Abstract
Here we show that cells lacking the heme-regulated inhibitor (HRI) are highly resistant to infection by bacterial pathogens. By examining the infection process in wild-type and HRI null cells, we found that HRI is required for pathogens to execute their virulence-associated cellular activities. Specifically, unlike wild-type cells, HRI null cells infected with the gram-negative bacterial pathogen Yersinia are essentially impervious to the cytoskeleton-damaging effects of the Yop virulence factors. This effect is due to reduced functioning of the Yersinia type 3 secretion (T3S) system which injects virulence factors directly into the host cell cytosol. Reduced T3S activity is also observed in HRI null cells infected with the bacterial pathogen Chlamydia which results in a dramatic reduction in its intracellular proliferation. We go on to show that a HRI-mediated process plays a central role in the cellular infection cycle of the Gram-positive pathogen Listeria . For this pathogen, HRI is required for the post-invasion trafficking of the bacterium to the infected host cytosol. Thus by depriving Listeria of its intracellular niche, there is a highly reduced proliferation of Listeria in HRI null cells. We provide evidence that these infection-associated functions of HRI (an eIF2α kinase) are independent of its activity as a regulator of protein synthesis. This is the first report of a host factor whose absence interferes with the function of T3S secretion and cytosolic access by pathogens and makes HRI an excellent target for inhibitors due to its broad virulence-associated activities.
Introduction
Greater knowledge of the mechanisms employed by microbial pathogens to overcome host defenses has allowed for the development of drug-like molecules that specifically target these pathogen virulence-associated structures. Since at least some of these virulence-associated structures are widely conserved among pathogens of animals and plants, such ‘virulence blocker’ compounds provide an attractive alternative to conventional antibiotics that typically target structures (e.g., ribosomes) or processes (e.g., cell wall synthesis) found in both pathogens as well as members of the microbiota. Several examples of broad-acting small molecule virulence blockers were originally identified as inhibitors of the type 3 secretion (T3S) system of the pathogenic yersiniae which delivers virulence factors directly into the host cell cytosol [1]. Subsequently it was shown that these compounds also inhibit T3SSs of other Gram-negative pathogens such as Chlamydia , Salmonella, and Pseudomonas [2]. Here we broaden this concept by identifying a host-encoded factor that is required by diverse pathogens to execute their respective cellular infection cycles.
In a yeast-based genetic screen using bacterial virulence factors as probes, we found that the stress-induced eIF2 signaling pathway plays a key role in the intracellular activities of both the Yersinia protein kinase A (YpkA) and Yersinia outer protein J (YopJ) [3]. In eukaryotes eIF2 signaling mediates the cellular responses to a variety of external and internal stress. Mammalian cells possess four different eIF2α kinases (GCN4, PERK, PKR, and HRI) that are activated by distinct stress conditions including nutritional deprivation (GCN4), endoplasmic reticulum stress (PERK), infection by viral-derived RNA (PKR) and heat/oxidative/heme-induced stresses (HRI). Phosphorylation eIF2α inhibits the formation of active ternary complexes thus leading to a reduction in protein synthesis. Our studies indicated that in yeast cells YpkA activated eIF2 signaling whereas YopJ, in contrast, negatively regulated eIF2 signaling [3].
Although the significance of the YpkA-induced eIF2 signaling during infection remains unknown, we subsequently showed that, like in yeast cells, YopJ negatively regulated eIF2 signaling in Yersinia -infected mammalian cells [4]. Additionally, we showed that an intact eIF2 signaling pathway was required for the infection-induced activation of NF-κB and pro-inflammatory cytokine expression [4]. In addition to its role in NF-κB activation and cytokine expression, we unexpectedly found that eIF2 signaling counteracts the host cell invasion of Yersinia as well as the intracellular pathogens Chlamydia and Listeria [4]. Cells that lacked a functional eIF2 pathway were highly invaded by these pathogens indicating that eIF2 signaling is important in protective anti-bacterial responses.
The heme-regulated inhibitor (HRI in humans, Hri in mice) was originally identified as the translation-level regulator (through its eIF2α kinase activity) that couples β-globin synthesis with heme levels during erythropoiesis and has more recently been shown to mitigate oxidative stress during erythroid differentiation [5,6]. HRI is also important for various stress responses in yeast and mammalian cells [7,8]. Here we investigated whether HRI plays a role in host cell infection by microbial pathogens. Unexpectedly, we found that HRI positively regulates specific virulence-related activities of diverse bacterial pathogens. Surprisingly, these HRI effects were independent of its canonical function as a translation regulator via eIF2α and thus identify a novel role for HRI in bacterial pathogenesis.
Materials and Methods
Host and pathogen strains
Hri and Pkr MEFs were provided by Randal J. Kaufman and Joan E. Durbin [4,8], respectively, and Hri knockout mice were generously provided by Jane-Jane Chen [9]. Mice were treated humanely in strict accordance with federal and state government guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health and their use was approved by the University of Miami institutional animal care and use committee (protocols 11-186).
The wild-type Yersinia pseudotuberculosis strain YPIII/pIB102 [10] was used except in the translocation assay a mutant YPIII strain (YPIII/pIB29MEKA), in which all 5 of the effector Yop-encoding genes were deleted [11], was transformed with a plasmid encoding a hybrid protein consisting of YopE (residues1-130) and a 40-residue Elk tag [12]. The Chlamydia trachomatis LGV-434, serovar L2 and the OVA-expressing Listeria monocytogenes strains were obtained from the American Type Culture Collection (ATCC) and DMX Corp. (DMX 09-082), respectively. The GFP-expressing L. monocytogenes strains were provided by Daniel Portnoy.
Infections
Transcript analysis was performed as previously described [4]. Hri +/+ and -/- MEFs were seeded on cover-slips and infected the next day. Overnight cultures of the Y. pseudotuberculosis strains were diluted to O.D. 0.1 in tissue culture media and subsequently propagated at 26 °C for 2 hrs and then 37 °C for 1 hr to induce expression of its type 3 secretion system prior to infection. Following infection, cells were first fixed with 2% paraformaldehyde in phosphate buffer saline (PBS, pH 7.4) for 30 minutes, then treated with permeabilization buffer (1% saponin and 3% bovine serum albumin in PBS) for 20 mins and then blocking buffer (0.3% bovine serum albumin and 0.1% Tween 20 in PBS) for 1 hour. Actin was visualized with Alexa-fluor-conjugated phalloidins (Molecular Probes), nuclei with DAPI (Molecular Probes) and vacuoles with LAMP1 (BD Bioscience). Cell images were captured with a Zeiss LSM 700 confocal microscope and analyzed using MacBiophotonics ImageJ software. The Yop translocation assay was performed as described [4]. To analyze if inhibition of protein synthesis of eukaryotic cells affected the kinetics of the cytotoxic response caused by the Y . pseudotubercolosis wild-type strain YPIII/pIB102, either 5 or 25 µg/ml of cycloheximide was added to cultured HeLa cells one hour prior and maintained during the infection. The morphology of the infected cells was analyzed by phase contrast microscopy up to 4 hrs post-infection. The C. trachomatis infections were performed as described [4] and initiated by adding the bacteria to cultured cells. Following the infection period, cells were either stained for C. trachomatis to microscopically determine direct counts of inclusions or alternatively cells were lysed and the resulting supernatants (which contain infectious C . trancomatis elementary bodies) were used to infect HeLa cell cultures to determine progeny yield. Quiescent (unstimulated) peritoneal macrophages were harvested from Hri +/+ and -/- aged/sex matched mice and seeded on coverslips in serum-containing media for a few hours and then unattached cells were removed. Next day cells were infected with C. trachomatis and processed as described above for MEFs.
Flow cytometry and imagining
For the antigen presentation and cytokine expression analyses, single-cell suspensions of splenocytes were prepared from Hri +/+ and -/- aged/sex matched mice, passed through a 70 micron filter and resuspended in serum-containing media. Cells were rested for 1 hr and then infected with either the OVA-expressing L. monocytogenes strain or the Y. pseudotuberculosis strain. For the OVA experiments, splenocytes were infected for 5 h in the presence of brefeldin A and then stained with anti-CD11c (eBioscience), anti-B220 (BD Bioscience), anti-MHC I Ova (Biolegend), anti-CD11b (BD Bioscience), anti-CD4 (eBioscience), anti-CD8 (Biolengend), F4/80 (Caltag Laboratories), and Live/dead near IR (Invitrogen) in fluorescence-activated cell sorting buffer (PBS containing 2% bovine serum albumin and 0.1% sodium azide) for 30 min at 4 °C and then fixed in 2% paraformaldehyde. A Cytofix/Cytoperm kit (BD-Pharmingen) was used to measure intracellular levels of TNFα. Data were acquired using a BD FACS LSRII flow cytometer (Becton Dickinson, San Jose, CA) and analyzed using FlowJo software (Tree Star, Inc.).
The microscopic-based L. monocytogenes infection assays were performed using techniques described above with the additional feature that the GFP-expressing bacteria were directly visualized. For the L. monocytogenes proliferation assay, MEFs were seeded in a 48-well plate (105 per well) and the next day infected with 4 x 106 cfus that were prepared from a stationary phase culture grown in brain heart infusium at 32 °C without shaking. After a 1.5 hr attachment period excess bacteria were removed and gentamicin was added at 2 µg/ml to kill non-internalized bacteria. After a 1.5 hr extracellular killing period internalized bacteria were enumerated at various time points by lysing the cells with water and determining the colony forming units (cfu) by plating.
Results
The host cellular factor HRI regulates infection-induced TNFα expression
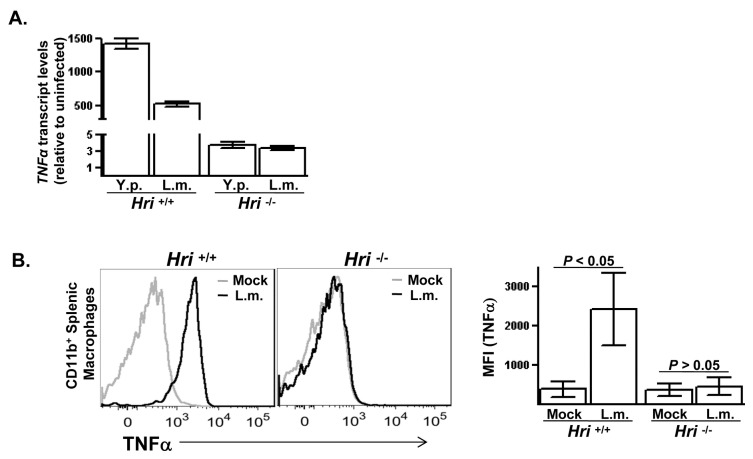
Prior studies have shown that LPS-induced expression of the proinflammatory cytokine TNFα is reduced in Hri -/- compared to Hri +/+ macrophages [13]. To determine whether this defective inflammatory response of Hri -/- macrophages to LPS reflects a role for HRI in the cellular response to infection with bacterial pathogens, TNFα expression was measured in Hri +/+ and Hri -/- primary macrophages infected with either Yersinia pseudotuberculosis or Listeria monocytogenes. In peritoneal macrophages from a Hri +/+ mouse infected in vitro with these pathogens, TNFα-encoding transcript levels increased >500-fold following a 3-hr infection period (Figure 1A). In contrast, there was only a modest increase (~3-fold) in the levels of TNFα-encoding transcript in infected Hri -/- peritoneal macrophages. Consistent with the transcript analysis, there was a several-fold increase in TNFα protein levels in Hri +/+ splenic macrophages following 5 hrs of infection with these two pathogens whereas there was essentially no changes in TNFα levels in similarly infected Hri -/- macrophages (Figure 1B ; data for Y. pseudotuberculosis not shown). Previously we showed that an intact eIF2 signaling pathway was required for infection-induced cytokine expression [4]. The findings presented here suggest that HRI is the primary eIF2α kinase responsible for this effect. However, upon closer examination it became apparent that the role HRI plays in the cellular infection cycle of these pathogens far exceeds its regulation of cytokine expression.
Figure 1. HRI is required for infection-induced cytokine expression.
(A) Peritoneal macrophages isolated from Hri +/+ and -/- mice were infected in vitro with either Yersinia pseudotuberculosis (Y.p.) or Listeria monocytogenes (L.m.) for 3 hr. Plotted are the levels of TNFα-encoding transcripts in infected Hri +/+ and -/- cells relative to their respective uninfected control cells. (B) Splenocytes isolated from Hri +/+ and -/- mice were either left uninfected or infected in vitro with L.m. for 5 hrs and then intracellularly stained for TNFα protein. Histrogram displays TNFa levels in macrophages (live, CD11b+-gated). The median fluorescence intensities (MFI) of 3 separate experiments is shown (right panel). P values calculated using student t test.
HRI is required for the function of Yersinia T3S system
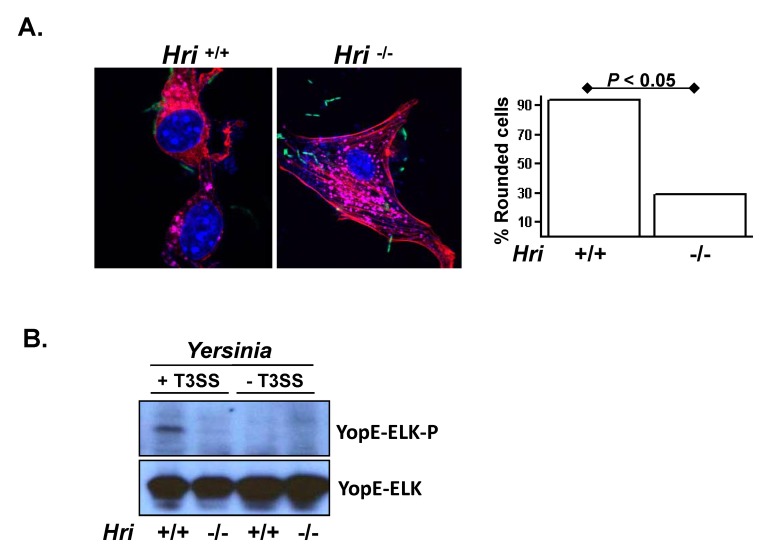
The pathogenicity of Yersinia is inextricably linked to its delivery of virulence factors (or effectors) directly into the host cell cytosol through the T3S system [14]. Upon their delivery into the cytosol, these effectors suppress the induction of host protective cytokine expression via YopJ-dependent mechanisms as well as cause a striking collapse of the cytoskeleton primarily by the actions of YopE [15]. The latter property serves as a readout for T3S activity [16] and can be readily observed in cultured wild-type (Hri +/+) mouse embryonic fibroblasts (MEFs) which transition from their normally flatten appearance to a partially detached ‘rounded’ morphology within an hour of infection (Figure 2). Unexpectedly, Hri -/- cells displayed almost no overt signs of this cytoskeletal disruption following infection (Figure 2). The resistance of Hri -/- cells to T3S-mediated disruption of the cytoskeleton could not be attributed to differential bacterial adhesion as determined both by direct microscopic examination and plating methods (Figure S1 ). Furthermore, there were no discernable generalized defects in the cytoskeletal function in the absence of HRI as both Hri +/+ and -/- cells migrated with comparable efficiency in Transwell migration assays (data not shown).
Figure 2. HRI facilitates the translocation of Yersinia virulence factors into the cytosol.
(A) Hri +/+ and -/- mouse embryonic fibroblasts (MEFs) were infected with a GFP-expressing strain Y. pseudotuberculosis for 2 hrs and then stained for nuclei, vacuoles, and actin (purple, pink, and red, respectively). Plotted is the percentage of rounded cells (resembling the cell shown in the left panel) of at least 5 microscopic fields of approximately 100 total cells. P value calculated using student t test. (B) MEFs were infected with a Y. pseudotuberculosis strain deleted for all six Yop effectors and possessing either an intact or a translocation-defective type 3 secretion (T3S) system and harboring a plasmid encoding a YopE-Elk translocation reporter. After 1 hr of infection the resulting whole cell lysates were examined by western analysis using antibodies specific for the Elk epitope tag either in its phosphorylated (YopE-Elk-P) or unphosphorylated (YopE-Elk) forms. YopE-Elk becomes phosphorylated exclusively within the eukaryotic cytosol and therefore the phosphorylation of the Elk epitope is readout for T3S-mediated translocation. For presentation purposes the lanes of a single blot were rearranged.
To directly test whether the lack of cytoskeletal disruption in Yersinia -infected Hri -/- cells was due to reduced functioning of the T3S system, we used a YopE-Elk translocation assay as a reporter of the delivery of Yersinia virulence factors into the host cell cytosol [12]. Compared to Hri +/+ cells, YopE translocated into Hri -/- cells was greatly reduected (Figure 2B ; lanes 1 and 2). The YopE translocation defect in Hri -/- MEF cytosol was similar to that observed for a Yersinia strain harboring a genetically inactivated T3S system (lanes 3 and 4; ref. [17]). Additionally, this translocation defect in the Hri -/- cells is independent of the Yop virulence factors themselves since this latter assay is performed in a Yersinia strain that is genetically deleted for all of the Yop virulence factors. These findings indicate that HRI is required for the T3S-mediated transfer of Yersinia -encoded virulence factors into the host cell cytosol. Although host cell processes have been described that promote T3S activity (e.g., the RhoA GTPases [18]), to the best of our knowledge this is the first description of a host factor that, when absent, renders a cell refractory to the translocation of T3S effectors.
To date all of the described activity of HRI is associated with it acting as a regulator of protein synthesis by virtue of its eIF2α kinase activity (see Introduction). Previously we reported that protein synthesis rates do not differ between cells infected with wild-type Y. pseudotuberculosis versus a T3S mutant derivative [15] and more recently we showed that there were no differences in Y. pseudotuberculosis T3S activity between infected wild-type host cells and cells expressing an eIF2α with an S51A replacement (the target residue of the eIF2α kinases) [4]. To further test whether T3S activity and protein synthesis in host cells are functionally linked, we treated cells with cycloheximide prior to (and during) their infection with Y. pseudotuberculosis. We observed no differences in either the kinetics or the severity of cytoskeletal disruption between untreated and cycloheximide-treated cells (Figure S2 ). Collectively these findings indicate that function of the Yersinia T3S system is not coupled to protein synthesis in host cells (or vice versa). Thus, the mechanism by which HRI positively impacts the T3S system in Yersinia infected cells is likely independent of HRI-mediated regulation of protein translation.
PKR and HRI independently regulate cellular infectivity of Chlamydia trachomatis
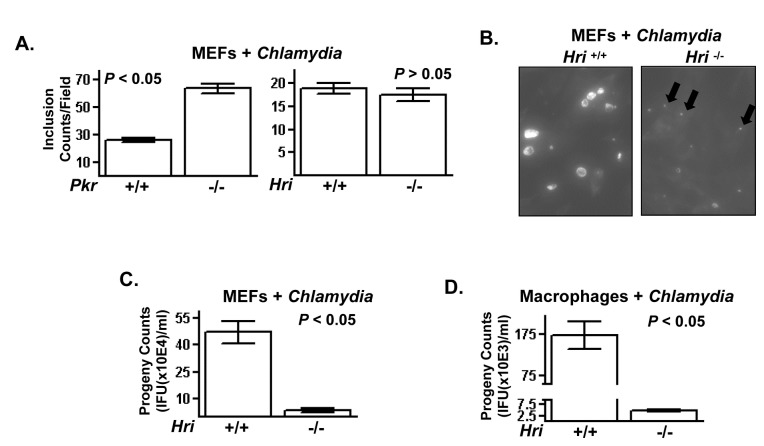
We recently showed that cells expressing an eIF2α with an S51A replacement (the target residue of the eIF2α kinases) were highly colonized by C. trachomatis indicating that this pathway plays a critical role in the complex cellular infection cycle of this bacterial pathogen [4]. Following invasion, C. trachomatis forms a membrane-bound inclusion body within which it differentiates from the invasive elementary bodies (EBs) into the proliferating reticulocyte bodies (RBs). To determine whether the eIF2α kinases PKR and/or HRI regulate the infection dynamics of this pathogen, wild-type (+/+) and knockout (-/-) cells were infected with C. trachomatis and the number of inclusion bodies (‘direct counts’) were determined. Pkr -/- cells were significantly more permissive for invasion compared to Pkr +/+ cells (Figure 3A). The higher level of invasion of Pkr -/- cells was similar to that we recently reported for cells expressing eIF2α(S51A) [4]. In contrast, there were comparable numbers of inclusion bodies observed in Hri +/+ and -/- cells following infection with C. trachomatis indicating that HRI does not substantially affect Chlamydial invasion (Figure 3A). The similar invasion phenotypes of the PKR null and eIF2α(S51A) cells suggest that PKR-mediated activation of eIF2 signaling opposes C. trachomatis invasion.
Figure 3. The eIF2α kinase PKR negatively regulates invasion and HRI positively regulates intracellular proliferation of Chlamydia .
(A) Pkr +/+ and -/- (left) and Hri +/+ and -/- (right) MEFs were infected with C. trachomatis L2 for 24 hr and then stained for C. trachomatis-containing inclusion bodies. The total number of inclusion forming units is plotted. (B) Infected Hri +/+ and -/- MEFs are shown with arrows denoting underdeveloped C . tranchomatis -containing inclusions in the Hri -/- cells. (C) Cells were infected as described and were then harvested and the resulting lysates used to infect cultured HeLa cells to determine their titer (plotted as ‘progeny counts’). (D) Unstimulated peritoneal macrophages were isolated from Hri +/+ and -/- mice and infected as described. P values calculated using student t test.
Although the number of inclusions that formed in Hri +/+ and -/- cells were comparable, there were, however, considerable difference in the sizes of these inclusions. The sizes of the inclusions in the Hri -/- cells were much reduced and these inclusions were populated by much fewer C. trachomatis compared to the Hri +/+ cells (Figure 3B). To quantitatively measure post-invasion proliferation a replating assay is performed in which infected cells are lysed and the yield of infectious units (‘progeny’) is determined. The yield progeny from Hri +/+ cells was ~10-fold greater than that recovered from Hri -/- cells (Figure 3C). Even greater differences (~50-fold) in progeny yields were observed in in vitro-infected peritoneal macrophages isolated from Hri +/+ and -/- mice (Figure 3D). These data show that HRI promotes the post-invasion intra-vacuolar proliferation of C. trachomatis.
Similar to Yersinia , pathogenic Chlamydia employs a T3S system to optimally infect eukaryotic cells [19]. The unaffected invasion rate but reduced intra-vacuolar growth phenotype of C. trachomatis in Hri -/- cells resembles that observed in cells treated with small molecules that disrupt Chlamydial T3SS functioning [20,21]. A high dose infection assay was therefore used to determine whether Chlamydial T3SS activity is reduced in Hri -/- cells. A relatively low multiplicity of infection (MOI) was used in the assays shown above such that cells were likely to be infected with a single EB. In cells infected with >1 EBs, each EB initially forms a single inclusion which eventually fuses with other EB-containing inclusions within the same cell. The fusion of multiple inclusions is abrogated by small molecule inhibitors of the T3S system and is dependent on IncA which is an effector of the C. trachomatis T3S system [22,23]. Cells were infected with a high MOI of C. trachomatis and then examined for the number of inclusions per infected cell. The majority of wild-type infected cells contained a single inclusion whereas Hri -/- cells contained small multiple inclusions consistent with a defect in IncA-mediated fusion activity (Figure S3 ). Collectively, these data indicate that HRI plays a positive role in the function of the C. trachomatis T3S system.
HRI is required for efficient trafficking of Listeria to the infected cell cytosol
The experiments described above indicate that HRI is required for effectors of the T3S system to gain access to the infected cell cytosol. It was next tested whether HRI regulates the cytosolic access of the Gram-positive pathogen Listeria monocytogenes. The intracellular infection cycle of L. monocytogenes consist of three distinct phases: (i) host cell invasion; (ii) vacuole escape of the bacterium to the cytosol; and (iii) intracytosolic association with actin and subsequent proliferation; each of these events is mediated by well-characterized virulence factors [24]. Previously we showed that, like C. trachomatis noted above, L. monocytogenes invaded cells expressing the non-phosphorylatable eIF2α(S51A) at a much higher level compared to wild-type cells [4]. This finding indicated that L. monocytogenes functionally interacts with the eIF2 signaling pathway during the cellular invasion phase of its infection cycle.
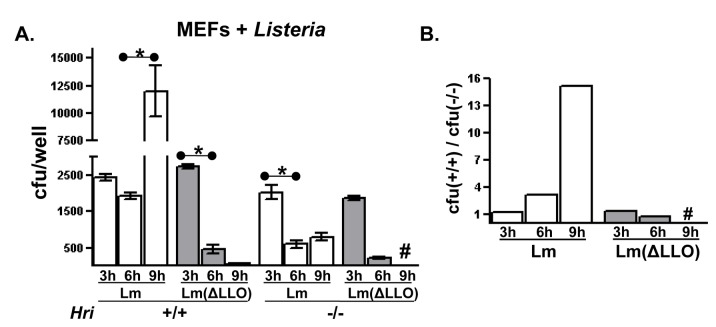
An in vitro infection assay was employed to determine whether HRI regulates the intracellular proliferation of L. monocytogenes. Using this assay the initial invasion phase of the infection does not appear to differ between Hri +/+ and -/- cells (Figure 4A ; compare the ‘3’ hr time points). Following invasion, L . monocytogenese readily proliferates within Hri +/+ cells increasing ~5-fold after an additional 6 hrs of infection. In contrast, there is a relatively rapid and sustained decrease in the number of viable L. monocytogenes recovered from Hri -/- cells (Figure 4A). This latter infection profile (i.e., normal levels of invasion but defective post-invasion proliferation) is similar to that observed in Hri +/+ cells infected with an attenuated L. monocytogenes mutant strain lacking listerolysin O (LLO) (Figure 4A). This mutant strain invades cells but is quickly killed due to its inability to gain access to the cytosol [25]. Additionally, the infection profile of the attenuated LLO mutant strain was similar in Hri +/+ and -/- cells (Figure 4B) indicating that HRI is not required to rapidly eliminate this attenuated strain. These data show that the infection dynamics of virulent L. monocytogenes is greatly impacted by HRI.
Figure 4. Absence of HRI reduces the intracellular proliferation of Listeria .
(A) Cultured Hri +/+ and -/- MEFs were infected with either wild-type L. monocytogenes (Lm) or a strain lacking the virulence factor listeriolysin O (ΔLLO). Following a 1.5 hr attachment and invasion period and a 1.5 hr treatment with an antibiotic to kill non-internalized Lm (3 hr total), internalized Lm were enumerated either immediately (3 hr) or at the indicated time points by lysing the cells and determining the colony forming units (cfus) by plating (Using two-tailed student t test: *, P < 0.05; #, below level of detection). (B) Derived from the data shown in (A), the ratio between cfus recovered from Hri +/+ and -/- cells infected with either Lm or Lm(ΔLLO) at each time point (#, below the level of detection).
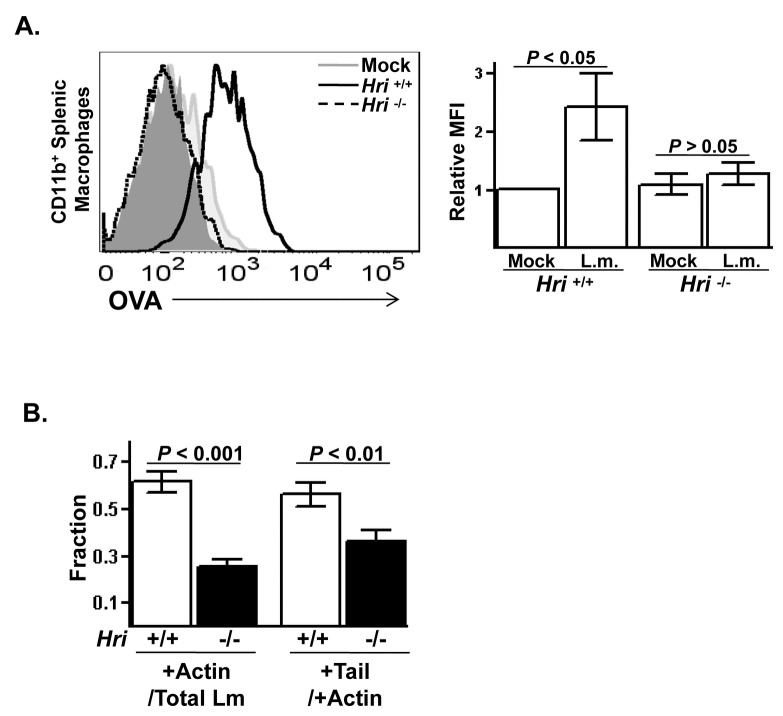
It was then tested whether HRI affects the delivery of Listeria -encoded factors to the cytosol. Splenocytes derived from Hri +/+ and -/- mice were infected in vitro with a L. monocytogenes strain expressing ovalbumin (OVA)-peptide. This peptide is a well-defined antigen that is loaded onto class I MHC molecules in the ER and presented on the surface of infected cells. Bacterial-derived OVA-peptide associated with MHC-I could readily be detected on Hri +/+ macrophages following a brief infection. In contrast, the MHC-I associated with OVA was not detected on Hri -/- macrophages (Figure 5A). These results suggest that Listeria -derived factors failed to access host cell cytosol in the absence of HRI.
Figure 5. Absence of HRI reduces the translocation of Listeria into the cytosol.
(A) Single-cell suspensions of splenocytes were prepared from Hri +/+ and -/- mice and either left uninfected or infected in vitro for 5 hrs with OVA-expressing L. monocytogenes. Histogram displays the levels of OVA surface staining on viable CD11b+-gated macrophages from a representative experiment and the relative median fluorescence intensities (MFI) of 3 separate experiments is shown in the right panel. The isotype control stained cells are shown in the grey filled plot. (B) Unstimulated peritoneal macrophages isolated from Hri +/+ and -/- mice were infected in vitro with GFP-expressing L. monocytogenes for 6 hr and then stained for actin. The fraction of L. monocytogenes (Lm) that were actin associated was determined by dividing the yellow and red-tailed green Lm by the total Lm (Hri +/+: N=418 Lm/168 macrophages; Hri -/-: N=542 Lm/167 macrophages). The fraction of cytosolic Lm that possessed actin tails was determined by dividing the red-tailed green Lm by the total number of yellow and red-tailed green Lm. (P values calculated using student t test.).
Next we directly determined whether HRI is required for L. monocytogenes to gain access to the cytosol. Unstimulated peritoneal macrophages derived from Hri +/+ and -/- mice were infected in vitro with L. monocytogenes. There was no appreciable differences between Hri +/+ and -/- macrophages in the levels of internalized L. monocytogenes following brief infection period indicating that, like for the experiment shown in Figure 4 using fibroblastic cells, the invasion phase of L. monocytogenes infection is not regulated by HRI. However, differences between Hri +/+ and -/- cells started to become evident upon prolonged infection periods. Actin-associated L. monocytogenes (which serves as a marker for cytosolic bacteria) are first observed in Hri +/+ macrophages after 2 hr of infection in contrast to Hri -/- macrophages in which L. monocytogenes is exclusively found in vacuoles. After 6 hr of infection there was a significantly higher fraction of actin-associated L. monocytogenes in Hri +/+ macrophages compared to Hri -/- macrophages (Figure 5B). However, a substantial fraction of the bacteria that did make it to the cytosol in the Hri -/- macrophages were competent to form actin tails (Figure 5B) indicating that this post-escape phase of the infection cycle of L . moncytogenes is not regulated by HRI. These observations indicate that L. monocytogenes requires HRI to efficiently traffic to the cytosol.
To determine how HRI impacts Listeria -host cell interactions over longer infection periods, Hri +/+ and -/- cells were infected for 18 hrs and then stained for intracellular vacuoles and actin. There was a much higher level of general cytotoxicity in the infected Hri +/+ cells as evidenced by a greater number of detached and rounded cells. Among the surviving Hri +/+ cells many contained large numbers of L. monocytogenes (Figure S4 ). Additionally, the vast majority of L. monocytogenes in Hri +/+ cells were within the cytosol. By all measures, Hri -/- cells were much less impacted by long-term infection with L. monocytogenes. There was very little evidence of cytotoxicity in that infected cells maintained their original morphology and a substantial fraction of L. monocytogenes did not co-localized with actin (Figure S4 , right panels). Collectively, three lines of evidence (bacterial proliferation, antigen processing, and direct observations) indicate that HRI specifically regulates the second phase of the L. monocytogenes cellular infection cycle: post-invasion trafficking to the cytosol.
Discussion
Here we demonstrate that HRI positively affects the cell-level infection dynamics of three dissimilar bacterial pathogens. The extracellular pathogen Yersinia , the vacuole-bound pathogen Chlamydia , and the cytosolic pathogen Listeria , all require HRI to efficiently complete their respective cellular infection cycles. A common denominator among these three pathogens is that they all require access to the infected cell cytosol: either for their virulence factors to manipulate host cell processes ( Yersinia and Chlamydia ) or for the bacterium itself to reach the compartment in which it proliferates ( Listeria ).
How could a HRI-mediated process promote pathogen access to the host cytosol? A commonality among the three pathogens used in our studies is that their respective infection cycles are dependent on forming pores in infected host cellular membranes. In Gram-negative pathogens such as Yersinia and Chlamydia , the T3S secrete translocators that assemble pore-forming structures in the host plasma membrane that mediate the transfer of effectors into the cytosol [14,17]. In some respects this process resembles that which occurs in Listeria -infected cells. Following its invasion of the host cell, Listeria secretes monomeric LLO that, following its activation by the host-encoded gamma-interferon-inducible lysosomal thiol reductase (GILT) [26], binds to and oligomerizes into pore-forming structures within the endosomal membrane. In addition to allowing leakage of antimicrobial factors, the resulting pores are also thought to allow the access of co-expressed and secreted phospholipases to the inner leaflet of the endosomal membrane [24,25]. It is possible that one or more of these events occur with reduced efficiency in HRI null cells.
We believe that the activities of HRI described here neither involve it acting as an eIF2α kinase nor otherwise affecting protein synthesis. That T3S secretion is not coupled to host cell protein translation was indicated by our finding that cycloheximide treatment did not affect YopE-mediated disruption of the host cell cytoskeleton. The most compelling data, in our opinion, supporting the model that the infection-specific activities of HRI are independent of its role as a regulator of protein synthesis is the fact that cells lacking the Ser51 residue of eIF2α (the phosphorylation site of HRI and the other eIF2α kinases) are just as competent as wild-type cells in supporting both the T3S activity of Yersinia as well as the T3S-dependent intracellular proliferation of Chlamydia [4]. However, eIF2α(S51A)-expressing cells are more permissive for bacterial invasion indicating that eIF2 signaling does impact the initial events of the pathogen–host cell interaction. For example, Chlamydia is much more efficient at forming inclusions in eIF2α(S51A) cells; however, the number of infectious EBs per inclusion (a measure of intra-vacuolar growth) is comparable between wild-type and eIF2α(S51A) cells [4]. This latter phenotype resembles that observed for PKR null cells (Figure 3) indicating that PKR-mediated eIF2 signaling acts to oppose bacterial invasion but does not affect the subsequent maturation of the Chlamydial inclusion. Our findings are consistent with the observation first noted by Alexander [27] of increased Chlamydial proliferation in cycloheximide-treated cells due to enhanced pathogen invasion.
Owing to its broad activity in promoting the intracellular proliferation of pathogens, HRI may be an excellent target for the development of anti-microbial compounds. HRI is an especially attractive target since it is not required for responses to non-pathogens but interferes with specific virulence mechanisms. Recently, it has been shown that HRI activity can be reduced by either direct targeting with small molecules or indirectly by using natural products that inhibit the HRI-cofactor Hsp90 [28–30]. The targeting of host factors may make it less likely that a pathogen would evolve drug-resistance since the pathogen would not have genetic control over the interaction between the compound and its target.
Supporting Information
The indicated MEFs were infected with Y. pseudotuberculosis at a MOI of 5 for 45 min. Cells were then washed several times, lysed, and the resulting lysates were plated on LB media. Two days later the resulting colonies were enumerated and plotted is the average number of colonies from three independently-infected wells.
(TIFF)
HeLa cells were treated were either treated or not with cycloheximide (25 µg/ml) one hour prior to the addition of Y. pseudotuberculosis as well as for 2 additional hours of infection at which time live cells were imagined.
(TIF)
MEFs were infected at an MOI of 5-10 and visualized 20 hrs post infection. On average one inclusion per cell was detected in Hri +/+ while multiple inclusions per cell routinely occurred in Hri -/- cells.
(TIF)
Hri +/+ and -/- MEFs were infected with GFP-expressing Lm for 18 hrs and then stained for actin (red), nuclei (purple), and vacuoles (pink). Shown in the enlarged images are numerous actin-associated Lm in Hri +/+ cells and in the Hri -/- cells either non-actin associated Lm (green) or Lm that are lightly associated with actin (yellow).
(TIFF)
Acknowledgments
This paper is dedicated to the memory of our co-author and long-standing colleague Roland Rosqvist who died on January 19, 2013. We thank Gregory V. Plano (Miami), Glen Barber (Miami), and Jane-Jane Chen (Harvard) for their generous assistance.
Funding Statement
This work was supported in part by Public Health Service grant AI53459 (KS) from the National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1. Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M (2003) Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem Biol 10: 241-249. doi:10.1016/S1074-5521(03)00046-2. PubMed: 12670538. [DOI] [PubMed] [Google Scholar]
- 2. Keyser P, Elofsson M, Rosell S, Wolf-Watz H (2008) Virulence blockers as alternatives to antibiotics: Type III secretion inhibitors against Gram-negative bacteria J Intern Med 264: 17-29. doi:10.1111/j.1365-2796.2008.01941.x. PubMed: 18393958. [DOI] [PubMed] [Google Scholar]
- 3. Wiley DJ, Shrestha N, Yang J, Atis N, Dayton K et al. (2009) The activities of the Yersinia protein kinase A YpkA) and outer protein J (YopJ) virulence factors converge on an eIF2alpha kinase. J Biol Chem 284:24744-24753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shrestha N, Bahnan W, Wiley DJ, Barber G, Fields KA et al. (2012) Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion J Biol Chem 287: 28738-28744. doi:10.1074/jbc.M112.375915. PubMed: 22761422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen JJ (2007) Regulation of protein synthesis by the heme-regulated eIF2alpha kinase: relevance to anemias. Blood 109: 2693-2699. PubMed: 17110456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Suragani RN, Zachariah RS, Velazquez JG, Liu S, Sun CW et al. (2012) Heme-regulated eIF2α kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood 119:5276-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhan K, Vattem KM, Bauer BN, Dever TE, Chen JJ et al. (2002) Phosphorylation of eukaryotic initiation factor 2 by heme-regulated inhibitor kinase-related protein kinases in Schizosaccharomyces pombe is important for fesistance to environmental stresses. Mol Cell Biol 22: 7134-7146. doi:10.1128/MCB.22.20.7134-7146.2002. PubMed: 12242291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McEwen E, Kedersha N, Song B, Scheuner D, Gilks N et al. (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280: 16925-16933. doi:10.1074/jbc.M412882200. PubMed: 15684421. [DOI] [PubMed] [Google Scholar]
- 9. Han AP, Yu C, Lu L, Fujiwara Y, Browne C et al. (2001) Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J 20:6909-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bölin I, Norlander L, Wolf-Watz H (1982). Temperature-inducible outer membrane protein of Yersinia pseudotuberculosis and Yersinia enterocolitica is associated with the virulence plasmid. Infect Immun 37:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Håkansson S, Galyov EE, Rosqvist R, Wolf-Watz H (1996) The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane Mol Microbiol 20: 593-603. doi:10.1046/j.1365-2958.1996.5251051.x. PubMed: 8736538. [DOI] [PubMed] [Google Scholar]
- 12. Day JB, Ferracci F, Plano GV (2003) Translocation of YopE and YopN into eukaryotic cells by Yersinia pestis yopN, tyeA, sycN, yscB and lcrG deletion mutants measured using a phosphorylatable peptide tag and phosphospecific antibodies. Mol Microbiol 47: 807-823. doi:10.1046/j.1365-2958.2003.03343.x. PubMed: 12535078. [DOI] [PubMed] [Google Scholar]
- 13. Liu S, Suragani RN, Wang F, Han A, Zhao W et al. (2007) The function of heme-regulated eIF2alpha kinase in murine iron homeostasis and macrophage maturation J Clin Invest 117: 3296-3305. doi:10.1172/JCI32084. PubMed: 17932563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Viboud GI, Bliska JB (2005) Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu Rev Microbiol 59: 69-89. doi:10.1146/annurev.micro.59.030804.121320. PubMed: 15847602. [DOI] [PubMed] [Google Scholar]
- 15. Rosqvist R, Forsberg A, Wolf-Watz H (1991) Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun 59: 4562-4569. PubMed: 1937815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Okan NA, Bliska JB, Karzai AW (2006) A Role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLOS Pathog 2: e6. doi:10.1371/journal.ppat.0020006. PubMed: 16450010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Håkansson S, Schesser K, Persson C, Galyov EE, Rosqvist R et al. (1996) The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity EMBO J 15: 5812-5823. PubMed: 8918459. [PMC free article] [PubMed] [Google Scholar]
- 18. Mejía E, Bliska JB, Viboud GI (2008) Yersinia controls type III effector delivery into host cells by modulating Rho activity. PLOS Pathog 4: e3. doi:10.1371/journal.ppat.0040003. PubMed: 18193942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Betts HJ, Wolf K, Fields KA (2009) Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal. Curr Opin Microbiol 12: 81-87. doi:10.1016/j.mib.2008.11.009. PubMed: 19138553. [DOI] [PubMed] [Google Scholar]
- 20. Muschiol S, Bailey L, Gylfe A, Sundin C, Hultenby K et al. (2006) A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis Proc Natl Acad Sci U S A 103: 14566-14571. doi:10.1073/pnas.0606412103. PubMed: 16973741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wolf K, Betts HJ, Chellas-Géry B, Hower S, Linton CN et al. (2006) Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol Microbiol 61: 1543-1555. doi:10.1111/j.1365-2958.2006.05347.x. PubMed: 16968227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Muschiol S, Normark S, Henriques-Normark B, Subtil A (2009) Small molecule inhibitors of the Yersinia type III secretion system impair the development of Chlamydia after entry into host cells. BMC Microbiol 9: e75. doi:10.1186/1471-2180-9-75. PubMed: 19383140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER (1999) The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol 1: 119-130. doi:10.1046/j.1462-5822.1999.00012.x. PubMed: 11207546. [DOI] [PubMed] [Google Scholar]
- 24. Cossart P (2011) Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes Proc Natl Acad Sci U S A 108: 19484-19491. doi:10.1073/pnas.1112371108. PubMed: 22114192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schnupf P, Portnoy DA (2007) Listeriolysin O: A phagosome-specific lysin Microbes Infect 9: 1176-1187. doi:10.1016/j.micinf.2007.05.005. PubMed: 17720603. [DOI] [PubMed] [Google Scholar]
- 26. Singh R, Jamieson A, Cresswell P (2008) GILT is a critical host factor for Listeria monocytogenes infection. Nature 455: 1244-1247. doi:10.1038/nature07344. PubMed: 18815593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alexander JJ (1968) Separation of protein synthesis in meningopneumonitisgent from that in L cells by differential susceptibility to cycloheximide. J Bacteriol 95: 327-332. PubMed: 5640375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rosen MD, Woods CR, Goldberg SD, Hack MD, Bounds AD et al. (2009) Discovery of the first known small-molecule inhibitors of heme-regulated eukaryotic initiation factor 2alpha (HRI) kinase Bioorg Med Chem Lett 19: 6548-6551. doi:10.1016/j.bmcl.2009.10.033. PubMed: 19854648. [DOI] [PubMed] [Google Scholar]
- 29. Zhao H, Brandt GE, Galam L, Matts RL, Blagg BS (2011) Identification and initial SAR of silybin: an Hsp90 inhibitor. Bioorg Med Chem Lett 21: 2659-2664. doi:10.1016/j.bmcl.2010.12.088. PubMed: 21273068. [DOI] [PubMed] [Google Scholar]
- 30. Davenport J, Manjarrez JR, Peterson L, Krumm B, Blagg BS et al. (2011) Gambogic acid, a natural product inhibitor of Hsp90. J Nat Prod 74: 1085-1092. doi:10.1021/np200029q. PubMed: 21486005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The indicated MEFs were infected with Y. pseudotuberculosis at a MOI of 5 for 45 min. Cells were then washed several times, lysed, and the resulting lysates were plated on LB media. Two days later the resulting colonies were enumerated and plotted is the average number of colonies from three independently-infected wells.
(TIFF)
HeLa cells were treated were either treated or not with cycloheximide (25 µg/ml) one hour prior to the addition of Y. pseudotuberculosis as well as for 2 additional hours of infection at which time live cells were imagined.
(TIF)
MEFs were infected at an MOI of 5-10 and visualized 20 hrs post infection. On average one inclusion per cell was detected in Hri +/+ while multiple inclusions per cell routinely occurred in Hri -/- cells.
(TIF)
Hri +/+ and -/- MEFs were infected with GFP-expressing Lm for 18 hrs and then stained for actin (red), nuclei (purple), and vacuoles (pink). Shown in the enlarged images are numerous actin-associated Lm in Hri +/+ cells and in the Hri -/- cells either non-actin associated Lm (green) or Lm that are lightly associated with actin (yellow).
(TIFF)