Abstract
Asexual blood stages of the malaria parasite, which cause all the pathology associated with malaria, can readily be genetically modified by homologous recombination, enabling the functional study of parasite genes that are not essential in this part of the life cycle. However, no widely applicable method for conditional mutagenesis of essential asexual blood-stage malarial genes is available, hindering their functional analysis. We report the application of the DiCre conditional recombinase system to Plasmodium falciparum, the causative agent of the most dangerous form of malaria. We show that DiCre can be used to obtain rapid, highly regulated site-specific recombination in P. falciparum, capable of excising loxP-flanked sequences from a genomic locus with close to 100% efficiency within the time-span of a single erythrocytic growth cycle. DiCre-mediated deletion of the SERA5 3' UTR failed to reduce expression of the gene due to the existence of alternative cryptic polyadenylation sites within the modified locus. However, we successfully used the system to recycle the most widely used drug resistance marker for P. falciparum, human dihydrofolate reductase, in the process producing constitutively DiCre-expressing P. falciparum clones that have broad utility for the functional analysis of essential asexual blood-stage parasite genes.
Introduction
The malaria parasite shares its life cycle between a vertebrate host and a mosquito vector, but all the clinical manifestations of malaria are caused by replication of the asexual blood stages within circulating erythrocytes. In the case of Plasmodium falciparum, responsible for the most life-threatening form of human malaria, asexual blood-stage forms can be maintained in continuous culture in human erythrocytes (Trager and Jensen, 1976), facilitating study of this important part of the life cycle. Advances in genetic tools for the modification of malarial genes have accelerated understanding of the biology of the parasite, aided within the last decade by the acquisition of the entire genome sequences of P. falciparum and several other Plasmodium species (e.g. Gardner et al., 2002; Pain et al., 2008). Because of the relative accessibility of asexual blood stages, these are always used for the introduction of targeting DNA constructs designed for genetic modification by ectopic transgene expression or homologous recombination. Since expression of many Plasmodium genes is stage-specific this allows disruption of genes with essential roles restricted to other parts of the life cycle, including the mosquito and exoerythrocytic life cycle stages. In contrast, disruption of parasite genes that are indispensable for blood-stage growth is lethal, preventing the establishment of null mutants and representing a bottleneck in the functional analysis of these genes. In attempts to overcome this, there has been great interest in developing conditional genetic tools suitable for exogenous control of gene expression in Plasmodium. One system in which the gene product of interest is fused to a destabilization domain that can be stabilized by small ligands, has been used in P. falciparum with some success (Armstrong and Goldberg, 2007; Russo et al., 2009; Dvorin et al., 2010; Muralidharan et al., 2011) but requires that the destabilization tag does not interfere with protein function. Moreover, since the destabilization domain mediates protein degradation by targeting it to the proteasome, the system has limited utility for proteins that are trafficked via the parasite secretory pathway. Tet repressor (TetR)-based regulation of transcription has proven difficult to develop in Plasmodium and there are no reports of efficient repression of transcription by TetR in these parasites. The most commonly used tetracycline-sensitive transactivator, a fusion between TetR and the Herpes simplex virus VP16 protein, does not activate minimal promoters in the group of apicomplexan parasites to which Plasmodium belongs (Meissner et al., 2001). An artificial transactivation domain (TATi), when fused to TetR has been successfully used in the related apicomplexan Toxoplasma gondii to generate conditional mutants (Meissner et al., 2002), and has been used with some success for inducible transgene expression in P. falciparum (e.g. Meissner et al., 2005; Gilson et al., 2008; O'Neill et al., 2011). Very recently, functional transactivation domains based on apicomplexan ApiAP2 transcription factors have been elegantly used to obtain stage-dependent tetracycline-dependent gene knock-down in T. gondii and Plasmodium (Pino et al., 1002). Though of great potential, these conditional approaches all require transactivator-responsive minimal promoters that accurately mimic the transcriptional profile of the gene of interest. Despite a promising report describing the use of peptide-morpholino oligomer conjugates in Plasmodium (Augagneur et al., 2012), most other widely used gene silencing approaches that affect transcript stability, translation or splicing, such as morpholino oligonucleotides, self-cleaving ribozymes and RNA interference (RNAi), have yet to be proven widely effective in Plasmodium (Agop-Nersesian et al., 2008), in the case of RNAi because of the absence of crucial components of the pathway in the parasite (Baum et al., 2009).
Site-specific recombination using phage or yeast-derived recombinases is a method of choice in many models for gene modification or deletion. Two recombinases, Cre and flippase (FLP), have been most widely used. Both recognize short, 34 bp sequences, respectively called loxP and FRT, and mediate either excision or inversion of intervening sequences depending on the relative orientation of the recognition motifs. FLP has been successfully used in the Plasmodium berghei rodent malaria model, exploiting an approach in which developmental stage-specific recombinase activity was obtained by placing FLP under the control of parasite promoters active only in insect stages (Combe et al., 2009; Falae et al., 2010; Giovannini et al., 2011; Lacroix et al., 2011). This system has only limited applicability for studying essential asexual blood stage-specific genes. Cre is active in both P. falciparum (O'Neill et al., 2011) and T. gondii (Brecht et al., 1999), but attempts to obtain robust regulation of this activity have been unsuccessful, resulting only in constitutive recombinase activity unsuitable for conditional gene modifications. Forms of Cre that can be regulated by hormones or small molecules have been described. In the DiCre system, Cre is expressed in the form of two separate, enzymatically inactive polypeptides, each fused to a different rapamycin-binding protein (either FKBP12 or FRB, the rapamycin-binding domain of the FKBP12-rapamycin-associated protein mTOR) (Chen et al., 1995; Choi et al., 1996; Liang et al., 1999). Rapamycin-induced heterodimerization of the two components restores recombinase activity (Jullien et al., 2003;2007). Recent work has shown that this technology functions efficiently in T. gondii, with recombination rates of up to 96% upon induction with rapamycin (Andenmatten et al., 2013).
Here we show that DiCre provides rapid, highly regulated Cre recombinase activity in P. falciparum, capable of excising loxP-flanked (floxed) sequences from a genomic locus with close to 100% efficiency within the time-span of a single erythrocytic growth cycle. As proof of principle, we have used the system to recycle one of the most widely used drug selectable markers, in the process producing DiCre-expressing P. falciparum clones that will be of great utility for conditional modification of P. falciparum genes, including those essential for asexual blood-stage growth.
Results
Design of a ‘single vector’ strategy for DiCre-mediated gene knock-down and selectable marker recycling in P. falciparum
X-ray crystal structure analysis of Cre recombinase has shown that it comprises two major helical domains linked by a short, relatively flexible segment (Guo et al., 1997). In earlier work, Jullien et al. (2007, 2003) showed that the rapamycin-mediated dimerization of two distinct, enzymatically inactive polypeptides approximately corresponding to the individual domains of Cre (residues Thr19–Asn59, called Cre59, and Asn60-343, called Cre60) each fused to a different rapamycin-binding protein (FKBP12 and FRB respectively), resulted in the reconstitution of Cre recombinase activity. Because induction of recombinase activity upon addition of rapamycin does not require de novo biosynthesis of the recombinase, induction is very rapid. The N-terminal FKBP12 and FRB fusion partners are linked to their partner Cre sequences through short Gly/Ser-rich linkers, and each protein possesses a nuclear localization signal at its extreme N-terminus. To adapt the DiCre system to P. falciparum, we first designed a DiCre cassette in which expression of the FKBP-Cre59 and FRB-Cre60 genes were placed under the control of the strong, constitutive P. falciparum BiP and hsp86 promoters, arranged in a head-to-head orientation (Fig. 1A). For introduction of the expression cassette into the parasite, we decided to incorporate this cassette into a larger targeting vector designed to integrate by homologous recombination into a P. falciparum locus in such a way that induction of recombinase activity would produce two distinct effects. First, we wanted to remove the drug selectable marker used to select for the initial integration event. This is because very few drug resistance markers are currently available for experimental genetic modifications in Plasmodium (Lin et al., 2011), and so the recycling of selectable markers increases flexibility for consecutive gene modifications. Second, we wished to explore the possibility of using a ‘single vector’ strategy to obtain downregulation or knockout of a selected gene of interest (GOI). The 3' UTR of eukaryotic genes generally regulates correct transcription termination and polyadenylation of the mRNA, important for mRNA stability and trafficking. Several previous reports (e.g. Yeoh et al., 2007; Combe et al., 2009; Giovannini et al., 2011) have shown that, whereas the endogenous 3' UTR of Plasmodium genes can usually be replaced with other Plasmodium 3' UTR sequences without deleterious effects on gene expression, complete removal of the 3' UTR can severely ablate expression levels, effectively resulting in gene knock-down. To investigate the use of DiCre as a means of obtaining conditional gene knock-down in a manner amenable to medium-throughput gene analysis, we sought to use it to obtain conditional removal of the 3' UTR of a GOI. As an initial target GOI we chose SERA5 (PlasmoDB ID PF3D7_0207600), a member of a family of nine SERA genes in P. falciparum (Arisue et al., 2011). Previous work (Miller et al., 2002; McCoubrie et al., 2007) has shown that whereas most members of this gene family can be individually disrupted with no phenotypic consequences, the SERA5 and SERA6 genes are refractory to disruption using conventional targeted homologous recombination, suggesting that they are indispensable in asexual blood stages of the parasite life cycle. SERA5 is expressed as an abundant, soluble parasitophorous vacuole protein of ∼ 126 kDa, and is thought to play a role in schizont rupture (egress) and/or erythrocyte invasion by released merozoites (reviewed in Blackman, 2008). However its precise function is unknown.
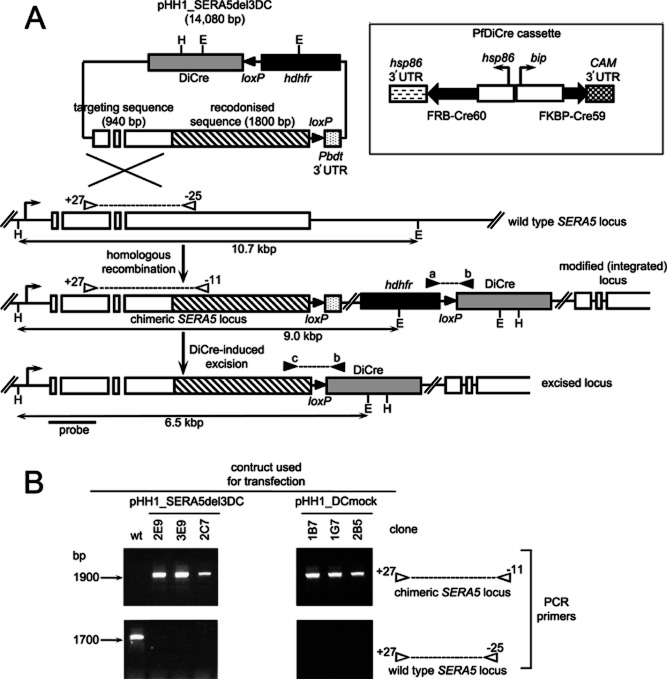
Fig. 1.
Design of the DiCre targeting vector, predicted homologous integration and recombinase-mediated excision events, and isolation of transgenic P. falciparum clones.
A. The boxed insert shows an expanded view of the PfDiCre expression cassette. Expression of the DiCre components (black block arrows) is driven by the P. falciparum BiP and hsp86 promoters, with transcription termination and polyadenylation being regulated by the 3' UTR sequences from the P. falciparum calmodulin (CAM) and hsp86 genes. Main figure: schematic (not to scale) showing the main features of the pHH1_SERA5del3DC plasmid construct. Targeting sequence derived from the native SERA5 gene was fused in-frame to recodonized sequence (hatched) derived from the SERA5synth gene. A single EcoNI restriction site (E) within the hdhfr cassette (black box), which confers resistance to the antifolate WR99210, is shown. The PfDiCre cassette contains another EcoNI site as well as a unique HindIII site (H). The P. berghei dhfr 3' UTR (Pbdt 3' UTR) lies just downstream of the SERA5synth sequence in the plasmid. Two loxP sites are indicated as black arrowheads in the plasmid. The endogenous SERA5 locus, which contains three introns, is shown below, as is the expected result of homologous integration of the entire plasmid and the architecture of the modified locus relative to flanking HindIII and EcoNI sites. Episomal plasmids are harboured as concatamers in P. falciparum, so integration of more than one copy can occur. However, because the plasmid contains only a partial SERA5 sequence not preceded by a promoter, the only SERA5 copy to be transcribed is the modified chimeric gene directly downstream of the endogenous promoter. DiCre-mediated recombination should result in excision of both the Pbdt 3' UTR from the modified SERA5 locus, and the hdhfr selectable marker, as well as removal of the entire pHH1_SERA5del3DC plasmid backbone. Note that, even if concatamers of pHH1_SERA5del3DC were to integrate, DiCre-mediated excision would still result in removal of all sequence between the duplicated loxP sites, leading to the terminal excised structure shown, in which only a single copy of the modified SERA5 coding sequence remains under the control of its endogenous promoter, and just a single genomic loxP site remains. Positions of primers +27, −11 and −25 used for diagnostic PCR of the expected integration of pHH1_SERA5del3DC and pHH1_DCmock (which is identical aside from the presence of a mock promoter in the place of the hsp86 and BiP promoters within the PfDiCre cassette) are shown by white arrowheads joined by dotted lines. Predicted sizes of the amplicons obtained with primer pairs +27 plus −11, and +27 plus −25, are 1911 bp and 1737 bp respectively. Primer +27 lies just upstream of the targeting sequence in the plasmid, so cannot produce a product from the transfection construct. Positions of primers CAM5‘_R4 (a), hsp86 3'_R1 (b) and sgs5seq5F (c) used for diagnostic PCR analysis of the modified and excised locus are indicated by black arrowheads joined by dotted lines. Predicted sizes of amplicons obtained with primers a plus b, and c plus b are 428 bp and 804 bp respectively. The relative position of the probe used for Southern analysis is indicated.
B. Diagnostic PCR analysis of genomic DNA from clones derived by limiting dilution of parasites transfected with constructs pHH1_SERA5del3DC or pHH1_DCmock, confirming integration into the SERA5 locus as expected. Only wild-type (wt) parasite DNA produced a product with primers +27 plus −25, while the amplicon produced with primer pair +27 plus −11, diagnostic of the expected integration event, was obtained only from the transgenic clones.
In initial preliminary work we produced construct pHH1SERA5chimWT, designed to integrate by single-crossover homologous recombination into the SERA5 locus to produce a chimeric gene still under the control of its endogenous promoter and encoding the unmodified SERA5 primary amino acid sequence, but using the 3' UTR from the P. berghei dihydrofolate reductase (dhfr) gene (Pbdt 3' UTR). The use of recodonized sequence within the construct was not essential for this work, but provided the option in future work of introducing desired mutations into this region in the knowledge that single-crossover recombination would preferably occur upstream of the recodonized sequence, as described previously (Child et al., 2010; Ruecker et al., 2012). Transfection of P. falciparum with pHH1SERA5chimWT and selection in the presence of the antifolate drug WR22910 resulted in rapid outgrowth of parasites in which integration of the construct had taken place in the expected manner. Parasite clones obtained by limiting dilution from these lines were phenotypically indistinguishable from wild-type parasites and expressed wild-type levels of SERA5 protein (R. Stallmach and M. Blackman, in preparation), showing that the chimeric SERA5 gene and appended Pbdt 3' UTR were fully functional in maintaining full SERA5 expression levels.
Rapid and efficient site-specific recombination at a genomic P. falciparum locus mediated by induction of DiCre activity
Encouraged by the above result, we produced constructs pHH1_DCmock and pHH1_SERA5del3DC (Fig. 1A), which used the same targeting sequence as pHH1SERA5chimWT for homologous integration into the SERA5 locus, but were designed to allow subsequent deletion of the Pbdt 3' UTR sequence downstream of the modified SERA5 gene, along with the human dhfr (hdhfr) selectable marker, upon induction of DiCre activity. While pHH1_SERA5del3DC contained the complete DiCre cassette shown in Fig. 1A, pHH1_DCmock contained a ‘mock’ cassette in which the hsp86 and BiP promoters were replaced by a single ∼ 930 bp stretch of bacterial coding sequence not expected to drive expression of the DiCre proteins, thereby acting as a negative control for DiCre expression. Transfection of both constructs into P. falciparum, followed by limiting dilution cloning of the resulting WR99210-resistant lines, resulted in the isolation of several parasite clones in which integration of pHH1_DCmock or pHH1_SERA5del3DC had taken place (Fig. 1B). To examine the effects of induction of DiCre activity, highly synchronous young ring-stage parasites of clones 2E9 and 3E9 (containing integrated pHH1_SERA5del3DC) and clone 1B7 (containing the integrated ‘mock’ construct pHH1_DCmock) were obtained by adding purified mature schizonts to fresh erythrocytes, allowing invasion to take place for a period of just 2 h, then removing residual schizonts using a combination of Percoll centrifugation and sorbitol-mediated lysis of the schizonts. The resulting cultures, now maintained in the absence of WR99210, were each divided into two and treated for exactly 4 h with either rapamycin (100 nM) or vehicle only (DMSO, 1% v/v). The parasites were then washed, returned to culture in medium lacking WR99210, and sampled at 24 h post invasion (mid-trophozoite stage) and 45 h post invasion (mature schizont stage). Genomic parasite DNA prepared from each time point was then analysed by PCR, using primers diagnostic for either the intact modified locus (primers a and b indicated in Fig. 1A), or the expected genomic product of DiCre-mediated site-specific recombination (primers c and b; Fig. 1A). As shown in Fig. 2A, rapamycin treatment resulted in the efficient conversion of the intact modified SERA5 locus to the expected excised form of the locus in which deletion of the floxed sequence had taken place (shown in Fig. 1A). Excision was clearly evident by 24 h post invasion, and by 45 h post invasion appeared almost complete, with very little of the intact locus detectable. No excision was detectable by PCR in the 1B7 clone irrespective of whether the parasites had been exposed to rapamycin treatment, indicating a requirement for DiCre expression in the parasite. Of particular importance, no background excision was detectable in 2E9 or 3E9 parasites that had not been exposed to rapamycin, demonstrating robust regulation of DiCre-mediated recombinase activity. To obtain a quantitative measure of excision efficiency, genomic DNA prepared from the 45 h time point was examined by Southern blot (Fig. 2B). This confirmed the high efficiency of the induced site-specific recombination event and allowed it to be calculated by phosphorimager analysis as ∼ 98% by comparison of the intensity of the signals corresponding to the excised and non-excised species in digests of genomic DNA from the rapamycin-treated parasites.
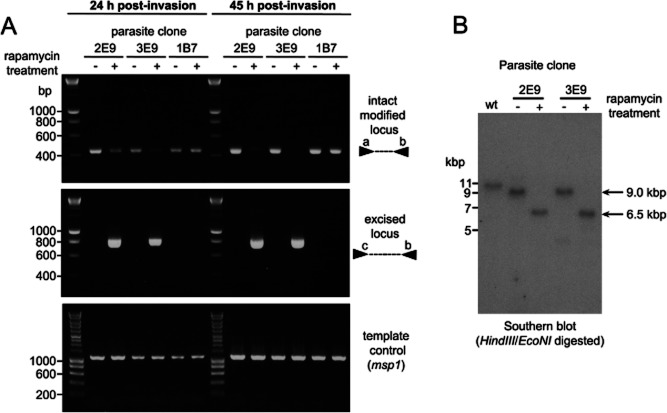
Fig. 2.
Rapid, regulated and highly efficient DiCre-mediated excision of a floxed genomic P. falciparum sequence.
A. PCR analysis showing detection of excision following a 4-h-long treatment of clones 2E9, 3E9 or 1B7 with rapamycin at ring stage. Genomic DNA was prepared from the treated parasites at 24 h and 45 h post invasion. Primer pairs a plus b detect the modified locus prior to excision, while primers c plus b detect the appearance of the rearranged locus resulting from site-directed recombination between the loxP sites (see Fig. 1A). Primers specific for the msp1 gene (MSP1_FOR and MSP1_REV; see Table S1) were used as a control to confirm the presence of genomic DNA in all the template samples.
B. Southern blots confirming efficient rapamycin-induced excision in the 2E9 and 3E9 transgenic P. falciparum clones. The genomic DNA used for the Southern blot was prepared at 45 h post invasion. Phosphorimager quantification of the 6.5 kb ‘excised locus’ signals in the rapamycin-treated 2E9 and 3E9 samples, compared with the position in the same tracks corresponding to the non-excised 9.0 kb species (where no band is visible by eye) showed that excision was 97.9% efficient in the case of clone 2E9, and 97.2% efficient in the case of clone 3E9. However, even upon prolonged exposure, no residual ‘non-excised’ signal was visually detectable in the genomic DNA digests from the rapamycin-treated parasites (not shown).
DiCre-mediated excision of the SERA5 3' UTR fails to silence gene expression due to transcription termination at an alternative cryptic polyadenylation site
The anticipated result of excision of the floxed sequence downstream of the modified SERA5 locus was to both remove the Pbdt 3' UTR, expected to reduce SERA5 expression levels, and also to excise the hdhfr cassette which provides resistance to WR99210 (Fig. 1A). To evaluate the first of these possible phenotypic outcomes, 45-h-old clone 2E9 and 3E9 schizonts from the experiment described above were examined by IFA using a mAb (24C6.1F1) specific for SERA5. Unexpectedly, no discernible difference in fluorescence intensity was evident between parasites that had been exposed to rapamycin at ring stage, and the parallel cultures that had not been rapamycin-treated (Fig. 3A). Given the efficiency of excision demonstrated by PCR and the Southern blot analysis (Fig. 2), this suggested that removal of the Pbdt 3' UTR from the modified SERA5 locus was not sufficient to significantly reduce SERA5 protein expression levels. This was confirmed by Western blot analysis (Fig. 3B).
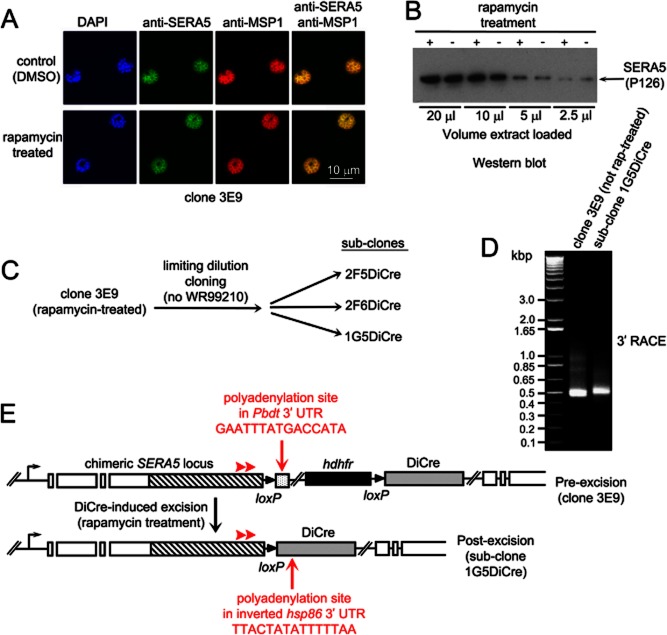
Fig. 3.
DiCre-mediated excision of the Pbdt 3' UTR does not reduce SERA5 expression levels due to the presence of an alternative polyadenylation site within the inverted hsp86 3' UTR.
A. IFA analysis of clone 3E9 schizonts derived from ring-stage parasites treated for 4 h with vehicle only (DMSO) or 100 nM rapamycin to induce DiCre-mediated excision of the Pbdt 3' UTR from the modified SERA5 locus (see Figs 1 and 2). Parasites were probed with mAbs specific for SERA5, or MSP1 as a control. No change in SERA5 expression levels was detected following rapamycin treatment. 4,6-Diamidino-2-phenylindol (DAPI) was used to detect parasite nuclei.
B. Western blot analysis of extracts of the same schizont populations. Different volumes of an SDS extract of intact schizonts were probed with the anti-SERA5 mAb 24C6.1F1. The results confirm no significant difference in SERA5 expression levels between the control and rapamycin-treated parasites.
C. Workflow showing isolation by limiting dilution of parasite subclones from rapamycin-treated P. falciparum clone 3E9, performed in order to obtain genetically homogeneous parasites harbouring the excised locus architecture. Subclone 1G5DiCre was used for subsequent 3' RACE and transfection analysis.
D. Agarose gel electrophoresis of 3' RACE products amplified from total RNA of clone 3E9 and subclone 1G5DiCre. A dominant RACE product was obtained in each case. No product was obtained in the absence of reverse transcriptase (not shown). Sizes in kb of DNA marker fragments (left-hand lane) are indicated.
E. Schematic (not to scale) showing the overall structure of the modified SERA5 locus in clones 3E9 (prior to excision) and subclone 1G5DiCre (following excision), depicted as in Fig. 1. Relative positions of polyadenylation sites identified by sequencing of the cloned 3' RACE products are indicated in red (see Fig. S1 for the aligned sequence data). Positions of SERA5synth gene-specific forward primers used for the semi-nested 3' RACE are shown (red arrowheads).
Although efficient knock-down of Plasmodium gene expression has previously been achieved through removal of 3' UTR regulatory sequences (e.g. Giovannini et al., 2011), the approach is not uniformly successful. In a recent in-depth investigation of such a case, Ecker et al. (1001) showed that FRT-mediated excision of the 3' UTR of the P. berghei chloroquine resistance transporter gene (pbcrt) failed to ablate gene expression due to the presence of alternative polyadenylation sites at the 3' end of the excised locus, that were efficiently used to stabilize the pbcrt mRNA. To address whether a similar phenomenon might explain the continued robust SERA5 expression observed following DiCre-mediated removal of the Pbdt 3' UTR in the present study, the rapamycin-treated clone 3E9 parasites were cloned by limiting dilution (in the absence of WR99210) to obtain genetically homogeneous subclones harbouring the excised genomic locus (Fig. 3C). A total of three subclones were expanded and examined by analytical PCR for the presence of the excised genomic architecture, as well as for the presence of the wild-type SERA5 coding sequence, which could potentially be reconstituted in these transgenic parasites through spontaneous reversion by homologous recombination between the excised modified locus (which lacks the hdhfr selectable marker) and the downstream partially duplicated SERA5 ORF (i.e. the reverse of the single-crossover recombination event depicted in Fig. 1). As expected, all three subclones exhibited the same excised genomic architecture as that in the rapamycin-treated clone 3E9 parasites, with no signs of the intact modified locus and no reversion to the wild-type locus, even after continuous culture for > 2 months (data not shown). One of the subclones (called 1G5DiCre) was then analysed by rapid amplification of cDNA ends (3' RACE), in parallel with the non-rapamycin-treated parental 3E9 clone, using nested forward primers specific for the SERA5synth gene to examine the 3' structure of the chimeric SERA5 mRNA transcript(s). As shown in Fig. 3D, there was a clear difference in the size on agarose gel electrophoresis of the dominant 3' RACE products from these two parasite clones, consistent with transcription termination occurring at distinct sites, as expected. To examine this in detail, the 3' RACE products were cloned and sequenced. A comparison of these sequences (Supporting Information Fig. S1) showed that mRNA transcribed from the non-excised chimeric SERA5 locus underwent polyadenylation as expected at a single major position ∼ 247 bp into the Pbdt 3' UTR. In contrast, in the excised locus polyadenylation occurred instead at a position within the inverted hsp86 3' UTR which lies immediately downstream of the SERA5 stop codon in the excised locus (Fig. 3E). No obvious sequence similarity was evident around the polyadenylation sites (Fig. 3E and Fig. S1). Note that the length of the sequenced RACE products was approximately in line with the observed size of the dominant signals observed on gel electrophoresis (∼ 460 bp and ∼ 490 bp for clone 3E9 and subclone 1G5DiCre respectively), suggesting that the sequenced clones were representative of the major RACE products in each case. These observations strongly suggest that the hsp86 3' UTR sequence (which was present in the modified locus in order to regulate transcription of the FRB-Cre60 component of the DiCre cassette; see Fig. 1 insert), possesses bidirectional transcription termination and polyadenylation functionality. Collectively, our results explain the observed lack of SERA5 knock-down upon removal of the Pbdt 3' UTR.
Efficient recycling of the hdhfr selectable marker through DiCre-mediated excision, and production of DiCre-expressing ‘recipient’ P. falciparum clones
To address the second predicted outcome of DiCre-mediated excision from the modified SERA5 locus – removal of the hdhfr selectable marker cassette – schizonts from all three clones 2E9, 3E9 and 1B7 treated with or without rapamycin as described in Fig. 2 were allowed to undergo invasion overnight in the absence of WR99210, to form a new generation of ring-stage parasites. Each culture was then divided into two, and culture continued in either the presence or absence of WR99210. As shown in Fig. 4A and B, whereas rapid expansion of the non-rapamycin-treated 2E9 and 3E9 clones took place in both the presence and absence of WR99210, those 2E9 and 3E9 parasites that had been rapamycin-treated at ring stage in the first cycle instead displayed extensive cell death in the presence of WR99210, consistent with the removal of the hdhfr cassette in the great majority of the population. In contrast, and again consistent with the excision results, the 1B7 clone parasites continued to grow well, irrespective of whether they had been rapamycin treated, or of the presence of WR99210. These data convincingly support the PCR and Southern blot results, showing efficient removal of the hdhfr resistance marker in the 2E9 and 3E9 parasites upon rapamycin-induced, DiCre-mediated excision of the floxed genomic sequence.
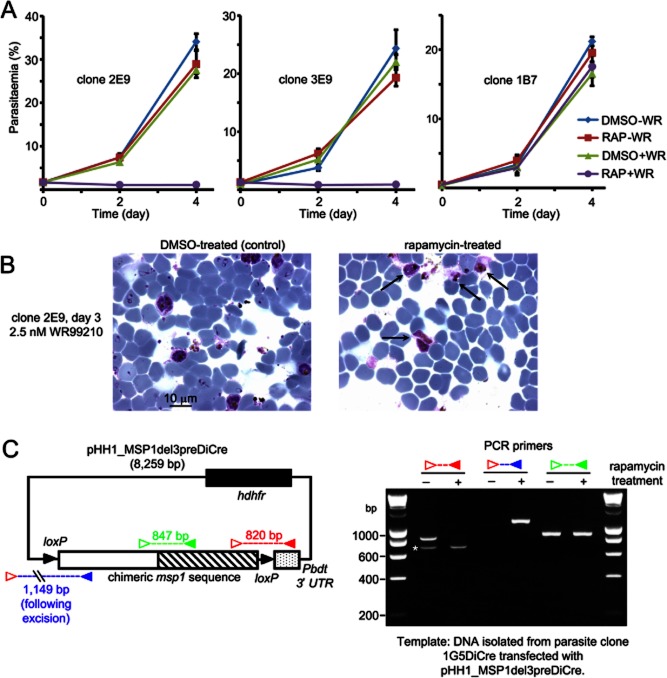
Fig. 4.
DiCre-mediated excision successfully recycles the hdhfr selectable marker and produces a P. falciparum recipient clone that constitutively expresses DiCre.
A. Typical growth assay showing the results of expanding the control (DMSO-treated) or rapamycin-treated clones 2E9, 3E9 and 1B7 in the presence or absence of WR99210. As predicted based on the efficient excision of the hdhfr cassette, the rapamycin-treated 2E9 and 3E9 clones did not replicate in medium containing WR99210. In contrast, rapamycin treatment of clone 1B7, which harbours the ‘mock’ DiCre cassette, had no effect on growth under any conditions. Day 0 corresponds to the point (∼ 60 h following treatment with rapamycin or DMSO) at which the cultures were transferred to fresh medium ± WR99210. Each data point represents the mean of triplicate microscopic counts, each of at least 500 cells. Error bars, ± 1 SD.
B. Giemsa-stained microscopic images of clone 2E9 at day 3 following the start of culture in the presence of WR99210. The appearance of the poorly staining, dysmorphic schizonts (arrowed) in the rapamycin-treated cultures is typical of that usually observed after WR99210 treatment of drug-susceptible wild-type parasites. By day 4, these had completely degenerated and disappeared from the cultures. In contrast, the DMSO-treated control cultures contain numerous healthy schizonts and young ring stages, indicative of normal levels of growth.
C. Induction of DiCre-mediated excision in the 1G5DiCre recipient subclone. Shown (left-hand side) is a schematic of reporter plasmid construct pHH1_MSP1del3preDiCre (not to scale) alongside results of PCR analysis of DNA isolated from 1G5DiCre parasites harbouring the plasmid, following treatment ± rapamycin. Rapamycin treatment induced efficient excision of the floxed Pbdt 3' UTR and hdhfr cassette from the plasmid, proving expression of DiCre from the genomic cassette incorporated into the modified SERA5 locus of the 1G5DiCre parasites. No excision was observed in the absence of rapamycin. The relative positions of primers used for the PCR are indicated as coloured arrowheads. Primers used were: 3D7synMSP1_FOR2 (open red); PbDT3'5’R1 (solid red); 3D7endoMSP1_REV4 (blue); 3D7endoMSP1FOR1 (open green); and 3D7synMSP1_REV3 (solid green). See Table S1 for primer sequences. The hatched region in the pHH1_MSP1del3preDiCre schematic indicates the synthetic (recodonized) msp1 sequence, and the expected sizes of the various amplicons are indicated. The extreme left-hand and right-hand tracks on the gel contain DNA size markers. The ∼ 700 bp fragment (asterisked) present in the 3D7synMSP1_FOR2 (open red) plus PbDT3'5’R1 (solid red) PCR tracks is a product of mis-priming, since this was also amplified from 1G5DiCre parasites that had not been transfected with plasmid pHH1_MSP1del3preDiCre (not shown). Rapamycin treatment results in loss of the 820 bp PCR product and the appearance of an excision-specific 1149 bp product. Note that under the PCR conditions used, no product was expected to be amplified from the intact pHH1_MSP1del3preDiCre with primers 3D7synMSP1_FOR2 (open red) and 3D7endoMSP1_REV4 (blue) due to the large size of that predicted product.
Prolonged culture (> 28 days) of the rapamycin-treated 2E9 and 3E9 parasites in the presence of WR99210 resulted in the eventual outgrowth of drug-resistant parasites (not shown). We reasoned that these surviving parasites (referred to as 2E9WR and 3E9WR) likely represented a very small minority of the original parasite clones that had either not undergone DiCre-mediated excision upon the first round of rapamycin treatment, or had undergone anomalous excision. To test this prediction, the expanded 2E9WR and 3E9WR parasite lines were synchronized then subjected to a second 4 h treatment ± rapamycin, and their genomic structure analysed again by PCR 40 h later. For comparison these experiments were performed in parallel with similar treatments of the original 2E9 and 3E9 clones that had not previously been exposed to rapamycin. As shown in Fig. S2, whereas the original 2E9 and 3E9 clones exhibited efficient rapamycin-dependent excision as seen previously, the WR99210-resistant parasite lines showed either poor excision (3E9WR) or good excision but with signs of pre-existing excision (2E9WR) which presumably had failed to remove the hdhfr cassette, leaving these parasites WR99210-resistant. While these results cannot be fully explained without detailed genomic sequencing of the modified loci, they were consistent with the recovered WR99210-resistant parasites being the results of very low-level anomalous DNA rearrangements that were not typical of the great majority of the rapamycin-treated 2E9 and 3E9 parasites.
To finally demonstrate the utility of our approach for producing a DiCre-expressing recipient parasite line with potential for further gene manipulation, we returned to a more detailed analysis of subclone 1G5DiCre, which was derived as described above by limiting dilution cloning (in the absence of WR99210) from clone 3E9 parasites that had been rapamycin-treated to induce excision of the hdhfr cassette. To test for expression of DiCre from the integrated genomic DiCre cassette in these parasites, subclone 1G5DiCre was transfected by electroporation with a reporter construct called pHH1_MSP1del3preDiCre (Fig. 4C). This construct contains a hdhfr selectable marker cassette and a chimeric msp1 gene fragment, followed by the Pbdt 3' UTR and flanked by loxP sites; importantly, it lacks a DiCre cassette, so excision of the floxed sequence can only take place if the 1G5DiCre parasites continue to express DiCre from the integrated genomic DiCre cassette. 1G5DiCre parasites not subjected to electroporation, or mock-transfected (i.e. subjected to electroporation in the absence of the transfection construct), rapidly died upon culture in the presence of WR99210, as expected (data not shown). In contrast, drug-resistant parasites appeared in the pHH1_MSP1del3preDiCre-transfected 1G5DiCre cultures within 6 days. These parasites were expanded, synchronized, then treated ± rapamycin for 4 h as described above and analysed 40 h later by PCR, using primers designed to detect excision of the floxed sequence, as well as control primers. As shown in Fig. 4C, rapamycin treatment resulted in efficient excision of the floxed sequence from the episomal pHH1_MSP1del3preDiCre construct, proving robust constitutive expression of DiCre in the 1G5DiCre parasites. These results confirm the usefulness of the 1G5DiCre parasite subclone as a recipient line for future DiCre-mediated mutational analysis of P. falciparum genes.
Discussion
Despite the medical importance of P. falciparum, and in particular of its asexual blood-stage life cycle, obtaining conditional regulation of gene expression in this organism has proven extremely challenging. The short erythrocytic life cycle of P. falciparum (∼ 48 h) raises particular problems in this regard; in order to observe developmental stage-specific phenotypic changes at the population level in a conditional mutant over the course of a single erythrocytic cycle – especially important if the outcome of mutagenesis is lethal – switch-off of gene expression needs to be both efficient and rapid. We have shown here that the DiCre system meets both those critical requirements, allowing rapid, highly regulated and remarkably efficient site-specific recombination within the course of a single erythrocytic growth cycle. We designed our strategy to simultaneously impose two measurable effects on gene expression in our transgenic clones. First we attempted knock-down of SERA5 expression by removal of its introduced Pbdt 3' UTR, anticipating that this would result in lowered protein expression levels by reducing mRNA stability. We were surprised to observe no detectable change in SERA5 expression levels. SERA5 is expressed at extraordinarily high levels in asexual blood-stage schizonts, and indeed is probably one of the most abundant schizont-stage parasite proteins (Lasonder et al., 2002; Le Roch et al., 2004). Small alterations in SERA5 mRNA stability may therefore have little effect on overall protein levels. We reasoned that an alternative explanation for the lack of alteration in SERA5 levels might be the presence of a cryptic 3' UTR in the apposed hsp86 3' UTR, which was shifted adjacent to the SERA5 stop codon following DiCre-mediated excision from the modified locus (Fig. 1A), even though this was in the opposite orientation to that previously known to mediate correct transcription termination. Examination of subclone 1G5DiCre, derived from the rapamycin-treated 3E9 parasites, confirmed this suspicion; 3' RACE analysis identified a polyadenylation site in the inverted hsp86 3' UTR at a position that shows no obvious sequence similarity to that identified in the authentic Pbdt 3' UTR, which is widely used to regulate transcription of transgenes in both P. falciparum and P. berghei. This suggests that the hsp86 3' UTR contains bidirectional transcription termination and polyadenylation signals. Interestingly, a recent report from Ecker et al. (1001) made a very similar observation; following unsuccessful attempts to obtain knock-down of the pbcrt gene in P. berghei those authors identified efficient polyadenylation sites in the inverted 3' UTR from the P. berghei thrombospondin related adhesive protein (trap) gene, which was translocated to a position immediately downstream of the pbcrt gene upon FLP-mediated excision. Together, these findings show that great caution should be exercised in the design of constructs intended for removal of 3' regulatory sequences as a route to gene knock-down in Plasmodium. Certainly, in the light of our results future work will focus on alternative approaches to DiCre-mediated SERA5 knockout, including truncation of the coding sequence by inserting the upstream loxP site into one of the 5'-proximal introns within the SERA5 gene, or indeed floxing the entire locus. This would likely require the use of a double-crossover strategy to incorporate the two loxP sites, a feasible approach in P. falciparum (Duraisingh et al., 2002).
The second intended outcome of DiCre-mediated excision of the modified SERA5 locus, removal of the hdhfr selectable marker cassette, resulted as expected in a dramatic reversal of resistance of the 2E9 and 3E9 clones to WR99210, consistent with the high efficiency of site-specific recombination determined by PCR and Southern blot analysis. Importantly, background excision was undetectable by PCR in the absence of rapamycin treatment, demonstrating stringent regulation of DiCre activity. The result demonstrates robust, conditional deletion of what is effectively an essential parasite transgene in the presence of the antifolate drug. It also demonstrates the ease with which the DiCre system can be used to recycle selectable markers, only a limited number of which are available for use in Plasmodium. This is crucial for enabling consecutive genetic manipulations, required for disruption of multiple genes at different loci, or complementation of knockouts. Three parasite subclones derived from the rapamycin-treated 3E9 clone were found to be genetically stable, and detailed examination of subclone 1G5DiCre showed it to be readily transformed back to WR99210-resistance by transfection with a reporter plasmid containing the hdhfr cassette, confirming recycling of the selectable marker. In addition we confirmed constitutive expression of DiCre in the parasites by demonstrating rapid excision of a floxed sequence from the input plasmid upon rapamycin treatment of the transfected parasites. The DiCre-expressing ‘recipient’ P. falciparum clones will be invaluable tools for further gene targeting studies, enabling the use of much smaller targeting constructs than that described here, since the constructs will no longer need to incorporate the ∼ 5.5 kb DiCre cassette (which is now integrated into the genome of the recipient parasite clones).
The DiCre strategy presented here will have important applications in other Plasmodium species, including the human pathogen Plasmodium knowlesi, which has recently been adapted to continuous culture in human erythrocytes (Moon et al., 2013). It also has great potential for use in rodent malaria models such as the widely used and highly genetically tractable species P. berghei. DiCre has previously been shown to be inducible in vivo using rapamycin (Jullien et al., 2007), and rapamycin is used clinically as an immunosuppressive agent, even accumulating strongly in erythrocytes following administration (Yatscoff et al., 1995; Trepanier et al., 1998). We would therefore expect that it should be possible to induce DiCre activity in circulating blood-stage rodent parasites by parenteral administration of rapamycin. As an alternative approach, and to avoid potential experimental complications introduced by the immunosuppressive and antiproliferative activities of rapamycin in mice, isolated parasites (which can be maintained transiently ex vivo) could be treated briefly in vitro with a ‘pulse’ of rapamycin to induce DiCre activity, then simply washed and reintroduced into the animal. Rapamycin does have antimalarial properties, possibly through binding the parasite FKBP homologue (Monaghan and Bell, 2005); however, the reported IC50 value of rapamycin against P. falciparum (2.6 μM; Bell et al., 1994) is much higher than the concentration used here to induce DiCre activation (100 nM), and in accord with that we observed no deleterious effects on parasite growth following brief treatment with the drug. The availability of a series of rapamycin analogues (‘rapalogues’) some of which bind certain FRB or FKBP12 mutants well, but have lowered affinity for endogenous mammalian FKBP12 or FRAP (Clackson et al., 1998; Pollock and Clackson, 2002; Bayle et al., 2006), provides even more potential flexibility for use of DiCre in vitro. Unfortunately several of these compounds are not suitable for use in mice as they appear to be rapidly cleared in vivo (Jullien et al., 2007). This limitation may be overcome by a second-generation ‘DiCre2’ system currently under development in which both DiCre components are fused to an FKBP mutant which can be dimerized by the rapalogue AP20187; preliminary studies have indicated that this is effective in vivo with no physiological side-effects (Herman and Jullien, 2012; also J.-P. Herman and C. Monetti, unpublished). We expect the DiCre strategy to have a marked impact on understanding of gene function in the malaria parasite and other apicomplexan pathogens.
Experimental procedures
Reagents and antibodies
Rapamycin was obtained from Sigma, UK (catalogue number R0395). Stock solutions (10 μM) were prepared in DMSO and stored in aliquots at −20°C. The antifolate drug WR99210 was from Jacobus Pharmaceuticals (New Jersey, USA). Monoclonal antibody (mAb) X509, which recognizes the parasite major merozoite surface protein MSP1, has been described previously (Blackman et al., 1991), as has the anti-SERA5 mAb 24C6.1F1 (Delplace et al., 1985), which was a kind gift of Jean-Francois Dubremetz (University of Montpellier 2, France).
Parasite cultures and transfections
Asexual blood-stage cultures of P. falciparum clone 3D7 were maintained in vitro and synchronized according to standard procedures (Blackman, 1994; Yeoh et al., 2007) in RPMI 1640 medium containing the serum substitute Albumax (Invitrogen). For introduction of transfection constructs, mature schizonts were enriched from highly synchronous cultures using Percoll (GE Healthcare) as described previously (Harris et al., 2005), and transfected by electroporation with 10 μg of circular plasmid DNA using the Amaxa 4D electroporator (Lonza) and the P3 Primary cell 4D Nucleofector X Kit L (Lonza) and program FP158, exactly as recently described for P. knowlesi (Moon et al., 2013). Selection for parasites harbouring the plasmid was performed by culture in medium containing 2.5 nM WR99210. Selection for parasites in which integration of transfected DNA into the genome had taken place was promoted by cycles of culture in the absence and presence of WR99210, as described previously (Harris et al., 2005). When integration was detected by diagnostic PCR, integrant clones were obtained by limiting dilution and maintained in medium containing WR99210. Parasite growth rates were assessed by microscopic examination of Giemsa-stained thin films at 2-day intervals, and expressed as percentage parasitaemia (percentage of parasitized erythrocytes in the population)
Production of P. falciparum transfection vector pHH1SERA5chimWT
A synthetic, recodonized SERA5 gene (SERA5synth), codon-optimized for expression in Escherichia coli and based on the predicted P. falciparum 3D7 sequence (PlasmoDB ID PF3D7_0207600) was synthesized by GeneArt AG (Regensburg, Germany) and provided with terminal 5’ SalI and 3’ XhoI sites. A 940 bp targeting sequence including the second and third intron of the SERA5 gene (PlasmoDB ID PF3D7_0207600) was amplified by PCR from P. falciparum 3D7 genomic DNA using the oligonucleotide pair +S5endogHpaI and −S5endogClaI (see Supporting information Table S1 for all primer sequences used in this work). A 3'-segment of the recodonized SERA5synth gene was amplified by PCR using the oligonucleotide pair +S5Seq1021 and −S5stopXhoI. The PCR amplicons, of 957 bp and 2007 bp respectively, were ligated into pCR-Blunt using the Zero Blunt PCR Cloning Kit (Invitrogen). Clones with a suitable insert orientation were selected and then the endogenous SERA5 targeting sequence was ligated in frame to the 5' end of the 3' segment of the SERA5synth gene, using restriction sites KpnI (derived from the pCR-Blunt multiple cloning site) and ClaI. The entire chimeric sequence was then excised with HpaI and XhoI and ligated into pHA3.HH1 (Yeoh et al., 2007) to produce pHH1SERA5chimWT. This final construct comprised 940 bp of authentic SERA5 sequence (the targeting sequence), fused in frame to 1800 bp of recodonized synthetic SERA5synth sequence encoding the remaining 3' region of the SERA5 ORF, followed by a stop codon, an XhoI site, then a 3HA epitope tag sequence, and another stop codon, all directly upstream of the 3' UTR from the P. berghei dihydrofolate reductase (dhfr) gene (Pbdt 3' UTR).
Production of the PfDiCre cassette
PCR amplification was conducted using AccuPrime™ Pfx SuperMix (Invitrogen, UK) and when required PCR products were subcloned into the pScB subcloning vector (Aligent Technologies, UK). A ‘mock’ promoter region was amplified from E. coli genomic DNA using primers EWmock1 and EWmock2. The resulting amplicon (corresponding to ∼ 930 bp of the E. coli pmba gene, GenBank/EMBL Accession No. X54152) was subcloned into pScB. Restriction digestion of the intermediate vector using PstI and HindIII released the mock promoter fragment which was subsequently cloned into the pBlueScript SK+ phagemid (GenBank/EMBL Accession No. X52328) pre-digested with PstI and HindIII, to form pmockINT. The FKBP-Cre59 gene was amplified using primers EWCRE59For and EWCRE59Rev from plasmid TUB8FKBP-Cre59-HX (Andenmatten et al., 2013), and subcloned into PstI and EcoRI-digested pmockINT to create pCre59INT. The 3' UTR of the P. falciparum calmodulin gene (PfCAM) was amplified from P. falciparum genomic DNA using primers EWCAM3For and EWCAM3Rev. The PCR fragment was subcloned into pScB. To excise the 3' CAM fragment, the intermediate vector was digested with NotI and PstI and inserted into pCre59INT to form the pCre59 vector. The FRB-Cre60 coding sequence was amplified by PCR using primers EWCRE60For and EWCRE60Rev from plasmid TUB8FRBCre60-HX (Andenmatten et al., 2013). The amplicon was subcloned into pScB, released using HindIII and KpnI and ligated into the pCre59 vector to form pCre59/Cre60INT. Finally, the hsp86 3' UTR was amplified by PCR from P. falciparum genomic DNA using primers EWHSP863For and EWHSP863Rev, cloned into pScB, released with ClaI and KpnI and ligated into pCre59/Cre60INT to create the ‘mock’ pCre59/Cre60 DiCre vector called (DiCre24A). To obtain expression of DiCre in P. falciparum, the mock promoter in this vector was replaced with the P. falciparum hsp86 and BiP promoters, arranged in a head-to-head orientation. To do this, the hsp86 5' flanking region was amplified from pA289-attP-BSD (a kind gift of Andy Osborne, University College London, UK) using primers hsp86 5'_F1 and hsp86 5'_R1. The resulting product was cloned into DiCre24A using EcoRI and HindIII restriction sites, replacing the mock promoter region and giving rise to plasmid pBS_DC_hsp86. The BiP 5' UTR was amplified from pHH4 using primers bip_F1 and bip_R1, and cloned into pBS_DC_hsp86 using AfeI and EcoRI, giving rise to the PfDiCre expression cassette vector pBS_DC_hsp86/Bip5'.
Production of P. falciparum transfection vector pHH1_SERA5del3DC
This construct was designed to integrate by single crossover homologous recombination into the SERA5 locus in such a manner that DiCre-mediated excision from the modified locus would remove its introduced 3' UTR (Pbdt 3' UTR), as well as removing the hdhfr selectable marker cassette from the modified locus. Cloning of other target gene sequences into the transfection plasmid can be carried out using a restriction site in the multiple cloning site (MCS) and the unique XhoI site immediately downstream of the recodonized SERA5 sequence (with or without a stop codon depending on whether the 3HA epitope tag is required) or the unique AvrII site.
PCR reactions were performed using pHH1SERA5chimWT as template, and primers sgS5seq4F and 3HA_AvrII_LoxP or AvrII_LoxP_PbDT3'_F and NotI_PbDT3'_R. Products from these reactions were mixed in equal amounts and an overlapping PCR carried out using primers sgS5seq4F and NotI_PbDT3'_R. The resulting amplicon was cloned into pHH1SERA5chimWT using the XhoI and NotI restriction sites, generating plasmid pHH1_sera5_LoxP1. This now contained an AvrII and a loxP site between the 3HA tag and Pbdt 3' UTR sequences. To incorporate the loxP site and a MCS (comprising SpeI, SnaBI, AflII and HpaI sites) upstream of the SERA5 targeting gene sequence in this plasmid, three sequential rounds of PCR amplification were carried out using forward primers (i) MCS_HpaI, (ii) U1_MCS and (iii) LoxP_MCS_XL and reverse primer EndoS5_R1. The resulting amplicon was blunt ended with T4 DNA polymerase then digested with BstZ171 and cloned into pHH1_sera5_LoxP1 pre-digested with HpaI and BstZ171, giving rise to plasmid pHH1_PreDiCre_A. This was digested with EcoRI, blunted with T4 DNA polymerase and re-ligated to remove the EcoRI site, giving rise to 3A_ΔEcoRI. This was digested with HindIII, blunted with T4 DNA polymerase and re-ligated to remove the HindIII site, giving rise to pHH1_PreDC_A_ΔH/ΔE. The entire DiCre cassette from plasmids DiCre24A or pBS_DC_hsp86/Bip5' was finally cloned into pHH1_PreDiCre_A and pHH1_PreDC_A_ΔH/ΔE respectively, using the SpeI and AflII restriction sites, to produce the final transfection constructs, pHH1_DCmock and pHH1_SERA5del3DC.
Production of P. falciparum reporter transfection vector pHH1_MSP1del3preDiCre
This construct was used to assay for inducible DiCre activity in the 1G5DiCre P. falciparum subclone. Briefly, it is identical to pHH1_PreDiCre_A described above except that the chimeric SERA5 sequence was replaced with ∼ 990 bp of msp1 sequence fused in frame to ∼ 1400 bp of recodonized msp1 sequence. The plasmid therefore contains two loxP sites flanking the Pbdt 3' UTR and hdhfr cassette. Full details of production of this construct will be provided in a later paper (S. Das and M. Blackman, in preparation).
Indirect immunofluorescence (IFA) and Western blot
IFA and Western blot analysis using mAbs 24C6.1F1 and X509 were performed as described previously, using SDS-extracts of mature intact Percoll-enriched schizonts for the Western blot analysis (Jean et al., 2003), and formaldehyde-fixed thin films of cultures containing mature schizonts for the IFA (Harris et al., 2005; Ruecker et al., 2012).
Southern blot
For Southern blot analysis, a 597 bp probe corresponding to endogenous SERA5 coding sequence lying just upstream of the targeting sequence in constructs pHH1SERA5chimWT, pHH1_A_DC_mock and pHH1_3A_H + B_ΔH/E_SERA5 (which was identical in all cases) was produced by PCR amplification from P. falciparum 3D7 genomic DNA with primers SERA5_US_F and SERA5_US_R (Table S1). Radiolabelling of the probe and hybridization to HindIII/EcoNI-digested genomic DNA from parasite clones of interest was performed as described previously (Ruecker et al., 2012). Quantification of the conversion of the signal corresponding to the non-excised modified SERA5 locus to the excised form was performed by phosphorimager analysis on a STORM 860 Molecular Imager (GE Healthcare) using ImageQuant software.
3' Rapid amplification of cDNA ends (3' RACE)
Total RNA from the 3E9 P. falciparum clone (non-rapamycin-treated) and the 1G5DiCre subclone was prepared using a TRIzol Plus RNA isolation kit. First-strand cDNA synthesis was conducted using 3 μg of total RNA, oligo dT adapter primers and SuperScript II reverse transcriptase, using a 3' RACE kit (Invitrogen) according to the manufacturer's protocol. Subsequently, semi-nested PCR reactions were performed to specifically amplify cDNA derived from polyadenylated SERA5 mRNA, using SERA5synth-specific primers SERA5_3R1 and SERA5_3R2 (Table S1) as first and second gene-specific primers. For both PCR reactions AUAP from the 3' RACE kit was used as the reverse primer. PCR products were purified (Qiagen MinElute kit) and cloned into the pGEM T-Easy vector (Promega) for nucleotide sequencing.
Acknowledgments
The authors are grateful to Malcolm Strath (NIMR) for excellent technical assistance with this work, and thank Andy Osborne, University College London, UK, for the gift of plasmid pA289-attP-BSD. This work was supported by the UK Medical Research Council (U117532063 to M.J.B.), and the European Community Seventh Framework Programme (FP7/2007–2013) under grant agreement No. 242095. C.R.C. was supported by a Wellcome Trust Project Grant (086550/Z/08/Z), and S.M. and M.M. by Wellcome Trust Senior Fellowships (WT061173MA and 087582/Z/08/Z respectively). S.D. was in receipt of a PhD studentship from the European Union Framework Programme 7-funded Network of Excellence EviMalAR.
Supplementary material
References
- Agop-Nersesian C, Pfahler J, Lanzer M, Meissner M. Functional expression of ribozymes in Apicomplexa: towards exogenous control of gene expression by inducible RNA-cleavage. Int J Parasitol. 2008;38:673–681. doi: 10.1016/j.ijpara.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Andenmatten N, Egarter S, Jackson AJ, Jullien N, Herman JP, Meissner M. Conditional genome engineering in Toxoplasma gondii uncovers alternative invasion mechanisms. Nat Methods. 2013;10:125–127. doi: 10.1038/nmeth.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisue N, Kawai S, Hirai M, Palacpac NM, Jia M, Kaneko A, et al. Clues to evolution of the SERA multigene family in 18 Plasmodium species. PLoS ONE. 2011;6:e17775. doi: 10.1371/journal.pone.0017775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CM, Goldberg DE. An FKBP destabilization domain modulates protein levels in Plasmodium falciparum. Nat Methods. 2007;4:1007–1009. doi: 10.1038/nmeth1132. [DOI] [PubMed] [Google Scholar]
- Augagneur Y, Wesolowski D, Tae HS, Altman S, Ben Mamoun C. Gene selective mRNA cleavage inhibits the development of Plasmodium falciparum. Proc Natl Acad Sci USA. 2012;109:6235–6240. doi: 10.1073/pnas.1203516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, Waters A, et al. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009;37:3788–3798. doi: 10.1093/nar/gkp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle JH, Grimley JS, Stankunas K, Gestwicki JE, Wandless TJ, Crabtree GR. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol. 2006;13:99–107. doi: 10.1016/j.chembiol.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Bell A, Wernli B, Franklin RM. Roles of peptidyl-prolyl cis-trans isomerase and calcineurin in the mechanisms of antimalarial action of cyclosporin A, FK506, and rapamycin. Biochem Pharmacol. 1994;48:495–503. doi: 10.1016/0006-2952(94)90279-8. [DOI] [PubMed] [Google Scholar]
- Blackman MJ. Purification of Plasmodium falciparum merozoites for analysis of the processing of merozoite surface protein-1. Methods Cell Biol. 1994;45:213–220. doi: 10.1016/s0091-679x(08)61853-1. [DOI] [PubMed] [Google Scholar]
- Blackman MJ. Malarial proteases and host cell egress: an ‘emerging’ cascade. Cell Microbiol. 2008;10:1925–1934. doi: 10.1111/j.1462-5822.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman MJ, Whittle H, Holder AA. Processing of the Plasmodium falciparum major merozoite surface protein-1: identification of a 33-kilodalton secondary processing product which is shed prior to erythrocyte invasion. Mol Biochem Parasitol. 1991;49:35–44. doi: 10.1016/0166-6851(91)90128-s. [DOI] [PubMed] [Google Scholar]
- Brecht S, Erdhart H, Soete M, Soldati D. Genome engineering of Toxoplasma gondii using the site-specific recombinase Cre. Gene. 1999;234:239–247. doi: 10.1016/s0378-1119(99)00202-4. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child MA, Epp C, Bujard H, Blackman MJ. Regulated maturation of malaria merozoite surface protein-1 is essential for parasite growth. Mol Microbiol. 2010;78:187–202. doi: 10.1111/j.1365-2958.2010.07324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combe A, Giovannini D, Carvalho TG, Spath S, Boisson B, Loussert C, et al. Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe. 2009;5:386–396. doi: 10.1016/j.chom.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Delplace P, Dubremetz JF, Fortier B, Vernes A. A 50 kilodalton exoantigen specific to the merozoite release-reinvasion stage of Plasmodium falciparum. Mol Biochem Parasitol. 1985;17:239–251. doi: 10.1016/0166-6851(85)90021-0. [DOI] [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Cowman AF. Negative selection of Plasmodium falciparum reveals targeted gene deletion by double crossover recombination. Int J Parasitol. 2002;32:81–89. doi: 10.1016/s0020-7519(01)00345-9. [DOI] [PubMed] [Google Scholar]
- Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS, et al. A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science. 2010;328:910–912. doi: 10.1126/science.1188191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker A, Lewis RE, Ekland EH, Jayabalasingham B, Fidock DA. Tricks in Plasmodium's molecular repertoire – escaping 3'UTR excision-based conditional silencing of the chloroquine resistance transporter gene. Int J Parasitol. 2012;42:969–974. doi: 10.1016/j.ijpara.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falae A, Combe A, Amaladoss A, Carvalho T, Menard R, Bhanot P. Role of Plasmodium berghei cGMP-dependent protein kinase in late liver stage development. J Biol Chem. 2010;285:3282–3288. doi: 10.1074/jbc.M109.070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson PR, O'Donnell RA, Nebl T, Sanders PR, Wickham ME, McElwain TF, et al. MSP1(19) miniproteins can serve as targets for invasion inhibitory antibodies in Plasmodium falciparum provided they contain the correct domains for cell surface trafficking. Mol Microbiol. 2008;68:124–138. doi: 10.1111/j.1365-2958.2008.06140.x. [DOI] [PubMed] [Google Scholar]
- Giovannini D, Spath S, Lacroix C, Perazzi A, Bargieri D, Lagal V, et al. Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa. Cell Host Microbe. 2011;10:591–602. doi: 10.1016/j.chom.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Guo F, Gopaul DN, van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, et al. Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog. 2005;1:241–251. doi: 10.1371/journal.ppat.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J-P, Jullien N. Regulation of Cre recombinase: use of ligand-regulated and dimerizable Cre for transgenesis. In: Morozov A, editor. Controlled Genetic Manipulations. Vol. 65. New York: Springer Protocols; 2012. pp. 1–28. Neuromethods 65. [Google Scholar]
- Jean L, Hackett F, Martin SR, Blackman MJ. Functional characterization of the propeptide of Plasmodium falciparum subtilisin-like protease-1. J Biol Chem. 2003;278:28572–28579. doi: 10.1074/jbc.M303827200. [DOI] [PubMed] [Google Scholar]
- Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien N, Goddard I, Selmi-Ruby S, Fina JL, Cremer H, Herman J. Conditional transgenesis using Dimerizable Cre (DiCre) PLoS ONE. 2007;2:e1355. doi: 10.1371/journal.pone.0001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix C, Giovannini D, Combe A, Bargieri DY, Spath S, Panchal D, et al. FLP/FRT-mediated conditional mutagenesis in pre-erythrocytic stages of Plasmodium berghei. Nat Protoc. 2011;6:1412–1428. doi: 10.1038/nprot.2011.363. [DOI] [PubMed] [Google Scholar]
- Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, et al. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419:537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Choi J, Clardy J. Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:736–744. doi: 10.1107/s0907444998014747. [DOI] [PubMed] [Google Scholar]
- Lin JW, Annoura T, Sajid M, Chevalley-Maurel S, Ramesar J, Klop O, et al. A novel ‘gene insertion/marker out’ (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS ONE. 2011;6:e29289. doi: 10.1371/journal.pone.0029289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoubrie JE, Miller SK, Sargeant T, Good RT, Hodder AN, Speed T, et al. Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design. Infect Immun. 2007;75:5565–5574. doi: 10.1128/IAI.00405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M, Brecht S, Bujard H, Soldati D. Modulation of myosin A expression by a newly established tetracycline repressor-based inducible system in Toxoplasma gondii. Nucleic Acids Res. 2001;29:E115. doi: 10.1093/nar/29.22.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M, Schluter D, Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- Meissner M, Krejany E, Gilson PR, de Koning-Ward TF, Soldati D, Crabb BS. Tetracycline analogue-regulated transgene expression in Plasmodium falciparum blood stages using Toxoplasma gondii transactivators. Proc Natl Acad Sci USA. 2005;102:2980–2985. doi: 10.1073/pnas.0500112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SK, Good RT, Drew DR, Delorenzi M, Sanders PR, Hodder AN, et al. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J Biol Chem. 2002;277:47524–47532. doi: 10.1074/jbc.M206974200. [DOI] [PubMed] [Google Scholar]
- Monaghan P, Bell A. A Plasmodium falciparum FK506-binding protein (FKBP) with peptidyl-prolyl cis-trans isomerase and chaperone activities. Mol Biochem Parasitol. 2005;139:185–195. doi: 10.1016/j.molbiopara.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Moon RW, Hall J, Rangkuti F, Shwen YH, Almond N, Mitchell GH, et al. Adaptation of the genetically tractable malaria pathogen Plasmodium knowlesi to continuous culture in human erythrocytes. Proc Natl Acad Sci USA. 2013;110:531–536. doi: 10.1073/pnas.1216457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan V, Oksman A, Iwamoto M, Wandless TJ, Goldberg DE. Asparagine repeat function in a Plasmodium falciparum protein assessed via a regulatable fluorescent affinity tag. Proc Natl Acad Sci USA. 2011;108:4411–4416. doi: 10.1073/pnas.1018449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MT, Phuong T, Healer J, Richard D, Cowman AF. Gene deletion from Plasmodium falciparum using FLP and Cre recombinases: implications for applied site-specific recombination. Int J Parasitol. 2011;41:117–123. doi: 10.1016/j.ijpara.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Pain A, Bohme U, Berry AE, Mungall K, Finn RD, Jackson AP, et al. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature. 2008;455:799–803. doi: 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino P, Sebastian S, Kim EA, Bush E, Brochet M, Volkmann K, et al. A tetracycline-repressible transactivator system to study essential genes in malaria parasites. Cell Host Microbe. 2012;12:824–834. doi: 10.1016/j.chom.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R, Clackson T. Dimerizer-regulated gene expression. Curr Opin Biotechnol. 2002;13:459–467. doi: 10.1016/s0958-1669(02)00373-7. [DOI] [PubMed] [Google Scholar]
- Ruecker A, Shea M, Hackett F, Suarez C, Hirst EM, Milutinovic K, et al. Proteolytic activation of the essential parasitophorous vacuole cysteine protease SERA6 accompanies malaria parasite egress from its host erythrocyte. J Biol Chem. 2012;287:37949–37963. doi: 10.1074/jbc.M112.400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo I, Oksman A, Vaupel B, Goldberg DE. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc Natl Acad Sci USA. 2009;106:1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Trepanier DJ, Gallant H, Legatt DF, Yatscoff RW. Rapamycin: distribution, pharmacokinetics and therapeutic range investigations: an update. Clin Biochem. 1998;31:345–351. doi: 10.1016/s0009-9120(98)00048-4. [DOI] [PubMed] [Google Scholar]
- Yatscoff RW, Wang P, Chan K, Hicks D, Zimmerman J. Rapamycin: distribution, pharmacokinetics, and therapeutic range investigations. Ther Drug Monit. 1995;17:666–671. doi: 10.1097/00007691-199512000-00020. [DOI] [PubMed] [Google Scholar]
- Yeoh S, O'Donnell RA, Koussis K, Dluzewski AR, Ansell KH, Osborne SA, et al. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell. 2007;131:1072–1083. doi: 10.1016/j.cell.2007.10.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.