Abstract
Purpose
Optimal triage of patients at risk of critical illness requires accurate risk prediction, yet little data exists on the performance criteria required of a potential biomarker to be clinically useful.
Materials and Methods
We studied an adult cohort of non-arrest, non-trauma emergency medical services encounters transported to a hospital from 2002–2006. We simulated hypothetical biomarkers increasingly associated with critical illness during hospitalization, and determined the biomarker strength and sample size necessary to improve risk classification beyond a best clinical model.
Results
Of 57,647 encounters, 3,121 (5.4%) were hospitalized with critical illness and 54,526 (94.6%) without critical illness. The addition of a moderate strength biomarker (odds ratio=3.0 for critical illness) to a clinical model improved discrimination (c-statistic 0.85 vs. 0.8, p<0.01), reclassification (net reclassification improvement=0.15, 95%CI: 0.13,0.18), and increased the proportion of cases in the highest risk categoryby+8.6% (95%CI: 7.5,10.8%). Introducing correlation between the biomarker and physiological variables in the clinical risk score did not modify the results. Statistically significant changes in net reclassification required a sample size of at least 1000 subjects.
Conclusions
Clinical models for triage of critical illness could be significantly improved by incorporating biomarkers, yet, substantial sample sizes and biomarker strength may be required.
Keywords: Biomarker, simulation, sample size, reclassification
INTRODUCTION
Over 4 million patients receive intensive care each year, and broad variation exists in critical care delivery. As such, the Institute of Medicine and multiple critical care professional societies called for a coordinated system of emergency and critical care. Similar to trauma care, one approach to improve coordination is to identify and systematically triage highest-risk patients to a higher level of care. This system of tiered regionalization could improve survival of high-risk patients while reducing costs.
The first challenge to effective regionalization is the accurate identification of patients who are most likely to benefit from triage to referral centers. Evidence suggests that emergency medical services (EMS) personnel could play a key role as well as clinicians evaluating new patients in the emergency department. Yet, critical illness risk prediction tools during emergency care of non-trauma patients demonstrate imperfect discrimination, as they would redistribute many patients without critical illness to regional centers while assigning high-risk patients with critical illness to hospitals without critical care resources. Without better tools, emergency care providers may inappropriately allocate thousands of low risk patients to referral centers while still overlooking patients at greatest risk.
The complex diagnoses and overlapping mechanisms of disease leading to critical illness among the non-injured are unlikely captured by clinical data alone. This fact is well recognized in the hospital, where clinicians often combine biomarkers with clinical data to improve assessments of risk and guide treatment. Now, as many biomarkers are measured using point-of-care platforms, the potential to move biomarker measurement to the forefront of emergency care - the prehospital phase - is close to reality. And yet, little empiric data helps guide which biomarkers could be most helpful or how they may improve classification beyond easily-measured clinical data. Nor has existing work employed state-of-the-art methods to measure incremental benefit for candidate markers.
In this study, we determined how strongly associated with outcome a biomarker must be in order to meaningfully improve classification of critical illness risk compared to clinical data alone. We hypothesized that in-silico biomarkers that are strongly associated with critical illness would provide incremental benefit over clinical data alone and that large studies would be needed to definitively document their value.
MATERIALS &METHODS
Conceptual approach
We sought to determine the strength of a biomarker necessary to meaningfully impact classification of emergency patients as high or low risk for critical illness. Emergency care personnel routinely combine physiological measurements (e.g. heart rate and blood pressure) with diagnostic aids (such as electrocardiograms) to make these critical triage decisions. In fact, physiologic measurements, diagnostic aids, or traditional blood tests could all be considered “biomarkers” of critical illness. We based our approach on a conceptual model where biomarkers could either improve triage accuracy by capturing otherwise unmeasured differences in inflammation and organ function, or only marginally improve triage accuracy if they are simply correlated with more easily measured clinical variables. We chose to study a cohort of EMS records, as the clinical data available during prehospital care is similar to initial, clinical exams for patients triaged at emergency department (ED) arrival.
Study Design, Setting, and Patients
We studied a population-based cohort of all adult, non- cardiac arrest, non-trauma EMS encounters in King County, Washington between 2002 and 2006. EMS records were linked to hospital discharge data to determine patient outcomes using a hierarchical, deterministic matching procedure. The details of cohort construction, data linking, quality assessments, and data abstraction are previously described. We restricted our analysis to patients in the validation cohort of the parent study (N=57,647) to reduce computational burden.
Primary outcomes and clinical variables
Our primary outcome was whether critical illness occurred anytime during hospitalization, defined as the presence of severe sepsis, delivery of mechanical ventilation, or death (heretofore referred to as “cases”). Hospitalizations without critical illness are termed “controls.” The components of our primary outcome were derived from hospitalization data including ICD-9-CM diagnosis and procedure codes, revenue codes, and discharge disposition for all hospitalized EMS encounters. The dataset also contains detailed demographics, incident characteristics, and initial prehospital vital signs.
Predicting critical illness risk with clinical data
For each EMS encounter, we calculated the predicted risk of critical illness using a multivariable logistic regression model including eight clinical variables from our previously published model: age, gender, heart rate, respiratory rate, systolic blood pressure, Glasgow Coma Scale score, pulse oximetry, and prehospital location (i.e. nursing home versus other location). We parameterized clinical variables as previously published in clinically relevant categories. We then grouped the predicted risk of critical illness for each EMS encounter using a priori categories: low (<0.05), intermediate (0.05–0.20), or high (>0.20).
Simulation procedure
We imagined a suite of biomarkers, among which the association between biomarker(s) and critical illness was variable. We began by informing the characteristics of these biomarkers using whole blood lactate, a well-studied, prognostic marker in critical illness. We identified the mean and standard deviation of lactate reported for patients with critical illness (4.0 ± 2.6) and without critical illness (2.5 ± 2.0 mmol/L) during emergency care. We used these parameters to simulate log normal random variables for cases and controls, respectively. We log-transformed all biomarker data and determined the unadjusted associations between the biomarker and observed critical illness in logistic regression models. No lactate measurements were prospectively collected in this dataset, and we used lactate characteristics to build in silico biomarker distributions.
We simulated many biomarker distributions (Additional File, Figure E1), each with an increasing association with critical illness (e.g. odds ratio ranging from 1.5 to 6.0). These odds ratios (OR) correspond to the increase in odds of critical illness for a one-unit change in the natural log of the biomarker. We show a crosswalk of biomarker OR with individual marker area under the receiver operating characteristic curve (AUCs) in the Additional File (Table E1). Because biomarkers likely correlate with clinical variables in our triage model, we also simulated biomarker data that correlated with initial prehospital systolic blood pressure (ρ = −0.2, Additional File Figure E2). Further detail of the derivation and assumptions of the biomarker distributions are provided in the Additional File: Supplemental Methods.
Primary data analysis
We added each simulated biomarker to the clinical prediction variables and refit the model. We assessed model performance in three ways: 1) overall discrimination and calibration, 2) risk reclassification tables, and 3) changes in proportions of cases and controls classified to the three risk categories.
First, we determined overall performance of models using the area under the receiver operating characteristic curve (AUC). We calculated the integrated discrimination improvement (IDI), an additional measure of the improved prediction performance gained with the biomarker when added to a clinical model. If the biomarker improves prediction, the IDI will be positive, while the IDI for a null biomarker is zero. Bootstrapped confidence intervals for the IDI which do not include zero are considered significant evidence of improvement in prediction. To assess calibration (model fit)we used a plot of observed vs. expected risk of critical illness over deciles of model based risk values.
Second, we evaluated risk reclassification using 3 × 3 tables of patients grouped by their risk for critical illness (low (<5%), intermediate (5–20%), high risk (>20%)). Risk reclassification tables cross-classify patients according to their risks of critical illness calculated according to the clinical model with and without inclusion of the biomarker. They illustrate how patients’ risk-group changes when the biomarker is included in the clinical model. Reclassification can be summarized using the net reclassification index (NRI), calculated as the sum of the net proportion of cases reclassified to a higher risk category and the net proportion of controls reclassified to a lower risk category. A marker that improves classification will have a positive NRI, while the NRI for a null marker is zero. We calculate the overall NRI, and for cases and controls separately. Reclassification can also be evaluated on the continuous scale, which avoids defining a priori risk groups. We illustrate individuals’ risk before and after addition of the biomarker(s) to the clinical model using scatter plots, and summarize changes with the continuous NRI.
We repeated these steps using biomarker data that included moderate correlation between the biomarker and systolic blood pressure. We also report results when using smaller cohorts (N=500, 1,000, 2,000, and 10,000 subjects) in order to determine sample sizes necessary to obtain statistical significance. We also tested how our results were sensitive to changes in risk categories (alternative parameterization: <10%, 10–50%, >50%). Further detail of performance and risk reclassification measures are provided in the online data supplement. We report 95% confidence intervals using bootstrap with 1000 replications. Tests of significance used a two-sided p-value with the comparison alpha error set at 0.05. The Institutional Review Boards for the Washington State Department of Health, King County Emergency Medical Services, and the University of Washington approved our study. A waiver of informed consent and HIPAA authorization was granted for this study of existing data. All analyses used STATA v11.0 (College Station, TX).
RESULTS
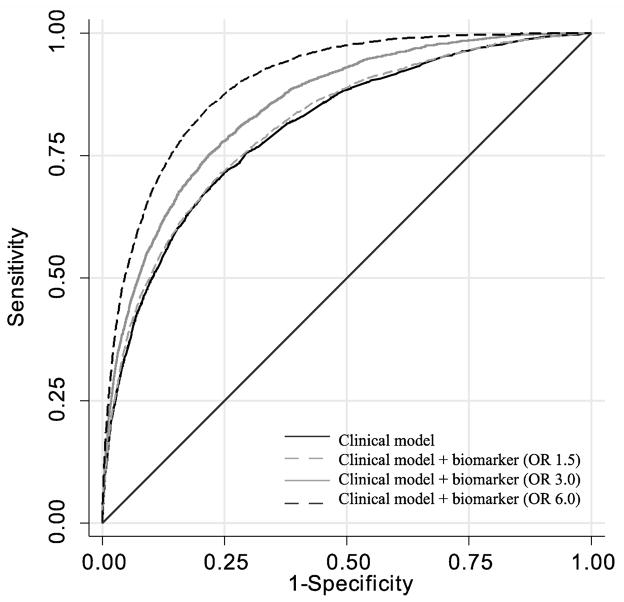
Of 57,647 encounters, 3,121 (5.4%) were hospitalized with critical illness (cases) and 54,526 (94.6%) without critical illness (controls). The clinical risk model alone had moderate discrimination between cases and controls (Table 1). When even weakest biomarkers (OR=1.5) were added to the clinical model, we observed statistically significant increases in area under the receiver operating characteristic curve (Figure 1) and integrated discrimination improvement (Table 1). The model discrimination steadily increased when we added stronger biomarkers, while model calibration was unchanged (Additional File, Figure E3).
Table 1.
Overall performance measures for the clinical model and models that include a biomarker of increasing strength.^
| Odds ratio (OR)of biomarker | Area under the ROC curve# | Integrated discrimination improvement (95%CI) |
|---|---|---|
| Clinical model | 0.800 (0.795, 0.811) | . |
| 1.5 | 0.812 (0.804, 0.820) | 0.007 (0.005, 0.009) |
| 2.0 | 0.823 (0.817, 0.831) | 0.019 (0.016, 0.022) |
| 2.5 | 0.834 (0.827, 0.842) | 0.029 (0.025, 0.033) |
| 3.0 | 0.850 (0.840, 0.860) | 0.047 (0.043, 0.053) |
| 3.5 | 0.857 (0.850, 0.864) | 0.056 (0.051, 0.062) |
| 4.0 | 0.873 (0.868, 0.879) | 0.074 (0.068, 0.080) |
| 4.5 | 0.878 (0.873, 0.883) | 0.081 (0.075, 0.088) |
| 5.0 | 0.887 (0.881, 0.892) | 0.095 (0.087, 0.010) |
| 5.5 | 0.890 (0.886, 0.896) | 0.104 (0.097, 0.111) |
| 6.0 | 0.898 (0.892, 0.903) | 0.111 (0.105, 0.119) |
95% confidence intervals for the AUC and IDI derive from the bootstrap (1000 replications)
All tests of the area under the curve (AUC) compare models enhanced with biomarkers vs. clinical model, and were statistically significant (p<0.05), where H0: (AUCenhanced − AUCclinical) = 0
Figure 1.
Receiver operating characteristic curve for clinical model compared to models that include biomarkers of increasing strength. Clinical model with a weak biomarker (OR 1.5) as shown as dashed grey line (OR=1.5), a moderate biomarker (OR=3.0) shown as grey line, and a strong biomarker (OR=6.0) shown as dashed black line.
To determine how strong a biomarker would be to meaningfully reclassify patients, we evaluated risk reclassification tables for cases and controls (example shown in Additional File, Table E2). In general, stronger biomarkers (e.g. greater OR for critical illness) increased the proportion of cases classified as higher risk, while controls were more likely to be classified as lower risk (Figure 2). For example, a biomarker with OR=3.0 increased the proportion of cases classified as high risk by 8.6% (95%CI: 7.5, 10.8%). The same biomarker decreased the proportion of cases classified as low risk by 5.4% (95%CI: 4.1, 7.2%) and intermediate risk by 3.2% (95%CI: 1.1, 5.7%). Among controls, a biomarker of OR=3.0 increased low risk classification by 1.0% (95%CI: 0.1, 2.3%) and reduced intermediate risk classification by 1.5% (95%: 0.6, 2.5%). In the extreme case, the strongest biomarker (OR=6.0) increased the proportion of cases deemed high risk by 20.7% (95%CI: 19, 22.9%), and increased the proportion of controls as classified as low risk by 4.4% (95%CI: 3.4, 5.7%).
Figure 2.
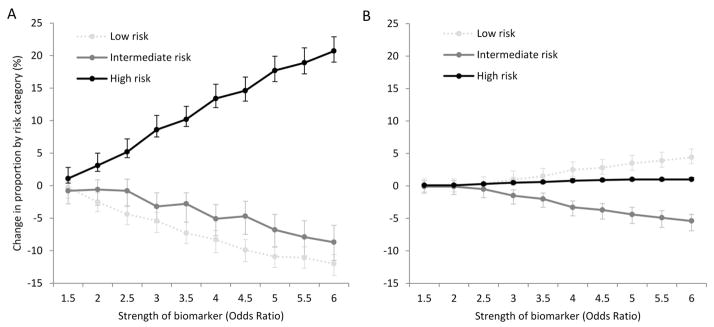
Proportional change in each risk category (low, intermediate, high) for critical illness when adding biomarkers of increasing strength to the clinical triage model. Cases (panel A) and controls (panel B) shown separately.
Interpretive example: When a moderate strength biomarker (OR=3.0) is added to a clinical risk model, the proportion of all cases classified as high risk increases by 9% (panel A), the proportion classified as intermediate risk decreases by 3%, and low risk decreases by 5%. In contrast, the proportion of controls classified as low risk increases by 1% (panel B) with no change in the proportion classified as high risk.
Taken together, we observed statistically significant NRI and NRI for cases when adding markers with OR ≥1.5. Only markers with OR >3.5 had statistically significant NRI among controls (Table 2). Reclassification on the continuous scale was significant even with weak markers (OR=1.5, NRI=0.23, 95%CI: 0.19, 0.26), and improved with marker strength (Figure 3).
Table 2.
Net reclassification results when comparing the clinical model to models with biomarkers of increasing strength (OR ranging from 1.5 to 6.0).
| Odds ratio (OR) of biomarker | Overall NRI (95%CI) | NRI for cases (95% CI) | NRI for controls (95% CI) |
|---|---|---|---|
| 1.5 | 0.014 (0.006, 0.036) | 0.014 (0.005, 0.039) | −0.001 (−0.010, 0.010) |
| 2.0 | 0.054 (0.042, 0.079) | 0.056 (0.041, 0.082) | −0.002 (−0.011, 0.010) |
| 2.5 | 0.095 (0.083, 0.123) | 0.095 (0.081, 0.124) | 0.000 (−0.009, 0.011) |
| 3.0 | 0.145 (0.132, 0.175) | 0.139 (0.123, 0.168) | 0.006 (−0.003, 0.018) |
| 3.5 | 0.182 (0.165, 0.207) | 0.173 (0.154, 0.196) | 0.010 (0.000, 0.021) |
| 4.0 | 0.227 (0.212, 0.259) | 0.210 (0.193, 0.239) | 0.018 (0.008, 0.030) |
| 4.5 | 0.258 (0.239, 0.285) | 0.237 (0.218, 0.261) | 0.021 (0.011, 0.033) |
| 5.0 | 0.300 (0.279, 0.329) | 0.273 (0.254, 0.299) | 0.027 (0.016, 0.039) |
| 5.5 | 0.317 (0.295, 0.343) | 0.285 (0.265, 0.310) | 0.031 (0.021, 0.043) |
| 6.0 | 0.341 (0.320, 0.370) | 0.310 (0.290, 0.330) | 0.040 (0.026, 0.048) |
The 95% confidence intervals for all NRI data derive from the bootstrap (1000 replications).
Interpretive example: Significant reclassification of critical illness risk was observed when including even a weak biomarker in a clinical risk model (OR 1.5, NRI=0.014 (95%CI: 0.006, 0.036)). However, reclassification of controls was significant only when adding a marker with strong association with outcome (OR ≥ 3.5).
Figure 3.
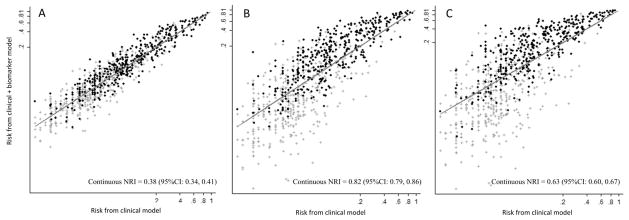
Reclassification plot of the risk of critical illness using a clinical model (X axis) compared to clinical model plus biomarkers of increasing strength (Y axis). Panel A corresponds to a weak biomarker (OR=1.5), Panel B a moderate strength marker (OR=3.0), and Panel C a strong marker (OR=6.0). Cases shown as dark circles and controls grey hashes. As biomarker strength increases, a greater proportion of cases are classified as higher risk (above the line) while more controls are reclassified as lower risk (below the line). All values plotted on the log scale from a random sample of 1000 cases and controls, respectively. Continuous net reclassification index (NRI) with 95% confidence intervals displayed on panel.
When we evaluated our models in smaller cohorts, we found that the statistical significance of the NRI was sensitive to sample size. For example, the overall NRI and NRI for cases was only significant for weak biomarkers when sample size was large (N>10,000), while a moderate biomarker (OR=3.0) required a sample size of at least 1000 subjects (Table 3). For biomarker distributions correlated with systolic blood pressure, we observed no changes to overall performance (Additional File, Table E3), reclassification (Additional File, Table E4), and changes by risk category (Additional File, Figure E4). Finally, we observed that our results were sensitive, in part, to the choice of risk thresholds. At higher risk cutoffs (<10%, 10–50%, >50%), we observed no change in overall reclassification or reclassification of cases, but reclassification of controls was not significantly improved by biomarkers of any strength (Additional file, Table E5).
Table 3.
Net reclassification results when comparing the clinical model to models with biomarkers of increasing strength (OR ranging from 1.5 to 6.0) in random samples of differing size.*
| Sample size | Odds ratio (OR) of biomarker | NRI overall | NRI among cases (95% CI) | NRI among controls (95%CI) |
|---|---|---|---|---|
| 500 | 1.5 | −0.04 (−0.08, 0.15) | −0.05 (−0.08, 0.15) | 0.006 (−0.03, 0.031) |
|
|
||||
| 3.0 | 0.04 (−0.04, 0.25) | 0.05 (−0.05, 0.27) | −0.004 (−0.04, 0.03) | |
|
|
||||
| 6.0 | 0.20 (−0.01, 0.42) | 0.19 (0.00, 0.36) | 0.006 (−0.02, 0.10) | |
|
| ||||
| 1,000 | 1.5 | 0.07 (−0.07, 0.22) | 0.07 (−0.08, 0.25) | 0.004 (−0.04, 0.04) |
|
|
||||
| 3.0 | 0.18 (0.05, 0.36) | 0.21 (0.05, 0.38) | −0.02 (−0.05, 0.05) | |
|
|
||||
| 6.0 | 0.26 (0.07, 0.43) | 0.27 (0.07, 0.43) | −0.02 (−0.04, 0.07) | |
|
| ||||
| 2,000 | 1.5 | 0.09 (−0.02, 0.20) | 0.11 (0.0, 0.20) | −0.02 (−0.04, 0.03) |
|
|
||||
| 3.0 | 0.11 (0.05, 0.26) | 0.13 (0.04, 0.27) | −0.02 (−0.02, 0.05) | |
|
|
||||
| 6.0 | 0.31 (0.20, 0.50) | 0.28 (0.18, 0.44) | 0.03 (−0.004, 0.09) | |
|
| ||||
| 10,000 | 1.5 | 0.02 (−0.02, 0.05) | 0.01 (−0.03, 0.06) | 0.004 (−0.02, 0.02) |
|
|
||||
| 3.0 | 0.16 (0.10, 0.22) | 0.14 (0.09, 0.21) | 0.01 (0.0, 0.03) | |
|
|
||||
| 6.0 | 0.36 (0.29, 0.41) | 0.31 (0.25, 0.37) | 0.04 (0.02, 0.06) | |
Gray boxes indicate statistical significance, defined as 95 % bootstrap confidence interval that does not include 0.0
Interpretive example: In modest samples sizes (N=1,000), we observed significant reclassification of critical illness risk with moderate and strongest biomarkers. Large samples sizes (N>10,000) are required for weak biomarkers (OR=1.5) to significantly improve reclassification.
DISCUSSION
We demonstrate that the addition of biomarkers could significantly improve clinical risk stratification for critical illness during emergency care. Even weak biomarkers reclassified cases at higher risk for critical illness, while only strong biomarkers could reclassify controls as lower risk. We found that our results required large cohort sizes to attain significance, but were robust to correlation between biomarkers and clinical predictors. These estimates inform future design of risk prediction studies during emergency and critical care, and highlight the need for combinations of biomarkers to significantly improve reclassification beyond clinical data.
Our simulations for critical illness risk prediction are broadly consistent with prior investigations in cardiovascular disease, cancer,, and acute lung injury. In these studies, biomarkers that are strongly associated with outcome often improve risk prediction. Though not universally reported, we observed almost linear improvement in reclassification as biomarker strength increased. However, our data uncovered differences in the incremental benefit for patients with and without critical illness. For example, biomarkers with strong associations with outcome (OR > 3.5) were required to move patients without critical illness into lower risk categories, while patients with critical illness were successfully reclassified as higher risk even with weakest markers (OR ≥ 1.5). This finding suggests that future studies testing empirical measurement of biomarkers in the emergency setting may need to study biomarker performance across different strata of baseline risk.
We created theoretical biomarkers that are generalizable to many existing markers in the literature. Although some biomarkers like procalcitonin, C-reactive protein, cardiac troponin, and multiple inflammatory cyto/chemokines have associations with patient outcomes that approach our simulated data, many are not as strongly associated with outcome. This may derive from negative bias resulting from absent or improper adjustment for matching covariates in nested case control studies. More importantly, though, we submit that odds ratio thresholds we study may be best reached using biomarker panels. Many such panels are under preliminary study in emergency care settings. Meanwhile, the growth of molecular phenotyping in critical illness has found multi-marker gene expression profiles with discrimination more similar to our simulated data (Table E1). Because significant cost and feasibility constraints limit biomarker selection when planning prospective studies, our simulations provide a framework for biomarker selection when improvements in either sensitivity or specificity are the desired goals.
Key to future studies of critical illness biomarkers will be adequate sample size. Our estimates were derived using existing data in a cohort much larger than traditional prospective studies. When we randomly sampled from this cohort, we observed that the precision of reclassification estimates were highly sensitive to cohort size. We found that cohorts exceeding 1000 subjects would be required even for studies of moderately strong biomarkers (OR ≥3.0) to attain significant reclassification beyond clinical data. This recipe assumes an outcome prevalence (~ 5%) and clinical model discrimination (AUC~ 0.8) similar to our dataset. Such sample size constraints are achievable, especially in multicenter and consortium studies of critical illness biomarkers.
Our study does not address practical limitations to using biomarkers in emergency critical care, including feasibility of measurement, the challenge of implementing decision support tools in austere clinical environments, or the broader limitations of coordinated critical illness triage. Our study was designed to address the scientific role of hypothetical biomarkers; future work should address their practical role. Second, we used a definition of critical illness which captures high risk patients most likely to require intensive care, yet may exclude some less acute patients admitted to the intensive care unit. Future validation studies of critical illness triage with and without biomarkers will be strengthened using non-administrative, generalizable definitions of critical illness. Third, we observed little impact of correlation between the biomarker and one clinical variable, but our simulation did not specify correlation coefficients between many variables (e.g. biomarker, heart rate, and pulse oximetry). Finally, we acknowledge that alternative risk thresholds would modify our reclassification analysis. We informed our thresholds using prior literature, and observed significant reclassification when analyzing NRI on the continuous scale. Some results in control reclassification were sensitive to our choice of risk threshold, and such category thresholds require validation and consensus across future studies.
CONCLUSIONS
In summary, clinical models for triage of critical illness could be significantly improved, especially by incorporating biomarker measurements. When designing such empirical cohort studies, substantial sample sizes may be required to uncover incremental benefit from candidate biomarkers or biomarker panels.
Supplementary Material
Acknowledgments
Acknowledgement of research support:
This study is supported in part by funding from the National Institutes of Health (KL2 RR025015, CWS; GM054438 & CA129934, MSP)
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Angus DC, Shorr AF, White A, et al. Critical care delivery in the United States: distribution of services and compliance with Leapfrog recommendations. Crit Care Med. 2006;34(4):1016–1024. doi: 10.1097/01.CCM.0000206105.05626.15. [DOI] [PubMed] [Google Scholar]
- 2.IOM. Committee on the Future of Emergency Care in the United States Health System. Washington, D.C: National Academies Press; 2007. Hospital based emergency care: at the breaking point. [Google Scholar]
- 3.Barnato AE, Kahn JM, Rubenfeld GD, et al. Prioritizing the organization and management of intensive care services in the United States: the PrOMIS Conference. Crit Care Med. 2007;35(4):1003–1011. doi: 10.1097/01.CCM.0000259535.06205.B4. [DOI] [PubMed] [Google Scholar]
- 4.Kahn JM, Linde-Zwirble WT, Wunsch H, et al. Potential value of regionalized intensive care for mechanically ventilated medical patients. Am J Respir Crit Care Med. 2008;177(3):285–291. doi: 10.1164/rccm.200708-1214OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seymour CW, Kahn JM, Cooke CR, et al. Prediction of critical illness during out-of-hospital emergency care. JAMA. 2010;304(7):747–754. doi: 10.1001/jama.2010.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wells PS, Anderson DR, Rodger M, et al. Excluding pulmonary embolism at the bedside without diagnostic imaging: management of patients with suspected pulmonary embolism presenting to the emergency department by using a simple clinical model and d-dimer. Ann Intern Med. 2001;135(2):98–107. doi: 10.7326/0003-4819-135-2-200107170-00010. [DOI] [PubMed] [Google Scholar]
- 7.Mills NL, Churchhouse AM, Lee KK, et al. Implementation of a sensitive troponin I assay and risk of recurrent myocardial infarction and death in patients with suspected acute coronary syndrome. JAMA. 2011;305(12):1210–1216. doi: 10.1001/jama.2011.338. [DOI] [PubMed] [Google Scholar]
- 8.Jones AE, Shapiro NI, Trzeciak S, et al. Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: a randomized clinical trial. JAMA. 303(8):739–746. doi: 10.1001/jama.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansen TC, van Bommel J, Mulder PG, et al. The prognostic value of blood lactate levels relative to that of vital signs in the pre-hospital setting: a pilot study. Crit Care. 2008;12(6):R160. doi: 10.1186/cc7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook NR, Ridker PM. Advances in measuring the effect of individual predictors of cardiovascular risk: the role of reclassification measures. Ann Intern Med. 2009;150(11):795–802. doi: 10.7326/0003-4819-150-11-200906020-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aufderheide TP, Hendley GE, Thakur RK, et al. The diagnostic impact of prehospital 12-lead electrocardiography. Ann Emerg Med. 1990;19(11):1280–1287. doi: 10.1016/s0196-0644(05)82288-7. [DOI] [PubMed] [Google Scholar]
- 12.Mikkelsen ME, Miltiades AN, Gaieski DF, et al. Serum lactate is associated with mortality in severe sepsis independent of organ failure and shock. Crit Care Med. 2009;37(5):1670–1677. doi: 10.1097/CCM.0b013e31819fcf68. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen HB, Rivers EP, Knoblich BP, et al. Early lactate clearance is associated with improved outcome in severe sepsis and septic shock. Crit Care Med. 2004;32(8):1637–1642. doi: 10.1097/01.ccm.0000132904.35713.a7. [DOI] [PubMed] [Google Scholar]
- 14.del Portal DA, Shofer F, Mikkelsen ME, et al. Emergency department lactate is associated with mortality in older adults admitted with and without infections. Acad Emerg Med. 2010;17(3):260–268. doi: 10.1111/j.1553-2712.2010.00681.x. [DOI] [PubMed] [Google Scholar]
- 15.Jones AE, Shapiro NI, Trzeciak S, et al. Lactate clearance vs central venous oxygen saturation as goals of early sepsis therapy: a randomized clinical trial. JAMA. 2010;303(8):739–746. doi: 10.1001/jama.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seymour CW, Band RA, Cooke CR, et al. Out-of-hospital characteristics and care of patients with severe sepsis: A cohort study. J Crit Care. 2010;25(4):553–562. doi: 10.1016/j.jcrc.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27(2):157–172. doi: 10.1002/sim.2929. discussion 207–112. [DOI] [PubMed] [Google Scholar]
- 18.Kerr KF, McClelland RL, Brown ER, Lumley T. Evaluating the incremental value of new biomarkers with integrated discrimination improvement. Am J Epidemiol. 2011;174(3):364–374. doi: 10.1093/aje/kwr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zethelius B, Berglund L, Sundstrom J, et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. N Engl J Med. 2008;358(20):2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 20.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98(8):529–534. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 21.Calfee CS, Ware LB, Glidden DV, et al. Use of risk reclassification with multiple biomarkers improves mortality prediction in acute lung injury. Crit Care Med. 2011;39(4):711–717. doi: 10.1097/CCM.0b013e318207ec3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pepe MS, Janes H, Longton G, et al. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159(9):882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 23.Huang DT, Weissfeld LA, Kellum JA, et al. Risk prediction with procalcitonin and clinical rules in community-acquired pneumonia. Ann Emerg Med. 2008;52(1):48–58. e42. doi: 10.1016/j.annemergmed.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaptoge S, Di Angelantonio E, Lowe G, et al. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: an individual participant meta-analysis. Lancet. 2010;375(9709):132–140. doi: 10.1016/S0140-6736(09)61717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrow DA, Cannon CP, Rifai N, et al. Ability of minor elevations of troponins I and T to predict benefit from an early invasive strategy in patients with unstable angina and non-ST elevation myocardial infarction: results from a randomized trial. JAMA. 2001;286(19):2405–2412. doi: 10.1001/jama.286.19.2405. [DOI] [PubMed] [Google Scholar]
- 26.Yende S, D’Angelo G, Kellum JA, et al. Inflammatory markers at hospital discharge predict subsequent mortality after pneumonia and sepsis. Am J Respir Crit Care Med. 2008;177(11):1242–1247. doi: 10.1164/rccm.200712-1777OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ioannidis JP, Panagiotou OA. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA. 2011;305(21):2200–2210. doi: 10.1001/jama.2011.713. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro NI, Trzeciak S, Hollander JE, et al. A prospective, multicenter derivation of a biomarker panel to assess risk of organ dysfunction, shock, and death in emergency department patients with suspected sepsis. Crit Care Med. 2009;37(1):96–104. doi: 10.1097/CCM.0b013e318192fd9d. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland A, Thomas M, Brandon RA, et al. Development and validation of a novel molecular biomarker diagnostic test for the early detection of sepsis. Crit Care. 2011;15(3):R149. doi: 10.1186/cc10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong HR, Cvijanovich NZ, Allen GL, et al. Validation of a gene expression-based subclassification strategy for pediatric septic shock. Crit Care Med. 2011;39(11):2511–2517. doi: 10.1097/CCM.0b013e3182257675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calfee CS, Ware LB, Glidden DV, et al. Use of risk reclassification with multiple biomarkers improves mortality prediction in acute lung injury. Crit Care Med. 2011;39(4):711–717. doi: 10.1097/CCM.0b013e318207ec3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenthal GE, Sirio CA, Shepardson LB, et al. Use of intensive care units for patients with low severity of illness. Arch Intern Med. 1998;158(10):1144–1151. doi: 10.1001/archinte.158.10.1144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.