Abstract
The Major histocompatibility complex (MHC) class I peptidome is thought to be generated mostly through proteasomal degradation of cellular proteins, a notion that is based on the alterations in presentation of selected peptides following proteasome inhibition. We evaluated the effects of proteasome inhibitors, epoxomicin and bortezomib, on human cultured cancer cells. Because the inhibitors did not reduce the level of presentation of the cell surface human leukocyte antigen (HLA) molecules, we followed their effects on the rates of synthesis of both HLA peptidome and proteome of the cells, using dynamic stable isotope labeling in tissue culture (dynamic-SILAC). The inhibitors reduced the rates of synthesis of most cellular proteins and HLA peptides, yet the synthesis rates of some of the proteins and HLA peptides was not decreased by the inhibitors and of some even increased. Therefore, we concluded that the inhibitors affected the production of the HLA peptidome in a complex manner, including modulation of the synthesis rates of the source proteins of the HLA peptides, in addition to their effect on their degradation. The collected data may suggest that the current reliance on proteasome inhibition may overestimate the centrality of the proteasome in the generation of the MHC peptidome. It is therefore suggested that the relative contribution of the proteasomal and nonproteasomal pathways to the production of the MHC peptidome should be revaluated in accordance with the inhibitors effects on the synthesis rates of the source proteins of the MHC peptides.
The repertoires and levels of peptides, presented by the major histocompatibility complex (MHC)1 class I molecules at the cells' surface, are modulated by multiple factors. These include the rates of synthesis and degradation of their source proteins, the transport efficacy of the peptides through the transporter associated with antigen processing (TAP) into the endoplasmic reticulum (ER), their subsequent processing and loading onto the MHC molecules within the ER, and the rates of transport of the MHC molecules with their peptide cargo to the cell surface. The off-rates of the presented peptides, the residence time of the MHC complexes at the cell surface, and their retrograde transport back into the cytoplasm, influence, as well, the presented peptidomes (reviewed in (1)). Even though significant portions of the MHC class I peptidomes are thought to be derived from newly synthesized proteins, including misfolded proteins, defective ribosome products (DRiPs), and short lived proteins (SLiPs), most of the MHC peptidome is assumed to originate from long-lived proteins, which completed their functional cellular roles or became defective (retirees), (reviewed in (2)).
The main protease, supplying the MHC peptidome production pipeline, is thought to be the proteasome (3). It is also responsible for generation of the final carboxyl termini of the MHC peptides (4), (reviewed in (5)). The final trimming of the n-termini of the peptides, within the endoplasmic reticulum (ER), is thought to be performed by amino peptidases, such as ERAP1/ERAAP, which fit the peptides into their binding groove on the MHC molecules (6) (reviewed in (7)). Nonproteasomal proteolytic pathways were also suggested as possible contributors to the MHC peptidome, including proteolysis by the ER resident Signal peptide peptidase (8, 9), the cytoplasmic proteases Insulin degrading enzyme (10), Tripeptidyl peptidase (11–13), and a number of proteases within the endolysosome pathway (reviewed recently in (14–17)). In contrast to the mostly cytoplasmic and ER production of the MHC class I peptidome, the class II peptidome is produced in a special compartment, associated with the endolysosome pathway (18–20). This pathway is also thought to participate in the cross presentation of class I peptides, derived from proteins up-taken by professional antigen presenting cells (21), (reviewed in (15–17, 22)).
The centrality of the proteasomes in the generation of the MHC peptidome was deduced mostly from the observed change in presentation levels of small numbers of selected peptides, following proteasome inhibition (3, 23). Even the location of some of the genes encoding the catalytic subunits of the immunoproteasome (LMP2 and LMP7) (24) within the MHC class II genomic locus, was suggested to support the involvement of the proteasome in the generation of the MHC class I peptidome (25). Similar conclusions were deduced from alterations in peptide presentation, following expression of the catalytic subunits of the immunoproteasome (26), (reviewed in (5)). Yet, although most of the reports indicated reductions in presentation of selected peptides by proteasome inhibition (3, 27–29), others have observed only limited, and sometimes even opposite effects (23, 30–32).
The matter is further complicated by the indirect effects of proteasome inhibition used for such studies on the arrest of protein synthesis by the cells (33–35), on the transport rates of the MHC molecules to the cell surface, and on their retrograde transport back to the vesicular system (36) (reviewed in (37)). Proteasome inhibition likely causes shortage of free ubiquitin, reduced supply of free amino acids, and induces an ER unfolded protein response (UPR), which signals the cells to block most (but not all) cellular protein synthesis (reviewed in (38)). Because a significant portion of the MHC peptidome originates from degradation of DRiPs and SLiPs (reviewed in (2)), arrest of new protein synthesis should influence the presentation of their derived MHC peptides. Taken together, these arguments may suggest that merely following the changes in the presentation levels of the MHC molecules, or even of specific MHC peptides, after proteasome inhibition, does not provide the full picture for deducing the relative contribution of the proteasomal pathway to the production of the MHC peptidome (reviewed in (7)).
MHC peptidome analysis can be performed relatively easily by modern capillary chromatography combined with mass spectrometry (reviewed in (39)). The peptides are recovered from immunoaffinity purified MHC molecules after detergent solubilization of the cells (40, 41), from soluble MHC molecules secreted to the cells' growth medium (42, 43) or from patients' plasma (44). The purified peptides pools are resolved by capillary chromatography and the individual peptides are identified and quantified by tandem mass spectrometry (40), (reviewed in (45–47)). In cultured cells, quantitative analysis can also be followed by metabolic incorporation of stable isotope labeled amino acids (SILAC) (48). Furthermore, the rates of de novo synthesis of both MHC peptides and their proteins of origin can be followed using the dynamic-SILAC proteomics approach (49) with its further adaptation to HLA peptidomics (50–52).
This study attempts to define the relative contribution of the proteasomes to the production of HLA class I peptidome by simultaneously following the effects of proteasome inhibitors, epoxomicin and bortezomib (Velcade), on the rates of de novo synthesis of both the HLA class I peptidome and the cellular proteome of the same MCF-7 human breast cancer cultured cells. The proteasome inhibitors did not reduce the levels of HLA presentations, yet affected the rates of production of both the HLA peptidome and cellular proteome, mostly decreasing, but also increasing some of the synthesis rates of the HLA peptides and cellular proteins. Thus, we suggest that the degree of contribution of the proteasomal pathway to the production of the HLA-I peptidome should be re-evaluated in accordance with their effects on the entire HLA class-I peptidome, while taking into consideration the inhibitors' effects on the synthesis (and degradation) rates of the source proteins of each of the studied HLA peptides.
EXPERIMENTAL PROCEDURES
Cell Culture
Cultured cells were maintained in Dulbecco's modified Eagles medium, supplemented with 10% fetal calf serum (FCS), 1% l-glutamine, 1% penicillin-streptomycin, in a humidified 5% CO2 incubator at 37 °C. The endogenous membranal HLA molecules, presented at the surface of the MCF-7 are HLA-A*0201, B*18, B*44, and Cw05 based on tissue typing performed in the Laboratory of Clinical Immunology and Tissue Typing, Rambam Hospital, Haifa.
MATERIALS AND METHODS
Epoxomicin, the specific irreversible proteasome inhibitor, was obtained from Peptide Institute (Minoh-shi, Osaka, Japan). Bortezomib (Velcade) was obtained from Selleckchem (Huston, TX). The antibodies used were: w6/32, a mouse anti-pan-HLA (native HLA A, B, C); bb7.2, a mouse anti native HLA-A2; and HB-149, a mouse anti β-2-microglobulin. The antibodies were all produced from the relevant hybridoma (obtained from the ATCC) grown in culture or from ascites fluid; rabbit monoclonal anti-HLA class I was obtained from Abcam (Cambridge, UK). The anti-ubiquitin was a rabbit polyclonal Ab, obtained from Dako (Glostrup, Denmark). Goat anti-rabbit IgG conjugated to alkaline phosphatase was obtained from Sigma-Aldrich (St. Louis, MO). Goat anti-mouse FITC was obtained from Jackson Immunoresearch (Pennsylvania, USA). Donkey anti-mouse IgG (H+L) Alexa Flour 594 was obtained from Invitrogen (Carlsbad, CA).
Metabolic Labeling of Cells with Stable Isotope Amino Acids
Simultaneously with the addition of 1 μm epoxomicin or 2 μm bortezomib, the growth medium of the cells was changed to Dulbecco's modified Eagles medium, lacking leucine, lysine, and arginine (purchased from Beit Haemek, Israel) and containing 10% dialyzed FCS. This medium was supplemented with heavy leucine (13C6,15N-Leu), heavy lysine (13C6,15N2-Lys), and arginine (13C6,15N4-Arg) at a final concentration of 52 mg/L, 147.6 μg/μl, and 87.3 μg/μl, respectively (Cambridge Isotope Laboratories, Andover, MA).
Trypsin Digestion of Protein Extracts
Proteins were dissolved in 8 m Urea containing 20 mm dithiotreitol and 400 mm ammonium bicarbonate and heated to 60 °C for 30 min. Next, the proteins were modified with 100 mm iodoacetamide at room temperature for 30 min, diluted with three volumes of water, and digested with modified trypsin (Promega, Madison, WI) at a 1:100 enzyme-to-substrate ratio, overnight at 37 °C.
Affinity Purification of the HLA Complexes
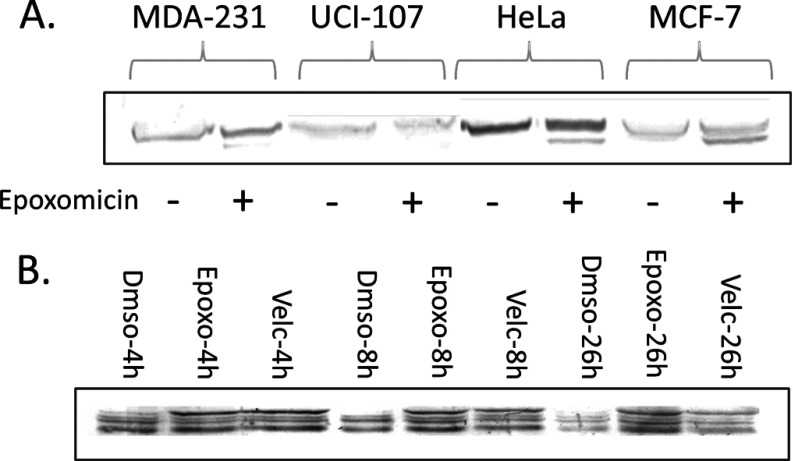
The HLA molecules were purified essentially as in (40) with minor modifications. The cells were released from the plates with trypsin, washed three times with cold PBS by centrifugation and then incubated for 1 h at 4 °C with mild stirring in lysis buffer, containing 0.25% sodium deoxycholate, 1% octyl-β-glucopyranoside, 33 μg/ml iodoacetamide, 1 mm EDTA, 1:200 Protease inhibitors mixture (Sigma), and 1:1000 Phenylmethanesulfonyl fluoride (PMSF). The cell lysate was spun at 18,000× g for 30 min and the supernatant was passed through a column containing the w6/32 antibody covalently bound to protein A Sepharose beads, as in (42) or to AminoLink beads (Pierce, Thermo-Fisher, Rockford, IL) as in (44). The columns were washed twice with ten column volumes of 150 mm NaCl, 20 mm Tris-HCl, once with ten volumes of 400 mm NaCl and with seven volumes of 20 mm Tris-HCl pH 8. The peptides were separated from the heavy subunits of the HLA molecules by elution with 1% trifluoroacetic acid (TFA) (Sigma) followed by their concentration and desalting on disposable Silica C-18 column (Harvard Apparatus, MA) in a modification (by Michal Bassani-Sternberg and Arie Admon) of the HLA peptide purification procedure described in (44), as follows: The disposable C-18 columns were washed with 0.1% trifluoroacetic acid (TFA) and the HLA peptides were eluted with 0.1% TFA and 30% acetonitrile, to separate the HLA peptides from the HLA α-chain and the β-2-microglubulin. Next, the protein subunits were eluted from these C18 disposable columns with 0.1% TFA and 80% acetonitrile. The recovery of the HLA molecules was evaluated by Western blotting (Fig. 3). The solvent was evaporated to dryness and the peptides were dissolved in 0.1% TFA and stored until use at −80 °C.
Fig. 3.
Validation by Western blot of the effects of epoxomicin and bortezomib on the levels of HLA-I in MCF7 cells before and after treatment with 1 μm epoxomicin or 2 μm bortezomib. The Western blots were developed with rabbit monoclonal anti-HLA class I and color development with alkaline phosphatase conjugated to polyclonal goat anti-rabbit IgG.
LC-MS/MS Analysis
The recovered HLA class I peptides (or tryptic peptides) were analyzed by μLC-MS/MS using an OrbitrapXL mass spectrometer (Thermo-Fisher) fitted with a capillary HPLC (Eksigent). The peptides were loaded onto a C18 trap column (0.3 × 5 mm, LC-Packings) connected on-line to a homemade capillary column (75 micron ID) packed with Reprosil C18-Aqua as in (53) and resolved using 7–40% acetonitrile gradients in the presence of 0.1% formic acid during 2 h. Top seven, (1–3 charged peptides for MHC peptides and 2–3 charged tryptic peptides) were selected for fragmentation by collision induced disintegration (CID) from each full mass spectrum according to the following criteria: Exclusion lists of up to 500 for 90 s;. the mass range of the selected peptides was 350–2000 Th, the AGC was set to target value of 107 ions for the full MS, and a target value of 5 × 105 for the full MS and 1 × 104 for the MS/MS. Ion selection threshold was set to 3 × 104 counts and the resolution was set to 7 × 104.
Data Analysis
Peptides were identified and the dynamic-SILAC data were quantified using the MaxQuant and the Proteome Discoverer software tools. Graphical representation of the bioinformatics results was performed with Perseus, version 1.3.0.4.
MaxQuant (54) version 1.3.0.5 was used with the Andromeda search engine (55) and the human section of Nov 2011 of UniProt containing 20257 entries and mass tolerance of 6 ppm for the precursor masses and 0.5 Da for the fragments. Methionine oxidation was accepted as variable modification for both tryptic and HLA peptides. Carbamidomethyl cysteine and n-acetylation were accepted as modification for the proteomics analyses. Minimal peptide length was set to seven amino acids and a maximum of two miscleavages was allowed for tryptic peptides. The false discovery rate (FDR) was set for tryptic peptides to 0.01 for protein identifications, and 0.05 for the MHC peptides. The resulting identified protein tables were filtered to eliminate the identifications derived from the reverse database, as well as common contaminants.
Proteome Discoverer version 1.3 (Thermo-Fisher) was used with UniProt of April 2012 for the Sequest search (containing 20220 entries) and July 2012 for the Mascot search (containing 20306 entries). Mass tolerance was set to 6 ppm for the precursors and 0.5 Da for the fragments. Methionine oxidation and n-acetylation were accepted as variable modification for both tryptic and HLA peptides. Carbamidomethyl cysteine was set as fixed modification for the proteomic analyses. Mass range of 350–5000 Da was used for tryptic peptides and mass range of 750–2500 Da was used for the MHC peptides. For both tryptic and MHC peptides the PSMs were filtered with at least 0.05 FDR (medium confidence), peptide maximum rank was set to 1. Minimal number of identified peptides per proteins was set to 2.
Flow Cytometry
Treated and untreated cells were released from the culture plates by short trypsin treatment and washed three times with phosphate-buffered saline (PBS) by centrifugation. Cells were incubated with the primary antibodies, w6/32 or bb7.2, for 1 h, followed by the anti-mouse FITC-labeled secondary antibody (Sigma), for 30 min, at 4 °C. Next, the cells were re-suspended in PBS for flow cytometry that was performed with a FACS-LSR-II (BD Biosciences).
Confocal Microscopy
Cells were grown on sterile cover slips in 6-well plates. The DNA was stained with 1:1000 Hoechst for 15 min. Cells were washed with PBS three times, fixed with 0.5% paraformaldehyde for 10 min, washed again with PBS three times and incubated with the primary antibody in the perforation buffer, containing 0.1% bovine serum albumin (BSA), 0.05% saponin, and 1% FCS in PBS, for 40 min. at 4 °C. The primary antibodies used were: w6/32 or bb7.2 and anti-ubiquitin. The cells were washed with PBS, 0.1% BSA and incubated with secondary antibodies labeled with Alexa-Fluor or with FITC in the perforation buffer for 20 min at 4 °C, washed and stored in PBS at 4 °C until use.
ELISA
The 96-well microtitre plates (Nunc, Rochester, NY) were coated overnight with 1 μg of the mAb, either the bb7.2 or the HB149 (anti-β2m) in 100 μl PBS. The plates were subsequently incubated with the solution containing the HLA complexes for 3 h, followed by incubation with biotinylated mAb w6/32 (0.2 μg/well), and next by streptavidin-HRP (Sigma, 1:2000, 100 μl/well). The color reaction was developed by adding a peroxidase colorimetric substrate—TMB Blotting Solution (Nalgene, Rochester, NY) and was stopped after color development with 1N H2SO4. The absorbance was measured at 450 nm with a Zenyth 3100 ELISA reader (Anthos).
Determining the Effectiveness of Proteasome Inhibitor
Cell extracts were incubated with increasing concentrations of epoxomicin and with 100 μm of the fluorogenic proteasome substrates Suc-LLVY-AMC or 400 μm Boc-LRR-AMC (Bachem Bioscience Inc.) in 25 mm Tris pH 7, 10 mm MgCl2, 10% glycerol, 1 mm ATP, and 1 mm DTT. The fluorescence was measured during the linear course of the peptidolytic reaction, following 10 min and 1 h of incubation. Excitation was at 380 nm and emission at 460 nm using an Aminco Bowman Series 2 fluorimeter.
Cell Proliferation Assay
The assay was performed with the Cell Proliferation Kit (XTT based) (Beit Haemek, Israel) according to the manufacturer's instructions. Briefly, the MCF-7 cells were cultivated in a flat 96-well plate with 100 μl of growth media. Fifty microliters of the reaction solution, containing activation solution and tetrazolium salt XTT in 1:50 ratio, were added to each well, which were treated with increasing concentration of the inhibitors. XXT is reduced by the metabolically active cells to an orange colored formazan. The absorbance ratio of the samples was measured by an ELISA reader at 450–500 nm, at different time points following the treatment.
RESULTS
Inhibition of the Proteasome did not Reduce HLA Class I Presentation
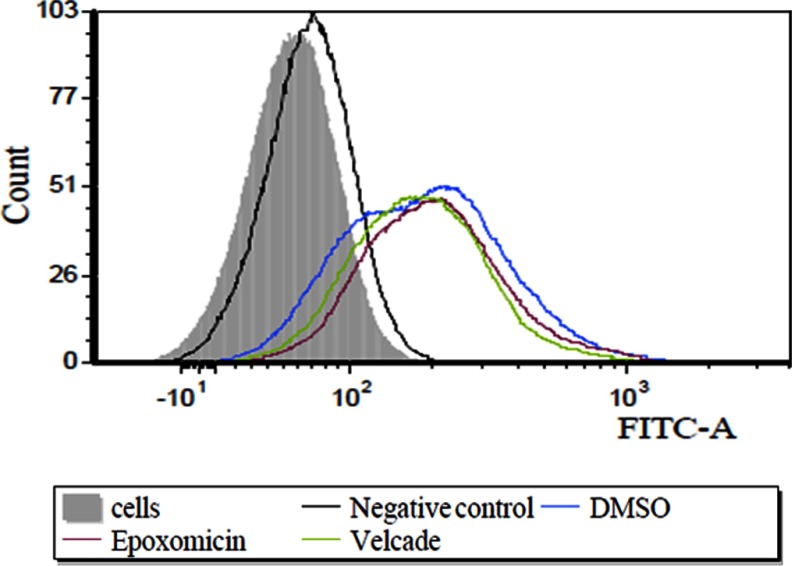
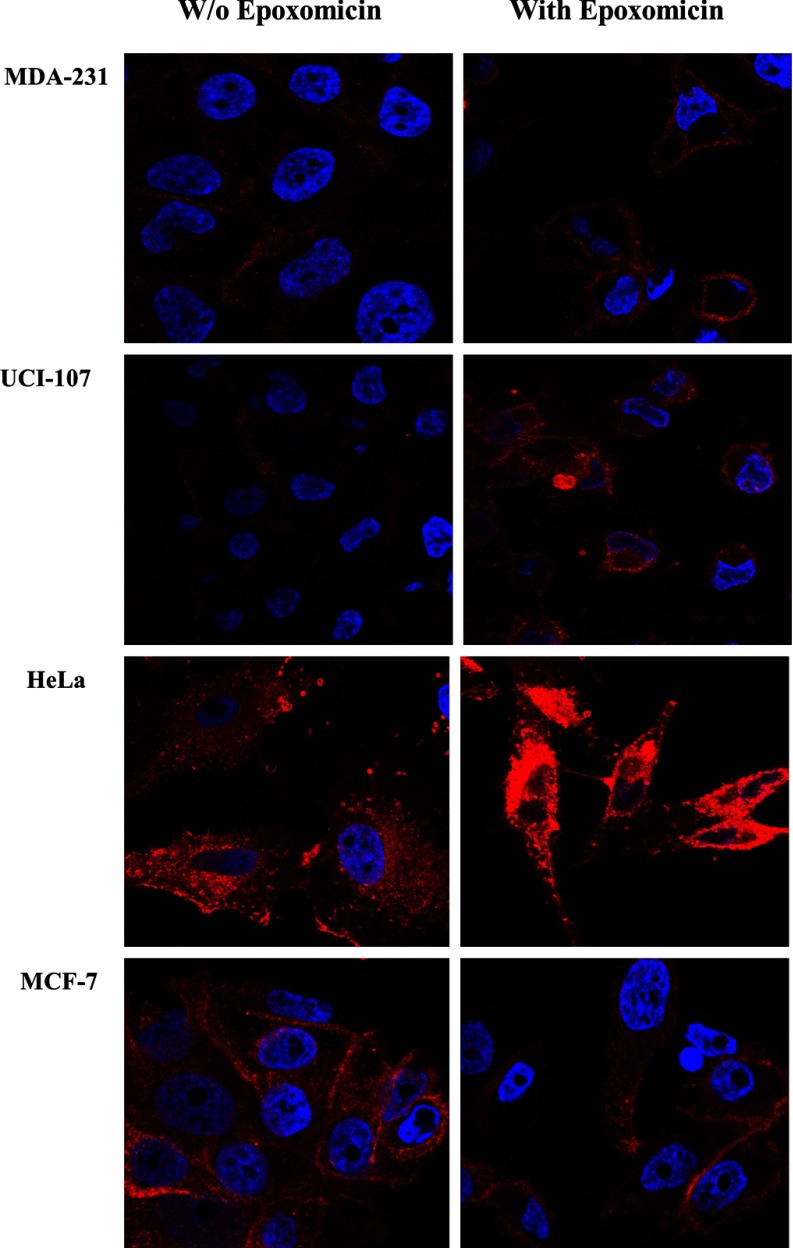
To assess the effect of the proteasome inhibitors on the production pipeline of the HLA-peptidome, we first tested the inhibitors effects on the levels of cell surface presentation of the HLA molecules, both by flow cytometry and by confocal microscopy. The levels of the mHLA class I molecules were not reduced by the proteasome inhibition in all of the tested cell lines, and in some cases the mHLA levels were even slightly increased. An example for the lack of effect of both epoxomicin and bortezomib on MCF-7 cells is shown in Fig. 1. Similar lack of effects of epoxomicin on other cell lines, such as UCI-107, MDA-231 and HeLa, and also the absence of effects by the proteasome inhibitors MG132 and lactacystin are shown in supplemental Fig. S1. The same phenomena were confirmed also by confocal microscopy, which suggested even that the inhibitors may have increased somewhat the intracellular levels of HLA molecules, possibly because of interference with their transport (Fig. 2). The effects of the inhibitors on the total levels of mHLA molecules were also assessed by Western blotting, confirming that the cellular pools of HLA molecules were not reduced and may have even been slightly elevated by the proteasome inhibition (Fig. 3).
Fig. 1.
The effect of proteasome inhibition on the levels of cell surface HLA molecules. Inhibition of the proteasomes by epoxomicin or bortezomib did not reduce the level of presentation of HLA class I molecules at the surfaces of the MCF7 cell line as observed by flow cytometry. Cells were decorated with w6/32 mAb and with secondary anti-mouse IgG FITC labeled antibodies Goat anti mouse FITC obtained or Donkey anti mouse IgG Alexa Flour 594.
Fig. 2.
Proteasome inhibition did not reduce the levels of the HLA class I. The levels of HLA-I of different cell lines were followed after treatment for 24 h with epoxomicin as detected by confocal microscopy. The DNA was stained with Hoechst (in blue) and the HLA class I was stained with the w6/32 mAb (in red).
Evaluating the Efficiency of the Inhibitors and Their Effect on the Cells' Viability
We first confirmed that the inhibitors were indeed effective at the tested concentrations for proteasome inhibition in the MCF-7 cells. Both inhibitors (bortezomib and epoxomicin) were similarly effective under these conditions. The inhibitors were first tested in vitro, focusing mostly on the chymotrypsin-like proteasome activity and using the fluorogenic substrate Suc-LLVY-AMC with cell extracts of MCF-7 cells. The treatment with epoxomicin and bortezomib reduced the chymotryptic-like proteasome activity to about 20% of its uninhibited activity and, as expected, had little effect on the proteasomes' trypsin-like activity with Boc-LRR-AMC as substrate (supplemental Fig. S2). Epoxomicin was also assessed in vivo for its effect on the viability of the cells, using the XTT assay (56). The lowest concentrations of epoxomicin and bortezomib used caused detectable reduction in the viability of the studied cells, already after 24 h of treatment (supplemental Fig. S3). This inhibition also caused significant elevation in the levels of ubiquitination of the cellular proteins, as seen by confocal microscopy with the anti-ubiquitin antibodies, demonstrating significant accumulation of ubiquitinated proteins in the cytoplasm (supplemental Fig. S4A). The same results were demonstrated by Western blotting, as indicated by the shift to the high molecular weight of the cellular ubiquitinated proteins (supplemental Fig. S4B). Thus, strong inhibition of the proteasomes was obtained in the studied cells by the tested concentration of the inhibitors. Yet, it became clear that these inhibitions affected many cellular processes. Most importantly, such high concentrations of the inhibitors used in this study could not have been used for extended periods of time because of their drastic effects on the cells' viability. It is still possible that the detected 10–20% residual proteasome activity was sufficient to supply the HLA peptidome production pipeline. These observations called for the use of a different approach for a comprehensive and unbiased evaluation of the inhibitor effects on the processes of HLA peptides production, while taking into consideration their impact on protein synthesis and degradation.
The Proteasome Inhibition Slowed the Synthesis Rates of Most Cellular Proteins
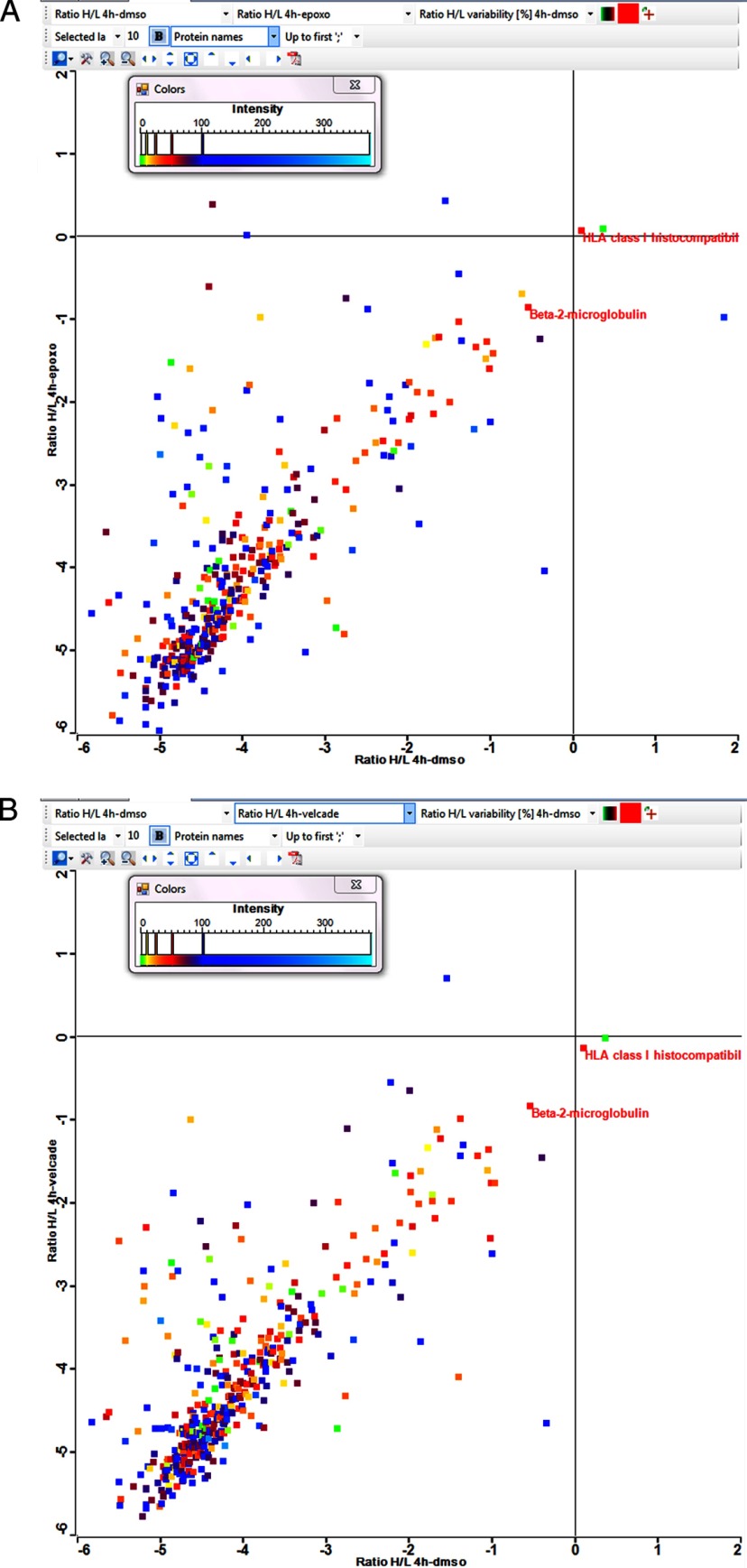
A combined HLA peptidomics and proteomics analysis was performed using the dynamic-SILAC methodology to simultaneously follow the effects of the proteasome inhibitors on the rates of synthesis of the cellular proteins and their derived proteolytic products, the HLA peptides. The effects of the inhibition on the cellular proteome and the HLA peptidome were first followed up to 26 h. However, when it became apparent that during such extended periods of treatment, and at the high concentration of inhibitors used, the synthesis of most proteins was severely slowed down (Fig. 4 and supplemental Table S1A and S1B) and many of the cells even became apoptotic (supplemental Fig. S3), the subsequent experiments were performed during shorter time scales. Samples were taken for proteomics and HLA peptidomics analyses at 2 and 4 h, just before the massive changes in protein synthesis took place (Fig. 4 and supplemental Table S2A and S2B). The proteasome inhibitors were tested in the MCF-7 cells using dynamic-SILAC after transferring the cells to growth media containing heavy lysine and arginine, simultaneously with the addition of the inhibitors. The synthesis rates of many of the cellular proteins, for whom rates could be defined, were significantly reduced or even stopped completely by the inhibitors. Yet, a few proteins were not affected by the inhibitors and the synthesis rates of a number of other proteins even increased. The data are shown for each inhibitor relative to the untreated cells (DMSO added instead of the inhibitor) as the ratio of heavy/light labeled peptides at 4 h after shifting the cells from the “light” to “heavy” amino acids medium (Figs. 5A-epoxomicin, and 5B-bortezomib). Importantly, the synthesis rates of the HLA proteins (both the heavy chain and the β-2-microglobulin) were not slowed down by the proteasome inhibitors (Fig. 6, Table I and supplemental Table S2A and S2B). The data set of about 2500 cellular proteins with their signal intensities and synthesis dynamics are listed in supplemental Tables S1A/B and S2A/B.
Fig. 4.
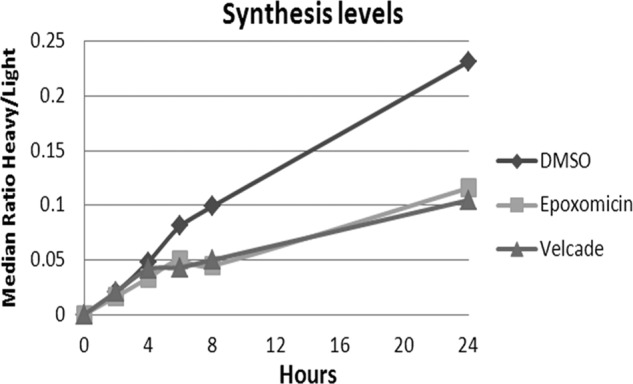
The rates of synthesis of most proteins were reduced by treatments with epoxomicin or bortezomib. Median synthesis rates of proteins detected with both heavy and lights forms in untreated, epoxomicin or bortezomib treated cells.
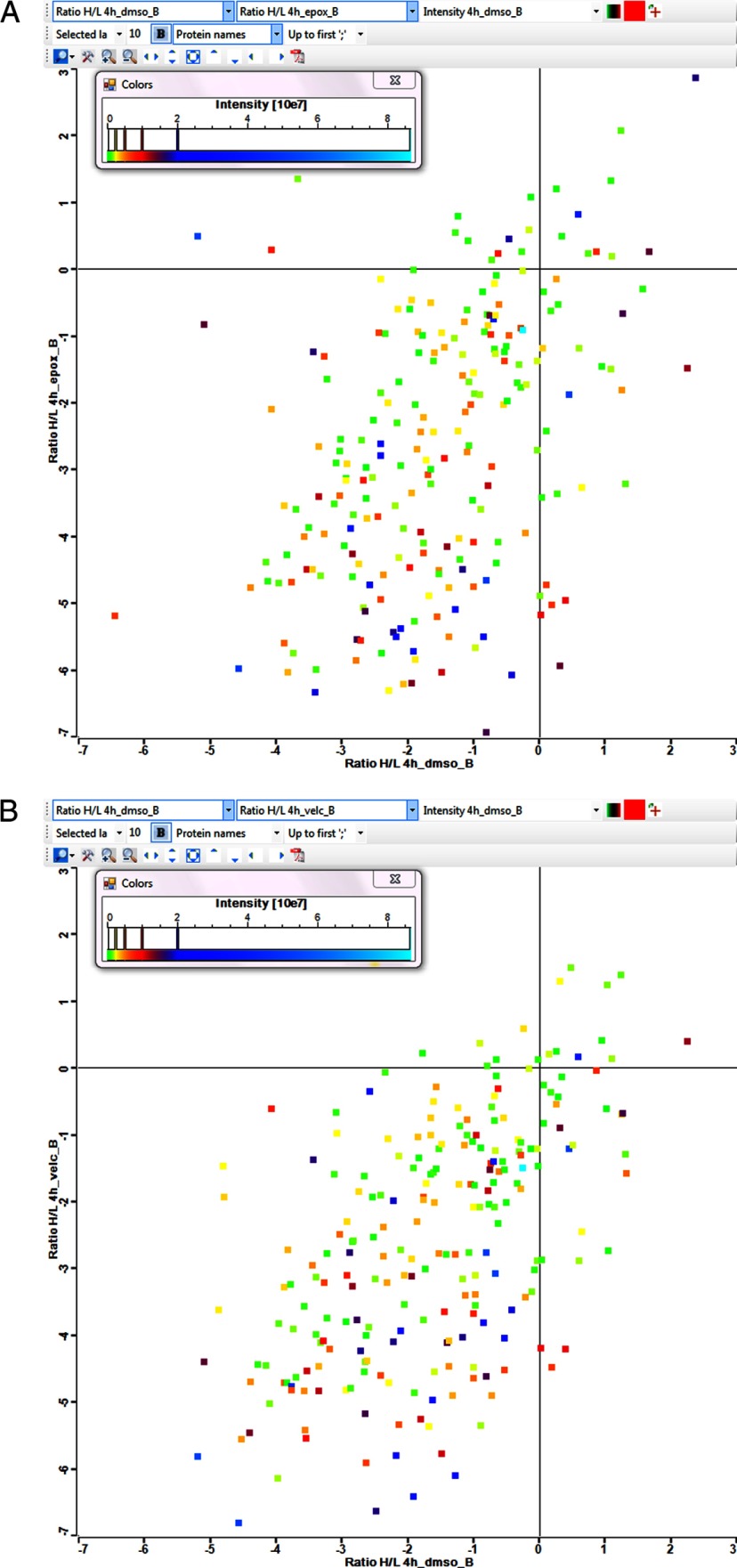
Fig. 5.
Examples for the degrees of labeling of the cellular proteins with heavy amino acids at the 4 h' time point of untreated cells (DMSO) relative to epoxomicin (A) or bortezomib (B) treated cells (log2 scales, results of the 2D-LC-MS/MS analysis).
Fig. 6.
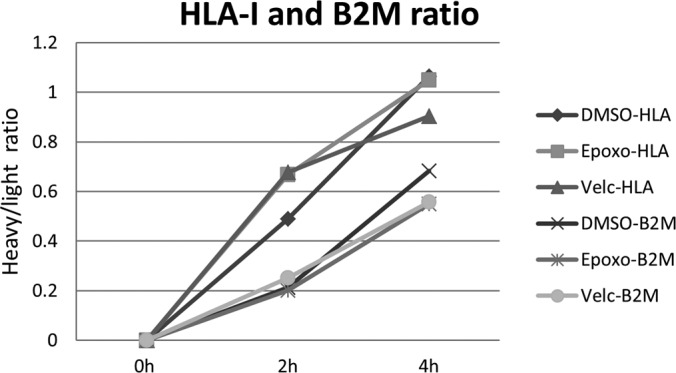
The rate of synthesis of the HLA heavy chain and β-2-microglobulin were not affected significantly by the proteasome inhibitors.
Table I. Examples for the effects of the inhibitors on the rates of synthesis of selected HLA peptides and their source proteins. The synthesis rates of HLA Peptides and tryptic (and thus the proteins) were defined by the rate of shift from their light to heavy forms. The entire data set is listed in supplemental Data Tables S1, S2, S3, and S4.
| Protein IDs | Protein names | Ratio H/L 0h-control | Ratio H/L 2h-dmso | Ratio H/L 4h-dmso | Ratio H/L 2h-epoxo | Ratio H/L 4h-epoxo | Ratio H/L 2h-bortezomib | Ratio H/L 4h-bortezomib | protein/HL A peptide |
|---|---|---|---|---|---|---|---|---|---|
| O75934 | splicing factor SPF27 | 0.22 | 0.38 | 0.23 | 0.49 | 0.25 | 0.50 | Protein | |
| O75934 | splicing factor SPF27 | 0.02 | 0.33 | 0.81 | 0.21 | 0.26 | 0.12 | HLA-p | |
| P01892 | HLA class I | 0.49 | 1.06 | 0.67 | 1.05 | 0.68 | 0.90 | Protein | |
| P01892 | HLA class I | 0.70 | 2.53 | 0.43 | 0.71 | HLA-p | |||
| P04155 | Trefoil factor 1 | 0.17 | 0.48 | 0.18 | 0.41 | 0.18 | 0.39 | Protein | |
| P04155 | Trefoil factor 1 | 0.41 | 0.90 | 1.41 | 0.57 | 0.70 | 0.96 | HLA-p | |
| P11802 | Cyclin-dependent kinase 4 | 0.26 | 0.16 | Protein | |||||
| P11802 | Cyclin-dependent kinase 4 | 0.05 | 0.13 | 0.05 | 0.09 | 0.10 | 0.17 | HLA-p | |
| P35244 | Replication protein A | 0.76 | 0.38 | 0.72 | 0.37 | Protein | |||
| P35244 | Replication protein A | 1.00 | 0.48 | 1.81 | 1.88 | HLA-p | |||
| P60660 | Myosin light polypeptide 6 | 0.16 | 0.32 | 0.21 | 0.43 | 0.22 | 0.43 | Protein | |
| P60660 | Myosin light polypeptide 6 | 0.01 | 0.18 | 0.51 | 0.02 | 0.02 | 0.01 | HLA-p | |
| P61769 | Beta-2-microglobulin | 0.21 | 0.68 | 0.20 | 0.55 | 0.25 | 0.56 | Protein | |
| P61769 | Beta-2-microglobulin | 0.43 | 1.16 | 0.44 | 0.34 | HLA-p | |||
| P62195 | 26S protease regulatory subunit 8 | 0.04 | 0.12 | 0.11 | 0.20 | 0.09 | 0.17 | Protein | |
| P62195 | 26S protease regulatory subunit 8 | 0.16 | 0.50 | HLA-p | |||||
| P84243 | Histone H3 | 0.09 | 0.21 | 0.15 | 0.26 | 0.11 | 0.68 | Protein | |
| P84243 | Histone H3 | 0.63 | 1.37 | 0.28 | 0.25 | 0.49 | 0.40 | HLA-p | |
| Q15008 | 26S proteasome subunit 6 | 0.16 | 0.39 | 0.19 | 0.42 | 0.20 | 0.40 | Protein | |
| Q15008 | 26S proteasome subunit 6 | 0.04 | 0.12 | 0.27 | 0.06 | 0.04 | 0.10 | 0.23 | HLA-p |
Inhibition of the Proteasome had Mixed Effect on the Levels and the Production Rates of the HLA Peptides
Similarly to their effects on protein synthesis, the proteasome inhibitors had variable effects on the levels and the synthesis rates of the HLA peptides. Although most of the HLA peptides were reduced in their amounts by the proteasome inhibition, the levels of many others were increased (supplemental Table S3A and S3B). Such results are expected, because the total level of the HLA molecules did not change because of the inhibition. This observation points to mixed effect of the inhibitors on the production pipeline of the individual HLA peptides, similarly to their complex effect on the synthesis rates of the cellular proteins. For the HLA peptidomic analysis, heavy leucine was added to the growth media, in addition to the heavy arginine and lysine normally used in proteomics SILAC experiments, to increase the number of labeled and quantifiable HLA peptides. Leucine is frequently found in the HLA peptides displayed by the MCF-7 cells' endogenous mHLA A2*01, B*18, B*44, and Cw05 allotypes. Moreover, peptides presented by these HLA alleles are good probes for the effects of these inhibitors on the chymotrypsin-like proteasome activity, which generates peptides with hydrophobic C termini residues, such as those presented by the allotypes of these cells. Proteome-Discoverer identified 1776 HLA peptides and 1224 unique HLA peptides. A few selected examples are listed in Table I and the entire data set is listed in supplemental Table S3A for the MaxQuant analysis and supplemental Table S3B for the Proteome-Discoverer analysis with the same raw LC-MS/MS data. Most peptide sequences fitted the known consensus binding motifs of the endogenous mHLA allotypes. The identified peptides were derived from a diversity of intracellular proteins, without significant preference to particular cellular functions or locations. The most striking observation was that even after treatment with epoxomicin or bortezomib, close to a third of the identified HLA peptides were detected in their heavy forms. About half of the identified peptides were detected in their heavy isotope isoforms, at multiple time points, in the uninhibited cells. Such labeled peptides were derived from proteins that were both synthesized and degraded after the treatment with the inhibitors. Different types of responses to the inhibitions were observed among the HLA peptides. The synthesis rates of most HLA peptides, whose kinetic data could be defined, were inhibited by the inhibitors, whereas others were unaffected and a small number of the HLA peptides even increased in their synthesis rates (Fig. 7). However, it should be stressed that the majority of the detected and identified HLA peptides shifted from the light to the heavy forms too slowly for determination of synthesis kinetics during the experiment timeframe, and not before the massive cell death took place. Peptides of this last group were observed only in their light forms throughout the experiments in both the treated and untreated cells. Selected examples for these groups of peptides are listed in Table I, whereas the entire data set is listed in supplemental Tables S3A and S3B.
Fig. 7.
The effect of epoxomicin and bortezomib on normalized LC-MS rates synthesis of HLA peptides in untreated relative to epoxomicin (A) and bortezomib (B) treated cells (log2 scales).
Relative Synthesis Rates of the HLA Peptides and Their Source Proteins
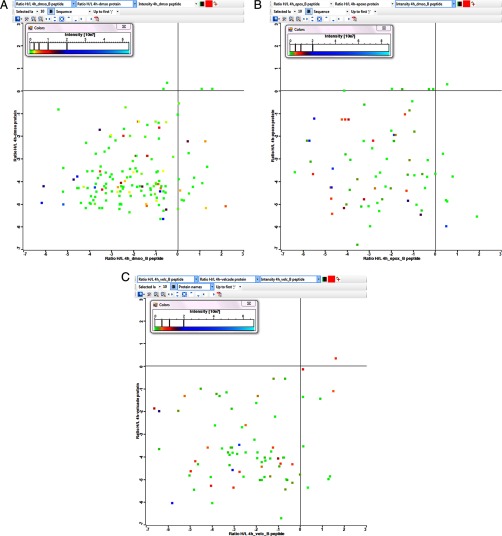
Combining the dynamic-SILAC data of the HLA peptidome and proteome of the same cultured cells enabled taking into consideration the inhibitor effects on the production rates of both the HLA peptides and their source proteins. Only a relatively small number of pairs of HLA peptides and their proteins of origin could be detected in this study with accurately definable synthesis dynamics. The detection of only a small numbers of HLA peptides and their source protein pairs is most likely caused by the normally low correspondence between the HLA peptidome and the proteome of the same cells. A few selected examples are listed in Table I and more are listed in supplemental Table S4. Indeed, complex effects were also observed on the relative synthesis rates of the cellular proteins and their derived peptides (Fig. 8). Yet, many of the detected HLA peptides shifted in their synthesis rates, to the heavy form, faster than their proteins of origin. The faster shifts from the light forms to the heavy forms of a number of HLA peptides, relative to their proteins of origin, were observed also in the epoxomicin and bortezomib treated cells.
Fig. 8.
The rates of synthesis of different proteins relative to their derived HLA peptides, as measured by the rates transformation from the light to the heavy forms at the 4 h' time point in untreated (A), or epoxomicin (B), or bortezomib (C) treated cells (log2 scales).
DISCUSSION
Dynamic SILAC Provides Rich Information About the HLA Peptidome Production Pipeline
If indeed the proteasome is the main protease responsible for degradation of cytosolic proteins, supplying the HLA class I peptidome pipeline (3, 29, 57, 58), its inhibition should have a profound effect on the HLA peptidome. One expected outcome is that the levels of the HLA molecules should be reduced because of the reduced supply of peptides for loading onto the HLA molecules in the ER, as happens in TAP deficient cells (T2 or RMAs); an outcome that was not observed in this study. Alternatively, the remaining residual proteasome activity is sufficient for producing excess amounts of peptides for loading onto the HLA molecules. In such case, no reductions in the level of the HLA are expected, even when the proteasomes are strongly inhibited. Another possibility is that other proteolytic activities produce some or most of the HLA peptides and supply sufficient amount of peptides for the HLA pipeline. Furthermore, it is also possible that such alternative proteolytic pipelines may become more active during proteasome inhibition.
Because the total levels of the peptidome remain relatively constant and individual peptides were influenced differently by the inhibitions, following changes in the levels of a few peptides by using specific T cells or TCR-like antibodies is not informative enough. Because we detected many peptides that responded differently in their levels and rates of synthesis to the inhibition, we believe that combining HLA peptidomics and proteomics with dynamic-SILAC, as described here, is essential for defining the effect of the inhibitors on the production pipelines of the HLA peptidome. The dynamic-SILAC methodology, used in this study, was previously used to follow individual rates of production of large sets of proteins (49, 59) and HLA peptides (50–52), reviewed in (39).
The modified method for the purification of MHC peptides that is described here (based on two step elutions from disposable reversed-phase columns, developed by M. B.-S. and A. A.) enables better yields of MHC peptides from limiting amounts of starting material. Better yield and higher purity of HLA peptide preparations are needed for analysis of the plasma soluble HLA, when quantities are limited by the available volumes of the patient's blood samples (44). After the immunoaffinity enrichment, the acid eluted peptides are collected directly onto a disposable C18 tip column and eluted with 30% acetonitrile. Next, the protein subunits are efficiently recovered with 80% acetonitrile, resulting in both pure peptides preparations in minimal volumes and sufficient amounts of the HLA a-chain and β-2-microglobulin for stained gel detection or for Western blot evaluation of the efficiency of the process (Fig. 3).
Faster Shifting Peptides, Relative to Their Source Proteins, may Indicate DRiPs Origin
A number of pairs of HLA peptides and their protein of origin were observed with measurable rates of shifts from their light to heavy forms. Interestingly, even among the few measurable dynamic-SILAC rates of proteins and their derived HLA peptides, a larger proportion of HLA peptides shifted from the light to the heavy forms faster than their source proteins. This observation supports a DRiPs origin for many of these HLA peptides. The HLA peptides that shift from the light to the heavy forms at the same or at slower rates relative to their proteins of origin are likely derived from “retiree” proteins. Yet, the observation that part of the peptides shifted to their heavy forms faster than their source proteins can only be explained by a significant contribution to the HLA peptidome from DRiPs sources. Furthermore, in the inhibited cells a few HLA peptides shifted from their light to the heavy forms faster than their source proteins. This observation suggests that the proteasome inhibitors increased the formation of DRiPs in these cells, possibly because of the deleterious effects of the inhibitors on the cellular metabolism. This phenomenon further complicates the interpretation of the contribution of the proteasome to the HLA peptidome. It hinders simple distinction between an increase in proteins misfolding and DRiPs production and increase in the activity of nonproteasomal proteolytic pathways, caused by the cellular stress induced by the proteasome inhibition. The existence of alternative production pipelines for MHC peptides, supplementing the peptide pool when the proteasomes are inhibited, was already suggested by (30), (reviewed in (7, 14)). It was also suggested that an alternative proteolytic pathway may be responsible for production of MHC peptides from DRiPs (60). It is possible that the data presented here fits such conclusions. Yet, the same observations can be explained by merely an increase in the misfolding of cellular proteins, which results in an increase in the production of MHC peptides.
Does an Alternative Proteolytic Pipeline Contribute to the Production of the HLA Peptidome?
A few reports have suggested that other, nonproteasomal, routes are significant in the generation of the HLA peptidomes (30, 61, 62) (reviewed in (7, 14)). The alternative pipeline for supplying the required variety of peptides needed for HLA display is likely the vacuolar/endolysosomal pathway (63). This pathway is supplied with cellular proteins for proteolysis mostly by autophagy. The vacuolar/endolysosomal pathway was proposed as a significant, nonproteasomal degradation pathway for a variety of cytosolic and membranal proteins (64). The endolysosomal pathway was studied more for its involvement in producing the HLA class II peptidomes (reviewed in (16, 17)) and for cross presentation of HLA class I peptides ((65), reviewed in (22, 66–68)). Furthermore, although the proteasome was proposed as the main protease responsible for degradation of newly synthesized proteins, including DRiPs and SLiPs, autophagy and lysosomal proteolysis were mostly associated with degradation of long-lived, house-keeping proteins and whole organelles at the end of their functional lifecycles (64). Of interest is a recent study that suggests that a significant fraction of the MHC peptidome is derived from proteins whose transcripts have micro-RNA elements, which may affect the DRiPs production rates of the encoded proteins (69). The quantitative data provided in this study, points to the possible large involvement of such nonproteasomal pathways with the production of the HLA class I peptidome, including contributions from newly synthesized proteins. Yet, it is also possible that such alternative pathways become more significant in supplying the HLA class I peptidome only when the proteasomes are inhibited.
CONCLUSIONS
Further studies are needed to evaluate the precise contribution of the different proteolytic pathways to the production pipelines of the HLA peptidome. Yet, relying on proteasome inhibition for such studies should be approached cautiously. The impact of these inhibitions should not be followed only by following their effects on the cell surface levels of individual MHC peptides presentations, but should also rely on analysis of their rates of synthesis, while correlating these with their indirect effects on the synthesis and degradation rates of their source proteins. The introduction of yet more powerful mass spectrometers and bioinformatics techniques are likely to provide larger coverage and better correlations between the dynamics of the proteomes and the MHC peptidomes of different cells and under different conditions. Elucidation of the role of the different proteases in production of the HLA peptidome is of value for its clinical implication, especially in light of the expanding use of proteasome inhibitors for treatment of different diseases (reviewed in (70)). Thus, the relative contributions of the proteasomal and nonproteasomal routes in shaping the HLA peptidome and their influence on the immune response to viruses, cancer, or self-antigens, should be further explored (71).
Supporting Information
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (72) with the data set identifier PXD000171.
Supplementary Material
Acknowledgments
We thank Ilana Navon for performing the mass spectrometry analyses and Dganit Melamed for help in data analysis.
Footnotes
* This research was funded by the Israel Science Foundation, Grant 916/05 and by the Greta Koppel SCLC Fund. The research described here is part of the Ph.D. thesis of E. M. and of the Master thesis of L. G.-K. The data deposition to the ProteomeXchange Consortium was supported by PRIDE Team, EBI.
 This article contains supplemental Tables S1 to S4 and Figs S1 to S4.
This article contains supplemental Tables S1 to S4 and Figs S1 to S4.
1 The abbreviations used are:
- HLA
- human leukocyte antigen
- MHC
- major histocompatibility complex
- DRiPs
- defective ribosome products
- SLiPs
- short lived proteins
- SILAC
- stable isotope labeling of amino acids in cell culture.
REFERENCES
- 1. Neefjes J., Jongsma M. L., Paul P., Bakke O. (2011) Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 [DOI] [PubMed] [Google Scholar]
- 2. Yewdell J. W. (2011) DRiPs solidify: progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 32, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 4. Mo X. Y., Cascio P., Lemerise K., Goldberg A. L., Rock K. (1999) Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J. Immunol. 163, 5851–5859 [PubMed] [Google Scholar]
- 5. Sijts E. J., Kloetzel P. M. (2011) The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol. Life Sci. 68, 1491–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saric T., Chang S. C., Hattori A., York I. A., Markant S., Rock K. L., Tsujimoto M., Goldberg A. L. (2002) An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 7. van Endert P. (2011) Post-proteasomal and proteasome-independent generation of MHC class I ligands. Cell Mol. Life Sci. 68, 1553–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Henderson R. A., Michel H., Sakaguchi K., Shabanowitz J., Appella E., Hunt D. F., Engelhard V. H. (1992) HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science 255, 1264–1266 [DOI] [PubMed] [Google Scholar]
- 9. Wei M. L., Cresswell P. (1992) HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature 356, 443–446 [DOI] [PubMed] [Google Scholar]
- 10. Parmentier N., Stroobant V., Colau D., de Diesbach P., Morel S., Chapiro J., van Endert P., Van den Eynde B. J. (2010) Production of an antigenic peptide by insulin-degrading enzyme. Nat. Immunol. 11, 449–454 [DOI] [PubMed] [Google Scholar]
- 11. Geier E., Pfeifer G., Wilm M., Lucchiari-Hartz M., Baumeister W., Eichmann K., Niedermann G. (1999) A giant protease with potential to substitute for some functions of the proteasome. Science 283, 978–981 [DOI] [PubMed] [Google Scholar]
- 12. Kloetzel P. M. (2004) Generation of major histocompatibility complex class I antigens: functional interplay between proteasomes and TPPII. Nat. Immunol. 5, 661–669 [DOI] [PubMed] [Google Scholar]
- 13. Endert P. (2008) Role of tripeptidyl peptidase II in MHC class I antigen processing - the end of controversies? Eur. J. Immunol. 38, 609–613 [DOI] [PubMed] [Google Scholar]
- 14. Del Val M., Iborra S., Ramos M., Lázaro S. (2011) Generation of MHC class I ligands in the secretory and vesicular pathways. Cell Mol. Life Sci. 68, 1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chemali M., Radtke K., Desjardins M., English L. (2011) Alternative pathways for MHC class I presentation: a new function for autophagy. Cell Mol. Life Sci. 68, 1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Münz C. (2010) Antigen processing via autophagy–not only for MHC class II presentation anymore? Curr. Opin. Immunol. 22, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Crotzer V. L., Blum J. S. (2009) Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. 182, 3335–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dörfel D., Appel S., Grünebach F., Weck M. M., Müller M. R., Heine A., Brossart P. (2005) Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood 105, 3199–3205 [DOI] [PubMed] [Google Scholar]
- 19. Nimmerjahn F., Milosevic S., Behrends U., Jaffee E. M., Pardoll D. M., Bornkamm G. W., Mautner J. (2003) Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur. J. Immunol. 33, 1250–1259 [DOI] [PubMed] [Google Scholar]
- 20. Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Münz C. (2005) Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307, 593–596 [DOI] [PubMed] [Google Scholar]
- 21. Li Y., Wang L. X., Yang G., Hao F., Urba W. J., Hu H. M. (2008) Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 68, 6889–6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joffre O. P., Segura E., Savina A., Amigorena S. (2012) Cross-presentation by dendritic cells. Nat. Rev. Immunol. 12, 557–569 [DOI] [PubMed] [Google Scholar]
- 23. Schwarz K., de Giuli R., Schmidtke G., Kostka S., van den Broek M., Kim K. B., Crews C. M., Kraft R., Groettrup M. (2000) The selective proteasome inhibitors lactacystin and epoxomicin can be used to either up- or down-regulate antigen presentation at nontoxic doses. J. Immunol. 164, 6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monaco J. J., McDevitt H. O. (1984) H-2-linked low-molecular weight polypeptide antigens assemble into an unusual macromolecular complex. Nature 309, 797–799 [DOI] [PubMed] [Google Scholar]
- 25. Fehling H. J., Swat W., Laplace C., Kühn R., Rajewsky K., Müller U., von Boehmer H. (1994) MHC class I expression in mice lacking the proteasome subunit LMP-7. Science 265, 1234–1237 [DOI] [PubMed] [Google Scholar]
- 26. Cresswell P., Bangia N., Dick T., Diedrich G. (1999) The nature of the MHC class I peptide loading complex. Immunol. Rev. 172, 21–28 [DOI] [PubMed] [Google Scholar]
- 27. Basler M., Lauer C., Beck U., Groettrup M. (2009) The proteasome inhibitor bortezomib enhances the susceptibility to viral infection. J. Immunol. 183, 6145–6150 [DOI] [PubMed] [Google Scholar]
- 28. Finn J. D., Hui D., Downey H. D., Dunn D., Pien G. C., Mingozzi F., Zhou S., High K. A. (2010) Proteasome inhibitors decrease AAV2 capsid derived peptide epitope presentation on MHC class I following transduction. Mol. Ther. 18, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harding C. V., France J., Song R., Farah J. M., Chatterjee S., Iqbal M., Siman R. (1995) Novel dipeptide aldehydes are proteasome inhibitors and block the MHC-I antigen-processing pathway. J. Immunol. 155, 1767–1775 [PubMed] [Google Scholar]
- 30. Vinitsky A., Anton L. C., Snyder H. L., Orlowski M., Bennink J. R., Yewdell J. W. (1997) The generation of MHC class I-associated peptides is only partially inhibited by proteasome inhibitors: involvement of nonproteasomal cytosolic proteases in antigen processing? J. Immunol. 159, 554–564 [PubMed] [Google Scholar]
- 31. Benham A. M., Grommé M., Neefjes J. (1998) Allelic differences in the relationship between proteasome activity and MHC class I peptide loading. J. Immunol. 161, 83–89 [PubMed] [Google Scholar]
- 32. Luckey C. J., Marto J. A., Partridge M., Hall E., White F. M., Lippolis J. D., Shabanowitz J., Hunt D. F., Engelhard V. H. (2001) Differences in the expression of human class I MHC alleles and their associated peptides in the presence of proteasome inhibitors. J. Immunol. 167, 1212–1221 [DOI] [PubMed] [Google Scholar]
- 33. Ding Q., Cecarini V., Keller J. N. (2007) Interplay between protein synthesis and degradation in the CNS: physiological and pathological implications. Trends Neurosci. 30, 31–36 [DOI] [PubMed] [Google Scholar]
- 34. Ding Q., Dimayuga E., Markesbery W. R., Keller J. N. (2006) Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J. 20, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 35. Rothman S. (2010) How is the balance between protein synthesis and degradation achieved? Theor. Biol. Med. Model 7, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boname J. M., Thomas M., Stagg H. R., Xu P., Peng J., Lehner P. J. (2010) Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic 11, 210–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Piper R. C., Lehner P. J. (2011) Endosomal transport via ubiquitination. Trends Cell Biol. 21, 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walter P., Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 39. de Verteuil D., Granados D. P., Thibault P., Perreault C. (2012) Origin and plasticity of MHC I-associated self peptides. Autoimmun Rev. 11, 627–635 [DOI] [PubMed] [Google Scholar]
- 40. Hunt D. F., Henderson R. A., Shabanowitz J., Sakaguchi K., Michel H., Sevilir N., Cox A. L., Appella E., Engelhard V. H. (1992) Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 255, 1261–1263 [DOI] [PubMed] [Google Scholar]
- 41. Falk K., Rötzschke O., Stevanovic S., Jung G., Rammensee H. (1991) Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 [DOI] [PubMed] [Google Scholar]
- 42. Barnea E., Beer I., Patoka R., Ziv T., Kessler O., Tzehoval E., Eisenbach L., Zavazava N., Admon A. (2002) Analysis of endogenous peptides bound by soluble MHC class I molecules: a novel approach for identifying tumor-specific antigens. Eur. J. Immunol. 32, 213–222 [DOI] [PubMed] [Google Scholar]
- 43. Prilliman K., Lindsey M., Zuo Y., Jackson K. W., Zhang Y., Hildebrand W. (1997) Large-scale production of class I bound peptides: assigning a signature to HLA-B*1501. Immunogenetics 45, 379–385 [DOI] [PubMed] [Google Scholar]
- 44. Bassani-Sternberg M., Barnea E., Beer I., Avivi I., Katz T., Admon A. (2010) Soluble plasma HLA peptidome as a potential source for cancer biomarkers. Proc. Natl. Acad. Sci. U.S.A. 107, 18769–18776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoppes R., Ekkebus R., Schumacher T. N., Ovaa H. (2010) Technologies for MHC class I immunoproteomics. J. Proteomics 73, 1945–1953 [DOI] [PubMed] [Google Scholar]
- 46. Admon A., Barnea E., Ziv T. (2003) Tumor antigens and proteomics from the point of view of the major histocompatibility complex peptides. Mol. Cell. Proteomics 2, 388–398 [DOI] [PubMed] [Google Scholar]
- 47. Mester G., Hoffmann V., Stevanović S. (2011) Insights into MHC class I antigen processing gained from large-scale analysis of class I ligands. Cell Mol. Life Sci. 68, 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 49. Pratt J. M., Petty J., Riba-Garcia I., Robertson D. H., Gaskell S. J., Oliver S. G., Beynon R. J. (2002) Dynamics of protein turnover, a missing dimension in proteomics. Mol. Cell. Proteomics 1, 579–591 [DOI] [PubMed] [Google Scholar]
- 50. Milner E., Barnea E., Beer I., Admon A. (2006) The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol. Cell. Proteomics 5, 357–365 [DOI] [PubMed] [Google Scholar]
- 51. García-Medel N., Sanz-Bravo A., Barnea E., Admon A., Lopez de Castro J. A. (2012) The origin of proteasome-inhibitor resistant HLA class I peptidomes: a study with HLA-A*68:01. Mol. Cell. Proteomics 11, M111.011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marcilla M., Cragnolini J. J., López de Castro J. A. (2007) Proteasome-independent HLA-B27 ligands arise mainly from small basic proteins. Mol. Cell. Proteomics 6, 923–938 [DOI] [PubMed] [Google Scholar]
- 53. Ishihama Y., Rappsilber J., Andersen J. S., Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A 979, 233–239 [DOI] [PubMed] [Google Scholar]
- 54. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 55. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 56. Hansen M. B., Nielsen S. E., Berg K. (1989) Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J. Immunol. Methods 119, 203–210 [DOI] [PubMed] [Google Scholar]
- 57. Dick L. R., Aldrich C., Jameson S. C., Moomaw C. R., Pramanik B. C., Doyle C. K., DeMartino G. N., Bevan M. J., Forman J. M., Slaughter C. A. (1994) Proteolytic processing of ovalbumin and beta-galactosidase by the proteasome to a yield antigenic peptides. J. Immunol. 152, 3884–3894 [PMC free article] [PubMed] [Google Scholar]
- 58. Niedermann G., Butz S., Ihlenfeldt H. G., Grimm R., Lucchiari M., Hoschützky H., Jung G., Maier B., Eichmann K. (1995) Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity 2, 289–299 [DOI] [PubMed] [Google Scholar]
- 59. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 60. Dolan B. P., Li L., Veltri C. A., Ireland C. M., Bennink J. R., Yewdell J. W. (2011) Distinct Pathways Generate Peptides from Defective Ribosomal Products for CD8+ T Cell Immunosurveillance. J. Immunol. 186, 2065–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Luckey C. J., King G. M., Marto J. A., Venketeswaran S., Maier B. F., Crotzer V. L., Colella T. A., Shabanowitz J., Hunt D. F., Engelhard V. H. (1998) Proteasomes can either generate or destroy MHC class I epitopes: evidence for nonproteasomal epitope generation in the cytosol. J. Immunol. 161, 112–121 [PubMed] [Google Scholar]
- 62. Yewdell J. W., Reits E., Neefjes J. (2003) Making sense of mass destruction: quantitating MHC class I antigen presentation. Nature reviews. Immunology 3, 952–961 [DOI] [PubMed] [Google Scholar]
- 63. English L., Chemali M., Duron J., Rondeau C., Laplante A., Gingras D., Alexander D., Leib D., Norbury C., Lippé R., Desjardins M. (2009) Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10, 480–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Klionsky D. J., Emr S. D. (2000) Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Houde M., Bertholet S., Gagnon E., Brunet S., Goyette G., Laplante A., Princiotta M. F., Thibault P., Sacks D., Desjardins M. (2003) Phagosomes are competent organelles for antigen cross-presentation. Nature 425, 402–406 [DOI] [PubMed] [Google Scholar]
- 66. Compeer E. B., Flinsenberg T. W., van der Grein S. G., Boes M. (2012) Antigen processing and remodeling of the endosomal pathway: requirements for antigen cross-presentation. Front Immunol. 3, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kreer C., Rauen J., Zehner M., Burgdorf S. (2011) Cross-Presentation: How to Get there - or How to Get the ER. Front Immunol. 2, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wagner C. S., Grotzke J. E., Cresswell P. (2012) Intracellular events regulating cross-presentation. Front Immunol. 3, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Granados D. P., Yahyaoui W., Laumont C. M., Daouda T., Muratore-Schroeder T. L., Côté C., Laverdure J. P., Lemieux S., Thibault P., Perreault C. (2012) MHC I-associated peptides preferentially derive from transcripts bearing miRNA response elements. Blood 119, e181–191 [DOI] [PubMed] [Google Scholar]
- 70. Bedford L., Lowe J., Dick L. R., Mayer R. J., Brownell J. E. (2011) Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Admon A., Bassani-Sternberg M. (2011) The Human Immunopeptidome Project, a suggestion for yet another postgenome next big thing. Mol. Cell. Proteomics 10, O111 011833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Vizcaíno J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 38, D736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.