Abstract
MYC-induced DNA damage is exacerbated in WRN deficient cells, leading to replication stress and accelerated cellular senescence. To determine if WRN deficiency impairs MYC driven tumor development, we utilized both xenograft and autochthonous tumor models. Conditional silencing of WRN expression in c-MYC overexpressing non-small cell lung cancer xenografts impaired both tumor establishment and tumor growth. This inhibitory effect of WRN knock-down was accompanied by increased DNA damage, decreased proliferation, and tumor necrosis. In the Eμ-Myc mouse model of B-cell lymphoma, a germline mutation in the helicase domain of Wrn (WrnΔhel/Δhel) resulted in a significant delay in emergence of lethal lymphomas, extending tumor free survival by >30%. Analysis of pre-neoplastic B cells from Eμ-Myc Wrn mutant mice revealed increased DNA damage, elevation of senescence markers, and decreased proliferation in comparison with cells from age-matched Eμ-Myc mice. Immunohistochemical and global gene expression analysis of overt Eμ-Myc WrnΔhel/Δhel lymphomas demonstrated a marked increase in expression of the CDK inhibitor, p16Ink4a, as well as elevation of TAp63, a known mediator of senescence. Collectively, these studies demonstrate that in the context of Myc-associated tumorigenesis, loss of Wrn amplifies the DNA damage response, both in pre-neoplastic and neoplastic tissue, engaging activation of tumor suppressor pathways. This leads to inhibition of tumor growth and prolonged tumor free survival. Targeting WRN or its enzymatic function could prove to be an effective strategy in the treatment of MYC-associated cancers.
Keywords: Werner helicase, therapeutic target, Myc-driven cancer, senescence, tumor suppressors
Introduction
We previously demonstrated that WRN, a gene encoding a RecQ DNA helicase, is a direct transcriptional target of c-MYC, and that the absence of WRN causes MYC overexpressing, h-Tert immortalized cells to undergo senescence (1). Mechanistically, the MYC/WRN co-dependence has been explained by the recent demonstration that MYC directly influences the DNA pre-replication machinery, and that MYC overexpression dramatically accelerates S-phase, thereby sensitizing cells to “replication stress” (2). Inhibition of WRN function in MYC overexpressing cells leads to excessive accumulation of DNA damage at sites of newly replicated DNA, triggering activation of the ATR-CHK1 pathway and, in turn, forcing the cells into a non-proliferative, senescent state (3). Thus, MYC transcriptional stimulation of the WRN gene provides a feed forward mechanism to limit MYC-associated DNA replication stress and enables continued cell proliferation.
Mutations in the WRN gene are associated with a progeroid syndrome in humans (Werner Syndrome, WS), which is characterized by accelerated aging, cellular senescence, genomic instability, and an increased incidence of otherwise rare cancers of mesenchymal origin (4, 5). WRN encodes a multifunctional protein with both DNA helicase and exonuclease activity, a property which sets WRN as the exception among other members of the RecQ family, which harbor only a DNA helicase domain (6, 7). WRN protein binds to and modifies DNA secondary structures that are likely to arise during DNA replication (8, 9). This property of WRN protein is consistent with its role in repair/recovery from replication-associated damage (10, 11). The role of WRN in maintaining DNA fidelity, coupled with the tumor predisposition of WS patients has suggested that WRN could function as a tumor suppressor gene. However, WRN mutations have not been reported in tumors, and we and others have found that WRN and other members of the RecQ helicase family are significantly overexpressed in cancer cell lines derived from Burkitts lymphoma, neuroblastoma, breast, ovarian and lung cancers (C. Grandori, unpublished results and (12)). Further, while fibroblasts from Wrn mutant mice exhibit enhanced sensitivity to DNA cross-linking agents, characteristic of human WS cells (13, 14), Wrn deficient mice have not shown a predisposition to spontaneous tumor development. These observations, together with the pro-survival role of WRN in MYC overexpressing cells suggest WRN could play a supporting role in the context of MYC dependent tumorigenesis and consequently, its deficiency might inhibit rather than accelerate tumor development. Here, we establish that acute WRN depletion in MYC overexpressing human lung cancer xenografts blocks tumor growth and further, that germline Wrn deficiency in mice causes a significant delay in Myc-induced lymphomagenesis and prolongs tumor free survival. Thus, in the context of Myc-driven cancers, Wrn provides a critical pro-survival function that is necessary for efficient tumor growth and constitutes a candidate druggable target in tumors driven by an “undruggable” oncogenic driver.
Materials and Methods
Mouse strains, genotyping, and tumor monitoring
The WrnΔhel/Δhel mutation (14) was backcrossed onto C57BL/6 mice to purity (N20). The C57BL/6 Eμ-Myc transgenic mouse strain (15) was interbred to C57BL/6 WrnΔhel/Δhel mice to generate Eμ-Myc WrnΔhel/+ and WrnΔhel/+ mice, which were subsequently intercrossed and backcrossed to parental lines to generate nontransgenic and Eμ-Myc transgenic Wrn+/+ and WrnΔhel/Δhel experimental animals (F1 generation). Mice were monitored daily for signs of morbidity and tumor development. Moribund animals were sacrificed and tumors and lymphoid organs were harvested for histopathological and molecular analysis. Germline transmission of the Eμ-Myc transgene, and the Werner helicase domain deletion (Wrn Δdel exon 3–4) was confirmed using conventional PCR–based genotyping strategies (14), (15). All animal protocols were approved by the Fred Hutchinson Cancer Research Center Laboratory Animal Care and Use Committee.
Immunoblotting of lymphomas
Whole cell protein extracts from primary pre-B cells and B-cell tumors from Eμ-Myc and Eμ-Myc WrnΔhel/Δhel transgenic mice were isolated as previously described (16). Equal amounts of clarified lysates (100–150μg per lane) were analyzed by western blotting with antibodies specific to mouse p19Arf (5-C3-1), p16Ink4a (M-156), Mdm2 (C-18) pAb, and Actin (I-19) from Santa Cruz Biotechnology, Mdm2 (MD-219- Abcam) mAb, Cyclin D1 (DCS6), pH2A.X (ser139), pAtr (ser424) p53 (ser18) and cleaved caspase-3 (Asp175) from Cell Signaling Technology, and p53 (CM5) from Novocastra. Detection was by enhanced chemiluminescence (Supersignal -Thermo Scientific).
Acidic β-galactosidase assay
The senescence-associated β-galactosidase assay (SA-β-gal) was performed as previously described with some modifications (17, 18). Briefly, murine tissues were harvested and frozen gradually (CO2) in cryomolds within OCT compound and stored at −80°C. Cryosections of tissues were cut using a cryotome at 6–12μm, fixed in 0.5% glutaraldehyde (6μm section /15 min) at room temperature (22°C), and rinsed with PBS (pH 7.3). Tissue sections were then covered with fresh SA-β-gal stain (40mM citric acid/sodium phosphate (pH 6.0), 1 mg/ml Xgal (DMF), 5 mM potassium ferricyanide, 5 mM ferrocyanide, 150 mM NaCl, 2 mM MgCl2) placed in humidity chambers, and incubated at 37°C in an ambient 21% O2 environment for 6–8 hours. After staining, tissue sections were rinsed with PBS and processed via standard immunohistochemical protocols.
Flow cytometric analysis of B-cell proliferation and apoptosis
Single-cell suspensions of bone marrow cells and splenocytes were prepared by harvesting marrow from femurs, disassociating the spleens between frosted microscope slides, and filtering both cell preparations through nylon cell strainers. For analysis of differentiation status, splenic cells were incubated with PE-conjugated anti-CD45R/B220, and PerCP-Cy5.5–conjugated anti-IgM antibodies (BD Biosciences Pharmingen). Proliferation rates were measured using a FITC BrdU Flow Kit (BD Biosciences Pharmingen) as described by the manufacturer. Animals were injected with BrdU (40 mg/gram bodyweight, i.p.), and bone marrow and spleen were harvested 2 h later. Apoptosis in pre-B (B220+/IgM−) and B-cell (B220+/IgM+) populations from spleen and bone marrow was measured using FITC-conjugated anti-active caspase-3 (BD Biosciences Pharmingen). All samples were analysed by a FACSCalibur (BectonDickinson).
Splenocyte isolation and immunoblotting
Single cell suspensions were generated from spleens of Eμ-Myc and Eμ-Myc WrnΔhel/Δhel mice as previously described (19). Cell suspensions were suspended in a hypotonic buffer to lyse red blood cells, and splenic lymphocytes and cellular debris were separated via centrifugation along a ficollhypaque (d=1.077) gradient (800× g at 4°C for 15'). Isolated splenocytes were then lysed in 50 mM Tris (pH 8.0), 200 mM NaCl, 5 mM EDTA, 1% Triton X-100, 10% Glycerol, 1 mM NaVO4, 2 mM DTT, 1.5 μg/ml aprotinin, 0.3 mg/ml pefabloc, 6 μg/ml leupeptin. Equal amounts of clarified protein lysates (50 μg per lane) were analyzed by Western blotting with antibodies to phospho-H2A.X (Serine 139), phospho-Atr (Serine 428), phospho-p53 (Serine18) from Cell Signaling Technology (CST), p53 (CM5) from Novocastra, and β-Actin from Biovision.
Sequence analysis of lymphomas
Total RNA was isolated from select lymphomas using Trizol (Invitrogen) followed by Qiagen RNeasy mini kit (Qiagen). The first strand cDNA was synthesized with dT oligos from Superscript III first strand kit (Invitrogen). p53 cDNA was amplified using the primer set for exons 2–11 (p53-fwd: 5'-GCTTCTCCGAAGACTGGATGACT-3' and p53-rev: 5'-GATTGTGTCTCAGCCCTGAAGTCA-3') (20). A second primer set for exons 5–8 (p53-fwd-Ex5: 5'-GTACCTTATGAGCCACCCGA-3' and p53-rev-Ex8: 5'TTTTCTTTTGCGGGGGA-3') was used for confirmation of sequence fidelity. PCR products were sequenced via ABI 3730xl DNA Analyser (Applied Biosystems) using the same primer sets. Sequencing data were aligned to Mus musculus p53 mRNA transcript sequences [ENSMUSP00000104298, …104297, …005371] (www.ensembl.org). Bi-allelic deletions of the Cdkn2a (Ink4a/ARF) locus were also examined from select lymphomas via PCR of genomic exons 1α, exon1β, exon 2, and exon 3.
Immunohistochemical staining of lymphomas
Tissues were fixed in normal buffered formalin (NBF), processed to paraffin and stained for H&E, specific proteins, BrdU (5-bromo-2'-deoxyuridine), or TUNEL (terminal deoxynucleotidyl transferase dUTP end labeling). Staining for p53 (Vector), p19Arf (Santa Cruz), Histone H3 (Serine10) (Cell Signaling Technology-CST), Histone H2A.X (Serine139) (CST), and cleaved caspase-3 (CST) were done using a standard three-step ABC method. Slides were developed using DAB/NiCl (Sigma) and counterstained with methylgreen. Sections for BrdU (DAKO) staining were treated with HCl and trypsin before incubating with primary antibody. For TUNEL staining, Trevigen TACS2 TdT kit was used as per manufacturer's instructions.
Expression analysis and in vitro assay of Wrn mediated knockdown
RNA was extracted from mouse tissue using the RNAeasy kit (Qiagen), reverse-transcribed using a Superscript II RT kit (Invitrogen). The following probe sets were used for qRT-PCR analysis: Wrn helicase domain (Mm00499253_m1), Wrn C-terminus (Mm01197916_m1), Bcl-2 (Mm00438070_m1), Prkdc (Mm01342967_m1), and Trp63 (Mm00495788_m1) (Applied Biosystems – Life Technologies Inc.). cDNAs were subjected to qRT-PCR analysis using a SYBR green PCR Master Mix (Applied Biosystems) with primers specific to TAp63 (fwd, 5'-TGCCCCGACCCTTACATCCA-3'; rev, 5'-GGAAGGACACATCGAAGCTGTG-3'); dNp63 (fwd, 5'-CTGGAAAACAATGCCCAGAC-3'; rev, 5'-GAGGAGCCGTTCTGAATCTG-3'); b-actin (fwd, 5'-GATCTGGCACCACACCTTCT-3, rev, 5'-GGGGTGTTGAAGGTCTCAAA-3') (21, 22). For in vitro assays, single cell suspensions were isolated from Eμ-Myc lymphomas as above and grown in B-cell media (DMEM/IMEM supplemented with 10% FBS and β-mercaptoethanol) on an NIH3T3 fibroblast feeder layer. Retroviral vectors were constructed using pLMP (gifted from Dr. Paddison) and siRNAs (Ambion) targeting the following sequences: shWRN1(CTGTGTGTGTCTGAGAGCAAAT); shWRN2(AACCCAGAACTTGACAGTTTAG), shControl (CAACAAGATGAAGAGCACCAA). Retroviral infections were performed using a phoenix ecotropic packaging cell line with helper plasmid pPEM5 (gifted from Dr. Miller). B-cell lymphoma cells were infected via “spinoculation”, 1500 RPM for 10 min at 32C, through four successive inoculations with retroviral containing supernatant from the phoenix cells supplemented with polybrene (Schmitt, C. pers. comm.). B-cell lymphoma cells were again cultured on an NIH3T3 feeder layer and selected with puromycin for three days, and harvested for gene expression at 6–8 days post-infection.
Microarray hybridization and data analysis
Total RNA was isolated from four lymphomas from each genotype using Trizol (Invitrogen) and RNeasy kit (Qiagen), labeled using the Message AMP kit (Ambion) and hybridized to Illumina BeadStudio Gene Expression microarray MouseRef 8v2 (Illumina Inc., San Diego, CA). Expression data were processed using the Illumina GenomeStudio software, using quantile normalization and background subtraction. Non-specific probe filtering of the array data was performed using a GenomeStudio-reported detection p-value cutoff of 0.1, reducing the number of genes to a filtered dataset of 7,865. The R/Bioconductor arrayQualityMetrics package was used to assess the quality of the BeadArray data as adequate for analysis. We used the R/Bioconductor stats package heatmap function to calculate hierarchical clustering dendrograms. In addition, the Bioconductor package lumi was used for affy-like background subtraction and to quantile normalize the full dataset. Negative control features were used to estimate signal-to noise ratios across the entire dataset. For a given gene, if s/n < 3 across all arrays, the gene was removed from further analysis. The Bioconductor package limma was used to perform modified t-statistic significance testing (23), and multiple testing corrections using Benjamini and Hochberg (24). All microarray data (MIAME compliant) are deposited in NCBI's Gene Expression Omnibus (25, 2002) and are accessible through GEO Series accession number (GSE25671).
Lentiviral shRNA construction and packaging
Lentiviral contructs for each shRNA and a scramble were designed and constructed in a pLenti6-tts system using the Gateway LR Clonase. Stbl3 chemically competent cells (Invitrogen) were transformed with 1–5ng of DNA, and clones were selected on blasticidin (50ug/ml) LB isolated, screened and propagated. 293FT cell line (derived from 293F cells, stably expressing the SV40 large T antigen for enhanced virus production) was cultured in DMEM/10%FBS/500ug/mlG418/pen/strep ViraPower Packaging Mix (Invitrogen) and resuspended in 195ul of sterile water to a concentration of 1ug/ul. When cotransfected with the pLenti-DEST expression construct into the 293FT producer cell line, this mixture of plasmids supplies the viral proteins in trans that are required to create viral products. Viral supernatants were generated using 293FT cells transfected with 10μg shRNA plasmid using Lipofectamine 2000 and the appropriate lentiviral packaging plasmid DNA (ViraPower). The media was changed 16 hr after transfection and the virus was harvested 48 to 56 hr later. Viral supernatant was filtered (0.40um), aliquoted and frozen at −80 for subsequent use.
Lentiviral mediated knockdown of WRN protein in A549 lung carcinoma cells
Human A549 small cell lung carcinoma cells (ATCC; Rockville, Md) cultured in DMEM/10%FBS/pen/strep were infected with lentiviral tet-mediated vectors with shRNA scramble control (19-mer - GUUGUUCUACUUCUCGUGG), shWRN-I (19-mer seq -GUCUAUCCGCUGUAGCAAU), shWRN-II (19-mer seq - GUACCUUAUCCACAUGGCA), or shWRN-III (19 mer seq - GAGACAAAUCAUCUUGUCU) 5'-3' target sequences and knockdown of the WRN protein was assessed via immunoblot 48 hours post-treatment with 0.01 μg/ml doxycycline (Sigma D9891) in culture media. Immunoblots were probed with WRN pAb (H-300), Actin (I-19), and c-Myc (N-262) antibodies purchased from Santa Cruz Biotechnology.
Xenograft tumor models
A549 cells were freshly infected with either the lentiviral tet-mediated shRNA scramble control or the shWRN-I (Fig. 1 E and F) or shWRN II (Fig. 1 G and H) and 2.5 × 106 cells in 0.2 mL PBS were inoculated subcutaneously into the left and right flanks of 10 eight week old NOD-SCID gamma null mice, respectively. For the experiment of Fig. 1 E and F (shWRN I), FOXN1 (nu/nu) nude mice were exposed to doxycycline from day 1. For the experiment of Fig. 1 G and H (shWRN II), NOD-SCID gamma null mice were given doxycycline (2mg/ml in 5% sucrose) ad libitum in their drinking water (n=5), only when tumors had reached a visible size, while an equal number of mice received only 5% sucrose water. The volume of the implanted tumor was measured every 2–3 days with a caliper, using the formula: V = L×W2/2; where V, volume (mm3); L, biggest diameter (mm); W, smallest diameter (mm). A tumor was defined as a palpable mass of ≥100 mm3 volume. Mice were sacrificed at 3–4 weeks.
Figure 1. WRN knock-down suppresses tumor growth.
A. Myc protein levels in A549 NSCLC cells with or without shWRN-I. HFF-Myc is shown as positive control for c-Myc expression, and IMR-32 cells are shown as a negative control for c-Myc expression. Actin was used as a loading control. B. WRN protein levels in A549 cells transduced with doxycycline (Dox)-conditional lentiviral vectors harboring WRN or control shRNAs (CTL) in the presence or absence of Dox. Actin was used as a loading control. C. Long-term colony forming assay of A549 cells with control (upper panels) or WRN specific shRNAs (lower panels) in the presence or absence of Dox. D. Time course of WRN mRNA knock-down measured by qRT-PCR in A549 cells after addition of doxycycline to induce expression of shWRN I. E. Representative image of an A549 xenograft at time of sacrifice showing a significant difference in tumor volume between negative control shRNA (left flank) and shWRN I (right flank). F. Tumor volume of xenografts expressing shWRN I and sh control over a 2-week interval while mice were exposed to Dox since the time of implantation. G. Representative image of the therapeutic xenograft model following induction of shWRN II or control shRNA with Dox in established A549 tumors. H. Tumor volume of xenografts indicating the growth inhibition of shWRN II expressing tumors. (*) Denotes a significant difference in mean tumor volume (n=5) between control and shWRN II at indicated times post-injection, p < 0.05 (unpaired t-test).
Statistical analysis
All images shown are representative of at least 10 fields viewed over two stained sections per animal. Quantitation was performed using ten fields per animal. All columns represent mean ± SEM, unless otherwise noted. All statistical analyses were performed using unpaired two-tailed t-test unless otherwise indicated. Statistical analysis of Kaplan-Meier survival curves was done via log-rank (Mantel-Haenszel) test (Graphpad Software, San Diego, CA) or SPSS (IBM, Worldwide).
Results
WRN is critical for establishment and growth of non-small cell lung cancer cells with MYC overexpression
To examine the role of WRN in a model of Myc associated cancer, the growth rates of A549 non-small cell lung carcinoma (NSCLC) xenografts were monitored after WRN knock-down. A549 cells express high levels of c-MYC, as confirmed via western blot analysis (Fig. 1A). Three shRNAs against WRN were first tested in transient assays in 293T cells. shWRN I and shWRN II indicated the best knock-down (data not shown). A549 cells were transduced with lentiviral vectors expressing doxycycline-conditional shWRN I or shWRN II specific to the WRN gene. Knockdown was measured by Western analysis (Fig. 1B) and through a time course of WRN mRNA following shWRN I (Fig. 1D). The latter indicated that optimal knock-down occurred several days after shRNA induction, consistent with the long half life of WRN protein and mRNA. To determine the effect of WRN depletion on the growth of A549 cancer cells, long-term colony assays were performed. In A549 cells harboring a control shRNA, growth was not affected by doxycycline treatment, while cells expressing the WRN specific shRNA demonstrated a dramatic inhibition of growth following addition of doxycycline (Fig. 1C). We next wished to test the role of WRN in tumor establishment and growth in two xenograft models. One, in which expression of the shRNA was initiated at the time of tumor cell injection, and a second, a therapeutic model in which tumors were established prior to WRN knockdown. Fig. 1F shows the tumor size over time in four mice, where each mouse was injected on one flank with A549 cells transduced with a control shRNA and on the other with shWRN I. Mice were exposed to doxycycline from the day of implantation to induce shRNA expression. A representative mouse showing inhibition of tumor growth by WRN knock-down is shown in Fig. 1E. The results obtained with the therapeutic model are shown in Fig. 1G and H where 10 mice (5 in each arm sh control and shWRN II) were exposed to doxycyline after tumors reached ~100mm3. In this therapeutic model, A549 tumor growth was markedly inhibited by shWRN II induction. Representative xenografts are shown in Fig 1G. Thus WRN depletion with two different hairpins in two settings markedly inhibited tumor establishment and growth. WRN depleted xenografts displayed a prominent increase in DNA damage, an increase in necrosis and a reduction in proliferative cells (Fig. 2A–B). However, no significant difference in caspase-3 dependent apoptosis compared to controls was observed (Fig. 2C). Together, these results establish that impaired tumor growth caused by WRN depletion is due to increased DNA damage and reduced proliferation.
Figure 2. Immunohistochemical profiling of A549 NSCLC xenografts indicates increased DNA damage and decreased proliferation following WRN knock-down.

A. Representative IHC images from A549 xenografts demonstrating decreased proliferation (H&E and Ki67), and increased DNA damage (γ-H2A.X, Serine139) in WRN depleted xenografts (shWRN I) vs. control xenografts (shCTL). In contrast no difference in apoptosis as judged by cleaved caspase-3 was detectable. H&E and Ki67 images show 10X field with 40X field inset. B. Quantitative scoring of DNA damage (H2A.X Serine 139) from control and shWRN xenografts. Bars represent mean+/−SD; n=3 mice per graft, ten 40x fields/mouse, ***p<0.0001 (unpaired t-test). C. Quantitative scoring of apoptosis (cleaved caspase-3) from control and shWRN xenografts. Bars represent mean+/−SD; n=3 mice per graft, ten 40x fields/mouse, no significant difference via unpaired t-test.
Wrn deficiency delays Eμ-Myc induced lymphomagenesis and prolongs tumor free survival
As the immune system and tumor microenvironment profoundly affect tumor development, we next addressed the role of Wrn in an autochthonous model of tumorigenesis in immunocompetent mice. To this end, we assessed the impact of a germline Wrn helicase mutation (WrnΔhel/Δhel) (14) on tumor latency in the Eμ-Myc mouse model. Eμ-Myc mice constitutively express c-myc in lymphoid precursor cells, leading to pre-B cell hyperplasia and subsequent lymphoma development (15). Cohorts of Eμ-Myc (n=41), Eμ-Myc WrnΔhel/Δhel (n=50), as well as wild type (n=30) and WrnΔhel/Δhel (n=30) mice were followed for >2 years and Kaplan Meier survival curves were generated. Virtually all Eμ-Myc mice developed lymphomas but the median latency in Eμ-Myc WrnΔhel/Δhel mice (151 days) was significantly delayed relative to Eμ-Myc mice (115 days) (Fig. 3A). Control wild type and WrnΔhel/Δhel mice did not develop any related pathology over the same time span (data not shown). To confirm the transcriptional induction of Wrn by c-Myc overexpression, previously observed in vitro (1), Wrn protein and mRNA expression was measured in normal spleen and lymphomas from Eμ-Myc mice. Wrn protein levels were indeed elevated in both pre-neoplastic and lymphoma derived cells from Eμ-Myc mice (Supplementary Fig. 1). In addition, expression of the truncated Wrn protein was verified in tissues from WrnΔhel/Δhel mice (Supplementary Fig. 2). Together, these results highlight a pro-tumorigenic role for the Wrn DNA helicase in a well-documented model of Myc-driven tumorigenesis.
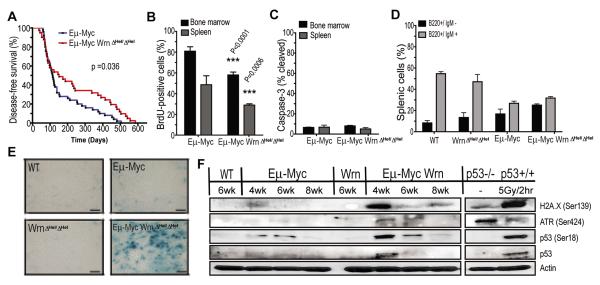
Figure 3. Werner deficiency (WrnΔhel/Δhel) delays lymphomagenesis.
A. Kaplan-Meier curves showing disease-free survival of Eμ-Myc (median survival of 115 days) and Eμ-Myc WrnΔhel/Δhel (median survival of 151 days) mice; significant difference via logrank test, p-value = 0.036. B. Flow-cytometry analysis of BrdU positive cells as a measure of proliferation; unpaired t-tests, bone marrow: ***p<0.0001, spleen: ***p=0.0006. C. Flow-cytometry analysis of cleaved caspase-3 as a measure of apoptosis. D. Splenic pre-B cells (B220+/IgM−) and mature B cells (B220+/IgM−) from Eμ-Myc and Eμ-Myc WrnΔhel/Δhel mice; matched littermates (n=2) from 4-week mice of all genotypes were analyzed in triplicate in panels b, c, d. E. Acidic B-galactosidase staining of germinal center lymphoid follicles from spleens of 4–6 week old mice of the indicated genotype. Images are representative of three mice of each genotype. Scale bar represents 40μm. F. Longitudinal analysis of protein levels from splenocytes isolated from mice at 4, 6, and 8 weeks. Antibodies to phospho-histone H2A.X (Serine 139), phospho-Atr (Serine 428), phospho-p53 (Serine 18), and p53 were used, and actin is shown as a loading control. WT refers to wild-type control, Wrn refers to WrnΔhel/Δhel, p53−/− and p53+/+ to splenocytes isolated from 6-week old mice, p53+/+ (p19−/−) mice were irradiated with 5Gy (137Cs) and splenocytes isolated 2 hours later.
Wrn deficiency triggers a DNA damage response leading to proliferation arrest in the preneoplastic stage of tumor development
To address the basis for the impaired lymphomagenesis in Eμ-Myc WrnΔhel/Δhel mice, we established another mouse cohort to examine markers of proliferation, apoptosis, and DNA damage in preneoplastic splenocytes prior to emergence of lymphomas. Preneoplastic B cells isolated from both bone marrow and spleen from Eμ-Myc WrnΔhel/Δhel mice showed reduced proliferation compared to Eμ-Myc mice (Fig. 3B). Quantification of cleaved caspase-3 positive cells indicated that apoptosis was not significantly elevated in Wrn deficient preneoplastic cells (Fig. 3C). Analysis of cell surface markers indicated that Wrn deficiency does not impinge on B cell homeostasis, as judged by the similar ratio of pre-B cell precursors versus mature B cells in Eμ-Myc mice from both Wrn genotypes (Fig. 3D). Consistent with reduced proliferation markers and the known role of Wrn in cellular senescence, a prominent increase in senescence-associated β-galactosidase (SA-β-gal) staining was observed in preneoplastic B-cell compartments (germinal center follicles of the spleen) from Eμ-Myc WrnΔhel/Δhel mice compared to Eμ-Myc mice (Fig. 3E). Longitudinal immunoblot analysis indicated an increase in the accumulation of γ-H2AX (Serine 139), Atr (Serine 428), and p53 (Serine18) in preneoplastic splenocytes from Eμ-Myc WrnΔhel/Δhel mice (Fig. 3F). Thus, Wrn deficiency blocks the expansion of preneoplastic cells by triggering a DNA damage response that impairs cell proliferation and favors senescence, thereby delaying the emergence of overt tumors.
Wrn deficiency delays lymphomagenesis by engaging tumor suppressor pathways
Previous research utilizing the Eμ-Myc mouse model has shown that development of lymphomas coincides with either bi-allelic deletions in the Cdkn2a locus or mutation in the p53 tumor suppressor (16, 26, 27). To further elucidate the mechanism by which Wrn deficiency delays lymphomagenesis, p53, p19Arf, p16Ink4a, cyclin D1, and cleaved caspase-3 were examined in Eμ-Myc and Eμ-Myc WrnΔhel/Δhel lymphomas. We observed a striking elevation of the cyclin dependent kinase (CDK) inhibitor p16Ink4a in the majority of Eμ-Myc WrnΔhel/Δhel (82%) compared to Eμ-Myc (33%) lymphomas (Fig. 4A, Supplementary Fig. 3A, *p=0.006). p16Ink4a normally functions to sequester Cdk4, allowing for the degradation of cyclin D1 and release of p27Kip1 from cyclin D1-Cdk4 complexes. Ultimately, this engages a G1 cell cycle arrest through Rb activation (28). Consistent with this role for p16Ink4a, cyclin D1 levels were found to be reduced in Eμ-Myc WrnΔhel/Δhel lymphomas, particularly late onset lymphomas with high p16Ink4a expression (Fig. 4A, asterisk indicates late onset). In addition, proliferation was significantly decreased in the Eμ-Myc WrnΔhel/Δhel (Fig. 4B and C) lymphomas. Consistent with our data from pre-neoplastic tissue, no elevation of caspase-3 dependent apoptosis was observed in Eμ-Myc WrnΔhel/Δhel lymphomas (Fig. 4A).
Figure 4. Molecular profiling of Eμ-Myc and Eμ-Myc WrnΔhel/Δhel lymphomas.
A. Representative immunoblot of Eμ-Myc (lanes 1–18) and Eμ-Myc WrnΔhel/Δhel (lanes 19–35) lymphomas showing levels of p53, p19Arf, p16Ink4a, cyclin D1, cleaved caspase-3 and actin (loading control). Late onset lymphomas are denoted with an asterisk and represent 8 lymphomas arising in Eμ-Myc mice (range 121–505 days), and 7 lymphomas arising in Eμ-Myc WrnΔhel/Δhel (range 122–495 days). Red and blue boxes refer to the mice used for IHC analysis in Supplementary Fig. 4A. B. Representative photomicrographs of Eμ-Myc and Eμ-Myc WrnΔhel/Δhel lymphomas with p53 over-expressed (ie. p53 mutation, ref. 16) and p19Arf over-expression, immunohistochemically stained for p53, p19Arf, BrdU, H2A.X (serine 139), and senescence-associated β-galactosidase (SA-β-gal); scale bar on all images represents 100μm. C. Immunohistochemical scoring of Eμ-Myc and Eμ-Myc WrnΔhel/Δhel lymphomas for proliferation (BrdU) and DNA damage (H2A.X Serine 139) of photomicrographs in B. Columns represent mean +/− sem (n=10, 40X and 100x fields, respectively; unpaired t-tests, BrdU: *** p<0.0001, H2A.X: ***p<0.0001).
We next assessed the frequency of p53 mutation in lymphomas. The results showed no significant difference in p53 mutation between Eμ-Myc lymphomas (35%) and Eμ-Myc WrnΔhel/Δhel lymphomas (45%) (Supplementary Fig. 3A, p=0.748). Furthermore, all tumors with stabilized p53 had mutations in p53, as previously reported (16) (Supplementary Fig. 3B). The tumor suppressor p19Arf was over-expressed in lymphomas with high p53, consistent with its known role in p53 stabilization (29) (Fig. 4A, Supplementary Fig. 3B). Proliferation and apoptosis were next assessed in late onset lymphomas, stratified for mutations in p53, anatomical location, and expression of p16Ink4a and p19Arf. These results indicate that in the context of a p53 mutant background, Wrn deficiency leads to a significant increase in DNA damage (γ-H2AX), and reduces proliferation (phospho-H3) and apoptosis (TUNEL) (Supplemental Fig. 4A and B). Upon closer examination of p53 mutant tumors, nuclear accumulation of p53 appears more pronounced in Eμ-Myc WrnΔhel/Δhel lymphomas with a concordant increase in senescence-associated β-galactosidase (Fig. 4B). These results confirm that a defect in Wrn activity leads to engagement of anti-proliferative/senescence pathways, likely through the combined effects of the tumor suppressors p16Ink4a/p19Arf and this does not require wild type p53.
Global gene expression analysis of Wrn deficient lymphomas identifies TAp63 as a possible mediator of senescence response
To globally define the pathways by which Wrn deficiency impairs tumor development, microarray gene expression analysis was performed on four lymphomas from each Wrn genotype (Supplementary Table 1, GSE25671). Ingenuity pathway analysis, based on known literature connections, of significantly up-regulated genes in Wrn deficient vs. Wrn wild type lymphomas identified genes that centered on the p53 homologue TAp63, the Cdkn2a (p16Ink4a/p19Arf) locus, and the p53-dependent stress-inducible nuclear protein, Trp53inp1 (Fig. 5A, Supplementary Table 1A). TAp63 has recently been recognized as an key mediator of senescence (21). Quantitative RT-PCR confirmed increased expression of TAp63 in additional Eμ-Myc WrnΔhel/Δhel lymphomas (Supplementary Fig. 3). Furthermore, induction of TAp63 was recapitulated in vitro by acute knockdown of Wrn in Eμ-Myc lymphoma-derived cells with p53 mutation (Fig. 5B and Supplementary Fig. 5). In addition, CDK inhibitors such as Cdkn1c (p57Kip2), and Cdkn1b (p27Kip1), were among the up-regulated genes in Eμ-Myc WrnΔhel/Δhel lymphomas (GSE25671). All together, the results obtained by global gene expression analysis correlate with the data derived from the immunohistochemical and biochemical assessment of lymphoma tissue, and delineate a senescence signature associated with Eμ-Myc WrnΔhel/Δhel lymphomas. In addition, they suggest that TAp63 could be the primary mediator of a senescence response in p53 mutant tumors.
Figure 5. Model of the impact of Wrn depletion in inhibiting Myc-driven tumorigenesis.

A. Ingenuity Pathway Analysis network based on known literature connections was built with genes differentially expressed in Wrn deficient and Wrn wild type lymphomas. Two major nodes emerged from this analysis: CDNK2A and TP63 and only genes linked to these nodes are shown here. Numbers below each icon indicate log2 fold changes expression values in Wrn deficient vs, Wrn wild type lymphoma. B. Relative mRNA expression of TAp63 in Eμ-Myc/p53 mutant cultured lymphoma cells in the presence of shControl or shWrn RNA. Bars represent the mean+/−SD (n=3), statistical analysis was performed using unpaired t-tests; shWrn#1 (*p=0.033), shWrn#2 (**p=0.009) vs. shControl. Additional data are shown in Supplementary Fig. 6. Inset: Immunoblot of WRN protein in same samples. C. Model of delayed Eμ-Myc lymphomagenesis caused by Wrn deficiency. Eμ-Myc expression triggers preneoplastic proliferation but also engages endogenous tumor suppressors p53 and p19ARF. In the absence of functional Wrn, Myc-induced replication stress is exacerbated, causing DNA damage, which hyperactivates multiple tumor suppressor pathways, impairs proliferation of tumor cells, and delays appearance of lymphomas.
Discussion
The RecQ DNA helicase WRN, has previously been associated with tumor suppression, as Werner Syndrome patients, in addition to premature aging, develop rare mesenchymal tumors late in life (5). In contrast, we hypothesized that WRN, by preventing senescence of cancer cells, could contribute to the tumorigenic process. This hypothesis was supported by our previous studies showing that the MYC oncoprotein directly stimulates transcription of the WRN gene, and in turn, loss of WRN function leads to senescence of MYC overexpressing cells (1). This co-dependence of WRN and MYC overexpression was attributed to the role of WRN in limiting replication-associated damage during S-phase, a process that is dramatically accelerated in MYC overexpressing cells through direct association of MYC with the pre-replication machinery (2, 3).
To determine if MYC-driven cancers depend on Wrn in an in vivo setting, we investigated two models of Myc-associated tumorigenesis, as well as the effect of both acute and chronic Wrn depletion on tumor growth. The acute depletion of WRN was carried out with NSCLC A549 cells which express high levels of c-MYC and have been optimized for the conditional knock-down of WRN in vivo. In addition, the utilization of a Myc-driven mouse model allowed us to test the consequences of chronically interfering with a helicase defective Wrn protein, much like one would envision in a therapeutic setting with small molecule inhibitors to the Wrn helicase domain. Both models demonstrated consistent results, that is impairment of WRN function profoundly effects tumor growth due to excessive accumulation of DNA damage and decreased proliferation with consequent senescence (observed in the Eμ-Myc lymphoma) or prominent necrosis (as observed in the A459 xenograft model). Wrn helicase deficiency effectively delayed the insurgence of lymphomas through activation of two tumor suppressors: p16Ink4a and TAp63 (see model in Fig. 5C). Importantly, we were able to recapitulate in lymphoma cells from Eμ-Myc mice that acute depletion of Wrn also induces TAp63, even in a p53 mutant setting (Fig. 5 and Supplemental Fig. 5). Our results are consistent with a recent report of increased senescence and delayed lymphomagenesis in Eμ-Myc p53515C/+ and Eμ-Myc p53515C/515C mutant mice relative to Eμ-Myc p53+/− mice (30) and highlight a mechanism of growth arrest and cellular senescence that has relevance for future therapeutic application of WRN inhibitors even in the context of p53 mutant cancers. In this view, the endogenous DNA damage generated by Wrn deficiency acts not unlike the damage generated via genotoxic treatment or telomere shorting to engage tumor suppressor pathways (31,32). The effect of Wrn depletion was lost in p53 deficient backgrounds (3), and Wrn deficiency increased the rate of cancer in a p53 deficient background (33), suggesting that complete loss of function of p53 could disable the senescence response in Wrn deficient cancer cells. Additional model systems will be needed to distinguish the impact of p53 deletion vs. mutation on Wrn function.
It is worthwhile to note that the Kaplan Meier curves comparing Eμ-Myc Wrn wild type with WrnΔhel/Δhel, indicated an age-dependent penetrance of tumor suppression caused by Wrn deficiency. While this requires further investigation, one hypothesis that is consistent with the observed elevation of p16Ink4a in Wrn deficient tumors, is that Wrn deficiency, superimposed with the natural aging of hematopoietic precursors (also characterized by an elevation of p16Ink4a) may additively provide a barrier for late onset tumor development (34). A second hypothesis is derived from the report that late onset lymphomas in Eμ-Myc mice are distinct from early onset tumors with respect to differentiation status and signaling pathways such as those regulated by NF-κB (35). Differential activation of these pathways may modify the impact of Wrn deficiency on engaging anti-proliferative pathways.
Our studies also highlight the differential sensitivity of cancer cells vs. normal cells to loss of WRN. This is reminiscent of other reports of tumor-specific sensitivity to loss of DNA repair function. For example, mice deficient in NHEJ activity due to mutation in Prkdc (the catalytic subunit of DNA-dependent protein kinase) are normal, barring a defect in V(D)J recombination. However, they are markedly resistant to development of squamous cell carcinoma, indicating active NHEJ is necessary for optimal tumor growth (36). Furthermore, a recent study indicates that Atr deficiency also protects mice from MYC-driven lymphomas (37). In contrast, Atm deficiency by eliminating a DNA damage induced apoptotic response accelerates MYC-driven tumorigenesis (38), highlighting differences in DNA damage pathways and their relation to MYC. This principle of differential sensitivity of cancer vs. normal cells to DNA repair deficiency has important therapeutic potential, as in the case of utilization of PARP inhibitors for cancers deficient in BRCA1 or BRCA2 function (39). Similarly, therapeutics directed against RecQ helicases may prove beneficial for cancers with amplification/alterations of MYC by marshaling the DNA damage response and intrinsic tumor suppressor pathways. Our experiments directly addressed the effects of a WRN helicase mutant in tumor growth, thus pinpointing not just the gene, but also the domain that could be targeted by small molecules. The catalytic activity of this domain necessitates both ATP binding and ATPase activity (4), and as such is highly druggable (40). Thus, targeting WRN or its enzymatic functions could prove to be an effective strategy to treat MYC associated cancers.
Supplementary Material
Acknowledgments
We thank Dr. Ray Monnat and Dr. Julia Sidorova for critically reviewing the manuscript and Dr. Denise Galloway and her laboratory for support and guidance during the course of this work. We also thank Dr. Patrick Paddison and Dr. Dusty Miller for viral vectors utilized for Wrn knock-down experiments, Dr. C.A. Schmitt for retroviral infection protocols and advice. JD and MM were supported in part by NIH P30 CA015704. This work was supported by R01 grants (NIA AG02661), (NCI CA099517) and the NCI Mouse Models of Human Cancer Consortium.
References
- 1.Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, Frank S, Amati B, Dalla-Favera R, Monnat RJ., Jr. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 2003;17:1569–1574. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 3.Robinson K, Asawachaicharn N, Galloway DA, Grandori C. c-Myc accelerates S-Phase and requires WRN to avoid replication stress. PLoS One. 2009;4:e5951. doi: 10.1371/journal.pone.0005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 5.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 7.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J Biol Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 8.Brosh RM, Jr., Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J Biol Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 9.Compton SA, Tolun G, Kamath-Loeb AS, Loeb LA, Griffith JD. The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J Biol Chem. 2008;283:24478–24483. doi: 10.1074/jbc.M803370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aggarwal M, Sommers JA, Morris C, Brosh RM., Jr. Delineation of WRN helicase function with EXO1 in the replicational stress response. DNA Repair (Amst) 2010;9:765–776. doi: 10.1016/j.dnarep.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidorova JM, Li N, Folch A, Monnat RJ., Jr. The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasnoselsky AL, Whiteford CC, Wei JS, Bilke S, Westermann F, Chen QR, Khan J. Altered expression of cell cycle genes distinguishes aggressive neuroblastoma. Oncogene. 2005;24:1533–1541. doi: 10.1038/sj.onc.1208341. [DOI] [PubMed] [Google Scholar]
- 13.Lombard DB, Beard C, Johnson B, Marciniak RA, Dausman J, Bronson R, Buhlmann JE, Lipman R, Curry R, Sharpe A, Jaenisch R, Guarente L. Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol Cell Biol. 2000;20:3286–3291. doi: 10.1128/mcb.20.9.3286-3291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc Natl Acad Sci U S A. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 16.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bandyopadhyay D, Gatza C, Donehower LA, Medrano EE. Analysis of cellular senescence in culture in vivo: the senescence-associated beta-galactosidase assay. Curr Protoc Cell Biol. 2005;Chapter 18(Unit 18):19. doi: 10.1002/0471143030.cb1809s27. [DOI] [PubMed] [Google Scholar]
- 18.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruisbeek AM. In vitro assays for mouse B and T cell function. Curr Protoc Immunology. 2000;Supplement 39(Unit 3.1.3) [Google Scholar]
- 20.Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell. 2009;16:33–43. doi: 10.1016/j.ccr.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, Vogel H, Mills AA. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth GK. `Bioinformatics and Computational Biology Solutions using R and Bioconductor'. 1 edition Springer; New York: 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 24.Benjamini Y. a. H., Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society B. 1995:289–300. [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eischen CM, Woo D, Roussel MF, Cleveland JL. Apoptosis triggered by Myc-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol. 2001;21:5063–5070. doi: 10.1128/MCB.21.15.5063-5070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 28.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 29.Sherr CJ, Bertwistle D, W DENB, Kuo ML, Sugimoto M, Tago K, Williams RT, Zindy F, Roussel MF. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–137. doi: 10.1101/sqb.2005.70.004. [DOI] [PubMed] [Google Scholar]
- 30.Post SM, Quintas-Cardama A, Terzian T, Smith C, Eischen CM, Lozano G. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene. 2010;29:1260–1269. doi: 10.1038/onc.2009.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 33.Lebel M, Cardiff RD, Leder P. Tumorigenic effect of nonfunctional p53 or p21 in mice mutant in the Werner syndrome helicase. Cancer Res. 2001;61:1816–1819. [PubMed] [Google Scholar]
- 34.Signer RA, Montecino-Rodriguez E, Witte ON, Dorshkind K. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev. 2008;22:3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori S, Rempel RE, Chang JT, Yao G, Lagoo AS, Potti A, Bild A, Nevins JR. Utilization of pathway signatures to reveal distinct types of B lymphoma in the Emicro-myc model and human diffuse large B-cell lymphoma. Cancer Res. 2008;68:8525–8534. doi: 10.1158/0008-5472.CAN-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kemp CJ, Vo K, Gurley KE. Resistance to skin tumorigenesis in DNAPK-deficient SCID mice is not due to immunodeficiency but results from hypersensitivity to TPA-induced apoptosis. Carcinogenesis. 1999;20:2051–2056. doi: 10.1093/carcin/20.11.2051. [DOI] [PubMed] [Google Scholar]
- 37.Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF, D'Artista L, Schleker T, Guerra C, Garcia E, Barbacid M, Hidalgo M, Amati B, Fernandez-Capetillo O. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol. 2011;18:1331–1335. doi: 10.1038/nsmb.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maclean KH, Kastan MB, Cleveland JL. Atm deficiency affects both apoptosis and proliferation to augment Myc-induced lymphomagenesis. Mol Cancer Res. 2007;5:705–711. doi: 10.1158/1541-7786.MCR-07-0058. [DOI] [PubMed] [Google Scholar]
- 39.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 40.Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr. Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci U S A. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.