Abstract
Mutation or common intronic variants in cardiac ion channel genes have been suggested to be associated with sudden cardiac death caused by idiopathic ventricular tachyarrhythmia. This study aimed to find mutations in cardiac ion channel genes of Korean sudden cardiac arrest patients with structurally normal heart and to verify association between common genetic variation in cardiac ion channel and sudden cardiac arrest by idiopathic ventricular tachyarrhythmia in Koreans. Study participants were Korean survivors of sudden cardiac arrest caused by idiopathic ventricular tachycardia or fibrillation. All coding exons of the SCN5A, KCNQ1, and KCNH2 genes were analyzed by Sanger sequencing. Fifteen survivors of sudden cardiac arrest were included. Three male patients had mutations in SCN5A gene and none in KCNQ1 and KCNH2 genes. Intronic variant (rs2283222) in KCNQ1 gene showed significant association with sudden cardiac arrest (OR 4.05). Four male sudden cardiac arrest survivors had intronic variant (rs11720524) in SCN5A gene. None of female survivors of sudden cardiac arrest had SCN5A gene mutations despite similar frequencies of intronic variants between males and females in 55 normal controls. Common intronic variant in KCNQ1 gene is associated with sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia in Koreans.
Keywords: Idiopathic Ventricular Arrhythmia; Death, Sudden, Cardiac; Mutation; Cardiac Ion Channel
INTRODUCTION
Ventricular tachyarrhythmia is predominant cause of sudden cardiac arrest (1). Ventricular tachyarrhythmia has been demonstrated to occur even in structurally normal heart (2). In this idiopathic ventricular tachyarrhythmia, its association with mutations in cardiac ion channels or associated proteins has been suggested, but it remains to be clarified (1). Cardiac ion channels are important in normal cardiac electrophysiology, and their mutations can lead to sudden cardiac arrest by generating ventricular arrhythmia (3). Mutations in genes encoding cardiac sodium channel such as SCN5A and potassium channel such as KCNQ1 and KCNH2 have been suggested to be associated with sudden cardiac death (4). It has been reported that common intronic variants (rs2283222 located in intron 11 in KCNQ1 and rs11720524 located in intron 1 in SCN5A) were significantly associated with sudden cardiac death (4). However, genetic studies in the Asian patients with sudden cardiac death are rare. Therefore, it is needed to demonstrate association between sudden cardiac arrest and mutations in cardiac ion channel in the Asian patients with idiopathic ventricular tachyarrhythmia.
This study aims to find mutations in cardiac ion channel genes of Korean sudden cardiac arrest patients with structurally normal heart and to demonstrate association of common variants in cardiac ion channel genes with sudden cardiac arrest in Koreans.
MATERIALS AND METHODS
Study populations were Korean subjects who survived sudden cardiac arrest caused by ventricular tachyarrhythmia in one institute. Exclusion criteria were as follows; any evidence of structural heart disease in transthoracic echocardiography (TTE), existence of abnormal findings in coronary angiography (CAG) or spasm provocation test with ergonovine if performed, inability to confirm the presence of ventricular tachyarrhythmia by routine electrocardiography (ECG) performed or electrophysiologic study, presence of typical ECG pattern suggestive of other disease which can cause ventricular tachyarrhythmia such as Brugada syndrome or long QT syndrome, and history of heart diseases such as coronary artery disease or other cardiomyopathy. Of these patients, we collected blood samples of the subjects who understood and agreed to our study.
Genomic DNA was extracted from peripheral blood leukocytes using the Wizard Genomic DNA Purification kit following the manufacturer's instructions (Promega, Madison, WI, USA). All coding exons and their flanking introns of the KCNQ1, KCNH2, and SCN5A gene were amplified using primer sets designed by the authors (Supplemental Tables 1~3). The polymerase chain reaction (PCR) was performed with a thermal cycler (model 9700, Applied Biosystems, Foster City, CA, USA) as follows: 32 cycles of denaturation at 94℃ for 30 sec, annealing at 60℃ for 30 sec, and extension at 72℃ for 30 sec. After treatment of the amplicon (5 µL) with 10 U shrimp alkaline phosphatase and 2 U exonuclease I (USB Corp., Cleveland, OH, USA), direct sequencing was performed with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems) on the ABI Prism 3100xl genetic analyzer (Applied Biosystems). To describe sequence variations, we followed the guidelines by the Human Genome Nomenclature Committee (HGVS) that 'A' of the ATG translation start site was numbered +1 for DNA sequence and the first methionine was numbered +1 for protein sequence.
We analyzed rs2283222 located at intron 11 in KCNQ1 and rs11720524 located at intron 1 in SCN5A to identify the presence of common variants reported to be associated with sudden cardiac death (4). We performed this analysis on the basis of the method described in the previous study (4). To inspect associations of these common variants with sudden cardiac arrest, we collected normal controls who have not experienced sudden cardiac arrest. These control subjects were collected randomly from gene bank of our institute. We could not get these patients' detailed information following guidelines of our institutional review board for genetic study. We also got blood samples of these control subjects and analyzed these intronic variants.
We also collected the data of patient's demographics, clinical presentation, and treatment from history taking and review of medical records. Echocardiographic images, CAG, and ECG were reviewed by experienced specialists.
Continuous variables were described as mean ± SD. Categorical variables were expressed as a number and as a percentage (%). Fisher's exact test was used to compare the frequency of aforementioned intronic variants in cardiac ion channel genes between case and control groups. Odds ratio (OR) and 95% confidence interval (CI) of these intronic variants in KCNQ1 and SCN5A genes for sudden cardiac arrest was calculated using logistic regression analysis. A P value < 0.05 was considered to be statistically significant. Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS) version 18.0 for Windows.
Ethics statement
Written informed consents were obtained from all the study subjects. Our institutional review board approved this study (Samsung Medical Center 2010-07-014).
RESULTS
Twenty-one Korean patients who survived sudden cardiac arrest caused by ventricular tachyarrhythmia and had no history of heart diseases were collected. Three patients were excluded because of abnormal findings on TTE. Two patients were excluded because their ECG showed typical pattern of Brugada syndrome. One patient showed positive finding in spasm provocation test during CAG and was excluded. Fifteen subjects remained eligible for analysis. These study population consisted of 11 males and 4 females, with a mean age of 39.7 ± 12.3 yr (range, 18 to 66 yr). Thirteen subjects of these took CAG and had normal findings. Two subjects who did not take CAG showed no regional wall motion abnormality on TTE and did not have any other abnormal findings on laboratory examination and history takings suggestive of the presence of ischemic heart disease. Eleven patients undertook spasm provocation test with ergonovine and showed negative findings. Ventricular tachyarrhythmia was confirmed with ECG performed when sudden cardiac arrest happened. Five patients had ventricular tachycardia, and seven patients had ventricular fibrillation. Three patients showed both ventricular tachycardia and fibrillation on their ECGs. Only one patient had family history of sudden cardiac death. All of 15 subjects received implantable cardioverter-defibrillator (ICD) therapy. Corrected QT interval was 0.448 ± 0.040 sec in female survivors and 0.421 ± 0.040 sec in male survivors, and its difference between genders was statistically insignificant (P value obtained by Mann-Whitney U test was 0.19). Clinical characteristics of these 15 subjects are presented in Table 1.
Table 1.
Clinical characteristics of patients who survived sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia
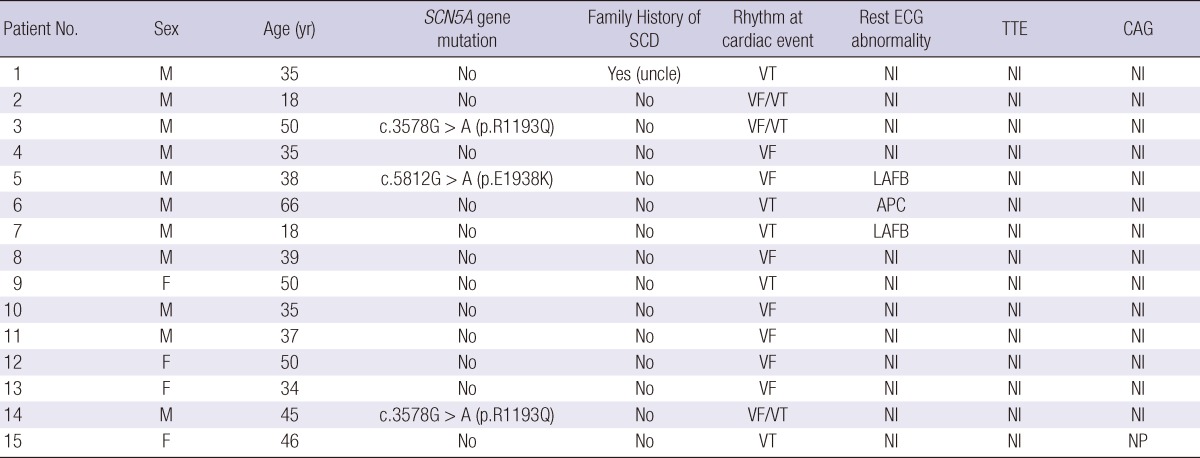
APC, atrial premature complex; CAG, coronary angiography; ECG, electrocardiography; LAFB, left anterior fascicular block; Nl, normal; NP, not performed; SCD, sudden cardiac death; TTE, transthoracic echocardiography; VF, ventricular fibrillation; VT, ventricular tachycardia.
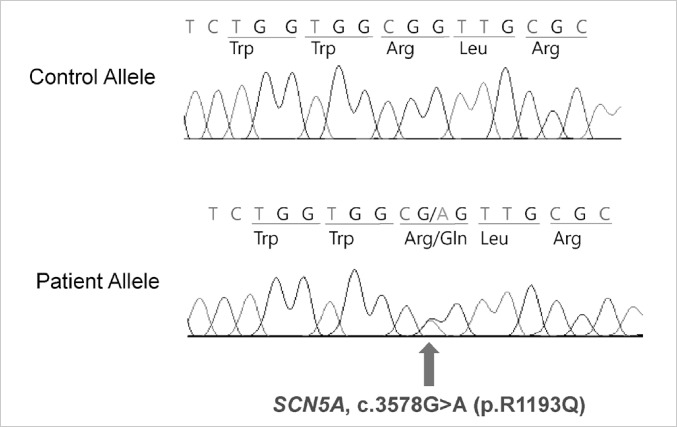
In the analysis of exons of SCN5A gene, mutations were found in three patients. Direct sequencing in genes encoding cardiac sodium channel of SCN5A revealed G-to-A mutation at position 3578 in exon 20 of SCN5A, which causes the substitution of arginine (R), a positively charged amino acid, for a glutamine (Q), a neutral amino acid, at position 1193 (Fig. 1). Also G-to-A mutation at position 5812 in exon 28 of SCN5A, which causes the substitution of lysine for glutamate at position 1938 was noted (Fig. 2). There was no mutation in the analysis of KCNQ1 and KCNH2 gene. Interestingly, all the three patients who had SCN5A gene mutation were male.
Fig. 1.
Direct sequencing in genes encoding cardiac sodium channel of SCN5A reveals G-to-A mutation at position 3578 in exon 20 of SCN5A, which causes the substitution of arginine (R), a positively charged amino acid, for a glutamine (Q), a neutral amino acid, at position 1193.
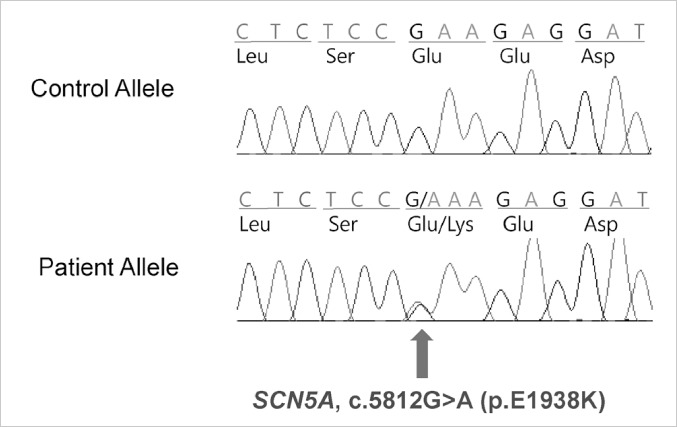
Fig. 2.
Direct sequencing in genes encoding cardiac sodium channel of SCN5A reveals G-to-A mutation at position 5812 in exon 28 of SCN5A, which causes the substitution of lysine for glutamate at position 1938.
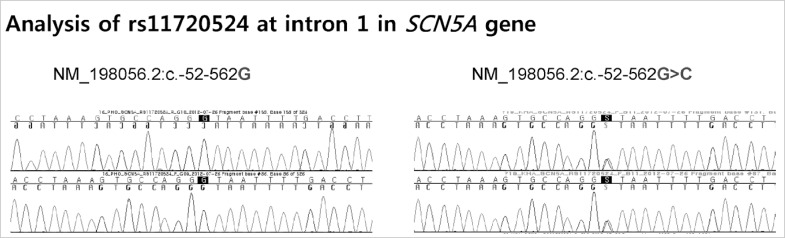
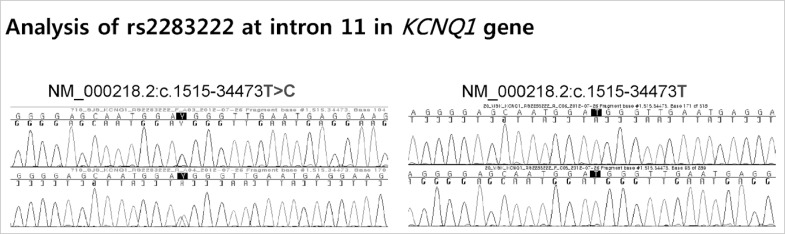
Blood samples from 14 of 15 patients were analyzed for common intronic variation in cardiac ion channel genes except one whose blood sample for this analysis was missing. Four patients of 14 patients had C-allele at rs11720524 in SCN5A gene (C-allele frequency was 0.143 [4/28]) (Fig. 3). Notably, all these four patients were male. All 14 subjects had T-allele at rs2283222 in KCNQ1 gene (T-allele frequency was 0.893 [25/28]) (Fig. 4). We collected 55 normal control subjects (21 males, 34 females) who have not experienced sudden cardiac arrest for analysis of intronic variants in KCNQ1 and SCN5A genes. Forty-eight subjects (18 males, 30 females) had T-allele at rs2283222 in KCNQ1 gene (T-allele frequency was 0.673 [74/110]). Eight subjects (3 males, 5 females) had C-allele at rs11720524 in SCN5A gene (C-allele frequency was 0.073 [8/110]). Frequencies of these variants in all the case and control subjects were 0.717 (99/138) at rs2283222 in KCNQ1 gene and 0.087 (12/138) at rs11720524 in SCN5A gene. Frequency of T-allele at rs2283222 in KCNQ1 gene was similar, and that of C-allele at rs11720524 in SCN5A gene was lower than those reported from previous studies (4). Associations between sudden cardiac arrest and these intronic variants in KCNQ1 and SCN5A genes in our study were compared with those reported previously (4) in Table 2. The frequency of T-allele at rs2283222 in KCNQ1 gene showed statistically significant differences between case and control groups (P value obtained by Fisher's exact test was 0.02). In this analysis, we numbered all the appearances of T-alleles at rs2283222 in KCNQ1 gene by counting two alleles per one subject. In additional recessive model, the presence of T-allele (TT) was also significantly different between two groups (P value obtained by Fisher's exact test was 0.04). In dominant model, the presence of T-allele (TT/TC) did not show significant differences between two groups (P value obtained by Fisher's exact test was 0.33). This discrepancy between two models probably results from higher percentage of TT homozygote in survivors of sudden cardiac arrest (11/14, 79%) compared with that in normal controls (26/55, 47%) (Table 3). In accordance with previous study (4), the presence of this intronic variant appeared to be associated with sudden cardiac arrest (OR [95% CI]) for sudden cardiac arrest was 4.05 [1.15-14.32]). The frequency of C-allele at rs11720524 in SCN5A gene did not show statistically significant difference between case and control groups (P value obtained by Fisher's exact test was 0.26), and its presence seemed to be associated with sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia, but did not show statistical significance (OR [95% CI] for sudden cardiac arrest was 2.13 [0.59-7.64]). We analyzed associations between sudden cardiac arrest and these intronic variants in KCNQ1 and SCN5A genes stratified by sex. The presence of T-allele at rs2283222 in KCNQ1 gene still had positive association with sudden cardiac arrest, but it did not show statistical significance in male patients (OR [95% CI] for sudden cardiac arrest in males = 2.27 [0.56-9.17], P = 0.25; T-allele frequency was 0.850 [17/20] and 0.714 [30/42] in male case and control groups, respectively). This association seemed to be slightly stronger in females although OR could not be calculated because T-allele frequency was 1.000 in female survivors of sudden cardiac arrest (T-allele frequency was 1.000 [8/8] and 0.647 [44/68] in female case and control groups, respectively). Association between sudden cardiac arrest and C-allele at rs11720524 in SCN5A gene became stronger in male patients, but did not reach statistical significance, either (OR [95% CI] for sudden cardiac arrest in males = 3.25 [0.65-16.20], P = 0.15).
Fig. 3.
Analysis of rs11720524 located at intron 1 in SCN5A gene shows the presence of common variants. Four patients of 14 patients had C-allele instead of G-allele at rs11720524 in SCN5A gene.
Fig. 4.
Analysis of rs2283222 located at intron 11 in KCNQ1 gene shows the presence of common variants. All 14 subjects have T-allele at rs2283222 in KCNQ1 gene (T-allele frequency was 0.893).
Table 2.
Associations between intronic variants in SCN5A or KCNQ1 gene and sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia
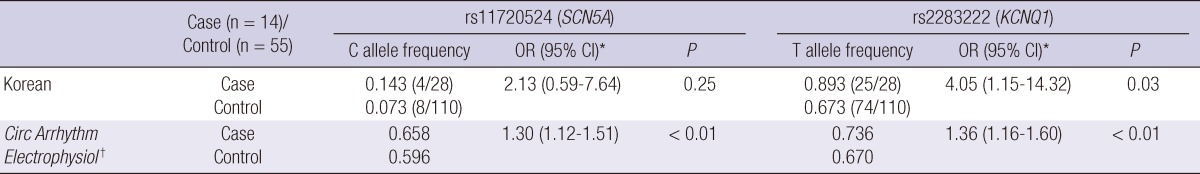
*Odds ratio (OR) and 95% confidence interval (CI) of intronic variants in SCN5A or KCNQ1 gene for sudden cardiac arrest was calculated with logistic regression analysis; †Associations between common intronic variants in SCN5A and KCNQ1 genes and sudden cardiac death reported by Albert CM, et al.(4).
Table 3.
The frequency of T-allele at rs2283222 in KCNQ1 gene and analysis using dominant and recessive model
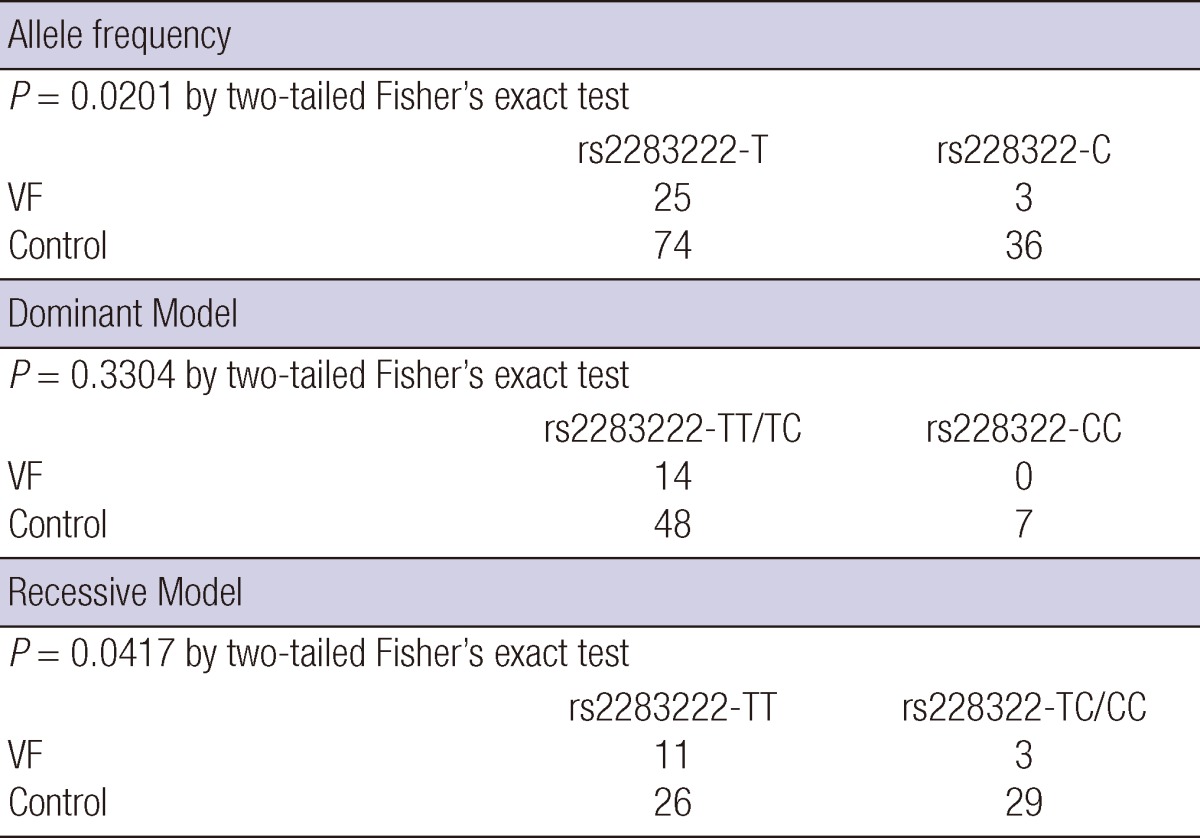
VF, ventricular fibrillation.
DISCUSSION
In this study, we found mutations of exons of SCN5A gene in Korean survivors of sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia. We have investigated common intronic variants in SCN5A and KCNQ1 genes reported to be associated with sudden cardiac death (4) in our study population and control groups and found its association with sudden cardiac arrest. As we know, this is the first report of mutation and common variants in cardiac ion channel genes for survivors of sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia in Koreans.
In our study, survivors of sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia showed male predominance (11/15, 73%), in accordance with previous studies that reported higher incidence of sudden cardiac death in males (5, 6). Spontaneous or induced ventricular tachyarrhythmia was more frequently noted in male patients who survived sudden cardiac death than females (7, 8). Gender difference in risk of sudden cardiac death can result from different incidences of other heart diseases such as coronary artery disease between males and females. Previous study reported that coronary artery diseases were more frequently observed in male survivors than female survivors of sudden cardiac death (9). However, our 15 study participants showed male predominance despite less likelihood of concomitant coronary artery disease considering the results of CAG and other diagnostic tests previously mentioned. Some studies have suggested sex differences in other properties such as ventricular repolarization or hormonal status may be other possible explanation for male predominance of sudden cardiac death (5, 10), but it remains to be clarified. It is noted that Ito-mediated action potential dome is a prerequisite for the development of phase 2 reentry dependent tachyarrhythmia and Brugada syndrome are predominantly seen in males whose Ito is significantly greater than that seen in females (11).
Mutations were noted at exons of SCN5A gene in three patients, but none in KCNQ1 and KCNH2 genes. Our study population showed significant association between sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia and T-allele at rs2283222 in KCNQ1 gene. The mechanism underlying possible strengthening of this association in females is unclear. A previous study has reported higher KCNQ1 mRNA level in female patients than male patients with long QT syndrome (12). Female gender has been suggested to be a risk factor for drug-induced long QT and cardiac arrhythmias in animal study (13). Gender difference in association between sudden cardiac arrest and genetic variants in KCNQ1 gene needs to be confirmed by further studies for idiopathic ventricular tachyarrhythmia patients. C-allele at rs11720524 in SCN5A gene seemed to be associated with sudden cardiac arrest in Koreans despite statistical insignificance. All the seven survivors of sudden cardiac arrest who had mutation at exons or intronic variant in SCN5A gene were male. None of female survivors had these mutation and intronic variants in SCN5A gene even though intronic variants in SCN5A gene showed similar frequencies between males and females in our normal control group. When stratified by sex, the presence of C-allele at rs11720524 in SCN5A gene had stronger association with sudden cardiac arrest in males, but statistically insignificant (OR [95% CI] for sudden cardiac arrest was 3.25 [0.65-16.20]). Although the mechanism underlying this result is uncertain, we can speculate that some factors in males may strengthen association between sudden cardiac arrest and this intronic variant at rs11720524 in SCN5A gene. Some authors have reported that difference in hormonal effects between males and females may be the answer to gender differences in many arrhythmias (5, 10). Male gonadal hormones have been reported to influence the susceptibility to reperfusion-induced sustained ventricular tachycardia in recent animal study (14). The explanation of the sex-related difference in sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia remains unknown (7, 8).
Our study has several limitations. First, two patients (one female patient aged 46 yr, one male patient aged 18 yr) of fifteen survivors of sudden cardiac arrest did not undertake CAG. Nevertheless, they showed low probability of ischemic heart disease considering other diagnostic tests. Two patients did not take spasm provocation test during CAG, but had no symptoms suggestive of variant angina. Comparison of common intronic variants was conducted with case-control design. Bias caused by nature of case-control study such as selection bias could be introduced. Second, we could not get detailed information about control subjects' clinical characteristics. Therefore, programmed selection and matching was unfeasible, and this is our study's limitation. Third, data on potential confounding factors influencing association between sudden cardiac arrest and genetic variants in cardiac ion channel were unavailable. An additional limitation of the study is that the number of survivors of sudden cardiac arrest was relatively small. Therefore further studies are needed to verify this association.
In conclusion, mutations in genes encoding cardiac sodium channel (SCN5A) were noted in Korean male survivors of sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia. Intronic variant in KCNQ1 gene was associated with sudden cardiac arrest in Koreans. Intronic variant in SCN5A gene seemed to be associated with sudden cardiac arrest especially in males. We suggest that sex should be considered when assessing associations between genetic variants and sudden cardiac arrest caused by idiopathic ventricular tachyarrhythmia, and further studies are needed to confirm these associations and their changes by sex.
Footnotes
This work was supported in part by the grant from the Korean Society of Cardiology (2012) and the Grant of Center for Genome Research, Samsung Biomedical Research Institute (#GRC D-B1-005-1).
The authors have no conflicts of interest to disclose.
Supplemental Material
References
- 1.Chopra N, Knollmann BC. Genetics of sudden cardiac death syndromes. Curr Opin Cardiol. 2011;26:196–203. doi: 10.1097/HCO.0b013e3283459893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai CF, Chen SA, Tai CT, Chiang CE, Ding YA, Chang MS. Idiopathic ventricular fibrillation: clinical, electrophysiologic characteristics and long-term outcomes. Int J Cardiol. 1998;64:47–55. doi: 10.1016/s0167-5273(98)00004-7. [DOI] [PubMed] [Google Scholar]
- 3.Napolitano C, Bloise R, Monteforte N, Priori SG. Sudden cardiac death and genetic ion channelopathies: long QT, Brugada, short QT, catecholaminergic polymorphic ventricular tachycardia, and idiopathic ventricular fibrillation. Circulation. 2012;125:2027–2034. doi: 10.1161/CIRCULATIONAHA.111.055947. [DOI] [PubMed] [Google Scholar]
- 4.Albert CM, MacRae CA, Chasman DI, VanDenburgh M, Buring JE, Manson JE, Cook NR, Newton-Cheh C. Common variants in cardiac ion channel genes are associated with sudden cardiac death. Circ Arrhythm Electrophysiol. 2010;3:222–229. doi: 10.1161/CIRCEP.110.944934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolbrette D, Naccarelli G, Curtis A, Lehmann M, Kadish A. Gender differences in arrhythmias. Clin Cardiol. 2002;25:49–56. doi: 10.1002/clc.4950250203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarnoz MJ, Curtis AB. More reasons why men and women are not the same (gender differences in electrophysiology and arrhythmias) Am J Cardiol. 2008;101:1291–1296. doi: 10.1016/j.amjcard.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 7.Freedman RA, Swerdlow CD, Soderholm-Difatte V, Mason JW. Clinical predictors of arrhythmia inducibility in survivors of cardiac arrest: importance of gender and prior myocardial infarction. J Am Coll Cardiol. 1988;12:973–978. doi: 10.1016/0735-1097(88)90463-9. [DOI] [PubMed] [Google Scholar]
- 8.Wigginton JG, Pepe PE, Bedolla JP, DeTamble LA, Atkins JM. Sex-related differences in the presentation and outcome of out-of-hospital cardiopulmonary arrest: a multiyear, prospective, population-based study. Crit Care Med. 2002;30:S131–S136. doi: 10.1097/00003246-200204001-00002. [DOI] [PubMed] [Google Scholar]
- 9.Albert CM, McGovern BA, Newell JB, Ruskin JN. Sex differences in cardiac arrest survivors. Circulation. 1996;93:1170–1176. doi: 10.1161/01.cir.93.6.1170. [DOI] [PubMed] [Google Scholar]
- 10.James AF, Choisy SC, Hancox JC. Recent advances in understanding sex differences in cardiac repolarization. Prog Biophys Mol Biol. 2007;94:265–319. doi: 10.1016/j.pbiomolbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Di Diego JM, Cordeiro JM, Goodrow RJ, Fish JM, Zygmunt AC, Pérez GJ, Scornik FS, Antzelevitch C. Ionic and cellular basis for the predominance of the Brugada syndrome phenotype in males. Circulation. 2002;106:2004–2011. doi: 10.1161/01.cir.0000032002.22105.7a. [DOI] [PubMed] [Google Scholar]
- 12.Moric-Janiszewska E, Głogowska-Ligus J, Paul-Samojedny M, Węglarz L, Markiewicz-Łoskot G, Szydłowski L. Age-and sex-dependent mRNA expression of KCNQ1 and HERG in patients with long QT syndrome type 1 and 2. Arch Med Sci. 2011;7:941–947. doi: 10.5114/aoms.2011.26604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu HR, Remeysen P, Somers K, Saels A, De Clerck F. Female gender is a risk factor for drug-induced long QT and cardiac arrhythmias in an in vivo rabbit model. J Cardiovasc Electrophysiol. 2001;12:538–545. doi: 10.1046/j.1540-8167.2001.00538.x. [DOI] [PubMed] [Google Scholar]
- 14.Lujan HL, Kramer VJ, DiCarlo SE. Sex influences the susceptibility to reperfusion-induced sustained ventricular tachycardia and beta-adrenergic receptor blockade in conscious rats. Am J Physiol Heart Circ Physiol. 2007;293:H2799–H2808. doi: 10.1152/ajpheart.00596.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.