Abstract
Although it has been established that all toxicoferan squamates share a common venomous ancestor, it has remained unclear whether the maxillary and mandibular venom glands are evolving on separate gene expression trajectories or if they remain under shared genetic control. We show that identical transcripts are simultaneously expressed not only in the mandibular and maxillary glands, but also in the enigmatic snake rictal gland. Toxin molecular frameworks recovered in this study were three-finger toxin (3FTx), CRiSP, crotamine (beta-defensin), cobra venom factor, cystatin, epididymal secretory protein, kunitz, l-amino acid oxidase, lectin, renin aspartate protease, veficolin, and vespryn. We also discovered a novel low-molecular weight disulfide bridged peptide class in pythonid snake glands. In the iguanian lizards, the most highly expressed are potentially antimicrobial in nature (crotamine (beta-defensin) and cystatin), with crotamine (beta-defensin) also the most diverse. However, a number of proteins characterized from anguimorph lizards and caenophidian snakes with hemotoxic or neurotoxic activities were recruited in the common toxicoferan ancestor and remain expressed, albeit in low levels, even in the iguanian lizards. In contrast, the henophidian snakes express 3FTx and lectin toxins as the dominant transcripts. Even in the constricting pythonid and boid snakes, where the glands are predominantly mucous-secreting, low-levels of toxin transcripts can be detected. Venom thus appears to play little role in feeding behavior of most iguanian lizards or the powerful constricting snakes, and the low levels of expression argue against a defensive role. However, clearly the incipient or secondarily atrophied venom systems of these taxa may be a source of novel compounds useful in drug design and discovery.
Venom secretory proteins evolve by a process in which the duplicate of a gene encoding for a key regulatory or bioactive protein is recruited for selective, tissue-specific expression in a venom gland (1, 2). Subsequently, the venom gland specific gene isoform is further duplicated, often explosively. Evidence of accelerated evolution and positive selection is characteristic of this “birth and death” model of venom protein evolution (1, 3–11). For example, the A-superfamily of conotoxin genes isolated from venomous cone snails is among the most rapidly evolving protein-coding gene family identified in metazoans to date (5). In particular, the bioactive residues on the surface of the toxic protein molecules undergo the highest level of accelerated evolution (4, 12–15). This diversification produces neo-functionalized forms with a myriad of new activities and potencies (1, 4, 5, 7, 16).
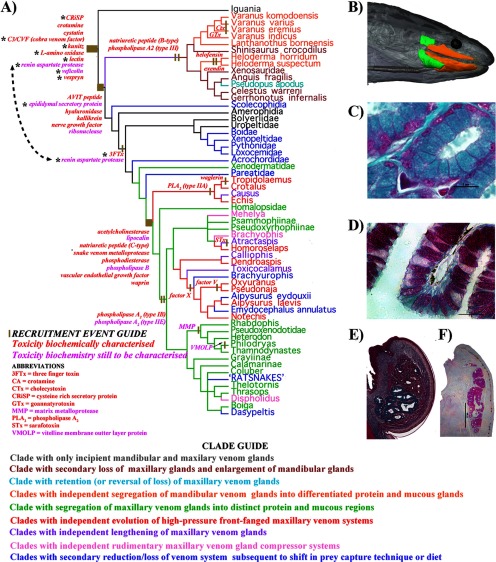
We have previously demonstrated that a core set of toxin-encoding genes was expressed in the oral glands of the common ancestor of all venomous reptiles (Toxicofera clade of squamates). This ancestral set was subsequently augmented by the recruitment of additional toxins and formed the substrate for the evolution of the complex venoms observed in modern venomous lizards and snakes. (Fig. 1A) (2, 7, 17–24). Although Toxicofera, which consists of three lineages (Anguimorpha lizards, Iguania lizards, and Serpentes) is well established as a monophyletic clade within the squamate reptiles (18, 25); phylogenetic studies based on nuclear genes have thus far failed to resolve the relative relationships of the snake/iguanian/anguimorph trichotomy [cf (2, 25, 26). However, other sources of evidence such as SINEs (short interspersed nuclear elements) and morphology favor the clustering of snakes with anguimorph lizards (27, 28). We therefore follow this arrangement here (Fig. 1A). The ancestral toxicoferan condition was the possession of thin, simple sero-mucous glands in both the mandibular and maxillary regions (18). These glands produced active substances that were the substrate for the evolution of toxins. Iguanian lizards apparently split off while this system was only in an incipient stage and little derivation has subsequently occurred within this clade. The apparent lack of specialization of the venom system within the Iguania is likely because of the fact that most species are insectivorous or vegetarian; however, we have shown that toxin genes continue to be expressed by the oral glands of these lizards (18). In the snakes and Anguimorpha lizards, by contrast, the venom system has been extensively diversified (18, 20, 22, 24, 29). In the anguimorph lizards, specialization of the mandibular gland was favored, whereas in snakes it was the maxillary gland that became most specialized; in both lineages highly derived forms exist, including encapsulated glands.
Fig. 1.
Cladogram of evolutionary relationships of toxicoferan reptiles (Fry et al., 2006; Lee, 2009; Piskurek et al., 2006; Vidal and Hedges 2002, 2004, 2005, 2009) showing relative timing of toxin recruitment events and derivations of the venom system. A, * indicates toxin type with relative gene recruitment event timing changed because of results from this study. B, Magnetic resonance imaging of C. ruffus showing the rictal (green) and maxillary/mandibular glands (orange). Histochemically stained in B Masson's Trichrome C. ruffus rictal gland and C, Periodic Acid Schiff's Aspidites melanocephalus mandibular gland; both with 5 μm histology scale bars. Mandibular glands stained with D Masson's Trichrome Anolis equestris showing the mixed sero-mucous gland or E Periodic Acid Schiff's Lepidophyma flavimaculatum showing the purely mucous secreting gland. n = nucleus; S.E. = secretory epithelial cells (S.E.); V = enlarged vesicles.
Snakes are known for their extensive development of oral glands, which may include a great variety of secretory structures mainly in the upper but also in the lower jaw, some of which develop in association with the dental lamina. Intriguing questions arise in relation to the number, volume, and diversification of these glands: why are there so many glands differing in general architecture, gross anatomy, and histochemistry, and how are these differences reflected in their function? In particular, within the complex of the circumlabial oral glands, those at the corner of the mouth (rictal glands) have received little attention except for a few detailed studies and some general investigations (30–37). McDowell (38) carried out a detailed study of the corner of the mouth of a large number of caenophidian snakes and described the rictal glands and muscles of the area together with a reinterpretation of the homology of the involved structures. Underwood (36) investigated the circumlabial oral glands of a series of henophidian snakes including Cylindrophis ruffus and the powerful constricting snake Aspidites melanocephalus and paid special attention to the glands of the rictal region. In C. ruffus, he found well-developed maxillary and mandibular glands as well as superior and inferior rictal glands. He described the lobules of the maxillary and mandibular glands as mixed: serous glands leading to mucous tubules that open into the oral cavity via short ducts along the margin of the lips. The C. ruffus superior and inferior rictal glands consisted of branched serous tubules opening into mucous tubules that led into wide mucous ducts opening directly into the corner of the mouth. Phisalix and Caius (39, 40) reported that the secretions of the rictal glands of Eryx conicus and six species of uropeltid snake were toxic to birds. These interesting observations do not appear to have been taken any further in the more than 80 years since they were reported. More recently, a test of oral secretions from a wide range of Australian snakes produced a curious cross-reactivity between oral secretions of the pythonid A. melanocephalus and tiger snake venom in the Australian Snake Venom Detection Kit (41). This result may be indicative of the fact that toxins are still being secreted from the oral glands of A. melanocephalus in detectable quantities.
The relative expression of genes in mandibular versus maxillary glands has remained uninvestigated. In addition, the molecular evolution of any associated secreted proteins in henophidian snakes has remained entirely neglected and that of the iguanian lizards has been hampered by only a single species having been investigated to-date and with only limited sequencing (18). Thus, the over-arching aim of this study was to examine the diversification of these glands and their secretory proteins in henophidian snakes and iguanian lizards to elucidate several intriguing aspects of reptile venom evolution. We also examined the genome of Anolis carolinensis for genes encoding putative toxin sequences.
In this study, iguanian lizards and henophidian snakes were both shown to express (at detectable levels) toxin types typically associated with the venoms of caenophidian snakes or anguimorph lizards. Our results revealed that the mandibular and maxillary glands are under a single system of genetic control, with identical transcripts being simultaneously expressed in both glands. Additionally, we showed that the rictal glands of snakes not only also secrete toxins and thus represent an unappreciated source of venoms but that this gland is under the same gene-expression control as the mandibular and maxillary glands.
MATERIALS AND METHODS
Species Examined (H = histology, M = MRI, T = transcriptome).
Snakes
Boidae
Boa constrictor imperator HMT (captive born stock of unknown geographical origin), Eunectes notaeus HM (captive born stock of unknown geographical origin), Corallus hortulanus HM (captive born stock of unknown geographical origin).
Cylindrophiidae
Cylindrophis ruffus HMT (Malaysia).
Pythonidae
Antaresia childreni HM (collected from Kununurra, Western Australia), Aspidites melanocephalus HMT (collected from Port Headland, Western Australia), Morelia spilota mcdowelli HMT (captive born stock of Brisbane, Queensland founder stock), Python regius HM (captive born stock of unknown geographical origin).
Lizards
Iguania
Agamidae
Amphibolurus longirostris H (collected from Newman, Western Australia), Chlamydosaurus kingii HT (collected from Kununurra, Western Australia), Ctenophorus nuchalis H (collected from Newman, Western Australia), Physignathus lesueurii HT (collected from Sydney, New South Wales), Uromastyx aegyptica HT (captive born stock of unknown geographical origin).
Chameleonidae
Chamaeleo calyptratus HT (captive born stock of unknown geographical origin).
Iguanidae
Anolis equestris HT (collected from Miami, Florida), Iguana iguana HT (captive born stock of unknown geographical origin).
Anguimorpha
Varanidae
Varanus glauerti T (collected from Kununurra, Western Australia), Varanus tristis T (collected from Perth, Western Australia).
Other
Cordylidae
Cordylus tropidosternum H (captive born stock of unknown geographical origin).
Gekkonidae
Gekko gecko H (captive born stock of unknown geographical origin).
Scincidae
Eumeces obsoletus H (captive born stock of unknown geographical origin).
Teiidae
Ameiva ameiva H (captive born stock of unknown geographical origin).
Xantusiidae
Lepidophyma flavimaculatum H (captive born stock of unknown geographical origin).
Histology
Histological sections were prepared from the intact head and the excised venom gland and delivery system. Whole heads were removed and a cut made to the underside to allow fast penetration of the fixative (10% neutral buffered formalin). After a minimum of 2 days excess tissue was removed and specimens were immersed in Kristensen's decalcification solution and placed on a rotor for up to 3 weeks (depending on the size of the head). Before processing the heads were bisected longitudinally for cutting transversely, at 3 microns, in two separate blocks. The processing schedule was: 10% neutral buffered formalin 2 h; absolute ethanol 4 × 1 h; histolene 3 × 1 h; followed by paraffin wax 2 × 90 min. The sections were taken every 100 microns and matching sections were stained with Periodic Acid Schiff's stain and Masson's Trichrome stain. In other specimens, the venom gland, venom duct, and the adjacent bony and muscular tissue were excised and placed in decalcifying solution (Cal-Ex, Fisher) for 72–168 h. Each sample was dehydrated and cleared through a progressive ethanol series and Cyto-Sol (Fisher) before embedding in Paraplast (Fisher). Serial sections were cut at 10–12 microns. All species were sectioned in the frontal plane; when available, the contralateral venom gland and delivery system were sectioned either parasagitally or transversely. Sections were stained using a variant of Van Gieson's stain, which provides clear distinction between connective tissue, muscle, and epithelium, or with hematoxylin and eosin.
Magnetic Resonance Imaging
Magnetic resonance imaging (MRI) was used to examine the three-dimensional shape and internal anatomy of the venom glands. Formalin-ethanol fixed heads were first submersed in Fomblin (Solvay Solexis) and placed under vacuum to prevent air artifacts. Depending on head size, imaging was performed on either 9.4 T (small/medium) or 17.6 T (large) vertical 89-mm-bore systems (Bruker BioSpin, Rheinstetten, Germany) with a Bruker Micro2.5 gradient system of 1 T/m and transmit/receive birdcage radiofrequency coil with diameter of 10–30 mm. Bruker ParaVision 4.0 software was used for image acquisition. Anatomical images were acquired using a 3D gradient echo sequence. The field-of-view and matrix were varied to fit the individual samples, resulting in voxel sizes between (40)3 mm3 and (70)3 mm3. Imaging parameters were: TE = 8 ms, TR = 40 ms, flip angle 200, 4–8 averages, total scan time 3–9 h per sample, depending on size and resolution. Image segmentation of glands was performed manually in Amira 4.1 (Mercury Computer Systems Inc.) and 3D surface renderings generated.
Transcriptomes
Mandibular venom glands of three specimens were pooled and total RNA extracted using the standard TRIzol Plus method (Invitrogen). Extracts were enriched for mRNA using standard RNeasy mRNA mini kit (Qiagen) protocol. mRNA was reverse transcribed, fragmented and ligated to a unique 10-base multiplex identifier (MID) tag prepared using standard protocols and applied to one PicoTitrePlate (PTP) for simultaneous amplification and sequencing on a Roche 454 GS FLX+ Titanium platform (Australian Genome Research Facility). Automated grouping and analysis of sample-specific MID reads informatically separated sequences from the other transcriptomes on the plates, which were then post-processed to remove low quality sequences before de novo assembly into contiguous sequences (contigs) using the MIRA software program. Assembled contigs were processed using CLC Main Work Bench (CLC-Bio) and Blast2GO bioinformatic suite (42–45) to provide Gene Ontology, BLAST, and domain/Interpro annotation. The above analyses assisted in the rationalization of the large numbers of assembled contigs into phylogenetic “groups” for detailed phylogenetic analyses outlined below. Sequence accession numbers are given in Table I and sequences are also available in supplementary File S1, nucleotide sequences.
Table I. Species recovered from (Genbank accession numbers).
| Toxin type | Species recovered from (Genbank accession numbers) |
|---|---|
| 3FTx | Aspidites melanocephalus (JX467122, JX467123), Cylindrophis ruffus (JX467124, JX467170) |
| CRiSP | Chameleo calyptratus (JX467127), Chlamydosaurus kingii (JX467128), Cylindrophis ruffus (JX467129), Iguana iguana (JX467125, JX467126) + Anolis genome (G1KVU3, G1KE85) |
| Crotamine | Uromastyx aegyptica (JX467130), Varanus glauerti (JX467131, JX467132, JX467133, JX467134), Varanus tristis (JX467135) |
| CVF | Anolis equestris (JX467140), Boa constrictor (JX467146, JX467147, JX467148), Chameleo calyptratus (JX467136, JX467137), Chlamydosaurus kingii (JX467138), Cylindrophis ruffus (JX467143, JX467144, JX467145), Iguana iguana (JX467139), Physignathus lesueurii (JX467142), Uromastyx aegyptica (JX467141) |
| Cystatin | Chameleo calyptratus (JX467152), Iguana iguana (JX467149, JX467150, JX467151), Physignathus lesueurii (JX467153) |
| ESP | Cylindrophis ruffus (JX467154) |
| Kunitz | Chameleo calyptratus (JX467155, JX467156), Heloderma suspectum (JX467158), Iguana iguana (JX467157) + Anolis genome (H9G8B3, H9GRC6, H9GQ81) |
| LAO | P. lesueurii fragment too small for database curation: |
| atgaaaagggacrgtctcctgatgaactctatgttggggaccttggacaaggtgacaaaaaactgcacaattttgaaggagaaatatgactcctattccaccaaggaatacttaattaaagaaggacatttgagcagaggagcagta | |
| Lectin | Aspidites melanocephalus (JX467159), Boa constrictor (JX467160), Chameleo calyptratus (JX467164), Cylindrophis ruffus (JX467161, JX467162), Morelia spilota mcdowelli (JX467163) |
| RAP | Boa constrictor (JX467165) + Anolis genome (G1KIV8, G1KH55) |
| Veficolin | Chlamydosaurus kingii (JX467166), Cylindrophis ruffus (JX467168), Morelia spilota mcdowelli (JX467167) |
| Vespryn | Boa constrictor (JX467169) + Anolis genome (G1KXL5) |
| Novel peptide | Aspidites melanocephalus (JX467494), Cylindrophis rufus (JX467494) |
Phylogenetics
Phylogenetic analyses were performed to allow reconstruction of the molecular evolutionary history of each toxin type for which transcripts were bioinformatically recovered. Toxin sequences were identified by comparison of the translated DNA sequences with previously characterized toxins using a BLAST search (46) of the UniProtKB protein database. Molecular phylogenetic analyses of toxin transcripts were conducted using the translated amino acid sequences. Comparative sequences from other venomous reptiles and physiological gene homologs identified from nonvenom gland transcriptomes were included in each data set as outgroup sequences. To minimize confusion, all sequences obtained in this study are referred to by their Genbank accession numbers (http://www.ncbi.nlm.nih.gov/sites/entrez?db = Nucleotide) and sequences from previous studies are referred to by their UniProtKB accession numbers (http://www.expasy.org/cgi-bin/sprot-search-ful). Resultant sequence sets were aligned using CLC Mainbench. When presented as sequence alignments, the leader sequence is shown in lowercase and cysteines are highlighted in black. > and < indicate incomplete N/5′ or C/3′ ends, respectively. Data sets were analyzed using Bayesian inference implemented on MrBayes, version 3.2 (47, 48). Run conditions were lset rates = invgamma with prset aamodelpr = mixed. The analysis was performed by running a minimum of 1 × 107 generations in four chains, and saving every 100th tree. The log-likelihood score of each saved tree was plotted against the number of generations to establish the point at which the log likelihood scores reached their asymptote, and the posterior probabilities for clades established by constructing a majority-rule consensus tree for all trees generated after completion of the burn-in phase.
Test for Recombination
Recombination can mislead phylogenetic and evolutionary selection interpretations (49). Hence, we evaluated the effect of recombination on all the toxins examined in this study (henophidian snake: lectin, three-finger toxin, epididymal secretory protein; iguanian lizard: cystatin, crotamine (beta-defensin), veficolin; crotaline snake: crotamine (beta-defensin); homalopsid snake: veficolin; caenophidian snake and anguimorph lizard: CRiSP) by employing Single Breakpoint Recombination implemented in the HyPhy package (50–52). Potential breakpoints were detected using the small sample Akaike information Criterion (AICc) and the sequences were compartmentalized before conducting the selection analyses.
Selection Analysis
Sophisticated likelihood models of coding-sequence evolution (53, 54) implemented in CODEML of the PAML (55) package were employed to evaluate the selection pressures on the major toxin transcripts recovered from henophidian snake and iguanian serous and mucus glands. We first employed the lineage-specific one-ratio model which assumes a single ω for the entire phylogenetic tree. This model tends to be very conservative and can only detect positive selection if the ω ratio averaged over all the sites along the lineage is significantly greater than one. Because such lineage-specific models assume a single ω for the entire tree, they often fail to identify regions in proteins that might be affected by episodic selection pressures and ultimately, underestimate the strength of selection. Hence, we employed site-specific models which estimate positive selection statistically as a nonsynonymous-to-synonymous nucleotide-substitution rate ratio (ω) significantly greater than 1. We compared likelihood values for three pairs of models with different assumed ω distributions as no a priori expectation exists for the same: M0 (constant ω rates across all sites) versus M3 (allows the ω to vary across sites within “n” discrete categories, n≥3); M1a (a model of neutral evolution) where all sites are assumed to be either under negative (ω <1) or neutral selection (ω = 1) versus M2a (a model of positive selection), which in addition to the site classes mentioned for M1a, assumes a third category of sites; sites with ω >1 (positive selection) and M7 (Beta) versus M8 (Beta and ω), and models that mirror the evolutionary constraints of M1 and M2 but assume that ω values are drawn from a beta distribution (56). Only if the alternative models (M3, M2a, and M8: allow sites with ω >1) show a better fit in Likelihood Ratio Test (LRT) relative to their null models (M0, M1a, and M8: do not show allow sites ω >1), are their results considered significant. LRT is estimated as twice the difference in maximum likelihood values between nested models and compared with the χ2 distribution with the appropriate degree of freedom—the difference in the number of parameters between the two models. The Bayes empirical Bayes (BEB) approach (Yang, 2005) was used to identify amino acids under positive selection by calculating the posterior probabilities that a particular amino acid belongs to a given selection class (neutral, conserved or highly variable). Sites with greater posterior probability (PP ≥ 95%) of belonging to the ‘ω > 1 class’ were inferred to be positively selected.
Further, Single Likelihood Ancestor Counting (SLAC), Fixed-Effects Likelihood (FEL), and Random Effects Likelihood models (57) implemented in HyPhy (58) were employed to further provide significant support to the aforementioned analyses and to detect sites evolving under the influence of positive and negative selection. The more advanced Mixed Effects Model Evolution (MEME) (59) was also used to detect episodic diversifying selection. MEME employs FEL along the sites and Random-effects likelihood (REL) across the branches to detect episodic diversifying selection.
For the effective comparison of iguanian and crotaline snake crotamines (beta-defensins); iguanian and homalopsid veficolins and anguimorph lizard and caenophidian snake cysteine-rich secretory proteins (CRiSPs)1, we employed Codeml Clade Model C analysis (60). The significance of the analysis was tested by comparison with site model M1a.
Structural Analyses
To depict the selection pressures shaping the evolution of toxins, we mapped the sites under positive selection on the homology models created using Phyre 2 webserver (61). Pymol 1.3 (62) was used to visualize and generate the images of homology models. Consurf webserver (63) was used for mapping the evolutionary selection pressures on the three-dimensional homology models whereas Selecton webserver (64, 65) was used to identify hypervariable regions on the toxins. Three-dimensional models and surface charge distributions were constructed as per (19).
Proteomics
Aspidites melanocephalus were obtained from Port Headland (Western Australia), the same population used as a “negative control” by Jelinek et al. (41). Specimens were anesthetized using zoletil (3 mg/kg) and then injected subcutaneously with pilocarpine (20 mg/kg) periglandular to the labial glands. Polyethylene equipment was used to collect and process samples in all cases. Samples were subsequently filtered using 20 Å syringe filters to remove large mucoidal strands and then lyophilized. Reduction and alkylation were undertaken by redissolving 3 μg of sample from each of the localities in 50 μl of 100 mm ammonium carbonate. After sonication and vortexing, 50 μl of 2% iodoethanol/0.5% triethylphosphine in acetonitrile was added to the re-dissolved sample. The reaction mixture was incubated for 2 h at 37 °C, before being dried and re-suspended in 15 μl of 2.5% acetonitrile and 0.5% formic acid. Additionally, 3 μl of reduced and alkylated sample was resuspended in 30 μl of 40 mm ammonium bicarbonate, before being incubated overnight with 750 ng trypsin. Digestion was stopped by addition of 1 μl of concentrated formic acid and 0.75 μg processed by liquid chromatography-tandem MS (LC-MS/MS). LC-MS/MS was performed using a Agilent Zorbax stable bond C18 column (2.1 mm × 100 mm, 1.8 μm particle size, 300 Å pore size) at a flow of 400 μl/min and a gradient of 1–40% solvent B (90% acetonitrile, 0.1% formic acid) in 0.1% formic acid over 15 min on a Shimadzu Nexera UHPLC coupled with an AB SCIEX 5600 Triple TOF mass spectrometer equipped with a Turbo V ion source heated to 450 °C. MS2 spectra were acquired at a rate of 20 scans/s, with a cycle time of 2.3 s, and optimized for high resolution. Precursor ions were selected between 80–1800 m/z, charge state of 2–5, and of an intensity of at least 120 counts per second, with a precursor selection window of 1.5 Da, and excluding isotopes within 2 Da for MS2. MS2 spectra were searched against all-reading frame translated A. melanocephalus mandibular and maxillary gland transcriptomes (16216 amino acid sequences) with Proteinpilot v4.5 (ABSciex) using a thorough identification search, specifying alkylation method (iodoethanol), tryptic digestion, and allowing for biological modifications and amino acid substitutions. This was performed to maximize the identification of protein sequences from the transcriptome despite the inherent variability of toxins, potential isoform mismatch with the transcriptomic data, and account for experimental artifacts leading to chemical modifications. For specific search parameters, modification probability and amino acid substitution probability settings set for the Paragon algorithm (version 4.5.0.0, 1654) see supplementary File S2. Spectra were inspected manually to eliminate false positives, excluding spectra with low signal to noise, erroneous modification assignments, and confidence values below 95% unless justifiable by presence of a-ions. The results were further analyzed using CLC Main Workbench v6.6.
1 Dimensional Gel Electrophoresis
SDS-PAGE of 20 μg A. melanocephalus secretion was carried out on 12% polyacrylamide gels as per the Laemmli-method, using a Bio-Rad Mini Protean setup. All reagents were purchased from Bio-Rad. Gels were stained overnight in Coommasie brilliant blue staining solution and de-stained again with 10% acetic acid.
RESULTS
Gland Arrangements
The rictal glands of the henophidian snake C. ruffus were large enough that they indented the mandibular and maxillary glands (Fig. 1B). In this anatomically simple and presumably plesiotypic condition, entirely serous tubules merge (Fig. 1C) to join a single duct descending vertically slightly anterior to the corner of the mouth. C. ruffus maxillary and mandibular glands were mixed glands, with the mandibular gland containing a greater proportion of serous cells than did the maxillary. The mandibular and maxillary glands of the constricting boid and pythonid snakes (Aspidites, Boa, Corallus, Eunectes, and Morelia) were also mixed but were predominantly composed of mucoid cells with only scattered serous cells (Fig. 1D). The iguanian lizards had mixed sero-mucous maxillary and mandibular glands, with small insectivorous or vegetarian species of any size having primarily mucous cells while species that feed on vertebrates, such as Anolis equestris, contained a greater proportion of serous cells at the base of the gland arrangements (Fig. 1E). In contrast, all nontoxicoferan lizards examined in this study (Ameiva ameiva, Cordylus tropidosternum, Eumeces obsoletus, Gekko gecko*, and Lepidophyma flavimaculatum) had mandibular mucous glands (e.g., Lepidophyma flavimaculatum in Fig. 1F) and had only extremely small maxillary glands or lacked them entirely.
Transcriptome Sequencing
Sequencing of transcriptomes of iguanian lizard and henophidian snake glands recovered transcripts encoding for proteins homologous to toxins previously characterized from anguimorph lizards or caenophidian snakes: 3FTx, CRiSP, crotamine (beta-defensin), CVF (cobra venom factor), cystatin, ESP (epididymal secretory protein), kunitz peptide, l-amino oxidase, lectin, RAP (renin aspartate protease), veficolin, and vespryn (Table I). Phylogenetic analyses showed all toxicoferan sequences to be monophyletic relative to endogenous body sequences, strongly suggestive of a single early recruitment, and in most cases the lizard and snake sequences were not reciprocally monophyletic, thus conclusively demonstrating a single early origin for that protein type. Intriguingly, whenever a particular toxin type was recovered, identical toxin transcripts were present in mandibular and maxillary glands within a species and in the case of C. ruffus, the identical transcripts were also recovered from the rictal gland in addition to the aforementioned glands.
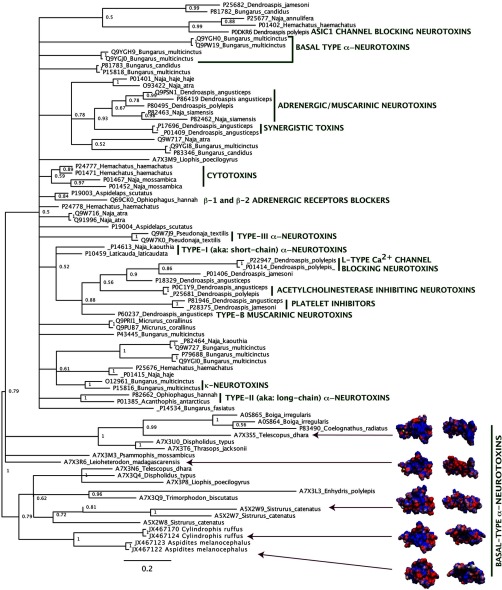
3FTx recovered from the henophidian snakes form a clade with the reptile and bird specific toxins (19, 66, 67) from nonfront fanged snakes along with the sequences from the viperid snake Sistrurus catenatus Fig. 2. The C. ruffus and A. melanocephalus isoforms shared a unique, newly evolved cysteine immediately before the doublet formed by the eighth and ninth ancestral cysteine. This results not only in a novel cysteine pattern but also in an odd number of cysteines (eleven). These changes would have structural implications not only for disulfide-bridging but also in the potential facilitation of dimerization. One C. ruffus variant has an exon deletion removing a significant N-terminal stretch of residues including five cysteines, resulting in a novel short peptide containing six cysteines (Fig. 3).
Fig. 2.
Molecular phylogeny of 3FTx toxins (nontoxin outgroups Q9WVC2 Mus musculus and Q9BZG9 Homo sapiens not shown). * indicates sequences obtained in this study. Clades with characterized activity are indicated.
Fig. 3.
3FTx sequence alignment: 1. Cruf RL12 rep c750, 2. CYRI0001D10, 3.ASLO Contig1, 4. ASUP Contig1, 5. O93422 Naja atra, 6. Q9PRI1 Micrurus corallinus, 7. Q9W717 Naja atra, 8. Q9W727 Bungarus multicinctus, 9. P25676 Hemachatus hemachatus, 10. O12961 Bungarus multicinctus, 11. Q9PW19 Bungarus multicinctus, 12. Q9YGH9 Bungarus multicinctus, 13. P43445 Bungarus multicinctus, 14. P01409 Dendroaspis angusticeps, 15. P22947 Dendroaspis polylepis, 16. P25677 Naja annulifera, 17. P24778 Hemachatus hemachatus, 18. P80495 Dendroaspis polylepis, 19. P14534 Bungarus fasiatus, 20. P82662 Ophiophagus hannah, 21. A7X3L3 Enhydris polylepis, 22. A7X3M3 Psammophis mossambicus, 23. A0S864 Boiga irregularis, 24. Q9W7J9 Pseudonaja textilis, 25. P15818 Bungarus multicinctus, 26. Q9YGI0 Bungarus multicinctus, 27. P0C1Y9 Dendroaspis angusticeps, 28. P25681 Dendroaspis polylepis, 29. P81946 Dendroaspis angusticeps, 30. P10459 Laticauda laticaudata, 31. P60237 Dendroaspis angusticeps. ‘+’ indicates N-terminal known only from peptide sequencing.
The iguanian crotamine (beta-defensin) sequences were also the most sequentially and phylogenetically divergent of all the iguanian transcript types (Figs. 4, 5). Some iguanian lizard crotamine (beta-defensin) sequences and the anguimorph lizard sequences unique proline-rich, highly charged N-terminal extensions not seen in the snake venom forms (Fig. 4). Sequence alignment and lack of phylogenetic affinity indicates that these novel proline rich regions are convergently evolved. A derived form from V. glauerti was notable in lacking two of the ancestral cysteines (Fig. 5).
Fig. 4.

Toxicofera crotamine (beta-defensin) peptide alignment 1. Q2XXN6 Pogona barbata, 2. Q2XXP0 Pogona barbata 3. Pogona barbata, 4. Pogona barbata 5. Uromastyx aegyptica, 6. Uromastyx aegyptica, 7. JX467135 Varanus tristis, 8. JX467132 Varanus glauerti, 9. JX467133 Varanus glauerti, 10. JX467134, Varanus glauerti, 11. JX467131 Varanus glauerti, 12. O57540 Crotalus durissus terrificus, 13. G9DCI6 Crotalus oreganus helleri, 14. G9DCI4 Crotalus oreganus helleri, 15. H2FLD8 Thamnodynastes strigatus.
Fig. 5.
Crotamine (beta-defensin) molecular phylogeny. * indicates sequences obtained in this study.
All lizard kunitz sequences had the same two-domain structure as the sequence from the venom of the caenophidian snake Austrelaps labialis (Fig. 6). Phylogenetic analyses showed that the two domain forms were basal to the derived mono-domain form (Fig. 7).
Fig. 6.
Kunitz peptide alignment 1. JX459935 Abronia graminea, 2. JX467158 Heloderma suspectum cinctum, 3. JX467155 Chameleo calyptratus, 4. JX467157 Iguana iguana, 5. H9G8B3 Anolis carolinensis, 6. H9GQ81 Anolis carolinensis, 7. H9GRC6 Anolis carolinensis, 8. B2BS84 Austrelaps labialis, 9. B5L5M7 Austrelaps superbus, 10. Q6T6S5 Bitis gabonica, 10 A7X3V4 Telescopus dhara. Gray blocks indicate kunitz peptide domains that are post-translationally cleaved from the precursor.
Fig. 7.
Molecular phylogeny of kunitz toxins and related nontoxin proteins. * indicates sequences obtained in this study.
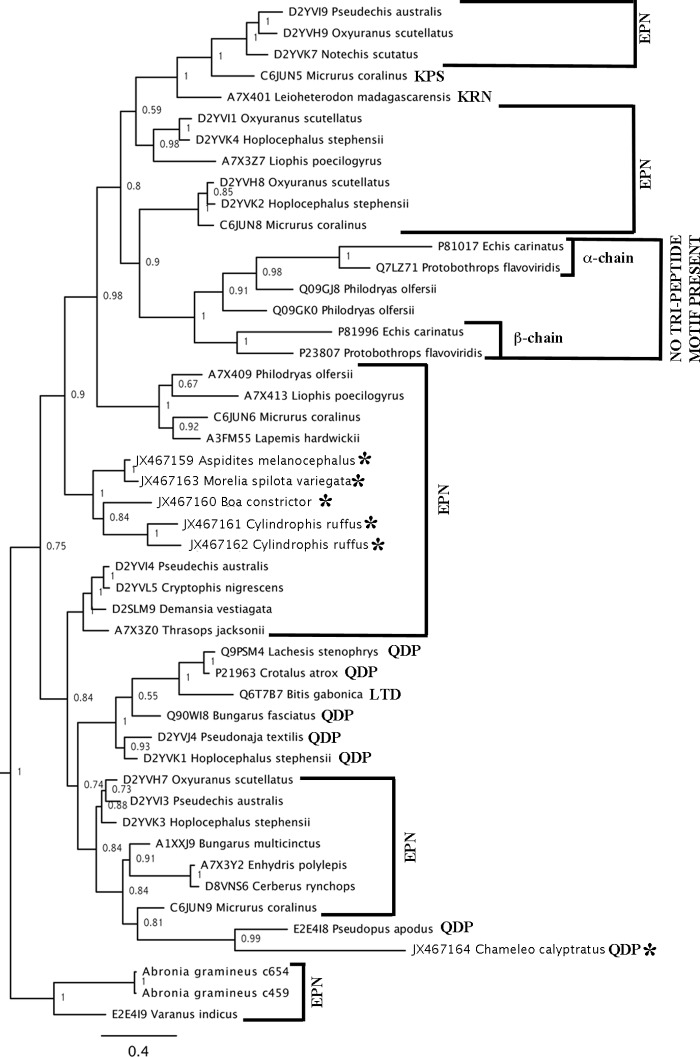
The henophidian snake lectin toxins had the galactose-binding EPN tripeptide functional motif whereas the C. calyptratus form is of the lactose-binding QPD motif. Phylogenetic analysis showed that the sequences containing the functional motifs EPN (glutamic acid + proline + asparagine) and QPD (glutamine + proline + aspartic acid) were not reciprocally monophyletic (Fig. 8).
Fig. 8.
Molecular phylogeny of lectin toxins (outgroups not shown). * indicates sequences obtained in this study. Functional motif triads are clade indicated for each clade.
Two variants of novel cysteine-rich peptide class were recovered from the A. melanocephalus and C. ruffus (Genbank accession numbers JX467493 and JX467494), which did not display homology to any previously characterized peptide class from any source, toxin or nontoxin.
Molecular Evolution Analyses
Examination using the highly conservative ORM (one ratio model) revealed considerable variability in molecular evolutionary rates in the proteins encoded by the transcripts we recovered (Supplemental Tables S1-S11). Estimated ratios of nonsynonymous codon substitutions over synonymous ones (ω) using maximum likelihood models ranged from indicating strong positive selection through to tremendous negative selection: crotaline snake crotamine (beta-defensin) = 1.42; henophidian snake lectin = 1.30; caenophidian snake CRiSP = 1.15; varanid crotamine (beta-defensin) 1.10; homalopsid snake veficolin = 0.86; anguimorph lizard CRiSP = 0.56; iguanian lizard veficolin = 0.52; iguanian lizard crotamine (beta-defensin) = 0.43; iguanian lizard cystatins = 0.42; henophidian snake 3FTx = 0.48; toxicoferan ESP = 0.34.
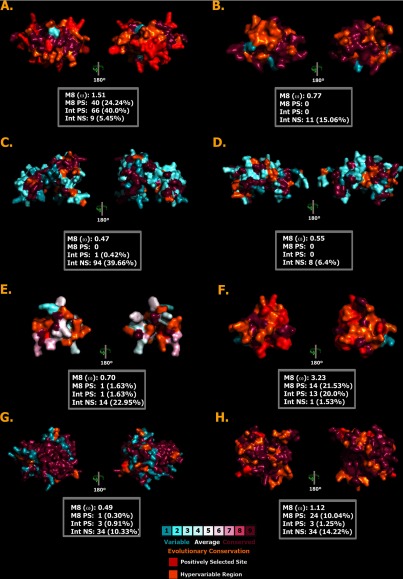
Because lineage-specific models of selection such as ORM often fail to detect episodic diversifying selection that affects only a few sites in the protein, we also employed site-specific models (Tables II, III, Fig. 9 and Supplementary Tables). Like ORM, site model M8 ω values also ranged widely (Tables II, III, and Fig. 9): crotaline snake crotamine (beta-defensin) = 3.23; henophidian snake lectin = 1.51; varanid lizard crotamine (beta-defensin) = 1.10; caenophidian snake CRiSP = 1.21; henophidian snake 3FTx = 0.77; anguimorph lizard CRiSP = 0.70; iguanian lizard crotamine (beta-defensin) = 0.70; iguanian lizard cystatin = 0.55; iguanian lizard veficolin = 0.49; toxicoferan reptile ESP = 0.47. The Bayes Empirical Bayes (BEB) method identified sites under positive selection for some toxin types (Tables II, III, and Fig. 9): 25% (40 sites) in the henophidian snake lectins; 24% (14 sites) in the crotaline snake crotamine; 10% (24 sites) in the homalopsid caenophidian snake veficolin; 8% (17 sites) in the caenophidian snake CRiSP; 2% (4 sites) in the anguimorph lizard CRiSP; while identifying only a single positively selected site each in iguanian crotamine (beta-defensin) and veficolin.
Table II. Selection analyses of basal snake and Iguania venom-encoding genes.
| SLACa | FELb | RELc | Integrative analyses | MEME Sitesd | PAMLg | |||
|---|---|---|---|---|---|---|---|---|
| SLAC + FEL + REL + MEME | ||||||||
| Basal Snake | M8 | M2a | ||||||
| ω>1e | 0 | 2 | 65 | 66 | 40 | 20 | ||
| Lectins | ω<1f | 1 | 9 | 0 | 9 | 5 | (16 + 24) | (3 + 17) |
| ω = | 1.30 | – | – | – | 1.51 | 1.52 | ||
| ω>1e | 0 | 0 | 0 | 0 | ||||
| 3FTx | ω<1f | 0 | 4 | 10 | 11 | 0 | 0 | 0 |
| ω = | 0.48 | – | – | – | 0.77 | 0.77 | ||
| ω>1e | 0 | 0 | 0 | 1 | ||||
| ESP | ω<1f | 3 | 30 | 94 | 94 | 1 | 0 | 0 |
| ω = | 0.34 | – | – | – | 0.47 | 0.57 | ||
| Iguania | ||||||||
| ω>1e | 0 | 0 | 0 | 0 | ||||
| Cystatins | ω<1f | 0 | 8 | 0 | 8 | 0 | 0 | 0 |
| ω = | 0.42 | – | 48 | – | 0.55 | 0.47 | ||
3FTx, Three-finger Toxin; ESP, Epididymal Secretory Protein.
a Single Likelihood Ancestor Counting.
b Fixed-effects likelihood.
c Random-effects likelihood.
d Sites detected as experiencing episodic diversifying selection (0.05 significance) by the Mixed Effects Model Evolution (MEME).
e Number of positively selected sites at 0.05 significance (for SLAC, FEL) or 50 Bayes factor (for REL).
f Number of negatively selected sites at 0.05 significance (for SLAC, FEL) or 50 Bayes factor (for REL).
g Positively selected sites detected using the Bayes Empirical Bayes approach implemented in M8 and M2a. Sites detected at 0.99 and 0.95 significance are indicated in the parenthesis; ω: mean dN/dS.
Table III. Comparative evolution of β-defensin, veficolins and cysteine-rich secretory proteins.
| SLACa | FELb | RELc | Integrative | MEME Sitesd | PAMLg | ||||
|---|---|---|---|---|---|---|---|---|---|
| SLAC + FEL + REL + MEME | M8 | M2a | Clade Model C (ω)h | ||||||
| Iguania, Varanidae and Crotalinae β-Defensin | |||||||||
| ω>1e | 0 | 1 | 0 | 1 | 1 | ||||
| Iguania | ω<1f | 3 | 9 | 9 | 14 | 0 | (0 + 1) | 0 | 3.61sig |
| ω = | 0.43 | – | 0.59 | – | 0.70 | 0.77 | |||
| ω>1e | 0 | 0 | 12 | 13 | 14 | 12 | |||
| Crotalinae | ω<1f | 1 | 0 | 0 | 1 | 2 | (6 + 8) | (6 + 6) | 4.43sig |
| ω = | 1.42 | – | 4.03 | – | 3.23 | 3.22 | |||
| ω>1e | 0 | 0 | 0 | 1 | |||||
| Varanidae | ω<1f | 0 | 5 | 14 | 14 | 1 | 0 | 0 | 1.97sig |
| ω = | 1.10 | – | – | – | 1.31 | 1.31 | |||
| Iguania and Homalopsidae Veficolins | |||||||||
| ω>1e | 0 | 1 | 10 | 17 | 1 | 1 | |||
| Iguania | ω<1f | 11 | 35 | 80 | 85 | 8 | (1 + 0) | (0 + 1) | 0.33sig |
| ω = | 0.52 | – | – | – | 0.49 | 0.49 | |||
| ω>1e | 0 | 0 | 0 | 3 | 24 | 14 | |||
| Homalopsidae | ω<1f | 2 | 5 | 34 | 34 | 3 | (8 + 16) | (2 + 12) | 3.06sig |
| ω = | 0.86 | – | – | – | 1.12 | 1.12 | |||
| Anguimorpha and Caenophidia CRiSPs | |||||||||
| ω>1e | 1 | 3 | 16 | 28 | 4 | 4 | |||
| Anguimorpha | ω<1f | 21 | 38 | 100 | 102 | 10 | (2 + 2) | (2 + 2) | 2.04sig |
| ω = | 0.56 | – | 0.77 | – | 0.70 | 0.83 | |||
| ω>1e | 3 | 13 | 62 | 67 | 17 | 26 | |||
| Caenophidia | ω<1f | 7 | 19 | 50 | 59 | 24 | (8 + 9) | (17 + 9) | 3.94sig |
| ω = | 1.15 | – | 4.13 | – | 1.21 | 1.21 | |||
a Single Likelihood Ancestor Counting.
b Fixed-effects likelihood.
c Random-effects likelihood.
d Sites detected as experiencing episodic diversifying selection (0.05 significance) by the Mixed Effects Model Evolution (MEME).
e Number of positively selected sites at 0.05 significance (for SLAC, FEL) or 50 Bayes factor (for REL).
f Number of negatively selected sites at 0.05 significance (for SLAC, FEL) or 50 Bayes factor (for REL).
g Positively selected sites detected using the Bayes Empirical Bayes approach implemented in the site models (M8 and M2a). Sites detected at 0.99 and 0.95 significance are indicated in the parenthesis.
h Mean dN/dS computed by the Clade Model C analysis for Iguania versus Crotalinae crotamines and Iguania versus Colubridae veficolins; ω: mean dN/dS; sig: Clade Model C analysis significant at 0.001.
Fig. 9.
Molecular evolution of lectin, 3FTx, epididymal secretory protein (A, B, and C, respectively), iguanian Cystatin (D), iguanian lizard and crotaline snake crotamines (beta-defensins) (E and F, respectively), iguanian lizard and Homalopsidae snake veficolins (G and H, respectively). The number of positively-selected sites (Codeml: Model 8, PP ≥ 0.95, Bayes-Empirical Bayes approach) and hypervariable regions are highlighted in red and orange, respectively. Various omega statistics (M8 omega value, total number of positively selected sites detected by M8 (PP ≥ 0.95, BEB)) and the number of positively and negatively selected sites detected by HyPhy integrative approach (SLAC and FEL: 0.05 significance; REL 50 Bayes factor) are also indicated.
Comparing the ω values estimated by the above models can be misleading, because the genes being compared may have a different proportion of sites under selection. Hence, we employed the Clade Model C approach for effectively comparing the evolution of anguimorph lizard, iguanian lizard, and crotaline snake crotamines; iguanian lizard and homalopsid snake veficolins and anguimorph lizard and caenophidian snake CRiSPs (Table III). Clade model analyses indicated that the crotaline snake crotamines (ω = 4.43) and homalopsid snake veficolins (ω = 3.06) have accumulated greater variations than their iguanian lizard crotamine (beta-defensin) (ω = 3.61) and veficolin (ω = 0.33) counterparts. The varanid crotamine (beta-defensin) peptides were less diverse (ω = 1.97) than either the snake or iguanian lizard counterparts. It is important to note that a lower omega value is only expected among closely related taxa versus more diverged taxa if for example, they share more ecological similarity to the extent that their toxins did not need to selectively diversify as much as between ecologically diverse taxa. The analysis also revealed that the caenophidian snake CRiSPs (ωω = 3.94) have evolved more rapidly than the anguimorph lizard CRiSPs (ω = 2.04). All these aforementioned analyses were significant (P≪0.001) in comparison with site model M1a. Single Likelihood Ancestral Counting (SLAC), Fixed Effects Likelihood (FEL), Random Effects Likelihood (REL), Mixed Effects Model of Evolution (MEME) and the identification of hypervariable regions, conclusively support these results (Tables II, III, and Fig. 9).
Surface Charge Molecular Modeling
Homology models were built using the NMR-structure of Bungatoxin from Bungarus Candidus venom as a template (PDB-code: 2JQP). Alternative templates (1ABT, 1CDT, 1KBA, and 2H7Z) were investigated, because not all of the sequences yielded a suitable similarity score to a single template structure. The bungatoxin structure used in this study was selected for its best fit to the cysteine sequence pattern. Regions of insertions and deletions were moved to loop structures rather than modeled in the core of the protein. Target template alignments were generated using SPDBV (68). Model structures were obtained from SwissModel server using the “project mode” option (68). Electrostatic surfaces were calculated using APBS (69) and visualized in visual molecular dynamics (70).
Proteomics
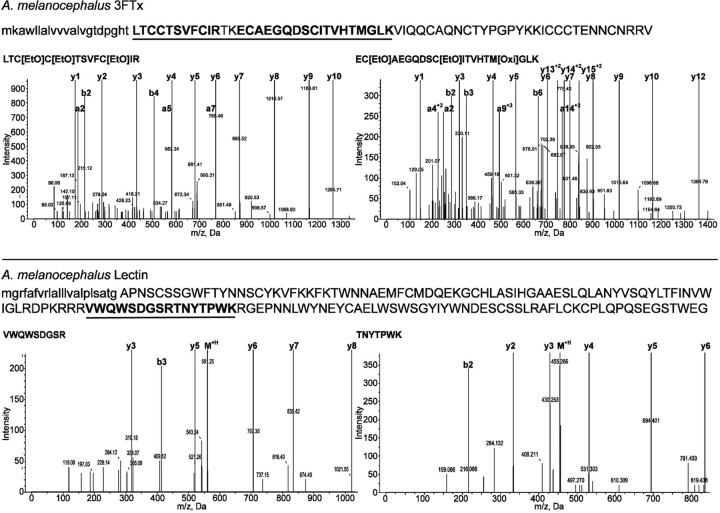
Proteomic analysis of the A. melanocephalus secretion was able to confirm the presence of the two dominant transcript types: 3FTx and lectin. Tandem MS spectra of tryptic fragments provided high confidence matches for each of the two toxin sequences (Fig. 10) (Supplemental Table S12). The low number of identified proteins, despite the large amount of analyzed sample (1 μg), supports the histological findings that the maxillary and mandibular glands of A. melanocephalus are primarily mucoidal but that low levels of protein secretion continue. The 3FTx present in the secretion was present in the identical <10kDa region in 1D gel analysis under reducing and non-reducing conditions (Fig. 11), demonstrating that the free cysteine (Fig. 3) is buried and thus does not promote dimerization.
Fig. 10.
Tandem MS spectra of 3FTx and lectin tryptic fragments from Aspidites melanocephalus secretion.
Fig. 11.
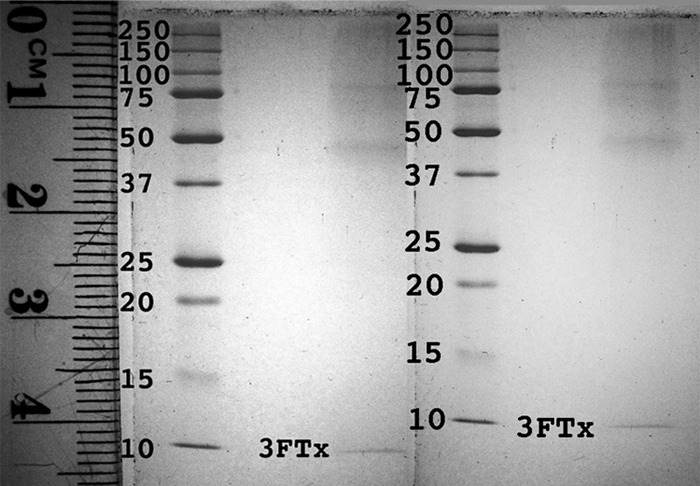
1D gel electrophoresis showing Aspidites melanocephalus oral secretion under A, reducing and B, nonreducing conditions with the 3FTx marked to show that the free-cysteine does not facilitate dimerization Supplementary Material.
DISCUSSION
Toxin gene recruitment events appear to have occurred in batches on three occasions: up to ten recruitments at the base of the toxicoferan tree; six in the common ancestor of anguimorph lizards and snakes; and eight more in the common ancestor of the caenophidian snakes (Fig. 1). Thus the Toxicofera venom system appears to have a punctuated molecular evolutionary history.
Our results significantly changed our perception of the relative timing of gene recruitment events for several toxin types (Fig. 1). 3FTx recruitment, previously thought to have occurred within the caenophidian snakes before the Viperidae split-off 54 million years ago (MYA), now appears to have occurred within the ancient Afrophidian snake clade at 104 MYA, a backwards shift of 50 million years (71). RAP was previously only known from Echis venoms within the Viperidae snakes and thus was considered to be the result of a recruitment event some 20 MYA. As we recovered it from B. constrictor, the recruitment may also have occurred within the ancestral Afrophidian snakes, a backwards shift of 84 million years. Intriguingly, a homologous renin aspartate protease sequence from the A. carolinensis genome was nested within the snake sequences, strongly suggestive of recruitment at the base of the toxicoferan reptile radiation some 166 MYA, which would be a shift of 146 million years from Echis. However, sequencing of homologous iguanian lizard mandibular or maxillary gland mRNA transcripts or proteins must first be accomplished before this timing can be absolutely confirmed. Other toxin types, however, were conclusively shown to be ancestral to the toxicoferan reptiles: CRiSP, CVF, kunitz, l-amino oxidase, lectin, and veficolin. CRiSP, CVF, lectin, and veficolin were previously sequenced from both snakes and anguimorph lizards but not from iguanian lizards, and thus this represents a backwards shift of 22 million years for the recruitment of this toxin type. Kunitz peptides and l-amino oxidase were considered exclusive to caenophidian snakes and thus our perception of the age of these toxin types shifts over 112 MYA.
This ancestral molecular diversity is particularly intriguing considering that the iguanian lizards have only poorly developed “incipient” venom glands. Despite these glands being of no obvious use in the feeding ecology of insectivorous or vegetarian species, and being predominantly mucoid in these lineages, they still secrete active peptides. Intriguingly, the highest expression level and greatest evidence of duplication and diversification within the iguanian transcripts was observed for the crotamine (beta-defensin) and cystatin peptides. All other transcript types were recovered in lower levels and typically with only one or two isoforms. It is noteworthy that the ancestral activity of these two peptide types is antimicrobial (72, 73)—an activity which is retained by at least some isoforms expressed in the venoms of highly toxic front-fanged snakes. Another ancestrally recruited protein type, l-amino oxidase, also has well-documented anti-microbial activity (74). Thus, the trigger for the development of the toxicoferan venom system may have been the rapid co-option of a newly developed antimicrobial peptide secreting system. However, it must be highlighted that the other toxin classes ancestrally recruited and still expressed even in the iguanian venom system (at very low levels) include those that have been developed into neurotoxins (CRiSP (75–79), vespryn (80) or hemotoxins: kunitz, lectin, and likely veficolin (81)). Thus, in the Iguania the evidence favors an antimicrobial function for the secretions of their incipient venom system. There is no known predatory function for the system within vegetarian or insectivorous species, although the significantly enlarged protein secreting region of species that predate on vertebrates (Fig. 1E) does point to a possible role in prey-capture. Conversely, the rapid accumulation of novel secretory frameworks appears to have underpinned the rapid radiation of the other toxicoferan reptile lineages (anguimorph lizards and the snakes). In particular, the venom system likely led to the explosive diversification of the caenophidian snakes (82).
The well-developed rictal glands in the henophidian snake C. ruffus (Fig. 1B), which are entirely serous in composition (Fig. 1C), suggest an active role for these structures in the life of these serpents. Similarly, the mandibular and maxillary glands of Cylindrophis are sero-mucous whereas those of the powerful constricting highly derived boid and pythonid snakes, stain histologically as predominantly (but not exclusively) mucoidal. Of particular note is the encoding of the same toxin transcripts in maxillary, mandibular, and rictal glands. This supports not only the hypothesized shared evolutionary history of the mandibular and maxillary glands (18) but also provides the first evidence that the rictal glands are derived from the same system.
The difference in gland structures and types between C. ruffus and the boid/pythonid snakes parallels the differences in their predatory ecology: with C. ruffus being representative of the presumably ancestral snake condition in being only weak constrictors and feeding upon prey that is typically not wider than their head; while boid/pythonid snakes represent a derived clade that has developed powerful constriction as a new adaptation to predate upon massive prey items often much wider than their head. Thus, alternate selection pressures operate on the oral glands of these snakes: C. ruffus would benefit from chemical assistance in prey capture; whereas the boid/pythonid snakes easily dispatch their prey using powerful constriction but have a need for appreciable amounts of mucous to lubricate the fur of mammalian or feathers of avian prey items. However, consistent with positive reaction generated by A. melanocephalus oral secretion in snake venom detection kit trials (41), our data demonstrates that the toxin genes are not only still transcribed but also continue to be translated and secreted as relics of the venom system that the boids/pythonids have down-regulated. Although it is noteworthy that A. melanocephalus is unusual among pythonids in that it feeds primarily on reptiles (slender prey items not dissimilar to those which C. ruffus preys on), it is probable that their oral secretions are of little to no use in prey capture as the snakes rely on constriction to kill the prey items. Despite this, however, they may be problematic for human bite victims as a result of their capacity to produce false-positives in the medical diagnosis of snake envenomation.
Our sequencing of transcripts that are homologous with well-characterized toxin types also provides new evidence for the continued diversification of the secreted proteins even within the glands of henophidian snakes and iguanian lizards. One example is the cysteine changes in the 3FTx of henophidian snakes that could have profound structural implications not only on the conformation of the peptide but also the facilitation of dimerization. However, our demonstration that the 3FTx in the A. melanocephalus secretion occupy the same <10kDa location under reducing and nonreducing conditions, demonstrates that the free cysteine (located as part of the cysteine-triplet near the C terminus) is buried in the molecular core and thus does not facilitate dimerization. Our results also provide evidence for ancient derivations such as the early loss of cysteine four in the lectins with replacement by glycine in one of the early duplicates. It had previously been unclear whether the EPN motif (which blocks platelets through mannose-binding sites), or the QPD motif (which blocks platelets through galactose-binding sites) was plesiotypic as the caenophidian snakes and anguimorph lizards each have both forms (20, 22). In this study, the QPD motif was present in the sole sequence recovered from an iguanian lizard (Chamaeleo calyptratus). Thus the QPD motif appears to be plesiotypic whereas the EPN is an ancient derivation. However, as neither motif is monophyletic, a degree of convergence in one or the other or both cannot be ruled out. Some henophidian snake isoforms have a glycine substituted for the fourth cysteine, a derivation shared with some caenophidian snake homologs but not present in the lizard forms. This indicates a secondary loss of this cysteine early in snake evolutionary history. In the kunitz peptides, the ancestral dual domain form previously sequenced from the caenophidian snake Austrelaps labialis was also recovered from the iguanian lizard C. calyptratus as well as the anguimorph lizards Abronia graminea and Heloderma suspectum cinctum (83). However, the derived mono-domain form favored in caenophidian snake venoms was also recovered from iguanian lizards (C. calyptratus and C. kingii). Thus the loss of the first domain was a derivation that occurred before the organismal diversification of the toxicoferan ancestor.
Evidence provided by various analyses of different henophidian snake and iguanian peptides/proteins and mapping of mutations on the homology models indicates that several of these have accumulated variation under the strong influence of positive selection (Tables II, III, and Fig. 9). The henophidian snake lectins in particular seem to have evolved rapidly (ω = 1.51; 40 positively selected (25%) sites). This estimated variation combined with the putative hemotoxic activity revealed by the conservation of functional motifs, may indicate a functional role for orally secreted lectins in the lives of these snakes, which are not known for using venom in prey capture. These analyses also revealed hypervariable regions in iguanian lizard crotamine (beta-defensin) and veficolin toxins. The observed variation in several regions could be a result of the putative antimicrobial function of crotamine (beta-defensin) peptides.
The recovery of novel protein scaffolds from the glands studied here reinforces how little is known about the protein composition of reptile venoms or the evolutionary history of the glands. This is underscored by the number and diversity of novel scaffolds recovered despite the relatively limited sampling employed, including an entirely new peptide class discovered in the pythonid libraries. The toxin types recovered in this study should not be considered as the full suite because the relatively limited sampling only recovered the dominant forms. More extensive sampling will no doubt recover novel isoforms of types identified to date as well as entirely new toxin classes. Of interest for follow-up investigations should be the relative secretion by the segregated mucous lobules in the independently derived mandibular systems of varanid and helodermatid lizards as well as the derived maxillary system of snakes. Both of these systems also have mucous and protein secreting cells divided into discrete glands, with the protein glands encapsulated. In the case of the anguimorph lizards, the varanids and helodermatids have independently derived the mandibular glands into complex compartmentalized glands. Further work is also needed to examine the relationship between transcription and translation, and thus the presence and representation of the proteins themselves in the secretory mixtures. These results highlight the relatively untapped potential of such complex mixtures as sources of novel investigational ligands or lead compounds for use in drug design and development. Of particular focus of follow-up work by us will be to investigate the bioactivity of iguanian and anguimorphan crotamine (beta-defensin) peptides to determine relative degree of antimicrobial activity versus more derived activities such as neurotoxicity. It is hoped that these results will stimulate further investigation of these neglected glands and their secretory proteins in an increasingly diverse range of toxicoferan reptile species.
Supplementary Material
Acknowledgments
We thank Professor Elazar Kochva for his helpful constructive criticism.
Footnotes
* This research was supported in part by the Portuguese Foundation for Science and Technology (FCT) through the PhD grant conferred to KS (SFRH/BD/61959/2009) and the project PTDC/AAC-AMB/121301/2010 (FCOMP-01-0124-FEDER-019490) to AA. EABU acknowledges funding from the University of Queensland (International Postgraduate Research Scholarship, UQ Centennial Scholarship, and UQ Advantage Top-Up Scholarship) and the Norwegian State Education Loans Fund. BGF was funded by an Australian Research Council Future Fellowship and by the University of Queensland. SAA was the recipient of postdoctoral fellowship (PDRF Phase II Batch-V) from Higher Education Commission (HEC Islamabad) Pakistan. TNWJ was funded by an Australian Postgraduate Award.
 This article contains supplemental Files S1 to S2 and Tables S1 to S12.
This article contains supplemental Files S1 to S2 and Tables S1 to S12.
1 The abbreviations used are:
- CRiSPs
- cysteine-rich secretory proteins
- CVF
- cobra venom factor
- ESP
- epididymal secretory protein
- RAP
- renin aspartate protease
- ORM
- one ratio model
- MYA
- million years ago.
REFERENCES
- 1. Kordis D., Gubensek F. (2000) Adaptive evolution of animal toxin multigene families. Gene. 261, 43–52 [DOI] [PubMed] [Google Scholar]
- 2. Fry B. G. (2005) From genome to “venome”: Molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 15, 403–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Binford G. J., Bodner M. R., Cordes M. H., Baldwin K. L., Rynerson M. R., Burns S. N., Zobel-Thropp P. A. (2009) Molecular evolution, functional variation, and proposed nomenclature of the gene family that includes sphingomyelinase D in sicariid spider venoms. Mol. Biol. Evolution 26, 547–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casewell N. R., Wagstaff S. C., Harrison R. A., Renjifo C., Wüster W. (2011) Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol. Biol. Evolution 28, 2637–2649 [DOI] [PubMed] [Google Scholar]
- 5. Chang D., Duda T. F., Jr. (2012) Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evolution 29, 2019–2029 [DOI] [PubMed] [Google Scholar]
- 6. Duda T. F., Jr., Palumbi S. R. (1999) Molecular genetics of ecological diversification: duplication and rapid evolution of toxin genes of the venomous gastropod Conus. Proc. Natl. Acad. Sci. U. S. A. 96, 6820–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fry B. G., Wüster W., Kini R. M., Brusic V., Khan A., Venkataraman D., Rooney A. P. (2003) Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evol. 57, 110–129 [DOI] [PubMed] [Google Scholar]
- 8. Nakashima K., Ogawa T., Oda N., Hattori M., Sakaki Y., Kihara H., Ohno M. (1993) Accelerated evolution of Trimeresurus flavoviridis venom gland phospholipase A2 isozymes. Proc. Natl. Acad. Sci. U. S. A. 90, 5964–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nei M., Gu X., Sitnikova T. (1997) Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. U. S. A. 94, 7799–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weinberger H., Moran Y., Gordon D., Turkov M., Kahn R., Gurevitz M. (2010) Positions under positive selection–key for selectivity and potency of scorpion alpha-toxins. Mol. Biol. Evolution 27, 1025–1034 [DOI] [PubMed] [Google Scholar]
- 11. Zhu S., Bosmans F., Tytgat J. (2004) Adaptive evolution of scorpion sodium channel toxins. J. Mol. Evolution 58, 145–153 [DOI] [PubMed] [Google Scholar]
- 12. Juarez P., Comas I., Gonzalez-Candelas F., Calvete J. J. (2008) Evolution of snake venom disintegrins by positive Darwinian selection. Mol. Biol. Evolution 25, 2391–2407 [DOI] [PubMed] [Google Scholar]
- 13. Gao B., Peigneur S., Dalziel J., Tytgat J., Zhu S. (2011) Molecular divergence of two orthologous scorpion toxins affecting potassium channels. Comparative Biochem. Physiol. 159, 313–321 [DOI] [PubMed] [Google Scholar]
- 14. Sunagar K., Johnson W. E., O'Brien S. J., Vasconcelos V., Antunes A. (2012) Evolution of CRiSPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol. Biol. Evolution 29, 1807–1822 [DOI] [PubMed] [Google Scholar]
- 15. Brust A., Sunagar K., Undheim E. A., Vetter I., Yang D., Casewell N. R., Ruder T., Jackson T. N., Koludarov I., Alewood P. F., Hodgson W. C., Lewis R. J., King G. F., Antunes A., Hendrikx I., Fry B. G. (2012) Differential Evolution of Domains:Comparative Evolution of Psammophis and Echis Snake Venom Metalloproteases (SVMP). Mol. Cell. Proteomics, 2, 651–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wong E. S. W., Belov K. (2012) Venom evolution through gene duplications. Gene 496, 1–7 [DOI] [PubMed] [Google Scholar]
- 17. Fry B. G., Lumsden N. G., Wüster W., Wickramaratna J. C., Hodgson W. C., Kini R. M. (2003) Isolation of a neurotoxin (alpha-colubritoxin) from a nonvenomous colubrid: evidence for early origin of venom in snakes. J. Mol. Evol. 57, 446–452 [DOI] [PubMed] [Google Scholar]
- 18. Fry B. G., Vidal N., Norman J. A., Vonk F. J., Scheib H., Ramjan S. F., Kuruppu S., Fung K., Hedges S. B., Richardson M. K., Hodgson W. C., Ignjatovic V., Summerhayes R., Kochva E. (2006) Early evolution of the venom system in lizards and snakes. Nature 439, 584–588 [DOI] [PubMed] [Google Scholar]
- 19. Fry B. G., Scheib H., van der Weerd L., Young B., McNaughtan J., Ramjan S. F., Vidal N., Poelmann R. E., Norman J. A. (2008) Evolution of an arsenal: structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Mol. Cell. Proteomics 7, 215–246 [DOI] [PubMed] [Google Scholar]
- 20. Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D. A., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., Rodríguez De La Vega R. C. (2009) The toxicogenomic multiverse: Convergent recruitment of proteins into animal venoms. 10, 483–511 [DOI] [PubMed] [Google Scholar]
- 21. Fry B. G., Roelants K., Winter K., Hodgson W. C., Griesman L., Kwok H. F., Scanlon D., Karas J., Shaw C., Wong L., Norman J. A. (2010) Novel venom proteins produced by differential domain-expression strategies in beaded lizards and gila monsters (genus Heloderma). Mol. Biol. Evolution 27, 395–407 [DOI] [PubMed] [Google Scholar]
- 22. Fry B. G., Winter K., Norman J. A., Roelants K., Nabuurs R. J., van Osch M. J., Teeuwisse W. M., van der Weerd L., McNaughtan J. E., Kwok H. F., Scheib H., Greisman L., Kochva E., Miller L. J., Gao F., Karas J., Scanlon D., Lin F., Kuruppu S., Shaw C., Wong L., Hodgson W. C. (2010) Functional and structural diversification of the Anguimorpha lizard venom system. Mol. Cell. Proteomics 9, 2369–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fry B. G., Wüster W. (2004) Assembling an arsenal: origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences. Mol. Biol. Evolution 21, 870–883 [DOI] [PubMed] [Google Scholar]
- 24. Fry B. G., Scheib H., de Junqueira L. M., Silva D. A., Casewell N. R. (2012) Novel transcripts in the maxillary venom glands of advanced snakes. Toxicon. 59, 696–708 [DOI] [PubMed] [Google Scholar]
- 25. Vidal N., Hedges S. B. (2005) The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. Comptes Rendus Biologies 328, 1000–1008 [DOI] [PubMed] [Google Scholar]
- 26. Townsend T., Larson A., Louis E., Macey J. R. (2004) Molecular phylogenetics of squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst. Biol. 53, 735–757 [DOI] [PubMed] [Google Scholar]
- 27. Piskurek O., Austin C. C., Okada N. (2006) Sauria SINEs: Novel short interspersed retroposable elements that are widespread in reptile genomes. J. Mol. Evolution 62, 630–644 [DOI] [PubMed] [Google Scholar]
- 28. Lee M. S. (2009) Hidden support from unpromising data sets strongly unites snakes with anguimorph ‘lizards’. J. Evolutionary Biol. 22, 1308–1316 [DOI] [PubMed] [Google Scholar]
- 29. Fry B. G., Scheib H., van der Weerd L., Young B., McNaughtan J., Ramjan S. F., Vidal N., Poelmann R. E., Norman J. A. (2008) Evolution of an arsenal: Structural and functional diversification of the venom system in the advanced snakes (Caenophidia). Molecular and Cellular Proteomics 7, 215–246 [DOI] [PubMed] [Google Scholar]
- 30. Gabe M., H S. G. (1969) Données histologiques sur les glandes salivaires des lépidosauriens. Mém. Mus. Natl. Hist. Nat. Paris 58, 1–118 [Google Scholar]
- 31. Kochva E. (1978) Oral Glands of the Reptilia In: Biology of the Reptilia, Gans C., Gans K. A. (Eds.). Academic Press, London and New York, 43–161 [Google Scholar]
- 32. Saint Girons H. (1988) Les glandes céphaliques exocrines des Reptiles. I. – Données anatomiques et histologiques. (The cephalic exocrine glands of reptiles. I. – Anatomical and histological data.). Annales des Sciences Naturelles, Zoologie, Paris. 9, 221–255 [Google Scholar]
- 33. Saint Girons H. (1989) Les glandes céphaliques exocrines des Reptiles. II.- Considérations fonctionnelles et évolutives. (The cephalic exocrine glands of reptiles. II. – Functional and evolutionary considerations.) Annales des Sciences Naturelles, Zoologie, Paris: 10, 1–17 [Google Scholar]
- 34. Underwood G., Kochva E. (1993) On the affinities of the burrowing asps Atractaspis (Serpentes: Atractaspididae). Zool J. Linn. Soc. 107 [Google Scholar]
- 35. Underwood G. (1997) An overview of venomous snake evolution. In: Venomous snakes, ecology, evolution and snakebite, p. 1–13 Thorpe R. S., Wüster W., Malhotra A. (eds.). Symposium of the Zoological Society of London, No. 70, Clarendon Press, Oxford, 1–13 [Google Scholar]
- 36. Underwood G. (2002) On the rictal structures of some snakes. Herpetologica 58, 1–17 [Google Scholar]
- 37. Wollberg M., Kochva E., Underwood G. (1998) On the rictal glands of some atractaspid snakes. Herpetological J. 8, 137–143 [Google Scholar]
- 38. McDowell S. B. (1986) The architecture of the corner of the mouth of colubroid snakes. J. Herpetology 20 [Google Scholar]
- 39. Phisalix M., Caius R. (1918) L'extension de la fonction venimeuse dans l'ordre entière des ophidiens et son existence chez des familles ou elle n'avait pas été soupçonnée jusqu'içi. Journal de Physiologie et de Pathologie Générale 17, 923–964 [Google Scholar]
- 40. Phisalix M. (1922) Animaux Venimeux et Venins. Masson, Paris [Google Scholar]
- 41. Jelinek G. A., Tweed C., Lynch D., Celenza T., Bush B., Michalopoulos N. (2004) Cross reactivity between venomous, mildly venomous, and non-venomous snake venoms with the Commonwealth Serum Laboratories venom detection kit. Emerg. Med. Australas 16, 459–464 [DOI] [PubMed] [Google Scholar]
- 42. Conesa A., Götz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 43. Conesa A., Götz S. (2008) Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genomics 2008, 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Götz S., Garcia-Gomez J. M., Terol J., Williams T. D., Nagaraj S. H., Nueda M. J., Robles M., Talon M., Dopazo J., Conesa A. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Götz S., Arnold R., Sebastian-Leon P., Martin-Rodriguez S., Tischler P., Jehl M. A., Dopazo J., Rattei T., Conesa A. (2011) B2G-FAR, a species-centered GO annotation repository. Bioinformatics 27, 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ronquist F., Huelsenbeck J. P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 [DOI] [PubMed] [Google Scholar]
- 48. Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., Larget B., Liu L., Suchard M. A., Huelsenbeck J. P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Posada D., Crandall K. A. (2002) The effect of recombination on the accuracy of phylogeny estimation. J. Mol. Evolution 54, 396–402 [DOI] [PubMed] [Google Scholar]
- 50. Frost S. D., Liu Y., Pond S. L., Chappey C., Wrin T., Petropoulos C. J., Little S. J., Richman D. D. (2005) Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J. Virol. 79, 6523–6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Delport W., Poon A. F., Frost S. D., Kosakovsky Pond S. L. (2010) Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26, 2455–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kosakovsky Pond S. L., Posada D., Gravenor M. B., Woelk C. H., Frost S. D. (2006) Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23, 1891–1901 [DOI] [PubMed] [Google Scholar]
- 53. Goldman N., Yang Z. (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evolution 11, 725–736 [DOI] [PubMed] [Google Scholar]
- 54. Yang Z. (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evolution 15, 568–573 [DOI] [PubMed] [Google Scholar]
- 55. Yang Z. (2007) PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evolution 24, 1586–1591 [DOI] [PubMed] [Google Scholar]
- 56. Nielsen R., Yang Z. (1998) Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kosakovsky Pond S. L., Frost S. D. W. (2005) Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol. Biol. Evolution 22, 1208–1222 [DOI] [PubMed] [Google Scholar]
- 58. Pond S. L., Frost S. D. W., Muse S. V. (2005) HyPhy: Hypothesis testing using phylogenies. Bioinformatics 21, 676–679 [DOI] [PubMed] [Google Scholar]
- 59. Kosakovsky Pond S. L., Murrell B., Fourment M., Frost S. D., Delport W., Scheffler K. (2011) A random effects branch-site model for detecting episodic diversifying selection. Mol. Biol. Evolution 28, 3033–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bielawski J. P., Yang Z. (2004) A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evolution 59, 121–132 [DOI] [PubMed] [Google Scholar]
- 61. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protocols 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 62. DeLano W. (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA [Google Scholar]
- 63. Armon A., Graur D., Ben-Tal N. (2001) ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 307, 447–463 [DOI] [PubMed] [Google Scholar]
- 64. Stern A., Doron-Faigenboim A., Erez E., Martz E., Bacharach E., Pupko T. (2007) Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 35, W506–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Doron-Faigenboim A., Stern A., Mayrose I., Bacharach E., Pupko T. (2005) Selecton: a server for detecting evolutionary forces at a single amino-acid site. Bioinformatics 21, 2101–2103 [DOI] [PubMed] [Google Scholar]
- 66. Fry B. G., Wüster W., Kini R. M., Brusic V., Khan A., Venkataraman D., Rooney A. P. (2003) Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J. Mol. Evolution 57, 110–129 [DOI] [PubMed] [Google Scholar]
- 67. Pawlak J., Kini R. M. (2008) Unique gene organization of colubrid three-finger toxins: complete cDNA and gene sequences of denmotoxin, a bird-specific toxin from colubrid snake Boiga dendrophila (Mangrove Catsnake). Biochimie 90, 868–877 [DOI] [PubMed] [Google Scholar]
- 68. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 69. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14, 33–38, 27–38 [DOI] [PubMed] [Google Scholar]
- 71. Vidal N., Rage J. C., Couloux A., Hedges S. B. (2009) Snakes (Serpentes). In “The Timetree of life”. Oxford University Press; (Hedges S. B., Kumar S., eds.), 390–397 [Google Scholar]
- 72. Semple F., Dorin J. R. (2012) beta-Defensins: Multifunctional Modulators of Infection, Inflammation and More? J. Innate Immunity 4, 337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shah A., Bano B. (2009) Cystatins in Health and Diseases. Int. J. Peptide Res. Therapeutics 15, 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Domingues Passero L., de Sousa K. (2010) Snake venoms - a perspective for development of drugs against microbes In: Venoms: Sources, toxicity and therapeutic uses Editors: Gjersoe J, Hundstad S. Advances Biol. Med., 77–95 [Google Scholar]
- 75. Brown R. L., Haley T. L., West K. A., Crabb J. W. (1999) Pseudechetoxin: a peptide blocker of cyclic nucleotide-gated ion channels. Proc. Natl. Acad. Sci. U. S. A. 96, 754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang J., Shen B., Guo M., Lou X., Duan Y., Cheng X. P., Teng M., Niu L., Liu Q., Huang Q., Hao Q. (2005) Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from Naja atra venom that targets the BKCa channel. Biochemistry 44, 10145–10152 [DOI] [PubMed] [Google Scholar]
- 77. Wang B., O'Malley T. M., Xu L., Vite C., Wang P., O'Donnell P. A., Ellinwood N. M., Haskins M. E., Ponder K. P. (2006) Expression in blood cells may contribute to biochemical and pathological improvements after neonatal intravenous gene therapy for mucopolysaccharidosis VII in dogs. Mol. Genet. Metab. 87, 8–21 [DOI] [PubMed] [Google Scholar]
- 78. Yamazaki Y., Brown R. L., Morita T. (2002) Purification and cloning of toxins from elapid venoms that target cyclic nucleotide-gated ion channels†,‡. Biochemistry 41, 11331–11337 [DOI] [PubMed] [Google Scholar]
- 79. Yamazaki Y., Koike H., Sugiyama Y., Motoyoshi K., Wada T., Hishinuma S., Mita M., Morita T. (2002) Cloning and characterization of novel snake venom proteins that block smooth muscle contraction. Eur. J. Biochem. 269, 2708–2715 [DOI] [PubMed] [Google Scholar]
- 80. Pung Y. F., Wong P. T., Kumar P. P., Hodgson W. C., Kini R. M. (2005) Ohanin, a novel protein from king cobra venom, induces hypolocomotion and hyperalgesia in mice. J. Biol. Chem. 280, 13137–13147 [DOI] [PubMed] [Google Scholar]
- 81. Yamazaki Y., Morita T. (2007) Snake venom components affecting blood coagulation and the vascular system: structural similarities and marked diversity. Current Pharmaceut. Design 13, 2872–2886 [DOI] [PubMed] [Google Scholar]
- 82. Pyron R. A., Burbrink F. T. (2012) Extinction, ecological opportunity, and the origins of global snake diversity. Evolution; International Journal of Organic Evolution 66, 163–178 [DOI] [PubMed] [Google Scholar]
- 83. Koludarov I., Sunagar K., Undheim E. A., Jackson T. N., Ruder T., Whitehead D., Saucedo A. C., Mora G. R., Alagon A. C., King G., Antunes A., Fry B. G. (2012) Structural and Molecular Diversification of the Anguimorpha Lizard Mandibular Venom Gland System in the Arboreal Species Abronia graminea. J. Mol. Evol. 75, 168–183 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.