Abstract
How bacteria coordinate cell growth with division is not well understood. Bacterial cell elongation is controlled by actin–MreB while cell division is governed by tubulin–FtsZ. A ring-like structure containing FtsZ (the Z ring) at mid-cell attracts other cell division proteins to form the divisome, an essential protein assembly required for septum synthesis and cell separation. The Z ring exists at mid-cell during a major part of the cell cycle without contracting. Here, we show that MreB and FtsZ of Escherichia coli interact directly and that this interaction is required for Z ring contraction. We further show that the MreB–FtsZ interaction is required for transfer of cell-wall biosynthetic enzymes from the lateral to the mature divisome, allowing cells to synthesise the septum. Our observations show that bacterial cell division is coupled to cell elongation via a direct and essential interaction between FtsZ and MreB.
Keywords: cell division, cell elongation, FtsZ, MreB, septum
Introduction
Actin and tubulin homologues are essential for the coordination of internal processes in all cells. These common and conserved proteins form structural elements that make up the cytoskeleton, a dynamic protein network that orchestrates many cellular processes, including protein recruitment and trafficking, cell motility and cell division in both eukaryotic and prokaryotic organisms (Erickson, 2007; Fletcher and Mullins, 2010). To gain a deeper insight into the cellular functions of cytoskeletons, it is important to understand how activities of both actin and tubulin families of proteins are coordinated. In eukaryotes, actin is regulated by tubulin microtubules that carry protein factors able to alter actin filament activities (Basu and Chang, 2007), while in prokaryotes such interactions through additional protein factors have not been described. The concept of a prokaryotic cytoskeleton arose from the discovery of two essential proteins: FtsZ and MreB, which are homologues of tubulin and actin capable of forming macro-molecular structures (Lutkenhaus et al, 1980; Wachi et al, 1987; Bork et al, 1992; De Boer et al, 1992). FtsZ and MreB are essential for cell division and shape maintenance respectively (Bi and Lutkenhaus, 1991; Jones et al, 2001). After a decade of research on MreB and over two on FtsZ in many bacterial species, it is clear that a general theme of organising cell morphology via an essential underlying cytoskeletal structure is an approach taken by most bacterial cells (Michie and Löwe, 2006; Typas et al, 2012). However, it is uncertain how the cytoskeleton interacts with the different components of the cell to carry out this function.
MreB is widely conserved among rod-shaped bacteria and interacts with the cell-wall elongation machinery through a membrane-spanning complex comprising of MreC, MreD and RodZ (Divakaruni et al, 2005; Kruse et al, 2005; Shiomi et al, 2008; Bendezú et al, 2009; White et al, 2010). This complex acts together with the peptidoglycan (PG) cell-wall biosynthetic enzymes to produce lateral PG, thereby giving bacterial rods their characteristic shapes. Pioneering work suggested that MreB formed a continuous helical pattern on the inside face of the cytoplasmic membrane (Jones et al, 2001). However, higher-resolution microscopy has indicated that MreB forms short filamentous structures in vivo (Domínguez-Escobar et al, 2011; Garner et al, 2011; Van Teeffelen et al, 2011). Biochemical characterisation of MreB has been difficult; although this family of proteins can form filaments in vitro, their dynamics are far from fully understood. However, solved crystal structures have provided insights into the biochemical properties of MreB including its establishment as a true actin homologue and defining the MreB–RodZ interaction surface (Van den Ent et al, 2001, 2010). MreB proteins from different species are known to directly interact with the cytoplasmic membrane, though the exact mechanism of binding varies between species (Salje et al, 2011). In E. coli, MreB has an N-terminal amphipathic helix that directly anchors MreB filaments to the inner leaflet of the cytoplasmic membrane (Salje et al, 2011).
FtsZ polymers, in conjunction with FtsA, ZapA, ZapB and other condensation factors, form a ring-like macro-molecular complex at mid-cell called the Z ring (De Boer, 2010). The regulated assembly of this structure is responsible for the spatial and temporal coordination of bacterial cell division. Recruitment of additional cell division factors to the Z ring occurs in an ordered mostly linear pattern, each protein requiring the localisation of upstream factors for its localisation forming a mature divisome complex (De Boer, 2010). Interestingly, the Z ring forms early in the cell cycle and persists without contracting until late in the cell cycle, indicating that maturation of the divisome occurs in two steps (Aarsman et al, 2005); the first being Z ring establishment, the second, downstream factor recruitment and Z ring contraction. The signal or events that trigger Z ring contraction are unknown.
When the divisome is complete, it provides the biochemical and mechanical activities required for septal PG synthesis, septation and separation (De Boer, 2010). In addition to cell division, the Z ring has an emerging secondary role in pre-septal PG synthesis, prior to synthesis of the septum (De Pedro et al, 1997; Aaron et al, 2007; Varma et al, 2007). The enzymatic requirements for pre-septal PG synthesis are not fully understood, although this process requires ZipA and seems to depend on the activity of penicillin-binding protein (PBP)2 rather than PBP3 (Wientjes and Nanninga, 1989; Varma and Young, 2009; Potluri et al, 2012).
The mreB gene is essential in E. coli under conditions that support rapid cell growth; however, it is possible to physiologically suppress the requirement for mreB by overexpression of FtsZ (Kruse et al, 2005; Bendezú and de Boer, 2008). Cells conditionally suppressed in this way grow as irregular spheres as they have lost the mechanism ensuring lateral PG incorporation (Kruse et al, 2005). The mechanism for this suppression is unclear, although it has been suggested that overexpression of FtsZ allows formation of Z rings, which will physically reach around the diameter of spherical cells (Kruse et al, 2005). Alternatively, additional FtsZ could help overcome membrane perturbation events brought about by uncoupling of membrane biosynthesis rates with cell volume in mreB mutants (Bendezú and De Boer, 2008).
Several indirect observations have raised the possibility that MreB plays a role in cell division. On a morphological level, compromising MreB function in bacterial cells gives both cell elongation and division phenotypes (Wachi and Matsuhashi, 1989; Fenton et al, 2010). The most direct evidence to suggest an involvement of MreB in division comes from Immuno-Fluorescence Microscopy (IFM) studies of Caulobacter crescentus, indicating that MreB forms ring-like structures at mid-cell that colocalise with the Z rings (Figge et al, 2004). In E. coli, MreB also localises as bands or rings, often positioned at mid-cell in pre-divisional cells (Vats and Rothfield, 2007; Vats et al, 2009). However, independent studies using IFM and GFP-labelled MreB did not observe ring-like MreB structures (Karczmarek et al, 2007; Swulius and Jensen, 2012). Moreover, different fusions between MreB and fluorescent proteins yield different subcellular patterns (Swulius and Jensen, 2012). Thus, the localisation pattern of MreB relative to FtsZ has not been firmly established in E. coli.
Here, we report a novel role of MreB in bacterial cell division using E. coli as the model organism. Microscopic observations validate previous suggestions that MreB is recruited to mid-cell and we comprehensively describe MreB dynamics in living cells. We show that MreB is recruited to the septum in virtually all cells via a direct interaction with FtsZ. We identify a mutation in MreB that removes the interaction with FtsZ and simultaneously blocks cell division. Remarkably, a single amino-acid (aa) change in FtsZ simultaneously restores the interaction with and suppresses the division defect of the MreB variant. Using fluorescently tagged cell-wall biosynthetic enzymes, we discovered that inhibition of cell division was correlated with the lack of recruitment of PBPs 1B and 2 to the Z ring. Our data support a model in which MreB delivers PBP1B and 2, and perhaps additional factors to the Z ring, thereby generating a link between cell elongation and division in bacteria.
Results
MreB is recruited to the Z ring
To study MreB protein dynamics, we generated a functional mYpet–MreB fusion protein (Supplementary Materials and methods) and expressed it in wild-type (wt) E. coli MG1655 cells at a level that did not affect growth rate or cell morphology (Supplementary Figure S1A). These cells had ∼6% of the total MreB pool labelled with mYpet (Supplementary Figure S1B). Addition of mYpet–MreB in this way had no impact on wt MreB protein levels and was therefore considered a phenotypically neutral cytoskeleton-labelling method (Supplementary Figure S1B).
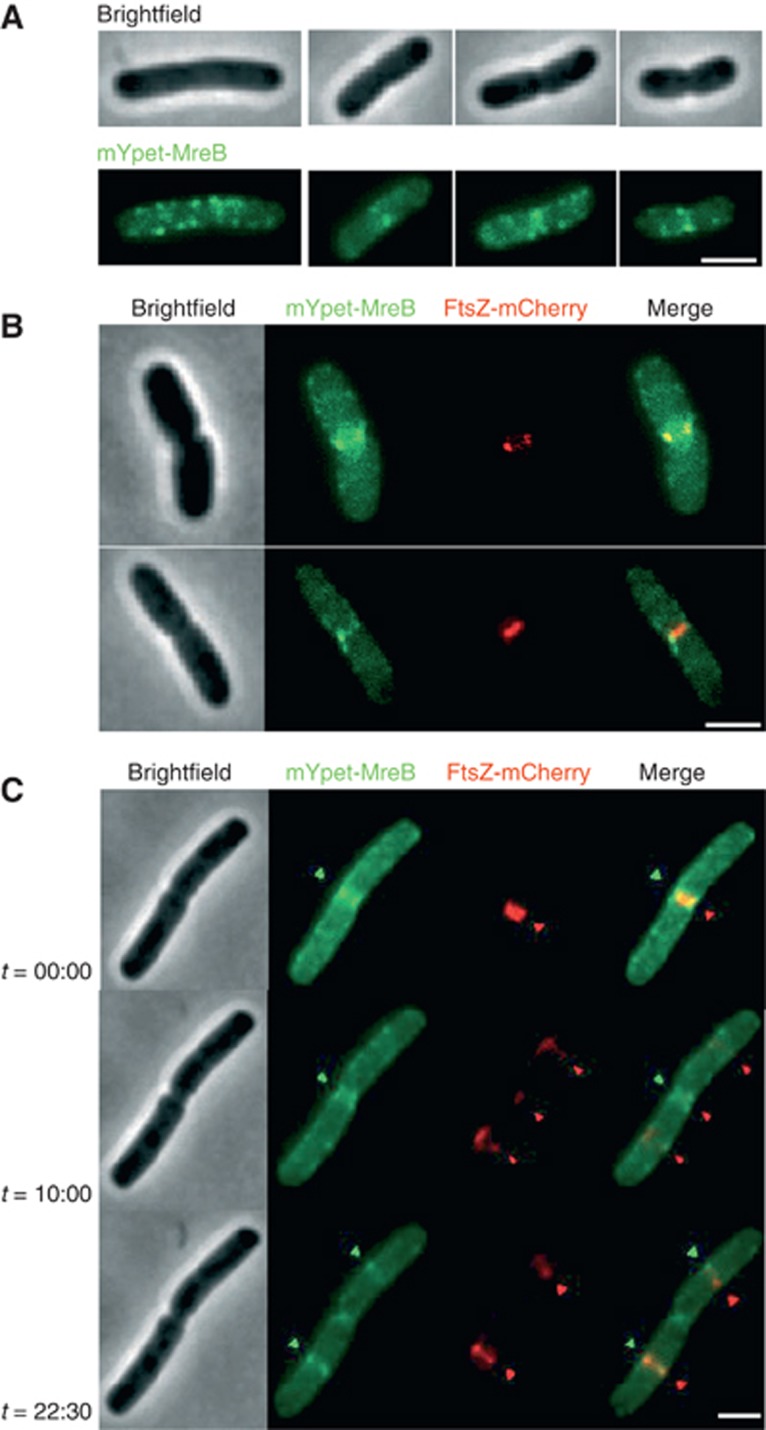
Our mYpet-labelling method revealed that MreB formed ring-like structures in addition to the punctate pattern present along the cell periphery (Figure 1A). The ring-like patterns only appeared at mid-cell in cells undergoing division. MreB structures at mid-cell colocalised with Z rings labelled with an FtsZ–mCherry fusion protein (Figure 1B). These MreB bands were present at all stages of cell invagination and were never observed independently of Z rings, raising the possibility that Z rings recruit MreB (see Movie in Supplementary Figure S9). Examining and scoring this colocalisation revealed that 75% of Z rings had overlapping MreB bands (449 Z rings scored, n=11).
Figure 1.
MreB is recruited to the FtsZ ring. (A) Representative bright field and fluorescent micrographs of MG1655 expressing the mYpet–MreB fluorescent protein from pAKF106 (PBAD::mYpet–mreBCD). Examples of both non-dividing and dividing cells at different stages of cell division are shown. (B) Dual-label fluorescence micrographs of MG1655 expressing both the mYpet–MreB fusion from pAKF106 and FtsZ–mCherry from pQW59 (Plac::ftsZ–mCherry). Merged images reveal signals colocalised at mid-cell. Using this technique, MreBwt–FtsZ colocalisation frequency was 75% (499 Z rings scored, n=11). (C) Selected images from a time-lapse series following a single MG1655 cell expressing the same fluorescent proteins shown in B. Arrows indicate the location and colocalisation of major MreB foci and FtsZ rings. The full time-lapse series and additional MreB–FtsZ colocalisation examples are shown in Supplementary Figures S9 and S10. All cells were grown in M9 glucose media with 0.05% arabinose inducer at 30°C 3 h prior to imaging (see Supplementary Figure S1A and B for growth curves and western blot). Induction of the pQW59 plasmid was not required to give Z rings labelled with FtsZ–mCherry. All images shown are representative of at least three independent repeats. Scale bars=2 μm.
To observe the localisation pattern of unlabelled native MreB and compare it to the mYpet–MreB observations we used IFM. E. coli cells were chemically fixed and polyclonal anti-MreB antibodies used to detect localisation patterns (see Supplementary Figure S1E for western blot). IFM revealed a very similar punctuated MreB localisation pattern along the cell periphery interrupted by MreB bands at mid-cell (Supplementary Figure S1C). Ring-like IFM signals colocalised with 68% of FtsZ–mCherry-labelled Z rings (Supplementary Figure S1D) and are thus consistent with the mYpet–MreB labelling (117 Z rings scored, n=3).
We also studied a functional fluorescent MreB sandwich fusion encoded by mreB–rfpSW, which had been successfully introduced into the E. coli chromosome by allelic replacement in strain FB76 (Bendezú et al, 2009). MreB–RFPsw showed localisation patterns at mid-cell very similar to those described above (Supplementary Figures S1F and G).
Using temperature-sensitive mutant strains revealed that mYpet–MreB could be recruited to Z rings consisting of only FtsZ and other essential condensation factors, without the additional downstream septation factors such as FtsA (Supplementary Figure S2). These findings were consistent with previous work; however, they did not reveal anything about interaction dynamics (Vats and Rothfield, 2007).
Time-lapse fluorescent microscopy showing mYpet–MreB behaviour revealed that MreB was rapidly recruited to newly formed Z rings (within 2.5 min) and frequently remained colocalised with Z rings throughout the entire cell division period (see Movie in Supplementary Figure S9, stills from the time lapse are shown in Figure 1C). Additionally in cells showing no previous Z-ring–MreB colocalisation, MreB was recruited to Z rings at the time of ring establishment (Supplementary Figure S10A). Spontaneous Z ring relocation events also led to MreB signal migration to the new division site (Supplementary Figure S10B). Finally, MreB bands were seen to disperse away from established Z rings in a minority of cells (Supplementary Figure S10C). Imaged in this way, virtually all newly formed Z rings showed initial MreB colocalisation; colocalisation either persisted through cell division or in a minority of cases dispersed away from the closing septum.
MreB interacts with FtsZ
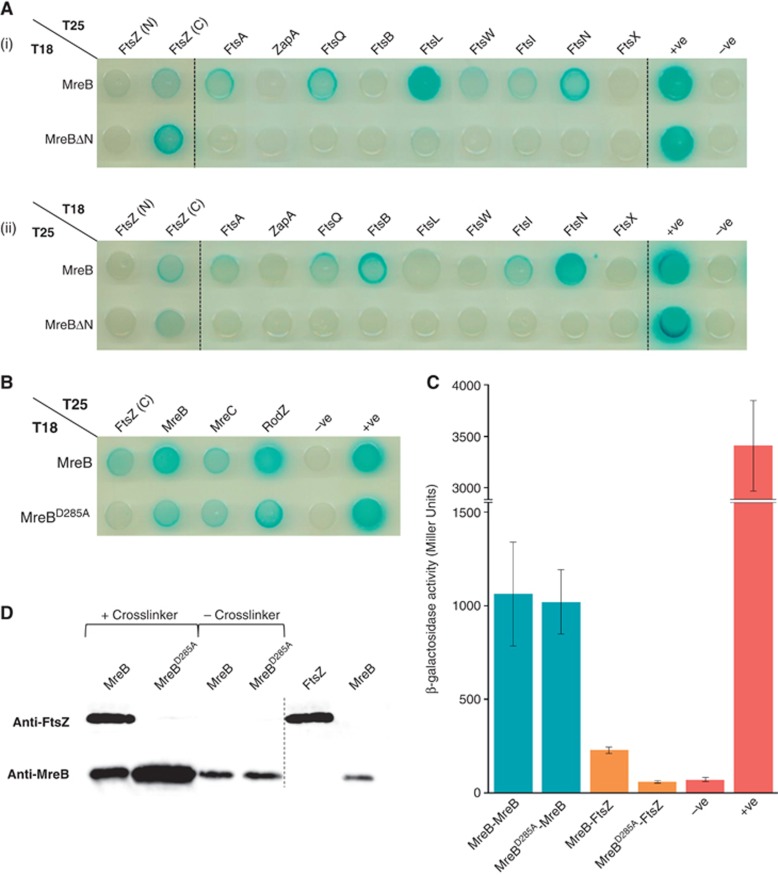
Using a Bacterial Two-Hybrid (BTH) approach (Karimova et al, 2005), we screened a series of known cell division factors for interaction with two full-length MreB fusions (T25–MreB and T18–MreB). This included all fusions reported by Karimova et al (2005) and full-length FtsZ labelled at both termini (Figure 2A). Initially, this approach revealed a cryptic pattern of tagged MreB protein interaction signals, some of which were strong (Figure 2A). The interaction pattern varied considerably between the two fusion series (compare panels (i) and (ii) in Figure 2A), raising the possibly that some of the signals were artefacts of the reporter technique.
Figure 2.
MreB interacts directly with FtsZ and this interaction is abolished in the MreBD285A variant. (A) Bacterial two-hybrid (BTH) screen of E. coli cell division factors against MreB. FtsZ has been labelled in both the N-terminus (N) and C-terminus (c). An MreB variant with a 9 aa N-terminal truncation (ΔN) is also included in this screen. Both T25- and T18-fusion orientations are shown for each BTH combination (i) and (ii). (B) BTH plate showing full-length T18-labelled MreB and MreBD285A against T25-labelled FtsZ (c) and known MreB interaction partners essential for cell elongation. The BTH101 strain was used for all BTH cloning, a full list of plasmid vectors used for this screen can be found in Supplementary Table SI. Cells were spotted onto NA plates containing ampicillin (50 μg/ml), kanamycin (25 μg/ml) and X-gal (40 μg/ml). Plates were incubated for 48 h at 30°C prior to imaging. (C) BTH β-galactosidase activity assays showing T18-labelled MreB and D285A variant against T25-labelled FtsZ and MreBwt. Error bars=s.d., n=4. In all cases, BTH −ve control strains contained empty pKNT25 and pUT18 vectors and +ve control strains contained the pKT25-zip and pUT18C-zip fusions. (D) Western blots of His-tagged MreB and MreBD285A formaldehyde crosslink and pull-down preparations, using both anti-FtsZ and anti-MreB primary antibodies. MC1000 cells containing either pTK500 (Plac::His8–mreB) or pAKF126 (Plac::His8–mreBD285A) were grown to an OD600 of ≈0.3 without induction, crosslinked with 1% formaldehyde and quenched after 10 min using 150 mM glycine. Resulting crosslinked complexes were affinity purified using the MagneHisTM suspension, crosslinks were reversed by incubation at 95°C for 1 h and complexes analysed. Equal volumes of protein complex preparations, un-crosslinked His–MreB and His–MreBD285A preparations and purified His–MreB (200 ng) and native FtsZ (100 ng) proteins were used as controls, n=4. MreB and FtsZ antibody affinity control western blots are shown in Supplementary Figure S3B.
The BTH assay is known to be particularly sensitive to detecting interactions between membrane-associated proteins (Karimova et al, 2005). MreB of E. coli interacts with the cell membrane via its N-terminal amphipathic helix. Deletion of this helix effectively abolished the ability of MreB to bind to the membrane (Salje et al, 2011). Therefore, we repeated the interaction study with MreB variants devoid of the N-terminal amphipathic helix (Figure 2A, MreBΔN panels of (i) and (ii)). A striking and similar effect was observed for both interaction series; only the single interaction signals between the soluble MreB and FtsZ were present in both cases (Figure 2A). This result was consistent with previous yeast two-hybrid studies suggesting a direct MreB–FtsZ interaction (Tan et al, 2011).
Using tap tagging, previous large-scale protein-interaction studies in E. coli identified potential interaction partners for FtsZ and MreB. Although other protein targets were found, these studies identified a putative MreB–FtsZ interaction (Butland et al, 2005; Hu et al, 2009). Here, we used in vivo crosslinking to validate the MreB–FtsZ interaction suggested by our BTH analysis. Wild-type cells expressing N-terminally His-tagged MreB were treated with formaldehyde, a chemical crosslinker with a very short spacer arm (2.3–2.7 Å) (Ishikawa et al, 2006). Crosslinked complexes were purified, dissociated and subjected to western blot analysis (Figure 2D). This revealed that His–MreB was able to pull out native FtsZ from wt cells, consistent with previous studies. Thus, two independent methods support that MreB and FtsZ interact.
MreB D285A does not interact with FtsZ and blocks cell division
By alignment of diverse MreB sequences, we identified a number of conserved residues located outside the five elements that form the canonical actin nucleotide-binding pocket (Bork et al, 1992) and changed them to alanine. These MreB variants were tested using the BTH assay. As seen from Figure 2B, changing the aspartate at position 285 to alanine severely reduced the interaction signal with FtsZ, whereas the interaction signals with wt MreB, MreC and RodZ were unchanged (Figure 2B and C). Using both MreBD285A fusions in the BTH assay showed that self-binding was maintained (Supplementary Figure S3A). Mapping of Asp285 on the Thermotoga maritima MreB crystal structure showed it to locate in subdomain IIA, near the predicted proto-filament interface and away from the RodZ interface (Van den Ent et al, 2001, 2010) (Supplementary Figure S3C). Together, these results showed that MreBD285A could still interact with the cell-wall elongation complex and that the reduced BTH interaction with FtsZ was not caused by reduced protein stability or misfolding.
To validate the BTH result, the MreBD285A variant was included in the in vivo crosslinking assay (Figure 2D). As seen, the MreBD285A –FtsZ interaction was severely reduced (>1000-fold). We conclude that the aspartate at +285 in MreB is essential for the MreB–FtsZ interaction.
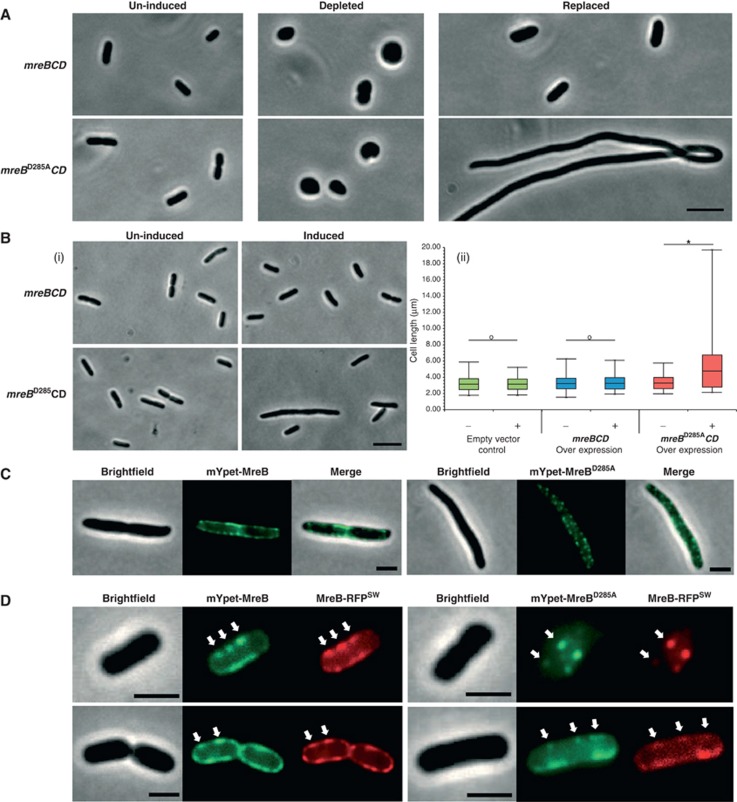
To assess the phenotype of the MreBD285A variant, we took two complementary approaches. In the first, we used an MreB depletion system to study the MreBD285A variant phenotype in the absence of wt MreB (Kruse et al, 2005). The depletion system utilises a ΔmreBCD strain complemented in trans by a low-copy R1 plasmid carrying Pmre::mreBCD. Depletion of the complementing plasmid was triggered by induction of the anti-sense RNA copA (under Plac control), which rapidly (within ≈10 min) blocks R1 plasmid replication (Supplementary Figure S4A). Both the plasmid and the MreBCD proteins were depleted by the natural turnover and dilution in growing E. coli populations with full loss of MreB signal on western blots taking ≈1 h (Supplementary Figure S4B). Complementation by MreBCD or MreBD285ACD was provided by a third plasmid carrying the test genes under control of the PBAD promoter (see Supplementary Figure S4A for a diagram of the experimental setup).
As expected, the R1 (Pmre::mreBCD) plasmid complemented the mreBCD deletion. Moreover, the PBAD::mreBwtCD or PBAD::mreBD285ACD plasmids had no effect on cell morphology or growth without induction (Figure 3A, left panels). Also as expected, depletion of MreBCD without the replenishment of MreBCD from the test plasmid led to the formation of rounded spherical cells (Figure 3A, middle panels), and rod-shape morphology was maintained by replacement with wt MreBCD, with a minor increase in cell width (Figure 3A, upper right panel). In contrast, cells expressing the MreBD285ACD variant in the absence of wt MreBCD exhibited extreme lengthening (Figure 3A, lower right panel). These experiments demonstrated that MreBD285A supported cell elongation but inhibited cell division. Also, under all conditions tested, the MreBD285A variant was unable to complement MreB depletion in liquid or on plates, and was thus incompatible with long-term cell viability.
Figure 3.
The MreBD285A variant inhibits cell division but not cell elongation. (A) Representative brightfield micrographs showing the MC1000 MreBCD depletion strain (MC1000 Δmre, pKG339, pTK554). Images show cells without MreB depletion ‘un-induced’, depleted of MreB ‘depleted’ or complemented with either mreBCD or mreBD285ACD expressed from pAKF128 (PBAD::mreBCD) or pAKF129 (PBAD::mreBD285ACD) ‘replaced’. Cells were grown in LB +tetracycline (20 μM), 2 mM IPTG and 0.2% arabinose for 3 h at 30°C prior to imaging. See Supplementary Figure S4A for diagram of depletion system. (B) MC1000 cells containing pAKF128 or pAKF129. Cells were grown in M9 medium with 0.000,05% arabinose inducer at 30°C for 5 h. (i) Representative bright field micrographs of cells with and without induction (ii) Box and Whisker plot showing cell length measurements with mreBCD and mreBD285ACD ‘overexpression’ compared to an empty vector control (pBAD24). Central line=mean length, box=± s.d. and whiskers=maximum/minimum lengths observed. *t-test=P>0.01, o=no significant difference between populations. 670 cells were sampled for each condition, n=3. For cell width measurements and a full series of control images, see Supplementary Figure S4C and D. (C) Representative brightfield and fluorescent micrographs of MC1000 carrying pAKF131 (PBAD::mypet–mreBCD; left panel) or pAKF132 (PBAD::mypet–mreBD285ACD; right panel), n=3. Note the septal localisation of the mYpet–MreB versus the punctate localisation pattern of mYpet–MreBD285A. (D) mYpet–MreB (pAKF131) and mYpet–MreBD285A (pAKF132) colocalise with the MreB–RFPSW in the FB76 strain. Arrows have been used to highlight selected colocalising foci, n=3. Scale bars=5 μm (A, B) and 2 μm (C, D).
In the second approach, we compared ‘low level’ overexpression of MreB and MreBD285A, together with MreCD, in wt cells. Overexpression was carefully controlled in these experiments to not perturb cell viability, cell width or growth rate (Supplementary Figures S3D and S4C, D). Interestingly, such mild expression of MreBD285A led to a 45% increase in average cell length, suggesting that cell division was inhibited (Figure 3B). In some 17% of such cases, this inhibition was very strong, resulting in cells greater than 18 μm in length (Figure 3B(i) and Supplementary Figure S4D). Statistical analysis showed that expression of MreBD285A but not wt MreB significantly increased the average cell length (Figure 3B(ii)). This supported the result from the depletion study and gave us a tractable genetic system to investigate the MreBD285A elongation phenotype.
MreB D285A is recruited to MreB assemblies but not to the Z ring
To investigate how the MreBD285A variant exerted its dominant-negative effect in cells also expressing wt MreB, we introduced the D285A allele into our mYpet–mreB construct. In wt cells, mYpet–MreBD285A exhibited a typical MreB localisation pattern with lateral patchy foci, suggesting that MreBD285A was capable of entering MreB assemblies (Figure 3C). Double labelling by expression of mYpet–MreBwt and mYpet–MreBD285A in FB76 (mreB–RFPsw) showed that the fusion proteins colocalised, thereby confirming that the D285A variant entered MreB assemblies (compare left and right panels of Figure 3D).
Next, we investigated the MreBD285A localisation pattern relative to FtsZ using an FtsZ–mCherry fusion protein and the same MreB-labelling strategy as shown in Figure 1B. Statistical analysis revealed a significant reduction in MreB–Z-ring recruitment from 75% for MreBwt to 28% for the MreBD285A variant (414 Z rings, n=7, P>0.01). Although this represents a large drop in colocalisation, the 28% proportion of colocalisation exhibited by MreBD285A is probably an overestimate of MreBD285A–Z-ring recruitment, as this variant could be recruited indirectly to the Z ring through its interaction with untagged wt MreB at the septum.
Mutations in ftsZ restore the FtsZ–MreB BTH interaction signal
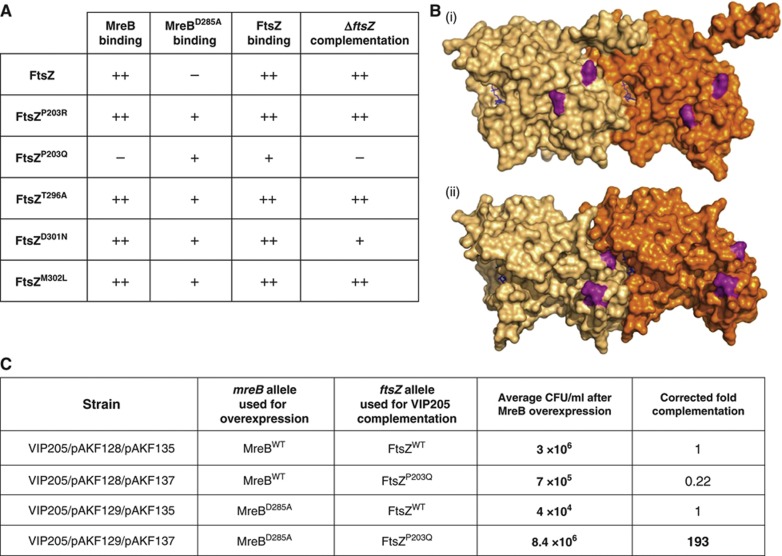
Using an error-prone variant of Pfu polymerase, Pfu(exo−) D473G, we PCR amplified the ftsZ open reading frame (ORF) and used the BTH assay to identify mutations in ftsZ that re-established the interaction with MreBD285A (Figure 4A, Supplementary Figure S5A) (Biles and Connolly, 2004). This screening generated five single aa changes in FtsZ that robustly restored the MreBD285A–FtsZ interaction signal. All five FtsZ variants interacted with wt FtsZ and all but one (FtsZP203Q) could still bind wt MreB (summarised in Figure 4A). When mapped onto the FtsZ crystal structure from Methanococcus jannaschii and Staphylococcus aureus all five aa changes clustered to a discrete patch on one face of FtsZ (Figure 4B).
Figure 4.
Single aa changes in FtsZ restore functional interaction with MreBD285A. (A) Table summarising the phenotypes of ftsZ point mutants identified in the ftsZ–mreBD285A PCR mutagenesis screen. Bacterial two-hybrid (BTH) scores represent signal strength from BTH101 double transformations, plated on NA plates with selective antibiotics and X-gal (see Supplementary Figure S5A for images of plates); ++=strong binding signal,+=weaker binding signal, −=no detectable signal. Details of plasmids used in these assays can be found in Supplementary Table SI. ‘ΔftsZ complementation scores’ are representative of CFU measurements shown in Supplementary Figure S5B; ++=full (comparable to ftsZwt complementation control),+=partial (100 drop in CFU compared to ftsZwt control) and −=no complementation (same CFU as −ve empty vector contol). Complementation was assayed using the VIP205 (MC1061: Ptac::ftsZ) strain complementing with Ptet::ftsZ (pAKF135) constructs containing the identified point mutations (pAKF136-140), the full list of plasmids can be found in Supplementary Table SI. Note that only the FtsZP203R variant has no detectable MreB binding and cannot complement the VIP205 ftsZ depletion strain. (B) Structural model of an FtsZ dimer of M. jannaschii (i) and S. aureus (ii) (Oliva et al, 2004; Matsui et al, 2012) showing the positions of the aas in FtsZ (purple), changes to which can restore the interaction with MreBD285A. The two monomers of the FtsZ dimer are shown in different shades of orange. GTP molecules are shown in blue. Identification of the equivalent aa positions in E. coli FtsZ were based on a sequence alignment of FtsZ (not shown). (C) FtsZP203Q can rescue MreBD285A expression. E. coli VIP205 (Ptac::ftsZ) cells were grown without IPTG inducer giving FtsZ depletion. This was complemented by expression of FtsZ from pAKF135 (Ptet::ftsZ) or pAKF137 (Ptet::ftsZP203Q) induced using 20 ng/ml chlorotetracycline. A FtsZ depletion system was used in this assay as overexpression of ftsZ can lead to cell elongation and thus CFU reduction. Strains contained either pAKF128 (PBAD::mreBCD) or pAKF129 (PBAD::mreBD285ACD) induced with 0.005% arabinose for controlled MreB overexpression. Cells were grown in M9 media for 8 h, serially diluted 1/10 and used for enumeration on NA plates with antibiotics and 30 μM IPTG, but without induction of the plasmid-borne ftsZ and mreBCD genes. These plates gave viable counts reflecting the effect of the mreB/ftsZ-mutant combinations in the liquid media. CFU measurements are an average of three independent repeats (n=3). Note that the expression of MreBD285A reduced CFU as cells elongate (compare CFU/ml when MreBwt or MreBD285A is overexpressed and the FtsZwt allele is used for VIP205 complementation). Using FtsZP203Q for VIP205 complementation in cells overexpressing MreBwt reduced cell viability four-fold (see corrected fold complementation). In contrast, FtsZP203Q expression in VIP205 cells also expressing MreBD285A yielded a near 200-fold higher CFU (in growing cultures). Images of serially diluted plates are shown in Supplementary Figure S5C along with plate images showing MreBD285A complementation for all the FtsZ variants identified in this study.
To test the functionality of the FtsZ variants in the absence of wt FtsZ, we used the FtsZ depletion strain VIP205 (Garrido et al, 1993). Depletion of FtsZ reproducibly generated a ≈106 reduction in cell viability, which could be fully complemented by inducing wt ftsZ in trans (Supplementary Figure S5B). Three of the five FtsZ variants: P203R, T296A and M302L fully complemented FtsZ depletion giving the same CFU as the wt ftsZ control. One variant, D301N, partially complemented the depletion giving a 100-fold drop in CFU. Only FtsZP203Q did not complement FtsZ depletion (Supplementary Figure S5B, summarised in Figure 4A). Strikingly, FtsZP203Q was the variant that no longer gave a positive-BTH interaction signal with MreBwt (Figure 4A, Supplementary Figure S5A). When used in a complementation assay, the FtsZP203Q variant elongated cells, raising the possibility that inhibition of cell division occurred through a mechanism similar to that of MreBD285A -induced elongation (Supplementary Figure S5D). Most importantly, the existence of aa changes in FtsZ that restore the interaction with MreB showed that the MreB–FtsZ interaction was direct.
FtsZ P203Q rescues the MreB D285A phenotype
Expression of mreBD285A in wt cells led to cell elongation through blockage of cell division (Figure 3A) and reduced the number of cells in a growing culture compared to a control culture expressing MreBwt (compare lines 1 and 3 in Figure 4C, see also Supplementary Figure S5C). Taking advantage of this reduction in viable counts, we tested the FtsZ variants for rescue of the MreBD285A phenotype in the FtsZ depletion strain VIP205. FtsZP203Q was a particularly important variant to test as it was the only one that interacted with MreBD285A but not with MreBwt (Figure 4A).
Cells were depleted of FtsZwt and complemented by induction of ftsZ from a plasmid (pAKF135-Ptet::ftsZ, or pAKF137-Ptet::ftsZP203Q) induced at a level that did not seriously alter cell morphology. Expression of MreB or MreBD285A was induced in these strains from pAKF128 (PBAD::mreBCD) or pAKF129 (PBAD::mreBD285ACD). After 8 h of growth in M9 media, cells were plated on solid medium containing antibiotics, IPTG (to replenish wt FtsZ), but without induction of the plasmid-borne ftsZ and mreBCD genes (Figure 4C, Supplementary Figure S5C).
Expression of FtsZP203Q plus MreBwt reduced viable counts four-fold (Figure 4C, compare MreBwt expression with FtsZ and FtsZP203Q complementation of VIP205). Critically, in cells expressing MreBD285A the pattern was reversed: FtsZP203Q expression increased viable counts by nearly 200-fold compared to FtsZwt (Figure 4C, compare FtsZwt or FtsZP203Q rescue of MreBD285A expression). Therefore, this assay demonstrated that the division-inhibition effect of MreBD285A could be counteracted by FtsZP203Q but not by FtsZwt.
Cells elongated by MreB D285A contain regularly spaced ‘locked’ Z rings
To investigate the elongation phenotype of cells expressing MreBD285A, we determined localisation patterns of fluorescently labelled cellular components. FtsZ–mCherry labelling revealed that Z rings were spaced regularly along the lengths of cells elongated by MreBD285A expression (Figure 5A, Supplementary Figure S7A). DNA staining using Hoescht showed that these Z rings formed between segregated nucleoids. Thus, cell elongation was not due to dysfunctional chromosome segregation (Supplementary Figures S6A and B). Together, these observations suggested that Z ring formation and positioning were unaffected in elongated cells, but raised the question of what protein activities were absent in the ‘locked’ Z rings.
Figure 5.
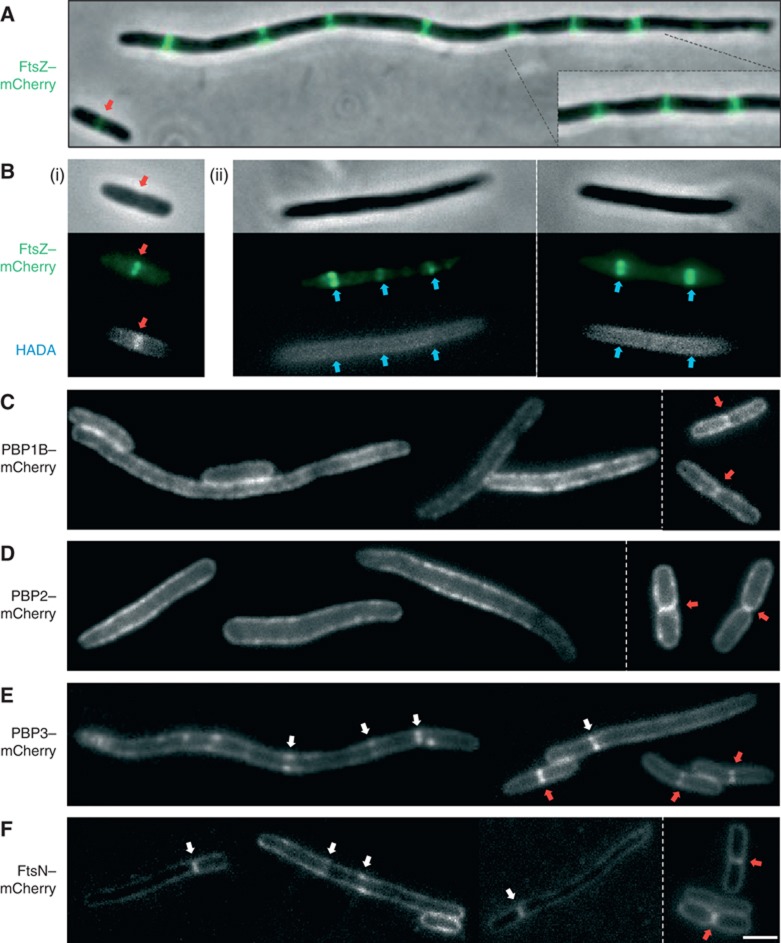
PBPs are mislocalised and septal PG synthesis is inhibited in MreBD285A-expressing cells. MC1000 cells elongated by expressing MreBD285A from pAKF129 (PBAD::mreBD285ACD) containing a second plasmid expressing GFP-tagged division and elongation factors: (A, B) FtsZ–mCherry (pAKF133, PBAD::ftsZ–mCherry), (C) PBP1B–mCherry (pAKF145, Plac::mrcB–mCherry), (D) PBP2–mCherry (pAKF146, Plac::mrdA–mCherry), (E) PBP3–mCherry (pAKF147, Plac::ftsI–mCherry) and (F) FtsN–mCherry (pAKF150, Plac::mCherry-ftsN). In most cases cells were grown in M9 media for 5 h at 30°C with 0.000,05% arabinose. For expression from pAKF146, cells were grown in M9 glucose with 0.1% arabinose inducer. Empty vector and MreB overexpression control images are shown in Supplementary Figure S7; contrasting elongated cells treated with PBP3-sepcific inhibitors are shown in Supplementary Figure S8. Elongated cells contain multiple evenly spaced Z rings (A). Fixed HADA fluorescent D-aa-labelled cells (B). MC1000 pAKF133 cells containing empty pBAD24 vector (i) or elongated through MreBD285ACD expression from pAKF129 (ii) were incubated with 1 mM HADA for 30 s, cells were fixed using 70% ethanol and washed in PBS before microscopy. Note HADA incorporation at mature Z rings in (i—red arrows) are absent from elongated MreBD285A cells (ii—blue arrows), see Supplementary Figure S6C for FtsI inhibitor controls. Note FtsZ, PBP1B, PBP2, PBP3 and FtsN are all recruited to the septum during normal cell division—red arrows (Supplementary Figure S7). In MreBD285A elongated cells, PBP3 and FtsN are recruited to inhibit septa (E, F—white arrows), whereas PBP1B and PBP2 are not (D, E). The behaviours of tagged PBP–mCherry fusions are shown in Supplementary Figure S11. Images are representative of at least four independent repeats. Scale bar=2 μm.
Locked Z rings do not support septal synthesis of peptidoglycan
To investigate the locked Z rings, we used a recently developed method that specifically labels sites that are actively synthesising PG. This technique exploits the tolerance of bacterial PG biosynthetic enzymes to accommodate D-amino acids with diverse sizes into PG. Addition of a fluorescent tag to these aas leads to progressive labelling of the cell wall over time, through a combination of growth and turnover (Kuru et al, 2012). In E. coli wt cells, pulse labelling the cell wall with the fluorescent D-aa HADA gives a very strong incorporation signal at Z rings, weaker incorporation at the cell periphery and very little incorporation at the relatively inert cell poles (described in Kuru et al (2012) and shown in Figure 5B (i)). Control cells treated with the FtsI inhibitor aztreonam show multiple sites of HADA incorporation colocalising with Z rings along their lengths (Supplementary Figure S6C). In contrast, in elongated MreBD285A-expressing cells, the cell-wall regions colocalising with the locked Z rings failed to incorporate HADA without affecting incorporation elsewhere (Figure 5B (ii)). This result showed that the locked Z rings did not actively synthesise PG, while the elongation enzymes remained functional.
Cells elongated by MreB D285A display mislocalisation of PBPs
To gain insight into the mechanism of MreBD285A-mediated cell elongation through inhibition of Z ring-dependent PG biosynthesis, we used a combination of fluorescent protein tagging and time-lapse microscopy. We focused on the localisation patterns of the PBP enzymes: PBP1A, PBP1B, PBP2 and PBP3, all of which are important for cell-wall and septum synthesis.
In wt cells, PBP1A is usually localised to the cell membrane and is thought to principally function in cell elongation (Vollmer and Bertsche, 2008; Banzhaf et al, 2012). In cells expressing MreBD285A, PBP1A–mCherry formed discrete foci at the cell periphery in both elongated and dividing cells, consistent with the expected pattern (Supplementary Figure S6D compared to Supplementary Figure S6E). In time-lapse imaging, these foci migrated along the long axis of elongating cells, similar to the behaviour of MreB (compare movies of Supplementary Figure S11A and Supplementary Figure S6D and E). Thus the localisation pattern of PBP1A–mCherry, which functions mainly in cell elongation (Vollmer and Bertsche, 2008), was not seriously affected by MreBD285A.
Both PBP1B and PBP2 are involved in cell elongation, but have putative roles in cell division (Den Blaauwen et al, 2003; Bertsche et al, 2006; Banzhaf et al, 2012; van der Ploeg et al, 2013). In wt cells both proteins localise as stable foci at the dividing septum, in addition to migrating peripheral foci along the cell length, reminiscent of MreB localisation (Figures 5C and D—red arrows in control cells). Discriminating between these two behaviours was technically challenging; here septum-associated foci were defined as those that stayed stationary at the cell periphery for more than three frames (7.5 min) and had a ‘partner’ focus on the opposing side of the membrane (Supplementary Figure S11 and stills in Supplementary Figures S7B and C). In MreBD285A elongated cells, such stable septum-like behaviours of PBP1B and PBP2 were not observed, with both proteins forming only longitudinally migrating foci (Supplementary Figure S11 and stills in Figure 5C and D). In contrast, using a PBP3-specific cell division inhibitor resulted in stationary PBP1B and PBP2 bands or ring-like foci in elongated cells (Supplementary Figures S8B and C). This suggested that both PBP1B and PBP2 were not successfully recruited to the locked Z rings present in MreBD285A elongated cells.
PBP3 is the penultimate essential factor known to be recruited to the Z ring (De Boer, 2010). Thus if PBP3 is present at the septa, it can be considered a marker for the localisation of all upstream cell division proteins (De Boer, 2010). In wt cells, PBP3 formed bands or rings at mid-cell immediately prior to cell division and remained associated with the constricting ring (Figure 5D, red arrows). In elongated MreBD285A cells, PBP3 rings were also detected, suggesting that all upstream septal proteins were present in these locked structures (Figure 5E, white arrows). Over time, the PBP3 rings dispersed instead of contracting (Supplementary Figure S11).
The presence of PBP3 in locked Z rings raised the possibility that these structures contained all the essential cell division factors. To investigate this, we studied the localisation pattern of a FtsN–mCherry fusion, which is the last essential cell division factor (De Boer, 2010). In wt cells, FtsN–mCherry is recruited immediately prior to septation (Figure 5F—red arrows in control cells, S7E). Expression of this fusion in MreBD285A elongated cells gave bands or rings of FtsN, similar to those observed for PBP3 (Figure 5F, white arrows).
In summary, our results demonstrated that locked Z rings recruited PBP3 and FtsN but failed to recruit PBP1B or PBP2. These observations indicated that the FtsZ–MreB interaction functions to deliver PBP1B and PBP2 to the Z ring.
Discussion
The discovery of a conserved bacterial cytoskeleton consisting of tubulin–FtsZ and actin–MreB was relatively recent and no mechanisms for coordination of these factors have been described—that is—each element has until recently been considered largely independent of the other. Here we show that, in E. coli, MreB interacts directly with FtsZ and that the interaction is essential for Z ring constriction, divisome maturation and PG septal synthesis.
Three separate MreB-labelling approaches showed that MreB colocalised with Z rings, in addition to adopting its previously established punctate localisation pattern. Unlike the patterns reported in Vats and Rothfield (2007), these ring-like patterns only appeared at mid-cell and in cells undergoing division. Time-lapse microscopy revealed that MreB was recruited early to the Z ring and remained colocalised throughout ring constriction. Our time lapses indicated that most if not all Z rings recruited MreB at the time of formation.
A single aa change in MreB (aspartate +285 to alanine) simultaneously reduced MreB recruitment to the Z ring and inhibited cell division. In cells devoid of wt MreB, inhibition of division by MreBD285A was severe, showing that recruitment of MreB to the Z ring was essential. MreBD285A was still capable of binding the known MreB-interacting partners MreC and RodZ, and supported cell elongation in an MreB depletion strain. Thus, MreBD285A was specifically defective in supporting cell division. The aspartate at position 285 in MreB is conserved among evolutionarily distant bacteria, raising the possibility that interactions through this residue could be a conserved phenomenon.
Crosslinking experiments showed that FtsZ and MreB interacted in vivo and that this interaction was abolished in MreBD285A. Single aa changes in FtsZ restored the interaction with MreBD285A, yielding strong support of a direct interaction. The aa changes in FtsZ were not located to the proto-filament interface, GTP-binding site, or C-terminal FtsA/ZipA-binding regions, suggesting that they would not disrupt the formation of functional Z rings (Ma and Margolin, 1999; Oliva et al, 2004; Matsui et al, 2012). One of the FtsZ variants (P203Q) had simultaneously lost its capability to complement depletion of wt FtsZ and its interaction with wt MreB but, strikingly, counteracted cell division inhibition exerted by ectopic expression of MreBD285A. This shows that the FtsZP203Q variant, although lethal when expressed as the sole copy of FtsZ, could support cell division in the right context. This lends further strong support to the finding that the MreB–FtsZ interaction is direct and essential.
Cells elongated by ectopic expression of MreBD285A formed regularly spaced, ‘locked’ Z rings, segregated their nucleoids and formed mature divisomes containing PBP3 and FtsN. However, these mature Z rings did not incorporate the fluorescent D-aa HADA, showing that no septal PG biosynthesis had taken place (see Figure 5B(ii) compared to Supplementary Figure S6C). PBP3 and FtsN are recruited late to the divisome, just before the onset of division (Addinall et al, 1997; De Boer, 2010) (Figure 6). Strikingly, even though the locked Z rings contained PBP3 and FtsN they did not recruit PBP1B and PBP2. This result was somewhat unexpected since PBP1B interacts directly with PBP3 (and FtsN) (Bertsche et al, 2006; Müller et al, 2007) and PBP2 seems to interact with PBP3 in vivo (Van der Ploeg et al, 2013). FtsN is the last known essential cell division protein recruited to the divisome (Müller et al, 2007; de Boer, 2010). Since FtsN has been proposed to function as a trigger of septation (Corbin et al, 2004; Gerding et al, 2009; Lutkenhaus, 2009), it was interesting to learn that locked Z rings contained FtsN, yet still did not divide. Taken together, our results show that PBP1B, PBP2 and possibly additional factors must be recruited to the divisome by MreB for cell division to occur. Our results therefore provide a mechanism for recruitment of essential PBP enzymes to the divisome from the elongation complex, namely PBP1B and/or PBP2 (Figure 6).
Figure 6.
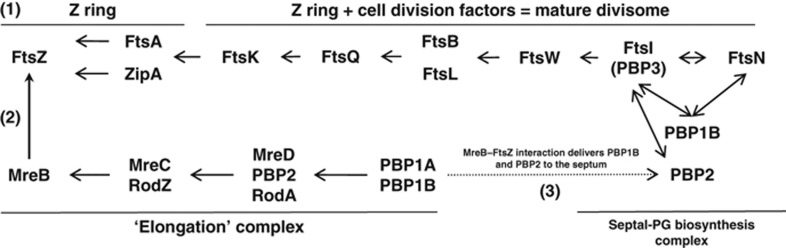
Model of the interaction network of cell elongation with cell division factors in E. coli. (1) FtsZ polymers at mid-cell assemble into the Z ring. (2) The direct interaction between MreB and FtsZ (this study) allows the downstream transfer of PBP1B and PBP2 into the mature divisome (3) (hatched arrow). This provides all the biosynthetic enzyme activities required for septal PG synthesis. Ordered assembly of the 10 essential cell division proteins was adapted from De Boer (2010). Binding structure of the elongation complex information was adapted from Kruse et al (2005); Bendezú et al (2009); White et al (2010); Banzhaf et al (2012). FtsI-, FtsN- and PBP1B-binding information was adapted from Müller et al (2007). FtsI-, PBP2-binding information was taken from Van der Ploeg et al (2013).
Based on these observations, we propose a model for the elongation to division transition of cell-wall biosynthesis. In the model, a proportion of the MreB pool is recruited to the Z ring to deliver protein factors from the cell-wall elongation complex (Figure 6). This recruitment depends on the MreB–FtsZ interaction that mediates transfer of protein factors between elongation and divisome complexes, in particular PBP2 and PBP1B identified in this study. However, other factors, such as MurG and MraY, are known to be present in both the elongation and divisome complexes (Aaron et al, 2007; Mohammadi et al, 2007). Further studies are required to determine if they are also recruited to the Z ring by an MreB-dependent mechanism.
MreB is a conditionally essential protein: during rapid growth, the cells become spherical, inflate and lyse. However, during slow growth, the cells propagate as spheres (Bendezú and De Boer, 2008). Thus our observations raise the question of how the spherical cells are able to divide in the absence of MreB that is clearly needed for division of wt cells. Spherical cells cannot synthesise lateral cell wall due to the lack of MreB; thus their cell wall is synthesised though the FtsZ-dependent septal and pre-septal cell-wall synthesis machinery (Varma and Young, 2009). Since, in the absence of MreB, there is no lateral cell wall, we infer that PBP1B and 2 by default are associated with the septal and/or pre-septal synthesis machinery, thus allowing the enzymes to perform their essential function. Alternatively, the absence of MreB changes the cell-wall machinery so dramatically that the requirement for PBP1B and/or 2 is bypassed.
This study has shown that MreB plays an essential role in cell division of E. coli under rapid growth conditions; previous work revealed that FtsZ plays a role in cell elongation through pre-septal synthesis (De Pedro et al, 1997). These observations expanded upon the established roles of both proteins and their associated protein complexes. The understanding that MreB and FtsZ interact during cell division to coordinate cell elongation with division opens a new window to study how cell division is controlled in bacteria. We suggest that the PBP1B and PBP2 proteins play essential roles in septal and/or pre-septal PG synthesis, respectively. It could be argued that MreB may not be involved in pre-septal PG synthesis because spherical cells of an mreB deletion indeed synthesised pre-septal PG (Potluri et al, 2012). However, as discussed above, the round cell morphology may bypass the requirement of MreB to deliver PBP 1B and 2 to the divisome, thereby resolving any such apparent paradox.
Materials and methods
Strains and growth media
For details of all strains and plasmids used in this study see Supplementary Table SI. All microscopic observations and growth curves were carried out using M9 growth media (Na2HPO4 0.6 g/l, KH2PO4 0.3 g/l, NaCl 0.05 g/l, NH4Cl 0.1 g/l, 0.2% w/v casamino acids, 0.001% w/v thiamine, 1 mM CaCl2, 1 mM MgS04, pH=7.4) made fresh from autoclaved components. Glycerol (0.5% v/v) was typically used as a carbon source; however, glucose (0.4% w/v) was occasionally used where indicated.
Epi-fluorescence microscopy
Epi-flourescence microscopy was carried out using an Olympus 1X71 inverted microscope through a 100X Zeiss Plan-NEOFLUAR oil objective (NA 1.3), held at 30°C using an environmental chamber.
For mYpet–MreB detection, MG1655 pAKF106 (PBAD::mYpet–mreB) stationary-phase cultures were diluted 1/100 into 25 ml M9 glucose media with 0.05% arabinose in a 125 ml conical flask. Under these conditions, induction of the PBAD promoter was minimal and did not affect growth rate or cell morphology (Supplementary Figures S1A and B). Resulting mixtures were incubated for 3 h at 30°C, typically giving OD600 values between 0.29 and 0.33. Culture samples were spotted onto pre-set 1% M9 glucose agarose pads, which had been pre-equilibrated to 30°C and imaged immediately. The same conditions were used for the MreB/FtsZ double-labelling strain: MG1655, pAKF106, pQW59 (Plac::ftsZ–mCherry) and all microscopy involving the FB76 (mreB–RFPSW) strain.
IFM was carried out using a standard protocol, the full details of which are given as Supplementary Data. Cells were fixed in 1% formaldehyde and 0.1% glutaraldehyde. MreB localisation was detected using rabbit polyclonal anti-MreB antibodies first reported in Kruse et al (2003), an antibody specificity western blot is shown in Supplementary Figure S1E. FITC-conjugated anti-rabbit-goat IgG antibody (Sigma-Aldrich) was used for secondary antibody binding and fluorescent detection.
Full details of the microscopic apparatus and software used for image acquisition, media recipes, IFM methods and details of the mYpet–MreB tag development can be found in the Supplementary Materials and methods.
MreBD285A ‘overexpression’ microscopy
Native mreB and mreBD285A was ‘overexpressed’ from plasmids pAKF128 and pAKF129 in MC1000 using empty vector (pBAD24) as a control. MC1000 (Δlac, ΔaraD) was used for microscopic studies as it allows fine control of expression from Plac and PBAD promoters. Stationary-phase cultures were diluted 1/100 into 25 ml M9 media with 0.000,05% (33.3 μM) arabinose. Resulting mixtures were incubated shaking at 30°C for 5 h giving OD600 values between 0.3 and 0.4. Cells were imaged and their dimensions measured using the MATLAB plug-in MicrobeTracker (Sliusarenko et al, 2012). The experiment was repeated in triplicate with at least 200 cells measured per repeat. A 670 cell sample cutoff was chosen as this equalled the smallest sample size collected.
For detection of PBP-fluorescent protein tags in mreBD285A-expressing cells the same protocol was used; except in the case of pAKF146 (Plac::mrdA–mCherry) where M9 glucose media with 0.1% arabinose inducer was used to repress expression. For chromosomal DNA staining 1 μg/ml Hoechst 33342 (Invitrogen) was used to stain cells immediately prior to imaging.
BTH analysis
Plasmids used for BTH analysis (Karimova et al, 2005) were co-transformed into BTH101 (cya-99). Five microlitre of stationary-phase culture of representative transformants was spotted onto nutrient agar (NA) plates containing selective antibiotics and 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal). After incubation at 30°C for 24–48h, plates were scanned on an Epson perfection V700 photo flatbed scanner. Measurements of β-galactosidase activity in liquid media were performed on 1 ml aliquots of growth-phase cultures as described in Miller (1972).
MreB–FtsZ in vivo formaldehyde crosslinking
E. coli MC100 cultures expressing His-tagged MreB from pTK500 or His–MreBD285A from pAKF126 were crosslinked using 1% formaldehyde for 10 min and quenched using 150 mM glycine. Crosslinked His–MreB complexes were purified under denaturing conditions as described in Ishikawa et al (2006). For full details see Supplementary Materials and methods. Equal volumes of protein were loaded onto SDS-PAGE gels and individual proteins identified by western blot using anti-FtsZ and anti-MreB antibodies (both used at a 1/10 000 dilution) (Kruse et al, 2003; Galli and Gerdes, 2012).
BTH ftsZ point mutagenesis screen
An error-prone PCR using a modified Pfu (exo−) D473G polymerase and primers: AKF_FtsZ_pKNT25_F and AKF_FtsZ_pKNT25_R were used to amplify the ftsZ ORF from pKNT25–ftsZ exactly as described in Biles and Connolly (2004). PCR products were digested with both HindIII and BamHI, ligated into pKNT25–ftsZ (cut with the same enzymes) and transformed into BTH101-containing pUT18–mreBD285A. Candidates that consistently gave blue cultures on NA plates containing X-gal (40 mg/ml) were selected and both plasmids re-isolated. Re-isolated pKNT25–ftsZ-mutant plasmids were co-transformed into BTH101 strains with either pUT18–ftsZ, pUT18C–mreB or pUT18C–mreBD285A to confirm the ‘re-binding’ phenotype and check protein variants could still bind FtsZ (Supplementary Figure S5A) before being sequenced. Primer sequences are given as Supplementary Data.
Other methods
Purification of native FtsZ protein was carried out exactly as described in Galli and Gerdes (2012). Affinity purification of His-tagged MreB was carried out using a HisTrap HP column (GE Healthcare). MreB depletion microscopy was carried out as described in Kruse et al (2005). Cell-wall HADA staining was carried out as described in Kuru et al (2012). In all cases, the full protocols are given as Supplementary Data.
Supplementary Material
Acknowledgments
We would like to thank Waldemar Vollmer and Romain Mercier for critical reading of the manuscript; Bernard Connolly for the generous gift of Pfu (exo-) D473G error-prone polymerase; Erkin Kuru, Yves Brun and Michael VanNieuwenhze for the gift of the HADA fluorescent aas; and Elisa Galli for purified FtsZ protein. We would also like to thank and acknowledge Chen Chen and Qing Wang who conducted essential preliminary work on this project. CC generated the mreB point mutant library and conducted preliminary work on mreB mutant-cell division phenotypes. QW engineered the pQW59 vector and the functional Ypet–linker–MreB fusion, which was further modified by AKF and used in this study.
Author Contributions: AKF carried out the majority of experiments, designed the experimental programme and co-authored the manuscript. KG designed parts of the experimental programme and co-authored the manuscript. This work was supported by the BBSRC and European Commission DIVINOCELL program.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C (2007) The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64: 938–952 [DOI] [PubMed] [Google Scholar]
- Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, Den Blaauwen T (2005) Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 55: 1631–1645 [DOI] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J (1997) FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol 25: 303–309 [DOI] [PubMed] [Google Scholar]
- Banzhaf M, Van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Distèche M, Den Blaauwen T, Vollmer W (2012) Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol Microbiol 85: 179–194 [DOI] [PubMed] [Google Scholar]
- Basu R, Chang F (2007) Shaping the actin cytoskeleton using microtubule tips. Curr Opin Cell Biol 19: 88–94 [DOI] [PubMed] [Google Scholar]
- Bendezú FO, De Boer PAJ (2008) Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J Bacteriol 190: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Hale CA, Bernhardt TG, De Boer PAJ (2009) RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman MEG, Kannenberg K, Von Rechenberg M, Nguyen-Distèche M, Den Blaauwen T, Höltje J-V, Vollmer W (2006) Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol 61: 675–690 [DOI] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- Biles BD, Connolly BA (2004) Low-fidelity Pyrococcus furiosus DNA polymerase mutants useful in error-prone PCR. Nucleic Acids Res 32: e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. PNAS 89: 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland G, Manuel J, Alvarez P, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A (2005) Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433: 531–537 [DOI] [PubMed] [Google Scholar]
- Corbin BD, Geissler B, Sadasivam M, Margolin W (2004) Z-Ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol 186: 7736–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Blaauwen T, Aarsman MEG, Vischer NOE, Nanninga N (2003) Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol Microbiol 47: 539–547 [DOI] [PubMed] [Google Scholar]
- De Boer PAJ (2010) Advances in understanding E. coli cell fission. Curr Opin Microbiol 13: 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer P, Crossley R, Rothfield L (1992) The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359: 254–256 [DOI] [PubMed] [Google Scholar]
- De Pedro MA, Quintela JC, Höltje JV, Schwarz H (1997) Murein segregation in Escherichia coli. J Bacteriol 179: 2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaruni AV, Ogorzalek Loo RR, Xie Y, Loo JA, Gober JW (2005) The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus. PNAS 102: 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R (2011) Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333: 225–228 [DOI] [PubMed] [Google Scholar]
- Erickson HP (2007) Evolution of the cytoskeleton. BioEssays 29: 668–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton AK, Lambert C, Wagstaff PC, Sockett RE (2010) Manipulating each MreB of Bdellovibrio bacteriovorus gives diverse morphological and predatory phenotypes. J Bacteriol 192: 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW (2004) MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Fletcher DA, Mullins RD (2010) Cell mechanics and the cytoskeleton. Nature 463: 485–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli E, Gerdes K (2012) FtsZ-ZapA-ZapB interactome of Escherichia coli. J Bacteriol 194: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 333: 222–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido T, Scnchez M, Palacios P, Aldea M, Vicente M (1993) Transcription of ftsZ oscillates during the cell cycle of Esherichia coli. EMBO J 12: 3957–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding Ma, Liu B, Bendezú FO, Hale CA, Bernhardt TG, De Boer PAJ (2009) Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol 191: 7383–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Janga SC, Babu M, Díaz-Mejía JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, Chandran S, Christopoulos C, Nazarians-Armavil A, Nasseri NK, Musso G, Ali M, Nazemof N, Eroukova V, Golshani A, Paccanaro A et al. (2009) Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol 7: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N (2006) A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol Microbiol 60: 1364–1380 [DOI] [PubMed] [Google Scholar]
- Jones LJ, Carballido-López R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Karczmarek A, Martínez-Arteaga R, RM-A Baselga, Alexeeva S, Hansen FG, Vicente M, Nanninga N, Den Blaauwen T (2007) DNA and origin region segregation are not affected by the transition from rod to sphere after inhibition of Escherichia coli MreB by A22. Mol Microbiol 65: 51–63 [DOI] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D (2005) Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187: 2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Bork-Jensen J, Gerdes K (2005) The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55: 78–89 [DOI] [PubMed] [Google Scholar]
- Kruse T, Møller-Jensen J, Løbner-Olesen A, Gerdes K (2003) Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J 22: 5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, De Pedro MA, Brun YV, Vannieuwenhze MS (2012) In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angewandte Chemie 51: 12519–12523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J (2009) FtsN--trigger for septation. J Bacteriol 191: 7381–7382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus JF, Wolf-Watz H, Donachie WD (1980) ftsA-envA region of the and identification of a new fts organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol 142: 615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Margolin W (1999) Genetic and functional analyses of the conserved C-Terminal core domain of Escherichia coli FtsZ. J Bacteriol 181: 7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Yamane J, Mogi N, Yamaguchi H, Takemoto H, Yao M, Tanaka I (2012) Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr 68: 1175–1188 [DOI] [PubMed] [Google Scholar]
- Michie KA, Löwe J (2006) Dynamic filaments of the bacterial cytoskeleton. Ann Rev Biochem 75: 467–492 [DOI] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics Cold Spring Harbor: Cold Spring Harbor Press, [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, Den Blaauwen T (2007) The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol 65: 1106–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Distèche M, Vollmer W (2007) The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem 282: 36394–36402 [DOI] [PubMed] [Google Scholar]
- Oliva MA, Cordell SC, Löwe J (2004) Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 11: 1243–1250 [DOI] [PubMed] [Google Scholar]
- Potluri L-P, Kannan S, Young KD (2012) ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J Bacteriol 194: 5334–5342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, Van den Ent F, De Boer P, Löwe J (2011) Direct membrane binding by bacterial actin MreB. Mol Cell 43: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Sakai M, Niki H (2008) Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J 27: 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C (2012) High-throughput, subpixel-precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol 80: 612–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swulius MT, Jensen GJ (2012) The helical MreB cytoskeleton in E. coli MC1000/pLE7 is an artifact of the N-terminal YFP tag. J Bacteriol 194: 6382–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Awano N, Inouye M (2011) YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol Microbiol 79: 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W (2012) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ent F, Amos LA, Löwe J (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413: 39–44 [DOI] [PubMed] [Google Scholar]
- Van den Ent F, Johnson CM, Persons L, De Boer P, Löwe J (2010) Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J 29: 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ploeg R, Verheul J, Vischer NOE, Alexeeva S, Hoogendoorn E, Postma M, Banzhaf M, Vollmer W, Den Blaauwen T (2013) Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol Microbiol 87: 1047–1087 [DOI] [PubMed] [Google Scholar]
- Van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z (2011) The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. PNAS 108: 15822–15827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, De Pedro MA, Young KD (2007) FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J Bacteriol 189: 5692–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A, Young KD (2009) In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2. J Bacteriol 191: 3526–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats P, Rothfield L (2007) Duplication and segregation of the actin (MreB) cytoskeleton during the prokaryotic cell cycle. PNAS 104: 17795–17800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats P, Shih Y-L, Rothfield L (2009) Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol Microbiol 72: 170–182 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Bertsche U (2008) Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta 1778: 1714–1734 [DOI] [PubMed] [Google Scholar]
- Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M (1987) Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J Bacteriol 169: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachi M, Matsuhashi M (1989) Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol 171: 3123–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Kitich A, Gober JW (2010) Positioning cell wall synthetic complexes by the bacterial morphogenetic proteins MreB and MreD. Mol Microbiol 76: 616–633 [DOI] [PubMed] [Google Scholar]
- Wientjes FB, Nanninga N (1989) Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J Bacteriol 171: 3412–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.