals grown in the presence and absence of NiCl2 conclusively identified the presence of nickel in the latter, but not in the former, crystals.
A three-wavelength MAD data set at 1.84 Å resolution was obtained on the crystals grown in the presence of NiCl2. The images were integrated and scaled with HKL2000 [22]. Selenium and nickel heavy atom sites were determined using the SHELX program suite [23–25] and HKL2MAP [26]. The peak wavelength data of the MAD data set, which contained useful anomalous signal out to 1.84 Å resolution, was used together with the program SOLVE [27] to produce an electron density map at 1.84 Å resolution. The majority of the structure was built automatically into the resolution map using RESOLVE [27–29] with the remainder built manually using coot [30] and refined with refmac5 [31] to 2.3 Å resolution. A final check on the stereochemical quality of the final model was assessed using the program MolProbity [32] and PROCHECK [16] and any conflicts addressed. MolProbity analysis indicated that the overall geometry of the final model ranked in the 45th percentile (MolProbity score of 2.22) where the 100th percentile is best among structures of comparable resolution. The clash score for “all-atoms” was 15.12 corresponding to a 45th percentile ranking for structures of comparable resolution. PROCHECK analysis showed that 98% of the psi/phi pairs were in most favored region and the remainder in additionally allowed regions. Overall, MolProbity and PROCHECK assessment indicated that the final model was a quality representation of the structure of cce_0567. The data collection and structure refinement statistics are given in Table 1 and the coordinates have been submitted to the Protein Data Bank (PDB ID 3CSX).
Table 1.
Summary of the diffraction data collection and refinement statistics for cce_05671
| X-ray source | X29 |
|---|---|
| Detector | ADSC Q315 CCD |
| X-ray wavelength (Å) | 0.9791 |
| Temperature (K) | 100 |
| Data set | SAD |
| Space group | P21 (P 1 21 1) |
| Unit-cell parameters | |
| a (Å) | 29.44 |
| b (Å) | 38.38 |
| c (Å) | 62.64 |
| α = γ | 90° |
| β | 96.70° |
| Resolution range (Å) | 30–1.84 (1.92–1.84) |
| Molecules per AU | 2 |
| Total number of reflections | 269,646 |
| Total number of unique reflections | 11,230 (625) |
| Completeness (%) | 92 (52) |
| Rmerge (%)a | 6.9 (26.1) |
| Mean I/σ(I) | 36.9 (2.6) |
| Wilson B (Å2) | 25.12 |
| Refinement | |
| R (%) | 21.6 |
| Rfree (%) | 29.4 |
| Mean B-value (Å2) | 28.14 |
| Δbonds (Å) | 0.015 |
| Δangles (°) | 1.50 |
| MolProbity Ramachandran analysis | |
| Most favored | 98% |
| Additionally allowed | 2% |
| MolProbity | |
| Clash score, all atoms | 15.1 (46th) |
| MolProbity score | 2.22 (45th) |
Values in parenthesis are the statistics for the highest resolution shell (1.92–1.84 Å).
Rmerge = ∑h∑j|Ih,j−<Ih>|/∑h∑j, where Ih,j is the intensity of the jth observation of unique reflection h.
3. Results and discussion
3.1. Crystal structure of cce_0567
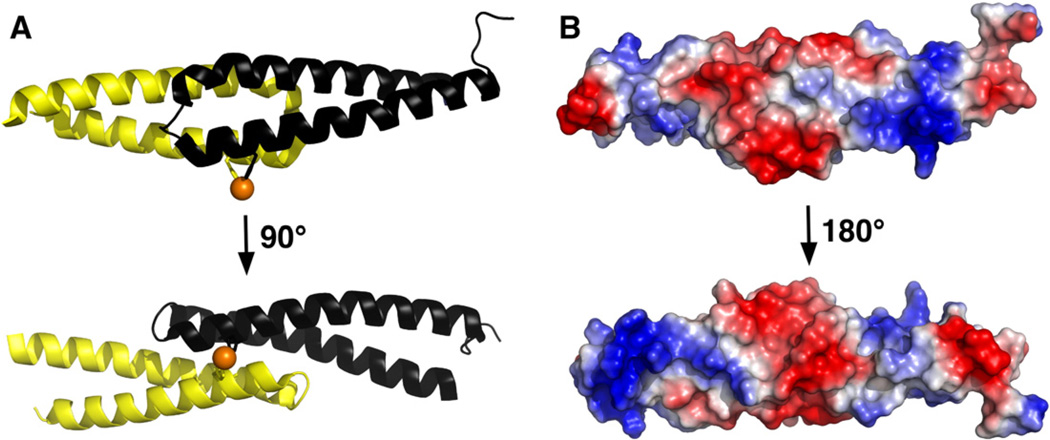
The asymmetric unit of the crystal contains two molecules of cce_0567 plus one nickel ion. Interpretable electron density was absent at the termini of both molecules suggesting the ends of the protein were disordered. Ten fewer residues could be mapped in molecule A (A15–E75)—then in molecule B (T5–E75), and therefore, the latter molecule will be discussed when referring to a single cce_0567 molecule. Aside from more missing electron density in one molecule, the structures of both molecules in the asymmetric unit are similar with a backbone RSMD of 0.96 Å and an all atoms RMSD of 1.79 Å (A15–E75) as determined using Superpose [33]. Indeed, the structure of both molecules are similar enough that previous NMR data, while suggesting cce_0567 was oligomeric in solution, generated only one set of amide cross peaks in the 1H–15N HSQC spectrum indicating that each subunit was in a magnetically similar environment [18]. Each individual cce_0567 molecule is composed of two α-helices, residues P11 to G41 (α1) and L49–E74 (α2), with the second α-helix containing a short 310-helix (Y46–N48). Four residues (L42–D45) connect the two helices and allow them to form an anti-parallel bundle. Fig. 1A shows that the two-helix bundle adopts an “X”-type shape as they cross over towards their termini.
Fig. 1.
(A) Two orientations of the likely cce_0567 dimer in solution that differ by ~90° about the horizontal axis. Each subunit is colored separately and the side chains of the H36 residue from each protein and the nickel (orange) highlighted. (B) The solvent accessible electrostatic surface for the cce_0567 dimer shown in two orientations. The upper orientation is similar to the orientation shown on the top of (A) with the lower orientation differing by ~180° about the horizontal axis.
Previous NMR data suggested that cce_0567 was oligomeric [18]. However, in the asymmetric unit of the crystal there clearly was little intermolecular contact between the two molecules as reflected in a calculated buried surface area of only 860 Å2 between them. A more likely interaction between two molecules of cce_0567 in solution is as shown in Fig. 1A. Such a dimer was observed upon expanding the symmetry related molecules of the asymmetric unit. The buried surface area due to the formation of such a dimer is 1860 Å2, a value larger than the value observed due to dimer formation in the asymmetric unit. Strong evidence in support of such a dimer is the recently determined solution structure for another protein that falls into the DUF683 family, Q60C73_METCA, from the methane-oxidizing bacterium M. capsulatus (PDB ID 2JS5). The crystal structure of the cce_0567 dimer shown in Fig. 1A (chain B, T10–A75) superimposes over the solution structure of the Q60C73_METCA dimer (model 1, chain B, M1–H66) with a backbone RMSD of 1.88 Å and an all-atoms RMSD of 2.32 Å (as determined using Superpose).
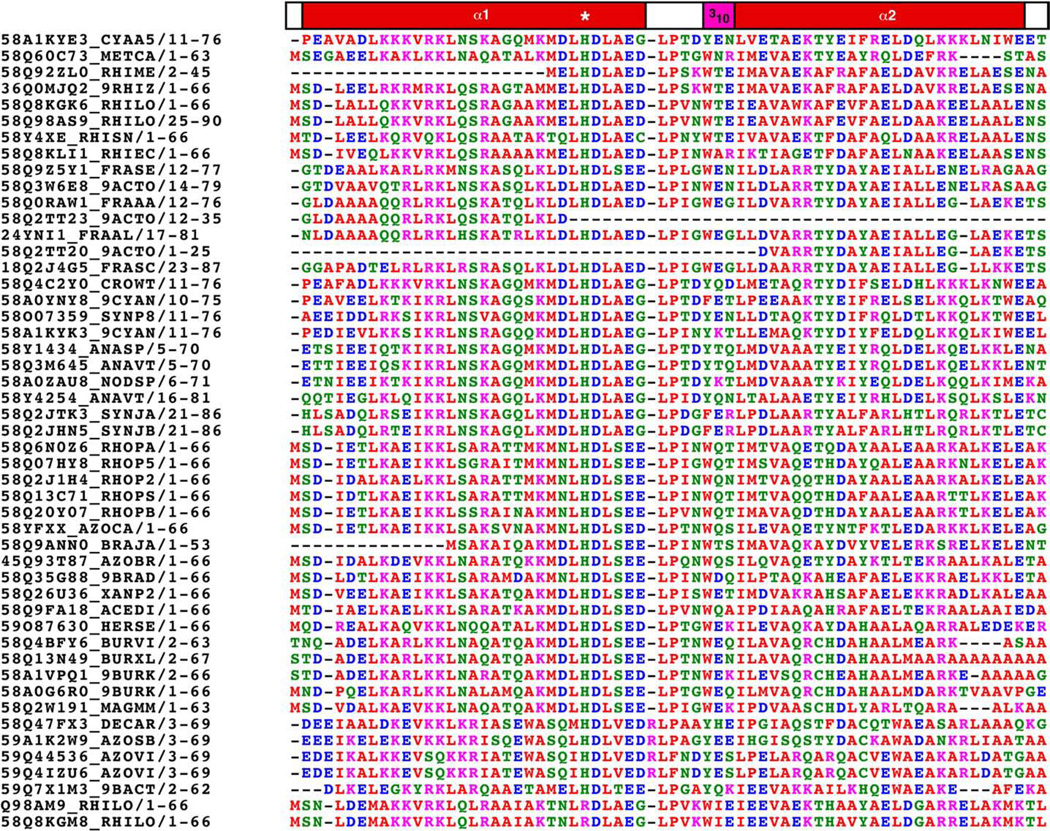
Fig. 1B shows an estimation of the electrostatic surface potential of the protein. There appears to be defined regions of alternating positive and negative charge. The biological importance of the electrostatic surface potential is unknown. However, sequence alignment of all 48 currently identified proteins in the DUF683 family using ClustalW2 [34], shown in Fig. 2, indicates that the charged residues are highly conserved at least in the N-terminal α-helix (α1). Such conservation of charged residues suggests that they may have a biological property.
Fig. 2.
ClustalW2 sequence alignment and color coding of cce_0567, Q60C73_METCA, and 46 other proteins listed by Pmaf in the DUF683 protein family (PF05082). Red = small and hydrodrophobic but not aromatic (A, V, F, P, M, I, L, W), blue = acidic (D,E); magenta = basic (R, K); green = hydrophilic (S, T, Y, H, C, N, G, Q).
3.2. Metal binding site
Crystals of SeMet-labeled cce_0567 that diffracted to 2.6 Å were original grown in the absence of divalent cation additives. However, it was not possible to generate an interpretable electron density map due to excessive disorder in the crystals. On the other hand, crystals of SeMet-labeled cce_0567 grown in the presence of NiCl2 diffracted to 1.84 Å resolution and these crystals generated excellent electron density maps. Nickel was unambiguously identified in the crystals grown in the presence of NiCl2 by X-ray fluorescence spectroscopy (data not shown) and the electron density observed for the metal ion in the electron density maps corroborated well for a Ni2+ ion. There was no evidence for nickel by X-ray fluorescence spectroscopy in the original crystals grown in the absence of NiCl2. Evidently, the nickel cation was necessary to provide additional order to the crystal lattice.
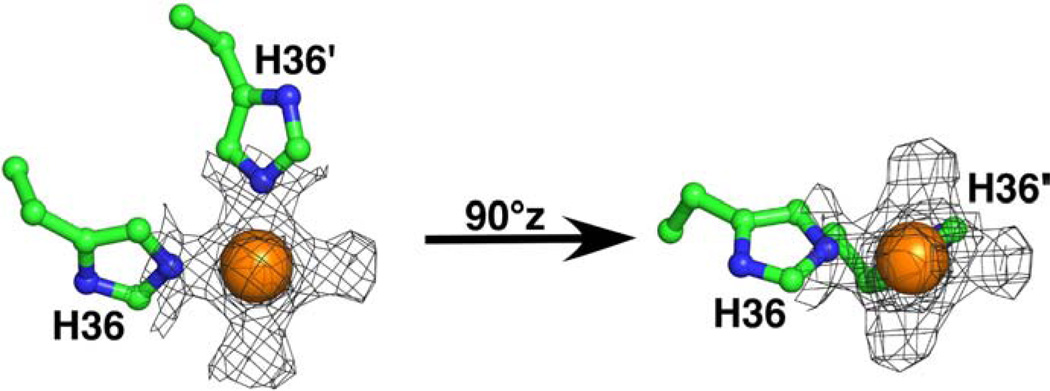
Fig. 3 illustrates the electron density at 1.5 sigma about the metal center and highlights the metal coordination to the protein side chains and water. The nickel is ligated to the N2 nitrogen atom of H36 from two cce_0567 molecules with a typical, average Ni–N bond length of 2.09±0.02 Å [35]. In addition to coordinating two nitrogen atoms, four additional “bulges” of electron density are observed around the nickel to form an overall pseudooctahedral geometry [35]. Two of the “bulges” are approximately in the plane of the two Ni–N bonds with the other two above and below this plane. Nickel (II) can exist in either the paramagnetic high-spin octahedral electronic configuration or the diamagnetic low-spin form. The latter form can adopt either a five-coordinate trigonal bipyramidal geometry or a four-coordinate square-planar geometry [36]. Consequently, the nickel appears to be in the paramagnetic high-spin state in the cce_0567 crystal structure, a configuration that differs from its low-spin state in solution as determined by NMR spectroscopy (to be discussed).
Fig. 3.
The electron density surrounding the Ni2+ ion in the crystal structure of cce_0567 illustrated at 1.5 σ. The N2 ring nitrogen from the histidine-36 side chain of two molecules coordinate the Ni2+ with water occupying the other sites around the metal. In the diagram on the right, H36′ has been rotated ~90° about the z-axis into the plane of the paper.
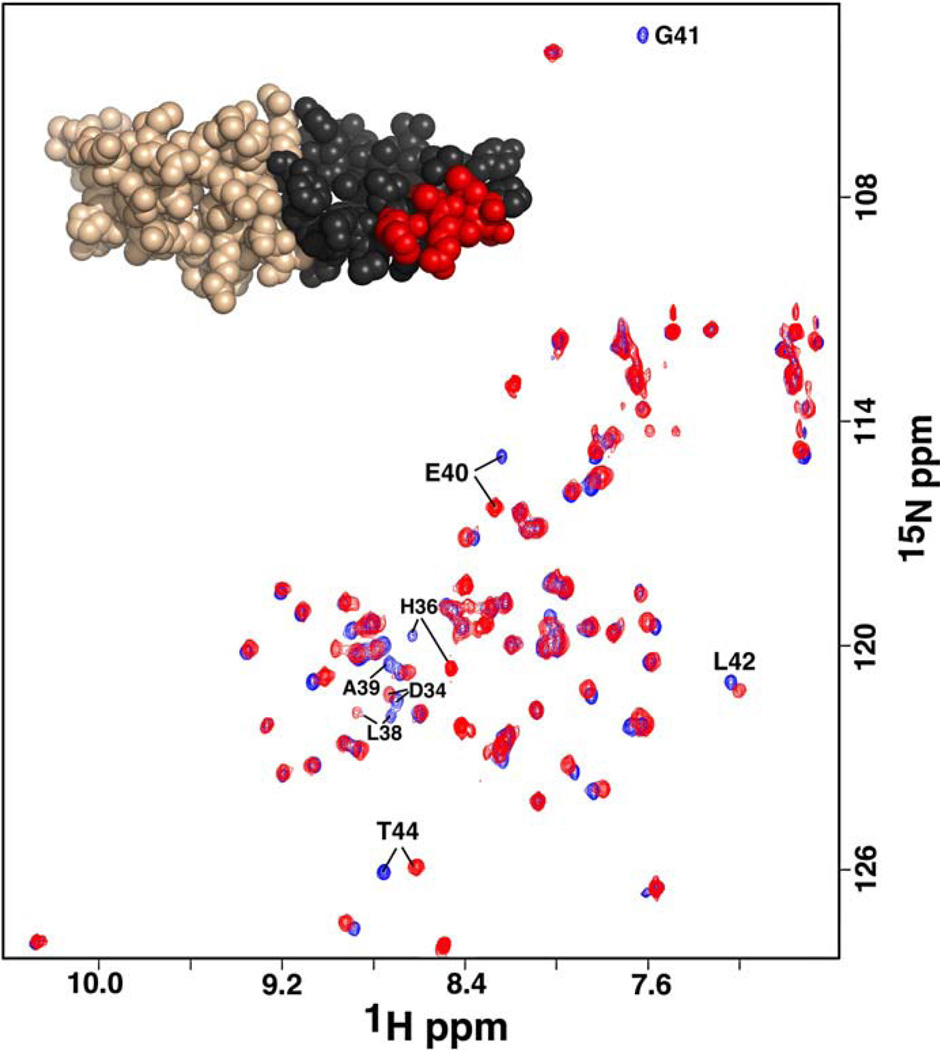
While nickel was necessary to solve the crystal structure of cce_0567 and is held in place by a histidine residue from each of the two molecules in the dimer, NMR studies indicate that nickel coordination is not necessary for dimer formation. Fig. 4 is an 1H–15N HSQC spectrum of cce_0567 obtained in the absence and presence of nickel. As illustrated, only a small subset of resonances in the 1H–15N HSQC spectrum are perturbed in the presence of nickel and these are at, or near, the nickel binding site or the turn between the two helices. Nickel associates with cce_0567 relatively tightly with a Kd of 10−6 M or less. Such a value is inferred from the observation of two subsets of cross peaks for residues about the metal-binding site in the presence of less than a 1:1 protein-to-metal molar ratios (data not shown) indicating that the equilibrium between the metal-free and metal-bound states are in slow exchange [37]. If nickel binding was responsible for dimer formation one would expect a larger subset of resonances to be perturbed in the 1H–15N HSQC spectrum with the addition of nickel to the solution. However, instead of chemical shift perturbations to residues through-out the dimer interface, colored black for one cce_0567 monomer in the inset of Fig. 4, perturbations are only observed for residues in a small part of α1, near the nickel binding site, and part of the linker region (colored red in the Fig. 4 inset). Furthermore, dimer formation would dictate a doubling of the protein's molecular weight. Such an event would manifest an increase in the visible line shape of the cross peaks in the 1H–15N HSQC spectrum (they would get broader with increasing molecular weight) and clearly this does not occur in Fig. 4. Dimer formation would also result in an increase in the estimated isotropic overall rotational correlation time (τc) for cce_0567, as inferred from 15N spin relaxation times [38]. There was no experimentally significant change in τc with the addition of nickel to cce_0567, measured at 17.1 ± 0.5 ns in the metal-free form [18]. Finally, our crystal structure for cce_0567 with nickel corroborates with the dimer structure determined by NMR-based methods for Q60C73_METCA deposited into the Protein Data Bank (2JS5) that did not contain a metal ligand. While the presence of Ni2+ would have been difficult, if not impossible, to identify using NMR methods alone, it is likely that the authors of the Q60C73_METCA structure analyzed their protein for metals using alternative methods.
Fig. 4.
Overlay of the 1H–15N HSQC spectrum of 15N-labeled cce_0567 (blue) over the spectrum collected in the presence a ~1:1 molar ratio of cce_0567:NiCl2 (red). The inset illustrates one molecule of cce_0567 with the residues in the dimer interface colored black and those residues with perturbed chemical shifts after the addition of Ni2+ colored red. Spectra were collected at a proton resonance frequency of 750 MHz, 25 °C, in the same buffer used for protein crystallization.
To establish that cce_0567 was initially expressed and purified in the metal-free form the metal chelator EDTA was added to the 15N-labeled NMR sample [39]. Ten fold molar excess EDTA had no effect on the 1H–15N HSQC spectrum suggesting that there was no significant level of metal bound to cce_0567 initially and that the chemical shifts deposited in the BioMagRes Bank database (BMRB-15539) are for the metal-free form of the protein. Indeed, after the addition of nickel to the solution, the 1H–15N HSQC spectrum could be converted back to the metal-free spectrum by the addition of EDTA (data not shown). Similar metal binding experiments were repeated with the divalent cations Zn2+ and Co2+. Similar effects were observed to the 1H–15N HSQC spectrum with the addition of Zn2+ as observed with Ni2+, while Co2+ had greater perturbations to the 1H–15N HSQC spectrum due to the paramagnetic effects of the ion [35, 40]. The one noticeable difference was that at molar ratios greater than 1:1, the protein was visibly observed to start precipitating out of solution in the presence of Zn2+ and Co2+ while excess Ni2+ had no visible effect on solubility.
3.3. Circular dichroism profile and thermal stability of metal-free cce_0567
Circular dichroism spectroscopy is a powerful tool to probe the conformation of proteins in solution [41, 42] because small changes in the backbone conformation can cause large changes in the CD spectrum [43]. Consequently, different elements of protein secondary structure each have distinct CD patterns and it is possible to estimate the secondary structure content for a protein from the deconvolution of its CD spectrum [42]. CD spectroscopy can also be used to follow the effect of many variables, such as pH, salt content, and temperature, on the structure of a protein.
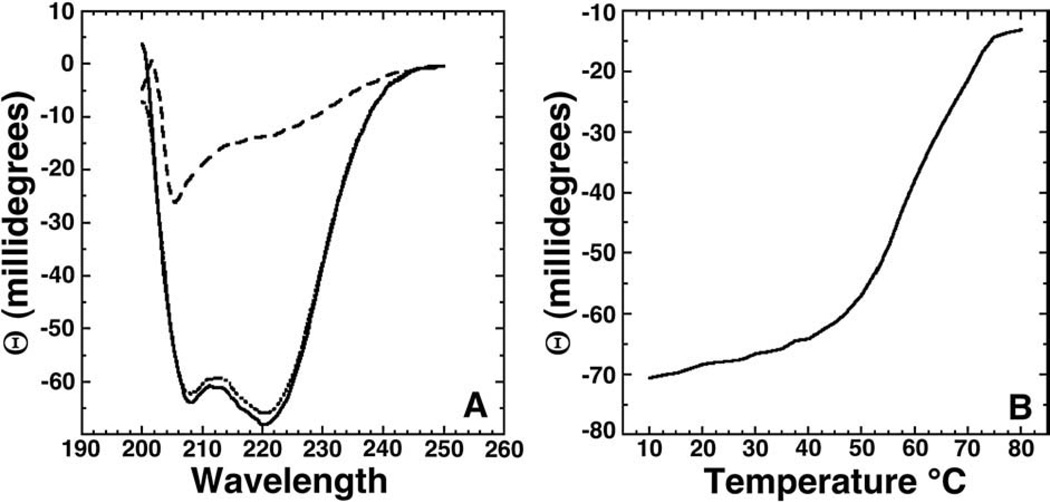
Fig. 5A contains the far-UV CD spectrum (solid line) for metal-free cce_0567 obtained in crystallization buffer at 25 °C. The dominant feature of the spectrum is the double minimumat 222 and 208–210 nm and maximum between 190 and 195 nm that are characteristic of a α-helical secondary structure [44]. Such an observation is expected given the crystal structure reported here for cce_0567. Fig. 5B assays the thermal stability of cce_0567 by monitoring the ellipticity at 220nmas a function of temperature between 10 and 80 °C. Avery gentle decrease in the ellipticity at 220 nmis observed from 10 to ~55 °C, upon which a steep decrease occurs up to ~75 °C at which point a plateau is reached. The inflection point for the transition is approximately 62 °C. Such a decrease in ellipticity with heating is typical of a phase transition from a folded to denatured state [45–48]. Indeed, the features of the CD spectrum of cce_0567 at 80 °C (dashed line) are characteristic of an unfolded protein [44]. However, after cooling the sample back to 25 °C (dotted line) the spectrum returns to essentially the identical pattern observed prior to heating the protein (solid line) indicating that denaturation is reversible. Such resilience to heating was confirmed by collecting 1H–15N HSQC spectra for 15N-labeled cce_0567 before and after heating for 30 min at 80 °C. The spectrum after heating was essentially identical to the spectrum collected prior to heating (data not shown).
Fig. 5.
(A) Circular dichroism spectrum of cce_0567 (30 µM) at 25 °C (solid line), 80 °C (dashed line), and again at 25 °C (dotted line) in buffer containing 500 mM NaCl, 20 mM Tris, 1 mM DTT, pH 7.2. (B) CD thermal melt for cce_0567 (30 µM) collected at 220 nm in 2.5 °C intervals between 10 and 80 °C.
3.4. Biological context
Circumstantial evidence that cce_0567 plays a role in nitrogen fixation is the gene's location in the nitrogen fixation operon. Physical evidence that cce_0567 may play a biochemical role in nitrogen fixation comes from transcriptomics microarray data for Cyanothece sp. 51142 cultures grown as previously described [49] for 48 h in alternating 12-hour periods of light and dark [50]. The expression profile for cce_0567 oscillates in sync with the known, essential nif genes, suggesting that cce_0567 may play some biochemical role in the nitrogen fixation process. Indeed, in Cyanothece it was observed that all 34 genes in the nitrogen fixation transcriptional regulon exhibited strong co-regulation in their expression [50]. The function of 16 of these 34 genes, including cce_0567 (DUF683), has not yet been determined.
Many of the nif proteins involved in nitrogen fixation contain metals and the crystal structure and NMR data presented here shows that cce_0567 binds the divalent cation nickel. However, these nif proteins bind the metals Fe, Mo, and S [51] and not nickel. On the other hand, uptake hydrogenases associated with nitrogenases in cyanobacteria [52] often contain nickel, and therefore, it may be speculated that cce_0567 is somehow associated with these uptake hydrogenases. However, nickel binding to cce_0567 may simply be serependitious as the NMR metal titration studies presented here shows that cce_0567 also binds to the divalent cations Zn2+ and Co2+. Further studies are necessary to determine if nickel is the biologically significant ligand for cce_0567 in vivo.
While the biologically significant metal that cce_0567 binds is not known, ClustalW2 sequence alignment of all 48 currently identified proteins in the DUF683 family strongly suggest that metal binding is an important property of the protein members in this family. As shown by such a sequence alignment in Fig. 2, the histidine residue is conserved in 46 of the 48 DUF683 family members. Furthermore, residues near this conserved histidine, and especially those participating in the linker region between the two helices, are also highly conserved through the family members. It is therefore very likely that divalent metal binding mediated through the conserved histidine residue plays a role in the biological function of cce_0567 and all other protein in the DUF683 family. Further biochemical studies are necessary to define the precise biological role cce_0567 has in the nitrogen fixation process. Given that proteins in the DUF683 family are primarily observed in cyanobacteria and cyanobacteria are the only bacteria known to have circadian clocks, perhaps the role of cce_0567 and other proteins in the DUF683 family is related to the circadian control of nitrogen fixation.
Acknowledgements
This work is part of a Membrane Biology EMSL Scientific Grand Challenge project at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by U.S. Department of Energy's Office of Biological and Environmental Research (BER) program located at Pacific Northwest National Laboratory (PNNL). PNNL is operated for the U.S. Department of Energy by Battelle. The assistance of the X29A beam line scientists at the National Synchrotron Light Source at Brookhaven National Laboratory is appreciated. Support for beamline X29A at the National Synchrotron Light Source comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. This manuscript has been authored by Battelle Memorial Institute, Pacific Northwest Division, under Contract No. DE-AC05-76RL0 1830 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Footnotes
The atomic coordinates have been deposited in the RCSB Protein Data Bank with accession code 3CSX.
References
- 1.Grey MW. The evolutionary origins of organelles. Trends Gen. 1993;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. Chloroplast evolution: secondary symbiogenesis and multiple-losses. Curr. Biol. 2002;12:R62–R64. doi: 10.1016/s0960-9822(01)00675-3. [DOI] [PubMed] [Google Scholar]
- 3.Kasting JF. The rise of atmospheric oxygen. Science. 2001;293:819–820. doi: 10.1126/science.1063811. [DOI] [PubMed] [Google Scholar]
- 4.Berman-Frank I, Lundgren P, Falkowski P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res. Microbiol. 2003;154:157–164. doi: 10.1016/S0923-2508(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 5.Sherman LA, Meunier P, Colon-Lopez MS. Diurnal rhythms in metabolism: a day in the life of a unicellular, diazotrophic cyanobacterium. Photosyn. Res. 1998;58:25–42. [Google Scholar]
- 6.Igarashi RY, Seefeldt LC. Nitrogen fixation: the mechanism of the Mo-dependent nitrogenase. Crit. Rev. Biochem. Mol. Biol. 2003;38:351–384. doi: 10.1080/10409230391036766. [DOI] [PubMed] [Google Scholar]
- 7.Sakurai H, Masukawa H. Promoting R and D in photobiological hydrogen production utilitzing mariculture-raised cyanobacteria. Mar. Biotechnol. 2007;9:128–145. doi: 10.1007/s10126-006-6073-x. [DOI] [PubMed] [Google Scholar]
- 8.Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 1992;56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Postgate JR. Nitrogen Fixation. Cambridge: Cambridge University Press; 1998. p. 112. [Google Scholar]
- 10.Zehr JP, Methe B, Foster R. In: New nitrogen-fixing microorganisms from the oceans: biological aspects and global implications. Wang Y-P, Lin M, Tian Z-X, Elmerich C, Newton WE, editors. Netherlands: Biological Nitrogen Fixation, Sustainable Agriculture and the Environment; 2005. pp. 361–365. [Google Scholar]
- 11.Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. U. S. A. 1993;90:5672–5681. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golden SS, Canales SR. Cyanobacterial circadian clocks—timing is everything. Nat. Rev., Microbiol. 2003;1:191–199. doi: 10.1038/nrmicro774. [DOI] [PubMed] [Google Scholar]
- 13.Woeffle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr. Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 14.Welsh EA, Liberton M, Stöckel J, Loh T, Elvitigala T, Wang C, Wollam A, Fulton RS, Clifton SW, Jacobs JM, Aurora R, Ghosh BK, Sherman LA, Smith RD, Wilson RK, Pakrasi HB. The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15094–15099. doi: 10.1073/pnas.0805418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskowski RA, Watson JD, Thornton JM. From protein structure to biochemical function? J. Struct. Func. Gen. 2003;4:167–177. doi: 10.1023/a:1026127927612. [DOI] [PubMed] [Google Scholar]
- 17.Kim S-H, Shin DH, Choi I-G, Schulze-Gahmen U, Chen S, Kim R. Structure-based functional inference in structural genomics. J. Struct. Funct. Genomics. 2003;4:129–135. doi: 10.1023/a:1026200610644. [DOI] [PubMed] [Google Scholar]
- 18.Buchko GW, Sofia HJ. Backbone 1H, 13C, and 15N NMR assignments for the Cyanothece 51142 protein cce_0567: a protein associated with nitrogen fixation in the DUF683 family. Biomol. NMR Assign. 2008;2:25–28. doi: 10.1007/s12104-007-9075-3. [DOI] [PubMed] [Google Scholar]
- 19.van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 20.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 21.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N Chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 24.Sheldrick GM. Kristallogr Z, editor. Macromolecular phasing with SHELXE. 2002;217:644–650. [Google Scholar]
- 25.Sheldrick GM. SHELXC. 2003 [Google Scholar]
- 26.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Crystallogr. 2004;37:843–844. [Google Scholar]
- 27.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. 1999;D55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terwilliger TC. Maximum-likelihood density modification using pattern recognition of structural motifs. Acta Crystallogr. 2001;D57:1763–1775. doi: 10.1107/S0907444901013737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terwilliger TC. Map-likelihood phasing. Acta Crystallogr. 2001;D57:1763–1775. doi: 10.1107/S0907444901013749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Pannu NJ, Murshudov GN, Dodson EJ, Read RJ. Incorporation of prior phase information strengthen maximum-likelihood structure refinement. Acta Crystallogr. 1998;D54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- 32.Lovell SC, Davis IW, Arendall WB, III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: π, σ, and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 33.Maiti R, van Domselaar GH, Zhang H, Wishart DS. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res. 2004;23:W590–W594. doi: 10.1093/nar/gkh477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson RJ, Higgins DG. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Pochapsky TC, Pochapsky SS, Ju T, Hoefler C, Liang J. A refined model for the structure of acireductone dioxygenase from Klebsiella ATCC 8724 incorporating dipolar couplings. J. Biomol. NMR. 2006;34:117–127. doi: 10.1007/s10858-005-5735-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciurli S, Mangani S. Nickel-containing Enzymes. New York, NY: Mercel Dekker Inc.; 2001. [Google Scholar]
- 37.Buchko GW, McAteer K, Wallace SS, Kennedy MA. Solution-state NMR investigation of DNA binding interactions in Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg): a dynamic description of the DNA/protein interface. DNA Repair. 2005;4:327–339. doi: 10.1016/j.dnarep.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Szyperski T, Yeh DC, Sukumaran DK, Moseley HNB, Montelione GT. Reduced-dimensionality NMR spectroscopy for high-throughput protein resonance assignment. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8009–8014. doi: 10.1073/pnas.122224599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buchko GW, Ni S, Holbrook SR, Kennedy MA. Solution structure of hypothetical Nudix hydrolase DR0079 from extremely radiation-resistant Deinococcus radiodurans bacterium. Proteins. 2004;56:28–39. doi: 10.1002/prot.20082. [DOI] [PubMed] [Google Scholar]
- 40.Buchko GW, Daughdrill GW, de Lorimier R, Rao BK, Isern NG, Lingbeck JM, Taylor JS, Wold MS, Gochin M, Spicer LD, Lowry DF, Kennedy MA. Interactions of human nucleotide excision repair protein XPA with DNA and RPA70ΔC327: chemical shift mapping and 15N NMR relaxation studies. Biochemistry. 1999;38:15116–15128. doi: 10.1021/bi991755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woody RW. Studies of Theoretical Circular Dichroism of Polypeptides: Contributions of β-turns. New York: John Wiley and Sons; 1974. [Google Scholar]
- 42.Kelly SM, Jess TJ, Price NC. How to study protein by circular dichroism. Biochim. Biophys. Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Manning CM, Illangaseke M, Woody RW. Circular dichroism studies of distorted α-helices, twisted β-sheets and β-turns. Biophys. Chem. 1988;31:77–86. doi: 10.1016/0301-4622(88)80011-5. [DOI] [PubMed] [Google Scholar]
- 44.Holzwarth GM, Doty P. The ultraviolet circular dichroism of polypeptides. J. Am. Chem. Soc. 1965;87:218–228. doi: 10.1021/ja01080a015. [DOI] [PubMed] [Google Scholar]
- 45.Galka JJ, Baturin SJ, Manley DM, Kehler AJ, O'Neil JD. Stability of the glycerol faciltator in detergent solution. Biochemistry. 2008;47:3513–3524. doi: 10.1021/bi7021409. [DOI] [PubMed] [Google Scholar]
- 46.Buchko GW, Hess NJ, Bandaru V, Wallace SS, Kennedy MA. Spectroscopic studies of zinc(II)- and cobalt(II)-associated Escherichia coli formamidopyrimidine-DNA glycosylase: extended X-ray absorption fine structure evidence for a metal-binding domain. Biochemistry. 2000;40:12441–12449. doi: 10.1021/bi001377k. [DOI] [PubMed] [Google Scholar]
- 47.Chang J-F, Hall BE, Tanny JC, Moazed D, Filman D, Ellenberger T. Structure of the coiled-coil dimerization motif of Sir4 and its interaction with Sir3. Structure. 2003;11:637–649. doi: 10.1016/s0969-2126(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 48.Kwok SC, Hodges RS. Clustering of large hydrophobes in the hydrophobic core of two-stranded α-helical coiled-coils controls protein folding and stability. J. Biol. Chem. 2003;278:35248–35254. doi: 10.1074/jbc.M305306200. [DOI] [PubMed] [Google Scholar]
- 49.Reddy KJ, Haskell JB, Sherman DM, Sherman LA. Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece . J. Bacteriol. 1993;175:1284–1292. doi: 10.1128/jb.175.5.1284-1292.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stöckel J, Welsch EA, Liberton M, Kunnvakkam R, Aurora R, Pakrasi HB. Global transcriptomic analysis of Cyanothece 51142 reveals robust diurnal oscillation of central metabolic process. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6156–6161. doi: 10.1073/pnas.0711068105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubio LM, Ludden PW. Biosynthesis of the iron–molybdenum cofactor of nitrogenase. Annu. Rev. Microbiol. 2008;62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 52.Tamagnini P, Axelsson R, Lindberg P, Oxelfelt F, Wünschiers R, Lindblad P. Hydrogenases and hydrogen metabolism in cyanobacteria. Microbiol. Mol. Biol. Rev. 2002;66:1–20. doi: 10.1128/MMBR.66.1.1-20.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]