Abstract
Current international interest in finding alternative sources of energy to the diminishing supplies of fossil fuels has encouraged research efforts in improving biofuel production technologies. In countries which lack sufficient food, the use of sustainable lignocellulosic feedstocks, for the production of bioethanol, is an attractive option. In the pre-treatment of lignocellulosic feedstocks for ethanol production, various chemicals and/or enzymatic processes are employed. These methods generally result in a range of fermentable sugars, which are subjected to microbial fermentation and distillation to produce bioethanol. However, these methods also produce compounds that are inhibitory to the microbial fermentation process. These compounds include products of sugar dehydration and lignin depolymerisation, such as organic acids, derivatised furaldehydes and phenolic acids. These compounds are known to have a severe negative impact on the ethanologenic microorganisms involved in the fermentation process by compromising the integrity of their cell membranes, inhibiting essential enzymes and negatively interact with their DNA/RNA. It is therefore important to understand the molecular mechanisms of these inhibitions, and the mechanisms by which these microorganisms show increased adaptation to such inhibitors. Presented here is a concise overview of the molecular adaptation mechanisms of ethanologenic bacteria in response to lignocellulose-derived inhibitory compounds. These include general stress response and tolerance mechanisms, which are typically those that maintain intracellular pH homeostasis and cell membrane integrity, activation/regulation of global stress responses and inhibitor substrate-specific degradation pathways. We anticipate that understanding these adaptation responses will be essential in the design of 'intelligent' metabolic engineering strategies for the generation of hyper-tolerant fermentation bacteria strains.
Keywords: Fermentation, Bioethanol, Lignocellulosic Inhibitors, Lignocellulolytic materials, Stress Response, Microbial Physiology, Phenolics.
Introduction
Rapid world industrialization has resulted in an overburdening demand for refined fossil fuels. This demand coupled with the continuous rise in cost of refined fossil fuels, their high contribution to greenhouse gas emissions and global warming, pose severe socio-economic challenges 1. There is thus an urgent need for the development of environmentally sustainable and affordable energy sources. One such promising environmentally friendly, affordable and sustainable alternative is bioethanol. One major advantage of bioethanol is that it results in far lower toxic gas emissions in comparison to fossil fuels such as gasoline, diesel and kerosene 2. With environmental protection laws in place in many countries for the implementation of bioethanol as an additive to fuel, the demand for bioethanol is rapidly increasing. Currently, ethanol blended with gasoline (e.g. gashol E5-E10 and gashol E80-85 containing 5-10% and 15-20% ethanol, respectively) is marketed in many developed economies including the USA and several European countries 3.
Presently, most technologies for bioethanol production make use of sugar cane juice and corn starch (1st generation). Around 10-15 billion gallons are currently produced per year, far short of the projected 60 billion gallons that would be required by the world economy 4. Furthermore, 1st generation bioethanol production places an extensive demand on the global food market for which these carbon substrates are destined, and the production costs may be as high as 40% of revenue derived from the bioethanol 5. 2nd generation bioethanol produced from inexpensive, renewable substrates such as lignocellulose is believed to be an attractive, affordable and sustainable alternative.
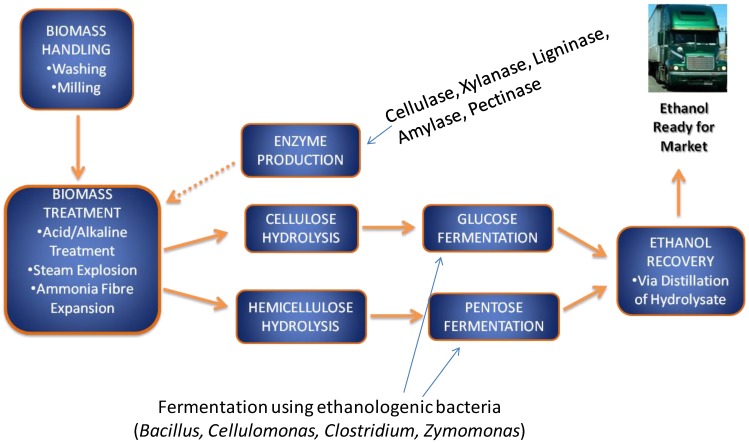
2nd generation bioethanol production involves several sub-processes, including a pre-treatment phase, where cellulosic substrates are extracted from the raw lignocellulosic products, followed by cellulose hydrolysis or hydrolysis into fermentable pentose and hexose sugars. Subsequently these sugars are fermented by ethanologenic microorganisms including bacteria, yeasts and fungi (Figure 1). Ethanologenic bacteria are of particular relevance as they have higher growth rate than fungi, which allow them to produce more fermentative enzyme, and they can utilize both pentose and hexose sugars (few exception such as Zymomonas mobilis), while fungi rarely use pentoses 6-11. An important advantage of some bacterial strain, such as Clostridium thermocellum, is the ability to ferment cellulose directly to ethanol 12, 13, 14. This ability opens an opportunity to use them in the consolidating bioprocessing of biomass to produce ethanol by 1 step (i.e without the need of breaking down the cellulose into its components). The alcohol by-products of microbial fermentation are subsequently distilled and dehydrated to produce an approximate 99.5% ethanol 15, 16, 17, 18.
Fig 1.
A flow chart of lignocellulose biomass conversion to bioethanol. The lignocellulose biomass is degraded into its constituent sugars by various pre-treatment methods and converted into ethanol using ethanologenic bacteria cells, which is distilled and ready for market.
There are several key challenges associated with the current 2nd generation bioethanol production methods that need to be addressed in order to develop sustainable lignocellulolytic material (LCM) bioconversion into ethanol. The key challenge is the development of a robust, sustainable, cost-effective, environmentally friendly and complementary alternative to 1st generation ethanol production and fossil fuels. These include many factors that relate to the microorganisms involved in sugar fermentation, and these may include their capacity to adequately ferment sugars into ethanol, their need for additional nutritional requirements, their sensitivity/tolerance towards ethanol and organic by-products of fermentation, as well as the high temperature and low pH associated with the pre-treatment, cellulose hydrolysis and fermentation phases of the LCM bioconversion process 6, 19-23. Furthermore, one of the major challenges associated with the current biological approach for bioethanol production is that during the pre-treatment and hydrolysis phases a number of by-products are produced that could inhibit the growth and metabolic capacity of ethanologenic microorganisms during the fermentation process 24-27. A major goal of current research relating to LCM bioconversion is the development of effective means to reduce or eliminate the fermentation inhibitors 28. Several detoxification protocols have been proposed and introduced, including physical (e.g. adsorption with activated carbon or ion exchange resins), chemical (e.g. lime or alkali treatment, ionic liquids; mixtures of cationic and anionic salts that melt mostly below 100°C) or biological (e.g. laccase or peroxidise) measures 26, 29-33. However, these methods come at an additional cost and frequently introduce further toxic waste products 30, 34. The screening and selection of microorganisms which are highly tolerant or resistant to fermentation inhibitors may represent a more sustainable and cost-effective strategy 35-38.
In this review, we discuss the fermentation inhibitors, as well as the means by which ethanologenic bacteria may have adapted to tolerate or resist these lignocellulosic inhibitory compounds. Microbial fermentation for ethanol production involves ethanologenic bacteria as well as fungi. Several publications covering and reviewing these topics with respect to ethanologenic fungi are available 6-10, 16- 18, 20, 39- 45. Microbial tolerance to organic solvents, other bio-products and chemicals from biorenewable fuels processes 46-48 were already discussed, however, not much of emphasis was laid on ethanolonogenic bacteria. Hence this review will focus on the effects of and tolerance/adaptation to inhibitors in ethanologenic bacteria, for which reviews are currently limited. Concise descriptions of lignocellulose as a potential source of biomass for bioethanol production and the 2nd generation bioconversion process are given. We discussed the inhibitory compounds generated during the pre-treatment and hydrolysis processes, how they affect the cellular activities of ethanologenic bacteria, as well as the mechanisms by which ethanologenic bacteria may be able to withstand and survive in the presence of these inhibitors. There have been a number of studies which have focused on the improvement of ethanol production through genetic engineering of ethanologenic bacteria. These have largely focused on yield, but not in means by which these bacteria can tolerate or resist the LCM bioconversion inhibitors 7-10. We anticipate that the topics covered in this review will be helpful in the future genetic engineering of ethanologenic bacteria with enhanced tolerance adaptation strategies towards these inhibitory substrates, thus contributing towards improvement of LCM-based ethanol production.
Lignocellulose: A potential feedstock for bioethanol production
Lignocellulose is an abundant natural biopolymer which accounts for 50% of the world's biomass. An estimated 10 - 50 billion tons of lignocelluloses is produced per annum 49. It can be obtained easily and inexpensively from various agricultural by products such as residual materials from grain crops, seeds, peels and shells of fruits and vegetables, vegetable oils, industrial and municipal waste, forestry residues and fast-growing energy grasses and trees 6, 49-51. One of the major advantages of using these wastes for bioethanol production, aside from their renewable nature, will be the lower energy, environmental and economic costs associated with their disposal, as they are still considered as waste products in many parts of the world and often disposed of by burning 52.
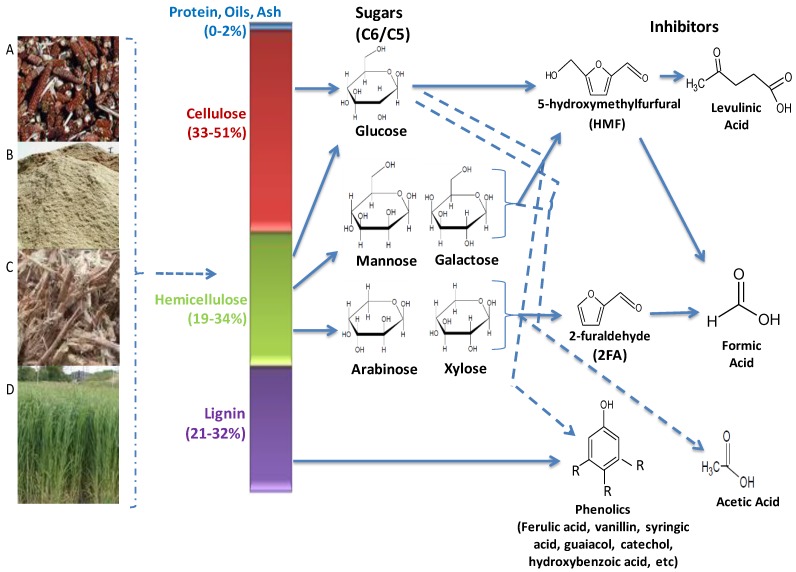
Lignocellulolytic materials (LCMs) consist of three main polymerized sugar components, cellulose, hemicellulose and lignin (Figure 2). Cellulose accounts for 33 - 51%, hemicellulose for 19 - 34% and lignin for 21 - 32% of the dry weight of LCM, while proteins, oils and ash make up the remaining fraction. Cellulose is the most abundant carbohydrate polymer in nature. It is composed of linear chains of hundreds to thousands of β-1,4-linked D- glucose molecules 53. It is closely linked with proteins, lignin, hemicelluloses and mineral elements, which makes it highly resistant to hydrolysis. However, it can be hydrolysed chemically (e.g. by acid treatment) or enzymatically (e.g. microbial cellulases) 39, 54, 55. Cellulose is of high industrial value, being used in the production of various foods, chemicals, textiles animal feeds, pulp and paper 53.
Fig 2.
Main hydrolytic components of lignocellulose biomasses and generated inhibitory compounds. Biomasses are generated from wastes such as (A) maize cobs (B) saw dust (C) sugar cane bagasse (D) fast growing grasses. Pre-treatment processes releases the sugars (C6/C5) and lignin, however these processes also cause the breakdown of lignin and dehydration of the sugars, producing the inhibitory compounds that greatly reduces the overall efficiencies of the ethanologenic cells in the lignocellulose hydrolysate.
Hemicellulose is a branched polysaccharides consisting of pentose (D-xylose and L-arabinose) and hexose (D-glucose, D-mannose, D-galactose) sugars, uronic acids (D-glucouronic, D-galactouronic) and various o-methylated sugars 56. Its composition is primarily dependent on the plant source, and unlike cellulose it is more readily hydrolyzed into its component sugars 56. However, some of these sugars can hamper the fermentation process as many microorganisms cannot utilize them as readily as glucose 57. Hemicellulose is also of high industrial value, as a major source of xylose and xylitol. The latter can be used as artificial sweetener and as antimicrobial agent in foods and other household products 58-60.
Lignin, a complex aromatic macromolecule, forms an integral component of the secondary plant cell wall 47. It is covalently linked with cellulose and hemicellulose and provides structural support to the plant cell, protects it from hydrolytic enzymes and pathogens entry, and plays a major role in the water conductivity of vascular tissues 61. It is formed by the polymerization and dehydration of three monolignols (ρ-coumaryl, coniferyl and sinapyl alcohols) to form phenylpropanoids (ρ-hydroxyphenyl, guaiacyl and syringal lignins, respectively) 62. Products of lignin degradation such as ferulic acid, vanillin, and catechol are used in the production of herbicides, pesticides, plastics, household products, food flavourants, anti-microbial and anti-foaming agents 39, 61, 62.
Inhibitory compounds produced during LCM conversion
The yield and productivity of LCM to ethanol bioconversion is greatly reduced due to the production of cytotoxic-inhibitory compounds generated from lignin degradation and/or sugars dehydration during the pre-treatment processes, which are subsequently released into the hydrolysate along with the fermentable sugars.
The pre-treatment phase is vital for LCM bioconversion. It allows delignification of cellulose-hemicellulose-lignin complex (CHLC) that then allows possible access to hemicellulose and cellulose for subsequent enzymatic depolymerisation into simple sugars (hexoses and pentoses) 19. A number of different pre-treatment methods have been introduced, including acid/alkaline hydrolysis 40, 63, steam/thermal explosion and hot water treatment 41-44 and ammonia fibre expansion 45, 64, 65 (Figure 1).
The type and quantity of these inhibitors is greatly dependent on the source of the biomass since lignin, one of the main sources of these inhibitory compounds, has different structural qualities and degrees of bonding/interaction with cellulose and hemicellulose depending on the plant, or waste LCM resource used for ethanol production 39, 47, 61, 62. Furthermore, the pre-treatment processes utilized as well as fermentation variables (e.g. oxygen concentration, pH of medium, etc.) can exacerbate the toxic effectors of the inhibitors 66. Regardless of the source or LCM preparation methods used, three main inhibitory compounds are produced during the pre-treatment and hydrolysis steps, namely organic acids, furan derivatives and phenolic compounds 27, 30, 67, 68.
Organic acids
Among the major organic acids produced during the pre-treatment and hydrolysis processes are lactate, succinate, formate and acetate 69. The latter, most abundant organic acid generated during pre-treatment and hydrolysis processes, is formed from the dehydration of released sugars and/or decomposition of acetylxylan, a byproduct of hemicellulose degradation (Figure 2) 63, 70.
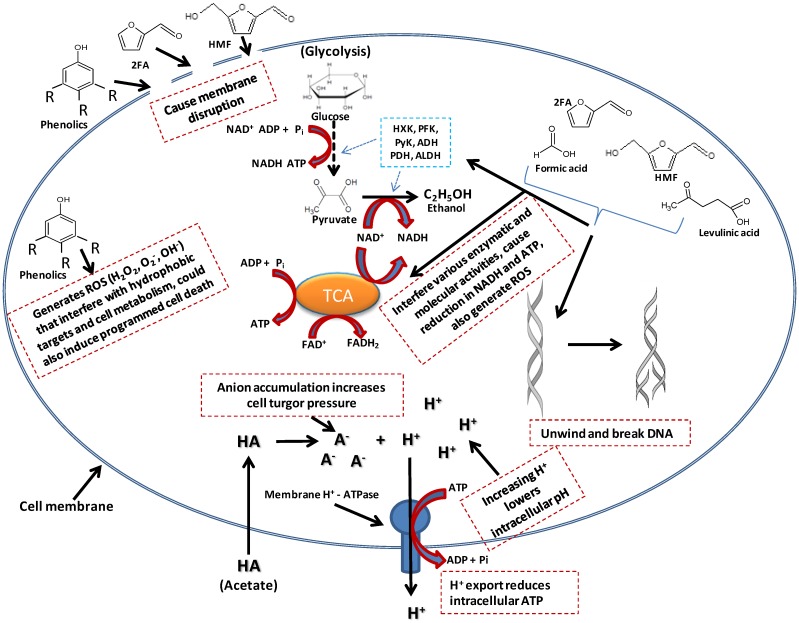
Acetate is liposoluble and therefore diffuses across the bacterial cell plasma membrane and dissociates into its anionic form, releasing protons into the cytoplasm (Figure 3) 71. This result in a drop in the intracellular pH, leading to disruption of the transmembrane pH potential, various damaging anion-specific effects on metabolism, protein/enzyme activity/stability, and higher turgor pressure within the cell 71. The dissociation of this weak acid inside the cytoplasm results from the higher intracellular pH of ethanologenic bacteria (pH = ~7.8) compared to that of acetate (pKa =4.75). While the dissociation of acetate in the cytoplasm has a net negative effect on the microorganismal cell growth and proliferation, it does not lead to a reduction in the production of ethanol 72. The cells tend to rapidly generate ATP in order to maintain the intracellular pH, forcing the microorganisms to switch into anaerobic respiration, consequently generating ethanol at the expense of biomass formation 72. Acetate concentrations as low as 0.5g/L were reported to inhibit Escherichia coli cell growth by 50% in a batch and fermenter culture respectively, but did not result in a reduction in the cell fermentation efficiency 73, 74. This can be linked to the bacterial cells generating more ATP in order to maintain the intracellular pH, forcing the bacteria to switch to anaerobic respiration, thereby generating ethanol, while at the same time exhausting the proton pumping capacity of the cell plasma membrane ATPase, resulting in depletion of the ATP content, dissipation of the proton motive force and acidification of the cytoplasm (Figure 3) 75, 76. The overall effect is a reduction in cell growth and proliferation 30.
Fig 3.
A model of effects of inhibitors presence in ethanologenic bacteria cells. As depicted in the illustration, inhibitory effect could range from membrane disruption, lowering of intracellular pH to interference with lots of cell metabolic targets/pathways.
Furan derivatives
2-furaldehyde (furfural) and 5-hydroxymethylfurfural (HMF) are dehydration products of pentose and hexose sugars, respectively, produced during acid pre-treatment and hydrolysis of LCMs (Figure 1 and 2) 77. The toxicity results from the inhibition of glycolytic and fermentative enzymes essential to central metabolic pathways (such as pyruvate, acetaldehyde and alcohol dehydrogenases) 78, protein-protein cross linking and DNA degradation into single strands (Figure 3) 79-82. Their high hydrophobicity allows furfural and HMF to compromise membrane integrity leading to extensive membrane disruption/leakage, which eventually will cause reduction in cell replication rate, ATP production, and consequently lower ethanol production 83. In-vitro incubation of furfural with double stranded lambda phage DNA led to single-strand breaks, primarily at sequence sites with three or more adenine or thymine bases 79-82. Furan derivatives are furthermore known to act synergistically with other inhibitors including phenolic and aromatic compounds as well as acetic, formic and levulinic acids 30, 83. The latter two acids also result as by-products of the acid degradation of HMF 79. Formic acid is more toxic than levulinic acid due to its smaller molecular size and undissociated form which facilitates its higher membrane permeability. Formic acid was shown to inhibit the synthesis of macromolecules, as well as DNA synthesis and repair 84, 85.
Phenolic compounds
Phenolic compounds formed during the degradation lignin and dehydration of sugars in the pre-treatment and hydrolysis stages are insoluble or partially soluble in the hydrolysate and include acids (ferulic acid, vanillic acids, 4-hydroxybenzoic acid and syringic acid), alcohols (guaiacol, catechol and vanillyl alcohol) and aldehydes (vanillin, syringic aldehyde and 4-hydroxylbenzaldehyde) 61, 62. These compounds are known to partition into biological membranes altering the permeability and lipid/protein ratio, which thus increases cell fluidity, leading to cell membrane disruption, dissipation of proton/ion gradients and compromising the ability of cellular membranes to act as selective barriers 86.This membrane disruption, allows the release of proteins, RNAs, ATP, Ions, out of the cytoplasm, consequently causing reduced ATP levels, diminished proton motive force and impaired protein function and nutrient transport 86. Furthermore, they enhance the generation of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), super oxides (O2-) and super hydroxyl (OH-) that interact with proteins/enzymes, which results in their denaturation, they damage cytoskeleton and other hydrophobic intracellular targets, cause DNA mutagenesis, and induce programmed cell death (Figure 3) 87. Phenolic compounds have been reported to be more toxic, even at low concentrations, than furfural and HMF 88, 89.
While the mechanism and extent of cytotoxicity of lignocellulose inhibitory compounds generally differ, they all result in gross physiological/metabolic changes in the ethanologenic microorganisms which concomitantly result in decreased cell viability and fermentation efficiency. One of the major determining factors of toxicity of these inhibitors is their hydrophobicity potentials. Hydrophobicity shows the extent to which a compound can accumulate in the cell cytoplasm. Table 1 shows the hydrophobicity potentials of the aforementioned inhibitory compounds. Log P, which is the partition coefficient of solvent in an equimolar mixture of octanol and water, is the measure of hydrophobicity (i.e the rate of interaction with non-polar molecules) 90, 91. A high value of Log P is an indication of high hydrophobicity (i.e the coumpound can readily translocate into the cell across the non-polar cell membrane), and as a consequent, the high inhibitory the compound 90-94.
Table 1.
Hydrophobicity potentials of organic acids, furans and phenolics on cell physiology.
| Inhibitor | IUPAC Name | Molecular Formular | Molecular Weight (g/L) | Log P |
|---|---|---|---|---|
| Acetic acid | Acetic acid | C2H4O2 | 60.05 | -0.32 |
| Formic acid | Formic acid | CH2O2 | 46.03 | -0.54 |
| Levulenic acid | 5-hydroxy-5-methyl-2-tetrahydrofuranone | C5H8O3 | 116.12 | 1.34 |
| HMF | 5-(hydroxymethyl)-2-furaldehyde | C6H6O3 | 126.11 | -0.37 |
| 2-furaldehyde | Furan-2-carbaldehyde | C5H4O2 | 96.08 | 0.41 |
| 4-Hydroxybenzoic acid | 4-Hydroxybenzoic acid | C7H6O3 | 138.12 | 1.56 |
| Ferulic acid | (E)-3-(4-hydroxy-3-methoxy-phenyl)prop-2-enoic acid | C10H10O4 | 194.18 | 1.641 |
| Syringic acid | 4-Hydroxy-3,5-dimethoxybenzoic acid | C8H10O5 | 198.17 | 1.129 |
| Vanillic acid | 4-Hydroxy-3-methoxybenzoic acid | C8H8O4 | 168.15 | 1.2014 |
| 4-Hydroxybenzaldehyde | 4-Hydroxybenzaldehyde | C7H6O2 | 122.12 | 1.392 |
| Syringaldehyde | 4-Hydroxy-3,5-dimethoxybenzaldehyde | C9H10O4 | 182.17 | 0.863 |
| Vanillin | 4-Hydroxy-3-methoxybenzaldehyde | C8H8O3 | 152.15 | 1.188 |
| Catechol | Pyrocatechol | C6H6O2 | 110.10 | 0.88 |
| 4-methyl catechol | 4-methyl catechol | C7H8O2 | 124.14 | 1.37 |
| Guaiacol | 2-methoxyphenol | C7H8O2 | 124.14 | 1.32 |
| Vanillyl alcohol | 4-Hydroxy-3-methoxybenzyl alcohol | C8H10O3 | 154.16 | 0.003 |
Log P is the partition coefficient of solvent in an equimolar mixture of octanol and water. It is a measure of hydrophobicity, which is the rate of interaction with non-polar molecules 90, 91. A high value of Log P is an indication of high hydrophobicity, and means the compound can readily translocate into the cell across the non-polar cell membrane, and as a consequent, the high inhibitory the compound. The Log P values of compounds were obtained from search on ChemSpider [Free Chemical Identifier Data Base; http://www.chemspider.com/].
Bacterial LCM bioconversion inhibitor tolerance and adaptation mechanisms
Efficient ethanologenic bacteria must be able to ferment a variety of sugars (pentoses and hexoses) and furthermore be capable of surviving, growing and replicating under the stressful conditions they will encounter during the LCM bioconversion process. This includes the ability to tolerate or adapt to inhibitors introduced during the LCM pre-treatment phases 19, 20, 95, 96. These factors form an integral component in the screening and selection of effective ethanologenic microorganisms, as well as representing important selectable markers in the genetic engineering of ethanologenic microorganisms, be they fungi, yeasts or bacteria. The molecular mechanisms employed by these ethanologenic bacteria to counteract these compounds are still largely unknown. However, a number of specific and global stress response mechanisms have been identified in bacteria which could be used by ethanologenic bacteria to provide tolerance, resistance or protection from the many of the above-mentioned fermentation inhibitors and these will be discussed below. These include mechanisms for the maintenance of pH homeostasis and cell membrane integrity, the activation of global stress responses and inhibitor degradation.
Maintenance of pH homeostasis
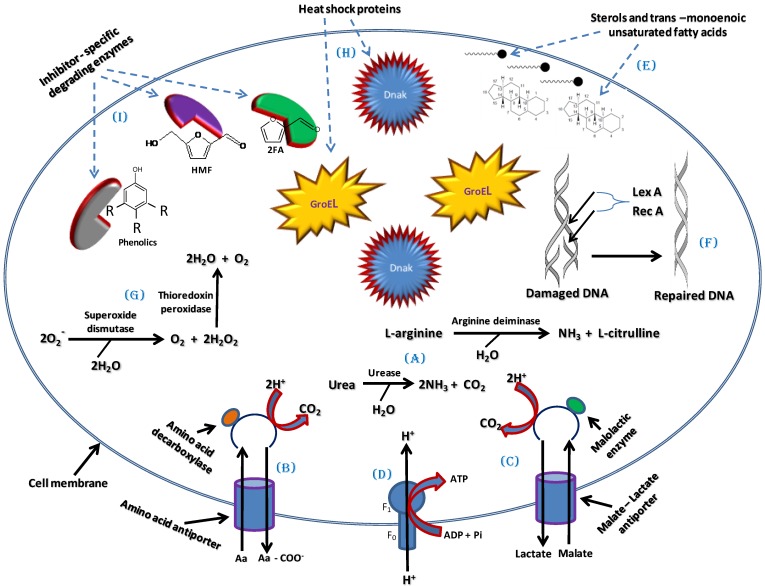
One of the main effects of the LCM bioconversion inhibitors is the intracellular acidification of ethanologenic microorganisms 97. The intracellular pH can be maintained by several means in ethanologenic bacteria. One means to achieve pH homeostasis in response to cellular acidification is through the increased production of ammonia (NH3) 98. NH3 will combine with the excess H+ ions present in the cell upon exposure to acids produced during LCM pre-treatment to form ammonium (NH4+) ions, consequently raising the intracellular pH 98. Several enzymes have been identified which enable the production of ammonium, including intracellular ureases and arginine deiminase. Urease converts urea into NH3 and CO2 while arginine deiminase converts L-arginine into NH3 and L-citrulline (Figure 4A) 98-100.
Fig 4.
A model of tolerance and adaptation mechanisms which could be employed by ethanologenic bacteria against the effects of lignocellulose inhibitors and which may involve maintenance of pH homeostasis and cell membrane integrity, activation and regulation of global cellular stress responses and degradation of Inhibitors.
Furthermore, the activation of an amino acid decarboxylase coupled with an antiporter, pumps in amino acids (arginine, glutamate or lysine) and pumps out decarboxylated products (agmatine, γ-amino butyrate or cadaverine) from the bacterial cell. This results in the expulsion of 2H+ molecules per decarboxylated product and leads to increase in intracellular pH (Figure 4B) 97, 100. This mechanism was well studied in Escherichia coli where glutamate-dependent decarboxylation was shown to be the most robust, in terms of pH stabilization, while the lysine-dependent decarboxylation is the least effective 97, 101. In addition to this effect, malolactic enzyme which converts mono-anionic malate into lactate by the addition of 2H+ can also contribute towards reducing the intracellular H+ concentration (Figure 4C) 97. The results of these decarboxylation processes concurrently generate a proton motive force (PMF) that is sufficient to drive ATP synthesis via F1-FO ATP Synthases activity, as a result providing ATP for metabolic functions (Figure 4D) 97.
Maintenance of cell membrane integrity
The hydrophobicity of the inhibitors results in the interference with fluidity and rigidity and concomitant instability of the bacterial cell membrane. One means to cope with this instability is by increasing sterol production and altering phospholipid fatty acids through synthesis of more trans-monoenoic than cis-monoenoic unsaturated fatty acids 102-104. This enhances membrane restructuring, conferring higher rigidity and resistance to disruption by external factors such as LCM bioconversion inhibitors (Figure 4E). This has been demonstrated in Pseudomonas putida P8 as well as several other bacteria belonging to the genera Pseudomonas and Vibrio which are resistant to high concentrations of phenolic compounds, and is linked to the constitutively expressed periplasm-localized enzyme cis-trans isomerase (Cti), which converts cis-unsaturated to trans-unsaturated fatty acids. 105,106. Sterols ensure that the bacterial cell membrane provide a greater hydrophobic barrier against polar molecules and rigidity barrier against non-polar molecules, consequently blocking non-specific translocation/permeation of toxic molecules into the cell 104, 105, 107. Bacterial Outer Membrane Proteins (OMPs) located in the outer membranes of Gram-negative bacteria and cell envelopes of Gram-positive bacteria may also play a vital role in providing a protective barrier against the influx of LCM bioconversion inhibitory compounds and/or facilitate their efflux through the plasma membrane, consequently protecting the cell 108-110. The role of OMPs in the extrusion of and protection against phenols has been described in both Pseudomonas species and E. coli 111 - 113.
Activation and regulation of global stress responses
Given the physiological stress introduced by LCM bioconversion inhibitors on bacterial cells, another means by which bacteria can tolerate these inhibitors is through the activation of global stress responses. Sigma factors (σS and σB) that regulate the general stress responses in bacteria play a major role in initiating the transcription of vital stress response genes 114-117. They form a complex with RNA polymerase that binds to the promoter regions of these response genes resulting in their transcription and subsequent translation. Activated response genes include those encoding SOS response proteins such as LexA and RecA which participate in various housekeeping functions including DNA repair and correction of mutation errors (Figure 4F) 118, 119, oxidative stress response proteins such as superoxide dismutase (which converts O2- to O2 and H2O2) and thioredoxin peroxidase (which converts H2O2 to H2O) thus relieving oxidative stress (Figure 4G) 117, 120, and heat shock proteins/chaperones (DnaK and GroESL complex) which are involved in the folding, renaturation and stability of cellular proteins or removal of damaged proteins during stress (Figure 4H) 117, 120, 121. Other regulators that could play similar role as the sigma factors are stress tolerance-related transcriptional factors such as Hfq, NhaA and HimA 122-124. These transcriptional factors were shown to be involved in the regulation of genes involved in resistance to lignocellulosic pre-treatment inhibitory compounds in Zymomonas mobilis. Over expression of the hfq, nhaA and himA genes resulted in increased resistance to these inhibitory compounds, while knock-out mutants were more sensitive 122-124.
Inhibitors degradation
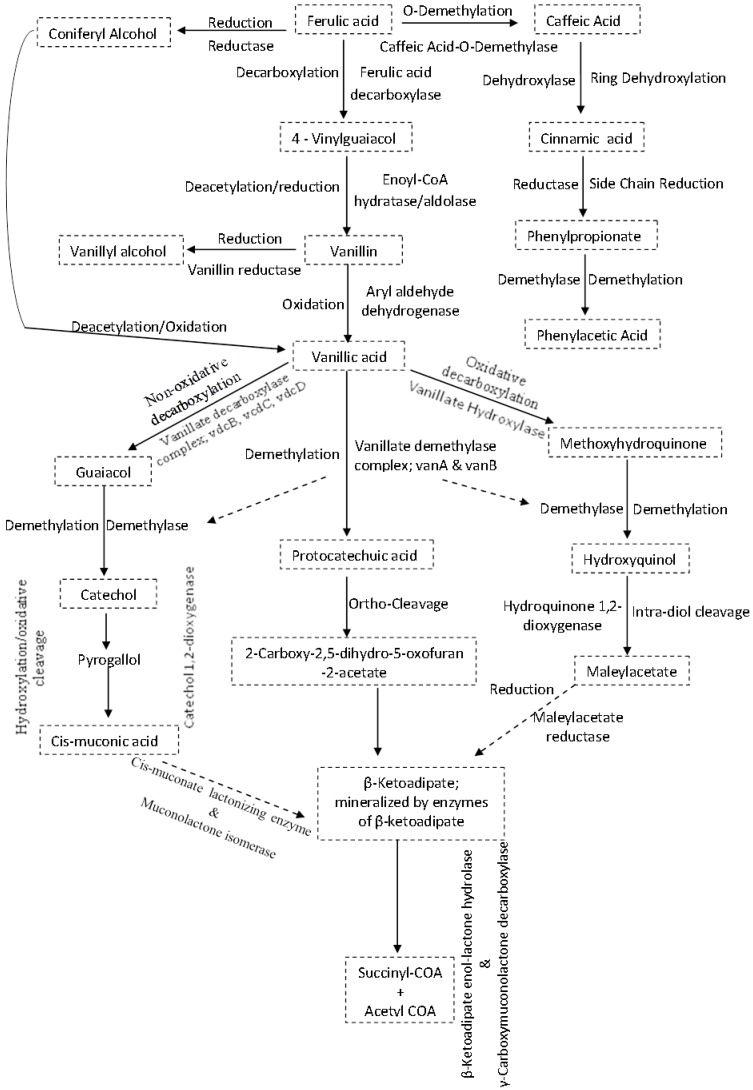
The synthesis and activation of proteins such as enzymes or co-enzymes in inhibitor-specific degradation pathways can contribute significantly towards alleviating the negative effects of the inhibitors on ethanologenic bacteria (Figures 4I and 5). The degradation of these inhibitory compounds has been widely demonstrated in many bacterial species, particularly for phenolics, where meta/ortho-cleavage and β-ketoadipate pathways are used for phenolic compound degradation 125-128. Phenolic degradation pathways have been previously described in Gram positive bacteria genera, such as Aerobacter 129 Arthrobacter 130, Bacillus 131-135, Lactobacillus 136, 137, Rhodococcus 138 and Paenibacillus 139, as well as in Gram negative bacteria genera such as Acinetobacter 140, Comamonas 141, 142, Enterobacter 143, Escherichia 144-148, Klebsiella 149, Pseudomonas 150-153 and Sphingomonas 154, 155. It has been shown that these microorganisms have evolved with abilities to degrade phenolics such as ferulic acid, vanillic acid, protocatechuic acid, and catechol, and be able to break them down into simple products via series of enzymatic processes, which can then channelled into the TCA and/or glyoxylate cycle(s) to produce energy (Figure 5) 125-128, 131, 139, 147, 150. Mechanisms such as decarboxylation, demethylation, dehydroxylation, oxidation, ring-cleavage, reduction, deacetylation are employed by several of the above-mentioned bacteria to degrade phenolic compounds. This degradation involves specific enzymes such as ferulic acid decarboxylase, vanillate demethylase, catechol-1,2-dioxygenase, maleylacetate reductase, muconolactone isomerise, β-ketoadipate enol-lactone hydrolase, γ-carboxymuconolactone decarboxylase (see Figure 5) 125-128, 131, 139, 147, 150.
Fig 5.
Inhibitor-specific degradative pathways in bacteria cells. Mechanisms often used by specific enzymes may involve decarboxylation, demethylation, dehydroxylation, oxidation, ring-cleavage, reduction, deacetylation e.t.c. The products formed are often less toxic to cells physiology and could easily be metabolised further to form products such as acetyl and succinyl COA that are easily assimilated by the cells.
The conversion of the aldehyde and/or carboxylic acid groups of phenolic compounds to alcohol group compounds were shown to be more beneficial to the cell physiology 76, 86, probably due to the reduced toxicity of the alcohol functional groups. The enzyme carboxylic acid reductase in Nocardia sp. was shown to convert the phenolic compound vanillic acid to vanillin aldehyde 156, while an aldehyde reductase in Gluconobacter oxydans can convert this aldehyde to vanillyl alcohol 157.
The enzyme furfural reductase produced by many different ethanologenic bacterial species can degrade furfural and HMF to the less toxic compounds furfuryl and hydroxymethyl furfuryl alcohol respectively 158, 159. It was shown that NADPH concentration plays a vital role in the activity of this enzyme, and that NADPH-dependent reduction of the furan compounds competes with normal cell metabolic biosyntheses that utilise NADPH 159. The silencing of the NADPH-dependent oxidoreductase genes yqhD and dkgA in E. coli EMFR9 resulted in greater tolerance towards furfural and HMF 160, 161. This was attributed to the increased availability of NADPH for use by furfural reductase, as over expression of glucose 6-phosphate dehydrogenase in S. cerevisiae that produces NADPH, was found to enhance tolerance to furfural 162. Genetic manipulation of four genetic traits (FucO, ucpA or pntAb and deletion of yqhD), were recently shown to increase furfural tolerance in ethanol producing E. coli LY180 (Strain W; ATCC derivative) and in succinate biocatalyst E. coli KJ122 (Strain C; ATCC8739 derivative) 163. These strains were reported to be highly resistant to furfural and mixture of other hemicellulose derived inhibitors, as equal yields of ethanol and succinate were produced when these strains were used in hemicelluloses hydrolysates (which contain furfural, HMF, formic and acetic acids) and in laboratory controlled fermentations 163.
The introduction of Laccase, a multicopper oxidase enzyme, in willow plant hemicellulose hydrolysate has also been found to greatly reduce the effect of lignocellulose inhibitory compounds (such as vanillic acid, catechol and 4-hydroxybenzoic acid) on fermentation 164. Laccases are produced by many bacterial species, including Azospirillum lipoferum 165, 166, Bacillus subtilis 167, 168, Marinomonas mediterranea 169, 170, Streptomyces griseus and Streptomyces cyaneus 171-173, where it participates in the biodegradation of polymers and ring cleavage of aromatic compounds.
Screening/engineering LCM inhibitor tolerant ethanologenic bacteria
The development of sustainable biofuel production will require an efficient utilization of alternative renewable and inexpensive biomass sources, such as LCMs. Presently, the large-scale use of bioconversion of LCMs to ethanol is hampered by the introduction of inhibitory compounds during the pre-treatment steps which negatively affect the ethanologenic microorganisms used for downstream ethanol production. There is thus an urgent need to improve and optimize the LCM pre-treatment and hydrolysis processes in order to overcome this technical challenge and thereby improve the fermentation efficiency 29, 54, 174. Existing chemical or physical strategies for inhibitor elimination or reduction are expensive and not entirely effective. Hence it has become important to look at means to improve the tolerance/resistance of ethanologenic microorganisms, including bacteria. This can be undertaken by screening for inhibitor tolerant/resistant microorganisms or through genetic engineering.
An important conventional biological approach which involves long-time course adaptation study could be employed. This could be done by culturing ethanologenic bacteria in specific medium containing high concentrations of LCM inhibitory compounds with low carbon (sugar) source, allowing them to adapt and develop in the new environment. This approach will lead to creation of inhibitor-tolerant bacterial strains, in a way that strains that have the ability to survive, grow and adapt within this medium, would have activated specific proteins/enzymes and mechanisms that would have facilitated the transformation of the LCM inhibitory compounds present in the hydrolysates, into less toxic compounds. This approach has resulted in increase in microbial biocatalysts efficiency in production of ethanol with increasing tolerance to LCM inhibitory compounds. For example, the Escherichia coli LY01 strain was found to show high tolerance to toxic aldehydes than its wild type KO11, by expressing high levels of genes involved in safeguarding osmolytic balance, stress response proteins and cell envelope components 76, 175. Furthermore, Escherichia coli strain LY168 engineered from wild type K011 from was shown to produce higher level of ethanol in a minimal nutritional supplement from various lignocellulose biomass containing LCM inhibitory compounds 176. It also shown that Methylobacterium extorquens, Pseudomonas sp., Flavobacterium indologenes, Acinetobacter sp., Arthrobacter aurescens could also degrade LCM inhibitory compounds when grown on them as the sole carbon and energy sources 177.The success of this approach infers that it could be applied to other ethanologenic bacteria to improve the overall tolerance to LCM inhibitory compounds and fermentation efficiencies. Resequencing the genome/proteome of these strains will also provide valuable information into the adaptation mechanism that may have been employed by these strains in adjusting to the new environment.
Another approach could be use of enzymatic detoxification. The use of laccase, phenoloxidase and/or lignin peroxidase enzymes could be a great potential, since treatment of LCM hydrolysate with these enzymes have led to degradation of phenolic compounds and increase the ethanol yield with a negligible loss of total sugars 178-180. Using appropriate genetic engineering to over-express the active production of these enzymes has been successfully carried out in S. cerevisiae 181-186. It will therefore be of better advantage if the same technology could be used for ethanologenic bacteria cells, since they have the abilities to grow quickly, producing more of these enzymes and they also could successfully ferment both C6 and C5 sugars in the LCM hydrolysate, unlike most fungi that ferment C6 and rarely utilize C5 sugars. Over-expression of homologous and heterologous potentially beneficial genes of dehydrogenases and reductases enzymes that are involved in numerous detoxification reactions and which are shown to alter the levels of co-factor NADPH and NADH could also confer resistance towards specific LCM inhibitory compounds 187, 188.
Furthermore, the use of genomics/metagenomic approach in biomining varieties of environmental and industrial niches for lignocellulolytic bacteria with genes that confers resistance or ability to degrade these inhibitory compounds, which could be cloned and expressed in existing industrial strains (that have high ethanol production capabilities) could further enhance the current fermentation technologies 189, 190. These bacteria are able to survive extreme habitats such as thermophilic, halophilic, acidophilic or alkaliphic environments as a result of their innate defence and adaptation mechanisms, and they often produce enzymes that are activity and potentially stable at these harsh conditions, which are often similar to extreme conditions present in lignocellulose degradation processes. Such bacteria could be used concurrently with the ethanologenic strains; helping to degrade the inhibitors while the ethanologenic bacteria ferments the hydrolysate 191. As example, the use of thermophilic bacterium, Ureibacillus thermosphaercus was found to potentially increase ethanol production when used concomitantly with S. cerevisiae for ethanol production in waste house wood hydrolysate 22. Phenolics and furan derivatives present in the hydrolysate were confirmed to be degraded by Chromatographic analysis, and the bacterium grows fast and utilizes below 5% of the released fermentable sugars.
Summary
It is anticipated that in the near future green energy such as biofuels (bioethanol in particular) will gradually replace fossil fuels as a global energy source 192 for home and industrial use. However, more vigorous research and focused approaches must be channelled into the development of robust and economical technologies that will utilize lignocellulose biomasses on a larger scale and improve lignocellulose fermentation efficiencies. This could be achieved by overcome the existing challenges facing the fermentation processes through better understanding of mechanisms of hydrolysate toxicity and engineering tolerance towards them. If this is actualized, lignocellulolytic biomass fermentation will be able to meet and exceed the productivity of sugar/food crop-based bioethanol bioprocesses that threatens world food security.
The further advancement of lignocellulose biotechnology will not only provide bioethanol for energy use, but will also create other opportunities to the world economies in terms of providing cheaply valuable raw materials for various industrial uses and reducing the burden of disposal of these biomasses, creating a cleaner, greener, and a comfortable, friendly environment 50, 51, 193. Moreover, the successes of USA and Brazil in food crop-based bioethanol production have proved that the same could be achieved with use of lignocellulosic biomasses if the appropriate technologies are put in place 194.
Acknowledgments
The authors wish to thank by the National Research Foundation (NRF) South Africa, the University of Western Cape and the Agricultural Research Council of South Africa, for their institutional and financial support.
References
- 1.The Economics of Global Climate Change. Jonathan MH, Brian R; http://www.ase.tufts.edu/gdae/education_materials/modules/The_Economics_of_Global_Climate_Change.pdf. [Google Scholar]
- 2.Hill J, Nelson E, Tilman D. et al. Environmental, economic and energetic cost and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006;30:11206–11210. doi: 10.1073/pnas.0604600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Energy Agency. Biofuels for Transport: Technology Roadmap; [2011]. http://www.iea.org/papers/2011/biofuels_roadmap.pdf. [Google Scholar]
- 4.Energy Information Administration: International Energy Outlook 2008: with projection to 2030. http://www.eia.doe.gov/oiaf/archive/ieo08/index.html.
- 5.Asif M, Muneer T. Energy Supply: its demand and security issues for developed and emerging economies. Renewable and Sustainable Energy Rev. 2007;11:1388–1413. [Google Scholar]
- 6.Chang T, Yao S. Thermophilic, lignocellulolytic bacteria for ethanol production: current state and perspectives. Appl Microbiol Biotechnol. 2011;92:13–27. doi: 10.1007/s00253-011-3456-3. [DOI] [PubMed] [Google Scholar]
- 7.Romero S, Merino E, Bolivar F. et al. Metabolic engineering of Bacillus subtilis for ethanol production: lactate dehydrogenase plays a key role in fermentative metabolism. Appl Environ Microbiol. 2007;16:5190–5198. doi: 10.1128/AEM.00625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AJ, Podkaminer KK, Desai SG. et al. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci USA. 2008;37:13769–13774. doi: 10.1073/pnas.0801266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cripps RE, Eley K, Leak DJ. et al. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab Eng. 2009;6:398–408. doi: 10.1016/j.ymben.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Yao S, Mikkelsen MJ. Metabolic engineering to improve ethanol production in Thermoanaerobacter mathranii. Appl Microbiol Biotechnol. 2010;1:199–208. doi: 10.1007/s00253-010-2703-3. [DOI] [PubMed] [Google Scholar]
- 11.Jarboe LR, Liu P Kautharapu K, Ingram LO. Optimization of enzyme parameters for fermentative production of biorenewable fuels and chemicals. Comp & Struct Biotechnol J. 2012;3(4):1–8. doi: 10.5936/csbj.201210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zverlov VV, Velikodvorskaya GA, Schwarz WH. A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum:investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose. Microbiol. 2002;148:247–255. doi: 10.1099/00221287-148-1-247. [DOI] [PubMed] [Google Scholar]
- 13.Zverlov VV, Schwarz WH. The Clostridium thermocellum cellulosome—the paradigm of a multienzyme complex. In: Ohmiya K, et al., editors. Biotechnology of lignocellulose degradation and biomass utilization. Tokyo, Japan: Uni Publishers; 2004. pp. 137–147. [Google Scholar]
- 14.Demain AL, Newcomb M, David Wu JH. Cellulase, Clostridia, and Ethanol. Microbiol. Mol Biol Rev. 2005;69(1):124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Béguin P, Aubert JP. The biological degradation of cellulose. FEMS Microbiol Rev. 1994;13:25–58. doi: 10.1111/j.1574-6976.1994.tb00033.x. [DOI] [PubMed] [Google Scholar]
- 16.Chandrakant P, Bisaria VS. Simultaneous bioconversion of cellulose and hemicellulose to ethanol. Crit Rev Biotechnol. 1998;18:295–331. doi: 10.1080/0738-859891224185. [DOI] [PubMed] [Google Scholar]
- 17.Sousa LD, Chundawat SPS, Balan V, Dale BE. 'Cradle-tograve' assessment of existing lignocellulose pretreatment technologies. Curr Opin Biotechnol. 2009;20:339–347. doi: 10.1016/j.copbio.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Perez J, Muñoz-Dorado J, de la Rubia T. et al. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Microbiol. 2002;5:53–63. doi: 10.1007/s10123-002-0062-3. [DOI] [PubMed] [Google Scholar]
- 19.Howard RL, Abotsi E, Jansen van Rensburg EL. et al. Lignocellulose biotechnology: issues of bioconversion and enzyme production. Afr J Biotechnol. 2003;2(12):602–619. [Google Scholar]
- 20.Zaldivar J, Nielsen J, Olsson L. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol. 2001;56:17–34. doi: 10.1007/s002530100624. [DOI] [PubMed] [Google Scholar]
- 21.Parawira W, Tekere M. Biotechnological strategies to overcome inhibitors in lignocelluloses hydrolysates for ethanol production: review. Crit Rev Biotechnol. 2011;1:20–31. doi: 10.3109/07388551003757816. [DOI] [PubMed] [Google Scholar]
- 22.Okuda N, Soneura M, Ninomiya K. et al. Biological detoxification of waste house wood hydrolysate using Ureibacillus thermosphaericus for bioethanol production. J Biosci Bioeng. 2008;2:128–133. doi: 10.1263/jbb.106.128. [DOI] [PubMed] [Google Scholar]
- 23.McCoy M. Biomass ethanol inches forward. Chemical Eng News. 1998;76:29–32. [Google Scholar]
- 24.Klinke HB, Thomsen AB, Ahring BK. Inhibition of ethanol producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol. 2004;66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- 25.Mosier N, Wyman C, Dale B. et al. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol. 2005;96:673–686. doi: 10.1016/j.biortech.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 26.Palmqvist E, Hahn-Hagerdal B. Fermentation of lignocellulosic hydrolysates. II: Inhibitors and mechanisms of inhibition. Bioresour Technol. 2000;74:25–33. [Google Scholar]
- 27.Mills TY, Sandoval NR, Gill RT. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol for Biofuels. 2009;2(1):26–36. doi: 10.1186/1754-6834-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding MZ, Wang X, Liu W, Cheng JS, Yang Y, Yuan YJ. Proteomic research reveals the stress response and detoxification of yeast to combined inhibitors. PLoS One. 2012;7(8):43474. doi: 10.1371/journal.pone.0043474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmqvist E, Hahn-Hägerdal B. Fermentation of lignocellulose hydrolysates. I: inhibition and detoxification. Biores Technol. 2000;74(1):17–24. [Google Scholar]
- 30.Mussatto SI, Roberto IC. Alternatives for detoxification of diluted-acid lignocellulosic hydrolyzates for use in fermentative processes: a review. Bioresour Technol. 2004;93:1–10. doi: 10.1016/j.biortech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 31.Tadesse H, Luque R. Advances on biomass pretreatment using ionic liquids: An overview. Energy Environ Sci. 2011;4:3913–3929. [Google Scholar]
- 32.Tang S, Baker GA, Ravula S, Jones JE, Zhao H. PEG-functionalized ionic liquids for cellulose dissolution and saccharification. Green Chem. 2012;14:2922–2932. [Google Scholar]
- 33.Brandt A, Gräsvik J, Halletta JP, Welton T. Deconstruction of lignocellulosic biomass with ionic liquids. Green Chem. 2013;15:550–583. [Google Scholar]
- 34.Liu ZL, Slininger PJ, Gorsich SW. Enhanced biotransformation of furfural and hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol. 2005;2:451–460. [PubMed] [Google Scholar]
- 35.Hahn-Hägerdal B, Wahlbom C, Gárdonyi M, van ZW, Otero R, Jönsson L. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Metab Eng. 2001;73:53–84. doi: 10.1007/3-540-45300-8_4. [DOI] [PubMed] [Google Scholar]
- 36.Liu ZL. Molecular mechanisms of yeast tolerance and in situ detoxification of lignocelluloses hydrolysates. Appl Microbiol Biotechnol. 2011;90(3):809–825. doi: 10.1007/s00253-011-3167-9. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Gellerstedt G, Toven K. Steam explosion lignins; their extraction, structure and potential as feedstock for biodiesel and chemicals. Bioresour Technol. 2009;100:2556–2561. doi: 10.1016/j.biortech.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Ostergaard S, Olsson L, Nielsen J. Metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol. 2000;64:34–50. doi: 10.1128/mmbr.64.1.34-50.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Chenj J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83:1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 40.Martinez A, Rodriguez ME, Wells ML. et al. Detoxification of dilute acid hydrolysate of lignocellulose with lime. Biotechnol Prog. 2001;2:287–293. doi: 10.1021/bp0001720. [DOI] [PubMed] [Google Scholar]
- 41.Hendriks AT, Zeeman G. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour Technol. 2009;1:10–18. doi: 10.1016/j.biortech.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 42.Alvira P, Tomas-Pejo E, Ballesteros M. et al. Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis: A review. Bioresour Technol. 2010;13:4851–4861. doi: 10.1016/j.biortech.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 43.Mosier N, Wymanb C, Dalec B. et al. Features of promising technologies for pretreatment of lignocellulosic biomass. Biores Technol. 2005;96:673–686. doi: 10.1016/j.biortech.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 44.Morjanoff PJ, Gray PP. Optimization of steam explosion as method for increasing susceptibility of sugarcane bagasse to enzymatic saccharification. Biotechnol Bioeng. 1987;29:733–741. doi: 10.1002/bit.260290610. [DOI] [PubMed] [Google Scholar]
- 45.Balan V, Bals B, Chundawat SP. et al. Lignocellulosic biomass pretreatment using Afex. Methods Mol Biol. 2009;581:61–77. doi: 10.1007/978-1-60761-214-8_5. [DOI] [PubMed] [Google Scholar]
- 46.Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals to biocatalysis and bioremediation. Metabolic Eng. 2010;12:307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Dunlop MJ. Engineering microbes for tolerance to next generation biofuels. Biotechnol for Biofuels. 2011;4:32–40. doi: 10.1186/1754-6834-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunlop MJ, Dossani ZY, Szmidt HL. et al. Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol. 2011;7:487. doi: 10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Classen PAM, van Lier JB, Lopez-Contreras AM. et al. Utilization of biomass for the supply of energy carriers. Appl Microbiol Biotechnol. 1999;52:741–755. [Google Scholar]
- 50.Wiselogel A, Tyson J, Johnson D. Wyman CE (ed) Handbook on bioethanol: production and utilization. Taylor and Francis, Washington, DC; 1996. Biomass feedstock resources and composition; pp. 105–118. [Google Scholar]
- 51.Wyman CE. Ethanol production from lignocellulosic biomass: overview. In: Wyman CE, editor. Handbook on bioethanol: production and utilization. Washington, DC: Taylor and Francis; 1996. pp. 1–18. [Google Scholar]
- 52.Levine JS. Biomass burning and global change. In: Levine JS, editor. Biomass burning and global change vol 1: Remote sensing and inventory development and biomass burning in Africa. Cambridge, Massachusetts, USA: The MIT Press; 1996. p. 35. [Google Scholar]
- 53.Malherbe S, Cloete TE. Lignocellulose biodegradation: fundamentals and application: A review. Environ Sci Biotechnol. 2003;1:105–114. [Google Scholar]
- 54.Olsson L, Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates for ethanol production. Enzyme Microb Technol. 1996;18:312–331. [Google Scholar]
- 55.Ghosh P, Singh A. Physicochemical and biological treatments for enzymatic/microbial conversion of lignocellulosic biomass. Adv Appl Microbiol. 1993;39:295–333. [Google Scholar]
- 56.Brigham JS, Adney WS, Himmel ME. Wyman CE (ed) Handbook on bioethanol: production and utilization. Taylor and Francis, Washington, DC; 1996. Hemicelluloses: diversity and applications; pp. 119–142. [Google Scholar]
- 57.Parajó JC, Domínquez HD, Domínquez JM. Biotechnological production of xylitol. Part 1: Interest of xylitol and fundamentals of its biosynthesis. Biores Technol. 1998;65:191–201. [Google Scholar]
- 58.Granström TB, Izumori K, Leisola M. A rare sugar xylitol. Part II: biotechnological production and future applications of xylitol. Appl Microbiol Biotechnol. 2007;74:273–276. doi: 10.1007/s00253-006-0760-4. [DOI] [PubMed] [Google Scholar]
- 59.Fry SC. Phenolic components of the primary cell wall. Biochem J. 1982;203:493–504. doi: 10.1042/bj2030493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alves LA, Felipe MGA. et al. Pretreatment of sugarcane bagasse hemicellulose hydrolysate for xylitol production by Candida guilliermondii. Appl. Biochem. Biotechnol. 1998;70:89–98. [Google Scholar]
- 61.Adler E. Lignin chemistry- past, present and future. Wood Sci Technol. 1977;11:169–218. [Google Scholar]
- 62.Zimmermann W. Degradation of lignin by bacteria. J Biotechnol. 1990;13(2-3):119–130. [Google Scholar]
- 63.Watanabe M, Aizawa Y, Lida T, Aida TM. et al. Glucose reactions with acid and base catalysts in hot compressed water at 473K. Carbohydrate Res. 2005;12:1925–1930. doi: 10.1016/j.carres.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 64.Kim Y, Hendrickson R, Mosier NS. et al. Enzyme hydrolysis and ethanol fermentation of liquid hot water and AFEX pretreatment distillers' grains at high solids loading. Bioresour Technol. 2008;99(12):5206–5215. doi: 10.1016/j.biortech.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 65.Lau MW, Dale BE. Cellulosic ethanol production from AFEX-treated corn stover using Saccharomyces cerevisiae 424A(LNH-ST) Proc Natl Acad Sci U.S.A. 2009;106(5):1368–1373. doi: 10.1073/pnas.0812364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taherzadeh MJ, Niklasson C, Liden G. On-line control of fed-batch fermentation of dilute-acid hydrolyzates. Biotechnol Bioeng. 2000;69:330–338. doi: 10.1002/1097-0290(20000805)69:3<330::aid-bit11>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 67.Liu ZL, Ma M, Song M. Evolutionary engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Genet Genomics. 2009;282(3):233–244. doi: 10.1007/s00438-009-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larsson S, Reimann A, Nilvebrant N-O, Jönsson LF. Comparison of different methods for the detoxification of lignocellulose hydrolyzates of spruce. Appl Biochem Biotechnol. 1999;77(1-3):91–103. [Google Scholar]
- 69.Rumbold K, van Buijsen HJJ, Overkamp KM. et al. Microbial production host selection for converting second-generation feedstocks into bioproduct. Micro Cell Fact. 2009;8:64. doi: 10.1186/1475-2859-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wende G, Fry SC. O-feruloylated, O-acetylated oligosaccharides as side-chains of grass xylans. Phytochemistry. 1997;6:1011–1018. doi: 10.1016/s0031-9422(96)00648-6. [DOI] [PubMed] [Google Scholar]
- 71.Roe AJ, McLaggan D, Davidson I, O'Byrne C, Booth IR. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Viegas CA, Sá-Correiq I. Activation of plasma membrane ATPase of Saccharomyces cerevisiae by octanoic acid. J Gen Microbiol. 1991;137:645–651. doi: 10.1099/00221287-137-3-645. [DOI] [PubMed] [Google Scholar]
- 73.Luli GW, Stohl WR. Comparison of growth, acetate production and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl Environ Microbiol. 1990;56:1004–1011. doi: 10.1128/aem.56.4.1004-1011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakano K, Rischke M, Sato S, Markl H. Influence of acetic acid on the growth of Escherichia coli K12 during high-cell-density cultivation in a dialysis reactor. Appl Microbiol Biotechnol. 1997;48:597–601. doi: 10.1007/s002530051101. [DOI] [PubMed] [Google Scholar]
- 75.Takahashi CM, Takahashi DF, Crvalhal ML, Alterthum F. Effects of acetate on the growth and fermentation performance of Escherichia coli KO11. Appl Biochem Biotechnol. 1999;81:193–203. doi: 10.1385/abab:81:3:193. [DOI] [PubMed] [Google Scholar]
- 76.Zaldivar J, Ingram LO. Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol Bioeng. 1999;66:203–210. doi: 10.1002/(sici)1097-0290(1999)66:4<203::aid-bit1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 77.Almeida JR, Bertilsson M, Gorwa-Grauslund MF. et al. Metabolic effects of furaldehyde and impacts on biotechnology processes. Appl Microbial Biotechnol. 2009;4:625–638. doi: 10.1007/s00253-009-1875-1. [DOI] [PubMed] [Google Scholar]
- 78.Modig T, Liden G, Taherzadeh MJ. Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J. 2002;363:769–776. doi: 10.1042/0264-6021:3630769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hadi SM, Shahabuddin H, Rehman A. Specificity of the interaction of furfural with DNA. Mutat Res. 1989;225:101–106. doi: 10.1016/0165-7992(89)90125-5. [DOI] [PubMed] [Google Scholar]
- 80.Shahabuddin H, Rahman A, Hadi SM. Reaction of furfural and methylfurfural with DNA: Use of single-strand-specific nucleases. Food Chem Toxicol. 1991;29:719–721. doi: 10.1016/0278-6915(91)90131-p. [DOI] [PubMed] [Google Scholar]
- 81.Khan QA, Hadi SM. Effects of furfural on plasmid DNA. Biochem Mol Biol Int. 1993;29(6):1153–1160. [PubMed] [Google Scholar]
- 82.Uddin S, Hadi SM. Reactions of furfural and methylfurfural with DNA. Biochem Mol Biol Int. 1995;35(1):185–195. [PubMed] [Google Scholar]
- 83.Zaldivar J, Martinez A, Ingram LO. Effect of selected aldehyde on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol Bioeng. 1999;65:24–33. doi: 10.1002/(sici)1097-0290(19991005)65:1<24::aid-bit4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 84.Cherrington CA, Hinton M, Chopra I. Effect of short-chain organic acids on macromolecular synthesis in Escherichia coli. J Appl Bacteriol. 1990;68:69–74. doi: 10.1111/j.1365-2672.1990.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 85.Sinha RP. Toxicity of organic acids for repair-deficient strains of Escherichia coli. Appl Environ Microbiol. 1986;51:1364–1366. doi: 10.1128/aem.51.6.1364-1366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heipieper HJ, Weber FJ, Sikkema J. et al. Mechanism of resistance of whole cell to toxic organic solvent. TIBTECH. 1994;12:409–415. [Google Scholar]
- 87.Mikuláasová M, Vodny S, Pekarovicová A. Influence of phenolics on biomass production by Candida utilis and Candida albicans. Biomass. 1990;23:149–154. [Google Scholar]
- 88.Philip TP, Zhang M. Role of pretreatment and conditioning processes on toxicity of lignocellulosic biomass hydrolysates. Cellulose. 2009;16:743–762. [Google Scholar]
- 89.García-Aparicio MP, Ballesteros I, González A, Oliva JM, Ballesteros M, Negro MJ. Effect of inhibitors released during steam-explosion pretreatment of barley straw on enzymatic hydrolysis. Appl Biochem Biotechnol. 2006;129:278–288. doi: 10.1385/abab:129:1:278. [DOI] [PubMed] [Google Scholar]
- 90.Partition coefficient. http://en.wikipedia.org/wiki/Partition_coefficient.
- 91.Bradley SP. Predicting models of toxic action from chemical structure: an overview. SAR QSAR Environ RES. 1994;2:89–104. doi: 10.1080/10629369408028842. [DOI] [PubMed] [Google Scholar]
- 92.Hansh C, Bonavida B, Jazireih AR, Cohen J, Milliron C, Kurup A. Quantitative structure-activity relationships of phenolic compounds causing apoptosis. Bioorg Medic Chem. 2003;11:617–620. doi: 10.1016/s0968-0896(02)00326-7. [DOI] [PubMed] [Google Scholar]
- 93.Selassie CD, Kapur S, Verma RP, Rosario M. Cellular apoptosis and cytotoxicity of phenolic compounds: a quantitative structure-activity relationship study. J Med Chem. 2005;48(23):7234–7242. doi: 10.1021/jm050567w. [DOI] [PubMed] [Google Scholar]
- 94.Hoelz LVB, Horta BAC, Araújo JQ, Albuquerque MG, de Alencastro RB, da Silva JFM. Quantitative structure-activity relationships of antioxidant phenolic compounds. J Chem Pharm Res. 2010;2(5):291–306. [Google Scholar]
- 95.Bothast RJ, Nichols NN, Dien BS. Fermentation with new recombinant organisms. Biotechnol Prog. 1999;15:867–875. doi: 10.1021/bp990087w. [DOI] [PubMed] [Google Scholar]
- 96.Ingram LO, Gomez PF, Lai X. et al. Metabolic engineering of bacteria for ethanol production. Biotechnol Bioeng. 1998;58:204–214. doi: 10.1002/(sici)1097-0290(19980420)58:2/3<204::aid-bit13>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 97.Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, Foster JW. Mechanism of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mobley HL, Island MD, Hausinger RP. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen YY, Burne RA. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol Lett. 1996;135:223–229. doi: 10.1111/j.1574-6968.1996.tb07993.x. [DOI] [PubMed] [Google Scholar]
- 100.Cunin R, Glansdorff N, Pierard A. et al. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gong S, Richard H, Foster JW. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol. 2003;185:4402–4409. doi: 10.1128/JB.185.15.4402-4409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hazel JR, Williams EE. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog Lipid Res. 1990;29:167–227. doi: 10.1016/0163-7827(90)90002-3. [DOI] [PubMed] [Google Scholar]
- 103.Guckert JB, Hood MA, White DC. Phospholipid ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholera: increase in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl Environ Microbiol. 1986;52:794–801. doi: 10.1128/aem.52.4.794-801.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Subczynski WK, Wisniewska A. Physical properties of lipid bilayer membranes: relevance to membrane biological function. Acta Biochimica Polonica. 2000;47(3):613–625. [PubMed] [Google Scholar]
- 105.Diefenbach R, Heipieper HJ, Keweloh H. The conversion of cis into trans unsaturated fatty acids in Pseudomonas putida P8: evidence for a role in the regulation of membrane fluidity. Appl Microbiol Biotechnol. 1992;38:382–387. [Google Scholar]
- 106.Hiepieper HJ, Meinhardt F, Segura A. The cis-trans isomerise of unsaturated fatty acids in Pseudomonas and Vibrio: biochemistry, molecular biology and psychological function of a unique stress adaptive mechanism. FEMS Microbiol Lett. 2003;229(1):1–7. doi: 10.1016/S0378-1097(03)00792-4. [DOI] [PubMed] [Google Scholar]
- 107.Eze Mo, McElhaney RN. The effect of alterations in the fluidity and phase state of the membrane lipids on the passive permeation and facilitated diffusion of glycerol in Escherichia coli. J Gen Microbiol. 1981;124:299–307. doi: 10.1099/00221287-124-2-299. [DOI] [PubMed] [Google Scholar]
- 108.Molloy MP, Herbert BR, Slade MB. et al. Proteomics analysis of the Escherichia coli outer membrane. Eur J Biochem. 2000;267:2871–2881. doi: 10.1046/j.1432-1327.2000.01296.x. [DOI] [PubMed] [Google Scholar]
- 109.Nikaido H. Molecular basis of bacteria outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bond PJ, Sansom MSP. The simulation approach to bacterial outer membrane proteins (Review) Mol Memb Biol. 2004;21:151–161. doi: 10.1080/0968760410001699169. [DOI] [PubMed] [Google Scholar]
- 111.Santos PM, Roma V, Benndorf D. et al. Mechanistic insights into the global response to phenol in the phenol-biodegrading strain Pseudomonas sp. M1 revealed by quantitative proteomics. OMICS. 2007;11(3):233–251. doi: 10.1089/omi.2007.0009. [DOI] [PubMed] [Google Scholar]
- 112.Roma-Rodrigues C, Santos PM, Benndorf D. et al. Response of Pseudomonas putida KT2440 to phenol at the level of membrane proteome. J Proteomics. 2010;73(8):1461–1478. doi: 10.1016/j.jprot.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 113.Zhang DF, Li H, Li XM. et al. Characterization of outer membrane proteins of Escherichia coli in response to phenol stress. Curr Microbiol. 2011;3:777–783. doi: 10.1007/s00284-010-9786-z. [DOI] [PubMed] [Google Scholar]
- 114.Haldenwang W G, Losick R. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci USA. 1980;77:7000–7004. doi: 10.1073/pnas.77.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boylan SA, Rutherford A, Thomas SM. et al. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Voelker U, Maul B, Hecker M. Expression of the sigmaB dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J Bacteriol. 1999;181:3942–3948. doi: 10.1128/jb.181.13.3942-3948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aertsen A, Michiels CW. Stress and how bacteria cope with death and survival. Crit Rev Microbiol. 2004;30:263–273. doi: 10.1080/10408410490884757. [DOI] [PubMed] [Google Scholar]
- 118.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 119.Cox MM, Goodman MF, Kreuzer KN. et al. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 120.Lim EM, Ehrlich SD, Maguin E. Identification of stress inducible proteins in Lactobacillus delbrueckii subsp. bulgaricus. Electrophoresis. 2000;21:2557–2561. doi: 10.1002/1522-2683(20000701)21:12<2557::AID-ELPS2557>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 121.Zou Y, Crowley DJ Van Houten B. Involvement of molecular chaperonins in nucleotide excision repair. DNaK leads to increased thermal stability of UvrA, catalytic UvrB loading, enhanced repair, and increased UV resistance. J. Biol. Chem. 1998;273:12887–12892. doi: 10.1074/jbc.273.21.12887. [DOI] [PubMed] [Google Scholar]
- 122.Yang S, Tschaplinski TJ, Engle NL. et al. Transcriptomic and metabolomic profiling of Zymomonas mobilis during aerobic and anaerobic fermentations. BMC Genomics. 2009;10:34. doi: 10.1186/1471-2164-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang S, Pelletier DA, Lu TY. et al. The Zymomonas mobilis regulator hfq contributes to tolerance against multiple lignocellulosic pretreatment inhibitors. BMC Microbiol. 2010;10:135. doi: 10.1186/1471-2180-10-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang S, Keller M, Brown SD. Genomics on pretreatment inhibitor tolerance of Zymomonas mobilis. Microbial Stress Tolerance for Biofuels. Microbiol Monographs. 2012;22:161–175. [Google Scholar]
- 125.Harwood CS, Parales RE. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 126.Masai E, Katayama Y, Fukuda M. Genetic and biochemical investigations on bacterial catabolic pathways for lignin-derived aromatic compounds. Biosci Biotechnol Biochem. 2007;71:1–15. doi: 10.1271/bbb.60437. [DOI] [PubMed] [Google Scholar]
- 127.Noor Hasyierah MS, Zulkali MMD, Ku Syahidah KI. Ferulic acid from lignocellulose biomass: Review. Proceedings of MUCET. 2008:1–8. [Google Scholar]
- 128.Rosazza JPN, Huang Z, Dostal L. et al. Biocatalytic transformation of ferulic acid: an abundant aromatic natural product. J Ind Microbiol. 1995;15:457–471. doi: 10.1007/BF01570016. [DOI] [PubMed] [Google Scholar]
- 129.Finkle BJ, Lewis JC, Corse JW. et al. Enzyme reaction with phenolic compounds: Formation of hydroxystyrenes through the decarboxylation of 4-hydroxycinnamic acids by Aerobacter. J Biol Chem. 1962;237:2926–2931. [PubMed] [Google Scholar]
- 130.Eaton RW. Plasmid-encoded phthalate catabolic pathway in Arthrobacter keyseri 12B. J Bacteriol. 2001;183:3689–3703. doi: 10.1128/JB.183.12.3689-3703.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Crawford RL. Novel pathway for degradation of protocatechuic acid in Bacillus species. J Bacteriol. 1975;121:531–536. doi: 10.1128/jb.121.2.531-536.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Crawford RL, Bromley JW, Perkins-Olson PE. Catabolism of protocatechuate by Bacillus macerans. Appl Environ Microbiol. 1979;37:614–618. doi: 10.1128/aem.37.3.614-618.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cavin J-F, Dartois V, Diviès C. Gene cloning, transcriptional analysis, purification and characterization of phenolic acid decarboxylase from Bacillus subtilis. Appl Environ Microbiol. 1998;64:1466–1471. doi: 10.1128/aem.64.4.1466-1471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karmakar B, Vohra RM, Nandanwar H. et al. Rapid degradation of ferulic acid via 4-vinylguaiacol and vanillin by a newly isolated strains of Bacillus coagulans. J Biotechnol. 2000;80:195–202. doi: 10.1016/s0168-1656(00)00248-0. [DOI] [PubMed] [Google Scholar]
- 135.Wolgel SA, Dege JE, Perkins-Olson PE. et al. Purification and characterization of protocatechuate 2,3-dioxygenase from Bacillus macerans: a new extradiol catecholic dioxygenase. J Bacteriol. 1993;175:4414–4426. doi: 10.1128/jb.175.14.4414-4426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cavin J-F, Barthelmebs L, Guzzo J. et al. Purification and characterization of an inducible P-coumaric acid decarboxylase from Lactobacillus plantarum. FEMS Microbiol Lett. 1997;147:291–295. doi: 10.1128/aem.63.5.1939-1944.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barthelmebs L, Divies C, Cavin J-F. Knockout of the P-coumarate decarboxylated gene from Lactobacillus plantarum reveals the existence of two other inducible enzymatic activities involved in phenolic acid metabolism. Appl Environ Microbiol. 2000;66:3368–3375. doi: 10.1128/aem.66.8.3368-3375.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Warhurst AM, Fewson CA. Biotransformations catalysed by the genus Rhodococcus. Crit Rev Biotechnol. 1994;14:29–73. doi: 10.3109/07388559409079833. [DOI] [PubMed] [Google Scholar]
- 139.Kasai D, Fujinami T, Abe T. et al. Uncovering the protocatechuate 2,3-cleavage pathway genes. J Bacteriol. 2009;191(21):6758–6768. doi: 10.1128/JB.00840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gerischer U, Segura A, Ornston LN. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J Bacteriol. 1998;180:1512–1524. doi: 10.1128/jb.180.6.1512-1524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Boon N, Goris J, de Vos P. et al. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline degrading Comamonas testosteroni strain, 12gfp. Appl Environ Microbiol. 2000;66(7):2906–2913. doi: 10.1128/aem.66.7.2906-2913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Provident MA, Mampel J, MacSween S. et al. Comamonas testosteroni BR6020 possessses a single genetic locus for extradiol cleavage of protocatechuate. Microbiology. 2001;147:2157–2167. doi: 10.1099/00221287-147-8-2157. [DOI] [PubMed] [Google Scholar]
- 143.Grbic-Galic D. Fermentation and oxidative transformation of ferulate by a facultatively anaerobic bacterium isolated from sewage sludge. Appl Environ Microbiol. 1995;50:1052–2057. doi: 10.1128/aem.50.4.1052-1057.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Garrido-Pertierra A, Cooper RA. Identification and purification of distinct isomerise and decarboxylase enzymes involved in the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. Eur J Biochem. 1981;117:581–584. doi: 10.1111/j.1432-1033.1981.tb06377.x. [DOI] [PubMed] [Google Scholar]
- 145.Abdulrashid N, Clark DP. Isolation and genetic analysis of mutations allowing the degradation of furans and thiophenes by Escherichia coli. J Bacteriol. 1987;169:1267–1271. doi: 10.1128/jb.169.3.1267-1271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Alam KY, Clark DP. Molecular cloning and sequence of the thdF gene, which is involved in thiophene and furan oxidation by Escherichia coli. J Bacteriol. 1991;173:6018–6024. doi: 10.1128/jb.173.19.6018-6024.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Diaz E, Ferrández A, Prieto MA. et al. Biodegradation of aromatic compounds by Escherichia coli. Microbiol Mol Biol Rev. 2001;65(4):523–569. doi: 10.1128/MMBR.65.4.523-569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zago A, Degrassi G, Bruschi CV. Cloning, sequencing and expression in Escherichia coli of the Bacillus pumilus gene for ferulic acid degradation. Appl Environ Microbiol. 1995;61:4484–4486. doi: 10.1128/aem.61.12.4484-4486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hashikado Y, Urashima M, Yoshida T. et al. Decarboxylative conversion of hydrocinnamic acids by Klebsiella oxytoca and Erwinia uredovora epiphytic bacteria of Polymnia sonchifolia leaf, possibly associated with formation of microflora on the damaged leaves. Biosci Biotech Biochem. 1993;57:215–219. doi: 10.1271/bbb.57.215. [DOI] [PubMed] [Google Scholar]
- 150.Ornston LN. The conversion of catechol and protocatechuate to β-keto-adipate by Pseudomonas putida II. Enzymes of the protocatechuate pathway. J Biol Chem. 1966;241:3787–3794. [PubMed] [Google Scholar]
- 151.Hinteregger C, Leitner R, Loidi M. et al. Degradation of phenol and phenolic compounds by Pseudomonas putida EKII. Appl Microbiol Biotechnol. 1992;37:252–259. doi: 10.1007/BF00178180. [DOI] [PubMed] [Google Scholar]
- 152.Huang Z, Dostal L, Rosazza JP. Microbial transformations of ferulic acid by Saccharomyces cerevisiae and Pseudomonas fluorescens. Appl Environ Microbiol. 1993;59(7):2244–2250. doi: 10.1128/aem.59.7.2244-2250.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maruyama K, Shibayama T, Ichikawa A. et al. Cloning and characterization of the genes encoding enzymes for the protocatechuate meta-degradation pathway of Pseudomonas ochraceae NGJ1. Biosci Biotechnol Biochem. 2004;68:1434–1441. doi: 10.1271/bbb.68.1434. [DOI] [PubMed] [Google Scholar]
- 154.Peng X, Egashira T, Hanashiro K. et al. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl Environ Microbiol. 1998;64(7):2520–2527. doi: 10.1128/aem.64.7.2520-2527.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hara H, Masai E, Katayama Y. et al. The 4-oxalomesaconate hydratase gene, involved in the protocatechuate 4,5-cleavage pathway, is essential to vanillate and syringate degradation in Sphingomonas paucimobilis SYK-6. J Bacteriol. 2000;182:6950–6957. doi: 10.1128/jb.182.24.6950-6957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.He A, Li T, Daniels L. et al. Nocardia sp. Carboxylic acid reductase: cloning, expression and characterization of new aldehyde oxidoreductase family. Appl Environ Microbiol. 2004;70(3):1874–1881. doi: 10.1128/AEM.70.3.1874-1881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schweiger P, Deppenmeier U. Analysis of aldehyde reductase from Gluconobacter oxydans 621H. Appl Microbiol Biotechnol. 2010;85(4):1025–1031. doi: 10.1007/s00253-009-2154-x. [DOI] [PubMed] [Google Scholar]
- 158.Gutierrez T, Buszko ML, Ingram LO, Preston JF. Reduction of furfural to furfuryl alcohol by ethanologenic strain of bacteria and its effect on ethanol production from xylose. Appl Biochem Biotechnol. 2002;98-100:327–340. doi: 10.1385/abab:98-100:1-9:327. [DOI] [PubMed] [Google Scholar]
- 159.Boopathy R, Bokang H, Daniels L. Biotransformation of furfural and 5-hydroxymethyl furfural by enteric bacteria. J Ind Microbiol. 1993;11:147–150. [Google Scholar]
- 160.Miller EN, Jarboe LR, Yomano LP. et al. Silencing of NADPH oxidoreductase genes (yqhD and dkgA) in furfural-resistance ethanologenic Escherichia coli. Appl Environ Microbiol. 2009;75:4315–4323. doi: 10.1128/AEM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Miller EN, Turner PC, Jaboe LR. et al. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol producing Escherichia coli LY180. Biotechnol Lett. 2010;32:661–667. doi: 10.1007/s10529-010-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gorsich SW, Dien BS, Nichols NN. et al. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2006;71:339–349. doi: 10.1007/s00253-005-0142-3. [DOI] [PubMed] [Google Scholar]
- 163.Wang X, Yomano LP, Lee JY, York SW, Zheng H, Mullinnix MT, Shanmugam KT, Ingram LO. Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. PNAS. 2013;110(10):4021–4026. doi: 10.1073/pnas.1217958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jönsson LJ, Palmqvist E, Nilverbrant NO. et al. Detoxification of wood hydrolysate with lacasse and peroxidase from the white-rot fungus Trametes versicolor. Appl Microbiol Biotechnol. 1998;49:691–697. [Google Scholar]
- 165.Alexandre G, Bally R. Emergence of laccase-positive variant of Azospirillum lipoferum occurs via a two-step phenotypic switching process. FEMS Microbiol Lett. 1999;174:371–378. doi: 10.1111/j.1574-6968.1999.tb13592.x. [DOI] [PubMed] [Google Scholar]
- 166.Givaudan A, Effosse A, Faure D. et al. Polyphenol oxidase in Azospirillum lipoferum isolated from rice rhizosphere: evidence for laccase activity in non-motile strains of Azospirillum lipoferum. FEMS Microbiol Lett. 1993;108:205–210. [Google Scholar]
- 167.Hullo M.-F, Moszer I, Danchin A. et al. CotA of Bacillus subtilis is a copper-dependent laccase. J Bacteriol. 2001;183:5426–5430. doi: 10.1128/JB.183.18.5426-5430.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Martins LO, Soares CM, Pereira MM. et al. Molecular and biochemical characterization of a highly stable bacterial laccase that occurs as a structural component of the Bacillus subtilis endospore coat. J Biol Chem. 2002;277:18849–18859. doi: 10.1074/jbc.M200827200. [DOI] [PubMed] [Google Scholar]
- 169.Sa´nchez-Amat A, Lucas-Elı´o P, Ferna´ndez E. et al. Molecular cloning and functional characterization of a unique multipotent polyphenol oxidase from Marinomonas mediterranea. Biochim Biophys Acta. 2001;1547:104–116. doi: 10.1016/s0167-4838(01)00174-1. [DOI] [PubMed] [Google Scholar]
- 170.Jimenez-Juarez N, Roman-Miranda R, Baeza A. et al. Alkali and halide-resistant catalysis by the multipotent oxidase from Marinomonas mediterranea. J Biotechnol. 2005;117(1):73–82. doi: 10.1016/j.jbiotec.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 171.Berrocal M, Rodrı´guez J, Ball AS. et al. Solubilisation and mineralisation of [14C]lignocellulose from wheat straw by Streptomyces cyaneus CECT 3335 during growth in solid-state fermentation. Appl Microbiol Biotechnol. 1997;48:379–384. [Google Scholar]
- 172.Endo K, Hosono K, Beppu T. et al. A novel extracytoplasmatic phenol oxidase of Streptomyces: it's possible involvement in the onset of morphogenesis. Microbiol. 2002;148:1767–1776. doi: 10.1099/00221287-148-6-1767. [DOI] [PubMed] [Google Scholar]
- 173.Arias ME, Arenas M, Rodr íguez J. et al. Kraft pulp biobleaching and mediated oxidation of a non-phenolic substrate by laccase from Streptomyces cyaneus CECT 3335. Appl Environ Microbiol. 2003;69(4):1953–1958. doi: 10.1128/AEM.69.4.1953-1958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Olsson L, Hahn-Hägerdal B. Fermentative performance of bacteria and yeasts in lignocellulosic hydrolysates. Process Biochem. 1993;28:249–257. [Google Scholar]
- 175.Gonzalez R, Tao H, Purvis JE, York SW, Shanmugam KT, Ingram LO. Gene array-based identification of changes that contribute to ethanol tolerance in ethanologenic Escherichia coli: comparison of KO11 (parent) to LY01 (resistant mutant) Biotechnol Prog. 2003;19:612–623. doi: 10.1021/bp025658q. [DOI] [PubMed] [Google Scholar]
- 176.Jarboe LR, Grabar TB, Yomano LP, Shanmugan KT, Ingram LO. Development of Ethanologenic Bacteria. Adv Biochem Engin/Biotechnol. 2007;108:237–261. doi: 10.1007/10_2007_068. [DOI] [PubMed] [Google Scholar]
- 177.López MJ, Nichols NN, Dien BS, Moreno J, Bothast RJ. Isolation of microorganisms for biological detoxification of lignocellulosic hydrolysates. Appl Microbiol Biotechnol. 2004;64:125–131. doi: 10.1007/s00253-003-1401-9. [DOI] [PubMed] [Google Scholar]
- 178.Jonsson LJ, Palmqvist E, Nilvebrant N-O, Hahn-Hagerdal B. Detoxification of wood hydrolysate with laccase and peroxidase from the white-rot fungus T. versicolor. Appl Microb Biotechnol. 1998;49:691–697. [Google Scholar]
- 179.Martin C, Galbe M, Wahlbom CF, Hahn-Hagerdal B, Johnsson LJ. Ethanol production from enzymatic hydrolysates of sugarcane bagasse using recombinant xylose-utilising Saccharomyces cerevisiae. Enzyme Microb Technol. 2002;31:274–282. [Google Scholar]
- 180.Chandel AK, Kapoor RK, Singh A, Kuhad RC. Detoxification of sugarcane bagasse hydrolysate improves ethanol production by Candida shehatae NCIM 3501. Bioresour Technol. 2007;98:1947–1950. doi: 10.1016/j.biortech.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 181.Kojima Y, Tsukuda Y, Kawai Y, Tsukamoto A, Sugiura J, Sakaino M, Kita Y. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J Biol Chem. 1990;265:15224–15230. [PubMed] [Google Scholar]
- 182.Bulter T, Alcade M, Sieber V, Meinhold P, Schlachtbauer C, Arnold FH. Functional expression of a fungal laccase in Saccharomyces cerevisiae by direct evolution. Appl Environ Biotechnol. 2003;69:987–995. doi: 10.1128/AEM.69.2.987-995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cassland P, Jönsson LJ. Characterization of a gene encoding Trametes versicolor laccase A and improved heterologous expression in Saccharomyces cerevisiae by decreased cultivation temperature. Appl Microbiol Biotechnol. 1999;52:393–400. doi: 10.1007/s002530051537. [DOI] [PubMed] [Google Scholar]
- 184.Kiiskinen LL, Saloheimo M. Molecular cloning and expression in Saccharomyces cerevisiae of a laccase gene from the ascomycete Melanocarpus albomyces. Appl Environ Microbiol. 2004;70:137–144. doi: 10.1128/AEM.70.1.137-144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Larsson S, Cassland P, Jönsson LJ. Development of a Saccharomyces cerevisiae strain with enhanced resistance to phenolic fermentation inhibitors in lignocellulose hydrolysates by heterologous expression of laccase. Appl Environ Microbiol. 2001;67:1163–1170. doi: 10.1128/AEM.67.3.1163-1170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Piscitelli A, Giardina P, Mazzoni C, Sannia G. Recombinant expression of Pleurotus ostreatus laccases in Kluyveromyces lactis and Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2005;69:428–439. doi: 10.1007/s00253-005-0004-z. [DOI] [PubMed] [Google Scholar]
- 187.Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2006;71:339–349. doi: 10.1007/s00253-005-0142-3. [DOI] [PubMed] [Google Scholar]
- 188.Nilsson A, Gorwa-Grauslund MF, Hahn-Hägerdal B, Lidén G. Cofactor dependence in furan reduction by Saccharomyces cerevisiae in fermentation of acid-hydrolyzed lignocellulose. Appl Environ Microbiol. 2005;71:7866–7871. doi: 10.1128/AEM.71.12.7866-7871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pace NR. A molecular view of microbial diversity and the biosphere. Sci. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 190.Rondon MR, August PR, Bettermann AD. et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganism. Appl Environ Microbiol. 2000;66:2541–2547. doi: 10.1128/aem.66.6.2541-2547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bader J, Mast-Gerlach E, Popović MK. et al. Relevance of microbial coculture fermentations in biotechnology. J Appl Microbiol. 2010;109:371–387. doi: 10.1111/j.1365-2672.2009.04659.x. [DOI] [PubMed] [Google Scholar]
- 192.Energy development. http://en.wikipedia.org/wiki/Energy_development.
- 193.Bungay H. Product opportunities for biomass refining. Enzyme Microb Technol. 1992;14:501–507. [Google Scholar]
- 194.Brazil vs United States Ethanol Industries. Sugar cane based ethanol production in Brazil; http://www.soybeansandcorn.com/Brazil-US-Ethanol-Production. [Google Scholar]