Abstract
Background
Little is known about the interaction between genetic and behavioral factors during lifecycle risk periods for obesity and how associations vary across race/ethnicity.
Objective
To examine joint associations of adiposity-related single nucleotide polymorphisms (SNPs) and moderate to vigorous physical activity (MVPA) with body mass index (BMI) in a diverse adolescent cohort.
Methods
Using data from the National Longitudinal Study of Adolescent Health (n=8,113: Wave II 1996; ages 12–21, Wave III; ages 18–27), we assessed interactions of 41 well-established SNPs and MVPA with BMI-for-age Z scores in European Americans (EA; n=5,077), African Americans (AA; n=1,736), and Hispanic Americans (HA; n=1,300).
Results
Of 97 assessed we found nominally significant SNP-MVPA interactions on BMI-for-age Z score in EA at GNPDA2 and FTO and in HA at LZTR2/SEC16B. In EA the estimated effect of the FTO risk allele on BMI-for-age Z score was lower (β=−0.13; 95% confidence interval (CI): 0.08, 0.18) in individuals with ≥ 5 versus < 5 (β=0.24; CI: 0.16, 0.32) bouts of MVPA per week (p for interaction 0.02). Race/ethnicity-pooled meta-analysis showed nominally significant interactions for SNPs at TFAP2B, POC5, and LYPLAL1.
Conclusions
High MVPA may attenuate underlying genetic risk for obesity during adolescence, a high-risk period for adult obesity.
Keywords: Genetics, genotype, BMI, physical activity, interaction, adolescence
INTRODUCTION
GWAS have successfully identified more than 41 genetic loci that influence BMI in European descent middle-aged adults [1]. Yet, little is known about the relevance of these variants across race/ethnicity groups, their importance across the life course, or how they interact with environmental and behavioral factors to influence obesity. Adolescence is a high-risk period for the development of adult obesity [2, 3]. Physical activity (PA) is one of the most promising behavioral candidates for preventing and reducing weight gain [4–6]. While there is evidence to support SNP by behavior interactions [7, 8], little is known about whether physical activity mitigates genetic susceptibility to obesity during lifecycle periods with the greatest potential for prevention and treatment.
The FTO locus is a well-studied common obesity variant that was among the first loci to show evidence for gene by PA interaction in obesity studies, with a larger estimated effect of the obesity risk allele at low levels of physical activity [9–12]. The majority of this literature, however, has been limited to middle-aged adult populations of European descent, although there has been work in African- and European-American adults [13], and a meta-analysis of ethnically diverse youth and adults in which PA significantly attenuated the association of FTO with obesity by 27% in adults but not youth [9].
We examined the interaction of 41 well-established obesity susceptibility SNPs with <5 versus ≥ 5 bouts MVPA per week in relation to BMI-for-age Z scores in a nationally representative sample of European American (EA), African American (AA) and Hispanic American (HA) adolescents. We hypothesized that physical activity would attenuate the association between genetic variants associated with BMI. In addition, we hypothesized that the joint role of moderate to vigorous physical activity and established obesity genetic variants on body mass would vary by race/ethnicity.
METHODS
Subjects
National Longitudinal Study of Adolescent Health
The National Longitudinal Study of Adolescent Health (Add Health) is a nationally representative, prospective cohort of ethnically diverse adolescents representative of the U.S. school-based population in grades 7 to 12 (11–22 years of age) in 1994–95 followed into adulthood. Add Health selected a systematic random sample of 80 high schools and 52 middle schools in the United States, stratified to ensure that the schools were representative of US schools with respect to region, urbanicity, school type, percentage of white students, and school size at Wave I (n=20,745)[14]. Respondents were followed through Wave II (n=14,738, 1996), Wave III (2001–2002, n=15,197) and most recently Wave IV (2008–2009, n=15,701), when DNA was first collected from all respondents, and consent given for banking and use in genetic studies (n=12,234). Add Health included a core sample plus subsamples of selected minorities, related adolescents (n=5,524), and other groups, including well-educated African Americans, collected under protocols approved by the Institutional Review Board at the University of North Carolina at Chapel Hill. The survey design and sampling frame have been discussed elsewhere [15–17].
Race/Ethnicity
Since genetic biomarkers to determine ancestry were unavailable, we used a race/ethnicity variable constructed from respondent and parental survey items on ancestral background and family relationship status, creating a race/ethnicity variable with priority for agreement between participant and parental report. We used a three-category classification: non-Hispanic European American (EA), non-Hispanic African American (AA), and Hispanic American (HA). Within HA, we also classified subpopulation: Cuban, Puerto Rican, Central/South Americans, Mexican, or Other Hispanic, as well as non-US born (first generation immigrants), and US-born (2nd or 3rd generation immigrants).
Sibling Relatedness
Add Health oversampled related adolescents (n= 5,524) [18]. Familial relatedness was classified according to participant and parental self-report. Twin zygosity was confirmed by 11 molecular genetic markers [19].
Genetic characterization
The 41 SNPs genotyped in the current study were selected based on 43 SNPs identified in published GWAS for their association with BMI at the time of our study (37 variants); obesity (4 variants), and central adiposity (2 variants) in European descent individuals [20–28].
Genotyping was performed using TaqMan assays and the ABI Prism 7900® Sequence Detection System (Applied Biosystems, Foster City, CA, USA). Sequences for primers and TaqMan probes are available upon request. The genotype call rate ranged from 97.8% to 98.2% and the discordance rate between blind duplicate pairs was (1/277, 0.3%). We excluded SNPs with too few observations (<10) in any combination of genotype by MVPA interaction (EA, n=1, rs11847697; HA, n=4, rs6548238, rs13107325, rs10508503, rs4788102), and (AA, n=8, rs6548238, rs13107325, rs12444979, rs228701, rs98723, rs2605100, rs545854, rs1805081). SNPs that failed tests for Hardy-Weinberg Equilibrium (HWE) within race/ethnicity groups (n=1, rs2922763) were excluded, resulting in 41 SNPs listed in Table S1.
Criteria for generalizability
Across all race/ethnicity groups generalizability was defined as similar direction of effect as reported in the literature (which is primarily European-descent populations) and nominal statistical significance (p<0.05) [29]. Thus, generalizability in the EA subpopulation is straightforward. A recent large GWA study in AA (Monda et al., submitted) suggests that some SNPs identified in EA populations fail to generalize in AA samples either due to limited power or linkage disequilibrium differences resulting in failure to capture the signal of the functional variant [30]. We excluded 14 SNPs with no evidence for generalization in AA (i.e., SNP effect estimates were directionally inconsistent and evidence for association p>0.20 in a large GWAS in African descent samples) (Monda et al., submitted). Given the lack of large GWA studies in Hispanics, all SNPs were considered for HA. In sum, we considered 41 SNPs in EA, 37 in HA, and 19 in AA for this analysis.
Body Mass Index (BMI)
Weight and height were measured during in-home surveys using standardized procedures. BMI (kg/m2) was calculated using measured height and weight assessed at Wave II or III when participants were aged 12–21 years, with priority for younger age at measurement (Wave II: n=14,646) unless the respondent was not seen at Wave II (Wave III: n=785) and was still between the ages of 12–21 years. Self-reported height and weight were substituted for those refusing measurement and/or weighing more than the scale capacity (Wave II n=119; Wave III n=34). Self-reported height and weight were substituted for those refusing measurement and/or weighing more than the scale capacity (Wave II n=119; Wave III n=34). The Add Health self-reported weight and height correlate strongly with measured weight (r=0.95) and height (r=0.94) [22]. Given the age range from 12 to 21 years there is substantial heterogeneity in growth. Thus, we used BMI-for-age Z scores in central analyses and weight- and height-for-age Z scores in sub-analyses.
Moderate-to-vigorous physical activity
Weekly frequency (bouts) of leisure-time MVPA (skating & cycling, exercise, and active sports) were queried at Waves II and III using a standard, interviewer administered activity recall based on validated questionnaires [31]. The questionnaire included activities relevant to adolescents at Wave II and was modified at Wave III to include age-appropriate activities; Wave III bouts were scaled for comparability with Wave II [32]. To explicitly test estimated effects for obesity SNPs relative to the recommendation for MVPA for adolescents at the time of study (i.e., ≥5 bouts of MVPA/wk American Cancer Society 2001), MVPA was dichotomized to high (≥5) versus low (<5) bouts per week using the Wave at which height and weight were measured (i.e. height and weight measured at Wave III then Wave III MVPA data was used).
Analytic sample
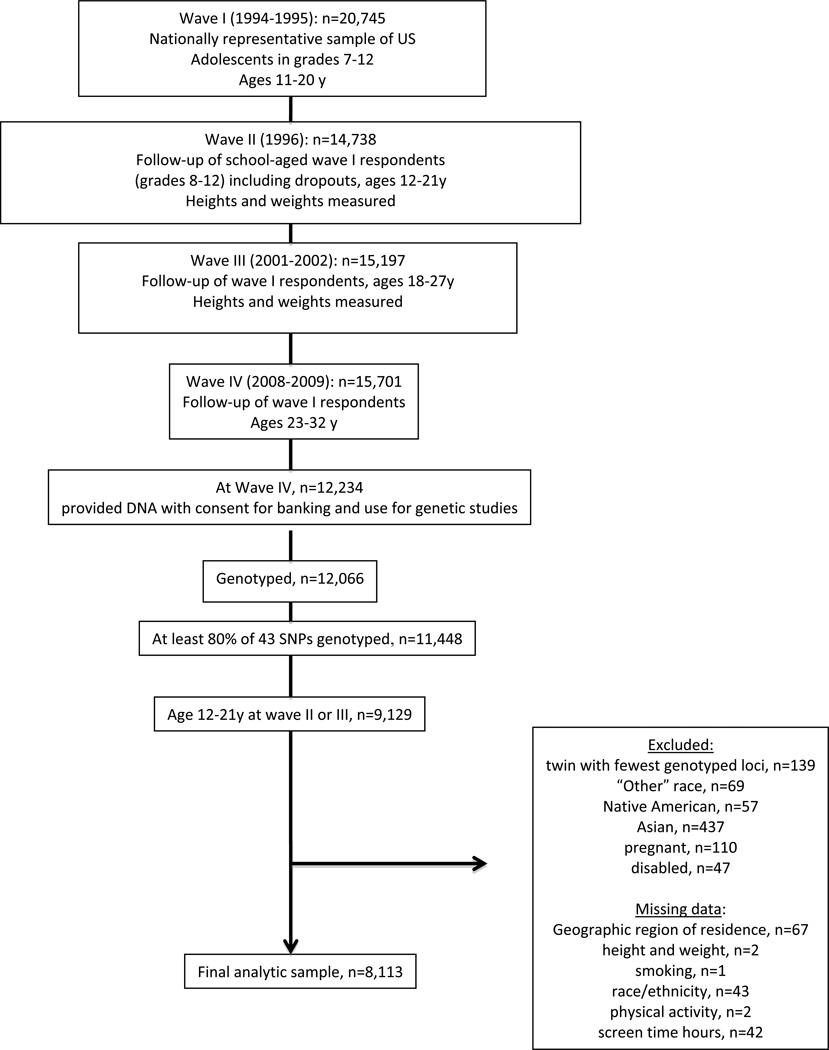
At Wave IV 59% (n=12,234) of the original Wave I (n=20,745) respondents provided samples with consent for banking and use in genetic studies from which DNA was extracted and genotyped (n=12,066). To be eligible for the current study, each individual had to have at least 80% of their 41 SNPs genotyped (n=11,448) and be between the ages of 12 and 21 years at either Waves II or III (n=9,129). Among the 9,129 eligible adolescents, we excluded the following participants: the monozygotic twin with fewer genotyped loci within each twin pair (n=139), individuals of Native American (n=57) or Asian (n=437) or unclassified (n=69) race/ethnicity due to insufficient sample size, pregnant (n=110), disabled (n=47), and those with missing data for: geographic region (n=67), BMI (n=2), current smoking (n=1), race/ethnicity (n=43), MVPA (n=2), or screen time (n=42). The final analytic sample included 8,113 individuals (Figure 1).
Figure 1.
Analysis Sample, derived from The National Longitudinal Study of Adolescent Health
Statistical analysis
Association analyses were conducted using Stata, version 12.1 (Stata Corp, College Station, Texas) and were stratified by race/ethnicity. We used means and proportions to describe the study population. In race/ethnicity-stratified, multivariable models, we controlled for age, sex, current smoking status (≥1 cigarette every day for 30 days), screen time (hours of screen time from television, video, and computer games per week), geographic region, and an indicator for self-reported heights and weights (n=86), with additional controls in AA models: oversampling of highly educated AAs (n=355), and in HA models: Hispanic ancestry: Cuban (n=193), Puerto Rican (n=224), Central/South American (n=120), Mexican (n=660), and other Hispanic (n=103), and foreign born (n= 268). Sample design effects and familial relatedness were accounted for in all multivariable linear models using separate random effects for school and family.
We assessed interactions between MVPA and each of the SNPs (41 in EA, 37 in HA; 19 in AA) using likelihood ratio tests comparing the log likelihoods between the main effects model and the models with a MVPA-by-SNP interaction variable. We also calculated betas for interaction, SNP and MVPA terms from nominally significant (p < 0.05) interaction models as well as risk allele frequencies and estimated main effects of each SNP on BMI-for-age, weight-for-age, and height-for-age Z score. To allow for inference of nominally significant interaction model results, we estimated the main effects of each SNP on BMI-for-age Z score stratified by high versus low MVPA. To test the hypothesis that MVPA may have a more potent attenuation effect in the more overweight subjects, we conducted a sensitivity analysis to determine whether interactions were more robust in the overweight and obese subjects using the (pediatric IOTF cut-off points that correspond to BMI>=25 kg/m2 in adults [33].
In addition, as we expected some effect estimates to be similar across race/ethnicity, we performed a race/ethnicity pooled inverse variance weighted meta-analysis using beta estimates from the models described above, implemented in the program METAL [34]. We included the AA in the meta-analysis to: 1) increase power by including all populations and 2) minimize bias that would occur if only the populations that had an effect were selected.
Multiple testing correction
To correct for multiple testing, we set the statistical significance threshold at α equal to 0.05/number of SNPs tested: 41 in EA; p=0.0012, 19 in AA; p= 0.0026, and 37 in HA; p= 0.0014. However, as this analysis is focused on the estimation of effects for established genetic variants, we interpreted all findings achieving nominal statistical significance.
RESULTS
Add Health includes a racially/ethnically diverse sample of American adolescents, as is reflected in the descriptive characteristics (Table 1). BMI, height, weight, current smoking, and MVPA varied across race/ethnicity, with about 60% of adolescents reporting high MVPA.
Table 1.
Study population characteristics, The National Longitudinal Study of Adolescent Health
| European American | African American | Hispanic American | Total | |
|---|---|---|---|---|
| N | 5,077 | 1,736 | 1,300 | 8,113 |
| Age, mean (SD), y | 16 (1.8) | 16 (1.9) | 17 (1.8) | 16.4 (1.8) |
| Male sex, n (%) | 2,404 (47.4) | 777 (44.8) | 641 (49.3) | 3,822 (47.1) |
| BMI1, mean (SD) | 23.0 (5.0) | 24.2 (5.8) | 23.9 (5.3) | 23.4 (5.3) |
| BMI-for-age Z scores, mean (SD) | 0.30 (1.1) | 0.53 (1.1) | 0.46 (1.1) | 0.38 (1.1) |
| Weight-for-age Z scores, mean (SD) | 0.53 (1.1) | 0.69 (1.1) | 0.42 (1.2) | 0.55 (1.1) |
| Height-for-age Z scores, mean (SD) | 0.43 (1.0) | 0.35 (1.1) | −0.12 (1.1) | 0.33 (1.1) |
| MVPA2, mean (SD) [range] | 6.2 (3.9) [0, 16.5] | 5.6 (3.6) [0, 16.5] | 5.7 (3.8) [0, 16.5] | 6 (3.8) [0, 16.5] |
| High MVPA3, n (%) | 3,324 (63.5) | 1,053 (58.8) | 782 (58.2) | 5,008 (61.7) |
| Screen time, mean (SD), h/wk | 19.1 (16.9) | 27.6 (22.2) | 21.3 (17.8) | 21.3 (18.6) |
| Current smoking, n (%) | 1,420 (27.1) | 166 (9.3) | 216 (16.1) | 1,750 (21.6) |
| Geographic region, n (%) | ||||
| West | 780 (15.4) | 246 (14.2) | 525 (40.4) | 1,551 (19.1) |
| Midwest | 1,862 (36.7) | 332 (19.1) | 96 (7.4) | 2,290 (28.2) |
| South | 1,668 (32.9) | 1,059 (61.0) | 495 (38.1) | 3,222 (39.7) |
| Northeast | 767 (15.1) | 99 (5.7) | 184 (14.2) | 1,050 (12.9) |
Abbreviation: MVPA moderate to vigorous physical activity, BMI Body Mass Index
Unless otherwise indicated, data are presented as number (column percentage).
Calculated as weight in kilograms divided by height in meters squared.
Bouts of MVPA per week
High: 5 or more bouts of MVPA per week
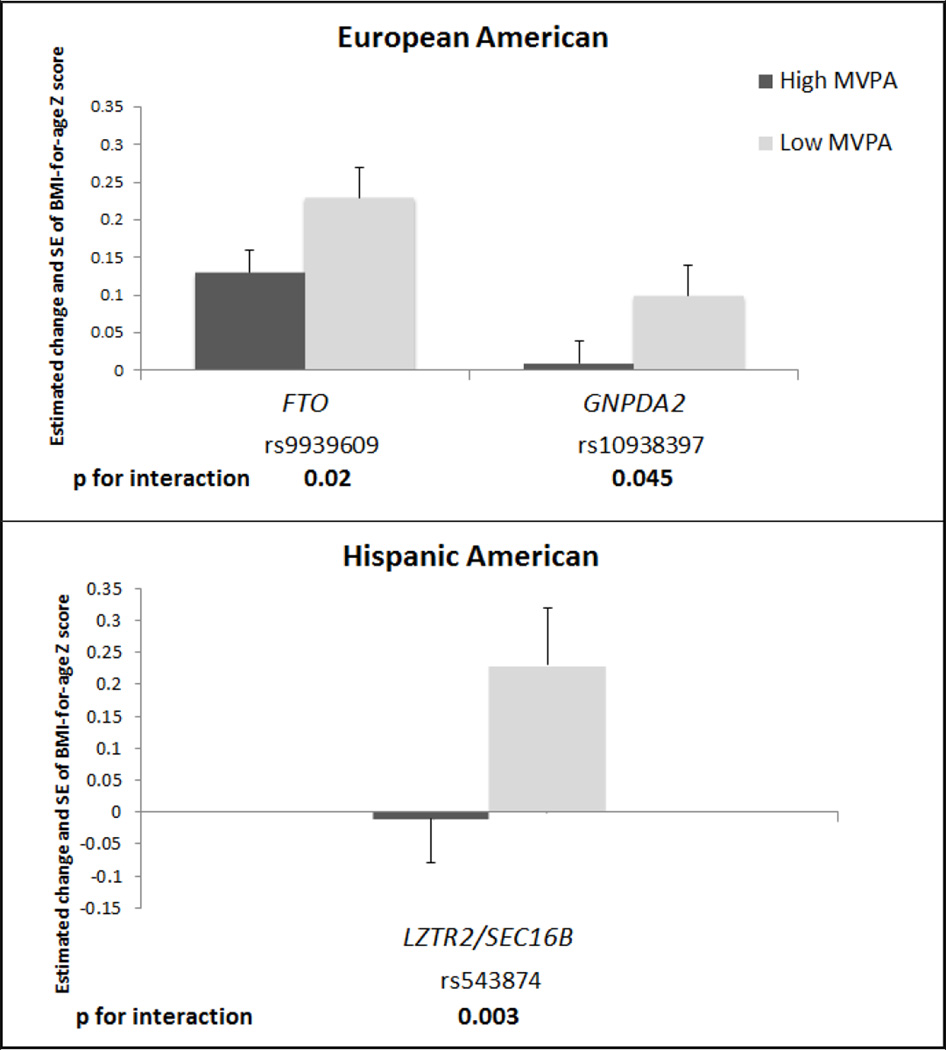
The SNPs considered (41 in EA; 37 in HA; 19 in AA) in the present study are shown in Table S1. Three SNP by MVPA interaction BMI effect estimates were nominally significant (P<0.05) in association with BMI-for-age Z score, with variation by race/ethnicity: two in EA (GNPDA2 (rs10938397) and FTO (rs9939609)), one in HA LZTR2/SEC16B (rs543874) and none in AA (Table 2). None reached significance with Bonferroni correction for the number of SNPs interrogated.
Table 2.
Model results for the three nominally significant interactions (p<0.05)1 between adiposity related loci by MVPA2 in association with BMI-for-age Z scores, by race/ethnicity and main effects on BMI-for-age Z scores
| β (95% CI) | |
|---|---|
| European American | |
| FTO (rs9939609 per copy of A allele) | 0.23 (0.16, 0.30) |
| MVPA (high versus low) | 0.12 (0.02, 0.21) |
| rs9939609 × MVPA | −0.10 (−0.19, 0.21) |
| p for interaction1 | 0.024 |
| GNPDA2 (rs10938397 per copy of G allele) | 0.11 (0.03, 0.18) |
| MVPA (high versus low) | 0.12 (0.02, 0.22) |
| rs10938397 × MVPA | −0.09 (−0.18, −0.00) |
| p for interaction1 | 0.045 |
| Main effects | |
| FTO (rs9939609 per copy of A allele) | 0.16 (0.12,0.21) |
| MVPA (high versus low) | 0.04 (−0.03, −0.10) |
| GNPDA2 (rs10938397 per copy of G allele) | 0.05 (0.00, 0.09) |
| MVPA (high versus low) | 0.04 (−0.02, 0.11) |
| Hispanic American | |
| LZTR2/SEC16B (rs543874 per copy of G allele) | 0.29 (0.13, 0.46) |
| MVPA (high versus low) | 0.02 (−0.13, 0.17) |
| rs5438749 × MVPA | −0.33 (−0.55, −0.11) |
| p for interaction1 | 0.003 |
| Main effects | |
| LZTR2/SEC16B (rs543874 per copy of G allele) | 0.10 (−0.01, 0.21) |
| MVPA (high versus low) | −0.10 (−0.23, 0.03) |
Abbreviations: SNP, single nucleotide polymorphism, BMI, body mass index, MVPA, moderate to vigorous physical activity, CI, confidence interval
Beta estimates are presented for the SNP, MVPA, SNP by MVPA interaction terms, and main effects in the interacted multivariable linear model of adolescent BMI-for-age Z scores regressed on SNP, MVPA, and SNP-by-MVPA interaction, controlling for age, sex, current smoking, screen time, region, indicator for self-reported heights and weights, oversampling of highly educated African Americans (AA stratum only), Hispanic subpopulation ancestry, as well as an indicator for foreign born (HA stratum only). Random effects allowed for individual, family and school with no sample weighting. Models were run separately for each SNP and race/ethnicity. Likelihood ratio tests were used to assess statistical interaction between the main effects (no interaction term) model and the interaction model, separately for each SNP by race/ethnicity. P values corrected for multiple testing are α equal to 0.05/number of SNPs tested (0.0012 in EA and 0.0014 in HA).
High MVPA: 5 or more bouts per week of MVPA, low MVPA: less than 5 bouts per week of MVPA
Allele frequencies (Tables S2) and main effects of SNPs for BMI-for-age Z-scores (Table S3) varied across race/ethnicity. Main effects for weight-for-age and height-for-age Z-scores are shown in Tables S4 and S5. To illustrate the effect of the risk allele in each strata of MVPA, we plotted the main effect of each SNP on BMI-for-age in race/ethnic groups separately in Figure 2. The estimated effect of the risk allele on BMI-for-age Z score was attenuated in individuals with high versus low levels of MVPA (Figure 2). Genotype frequencies are shown in Table S6. For SNP by MVPA associations in the overweight/obese subsample [n= 2,548 (EA: 1,457; AA: 636; HA: 455)] interaction effect sizes were moderately attenuated compared to the central analysis and none were nominally significant (p>0.05) Table (S7).
Figure 2.
Estimated effects1 for BMI-for-age Z score per one copy of risk allele in the 3 SNPs with nominally significant interactions2 by high versus low MVPA3 and across European American and Hispanic American subpopulations
Abbreviation: MVPA moderate to vigorous physical activity, EA, European American, AA, African American, HA, Hispanic American
1 MVPA stratified model: Multi-level model of adolescent BMI-for-age Z score regressed on SNP, controlling for age, sex, current smoking (at least one cigarette every day for 30 days), screen time (hours of screen time from television, video, and computer games per week), region, an indicator for self-reported heights and weights where necessary. Additional race/ethnicity information was controlled for in the non-EA subpopulations: oversampling of highly educated African Americans (AA stratum only), Hispanic subpopulation ancestry, as well as an indicator for foreign born (HA stratum only). Separate random effects allowed for individual, family and school with no sample weighting. Models run separately for each SNP, by high and low MVPA and race/ethnicity. P values corrected for multiple testing are α equal to 0.05/number of SNPs tested (0.0012 in EA and 0.0014 in HA).
2 Likelihood ratio tests were used to assess statistical interaction between the main effects (no interaction term) model and the interaction model, separately for each SNP by race/ethnicity.
3 High MVPA: 5 or more bouts per week of MVPA, low MVPA: less than 5 bouts per week of MVPA
In the race/ethnicity pooled meta-analysis (Table 3) we detected directionally consistent effect estimates for the FTO and GNPDA2 SNPs observed in the EA sample. Two additional SNPs, POC5 (rs2112347) and TFAP2B (rs987237), showed attenuation in BMI-for-age Z score with high MVPA for each copy of the risk allele. An additional SNP, LYPLAL1 (rs2605100), showed higher BMI-for-age Z score with high MVPA for each copy of the risk allele. Findings for FTO and GNPDA2 showed larger estimated effect size than in the EA stratum (Tables 2 and 3).
Table 3.
Model results for the five nominally significant interactions (p<0.05)1 between adiposity related loci by MVPA2 in association with BMI-for-age Z scores in the race/ethnicity pooled meta-analysis sample3
| β (95% CI) | |
|---|---|
| FTO (rs9939609 per copy of A allele) | −0.43 (−0.76, −0.42) |
| p for interaction3 | 0.009 |
| GNPDA2 (rs10938397 per copy of G allele) | −0.42 (−0.08, −0.45) |
| p for interaction3 | 0.01 |
| POC5 (rs2112347 per copy of T allele) | −0.41 (−0.77, −0.35) |
| p for interaction3 | 0.03 |
| LYPLAL1 (rs2605100 per copy of the G allele) | 0.42 (0.80, 0.36) |
| p for interaction3 | 0.03 |
| TFAP2B (rs987237 per copy of G allele) | −0.44 (−0.01, −0.53) |
| p for interaction3 | 0.045 |
Abbreviations: SNP, single nucleotide polymorphism, BMI, body mass index, MVPA, moderate to vigorous physical activity, CI, confidence interval
Beta estimates are presented for the SNP, MVPA and SNP by MVPA interaction terms in the interacted multivariable linear model of adolescent BMI-for-age Z scores regressed on SNP, MVPA, and SNP-by-MVPA interaction, controlling for age, sex, current smoking, screen time, region, indicator for self-reported heights and weights, oversampling of highly educated African Americans (AA stratum only), Hispanic subpopulation ancestry, as well as an indicator for foreign born (HA stratum only). Random effects allowed for individual, family and school with no sample weighting. Models were run separately for each SNP and race/ethnicity. Likelihood ratio tests were used to assess statistical interaction between the main effects (no interaction term) model and the interaction model, separately for each SNP by race/ethnicity. P- value corrected for multiple testing is 0.0012.
High MVPA: 5 or more bouts per week of MVPA, low MVPA: less than 5 bouts per week of MVPA
Beta estimates are presented for the meta-analysis of race/ethnicity pooled sample estimates shown for SNP by MVPA interaction models in METAL [34]. The Z test was used to assess statistical interaction using comparison of SNP by MVPA interaction estimates.
DISCUSSION
In this large and nationally representative sample of US adolescents, we found that high levels of MVPA appear to attenuate the influence of three genetic variants on BMI-for-age Z score during a major risk period for the development of adult obesity [2, 3]. In addition, there was variation in these effect estimates across race/ethnicity. The estimated effect sizes differed between high and low MVPA by approximately 0.2 BMI-for-age Z score units observed across the 2 loci (FTO (rs9939609), GNPDA2 (rs10938397)) in the EA subpopulations.
The different allele frequencies by race/ethnicity reflect the different patterns of population histories in EA, AA, and HA. In general, main SNP effects on BMI-for-age Z scores were very similar in direction and statistical significance as those estimated on BMI across race/ethnicity in a separate analysis [29]. We observed nominally significant (p<0.05) main SNP effects on BMI-for-age Z score for two SNPs (EA, n=1; rs12444979) and (AA, n=1; rs7138803) that did not have nominally significant associations with BMI. Conversely we observed two SNPs (AA, n=1; rs2241423) and (HA, n=1; rs571312) with main effects on BMI-for-age Z score that did have nominally significant associations with BMI. In the race/ethnicity pooled meta-analysis, the increase in BMI-for-age Z score per copy of the FTO, GNPDA2, POC5, and TFAPB2 risk alleles was 0.4 BMI-for-age Z score units greater in individuals with low versus high MVPA. We also observed a nominally significant interaction between MVPA and a SNP in/near LYPLAL1 (associated with WC [24]), although the effect of the BMI increasing allele was heightened in the high MVPA group. While GWAS in populations of European descent have identified several common BMI candidates for adults, little is known about adolescence, a lifecycle period with great potential for obesity prevention and treatment. Even less is known about how obesity susceptibility loci interact with modifiable behaviors, such as MVPA [4–6], which are known to prevent weight gain. The few gene-by-environment interactions investigated in adolescents have mainly interrogated the single FTO locus [9, 35–38]. Although associated variants typically implicate genomic regions rather than individual genes, we note that some of the loci presented in this study underlie positional candidate genes with interesting and established connections to obesity [39–42].
The risk allele of FTO (rs9939609) has been reported to interact with PA [9, 35, 37, 38], possibly increasing weight gain susceptibility through energy balance regulation via the central nervous system through expression in the arcuate nucleus of the hypothalamus [43]. Our findings in the EA subpopulation and in the race/ethnicity pooled meta-analysis support the previously reported FTO by PA association.
We also observed nominally significant interactions (attenuation of BMI in combination with MVPA) in EA for SNPs in or near the GNPDA2 gene that encodes Glucosamine-6-phosphate deaminase, involved in the biosynthesis of glucosamine and expressed in the hypothalamus [44]. In the overweight/obese subsample analysis, the nominally significant SNP by MVPA interactions in the EA and HA lost statistical significance most likely because of lack of power or possibly due to timing of MVPA relative to weight gain, which could have obscured interaction effects. There is clearly a need for additional study of SNP by MVPA interactions in longitudinal samples with longer periods of follow-up to fully understand the nature of the temporal relationships that lead to weight gain.
Our findings may lend further support to the role of neuronal pathways in relation to BMI through energy balance, potentially via appetitive and satiety mechanisms [45]. We observed a nominally significant SNP by MVPA interaction for TFAPB2 (which may play a role in energy balance and inflammation [46]) in association with BMI-for-age Z score in the race/ethnicity pooled meta-analysis. The fact that physical activity can lead to temporary appetite suppression [47], suggests some biologically plausible support for the observed attenuation of obesity risk allele associations by MVPA near the TFAPB2, GNPDA2 and FTO genes.
We also observed a nominally significant SNP by MVPA interaction for the POC5 and LYPLAL1 loci in association with BMI-for-age Z score in the race/ethnicity pooled meta-analysis. LYPLAL1 supports potentially important mitigating pathways related to fatty acid metabolism and glucose uptake by adiponectin [48] and POC5 is a gene necessary for building the distal half of centrioles [49]. In HA, we observed evidence for an interaction between a SNP in or near SEC16B/LZTR2 in relation to BMI-for-age Z score.
Obesity genetics research has largely focused on European descent populations limiting the ability for comparative research in minority populations at greatest risk for obesity [50]. Furthermore, the small number of racially/ethnically diverse cohorts with high quality phenotype, behavior and genetic data prohibits the investigation of gene-by-environment interactions in many minority populations. It is notable, however, that the few gene-by-environment interaction studies that have been conducted in minority populations have observed differential findings by race/ethnicity [35, 48]. One challenge is that different linkage disequilibrium patterns across ancestral populations reduce the ability to detect associations. For example, ancestry specific linkage disequilibrium patterns may underlie the attenuated estimates of well-established FTO obesity related variants originally identified in Europeans in African-descent populations, despite adequate statistical power [51].
We recognize multiple testing is a substantial issue in the analyses of gene-environment interactions. We attempted to limit the impact of this problem by focusing extensive analyses only on those SNPs with well-established main effects in European descent populations as well as SNPs that have been shown to generalize in African ancestry populations. Although this strategy led to fewer statistical tests, it may have introduced some bias. Furthermore, while we interpret the findings of nominally significant interactions as likely to represent significant genotype-by MVPA interactions, another and perhaps more simple explanation may be that there is insufficient power to detect association in the high/or low activity strata. Clearly further independent research is needed to confirm and refine this work.
While our study capitalizes upon a racially/ethnically diverse nationally representative cohort measured during a unique period of the lifecycle, there are limitations to consider. First, the lack of established obesity loci in all race/ethnicity groups is a limitation, particularly in HA, where there have not been large GWAS. As larger genome-wide association and sequencing studies of African-and Hispanic-descent individuals are published, more information will clarify the population differences in the genetic architecture of obesity. Second, we rely on self-reported physical activity behaviors, where objective and precise measures of energy expenditure might have increased power to detect associations of interest. However, there are no large, ethnically diverse population studies with such data. Third, we lack ancestry informative markers to account for population admixture. However, as these loci are well established, our inferences are unlikely to be unduly influenced by cryptic population stratification. Fourth, there may be more complex interactions such as gene-by-gene and/or gene-by-unmeasured environmental influences that might influence BMI, such as dietary patterns. Lastly, in our cross-sectional analysis we are unable to assess temporal relationships with MVPA behavior patterns and BMI change over time. Further longitudinal research is needed. Despite these limitations, our study uses well-established obesity loci to investigate the joint role of MVPA and established obesity variants on BMI during adolescence.
Conclusions
Our findings suggest that higher levels of MVPA may attenuate the influence of obesity susceptibility variants on BMI during adolescence. Whether the observed race/ethnicity differences indicate true differences across populations or limited power to detect effects remains to be seen. As more studies become available, replication of these findings in large, ethnically diverse samples, and broadly across the lifecycle, will be important.
Supplementary Material
What is already known about this subject.
-
-
GWAS have successfully identified numerous genetic loci that influence BMI in European descent middle-aged adults.
-
-
Adolescence is a high-risk period for the development of adult obesity and severe obesity.
-
-
Physical activity (PA) is one of the most promising behavioral candidates for preventing and reducing weight gain, particularly among youth
What this study adds.
-
-
An examination of the joint association between 41 of the well-established obesity susceptibility SNPs with <5 versus ≥ 5 bouts MVPA per week in relation to BMI-for-age Z score in a nationally representative sample of European American, African American and Hispanic American adolescents.
-
-
Three nominally significant interactions (p<0.05) varied by race/ethnicity.
-
-
Overall, the estimated effect of the risk allele on BMI-for-age Z score was greater in individuals with < 5 than those with ≥ 5 bouts MVPA per week.
ACKNOWLEDGMENT
We thank Amy Perou of the BioSpecimen Processing facility and Jason Luo of the Mammalian Genotyping Core at University of North Carolina at Chapel Hill. This work was funded by National Institutes of Health grant R01HD057194. P.G.L., K.E.N., E.L. and A.S.R. contributed to study design, A.S.R. and M.I.G. to data analysis, and A.S.R., K.E.N., and P.G.L. contributed to writing of the manuscript, all other authors provided critical evaluation of the manuscript. A.S.R., K.E.N., and P.G.L. had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. This research uses data from Add Health, a program project directed by Kathleen Mullan Harris and designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill, and funded by grant P01-HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 23 other federal agencies and foundations. Special acknowledgment is due Ronald R. Rindfuss and Barbara Entwisle for assistance in the original design. Information on how to obtain the Add Health data files is available on the Add Health website (http://www.cpc.unc.edu/addhealth). No direct support was received from grant P01-HD31921 for this analysis. We are grateful to the Carolina Population Center (R24 HD050924) for general support.
ABBREVIATIONS
- MVPA
Moderate to Vigorous Physical Activity
- PA
Physical Activity
- SNP
Single Nucleotide Polymorphism
- GWA
Genome-Wide Association
- EA
non-Hispanic European American
- AA
non-Hispanic African American
- HA
Hispanic American
Footnotes
CONFLICT OF INTEREST
There were no potential or real conflicts of financial or personal interest with the financial sponsors of the scientific project.
References
- 1.Hebebrand J, Volckmar AL, Knoll N, Hinney A. Chipping away the 'missing heritability': GIANT steps forward in the molecular elucidation of obesity - but still lots to go. Obes Facts. 2010;3(5):294–303. doi: 10.1159/000321537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The NS, Suchindran C, North KE, Popkin BM, Gordon-Larsen P. Association of adolescent obesity with risk of severe obesity in adulthood. JAMA. 2010;304(18):2042–2047. doi: 10.1001/jama.2010.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh AS, Mulder C, Twisk JW, van Mechelen W, Chinapaw MJ. Tracking of childhood overweight into adulthood: a systematic review of the literature. Obes Rev. 2008;9(5):474–488. doi: 10.1111/j.1467-789X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 4.Hankinson AL, Daviglus ML, Bouchard C, et al. Maintaining a high physical activity level over 20 years and weight gain. JAMA. 2010;304(23):2603–2610. doi: 10.1001/jama.2010.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowler WC, Barrett-Connor E, Fowler SE, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346(6):393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waller K, Kaprio J, Kujala UM. Associations between long-term physical activity, waist circumference and weight gain: a 30-year longitudinal twin study. Int J Obes (Lond) 2008;32(2):353–361. doi: 10.1038/sj.ijo.0803692. [DOI] [PubMed] [Google Scholar]
- 7.Rankinen T, Bouchard C. Gene-physical activity interactions: overview of human studies. Obesity. 2008;16(Suppl 3):S47–S50. doi: 10.1038/oby.2008.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agurs-Collins T, Bouchard C. Gene-nutrition and gene-physical activity interactions in the etiology of obesity. Introduction. Obesity. 2008;16(Suppl 3):S2–S4. doi: 10.1038/oby.2008.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilpelainen TO, Qi L, Brage S, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Medicine. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreasen CH, Stender-Petersen KL, Mogensen MS, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- 11.Rampersaud E, Mitchell BD, Pollin TI, et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med. 2008;168(16):1791–1797. doi: 10.1001/archinte.168.16.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vimaleswaran KS, Li S, Zhao JH, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90(2):425–428. doi: 10.3945/ajcn.2009.27652. [DOI] [PubMed] [Google Scholar]
- 13.Demerath EW, Lutsey PL, Monda KL, et al. Interaction of FTO and physical activity level on adiposity in African-American and European-American adults: the ARIC study. Obesity (Silver Spring) 2011;19(9):1866–1872. doi: 10.1038/oby.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris KM, Halpern CT, Whitsel E, et al. The National Longitudinal Study of Adolescent Health: Research Design. 2009 Available from: http://www.cpc.unc.edu/projects/addhealth/design. [Google Scholar]
- 15.Miller WC, Ford CA, Morris M, et al. Prevalence of chlamydial and gonococcal infections among young adults in the United States. JAMA. 2004;291(18):2229–2236. doi: 10.1001/jama.291.18.2229. [DOI] [PubMed] [Google Scholar]
- 16.Resnick MD, Bearman PS, Blum RW, et al. Protecting adolescents from harm. Findings from the National Longitudinal Study on Adolescent Health. JAMA. 1997;278(10):823–832. doi: 10.1001/jama.278.10.823. [DOI] [PubMed] [Google Scholar]
- 17.Harris KM. An integrative approach to health. Demography. 2010;47(1):1–22. doi: 10.1353/dem.0.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris KM, Halpern CT, Smolen A, Haberstick BC. The National Longitudinal Study of Adolescent Health (Add Health) twin data. Twin Res Hum Genet. 2006;9(6):988–997. doi: 10.1375/183242706779462787. [DOI] [PubMed] [Google Scholar]
- 19.The Add Health Biomarker Team. Biomarkers in Wave III of the Add Health Study. 1999 [Google Scholar]
- 20.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 21.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41(1):25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindgren CM, Heid IM, Randall JC, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyre D, Delplanque J, Chevre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 26.Okada Y, Kubo M, Ohmiya H, et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet. 2012;44(3):302–306. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen W, Cho YS, Zheng W, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44(3):307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Grant SF. Genetics of childhood obesity. J Obes. 2011;2011:845148. doi: 10.1155/2011/845148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graff M, North KE, Mohlke KL, et al. Estimation of genetic effects on BMI during adolescence in an ethnically diverse cohort: The National Longitudinal Study of Adolescent Health. Nutr Diabetes. 2012;2:e47. doi: 10.1038/nutd.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang SJ, Chiang CW, Palmer CD, et al. Genome-wide association of anthropometric traits in African- and African-derived populations. Hum Mol Genet. 2010;19(13):2725–2738. doi: 10.1093/hmg/ddq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sallis JF, Strikmiller PK, Harsha DW, et al. Validation of interviewer- and self-administered physical activity checklists for fifth grade students. Med Sci Sports Exerc. 1996;28(7):840–851. doi: 10.1097/00005768-199607000-00011. [DOI] [PubMed] [Google Scholar]
- 32.Willett WC. Nutritional Epidemiology. 2nd ed. New York: Oxford University Press; 1998. [Google Scholar]
- 33.Cole TJ, Lobstein T. Extended international (IOTF) body mass index cut-offs for thinness, overweight and obesity. Pediatr Obes. 2012;7(4):284–294. doi: 10.1111/j.2047-6310.2012.00064.x. [DOI] [PubMed] [Google Scholar]
- 34.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmad T, Lee IM, Pare G, et al. Lifestyle interaction with fat mass and obesity-associated (FTO) genotype and risk of obesity in apparently healthy U.S. women. Diabetes Care. 2011;34(3):675–680. doi: 10.2337/dc10-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallman DM, Friedel VC, Eissa MA, et al. The association of variants in the FTO gene with longitudinal body mass index profiles in non-Hispanic white children and adolescents. Int J Obes (Lond) 2012;36(1):61–68. doi: 10.1038/ijo.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz JR, Labayen I, Ortega FB, et al. Attenuation of the effect of the FTO rs9939609 polymorphism on total and central body fat by physical activity in adolescents: the HELENA study. Arch Pediatr Adolesc Med. 2010;164(4):328–333. doi: 10.1001/archpediatrics.2010.29. [DOI] [PubMed] [Google Scholar]
- 38.Scott RA, Bailey ME, Moran CN, et al. FTO genotype and adiposity in children: physical activity levels influence the effect of the risk genotype in adolescent males. Eur J Hum Genet. 2010;18(12):1339–1343. doi: 10.1038/ejhg.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loos RJ. Recent progress in the genetics of common obesity. Br J Clin Pharmacol. 2009;68(6):811–829. doi: 10.1111/j.1365-2125.2009.03523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volckmar AL, Bolze F, Jarick I, et al. Mutation screen in the GWAS derived obesity gene SH2B1 including functional analyses of detected variants. BMC Med Genomics. 2012;5(1):65. doi: 10.1186/1755-8794-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy RA, Nalls MA, Keller M, et al. Candidate Gene Association Study of BMI-Related Loci, Weight, and Adiposity in Old Age. J Gerontol A Biol Sci Med Sci. 2012 doi: 10.1093/gerona/gls227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wing MR, Ziegler J, Langefeld CD, et al. Analysis of FTO gene variants with measures of obesity and glucose homeostasis in the IRAS Family Study. Hum Genet. 2009;125(5–6):615–626. doi: 10.1007/s00439-009-0656-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmid PM, Heid IM, Buechler C, et al. Expression of fourteen novel obesity-related genes in Zucker diabetic fatty rats. Cardiovasc Diabetol. 2012;11(1):48. doi: 10.1186/1475-2840-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llewellyn CH, van Jaarsveld CH, Plomin R, Fisher A, Wardle J. Inherited behavioral susceptibility to adiposity in infancy: a multivariate genetic analysis of appetite and weight in the Gemini birth cohort. Am J Clin Nutr. 2012;95(3):633–639. doi: 10.3945/ajcn.111.023671. [DOI] [PubMed] [Google Scholar]
- 46.Ugi S, Nishio Y, Yamamoto H, et al. Relation of the expression of transcriptional factor TFAP2B to that of adipokines in subcutaneous and omental adipose tissues. Obesity. 2010;18(7):1277–1282. doi: 10.1038/oby.2009.442. [DOI] [PubMed] [Google Scholar]
- 47.Martins C, Morgan L, Truby H. A review of the effects of exercise on appetite regulation: an obesity perspective. Int J Obes (Lond) 2008;32(9):1337–1347. doi: 10.1038/ijo.2008.98. [DOI] [PubMed] [Google Scholar]
- 48.Edwards TL, Velez Edwards DR, Villegas R, et al. HTR1B, ADIPOR1, PPARGC1A, and CYP19A1 and obesity in a cohort of Caucasians and African Americans: an evaluation of gene-environment interactions and candidate genes. Am. J. Epidemiol. 2012;175(1):11–21. doi: 10.1093/aje/kwr272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azimzadeh J, Hergert P, Delouvee A, et al. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol. 2009;185(1):101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999–2008. JAMA. 2010;303(3):235–241. doi: 10.1001/jama.2009.2014. [DOI] [PubMed] [Google Scholar]
- 51.Bressler J, Kao WH, Pankow JS, Boerwinkle E. Risk of type 2 diabetes and obesity is differentially associated with variation in FTO in whites and African-Americans in the ARIC study. PLoS One. 2010;5(5):e10521. doi: 10.1371/journal.pone.0010521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.