Abstract
RMP has been shown to function in the transcription regulation through association with RNA polymerase (RNAP) II subunit RPB5. It also has been shown to be required for the proliferation of hepatocellular carcinoma (HCC) cells with an antiapoptotic property. In this article, we further demonstrate that RMP displays distinct features in HCC cells compared with normal hepatic cells. RMP expression is remarkably increased in various cancer cell lines including HCC cells when compared with normal cells. Depletion of RMP could inhibit the proliferation of HCC cells, but not the normal hepatic cells. RMP significantly prevented apoptosis of HCC cells in SMMC-7721 and HepG2, but had little effect on apoptosis in the normal hepatic cells. The mechanisms of RMP's distinct features rely on different responsive expressions of apoptosis factors induced by RMP in HCC and hepatic cells. Either overexpression or depletion of RMP significantly affected the expression of apoptosis factors in HCC cells. However, normal hepatic cells showed a tendency to resist RMP for the regulation of apoptosis. In the clinical samples, the increased expression of RMP in HCCs was also observed when compared with the matched non-tumor tissues from 30 HCC patients. The different expression levels of and distinct responses to RMP between HCC and hepatic cells suggest that RMP might serve as not only a biomarker for the diagnosis of HCC, but also a potential target for the HCC therapy.
Keywords: RMP, hepatocellular carcinoma cells
Introduction
The transcription of structure genes is carried out by the RNA polymerase (RNAP) II 1-3. There are 12 subunits of RNAP II, with RPB5, RPB6 and RPB8 shared by RNAP I, RNAP II and RNAP III 4. RPB5 is also an active communicating subunit which serves as targets of transcription factors 5-7. Hepatitis H virus (HBV) encodes a small X oncoprotein (HBx), which is responsible for the transformation of infected hepatic cells into hepatocellular carcinoma (HCC). HBx has been identified to target RPB5 to activate the transcription 8. Later on, a novel gene which encodes the RPB5-mediating protein (RMP) was isolated from a human HCC HepG2 cDNA library 9. An unusually large member of prefoldins (PFDs), termed as unconventional prefoldin RPB5 interactor (URI), was also identified as an alternative form of RMP, which associates with RPB5 and other small PFDs 10.
Although the majority of endogenous RMP/URI proteins are cytoplasmic 11-13, both human and yeast URI proteins were identified as a transcriptional factor 9, 10, 14, 15. The interactions between RMP/URI and RNA polymerase II enable some RMP/URI protein associated with chromatin. Polytene chromosome staining revealed that the majority of the RMP/URI positive bands are sites of active transcription 13.
As a transcriptional regulator, RMP represses the activated transcription by VP16 and HBx 9. A large fraction of genes activated in RMP/URI depletion cells contained a consensus-binding site for Gcn4p, a transcription factor related to amino acid biosynthesis. Functional analysis revealed that URI is a target of nutrient signalling and involved in controlling of transcription of nutrient-sensitive genes 10.
Interestingly, recently both groups of Dr. Krek and ours simultaneously uncovered that RMP/URI plays active roles in carcinogenesis 16, 17. RMP/URI was amplified and overexpressed in tissues and cell lines of human ovarian carcinomas 16. The increased copy number of URI genes is selectively required for the survival of ovarian cancer cells. By constitutive binding to PP1γ, URI detains PP1γ in an inactive state and thus enhances the S6K1 survival signalling 12. The release of PP1γ by the phosphorylation of URI results in apoptosis and cell death.
Almost at the same time, we find that RMP actively participates in the proliferation of HCC cells 17. Depletion of RMP in the HCC cells resulted in apoptosis which was enhanced by ionizing radiation. An antiapoptotic role of RMP was identified, which supports the proliferation and growth of HCC cells. Further in vivo investigation has shown that RMP is required for development and growth of HCC xenograft tumours.
RMP has been shown to be required for both the proliferation of HCC cells and the survival of ovarian cancer cells. Then our concerns are: is RMP/URI an oncogene specifically in HCC and ovarian cancer, or is RMP/URI simply a gene required for the carcinogenesis of multiple tumours? Is RMP/URI required for the growth of not only HCC cells, but also normal hepatic cells? Or is RMP/URI virtually a gene generally required for the proliferation and development of various normal cells? Therefore it is important to address the effect of RMP on the growth of both normal and cancer cells.
In this article, we show that RMP possesses distinct features in HCC cells compared with normal hepatic cells. RMP expression is increased in HCC cell lines as well as in the HCC tissues from patients, compared with normal cell lines or non-tumour tissues. Depletion of RMP inhibited the proliferation of HCC cells, but not the normal hepatic cells. The expressions of apoptosis factors respond differently to RMP, which suggests a different mechanism of RMP regulation pathway in the normal and HCC cells.
Materials and Methods
Plasmids, Cell Lines and Reagents. The plasmid pFlagCMV4 was purchased from Sigma. NKCflag-RMP was obtained from Dr. Seishi Murakami (Kanazawa University, Japan). pGPU6-Neo was purchased from Jima Co. (Shanghai, China). The plasmids pGPU6/Neo-SCR, pGPU6/Neo-RMPi for RMP depletion and pFlagCMV4-RMP for RMP overexpression were constructed as described previously 17. All cell lines used in this article were obtained from our own laboratory of the Department of Cell Biology at Soochow University. Cells were maintained in MEM medium (Gibco-BRL, Shanghai, China) supplemented with 10% Fetal Bovine Serum (FBS) (Sino-American Biotechnology Co, Shanghai, China). The Annexin-V-FITC/PI apoptosis detection kit was purchased from BD Biosciences (Shanghai, China). Methyl-thiazolyl-tetrazolium (MTT) was purchased from Sigma (Shanghai, China).
Antibodies against Bax, Bcl-2, p53 and β-actin were purchased from Boster Company (Wuhan, China). The RMP antibody was a product from AppTec Pharmaceutical (Suzhou, China).
Cell Culture, Transfection and Western Blot Analysis. Twenty four hours prior to transfection, a total of 2×105 cells were seeded into each well of a 24-well plate. Cell numbers were chosen to finally reach 90% confluency at the time of transfection. These cells were transfected with lipofectamaine 2000 (Invitrogen, Shanghai, China) according to the manufacturer's protocol. LF2000/DNA complexes were allowed to form in OptiMEM with final concentration of 10 μg/ml LF2000. Selection of stably transfected cells and Western blot analysis were conducted as previously described 17, 18.
Cell Viability Assay. MTT assays were carried out as described previously 17, 18. Briefly, after SMMC-7721 and HepG2 cells were cultured for 24 hours, 30 μl of MTT (5 mg/ml) was added to each well. Crystals were formed and then dissolved by adding 300 μl isopropanol acidified with HCl (0.04 N) containing 10% Triton X 100. Then, 200 μl of the blue formazan mixtures was transferred to 96-well plates. The plates were read at 570 nm using a Microplate Reader (Model 550, BIO-RAD, Shanghai, China).
Analysis of Apoptosis by Flow Cytometry. Cells were exposed to ionising radiation (IR) using a 60Co γ-irradiator at a dose of 2 or 6 Gy. Cells radiated or unradiated were then stained with Annexin V-FITC and propidium iodide (PI) and analyzed for apoptosis according to the manufacturer's instructions (BD Biosciences, Shanghai, China). Briefly, after two treatments with washing buffer, cells were resuspended in 400 μl of Dulbecco's phosphate-buffered saline (PBS). Then, 100 μl of this cell suspension was incubated with 10 μl of PI (50 μg/ml) and 5 μl Annexin V-FITC for 15 minutes at room temperature (RT) in the dark. Cells were then analyzed by flow cytometry. Cells that stained positive for only Annexin V-FITC were in the early stage of apoptosis, whereas cells that stained positive for both Annexin V-FITC and PI were in the stage of late apoptosis or primary necrosis.
Tumor Formation Assay in Nude Mice. For the in vivo tumor formation assay, 5×106 HCC cells of SMMC-7721 in 0.1 ml of PBS were injected subcutaneously into the right flank of 15-20 g female nude mice (Animal Centre of Soochow University). Xenograft tumors developed in the nude mice two weeks later after injection and then the mice were divided into 4 groups, each consisting of 5 mice. The xenograft tumors of each group mice were subjected to treatment by vectors of RMP overexpression, RMP depletion. Briefly, A suspension (25 μl/mouse) of Lipofectamine 2000 (20 μg) was mixed with DNA vectors (10 μg) in a final concentration of 100 μg /ml and incubated for 20 min to allow them to complex. Then the complex was delivered by multiple intratumor injection every other day for 2 weeks. Mice were monitored daily, and tumor growth was measured by caliper every 3 days. At the end of the fourth week, tumors were dissociated and evaluated. The animal operations and procedures were approved by the Committee on the Use of Live Animals in Teaching and Research of Soochow University. The tumor inhibition rates were calculated as follows: tumor inhibition rate (%)=(1-weight of experimental group tumor/weight of control group tumor)×100%.
Human Tissue Samples. Hepatocellular carcinoma and the matched non-tumor hepatic tissues as well as clinical data were obtained from Sun Yat-Sen University Cancer Center, with the approval of institutional review boards and the ethics committee of Yat-Sen University. Informed consent was obtained from all subjects.
Reverse transcription and Quantitative real time Polymerase Chain Reaction (qRT-PCR). Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Shanghai, China) following the manusfactures instructions and was reverse-transcribed using the Thermoscript RT system (Invitrogen, Shanghai, China), according to the manufacturer's protocol. The reaction was performed in triplicate in a total volume of 20 μl containing 10 μl SsoFast EvaGreen supermix (Bio-Rad) with SYBR Green, 2 μl cDNA, 2 μl each of the primers, and 4 μl RNase-free water. The PCR program was 94 °C for 2 min, followed by 40 cycles at 94°C for 15 s, 60 °C for 15 s, and 72 °C for 30 s. Quantitative real-time PCR assay was run on a Mini OpticonTM Real-time PCR instrument. Each reaction contains 3 technical replicates for qRT-PCR analysis. Primers for RMP were 5'- TCC GAA TAA ATA CTG GAA AG -3' and 5'-AAG GCT CTG TAA ATG TCT GC -3'. Primers for Bax were 5'- TTT TGC TTC AGG GTT TCA TC -3' and 5'- GAC ACT CGC TCA GCT TCT TG -3'. Primers for Bcl-2 were 5'- GGT GGG AGG GAG GAA GAA -3' and 5'- CGC AGA GGC ATC ACA TCG -3'. Primers for caspase-3 were 5'- AGA GCT GGA CTG CGG TAT TGA G -3' and 5'- GAA CCA TGA CCC GTC CCT TG -3'. Primers for GAPDH were 5'-GAC CTG ACC TGC CGT CTA-3' and 5'- AGG AGT GGG TGT CGC TGT -3'.
Immunohistochemistry Detection. Tissues from HCC patients and nude mice were subjected to formalin fixation, paraffin embedding and sectioning for immunohistochemistry assays as describe previously 17. The slides were blocked for 30 minutes in PBST containing 3% BSA (PBST-BSA) at 37°C and incubated overnight at 4°C with mouse monoclonal antibodies (1:500 in PBST-BSA). Slides were then washed 3 times in PBST (10 minutes at RT) and incubated for 2 hours with the secondary antibody (anti-mouse HRP, 1:200) in PBST at RT. After incubation, slides were washed three times (10 minutes each) in PBST and mounted with 3 μl of Vectashield for further analysis. Sections were examined at high power (x 400) under a standard light microscope. Cell staining was regard as positive if nuclear was homogeneous stained or 10% or more of cytoplasm was heterogeneously stained.
Statistical analysis- Values are expressed as the mean +/- standard error (SE). Student's t-test was used to determine the significance of the difference between compared groups. P < 0.05 indicated significant differences.
Results
RMP expression is increased in cancer cells and HCC tissues. In our previous work, it was observed that overexpression of RMP promoted the proliferation of HCC cells, while depletion of RMP inhibited the proliferation of HCC cells 17. Wondering if RMP expression is increased in multiple cancer cells, we decided to study the RMP expression in various cancer and normal cell lines.
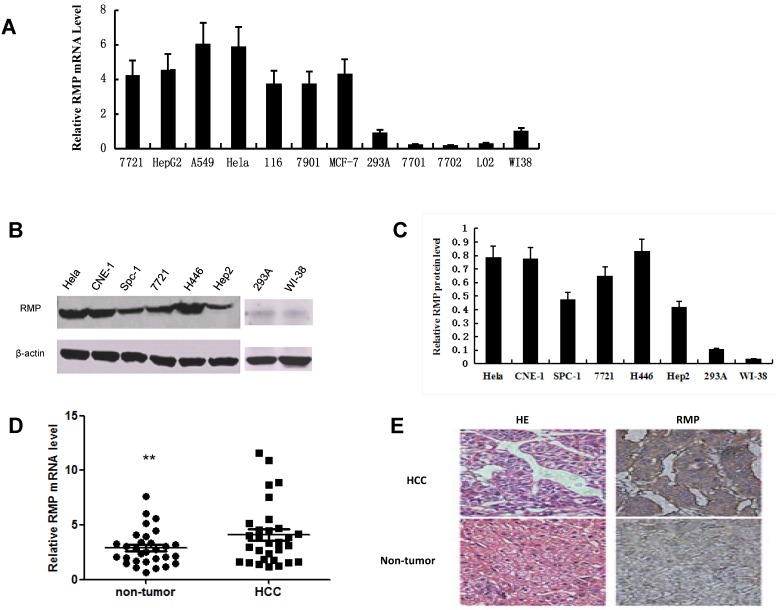
We examined the mRNA expression of RMP in 7 human cancer cell lines: HCC cells of SMMC-7721 and HepG2 17, lung adenocarcinoma A-549 cells 19, the commonly used HeLa cells of cervical carcinoma, colon carcinoma HCT-116 cells 20, SGC-7901 cells, a perpetual cell line which was derived from the gastric adenocarcinoma with lymphatic metastasis21 and the breast carcinoma MCF-7 cells 22. The mRNA expression of RMP was also examined in human normal cell lines including kidney QBI-293A cells 23, WI-38 cells of a diploid human cell culture line composed of fibroblasts from normal embryonic lung of female 24 and three normal hepatic cell lines of QSG-770125, HL-7702 26 and L02 27. Apparently, all 7 cancer cell lines expressed higher RMP mRNA level than that in 5 normal cell lines (Fig. 1A).
Fig 1.
RMP expression is increased in cancer cells and HCC tissues. (A) The mRNA expression of RMP in normal and cancer cell lines was examined by qRT-PCR as described in Materials and Methods. The relative mRNA levels of RMP was normalized against GAPDH and depicted graphically. (B) The RMP protein expression was examined by Western blot analysis as described in Materials and Methods. The relative levels of RMP protein expression was normalized against β-actin and depicted graphically in (C). Results are reported as mean +/-S.D. of three independent experiments. (D) Higher expression of RMP in HCC. Total RNA was extracted from 30 pairs of tumor and matched non-tumor tissues of HCC patients. Quantitative real-time RT-PCR was performed for the detection of RMP mRNA. The expression levels RMP mRNA were shown in close dots (Non-tumor) and squares (HCC). The relative expression levels of RMP mRNA was normalized against β-actin. Student's t test, **, p<0.01, relative to controls. (E) The tumor and matched non-tumor tissues from HCC patients were sectioned and subjected to H-E staining and immunostaining with antibody against RMP. The sections were observed in a microscope at 400 magnifications. Cell line abbreviation: 7721 and HepG2: Human Hepatocellular carcinoma cells; A-549: Human lung adenocarcinoma A-549 cells; HeLa: Human cervical carcinoma HeLa cells; 116: Human colon carcinoma HCT-116 cells; 7901: Human gastric carcinoma SGC-7901 cells; MCF-7: Human breast carcinoma MCF-7 cells; 293A: Normal human kidney QBI-293A cells; WI-38: Normal human diploid embryonic lung fibroblasts WI-38 cells; 7701, 7702 and L02: Normal hepatic cell lines of QSG-7701, HL-7702 and L02; CNE-1: Human nasopharyngeal carcinoma CNE-1 cells; Spc-1: Human lung adenocarcinoma Spc-1 cells; H446: Human small cell lung cancer H446 cells; Hep2: Human caucasian larynx carcinoma squamous Hep2 cells.
We next determined if RMP protein expression was consistent with its mRNA expression. Western blot analysis was carried out with extract of cancer and normal cell lines. As shown in Fig. 1B and 1C, the protein expression of RMP were increased in cervical carcinoma HeLa cells, human nasopharyngeal carcinoma CNE-1 cells 28, lung adenocarcinoma Spc-1 cells 29, HCC cell line SMMC-7721, small cell lung cancer H446 cells 30and caucasian larynx carcinoma squamous Hep2 cells 31, compared with human normal cell lines of QBI-293A and WI-38.
We also examined the expression of RMP in HCC tissues from 30 patients and the matched non-tumor hepatic tissues which are adjacent to carcinoma from same patients. The results by qRT-PCR showed that RMP expression in HCC was significantly higher than that in the matched non-tumor hepatic tissues (Fig.1D), which is consistent with the in vitro experiments. Immunostaining with antibody against RMP also showed elevated expression of RMP in HCCs compared with non-tumor hepatic tissues (Fig.1E).
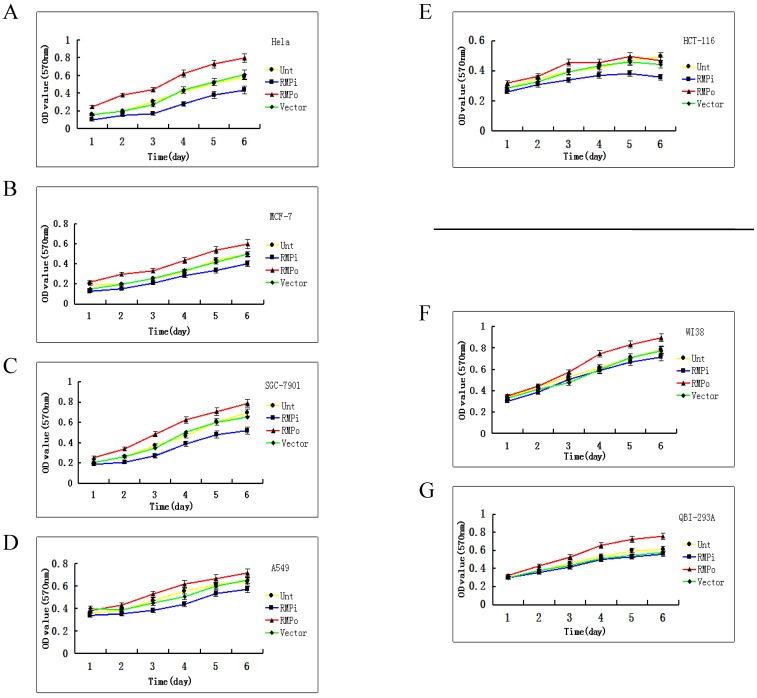
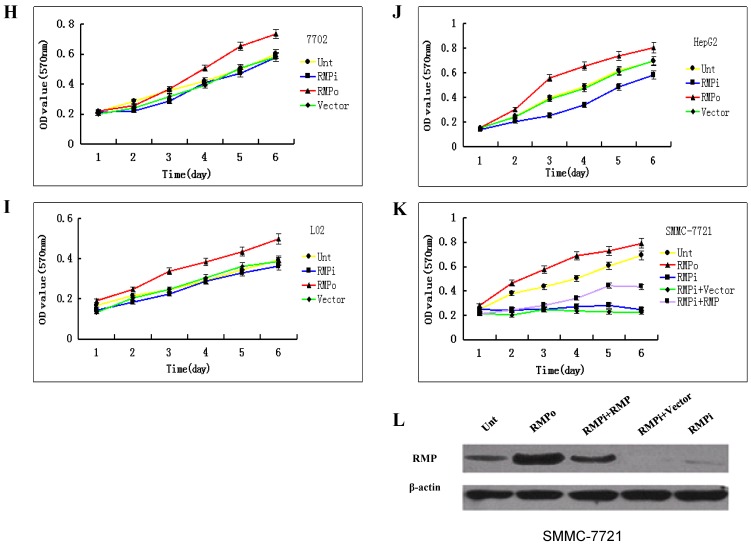
The cell growth is sensitive to RMP in cancer cells, but not in normal cells. As stated above, the expression of RMP is higher in cancer cells than in normal cells. Then we explored the relationship between the RMP expression level and cell growth of cancer and normal cells. The effect of RMP on the cell proliferation was examined in various human cancer cell lines of HeLa (Fig.2A), MCF-7 (Fig.2B), SGC-7901 (Fig.2C), A-549 (Fig.2D) and HCT-116 (Fig.2E). Interestingly, the growth of cancer cells was promoted by the overexpression of RMP (RMPo), while depletion of RMP by RNA interference (RMPi) dramatically reduced the proliferation of all five cancer cell lines (Fig.2A-2E), compared with the cells untransfected (Unt) or transfected with vector alone (Vector).
Fig 2.
The growth of cancer cells, but not normal cells is sensitive to RMP depletion. The plasmids for RMP overexpression (RMPo), RMP interference (RMPi), or vector alone (Vector) were introduced into various cancer cell lines (A-E) and normal cell lines as indicted (F, G). Untransfected (Unt) parent cells were applied as a control. The growth of cells was determined by MTT assay as described in Material and Methods. Human normal hepatic cell lines HL-7702 (H), L02 (I) and HCC cell lines of HepG2 (J) were transfected with expression plasmids as indicated. (K) HCC cell lines of SMMC-7721 stably depleted of RMP (RMPi), were transiently transfected RMP expression plasmid (RMPi+RMP) or vector alone (RMPi+Vector). The SMMC-7721 cells stably overexpressing RMP (RMPo), cells transfected with vector alone (Vector) and untransfected (Unt) parent cells were applied as controls. The growth of cells was determined by MTT assay everyday starting 24 hours after transfection as described in Material and Methods. (L) The expression of RMP in the transfected SMMC-7721 cells was determined by Western blot analysis as described in Materials and Methods. Results are reported as mean +/-S.D. of three independent experiments.
We also examined if RMP was also required for the growth of normal cells. The overexpression of RMP promotes the proliferation of normal cell lines of WI-38 cells (Fig.2F) and QBI-293A cells (Fig.2G). However, depletion of RMP did not significantly affect the proliferation of either cell line, compared with the untransfected parent cells and cells with vector alone.
We then studied the relation between the growth of HCC and normal hepatic cells and the RMP expression level. As shown in Fig.2H and 2I, although the overexpression of RMP promoted the growth of normal hepatic cells, RMP depletion had little effect on the proliferation of normal hepatic cells HL-7702 and L02. There was no significant difference between the growths of normal hepatic cells with or without RMP depletion. In contrast, the proliferation of HCC cells of HepG2 and SMMC-7721 increased significantly when RMP was overexpressed, while depletion of RMP dramatically reduced the growth rate of HCC cells (Fig.2J and 2K).
To confirm the effect of RMP on the growth HCC cells, SMMC-7721 cells stably depleted of RMP (RMPi) were transfected back with RMP expression vector (RMPi+RMP) (Fig. 2K). The rescue transfection restored the expression of RMP in the SMMC-7721 cells, as shown by the Western blot analysis (Fig.2L). Expectedly, consistent with the restoration of RMP expression, the proliferation of HCC cells originally depleted of RMP (RMPi) was significantly enhanced by the RMP restoration (RMPi+RMP) (Fig. 2K).
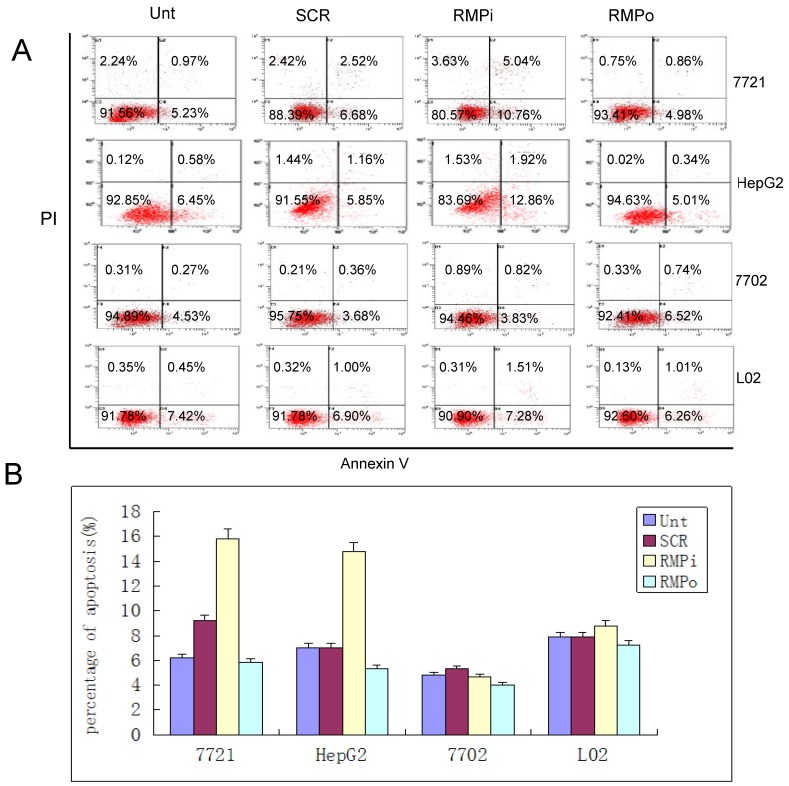
RMP depletion induces apoptosis in HCC cells, but not in normal hepatic cells. As apoptosis is closely related to the control of cell proliferation, we examined the effect of RMP on the apoptosis of HCC cells and normal hepatic cells as well. The overexpression of RMP inhibited the apoptosis of HCC cells, but did not significantly reduce the apoptosis of normal hepatic cells (Fig.3A and 3B). The depletion of RMP dramatically induced the apoptosis of both HCC cells of HepG2 and SMMC-7721. However, the depletion of RMP had little effect on the apoptosis of normal hepatic cells HL-7702 and L02 (Fig.3A and 3B). These results indicate that HCC cells are sensitive to RMP in apoptosis, while normal hepatic cells are resistant to RMP in apoptosis, which is consistent with the effect of RMP on the cell proliferation in HCC and normal hepatic cells, as shown in Fig.2.
Fig 3.
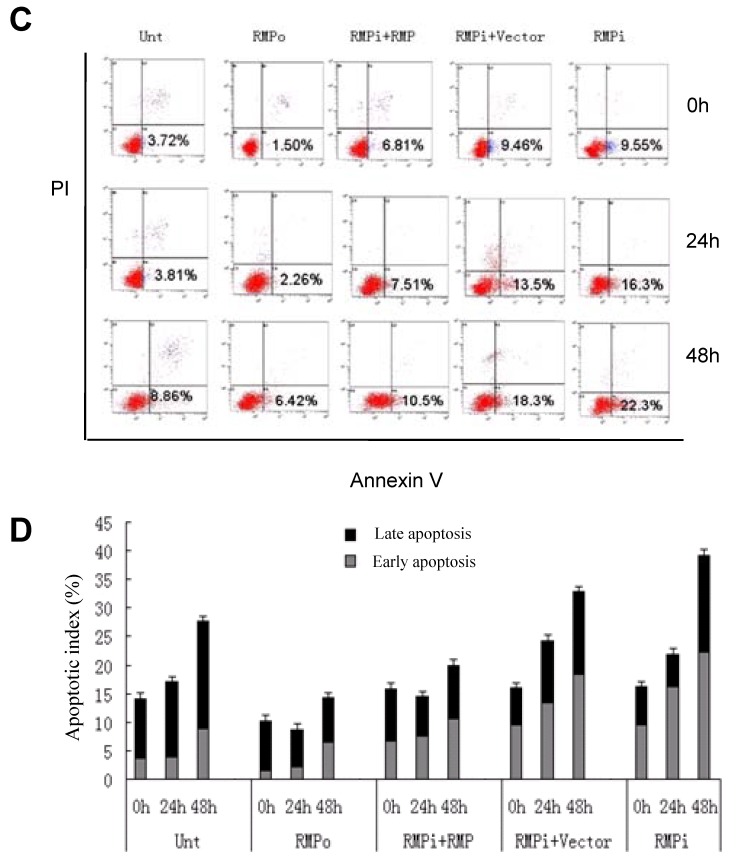
RMP depletion induces apoptosis in HCC cells, but not in normal hepatic cells. (A) HCC cell lines (SMMC-7721 and HepG2) and normal hepatic cell lines (HL-7702 and L02) were transfected with expression plasmids as indicated and subjected to apoptosis analysis by flow cytometry 72 hours after transfection. (B) The percentage of apoptotic cells were scored and represented in histogram with bar as mean +/-S.D. of three independent experiments. (C) HCC cells SMMC-7721 were transfected with expression plasmids as indicated. 24 hours later the transfected cells were subjected for irradiation at 6 Gy and then cultured for additional times as indicated. Finally cells were harvested and subjected to apoptosis analysis by flow cytometry 48 hours after irradiation. (D) The percentage of apoptotic cells were scored and represented in histogram with bar as mean +/-S.D. of three independent experiments.
To confirm the sensitivity of HCC cells to RMP in apoptosis, we also investigated the induced apoptosis by 60Co γ-irradiation (Fig.3C and 3D). Irradiation increased the apoptotic population of HCC cells after 24h and 48h. The HCC cells with RMP overexpression (RMPo) demonstrated less apoptotic cells compared with the untransfected parent cells, while RMP depletion (RMPi) greatly enhanced the induced apoptosis. The rescued expression of RMP in the cells depleted of RMP (RMPi+RMP) partially restored the resistance of SMMC-7721 to the apoptosis induced by irradiation (Fig.3C and 3D).
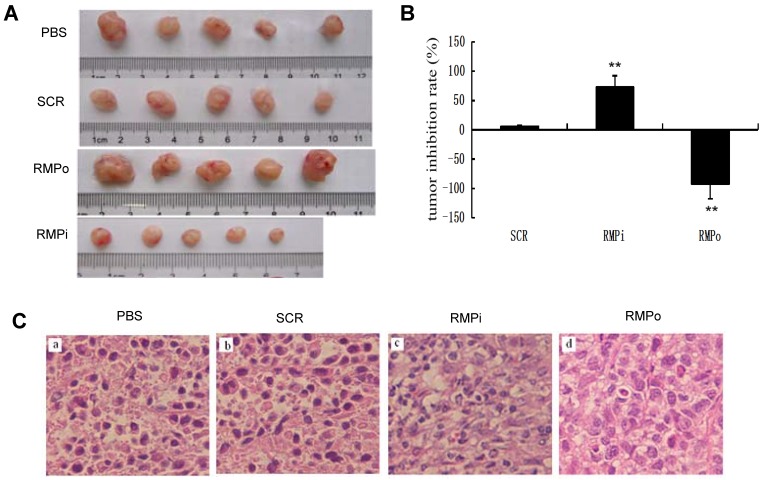
RMP promotes the growth of HCC cells in vivo. Then we checked the effect of RMP on the growth of xenograft tumors. As shown in Fig. 4A, The HCC xenograft tumors were formed in the nude mice inoculated with SMMC-7721 cells, while there was no tumor formed after inoculation with HL-7702 hepatic cells. The developed tumors were then treated with the various plasmids for RMP overexpression or RMP depletion (Fig. 4A). The results demonstrated that the overexpression of RMP (RMPo) significantly promoted the growth of HCC xenograft tumors, while the growth of tumors treated with RMP depletion vector (RMPi) was inhibited compared with controls (Fig. 4A and 4B). The tumor tissues were then examined. Interestingly, the nucleus size of RMPo tumor cells was significantly larger than that of RMPi and controls. Also more mitotic figures could be seen in RMPo tumors than in RMPi group (Fig. 4C).
Fig 4.
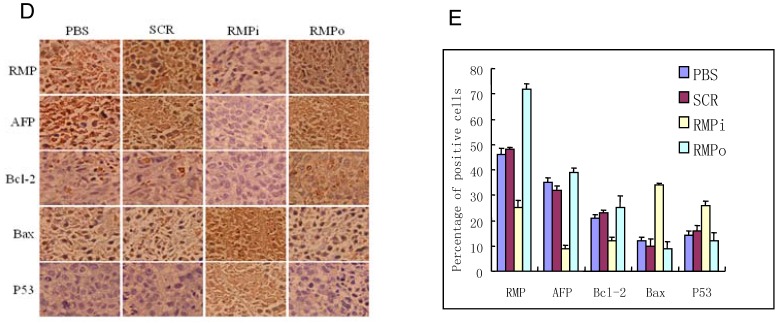
RMP promotes the growth of HCC xenograft tumors in vivo. (A) SMMC-7721 HCC cells were inoculated subcutaneously for the tumor formation in nude mice as described in the Materials and Methods. The developed tumors in the mice were then treated with expression plasmids as indicated, or with PBS as a control. Finally tumors were dissected and photographs from each group are shown. (B) Tumor inhibition rate was indicated as percentage of growth reduction under each treatment relative to cells treated with PBS and represented by histogram. Student's t test, **, p<0.01, relative to controls. (C) The HCC xenograft tumors treated with expression plasmids as indicated or with PBS as a control were sectioned and subjected to H-E staining for pathological examination. (D) Immunohistochemical examination of RMP, AFP, Bax, Bcl-2 and p53 in the HCC xenograft tumors treated with expression plasmids as indicated, or with PBS as a control. (E) The percentage of positive cells expressing RMP, AFP, Bax, Bcl-2 and p53 was scored and represented by histogram.
Immunohistochemical analysis demonstrated that the expression of RMP was enhanced in the xenograft tumors from RMP-overexpressing cells (RMPo), while decreased in tumors from cells depleted of RMP (RMPi), compared with control tumors treated with PBS and SCR (Fig. 4D and 4E). The expression of HCC marker AFP and antiapoptotic factor Bcl-2 were also elevated in RMPo tumors, while decreased in RMPi cells. In contrast, the expression of p53 and apoptotic factor Bax increased in RMPi tumors, but decreased in the RMPo tumors (Fig. 4D and 4E).
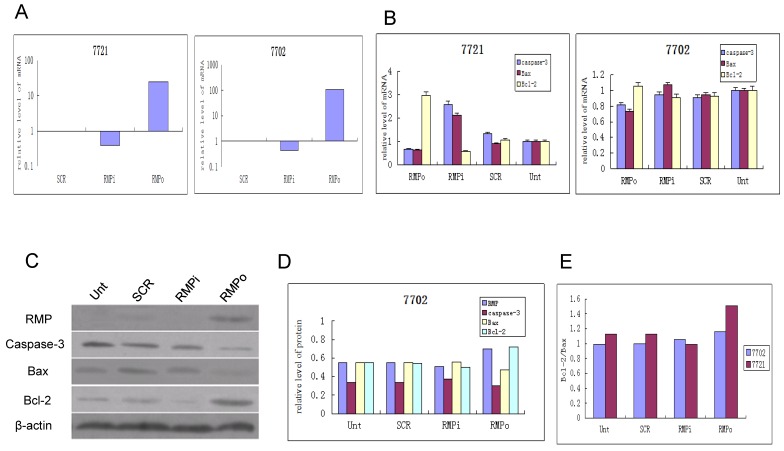
The expression of apoptosis factors is differentially regulated by RMP in HCC and normal hepatic cells. To investigate the mechanism of distinct responses to RMP in HCC and hepatic cells, we examined the expression responses of apoptotic factors in SMMC-7721 and HL-7702 cells. As shown in Fig. 5A, RMP was highly overexpressed in both HCC and hepatic cells when transfected with RMP expression vector (RMPo). RMPi significantly reduced RMP expression level in SMMC-7721 and HL7702 cells.
Fig 5.
The expression of apoptosis factors is affected by RMP differently in HCC and normal hepatic cells. HCC cell SMMC-7721 and normal hepatic cells HL-7702 were transfected with expression plasmids as indicated. qRT-PCR was performed to determine the expression level of RMP (A) and apoptosis factors (B). The relative levels of mRNA expression was normalized against GAPDH and represented by histogram. Results are showed as mean +/-S.D of three independent experiments. (C) Normal hepatic cells HL-7702 were transfected with vectors of RMP expression (RMPo), RMP depletion or scrambled sequence (SCR). The untrasfected (Unt) cells were applied as an additional control. The Western blot analysis was carried out with antibodies as indicated. The relative protein levels of Caspase-3, Bax and Bcl-2 were normalized against β-actin and plotted in (D). The protein ratio of Bcl-2 against Bax (Bcl-2/Bax) in hepatic cells HL-7702 and HCC cells SMMC-7721 were calculated and plotted in (E).
The mRNA expression of apoptosis factor of both caspase-3 and Bax significantly increased in the SMMC-7721 cells with RMP depletion (RMPi), but decreased in the cells with RMP overexpression (RMPo) (Fig.5B, left panel). In contrast, the expression of antiapoptotic factor Bcl-2 dramatically increased in the HCC cells with RMPo, but decreased in the HCC cells with RMPi. Interestingly, little effect of RMP could be detected on the expression of Bax, caspase-3 and Bcl-2 in the normal hepatic cells with either RMPo or RMPi (Fig. 5B, right panel).
In our previous work, it was also observed that the protein expression of Bax and Bcl-2 efficiently regulated by RMP in HCC cells, as demonstrated by Western blot analysis and immunohistochemical assay 17. We further examined the effect of RMP on the protein expression of apoptosis and antiapoptosis factors in the normal hepatic cells HL7702 by Western blot analysis (Fig. 5C). The results showed that although RMPo or RMPi also affected the expression of Caspase-3, Bax and Bcl-2, the degree of effect of RMP on these factors is slight (Fig. 5C and Fig. 5D). The protein ratio of Bcl-2 and Bax is much lower in the normal hepatic cells transfected with RMP expression or depletion vectors, when compared with the ratio in HCC cells of our previous work 17 (Fig. 5E).
Discussion
RMP/URI is involved in nutrient availability with cell growth and proliferation by controlling nutrient metabolism genes 10. Rapamycin treatment and amino acid starvation in yeast results in a similar transcription response, which overlaps the expression of genes involved in the amino acid biosynthesis in favor of cell survival. Yeast URI-1 has been identified to be involved in TOR signaling pathway through nutrient utilization 10. In C. Elegans, URI-1 was also found to be required in the proliferation, genome stability and survival of germ cells32.
RMP/URI was previously reported to be overexpressed or amplified in ovarian cancer cells 16. In this article, we found that RMP is overexpressed virtually in multiple cancer cell lines. The exogenous overexpression of RMP in HCC cells accelerated the cell growth and division, while depletion of RMP significantly reduced the growth rate of cultured cancer cells including HCC cells. In vivo, RMP promoted the HCC xenograft tumors in the nude mice, while depletion of RMP resulted in a retarded growth of the xenograft tumors. All these evidence support a conclusion of RMP as an oncogene in the carcinogenesis of HCC.
The oncogenicity of RMP results from a fact that RMP actually is an antiapoptic factor as we reported previously 17. Overexpression of RMP inhibited the apoptosis of HCC cells; while depletion of RMP significantly induced the apoptosis of HCC cells in vitro and in vivo. The expression of apoptotic factors, such as Bax was induced by RMP depletion, and inhibited by the overexpression of RMP. In contrast, the expression of antiapoptotic factors, such as Bcl-2 was increased by the RMP overexpression, and inhibited by RMP depletion. The antiapoptotic effect of RMP might be, at least partially, through regulating the expression of both apoptotic and antiapoptotic factors.
In this work, we identified the distinct sensitivity of normal hepatic and HCC cells to RMP. Overexpression or depletion of RMP in cancer cells significantly increased or decreased the growth rate of HCC cells. However, little effect was observed in normal hepatic cells. It was also observed that overexpression or depletion of RMP significantly inhibited or promoted the apoptosis of HCC cells. RMP expression in HCC was also confirmed to be significantly higher than that in non-tumor tissues. All these observation indicates that cancer cells, including HCC cells are more sensitive to RMP than normal hepatic cells, which might also be true for other cancer cells with their normal counterparts.
The detailed mechanism still remains unclear about the distinct sensitivity of HCC and normal hepatic cells to RMP. The distinct sensitivity could result from a different modification system of RMP in normal and cancer cells. However, it is clear that the expression of apoptotic factors is induced by RMP differently in normal hepatic and HCC cells. Either overexpression or depletion of RMP in HCC cells and in the tissues of xenograft tumors significantly decreased or increased the expression of apoptotic factors, such as Bax, Caspase-3 and antiapoptotic factor Bcl-2, which was consistent with our previous work by Western blot analysis 17. However, as presented in this work, the expression of apoptosis and antiapoptosis factors is resistant to the regulation by RMP in normal hepatic cells with an unknown mechanism.
The distinct RMP responses of apoptosis in normal hepatic and HCC cells should be further explored and applied in the clinic investigation and practice. The different expression levels of RMP in cells of normal hepatic and HCC cells indicate that RMP might serve as a potential biological marker for the diagnosis of HCC. The distinct sensitivity of normal hepatic and HCC cells also makes RMP a potential and exciting target for HCC therapy. Specifically targeting RMP with RNA interference would effectively destroy the HCC cells with increased expression of RMP and high sensitivity to RMP, but may barely damage the normal hepatic cells and tissues, although further exploration and medical trial are required for.
We found a tendency of increased expression of RMP not only in HCC cells, but also in multiple cancer cells. In contrast, the expression of RMP is low in multiple normal cells, besides the normal hepatic cells. These finding suggested that RMP might also be a general oncogene for multiple cancers and thus a potential marker for the diagnosis of various cancers. The fact that the depletion of RMP significantly reduced the proliferation of many kinds of cancer cells, while had little effect on the growth of normal cells, suggests that RMP might also potentially be a specific target for the treatment of multiple cancers, although more number and varieties of cells and tissue samples require confirmation.
Acknowledgments
This study was partly supported by Nature Science Foundation of China (81172347, 81272301), Open Grant from State Key Laboratory of Oncology in South China (2010) and the Graduate Innovation Project of Jiangsu Province (CXZZ12-0834 and CXZZ13-0828).
References
- 1.Petesch SJ, Lis JT. Overcoming the nucleosome barrier during transcript elongation. Trends in genetics: TIG. 2012;28:285–94. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palangat M, Larson DR. Complexity of RNA polymerase II elongation dynamics. Biochimica et biophysica acta. 2012;1819:667–72. doi: 10.1016/j.bbagrm.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcello A. RNA polymerase II transcription on the fast lane. Transcription. 2012;3:29–34. doi: 10.4161/trns.3.1.19147. [DOI] [PubMed] [Google Scholar]
- 4.Zaros C, Briand JF, Boulard Y, Labarre-Mariotte S, Garcia-Lopez MC, Thuriaux P. et al. Functional organization of the Rpb5 subunit shared by the three yeast RNA polymerases. Nucleic acids research. 2007;35:634–47. doi: 10.1093/nar/gkl686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye X, Xiao P, Hu X, Chen Y, Zhang L, Xie W. et al. Crystallization and preliminary X-ray analysis of the RPB5 subunit of human RNA polymerase II. Acta crystallographica Section F, Structural biology and crystallization communications. 2011;67:1391–3. doi: 10.1107/S1744309111033288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le TT, Zhang S, Hayashi N, Yasukawa M, Delgermaa L, Murakami S. Mutational analysis of human RNA polymerase II subunit 5 (RPB5): the residues critical for interactions with TFIIF subunit RAP30 and hepatitis B virus X protein. Journal of biochemistry. 2005;138:215–24. doi: 10.1093/jb/mvi119. [DOI] [PubMed] [Google Scholar]
- 7.Wei W, Dorjsuren D, Lin Y, Qin W, Nomura T, Hayashi N. et al. Direct interaction between the subunit RAP30 of transcription factor IIF (TFIIF) and RNA polymerase subunit 5, which contributes to the association between TFIIF and RNA polymerase II. J Biol Chem. 2001;276:12266–73. doi: 10.1074/jbc.M009634200. [DOI] [PubMed] [Google Scholar]
- 8.Cheong JH, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. The EMBO journal. 1995;14:143–50. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorjsuren D, Lin Y, Wei W, Yamashita T, Nomura T, Hayashi N. et al. RMP, a novel RNA polymerase II subunit 5-interacting protein, counteracts transactivation by hepatitis B virus X protein. Molecular and cellular biology. 1998;18:7546–55. doi: 10.1128/mcb.18.12.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gstaiger M, Luke B, Hess D, Oakeley EJ, Wirbelauer C, Blondel M. et al. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–12. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- 11.Delgermaa L, Hayashi N, Dorjsuren D, Nomura T, Thuy le TT, Murakami S. Subcellular localization of RPB5-mediating protein and its putative functional partner. Molecular and cellular biology. 2004;24:8556–66. doi: 10.1128/MCB.24.19.8556-8566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R. et al. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Molecular cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Kirchner J, Vissi E, Gross S, Szoor B, Rudenko A, Alphey L. et al. Drosophila Uri, a PP1alpha binding protein, is essential for viability, maintenance of DNA integrity and normal transcriptional activity. BMC molecular biology. 2008;9:36. doi: 10.1186/1471-2199-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenxiang W, Jun Xia G, Cui Qing Z, Feng Yan S, Dorjbal DORJSUREN YL. Interaction with general transcription factor IIF (TFIIF) is required for the suppression of activated transcription by RPB5-mediating protein (RMP) Cell research. 2003;13:111–20. doi: 10.1038/sj.cr.7290155. [DOI] [PubMed] [Google Scholar]
- 15.Mita P, Savas JN, Djouder N, Yates JR 3rd, Ha S, Ruoff R. et al. Regulation of androgen receptor-mediated transcription by RPB5 binding protein URI/RMP. Molecular and cellular biology. 2011;31:3639–52. doi: 10.1128/MCB.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK. et al. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer cell. 2011;19:317–32. doi: 10.1016/j.ccr.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Gu J, Zheng Q, Li M, Lian X, Miao J. et al. RPB5-mediating protein is required for the proliferation of hepatocellular carcinoma cells. J Biol Chem. 2011;286:11865–74. doi: 10.1074/jbc.M110.136929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu J, Sun D, Zheng Q, Wang X, Yang H, Miao J. et al. Human Elongator complex is involved in cell cycle and suppresses cell growth in 293T human embryonic kidney cells. Acta biochimica et biophysica Sinica. 2009;41:831–8. doi: 10.1093/abbs/gmp072. [DOI] [PubMed] [Google Scholar]
- 19.Lin CC, Kuo CT, Cheng CY, Wu CY, Lee CW, Hsieh HL. et al. IL-1 beta promotes A549 cell migration via MAPKs/AP-1- and NF-kappaB-dependent matrix metalloproteinase-9 expression. Cellular signalling. 2009;21:1652–62. doi: 10.1016/j.cellsig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Hu R, Chen N, Yao J, Zhao Q, Zhang F, Li ZY. et al. The role of Nrf2 and apoptotic signaling pathways in oroxylin A-mediated responses in HCT-116 colorectal adenocarcinoma cells and xenograft tumors. Anti-cancer drugs. 2012;23:651–8. doi: 10.1097/CAD.0b013e3283512703. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Z, Zhu JS, Xu ZP, Zhang Q. Lentiviral vector-mediated siRNA knockdown of the YAP gene inhibits growth and induces apoptosis in the SGC7901 gastric cancer cell line. Mol Med Rep. 2011;4:1075–82. doi: 10.3892/mmr.2011.543. [DOI] [PubMed] [Google Scholar]
- 22.Spink BC, Bennett JA, Lostritto N, Cole JR, Spink DC. Expression of the aryl hydrocarbon receptor is not required for the proliferation, migration, invasion, or estrogen-dependent tumorigenesis of MCF-7 breast cancer cells. Molecular carcinogenesis. 2012. [DOI] [PMC free article] [PubMed]
- 23.Arguin G, Caron AZ, Elkoreh G, Denault JB, Guillemette G. The transcription factors NFAT and CREB have different susceptibilities to the reduced Ca2+ responses caused by the knock down of inositol trisphosphate receptor in HEK 293A cells. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2010;26:629–40. doi: 10.1159/000322330. [DOI] [PubMed] [Google Scholar]
- 24.Cmielova J, Havelek R, Jiroutova A, Kohlerova R, Seifrtova M, Muthna D. et al. DNA damage caused by ionizing radiation in embryonic diploid fibroblasts WI-38 induces both apoptosis and senescence. Physiological research / Academia Scientiarum Bohemoslovaca. 2011;60:667–77. doi: 10.33549/physiolres.932083. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F, Xu R, Zhao MJ. QSG-7701 human hepatocytes form polarized acini in three-dimensional culture. Journal of cellular biochemistry. 2010;110:1175–86. doi: 10.1002/jcb.22632. [DOI] [PubMed] [Google Scholar]
- 26.Zhang ZY, Pan LJ, Zhang ZM. Functional interactions among STIM1, Orai1 and TRPC1 on the activation of SOCs in HL-7702 cells. Amino acids. 2010;39:195–204. doi: 10.1007/s00726-009-0398-5. [DOI] [PubMed] [Google Scholar]
- 27.Cao MM, Xu WX, Li CY, Cao CZ, Wang ZD, Yao JW. et al. Hepassocin regulates cell proliferation of the human hepatic cells L02 and hepatocarcinoma cells through different mechanisms. Journal of cellular biochemistry. 2011;112:2882–90. doi: 10.1002/jcb.23202. [DOI] [PubMed] [Google Scholar]
- 28.Shao J, Tang J, Ji J, Zhou W. Therapy effects of gold nanorods on the CNE-1 nasopharyngeal carcinoma cell line. Drug design, development and therapy. 2012;6:297–301. doi: 10.2147/DDDT.S36706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang MH, Hu YD, Xu Y, Xiao Y, Luo Y, Song ZC. et al. Human mesenchymal stem cells enhance autophagy of lung carcinoma cells against apoptosis during serum deprivation. International journal of oncology. 2013;42:1390–8. doi: 10.3892/ijo.2013.1810. [DOI] [PubMed] [Google Scholar]
- 30.Sun MZ, Guan Z, Liu S, Zhou X, Wang N, Shao S. et al. Caveolin-1 interferes cell growth of lung cancer NCI-H446 cell through the interactions with phospho-ERK1/2, estrogen receptor and progestin receptor. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2012;66:242–8. doi: 10.1016/j.biopha.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Jeong YJ, Lee SW, Kim D, Oh SJ, Lim HS. et al. EGCG induces apoptosis in human laryngeal epidermoid carcinoma Hep2 cells via mitochondria with the release of apoptosis-inducing factor and endonuclease G. Cancer letters. 2010;290:68–75. doi: 10.1016/j.canlet.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 32.Parusel CT, Kritikou EA, Hengartner MO, Krek W, Gotta M. URI-1 is required for DNA stability in C. elegans. Development. 2006;133:621–9. doi: 10.1242/dev.02235. [DOI] [PubMed] [Google Scholar]