Abstract
The most important indoor allergens for humans are house dust mites (HDM). Fourteen Dermatophagoides farinae allergens (Der f 1–3, 6, 7, 10, 11, 13–18, and 22) are reported although more than 30 allergens have been estimated in D. farinae. Seventeen allergens belonging to 12 different groups were identified by a procedure of proteomics combined with two-dimensional immunoblotting from D. farina extracts. Their sequences were determined by Edman degradation, mass spectrometry analysis, and cDNA cloning. Their allergenicities were assayed by enzyme-linked immunosorbent assay inhibition tests, immunoblots, basophil activation test, and skin prick tests. Eight of them are the first report as D. farinae allergens. The procedure of using a proteomic approach combined with a purely discovery approach using sera of patients with broad IgE reactivity profiles to mite allergens was an effective method to investigate a more complete repertoire of D. farinae allergens. The identification of eight new D. farinae allergens will be helpful for HDM allergy diagnosis and therapy, especially for patients without response for HDM major allergens. In addition, the current work significantly extendedthe repertoire of D. farinae allergens.
The house dust mites (HDM)1 are major sources of indoor allergens for humans, which induce asthma, rhinitis, dermatitis, and other allergic diseases (1). Extensive studies have been conducted to understand the biological, chemical, and structural properties of dust mite allergens. Most of the best characterized allergens are from dust mites Dermatophagoides pteronyssinus and D. farinae (Acari: Pyroglyphidae). Twenty-three groups of dust mite allergens are listed in the (IUIS) nomenclature data set, and 21 of them have been identified from Dermatophagoides spp (http://www.allergen.org/). There is an extreme diversity of dust mite allergens. Western blotting studies with human sera containing high levels of anti-mite IgE showed more than 32 bands with molecular weights ranging from 11 to greater than 100 kDa (2). Two groups of mite allergens (group 1 and 2) have been extensively studied. They are a 25-kDa cysteine protease and a 14-kDa epididymal protein, respectively. More than 80% of humans with house dust mite allergy mount an IgE response to the group 1 and more than 90% to the group 2 (3–6).
The group 1 and 2 molecules are major allergens in HDMs but about 20% of patients do not have IgE antibody to the two group allergens (3). It has been found that there are also many other HDM allergens containing high IgE binding activity although these are present in low and variable concentrations in mite extracts (minor allergens), usually at less than 1% of the group 1 and 2 allergens (3). Allergens present in low amount in mite extracts, which can induce high titers of IgE, suggest that they are potent at low concentration. Another possibility is that the amount of allergen required to induce allergic responses in the airways is more than that required to induce IgE. It has been estimated that there are at least 30 allergens in the extracts of D. farinae by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) combined with autoradiography analysis (7). Two-dimensional (2-D) immunoblotting has been applied to study mapping of D. farinae mite allergens (7). Seven allergens including Der f 1, Der f 2, Der f 3, Der f 4, Der f 5, and 2 high molecular mass allergens, which share significant homologies with allergen Mag 3 from D. farinae and with a chitinase from prawn Penaeus japonicus, have been identified from the 2-D immunoblotting analysis (7). Up to now, 14 allergens from D. farinae have been named. Most of them are in the molecular weight range of 14 to 60 kDa. Given the extreme diversity of mite allergens, many investigations with novel allergen identification are still in progress or are yet to be undertaken. It is well known that many mite allergens are not identified on the basis of two possible reasons: (1) it is difficult to purify and characterize minor allergens because they present in low concentration in mite extracts; (2) some minor allergens are neglected because of their minor amount or abilities to only induced allergy to a minor population. It is necessary to develop efficient procedure with high accuracy and resolution to purify and characterize allergens from mite extracts. In this work, 17 allergens or their isoforms have been identified from the mite extracts of D. farinae by a procedure of proteomics combined with two-dimensional immunoblotting. Eight of them are the first to be reported as mite allergens.
MATERIALS AND METHODS
Patient Selection and Skin Prick Test (SPT)
Sera were collected from 41 asthmatic patients who had a positive skin reaction to crude D. farinae extract and the positive skin tests were further confirmed by measuring D. farinae-specific IgE antibodies with the CAP System (Pharmacia & Upjohn Diagnostics AB, Uppsala, Sweden). Of these patients, 15 (42%) are children from 10 to 18 years of age (mean 14.3 years) and 21 (58%) are adults from 20 to 55 years of age (mean 41.6 years). In addition, sera from 16 healthy individuals with a negative SPT response to crude D. farinae extract were collected as negative controls. SPTs with D. farinae extract or purified allergens were performed by using the single-prick technique according to the method of Dreborg (8). Negative (sodium chloride) and positive (histamine, 5 mg/ml) controls were used for the comparison. Skin response was observed after 15 min and defined as positive when the presence of a wheal was 3 mm larger than the negative control.
Approval to conduct these studies was obtained from the ethics committee of the Institutional Review Board of the Kunming Institute of Zoology, Chinese Academy of Sciences. All participants were provided written informed consent for the use of blood samples and skin test before study entry (supplemental Fig. S3).
D. farinae Culture and D. farinae Extract Preparation
According to Sasa's method (9), dust mites were reared at 25°C with a relative humidity of 80%. The culture medium consisted of yeast extract and mouse diet. Mites were isolated from the medium by using a modified heat-escape method. About 8.0 grams of dust mites were homogenized in 0.05 m Tris-HCl, pH 8.0 and centrifuged at 10,000 rpm for 30 min under refrigeration. The supernatant was termed dust mite extract (DME) and lyophilized.
Allergens Purification from DME
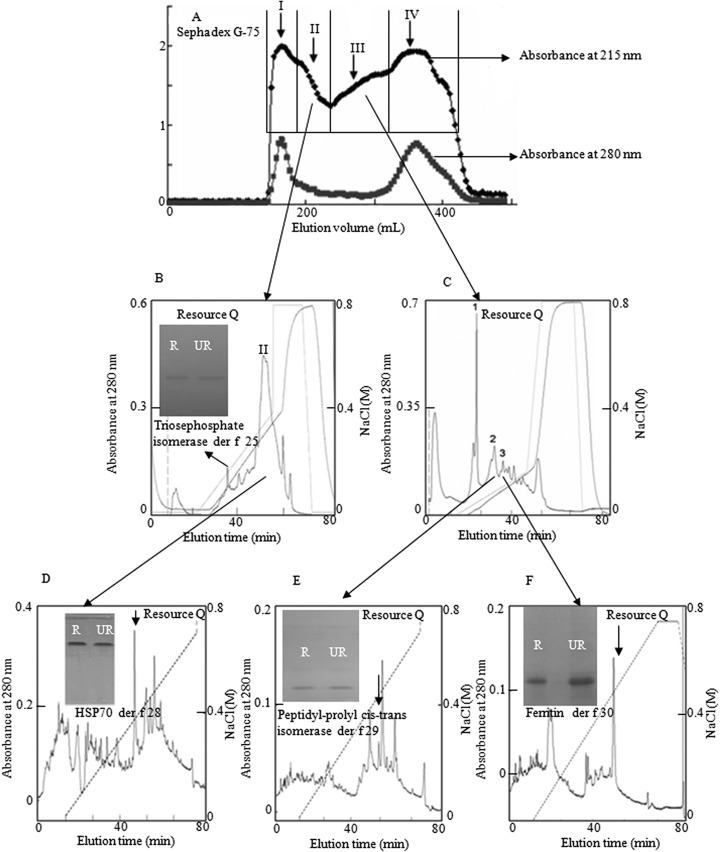
The lyophilized DME sample was dissolved in 100 ml of 0.05 m Tris-HCl, pH 8.0. Aliquot of 10 ml was loaded on a Sephadex G-75 (Superfine, Amersham Biosciences; 26×100 cm) gel filtration column pre-equilibrated with 0.05 m Tris-HCl pH 8.0. The elution was performed with the same buffer at a flow rate of 0.3 ml/min, with fractions collected of 3.0 ml. The eluted fractions were monitored at 280 nm and assayed for the allergenicity. Fractions with allergenicity were pooled and further purified by Resource Q anionic exchange column (1 ml volume, GE Healthcare) on a AKTA explorer fast protein liquid chromatography (FPLC) system (GE Healthcare) as illustrated in Fig. 1.
Fig. 1.
Purification of allergens from DME. A, 380 mg of DME was divided into four fractions by Sephadex G-75 (Superfine, Amersham Biosciences; 26 × 100 cm), the absorbance of eluted fractions were monitored at 280 and 215 nm. B, C, Pooled fraction II and III was dialyzed against 20 mm Tris-HCl, pH 8.0 and subjected to further purification by Resource Q anionic exchange column equilibrated with 0.02 m Tris-HCl, pH 8.0 on an AKTA FPLC system, and eluted at a flow rate of 1 ml/min with the indicated NaCl gradient in 0.02 m Tris-HCl pH 8.0, respectively. D–F: peak II from Fig. 1B, peak 1, and peak 2 were subjected to further purification by Resource Q anionic exchange column equilibrated with 0.02 m Tris-HCl, pH 8.0 on an AKTA FPLC system, and eluted at a flow rate of 1 ml/min with the indicated NaCl gradient in 0.02 m Tris-HCl pH 8.0, respectively. In every purification step, the absorbance of elution peaks was monitored at 280 nm. Inserts: Purified proteins were subjected to SDS-PAGE analysis on a 15% gel. R, reduced; UR, unreduced.
2-D Electrophoresis (2-DE) and Immunoblotting Analysis with Sera from Patients with Dust Mite Allergy
Fractions with allergencity from the Sephadex G-75 gel filtration were pooled and treated further with 2-D Clean-Up Kit (GE Healthcare BioScience) following manufacturer's instructions. Briefly, 50 μl sample was mixed with 300 μl precipitant and co-precipitant, and then incubated for 15 min on ice. The mixture was centrifuged at 12,000 × g for 5 min. After removing the supernatant, the precipitate was washed with 1 ml washing buffer and 5 μl washing additive until the pellets are fully dispersed, and then incubated at - 20°C for 30 min. The mixture was further centrifuged at 12,000 × g for 5 min, and the supernatant was discarded. After air drying the pellet briefly, the precipitate was resuspended in 125 μl rehydration buffer (8 m Urea, 0.5% w/v 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, 0.2% w/v dithiotreitol (DTT), 0.2% v/v IPG stands for immobilized pH gradient buffer pH 3–10).
Isoelectric focusing (IEF) was performed using 7 cm, PI 3–10 nonlinear IPG strips (GE Healthcare BioSciences). The rehydration buffer (125 μl) containing about 50 μg sample was loaded onto the IPG strip, and then the IPG strips were rehydrated overnight at room temperature in Strip Holders (GE Healthcare BioSciences), and focused at 50 μA/IPG strip for a total of 6.0 kVh at 20°C using the Ettan IPGphor III system (GE Healthcare BioSciences). After focusing, the strips were washed in equilibration solution (ES) containing 6 m urea, 30% v/v glycerol, 2% w/v SDS and 0.1% w/v bromphenol blue. The solution used in the first equilibration step was ES plus 1% DTT. ES containing 4.8% w/v iodoacetamide was used for the second equilibration. Each equilibration lasted for 15 min. After two steps equilibration, IPG strips were washed with SDS-PAGE running buffer (25 mm Tris, 192 mm glycine, and 0.1% w/v SDS) and applied onto the top of 12% SDS-PAGE gels using S.E. 260 vertical electrophoresis systems (GE Healthcare BioSciences). Gels were run at 20 mA/gel at 10 °C until bromphenol blue dye reached the bottom of each gel. Gels were either stained with Coomassie or transferred to polyvinylidene difluoride membranes. Coomassie blue stained gels were scanned and analyzed by ImageScannerTM and ImageMasterTM 2D platinum.
Proteins in 2-D gels were transferred to polyvinylidene difluoride membranes (Millipore). Membranes were blocked with 3% bovine serum albumin (BSA) for 2 h at room temperature and incubated overnight at 4 °C with the pooled serum from dust mite allergic patients diluted at a ratio of 1:20 v/v, followed by 1:2500 diluted peroxidase-labeled anti-human IgE monoclonal antibody (KPL, Inc., Gaithersburg, MD). After agitation for 1 h at room temperature and washing three times with PBS, IgE binding proteins were visualized by using the enhanced chemiluminescence ECL reaction (Amersham Biosciences) and exposed to Kodak Imaging film. Normal human serum (detected by Pharmacia Unicap 100 system) from healthy subjects with no history of dust mite allergy was used as a negative control.
Structural Analysis
The N-terminal peptide and partial interior peptide fragments produced by trypsin hydrolysis were undertaken by automated Edman degradation on an Applied Biosystems pulsed liquid-phase sequencer, model 491 (Applied Biosystems, Foster City, CA). The mass spectra were obtained by using nano-electrospray quadripole time of flight (ESI-QUAD-TOF) mass spectrometry (QTOF II, Micromass, Manchester, UK). The instrument was operated in a positive ion mode under the following parameters: capillary voltage, 3.0 Kv; nebuliser gas (N2) flow rate, 80 L/h; desolvation gas (N2) flow rate, 400 L/h. A range of different collision energies were used depending on the m/z value of the selected precursor to produce optimal fragmentation. The peaklist-generating software was Mascot.dll v1.6b23 (Applied Biosystems), and the search engine used was Mascot Version 2.1.0 (Matrix Science, London, UK), the database searched was NCBI nr (2011. 11. 09; 16044589 sequences). The following search parameters were used: trypsin was used as the cutting enzyme for cutting the C-term side of arginine and lysine unless next residue is proline, and the one missed cleavage was permitted. Carbamidomethyl (C) and Oxidation (M) were chosen as fixed and variable modifications, mass tolerance for monoisotopic peptide was set to 500 ppm, fragment mass tolerance was set to 0.8 Da, The proteins identified with the probability greater than 95% were taken as acceptable when their mascot scores were greater than 50.
SDS-PAGE and Protein Quantification
SDS-PAGE was performed with a Bio-Rad Mini Protean II apparatus under reduced and/or unreduced conditions. Protein samples were boiled for 5 min at the presence of 5 × loading buffer before their application to polyacrylamide gels (15%). After the electrophoresis, protein bands were observed using a standard Coomassie Blue R250 stain. The protein concentration was determined by a protein assay kit (Bio-Rad, Hercules, CA, USA) with Bovine serum albumin (BSA) as a standard.
SMART cDNA Synthesis and cDNA Library Construction
Total RNA was extracted using TRIzol Reagent (Invitrogen) from 80 mg dust mites. mRNA was purified by an Oligotex mRNA Mini kit according to the protocols provided (Qiagen). cDNA was synthesized by SMARTTM techniques using a CreatorTM SMARTTM cDNA Library Construction Kit (Clontech, Palo Alto, CA). The first strand of cDNA was synthesized by using 3′SMART CDS III/3′ PCR primer, 5′-ATTCTAG AGGCCGAGGCGGCCGACATG-d(T)30N_1N-3′ (n = A, G, C, or T; N_1 = A, G, or C), SMART IV oligonucleotide 5′-AAGCAGTGGTATCAAC GCAGAGTGGCCATTACGGCCGGG-3′ and SMART MMLV reverse transcriptase. The second strand was amplified using Advantage polymerase by 5′ PCR primer, 5′-AAGCAGTGGTATCAACGCAGAGT-3′ and CDS III/3′ PCR primer, 5′-ATTCTAGAGGCCGAGGCGGCCGACATG-3′. A directional cDNA library was constructed with a plasmid cloning kit (SuperScriptTM plasmid System; Invitrogen) following manufacturer's instructions, producing a library of about 2.5×105 independent colonies.
Screening of cDNA library
The specific primers (Table S1), which were designed according to the peptide sequences determined by Edman degradation or ESI-QUAD-TOF mass spectrometry, and 3′ PCR primer CDS III as mentioned above were used for DNA amplification. The PCRs were performed using Advantage polymerase from Clontech (Palo Alto, CA) as follows: 2 min at 94°C followed by 30 cycles of 10 s at 92°C, 30 s at 50°C, and 40 s at 72°C. The PCR products were recovered by DNA Gel Extraction Kit (Tiangen China) and ligated into pMD19-T vector (TaKaRa Biotechnology Dalian Co., Ltd., China) following manufacturer's instructions. DNA sequencing was performed on an Applied Biosystems DNA sequencer, model ABI PRISM377.
Immunoblotting Analysis with Sera from Allergic Patients to Dust Mites
SDS-PAGE of the purified allergens was performed using a gel concentration of 15%. Protein bands on the gel were electro-transferred to nitrocellulose membranes (Bio-Rad, Hercules, CA, USA) for immunoblotting analysis. Membranes were blocked for 2 h with 5% BSA in room temperature. The membranes were incubated with patients' sera 1:10 (v/v) diluted in 5% BSA-PBST (PBST: PBS containing 0.05% Tween 20) overnight at 4°C under constant agitation on a rotary shaker. After washing three times with PBST, the membranes were incubated with 1:4000 dilution of horseradish peroxidase (HRP)-labeled goat anti-human IgE monoclonal antibody (KPL Inc., Gaithersburg, MD) in 5% BSA-PBST. After washing with PBST for three times, fluorescence was visualized by incubating the membrane with the substrate of HRP.
Enzyme-linked Immunosorbent Assay (ELISA) and ELISA Inhibition
Sera IgE antibodies specific for purified allergens were measured by indirect ELISA as described (10, 11). Briefly, 96-well microtiter plates (Nunc, Roskilde, Denmark) were coated with 2 μg purified allergen in 100 μl carborate-biocarborate buffer (15 mm Na2CO3 and 35 mm NaHCO3, pH 9.6) overnight at 4°C and then blocked for 30 min at 37°C with 200 μl 3% BSA in PBS. After blocking, the plates were incubated for 40 min at 37°C with 50 μl 1:10 (v/v) diluted serum in 0.1% BSA-PBST. After IgE binding, the plates were incubated with peroxidase-labeled goat anti-human IgE (Kpl InC., Gaithersburg, MD,1:2000) for 30 min at 37°C. Each incubating step was followed by 3 washes with PBST. The color was developed by adding 100 μl chromogenic substrate to each well and stopped by the addition of 50 μl 2 m sulfuric acid. The plates were read with a microplate reader at 450 nm (Epoch Etock, BioTek). Two patients (6 and 8) whose sera recognized purified allergens were chosen for ELISA inhibition assay according to previously described method (11). Patients sera (diluted 1:20 in 2% BSA, 0.05% Tween 20 phosphate buffer saline) were pre-incubated with purified allergen or DME (final concentration: 0.0003–30 μg/ml) at 37°C for 1 h and then were added to the microtiter plates previously coated with 100 μl per well of 20 μg/ml DME. The subsequent steps were the same as those for direct ELISA described above. All ELISAs were tested in triplicate and the data were presented as the mean ± S.D.
Basophil Activation Test (BAT)
After in vitro incubation with allergen, the up-regulation of CD63 on the basophil surface has been considered as the marker of basophil activation (12). Venous blood was collected from six dust-mite-allergic patients and six nonallergic subjects. Peripheral blood mononuclear cells (PBMCs) were separated by Ficoll-Paque density gradient and resuspended in stimulation buffer (20 mm HEPES,133 mm NaCl, 7 mm KCl, 3.5 mm MgCl2, 1 mg/ml BSA, 2 ng/ml IL-3, pH 7.4). Then 6 aliquots of PBMCs from a dust-mite-allergic patient or a nonallergic patient were incubated with 1.0 μg/ml testing allergen, goat anti-human IgE antibody (Kpl InC.) and stimulation buffer at 37°C for 40 min, respectively. The goat anti-human IgE antibody was taken as positive control and the stimulation buffer was used to evaluate the basal CD63 level. The reaction was stopped on ice and the basophils were incubated with anti-human CD63-FITC antibody and anti-human CD193 (CCR3)-PE antibody (Biolegend, CA, USA) at 4°C for 15 min. The CCR3 and low granularity were used for gating basophils (13). Basophil activation was quantified as the percentage of CD63-positive basophils. Basophil surface markers were analyzed at 488 nm on a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ) using FACSDivaTM software.
Statistical Analysis
Statistical significance was analyzed by using the SPSS 13.0 version for t test. Data were shown as mean ± S.D. (Standard deviation) and the data for direct ELISA were plotted in a scatter plot. Mann-Whitney U test was employed to analyze the nonparametric data. p value of less than 0.05 was taken as statistically significant.
RESULTS
Allergens Purification from D. farinae Mite Extracts (DME)
As indicated in Fig. 1A, the supernatant of DME was divided into four fractions by Sephadex G-75 gel filtration. These fractions were analyzed by SDS-PAGE and IgE immunoblot. Allergenic proteins were found in fractions I, II, and III. Fractions II and III were subjected to further purification by Resource Q anionic exchange column on an AKTA FPLC system (Figs. 1B-1F) and two-dimensional electrophoresis analysis (Fig. 2). Four allergens were purified. SDS-PAGE analysis indicated that they are homogenized protein (inserts in Figs. 1B-1F). They were determined as triosephosphate isomerase, heat shock protein 70, peptidyl-prolyl cis-trans isomerase, and ferritin, respectively, as described below.
Fig. 2.
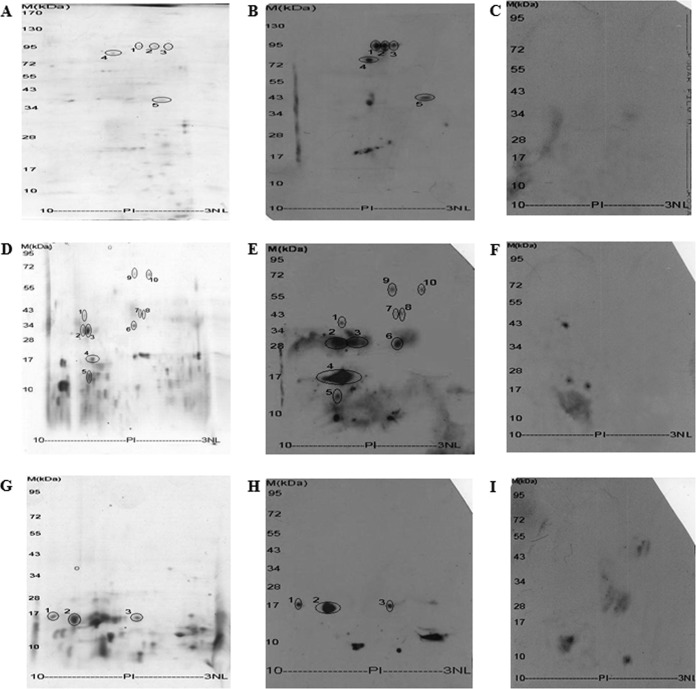
D. farinae allergens identification by coupling 2-DE with 2-D immunoblotting. Protein fraction I, II and III from Sephadex G-75 gel filtration were further separated by 2-DE and stained with Coomassie brilliant blue (A, C, and E) or transferred to PVDF followed by IgE immunoblotting (B, D, and F), respectively.
Analysis of Dust Mite Allergen Components by 2-DE and IgE Immunoblotting
As mentioned above, three fractions (fraction I, II, and III) from Sephadex G-75 gel filtration have been found to contain allergen proteins. To understand allergen mapping of the dust mite, these three fractions were further separated by 2-DE and their IgE-binding abilities was detected by IgE immunoblotting as illustrated in Fig. 2. There were four, ten, and three IgE immune-reactive spots that matched well with protein spots in 2-DE of fraction I, II, and III, respectively. As listed in Table I and Table II, these protein spots were found to belong to 12 different protein families by ESI-QUAD-TOF-based mass spectrometry analysis (supplemental Fig. S1). Among them, four proteins belong to known allergens (Der f 2, 10, 15, and 22) of D. farinae, and the other 8 proteins are new allergens. Three proteins with 2–3 spots may represent their different isoforms. The eight new IgE-binding proteins are arginine kinase (Figs. 2C and 2D, spot No.1), glutathione S-transferase (Figs. 2C and 2D, spots No.2 and No.3), triosephosphate isomerase (TIM) (Figs. 2C and 2D, spot No.6), translation elongation factor 2 (Figs. 2C and 2D, spot No.7), serpin (Figs. 2C and 2D, spot No.8), heat shock protein 70 (HSP 70) (Figs. 2C and 2D, spots No.9 and No.10), peptidyl-prolyl cis-trans isomerase (Figs. 2E and 2F, spot No.1), and ferritin (Figs. 2E and 2F, spot No.3). These eight new allergens were named Der f 20, Der f 8, Der f 25 to Der f 30, respectively, as listed in Tables I and II.
Table I. The identification of specific IgE-binding proteins of D. farina by ESI-QUAD-TOF mass spectrometry. The proteins identified with the probability greater than 95% were taken as acceptable when their mascot scores were greater than 50.
| Spot No. | Accession No. | Protein name | Species | Identical peptide span | Mascot score | Seq coverage (%) |
|---|---|---|---|---|---|---|
| Fraction I (Figs. 2A and 2B, supplemental Figs. S1A–S1D) | ||||||
| 1 | gi|5815436 | Chitinase | D. farinae | 126∼136 171∼181 273∼284 294∼301 397∼406 | 388 | 11 |
| 2 | gi|5815436 | Chitinase | D. farinae | 24∼34 48∼70 126∼136 171∼181 273∼291 397∼406 | 500 | 15 |
| 3 | gi|5815436 | Chitinase | D. farinae | 24∼34 48∼60 126∼136 171∼181273∼291 397∼406 | 442 | 13 |
| 5 | gi|1359436 | Tropomyosin | D. farinae | 28∼43 90∼116 126∼164 168∼175 183∼197 205∼241 254∼259 267∼281 | 1538 | 53 |
| Fraction II (Figs. 2C and 2D, supplemental Fig. S1E-S1N) | ||||||
| 1 | gi|156938897 | Argentines kinase | D. pteronyssinus | 236∼245 296∼303 310∼329 | 173 | 10 |
| 2 | gi|60920899 | Glutathione S-transferase | D. pteronyssinus | 2∼12 20∼33 168∼182 | 257 | 18 |
| 3 | gi|60920899 | Glutathione S-transferase | D. pteronyssinus | 19∼43 56∼122 | 161 | 28 |
| 4 | gi|109629736 | Unknown | D. farinae | 36∼49 61∼74 | 109 | 15 |
| 5 | gi|61679632 | NPC2 family | D. farinae | 1∼48 56∼77 97∼126 | 593 | 77 |
| 6 | gi|262305867 | triosephosphate isomerase | Lynceus sp | 56∼68 83∼96 | 277 | 18 |
| 7 | gi|161661017 | translation elongation factor 2 | Lycosa singoriensis | 293∼302 371∼405 | 248 | 9 |
| 8 | gi|37958175 | Serpin | D. farinae | 159∼174 101∼123 289∼296 | 94 | 8 |
| 9 | gi|325303326 | HSP70 | Amblyomma variegatum | 37∼49 103∼112 127∼137 160∼188 221∼247 | 589 | 17 |
| 10 | gi|209867652 | HSP70 | Philodina roseola | 35∼47 125∼135 170∼186 219∼234 324∼340 | 249 | 11 |
| Fraction III (Figs. 2E and 2F, supplemental Figs. S1O-S1Q) | ||||||
| 1 | gi|37958141 | Peptidyl-prolyl cis-trans isomerase | D. farinae | 6∼19 26∼76 83∼164 | 1007 | 89 |
| 2 | gi|61679632 | NPC2 family | D. farinae | 1∼48 56∼77 101∼126 | 619 | 61 |
| 3 | gi|15072346 | ferritin heavy chain | D. farinae | 130∼163 | 290 | 18 |
Table II. D. farinae allergens identified by Two-dimensional electrophoresis. The allergens underlined represent the new allergens identified from D. farinae.
| Spot No. | Name | Molecule | MW(KDa) | pI | Similar allergens (Resource, Reference) |
|---|---|---|---|---|---|
| Peak I (Figs. 2A and 2B, supplemental Figs. S1A-S1D) | |||||
| 1 | Der f 15 | Chitinase | 95 | 6.18 | Der p 15 (D. pteronyssinus, Reference 14) |
| 2 | Der f 15 | Chitinase | 95 | 5.3 | Der p 15 (D. pteronyssinus, Reference 14) |
| 3 | Der f 15 | Chitinase | 95 | 4.67 | Der p 15 (D. pteronyssinus, Reference 14) |
| 5 | Der f 10 | Tropomyosin | 36 | 5.0 | Der p 10 (D. pteronyssinus, Reference 20) |
| Peak II (Figs. 2C and 2D, supplemental Figs. S1E-S1N) | |||||
| 1 | Der f 20 | Arginine kinase | 40 | 8.0 | Der p 20 (D. pteronyssinus, Reference 36) |
| 2 | Der f 8 | Glutathione S-transferase | 32 | 8.25 | Der p 8 (D. pteronyssinus, Reference 37) |
| 3 | Der f 8 | Glutathione S-transferase | 32 | 7.8 | Der p 8 (D. pteronyssinus, Reference 37) |
| 4 | Der f 22 | Unknown | 17 | 6.7 | Der f 2 (D. farinae) |
| 5 | Der f 2 | NPC2 family | 13 | 7.8 | Der p 2 (D. pteronyssinus, Reference 15) |
| 6 | Der f 25 | Triosephosphate isomerase | 34 | 6.2 | Cra c 8, etc (Crangon crangon, etc, Reference 23, 24) |
| 7 | Der f 26 | Translation elongation factor 2 | 43 | 6.0 | EF-2 (Cynodon dactylon, Reference 25, 26) |
| 8 | Der f 27 | Serpin | 43 | 5.8 | Gal d 2, Tri a 33, etc (Triticum aestivum, Gallus domesticus, etc, Reference 21, 22) |
| 9 | Der f 28 | Hot shock protein 70 | 70 | 6.2 | HSP70 (Hordeum vulgare, Malassezia globosa, Reference 27–29) |
| 10 | Der f 28 | Hot shock protein 70 | 70 | 5.6 | HSP70 (Hordeum vulgare, Malassezia globosa, Reference 27–29) |
| Peak III (Figs. 2E and 2F, supplemental Figs. S1O-S1Q) | |||||
| 1 | Der f 29 | Peptidyl-prolyl cis-trans isomerase | 16 | 9.6 | Asp f 11, Bet v 7,etc (Aspergillus fumigatus, Betula verrucosa,etc, Reference 30–34) |
| 2 | Der f 2 | NPC2 family | 15 | 8.7 | Der p 2 (D. pteronyssinus, Reference 15) |
| 3 | Der f 30 | Ferritin | 16 | 5.8 | Ferritin (Pacifastacus leniusculus, Reference 35) |
Structural Characterization of New Allergens from Dust Mites
Amino acid sequences of N terminus and partial interior peptide fragments of eight new allergens were obtained by Edman degradation and/or ESI-QUAD-TOF-based mass spectrometry analysis (supplemental Fig. S1). Based on the sequences of N-terminuses and interior peptide fragments, degenerate primers were designed to clone the cDNA sequences encoding these allergens as illustrated in supplemental Fig. S2.
Triosephosphate isomerase (Der f 25), heat shock protein 70 (HSP70, Der f 28), peptidyl-prolyl cis-trans isomerase (Der f 29), and ferritin (Der f 30) also purified from DME (Fig. 1) and identified by 2-D immunoblotting (Fig. 2). cDNAs encoding their precursors (GenBank accession numbers KC305500, KC305502, AY283280.1, KC305503) were cloned from the cDNA library of D. farinae (supplemental Fig. S2). The precursor of Der f 25, Der f 28–30 is composed of 247, 659, 164, and 171 aa as illustrated in supplemental Fig. S2C, supplemental Fig. S2F-S2H, respectively.
The complete cDNA sequence encoding the precursor of Der f 8 (KC305499), Der f 20 (EU106619.1), Der f 26 (KC305501), and Der f 27(AY283297.1), which were identified by 2-D immunoblotting, was also cloned from the cDNA library D. farina, respectively. The precursor of Der f 8 (supplemental Fig. S2B), Der f 20 (supplemental Fig. S2A), Der f 26 (supplemental Fig. S2D), and Der f 27 (supplemental Fig. S2E) is composed of 197, 362, 429, and 427 aa, respectively.
Immunoreactivity to IgE
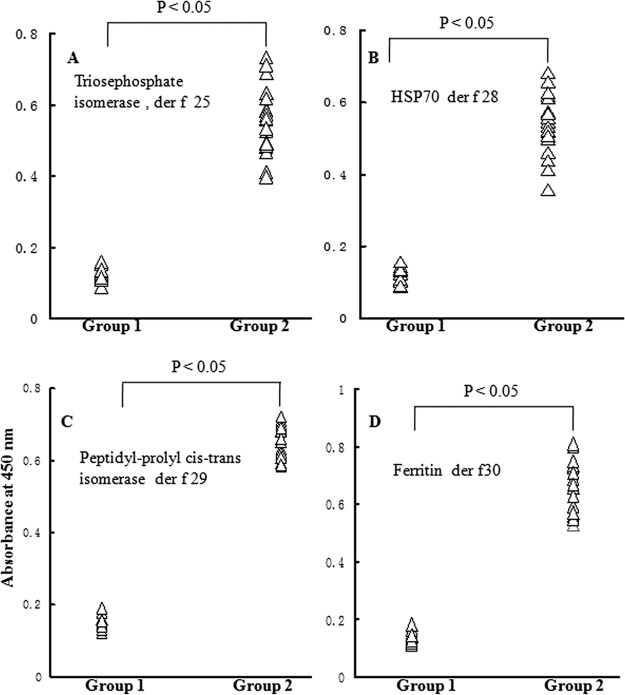
To determine the allergenicity of four purified proteins, immunoblottings were performed using individual sera from 41 dust mite allergic patients. The results demonstrated that sera IgE from 31, 28, 35, and 26 of 41 (75.6%, 68.3%, 85.6% and 63.4%) dust mite allergic patients reacted to Der f 25, Der f 28, Der f 29, and Der f 30, respectively. IgE-binding ability of these four purified allergens in a representative group of nine patients and one control is illustrated in Fig. 3. Most of IgE-binding bands with molecular weight around 34 kDa (Der f 25), 70 kDa (Der f 28), and 15 kDa (Der f 29 and Der f 30) were positive for patients, but completely negative in healthy subjects. The specific immunoreactivity of IgE against four new purified allergens was further confirmed by direct ELISA (Fig. 4). In comparison with the sera from healthy control subjects, the IgE-reactivity of four purified allergens in the sera from positive patients increased by 4.1, 3.7, 4.6, and 5.1-fold, respectively (Fig. 4).
Fig. 3.
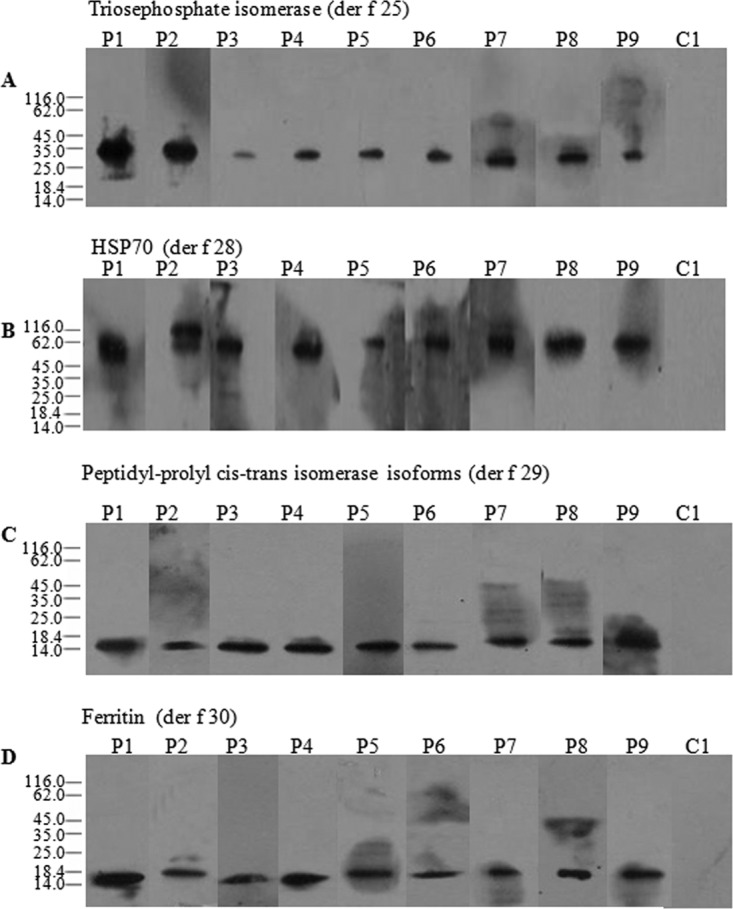
Immunobloting analysis of specific IgE reactivity to allergen der f 25 (A), der f 28 (B), der f 29 (C), and der f 30 (D) in the sera from the patients with dust mite allergy. Lanes 1–9, representatives of sera from allergic subjects. Lane 10 marked as C, serum from healthy individual as negative control.
Fig. 4.
Evaluation of specific IgE reactivity to allergen der f 25 (A), der f 28 (B), der f 29 (C), and der f 30 (D) by direct ELISA. Group 1: the sera from healthy control subjects, group 2: the sera from der f 25, der f28, der f 29 or der f 30-positive patients, respectively.
For ELISA inhibition assay, two patients' sera (6 and 8), which had positive reactions to Der f 25, Der f 28–30, were chosen. Different concentrations of these allergens or DME were incubated with the serum. These allergens inhibited the patients' serum IgE binding to the coated DME in a dose-dependent manner (Fig. 5).
Fig. 5.
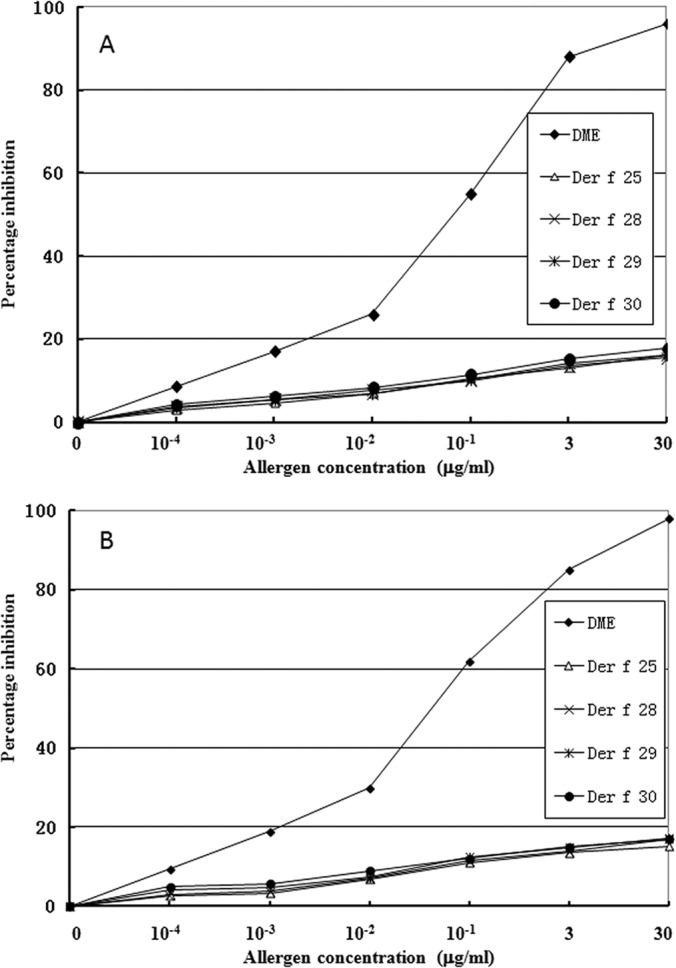
Purified allergens inhibited the patients' serum IgE binding to the coated DME in a dose-dependent manner. A and B represent the serum from dust mite allergy patients 6 and 8, respectively. Allergen concentrations ranged from 0 to 30 μg/ml. DME: dust mite extracts.
Skin Prick Testing
The allergic activity of Der f 25, Der f 28–30 was evaluated by SPT. Six (60%), 7 (70%), 7 (70%), and 6 (60%) of 10 dust mite allergic patients showed positive reaction to Der f 25 and Der f 28–30, respectively (supplemental Table S2).
Basophil Activation Analysis
To analyze basophil activation of four purified allergens, the optimal concentration 1 μg/ml of allergen was applied to this experiment. In comparison with healthy control, Der f 25, Der f 28, Der f 29, and Der f 30 induced approximately up to 5.2, 4.8, 7.1, and 5.9-fold increase, respectively, in CD63 and CCR3 double-positive cells following incubating the allergens with PBMC from patients with dust mite allergy (Fig. 6).
Fig. 6.
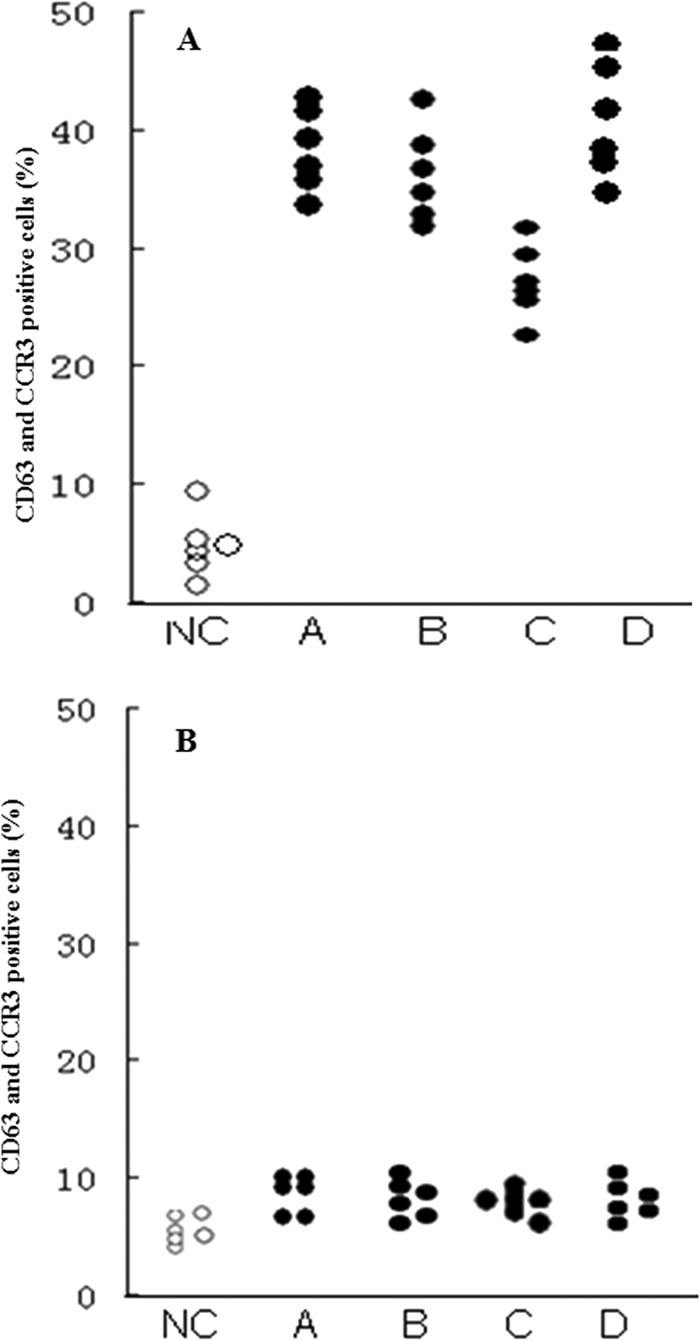
Induction of basophil activation by 4 purified allergens. Allergen was incubated with six dust mite allergenic patients' PBMC (A) or 6 healthy subjects' PBMC (B). NC: using the stimulation buffer to evaluate the basal CD63 level. A: der f 25, B: der f 28, C: der f 29, D: der f 30.
DISCUSSION
An extended repertoire of D farinae mite allergens recognized by specific IgE antibodies from human sera of patients with dust mite allergy was presented here. The repertoire was well defined by the high-resolution 2-D technique combined with IgE immunodetection. Eight new allergens not previously described in the literature were identified (Table I and Table II). Four of them were purified from the mite extracts. Their primary structures and allergenicities were characterized by Edman degradation, cDNA cloning, mass spectrometry, competitive ELISA, BAT, and SPT.
To enrich minor allergens in total proteins and get rid of nonprotein components, the mite extract was fractionated by a sephadex G-75 gel filtration (Fig. 1). All proteins with molecular weight larger than 10 kDa are concentrated at fractions I-III, which were then subjected to 2-D immunobloting analysis (Fig. 2) and further purification. Four IgE-binding proteins including three chitinases and a tropomyosin were identified from fraction I (Figs. 2A and 2B). Chitinases from D. farinae mite have been identified as major allergens (Der f 15) of dogs (14). The current work also has been indicated that D. farinae chitinases (Der f 15) also have ability to bind IgE of human allergic sera (Fig. 2B). Three chitinases reported here have pI value of 6.18, 5.3, and 4.67, respectively. Some studies have shown that most of the important mite allergens can occur as isoforms with varying amino acid sequences or glycosylation. Ten different isoforms of Der p 2 allergen have been shown by 2-DE (15). Other allergens including groups 1, 3, 5, and 7 have also been shown to be polymorphic (16–19). Three chitinase isoforms reported here have the similar molecular weight but different pI, which may be a result of varying amino acid sequences or glycosylation. Tropomyosin has been identified as Der f 10 in D. farinae (20).
As illustrated in Fig. 2B and listed in Table I and Table II, ten IgE-binding proteins including Der f 20, Der f 8 (two isoforms), Der f 22, Der f 2, Der f 27, Der f 25, Der f 26, and two isoforms of Der 28 were identified from fraction II. The cDNA encoding Der f 22 has been cloned from D. farinae (DQ643992) but native or recommbinant Der f 22 allergen has not been reported. Native Der f 22 was identified from the mite extracts in the current work (Fig. 2C and 2D). Der f 27 is a serpin, which shares significant similarity with serpin allergens (Tri a33 and Gal d 2) from Triticum aestivum and Gallus domesticus (21, 22). The current serpin (Der f 27) from DME was demonstrated to bind with serum IgE from patients with allergy to D. farinae (Fig. 2). Triosephosphate isomerases (TIMs) have been identified as allergens in sea shrimp and wheat flour (23, 24). The TIM (Der f 25) was purified and characterized from the mite extracts of D. farinae (Fig. 1B and Fig. 3A). Competitive ELISA, BAT, and SPT indicated that it was an allergen (Fig. 4A, Figs. 5 and 6 and supplemental Table S2). Translation elongation factor 2 from grass pollens has been identified as an allergen (25, 26). The current work demonstrated that there is the presence of translation elongation factor 2 (Der f 26) in D. farinae extracts and it has the ability to bind IgE (Fig. 2D). HSP70 has been identified as an allergen in several species including barley, corn, German cockroach, and Malassezia globosa (27–29). Our current work identified two isoforms of HSP70 with IgE-binding ability from the mite extracts and one of them was purified and characterized as a new D. farinae allergen (Der f 28, Fig. 1D, Fig. 3B, Fig. 4B, and Figs. 5 and 6).
Three IgE-binding proteins including Der f 2, Der f 29, and Der f 30 were identified from fraction III (Figs. 2E and 2F and Table I and 2). The current Der f 2 might be an isoform of the der f 2 found in fraction II because they have different molecular weight and pI value. Der f 29 is a peptidyl-prolyl cis-trans isomerase or cyclophilin (Fig. 1E and supplemental Fig. S1N). Several allergens of cyclophilins from plants or fungi have been determined (30–34). No allergens of animal cyclophilin have been reported. The native cyclophilin (Table I and II) purified from the dust mite extracts has been found to be an allergen (Figs. 2E and 2F, Fig. 3C, Fig. 4C, Figs. 5 and 6, supplemental Fig. S2, and supplemental Table S2) and it was named Der f 29. Ferritin was reported as allergens in Procambarus clarkia (35). No other ferritin allergens were identified. Native ferritin (der f 30, Tables I & II) was purified from D. farinae allergen extracts (Fig. 1G) and demonstrated to be a new mite allergen (Fig. 2E & 2F, Fig. 3D, Fig. 4D, Fig. 5 and 6, and supplemental Table S2).
The development of effective diagnostic and therapeutic approaches depends on the continuity of research of HDM allergens and the identification of allergen diversity. Seventeen allergens belonging to 12 different groups were identified from the D. farinae mite extracts by a procedure of proteomics combined with 2-D immunoblotting. Nine of them are the first report as D. farinae allergens. These allergens will be helpful for HDM allergy diagnosis and therapy, especially for patients without response for HDM major allergens. The current work indicated that many novel D. farinae allergens remain to be investigated. The approach with high resolution is necessary to detect the complete repertoire of D. farinae allergens.
Supplementary Material
Footnotes
* This work was supported by Chinese National Natural Science Foundation (31070701, 31000962, 31025025, 31200590, 30971179, 30972848, 31028006), the Ministry of Science and Technology (2010CB529800, 2009ZX09103-1/091, 011ZX09102-002-10, 2013CB911300).
 This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
1 The abbreviations used are:
- HDM
- house dust mite
- 2-D
- two-dimensional
- SPT
- skin prick test
- DME
- dust mite extract.
REFERENCES
- 1. Tovey E.R., Chapman M.D., Platts-Mills T.A. (1981) Mite faeces are a major source of house dust mite allergens. Nature 289, 592–593 [DOI] [PubMed] [Google Scholar]
- 2. Hong C.S., Lee M.K., Oh S.H. (1991) Identification of major allergens from the house dust mites, Dermatophagoides farinae and Dermatophagoides pteronyssinus, by electroblotting. Yonsei. Med. J. 32, 24–32 [DOI] [PubMed] [Google Scholar]
- 3. Thomas W.R., Smith W.A., Hales B.J. (2004) The allergenic specificities of the house dust mite. Chang. Gung. Med. J. 27, 563–569 [PubMed] [Google Scholar]
- 4. Van Der Veen M.J., Jansen H.M., Aalberse R.C., Van der Zee J.S. (2001) Der p 1 and Der p 2 induce less severe late asthmatic responses than native Dermatophagoides pteronyssinus extract after a similar early asthmatic response. Clin. Exp. Allergy 31, 705–714 [DOI] [PubMed] [Google Scholar]
- 5. Trombone A.P., Tobias K.R., Ferriani V.P., Schuurman J., Aalberse R.C., Smith A.M., Chapman M.D, Arruda L.K. (2002) Use of a chimeric ELISA to investigate immunoglobulin E antibody responses to Der p 1 and Der p 2 in mite-allergic patients with asthma, wheezing and/or rhinitis. Clin. Exp. Allergy 32, 1323–1328 [DOI] [PubMed] [Google Scholar]
- 6. Meyer C.H., Bond J.F., Chen M.S., Kasaian M.T. (1994) Comparison of the levels of the major allergens Der p I and Der p II in standardised extract of the house dust mite, Dermatophagoides pteronyssinus. Clin. Exp. Allergy 24, 1041–1048 [DOI] [PubMed] [Google Scholar]
- 7. Le Mao J., Mayer C.E., Peltre G., Desvaux F.X., David B., Weyer A., Sénéchal H. (1998) Mapping of Dermatophagoides farinae mite allergens by two-dimensional immunoblotting. J. Allergy. Clin. Immunol. 102, 631–636 [DOI] [PubMed] [Google Scholar]
- 8. Dreborg S., Foucard T. (1983) Allergy to apple, carrot and potato in children with birch pollen allergy. Allergy 38, 167–172 [DOI] [PubMed] [Google Scholar]
- 9. Sasa M., Miyamoto J., Shinohara S., Suzuk i H., Katsuhata A. (1970) Studies on mass culture and isolation of Dermatophagoides farinae and some other mites associated with house dust and stored food. Jpn. J. Exp. Med. 40, 367–372 [PubMed] [Google Scholar]
- 10. Ma D., Li Y., Dong J., An S., Wang Y., Liu C., Yang X., Xu X., Lin D., Lai R. (2011) Purification and characterization of two new allergens from the salivary glands of the horsefly, Tabanus yao. Allergy 66, 101–109 [DOI] [PubMed] [Google Scholar]
- 11. An S., Ma D., Wei J.F., Yang X., Yang H.W., Yang H., Xu X., He S., Lai R. (2011) A novel allergen Tab y 1 with inhibitory activity of platelet aggregation from salivary glands of horseflies. Allergy 66, 1420–1427 [DOI] [PubMed] [Google Scholar]
- 12. Schuerwegh A.J, Ebo D.G., Bridts C.H., De Clerck L.S., Stevens W.J. (2001) CD63 expression on basophils of nonallergic controls and patients allergic to wasp. J. Allergy. Clin. Immunol. 108, 150–152 [DOI] [PubMed] [Google Scholar]
- 13. Pruzansky J.J., Grammer L.C., Patterson R., Roberts M. (1983) Dissociation of IgE from receptors on human basophils. I. Enhanced passive sensitization for histamine release. J. Immunol. 131, 1949–1953 [PubMed] [Google Scholar]
- 14. McCall C., Hunter S., Stedman K., Weber E., Hillier A., Bozic C., Rivoire B., Olivry T. (2001) Characterization and cloning of a major high molecular weight house dust mite allergen (Der f 15) for dogs. Vet. Immunol. Immunopathol. 78, 231–247 [DOI] [PubMed] [Google Scholar]
- 15. Chua K.Y., Huang C.H., Shen H.D., Thomas W.R. (1996) Analysis of sequence polymorphism of a major mite allergen, Der p 2. Clin. Exp. Allergy 26, 829–837 [PubMed] [Google Scholar]
- 16. Chua K.Y., Kehal P.K., Thomas W.R. (1993) Sequence polymorphism of cDNA clones encoding the mite allergen Der p 1. Int. Arch. Allergy. Immunol. 101, 364–368 [DOI] [PubMed] [Google Scholar]
- 17. Smith W.A., Thomas W.R. (1996) Sequence polymorphism of the Der p 3 house dust mite allergen. Clin. Exp. Allergy 26, 571–579 [PubMed] [Google Scholar]
- 18. Lin K.L., Hsieh K.H., Thomas W.R., Chiang B.L., Chua K.Y. (1994) Characterization of Der p V allergen, cDNA analysis, and IgEmediated reactivity to the recombinant protein. J. Allergy. Clin. Immunol. 94, 989–996 [DOI] [PubMed] [Google Scholar]
- 19. Shen H. D., Chua K.Y., Lin W.L., Hsieh K.H., Thomas W.R. (1995) Characterization of the house dust mite allergen Der p 7 by monoclonal antibodies. Clin. Exp. Allergy 25, 416–422 [DOI] [PubMed] [Google Scholar]
- 20. Aki T., Kodama T., Fujikawa A., Miura K., Shigeta S., Wada T., Jyo T., Murooka Y., Oka S., Ono K. (1995) Immunochemical characterization of recombinant and native tropomyosins as a new allergen from the house dust mite, Dermatophagoides farinae. J. Allergy. Clin. Immunol. 96, 74–83 [DOI] [PubMed] [Google Scholar]
- 21. Sander I., Flagge A., Merget R., Halder T.M., Meyer H.E., Baur X. (2001) Identification of wheat flour allergens by means of 2-dimensional immunoblotting. J. Allergy. Clin. Immunol. 107, 907–913 [DOI] [PubMed] [Google Scholar]
- 22. Hoffman D.R. (1983) Immunochemical identification of the allergens in egg white. J. Allergy. Clin. Immunol. 71, 481–486 [DOI] [PubMed] [Google Scholar]
- 23. Bauermeister K., Wangorsch A., Garoffo L.P., Reuter A., Conti A., Taylor S.L., Lidholm J., Dewitt A.M., Enrique E., Vieths S., Holzhauser T., Ballmer-Weber B., Reese G. (2011) Generation of a comprehensive panel of crustacean allergens from the North Sea Shrimp Crangon crangon. Mol. Immunol. 48, 1983–1992 [DOI] [PubMed] [Google Scholar]
- 24. Sander I., Rozynek P., Rihs H.P., van Kampen. V, Chew F.T., Lee W.S., Kotschy-Lang N., Merget R., Brüning T., Raulf-Heimsoth M. (2011) Multiple wheat flour allergens and cross-reactive carbohydrate determinants bind IgE in baker's asthma. Allergy 66, 1208–1215 [DOI] [PubMed] [Google Scholar]
- 25. Kao S.H., Su S.N., Huang S.W., Tsai J.J., Chow L.P. (2005) Sub-proteome analysis of novel IgE-binding proteins from Bermuda grass pollen. Proteomics 5, 3805–3813 [DOI] [PubMed] [Google Scholar]
- 26. Dai S., Wang T., Yan X., Chen S. (2007) Proteomics of Pollen Development and Germination. J. Proteome. Res. 6, 4556–4563 [DOI] [PubMed] [Google Scholar]
- 27. Chiung Y.M., Lin B.L., Yeh C.H., Lin C.Y. (2000) Heat shock protein (hsp 70)-related epitopes are common allergenic determinants for barley and corn antigens. Electrophoresis 21, 297–300 [DOI] [PubMed] [Google Scholar]
- 28. Chuang J.G., Su S.N., Chiang B.L., Lee H.J., Chow L.P. (2010) Proteome mining for novel IgE-binding proteins from the German cockroach (Blattella germanica) and allergen profiling of patients. Proteomics 10, 3854–3867 [DOI] [PubMed] [Google Scholar]
- 29. Ishibashi Y., Kato H., Asahi Y., Sugita T., Nishikawa A. (2009) Identification of the major allergen of Malassezia globosa relevant for atopic dermatitis. J. Dermatol. Sci. 55, 185–192 [DOI] [PubMed] [Google Scholar]
- 30. Cadot P., Diaz J.F., Proost P., Van Damme J., Engelborghs Y., Stevens E.A., Ceuppens J.L. (2000) Purification and characterization of an 18-kd allergen of birch (Betula verrucosa) pollen: identification as a cyclophilin. J. Allergy. Clin. Immunol. 105, 286–291 [DOI] [PubMed] [Google Scholar]
- 31. Flückiger S., Fijten H., Whitley P., Blaser K., Crameri R. (2002) Cyclophilins, a new family of cross-reactive allergens. Eur. J. Immunol. 32, 10–17 [DOI] [PubMed] [Google Scholar]
- 32. Glaser A.G., Limacher A., Flückiger S., Scheynius A., Scapozza L., Crameri R. (2006) Analysis of the cross-reactivity and of the 1.5 A crystal structure of the Malassezia sympodialis Mala s 6 allergen, a member of the cyclophilin pan-allergen family. Biochem. J. 396, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marzban G., Herndl A., Kolarich D., Maghuly F., Mansfeld A., Hemmer W., Katinger H., Laimer M. (2008) Identification of four IgE-reactive proteins in raspberry (Rubus ideaeus L.). Mol. Nutr. Food. Res. 52, 1497–1506 [DOI] [PubMed] [Google Scholar]
- 34. Ortona E., Vaccari S., Margutti P., Delunardo F., Rigano R., Profumo E., Buttari B., Rasool O., Teggi A., Siracusano A. (2002) Immunological characterization of Echinococcus granulosus cyclophilin, an allergen reactive with IgE and IgG4 from patients with cystic echinococcosis. Clin. Exp. Immunol. 128, 124–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. González-de-olano D., Pastor-Vargas C., Gandolfo-Cano M., González-Mancebo E., Meléndez-Baltanás A., Morales-Barrios M.P., Pérez-Gordo M., Vivanco F, Bartolomé B. (2011) Allergy to crayfish. J. Investig. Allergol. Clin. Immunol. 21, 318–319 [PubMed] [Google Scholar]
- 36. Hales B.J., Martin A.C., Pearce L.J., Laing I.A., Hayden C.M., Goldblatt J., Le Souëf P, N., Thomas W.R. (2006) IgE and IgG anti-house dust mite specificities in allergic disease. J. Allergy. Clin. Immunol. 118(2), 361–367 [DOI] [PubMed] [Google Scholar]
- 37. O'Neill G.M., Donovan G.R., Baldo B.A. (1994) Cloning and characterization of a major allergen of the house dust mite, Dermatophagoides pteronyssinus, homologous with glutathione S-transferase. Biochim. Biophys. Acta 1219, 521–528 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.