Abstract
Gaining understanding of common complex diseases and their treatments are the main drivers for life sciences. As we show here, comprehensive protein set analyses offer new opportunities to decipher functional molecular networks of diseases and assess the efficacy and side-effects of treatments in vivo. Using mass spectrometry, we quantitatively detected several thousands of proteins and observed significant changes in protein pathways that were (dys-) regulated in diet-induced obesity mice. Analysis of the expression and post-translational modifications of proteins in various peripheral metabolic target tissues including adipose, heart, and liver tissue generated functional insights in the regulation of cell and tissue homeostasis during high-fat diet feeding and medication with two antidiabetic compounds. Protein set analyses singled out pathways for functional characterization, and indicated, for example, early-on potential cardiovascular complication of the diabetes drug rosiglitazone. In vivo protein set detection can provide new avenues for monitoring complex disease processes, and for evaluating preclinical drug candidates.
The application of reductionism and experimental manipulation in the 20th century biological research has generated important insights into functional processes of life. Based on this successful paradigm, researchers rationally dissected multiple underlying molecular mechanisms of “living systems” and efficiently developed drugs. However, drugs or dietary interventions can interfere with numerous proteins in hundreds of different cell types in various tissues, not to mention potential crosstalk on various levels of biological organization. Not surprisingly, conventional in vitro and lengthy preclinical studies that target only specific marker molecules often missed out important but unexpected physiological effects of drug treatment. Although complex biological phenomena such as physiological outcomes of disease treatment depend on various individual molecules, they are based on in vivo network properties, which cannot be adequately described or explained by “parts of the sum” of mechanistic events.
Soft-ionization mass spectrometry (MS) has been widely validated as a tool for precise quantitative analysis of biomolecules (1, 2), and isotope-labeling procedures were introduced to detect protein expression, primarily in cell culture models (3, 4). Previous attempts of using mass spectrometry for protein quantification in mammalian disease models were limited to analysis of a small number of usually abundant proteins, which made comprehensive pathway analysis and physiological outcome prediction impossible (5, 6). Recent technical pilot studies provided extensive information on the protein inventories of different mouse tissues (7, 8), and isotope-labeled mice have been introduced as a resource for accurate protein quantification (9).
The development of diet-induced obesity and diabetes is a complex pathophysiological process involving a number of interacting organs, in which chronic hyperglycemia and hyperlipidemia lead to cumulative damaging effects on metabolic tissues such as skeletal muscle, liver, and adipose tissues. As we show here, disease processes and in particular physiological effects of drug treatment are largely determined by the actual cellular protein expression levels and post-translational modifications of proteins. Whereas analyses of single protein changes were mostly uninformative, quantitative protein set enrichment analysis was an efficient tool to monitor tissue-specific responses of anti-diabetic treatments. This approach allows for investigation of interacting molecular and physiological processes that occur on the pathway level, and enables sensitive, unbiased and robust diagnostic detection of treatments in vivo.
In this pilot study, we compared the effects of the drug rosiglitazone (RSG)1, which has been associated with a number of undesirable side effects (10), and the plant-derived amorfrutin A1 (A1) (11) in diet-induced obesity (DIO) mice. Both compounds' antidiabetic effects appear to be derived from activation of the peroxisome proliferator-activated receptor gamma (PPARγ).
EXPERIMENTAL PROCEDURES
Animal studies were carried out according to internationally approved standards as described recently (11), and have been validated and approved by the State Office of Health and Social Affairs Berlin (LAGeSo). The animals were maintained one per cage under temperature-, humidity- and light-controlled conditions (22 °C, 50% humidity, 12 h light/12 h dark-cycle). The health status and behavior of mice were examined daily. Mice had ad libitum access to food and water. Mice and food were weighed regularly to determine changes in body weight and food intake. Low-fat diet (LFD, D12450B, 10 kcal% fat, 18.0 MJ/kg, ssniff, Soest, Germany) was composed of: 4.1% crude fat, 18.1% crude protein, 26.6% starch, 35.5% sugar/dextrines, 4.7% crude fiber. High-fat diet (HFD, D12492, 60 kcal% fat, 25.3 MJ/kg, ssniff) was composed of: 34.0% crude fat, 24.1% crude protein, 1.1% starch, 23.8% sugar/dextrines, 6.0% crude fiber.
For the therapeutic study, we subjected DIO mice to a short-term treatment. Therefore, 6-week-old male C57BL/6 mice were fed with a HFD for 12 weeks. The mice were then weighed and randomly distributed equally to three groups (n = 13 each). DIO mice were then treated over 3 weeks with 4 mg/kg/d RSG or 100 mg/kg/d A1 or vehicle only. A number of physiological assays such as glucose tolerance or insulin sensitivity tests were performed as described recently. Mice of similar age treated only with LFD were served as healthy controls. Plasma and tissues were collected and stored at −80 °C before use.
For the A1 prevention study, 9-week-old male C57BL/6 mice were weighed and randomly assigned to each treatment. Then the mice were fed over 15 weeks with either LFD, HFD, or HFD with low-dose (37 mg/kg/d) of A1 (HFD+A1prev). A number of physiological tests were performed as described recently (11). After 15 weeks of dosing, fasted mice were sacrificed by cervical dislocation. Plasma and tissues were collected and stored at −80 °C before use.
Tissue Harvest for MS Analysis
To discover reliable changes in proteome expression of important metabolic peripheral target tissues such as visceral adipose-, heart-, and liver-tissue, we used pools of eight mice per treatment cohort. Tissues were dissected, washed in phosphate buffered saline (PBS, pH7.4) and shock frozen in liquid nitrogen. Instead of labeled cell culture references, we preferred 13C6 lysine labeled reference tissues (Silantes, Martinsried, Germany) to certainly compare animal tissues with each other (K and R labeled mice we unfortunately not available). The only exception was the reference for the phosphoproteome, for which we used 13C615N2 lysine and 13C615N4 arginine labeled murine hepatoma cells (Hepa 1–6) obtained from ATCC (Manassas, VA). As recent method developments showed this method can produce a much higher number of phosphopeptides with a tryptic digest versus Lys-C digest (12).
Sample Preparation for MS analysis
Tissues from eight mice were pooled in frozen condition and a lysis buffer containing 4% SDS, 0.1 m DTT, 0.1 m Tris pH 8.0 was added. In general, we analyzed tissue proteins in duplicates. Tissues were homogenized with a FastPrep (3 × 6.5 m/s for 60 s) immediately after lysis to avoid any proteolytic activities. SILAC mouse reference tissues were lysed in the same way. The direct comparison of labeled reference and unlabeled mouse tissues under investigation allows for straightforward quantitative protein analysis, as both tissues types are very similar to each other. Lysates were sonicated for 1 min at the lowest intensity, centrifuged at 15,000 × g and boiled for 5 min. Supernatants were transferred to low protein binding tubes (Eppendorf, Germany).
For protein separation, samples were mixed 1:1 with the SILAC reference samples and about 200 μg protein was loaded onto a 12% SDS gel. After destaining the Coomassie blue stained gel, 18 gel slides from high to low molecular weight were excised and cut into small pieces of no larger than 1 mm3.
The in-gel Lys-C digestion was done as described (13). Each sample was dissolved in 5% acetonitrile, 2% formic acid and (5 μl of 19 μl) were used for LC-MS analysis. Every sample was analyzed in duplicates.
For the liver phosphoproteome 90% of the peptides were sequentially separated and enriched with SCX (strong cation exchange, (3M Purification, USA)) and TiO2 (GL Sciences, Japan): SILAC labeled Hepa 1–6 cells were mixed with equal amounts of protein from liver tissues of LFD, HFD, or HFD +A1prev groups, each contained a total of 15 mg of proteins and precipitated in acetone overnight at −20 °C. Pelleted precipitates were lyophilized and dissolved in 8 m urea with 10 mm Tris pH 8.0. Lys-C digestion (2.5 μg/sample) was performed for 4 h followed by a trypsin digestion (50 μg/sample) in 2 m urea overnight at 37 °C. Peptides were desalted with C18 StepPack columns.
The remaining 10% of the peptides were sequentially separated on a SCX and SAX (strong anion exchange) column (3M Purification). SCX separation was performed according to (14), TiO2 enrichment for phosphopeptides (90% fraction) according to (12), and SAX separation (10% fraction) according to (15). The use of labeled cells allows to up-scale the method in a cost-efficient way (as SILAC mouse tissues are still very expensive). On the other hand, cells from culture do not completely reflect the protein inventory of cells in tissue. For example, as detected by mass spectrometry, FABP1 is highly expressed in liver but only to a very limited degree in Hepa 1–6 cells, rendering normalization for quantitative analysis difficult.
Liquid Chromatography, Tandem Mass Spectrometry, and Data Processing
Liquid chromatography, tandem mass spectrometry (LC-MS/MS) was carried out by nanoflow reverse phase liquid chromatography (RPLC) (Agilent, Santa Clara, CA) coupled online to a Linear Ion Trap (LTQ)-Orbitrap XL mass spectrometer (Thermo-Electron Corp). Briefly, the LC separation was performed using a PicoFrit analytical column (75 μm ID × 150 mm long, 15 μm Tip ID (New Objectives, Woburn, MA)) in-house packed with 3-μm C18 resin (Reprosil-AQ Pur, Dr. Maisch, Germany). Peptides were eluted using a nonlinear gradient from 2 to 40% solvent B over 160 min at a flow rate of 200 nL/min (solvent A: 97.9% H2O, 2% acetonitrile, 0.1% formic acid; solvent B: 97.9% acetonitrile, 2% H2O, 0.1% formic acid). A 1.8kV voltage was applied for nanoelectrospray generation. A cycle of one full FT scan mass spectrum (300–2000 m/z, resolution of 60,000 at m/z 400) was followed by 10 data-dependent MS/MS scans acquired in the linear ion trap with normalized collision energy (setting at 35%). Target ions already selected for MS/MS were dynamically excluded for 60 s.
Primary Mass Spectrometry Data Analysis
Raw files for each LC-MS/MS experiment were submitted to MaxQuant v1.0.13.13 for database searching (16), based on Mascot v2.2.2. For the liver phosphoproteome (in combination with all other liver samples), we used MaxQuant version 1.2.2.5, based on Andromeda. The mouse database (ipi.MOUSE.v3.87 from 20 Sep. 2011, 91.464 entries) was used for database searching. The cutoff value was set to 1% false discovery rate (FDR) to be 99% confident at the peptide level. As SILAC modification we used 13C6-labeled lysine, but in the case of the phospho proteome we used: 13C615N2 lysine and 13C615N4 arginine. Following chemical modifications were selected as variable modifications during database search: protein N-terminal acetylation and methionine oxidation, for the phospho proteome: methionine oxidation and phospho STY. Carbamidomethyl C was used as fixed modification. Lys-C or trypsin was set with a maximum of two missed cleavage sites. Mass tolerance for precursor and fragment ions was 0.5 Da and 7 ppm (for MaxQuant v1.0.13.13) and 0.5 Da and 6 ppm (MaxQuant version 1.2.2.5). Known contaminants are indicated in MaxQuant result lists. Quantification was performed with at least two identified peptides. Protein ratios were calculated as the exponent of the median of the log-transformed evidence ratios by MaxQuant, no minimum thresholds were set and no outliers were removed. Thereby, SILAC protein ratios were determined as the median of all peptide ratios assigned to the protein. Scatter plot analysis of each treatment using the results derived from two samples revealed in general high data correlation of the duplicates (Fig. S1), as was also observed in previous studies (17). In general, MaxQuant normalized H/L SILAC ratios were used, except data for adipose tissue, which were manually normalized. MaxQuant Viewer v1.2.2.5 was used to extract annotated spectra of all phosphorylated peptides of the liver tissue.
Protein expression changes between samples were calculated using this formula:
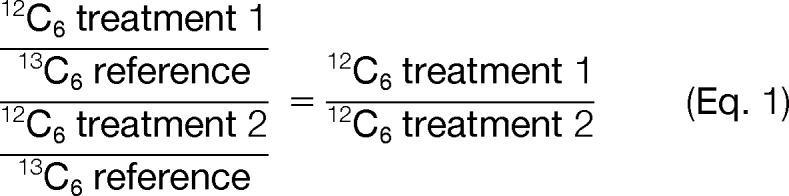 |
Raw- and MaxQuant processed data including annotated phoshopeptides can be downloaded via: ftp://PASS00201:BG7335ub@ftp.peptideatlas.org/
Secondary Protein Data Analyses
For fine-tuning of pathways analyses, we performed protein set enrichment analysis (PSEA) using the GSEA tool ((18, 19), v2.07, http://www.broadinstitute.org/gsea/index.jsp) to analyze whether an a priori defined set of proteins revealed statistical significance and concordant differences between two diet regimes or treatments. For PSEA, the following parameters were chosen if not otherwise noted: 1000 protein set permutations, weighted enrichment statistics, minimal gene set size of 5, and log2 ratio metric with preranking. RNA expression data and protein SILAC ratios were analyzed using the Reactome database (version 3.0, 430 pathways) from the Molecular Signature Database (MSigDB). We considered regulated pathways only as statistically significant, if the FDR was ≤0.25. Heatmaps were carried out with Mayday 2.8 (20). For presenting different treatment effects in heatmaps, the normalized enrichment score (NES) for a pathway was adjusted with the appropriate FDR as follows: adjustedNES = (1-FDR) × NES.
Protein distance matrix (PDM) analyses included the protein expression data of every treatment, and the expression value for each protein was translated to a vector in Euclidean space, thus, the complete expression profile was collapsed to a high-dimensional vector sum. Pairwise distances were calculated for comparison of two treatments. The Euclidean distance between the vector sums of two different treatments was therefore a measure of similarity between the protein expression profiles. PDM analyses were conducted using the MeV 4.3 software tool (21).
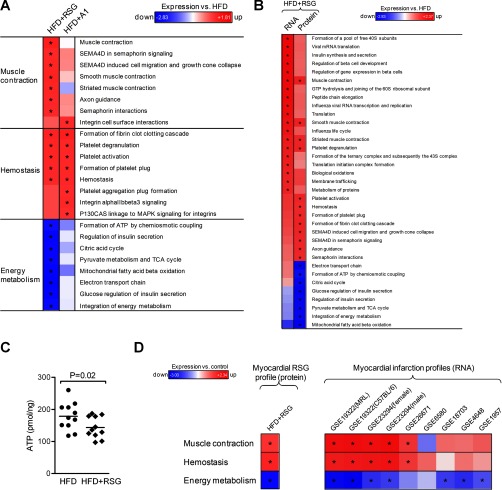
Meta-analysis on the SILAC data of RSG-treated heart samples were performed with publicly available expression data of heart diseases. We therefore created lists of proteins that contributed to the enrichment of the pathway clusters “muscle contraction,” “hemostasis,” and “energy metabolism” as detected by PSEA (Fig. 4A). Expression of these three pathways was analyzed in gene expression data of studies related to myocardial infarction in rodents (GSE1957, GSE4648, GSE6580, GSE18703, GSE19322, GSE23294, and GSE26671), which were extracted from the NCBI Gene Expression Omnibus (GEO) database. Additionally, our SILAC data of RSG-treated heart samples were tested for connection with expression signatures of known drugs using the Connectivity Map (22), which is a collection of gene expression profiles from cultured cells treated with small molecules in combination with a pattern-matching software. We therefore used a merged protein list of the three pathway clusters “muscle contraction,” “hemostasis,” and “energy metabolism” detected by PSEA (Fig. 4A) to create a query signature for the Connectivity Map tool. This SILAC-based RSG profile was then tested for correlation with gene expression signatures of 1310 small molecules. To investigate if these connected drugs have been previously reported to induce myocardial defects, we extracted a list of drugs that are linked to “cardiac failure” or “myocardial infarction” from the SIDER database (23).
Fig. 4.
Protein and RNA pathway analysis of heart tissue. A, Pathway-level regulation of protein expression in the heart after treatment of HFD-fed mice with rosiglitazone (HFD+RSG) or amorfrutin A1 (HFD+A1). Regulation is displayed as FDR-adjusted enrichment score relative to HFD-fed mice. Protein sets were filtered with FDR ≤ 0.05 for one condition. B, Comparison of regulated pathways on RNA and protein level in the heart of RSG-treated mice. C, Cellular ATP concentration (-19% with RSG, n = 11 each group), normalized to total DNA. D, Comparison of the RSG-induced myocardial protein expression profile (left) with published RNA expression data related to myocardial infarction (right). The RSG protein profile (left) was determined in mice treated for 3 weeks with RSG by mass spectrometry and subsequently subjected to PSEA, whereas the RNA myocardial infarction profiles derived from severely diseased animals (right) were extracted from the NCBI gene expression omnibus (GEO) database. Regulation is presented relative to HFD-fed or uninfarcted control mice, respectively. *, p ≤ 0.05.
Kinase enrichment analysis (KEA) (24) was performed using a web-based tool with an underlying database to link lists of mammalian proteins with the kinases that phosphorylates them. KEA considers several kinase-substrate databases to calculate kinase enrichment probability based on the distribution of kinase-substrate proportions in the respective background database compared with kinases found to be associated with a user input list of proteins. We performed KEA using data of two-fold regulated peptides with one phospho-site.
To extract phosphorylation motifs from our large tandem mass spectrometry-based data sets, we performed kinase motif search (motif X search (v1.2 10.05.06, http://motifx.med.harvard.edu/motif-x.html)).
For investigating and visualizing of enriched pathways in the phosphoproteome of HFD-fed versus LFD-fed mice, we mapped hypo- and hyperphosphorylated proteins (ratio ≥ 1.33) with the Ingenuity Pathway Analysis (IPA) Software (Ingenuity Systems, CA).
RNA Expression Analysis
Total RNA was isolated and purified using TRIzol reagent (Invitrogen) with subsequent usage of the RNeasy Mini Kit (Qiagen, Germany) according to the manufacturers. Tissues were lysed and homogenized in TRIzol reagent with 5 mm steel beads at 20 Hz for 4 min (TissueLyser, Qiagen). Genomic DNA was digested on column using the DNase-Set (Qiagen, Valencia, CA). RNA quality was determined by the Bioanalyzer 2100 (Agilent, Santa Clara, CA). Biotin-labeled cRNA was generated from the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX) following the manufacturer's instructions. Cy3-stained cRNA was hybridized onto MouseWG-6 v2.0 Expression BeadChips (Illumina, Eindhoven, The Netherlands). Scanning was executed on Illumina BeadStation 500 platform. Reagents were applied according to the manufacturer's protocols. Samples were hybridized in biological triplicates. All basic expression data analyses were carried out using GenomeStudio V2011.1 (Illumina). Raw data were background-subtracted and normalized applying the cubic spline algorithm. Processed data were subsequently filtered for significant detection (p value ≤ 0.01) and differential expression versus vehicle treatment according to the Illumina t test error model, and were corrected according to the Benjamini-Hochberg method (p value ≤0.05) of the GenomeStudio software. Gene expression data were submitted in MIAME-compliant form to the NCBI Gene Expression Omnibus database (GSE38856).
Qualitative correlation was calculated as the part of the genes that were regulated in the same direction by RNA and protein expression. A correlation of 50% was thus considered as correlated only by chance. For correlation analyses between RNA and protein expression on single gene level, differentially expressed genes or proteins that were detectable in both transcriptome and proteome were filtered for candidates with fold change ≥1.33 or ≤0.75 versus vehicle control treatment. For correlation analyses on the pathway level, we compared the Reactome pathway regulation determined by GSEA for RNA and PSEA for protein expression, and included only detectable regulated pathways with FDR ≤ 0.05 for RNA or protein. Correlation analyses were done for each treatment and tissue.
Mitochondrial DNA
DNA of heart samples was isolated with the QIAamp Mini Kit (Qiagen). Mitochondrial DNA content was subsequently determined by quantitative real-time PCR as described recently (11). Relative mitochondrial DNA content was quantified by the 2−ΔΔCt method and adjusted with primer efficiency. Primers of the following sequences were used: mtDNA specific, COX1, forward 5′-TGCTAGCCGCAGGCATTAC-3′, reverse 5′-GGGTGCCCAAAGAATCAGAAC-3′, and ND1, forward 5′-CCGCAAGGGAAAGATGAAAGAC-3′, reverse 5′-TCGTTTGGTTTCGGGGTTTC-3′; nuclear DNA specific, HK2, forward 5′-GCCAGCCTCTCCTGATTTTAGTGT-3′, reverse 5′-GGGAACACAAAAGACCTCTTCTGG-3′, and NDUFV1, forward 5′-CTTCCCCACTGGCCTCAAG-3′, reverse 5′-CCAAAACCCAGTGATCCAGC-3′.
Physiological Parameters
To measure liver triglyceride levels, tissues were weighed and disrupted at a concentration of 44 mg/ml in 100% isopropanol. Disruption was performed with 5 mm steel beads at 20 Hz for 4 min (TissueLyser, Qiagen). After centrifugation for 10 min at 20,000 ×g at 4 °C, the supernatants were collected and measured in the colorimetric triglyceride assay (BioVision, Mountain View, CA). To measure ATP in hearts, tissues were weighed and disrupted at a concentration of 50 mg/ml in ATP assay buffer (BioVision). Disruption was performed with 5 mm steel beads at 20 Hz for 4 min. After centrifugation for 10 min at 20,000 × g, 4 °C, the supernatants were collected and deproteinized using the perchloric acid precipitation method (BioVision) according to the manufacturer's instructions. Finally, ATP levels in heart lysates were measured in a colorimetric quantification kit (BioVision) and were normalized by DNA as quantified by PicoGreen assay (Quant-iT, Invitrogen, USA).
For determination of liver glycogen, tissues were weighed and disrupted at a concentration of 28 mg/ml in 200 mm sodium acetate (pH 4.8) using the TissueLyser (Qiagen), heated to 70 °C for 10 min, and then centrifuged for 10 min at 6000 × g and 4 °C. Three μl of sample supernatants were added to 57 μl of 200 mm sodium acetate (pH 4.8) without or with 27 U/ml amyloglucosidase (A1602, Sigma-Aldrich) and incubated at 41 °C for 2 h. Afterwards samples were neutralized with 15 μl of 280 mm sodium hydroxide, and free glucose was measured with a colorimetric glucose assay kit (Invitrogen). For determination of TNFα concentrations in liver, tissues were weighed and disrupted at a concentration of 100 mg/ml in a tissue lysis buffer containing 20 mm Tris, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm EDTA, 0.1% SDS and protease inhibitor mixture (Roche), using disruption with 5 mm steel beads at 20 Hz for 4 min (TissueLyser). Samples were centrifuged for 10 min at 20,000 g and 4 °C, and supernatants were measured in a TNFα ELISA (TNFα ELISA Ready-SET-Go, eBioscience, NatuTec, Frankfurt, Germany) and normalized against DNA content measured with the PicoGreen assay (Quant-iT, Invitrogen). Plasma alanine transaminase (ALT) was quantitatively measured using a colorimetric quantification kit (Biovision, BioCat), according to the manufacturer's instructions.
Mitochondrial Sample Preparation for Western Blot Analysis and Enzyme Measurements
Mouse heart tissues (10–30 mg) from 13 mice fed either a high-fat diet (HFD) or a HFD plus rosiglitazone (4 mg/kg/d) were homogenized with a tissue disintegrator (Ultraturrax, IKA, Staufen, Germany) in extraction buffer (20 mm Tris-HCl, pH 7.6, 250 mm sucrose, 40 mm KCl, 2 mm EGTA) and finally homogenized with a motor-driven Teflon-glass homogenizer (Potter S, Braun, Melsungen, Germany). The homogenate was centrifuged at 600 × g for 10 min at 4 °C. The supernatant containing the mitochondrial fraction was diluted 1/100 for measuring enzyme activities and Western blot analysis.
Western Blot Analysis
Ten microgram protein/lane from the previous step were loaded onto 15% polyacrylamide gels. Nitrocellulose membranes were blocked with 1% blocking solution and developed with the Lumi-LightPLUS Western blotting Kit (Roche). Western blot analysis was performed with a rabbit polyclonal antibody against the mitochondrial protein ATP5A1 (14676–1-AP, Proteintech Group, IL). A mouse monoclonal antibody against β-actin (β-Actin (C4) sc-47778, Santa Cruz Biotechnology, TX) was applied as loading control. Western blot images were analyzed using GelAnalyzer 2010a (www.gelanalyzer.com).
Mitochondrial Enzyme Measurements
Enzymatic activity of the citrate synthase was determined according to Srere et al. (24), with the following modifications to the assay buffer: 50 mm HEPES pH 7.6, 2 mm MnCl2, 4 mm dl-isocitrate and 0.1 mm NADP (nicotinamide adenine dinucleotidephosphate). Oligomycin sensitive ATPase activity of complex V was determined using buffer conditions described by Rustin et al. (26), but by applying sonification of the whole reaction mixture for 10 s with an ultra-sonifier (Bio cell disruptor 250, Branson, Vienna, Austria) at the lowest energy output (27). The concentration of oligomycin was 3 μm. All spectrophotometric measurements (Uvicon 922, Kontron, Milano, Italy) were assayed in duplicates and performed at 37 °C.
Statistical Analyses
If not stated otherwise, statistical significance was determined by unpaired two-tailed Student's t test for single comparisons and one-way ANOVA with Dunnett's post-hoc test for multiple comparisons. Pearson correlation analyses were carried out using GraphPad Prism 5.0. A p value ≤ 0.05 was defined as statistically significant. For gene and protein set enrichment analyses a FDR ≤ 0.25 was considered as statistically significant.
RESULTS
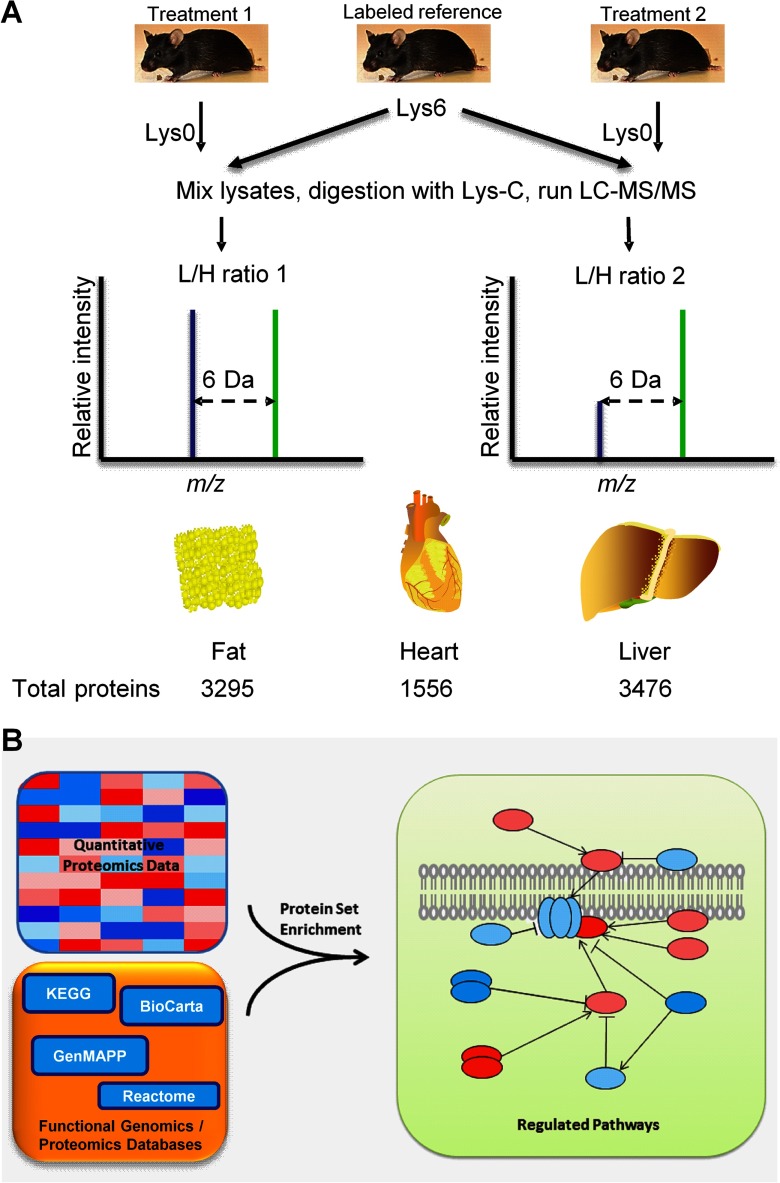
We analyzed protein expression from tissues of mice fed with LFD or HFD, as well as HFD supplemented with either RSG or A1. As a reference for quantitative mass spectrometry, we used isotope-labeled mouse tissues or cells in combination with high precision mass spectrometry. In general, we detected several thousands of proteins per tissue and identified several hundreds of up- or down-regulated proteins per experiment (Fig. 1A).
Fig. 1.
General work scheme for sample- and data analysis. A, Proteomic workflow. Lysates of tissues of interest from an isotope-labeled reference mouse are equally mixed with unlabeled treated or mock-treated tissues. Proteins are extracted, separated via gel electrophoresis, and digested with the protease Lys-C. Resulting peptides are analyzed on a LC-MS/MS system. MaxQuant software is used to calculate differentially expressed proteins (16). Using 200 μg amounts of protein extracts derived from tissues and simple gel-based protein fractionation, we could detect 3295 proteins in the adipose tissue, 1556 proteins in the heart tissue, and 3476 proteins in the liver tissue. Not surprisingly, the number of identified proteins in the heart tissue was comparably low, as we detected many abundant large structural proteins. Depending on treatment conditions, we observed differential expression of several hundreds of up- or down-regulated proteins per experiment. Although individual proteins showed subtle variation, sets of functionally related proteins revealed insights in physiology and metabolic regulation in the differentially treated DIO mice. We further investigated the phosphoproteome of fatty and chronically inflamed liver of DIO mice. These sophisticated analyses required 15 mg of protein and column-based peptide enrichment. In total, 6956 liver proteins were identified, 3862 belonging to the phosphopeptide enrichment fraction. B, We adopted the widely accepted functional genomics principle of gene set enrichment to explain physiological effects in mice derived from concerted actions of individual proteins. Protein sets or pathways can be extracted from quantitative mass spectrometry results using appropriate maintained databases, as is exemplified in the figure.
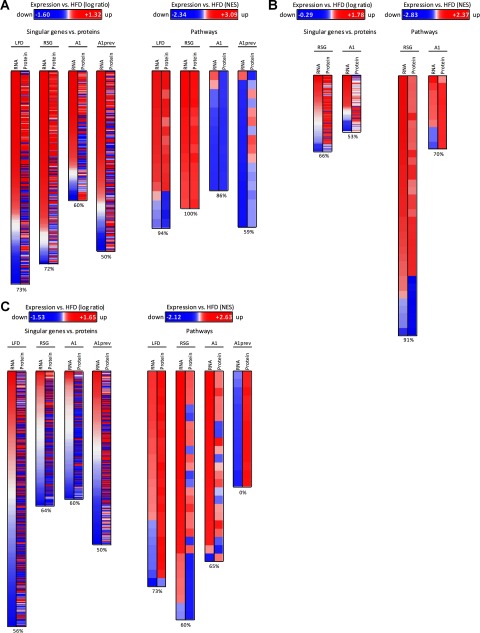
To get insight into the potentially differential dynamics of RNA and protein expression, we first analyzed the correlation between genes and proteins or pathways that were either both up-regulated or both down-regulated. Comparing the expression of single RNAs with the corresponding proteins, we observed a mean correlation of 60% from all analyzed tissues and treatments (Fig. 2), suggesting that the level of expressed proteins only marginally correlated with the level of RNA transcripts (28).
Fig. 2.
Correlation between gene and protein expression from different treatments on singular gene/protein or pathway level in visceral white adipose tissue (A), heart (B), and liver (C). Mice were treated with low-fat diet (LFD) or high-fat diet (HFD) and with rosiglitazone (RSG) or amorfrutin A1 (A1) after HFD feeding or amorfrutin A1 (A1prev) during the HFD feeding. In heart, we focused on differential effects of RSG and A1 treatment. Regulation is presented relative to HFD-fed mice. Differentially expressed genes/proteins, which were found in both transcriptome and proteome data sets, are displayed as relative change in expression in logarithmic scale (change≥1.33 for RNA or protein). Pathway regulation was analyzed using PSEA and is displayed as normalized enrichment score (FDR≤0.05 for RNA or protein). Percentage values represent qualitative correlation between gene and protein expression. Whereas on single gene level treatment-induced expression of RNA and protein only marginally correlated to each other, collapsing to biological pathways revealed high correlation between RNA and protein regulation, partially up to 100% in white adipose tissues (A). However, in the heart (B) RSG-treatment led to pathway down-regulation exclusively on the protein level, which was subsequently biochemically validated. In the liver (C), for A1prev we also observed inverse expression tendencies of RNA and respective protein pathways. However, only protein expression data were statistically significant (14 Reactome pathways below FDRs of 0.05), whereas RNA expression data indicated no statistical significance (FDRs ranging from 0.64–1). Most proteins detected contributed to ribosomal biogenesis and translation pathways, suggesting that A1prev led to increased translational processes in the liver during HFD-feeding.
As is summarized for the various treatments in supplemental Table S1, a large fraction of detected proteins was slight regulated (ratio 0.75 to 1.33); only few proteins were three-fold up- or down-regulated. To extract relevant molecular pathways from protein expression data of slightly regulated individual proteins, we applied protein set enrichment analysis (PSEA, Fig. 1B) (29, 30), an extension of gene set enrichment analysis (GSEA) (18). This approach allows detecting the effects of coordinated differential expression of groups of functionally related molecules, which show only subtle changes at the level of individual proteins. Applying this rationale, protein pathways correlated better with respective transcriptomic pathways than individual genes and proteins, leading to a mean correlation of 70% (partly even 100%) over all analyzed tissues and treatments (Figs. 2 and supplemental Fig. S2). The nonetheless rather small correlation in expression of corresponding sets of RNAs and proteins indicated different dynamics of production and degradation of these biomolecules.
Visceral Adipose Tissue
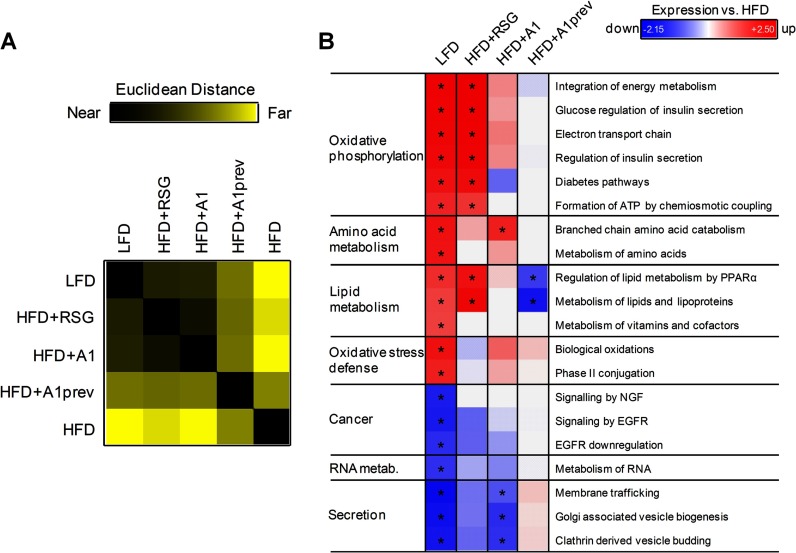
We started our protein set analyses with visceral adipose tissue, because physiological effects of ligand-based PPARγ activation are triggered in white fat cells (31). To determine the treatment effects of RSG and A1 on protein expression, we performed protein distance matrix (PDM) analyses. The PDM of our adipose tissue samples clearly showed a large difference in protein expression between nondiabetic (LFD) and obese (HFD) mice (Fig. 3A, supplemental Table S2). Treatment of obese mice with RSG or A1 strongly shifted the expression profile toward that of nondiabetic (LFD) mice. PSEA revealed a strong relative down-regulation of oxidative phosphorylation signaling after HFD feeding (FDR<0.25, Fig. 3B), but which was reconstituted by treatment with RSG and also, less efficiently, with A1. Interestingly, HFD feeding also resulted in a significant decrease in the protein expression of key enzymes involved in the degradation of branched-chain amino acids (BCAAs) (supplemental Fig. S3A). Elevated BCAA levels can induce insulin resistance in fat cells via mTOR signaling (32). Treatment of DIO mice with A1 effectively counteracted the HFD-induced down-regulation of catabolic BCAA protein pathways.
Fig. 3.
Protein pathway analysis of visceral white adipose tissue. A, Protein distance matrix of protein expression profiles in white adipose tissue from low-fat diet (LFD) or high-fat diet (HFD)-fed mice treated with rosiglitazone (RSG) or amorfrutin 1 (A1) or treatment by amorfrutin 1 during HFD feeding (A1prev). Squares show the distance of two conditions in Euclidean space, ranging from exactly the same profile (black) to completely different (yellow). Mass spectrometry ratios were filtered with a minimal change in protein expression of 1.33. B, Regulation of pathways on protein level in white adipose tissue relative to HFD-fed mice. Mass spectrometry ratios from (A) were used for protein expression analysis and pathway regulation was explored with subsequent PSEA against the Reactome database. PSEA is applied to determine whether any defined protein sets are enriched from a list of proteins ordered according to expression differences between two classes using Kolmogorov-Smirnov running sum statistics as described in Supplemental Material. Regulation is displayed as FDR-adjusted enrichment score and was normalized to HFD-fed mice for technical reasons. Protein sets were filtered with FDR ≤ 0.25 for LFD treatment.
Notably, HFD-mice exhibited reduced expression of oxidative stress defense pathways (Fig. 3B and supplemental Fig. S3B), including lowered expression of glutathione-S-transferases and cytochrome P450 enzymes. Strikingly, only A1 (but not RSG) treatment rescued in part these changes to the oxidative stress defense system, which was accompanied by a decrease in body weight (supplemental Fig. S4A). Additionally, the proteins of secretory pathways were significantly up-regulated in HFD-fed DIO mice (Fig. 3B). Reversal of these effects was achieved by A1, and correlated again with reduction of body weight of A1-treated mice (supplemental Figs. S4B, C). Furthermore, A1 treatment of DIO mice exclusively led to down-regulation of ribosomal biogenesis and translation (supplemental Fig. S5A) via reduced expression of ribosomal proteins (supplemental Fig. S5B). Since increased ribosomal biogenesis has been correlated with elevated nutrient availability and tumorigenesis (33), these results may contribute to understanding the mechanisms of potential antiproliferative effects attributed to selective PPARγ activation (34).
We observed strong effects of PPARγ ligand-induced regulation. Characteristically, the full agonist RSG was efficient in shaping gene and protein expression profiles during HFD to a LFD status (Fig. 3B), but with the concomitant disadvantage of unspecific expression of proteins that can contribute to increased weight gain such as fatty acid binding proteins as FABP1 (fold up-regulation in RSG: 2.54; A1: 0.59), fatty acid transporters as FATP (fold up-regulation in RSG: 2.97; A1: 1.48) or fat storing proteins as FACL2 (fold up-regulation in RSG: 1.47; A1: 0.57).
In summary, treatment of obese mice with RSG and A1 showed in visceral white adipose tissue that protein and RNA expression profiles shifted back to the state of nondiabetic mice. Both treatments displayed beneficial effects. In contrast, preventive A1 treatment had no significant effect in this tissue.
Heart Tissue
RSG was withdrawn from the pharmaceutical market in 2011, several years after its release by the FDA because of an increased cardiovascular risk, in part as a result of fluid retention caused by impaired kidney function, which can result in chronic stress of the heart (35, 36). To demonstrate the potential diagnostic strengths of detecting protein pathways, we investigated whether our approach would enable early prediction of adverse effects in the heart tissues of RSG- or A1-treated DIO mice. In RSG-treated DIO mice, hemostasis, muscle contraction, and cytoskeletal pathways were remarkably impaired (Fig. 4A). For example, RSG strongly induced the expression of myosins and tropomyosins (Fig. 5A) as well as axon guidance pathways and semaphorin interactors. These changes were indicative for cardiac hypertrophy and may provide another link to cardiovascular disease phenotypes (37). In contrast to RSG, in general A1 did not induce significant changes in the heart muscle contraction protein expression data, consistent with previous reports that A1 treatment did not induce fluid retention (11) (Fig. 4A and Fig. 5A). Only the hemostasis pathways were significantly up-regulated in both, RSG and A1 treated mice (Fig. 4A and Fig. 5B).
Fig. 5.
Expression of heart proteins involved in muscle contraction (A) or hemostasis (B) after treating mice with high-fat diet with rosiglitazone (HFD+RSG) or amorfrutin A1 (HFD+A1). Protein expression is presented relative to HFD-fed mice. C, Comparison of RNA and protein expressions in energy metabolism of RSG-treated mice.
Although correlation between RNA and protein expression remained high at the pathway level, we detected several striking differences between the heart proteome and its transcriptome (Fig. 4B). We observed strongly down-regulated pathways that are involved in the citric acid cycle and oxidative phosphorylation after RSG (but not A1) treatment of HFD mice only in the proteome, not at the transcriptome level (Fig. 4A and Fig. 5C).
A series of Western blot and enzymatic experiments confirmed the mitochondrial protein sets down-regulation detected by mass spectrometry, which in part resulted from the reduced number of mitochondria in the heart tissue (supplemental Figs. S6A-6C), leading to 19% reduction of ATP in the heart of RSG-treated HFD-mice (Fig. 4C).
Using the connectivity map approach (22) we further compared the RSG-induced regulation of the characteristic protein pathway sets with gene expression profiles of drugs with side effects as “cardiac failure” or “myocardial infarction.” Interestingly, we found striking overlap between data from our proteomic analysis of mice subjected to only 3 weeks of RSG treatment with the transcriptomic data reported for severely heart-diseased rodents (Fig. 4D and supplemental Table S3). Concordantly, 8 of 10 drugs were significantly correlated to our RSG-induced protein expression data from the murine heart (supplemental Table S3).
In summary, heart tissue showed pathway regulations upon RSG treatment, which were indicative for heart failure, like up-regulations of hemostasis and cytoskeleton and down-regulation of mitochondrial energy metabolism. These affected pathways were unchanged at the RNA expression level. Thus, protein set analysis in the heart was predictive for potential systemic cardiovascular complications of RSG treatment at an early preclinical stage and can therefore be used as a method for drug testing. Interestingly, the natural A1 compound showed no maleficent changes of the cytoskeleton and mitochondrial energy metabolism in the heart.
Liver Tissue
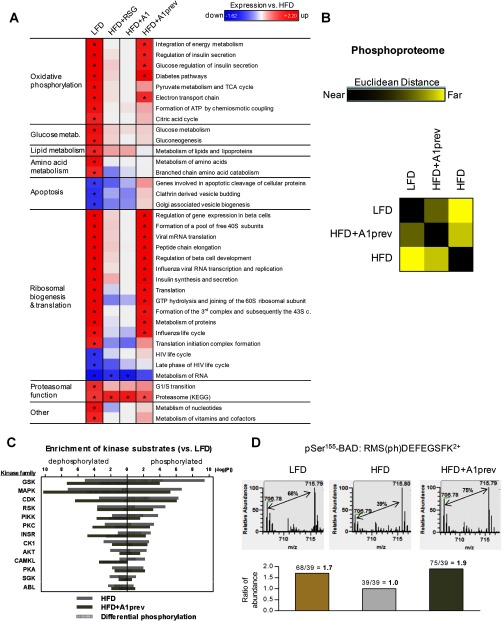
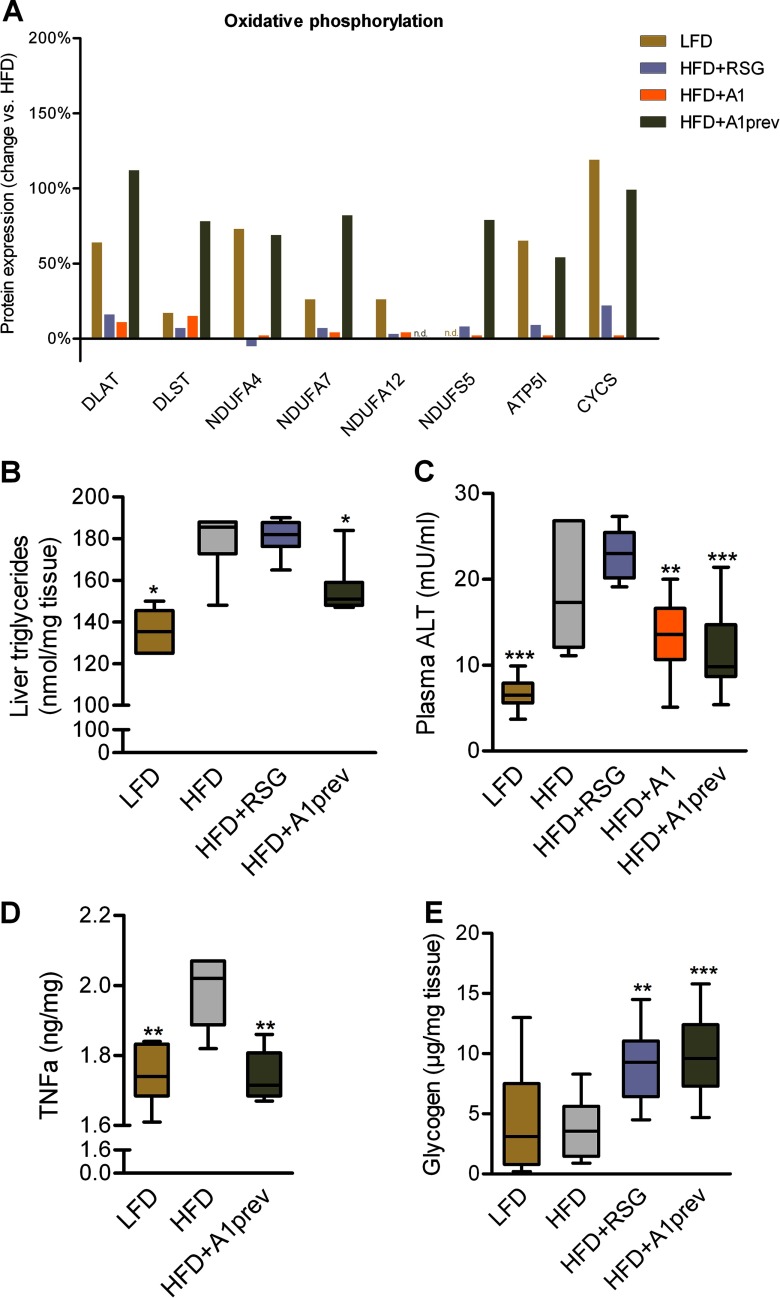
Diet-induced obesity usually leads to liver steatosis because of excessive storage of fat in central organs (37). In the livers of HFD-fed mice, we observed down-regulation of proteins involved in oxidative phosphorylation and citric acid cycle (Figs. 6A and 7A). Whereas RSG and A1 treatment showed no significant influence on protein expression, preventive application of A1 during HFD feeding minimized the impairment of these key metabolic pathways. HFD-induced obesity led amongst others to an up-regulation of apoptosis proteins and concomitant reduction of proteins involved in ribosomal biogenesis and translation, indicating liver injury as observed in nonalcoholic steatohepatitis (NASH) (39), which was consistent with detected pertinent physiological liver parameters (Figs. 7B–7E). HFD-induced obesity further led to significant down-regulation of proteins involved in proteasomal function (Fig. 6A), in agreement with recent studies that reported the role of proteasomal impairment in obesity (40).
Fig. 6.
Protein pathway analysis of liver tissue and its phosphoproteome. A, Regulation of protein pathways in the liver. Pathway regulation was analyzed by PSEA. Regulation is displayed as FDR-adjusted enrichment score and was normalized to HFD-fed mice. Protein sets were filtered with FDR ≤ 0.25 for LFD treatment. As shown in B–D, quantitative mass spectrometry analysis can further provide valuable insights into phosphorylation dynamics. B, Phosphopeptide distance matrix (PPDM) of the phosphoproteome of liver samples. Squares show the distance of two conditions in Euclidean space, ranging from exactly the same profile (black) to completely different (yellow). Ratios of phosphorylated and nonphosphorylated peptides were determined by mass spectrometry and restricted to peptides with a ratio ≥2 or ≤0.5 for one condition (528 peptides final). C, Kinase enrichment analysis of the liver phosphoproteome from HFD-fed mice with or without A1 preventive treatment versus healthy LFD-fed mice. Measured kinase substrates were distinguished between dephosporylated peptides and phosphorylated peptides on treatment. Displayed differential phosphorylation is the difference between hyper- and hypophosphorylation. Only kinase families with an enrichment p value ≤ 0.01 are shown. D, Mass spectra from a phosphorylated BAD peptide (Uniprot ID: Q61337). Differentially down-regulated phosphorylation of BAD at Ser-155 during HFD could be reversed by amorfrutin supplementation leading to phosphorylation levels similar as during LFD feeding. The light peak at m/z 706.78 is originated from the unlabeled mouse, whereas the heavy peak at m/z 715.79 represents the 13C-lysine labeled reference mouse. Thus, ratios between LFD/HFD and HFD+A1prev/HFD could be calculated.
Fig. 7.
Effects of HFD feeding and treatment on different metabolites. A, Expression of proteins involved in oxidative phosphorylation in liver after treatment of mice with low-fat diet (LFD) or high-fat diet (HFD) with rosiglitazone (HFD+RSG) or amorfrutin A1 after HFD feeding (HFD+A1) or amorfrutin A1 during HFD feeding (HFD+A1prev) as displayed in Fig. 6A. B, Effect of HFD feeding and treatment on liver triglycerides. Data are expressed as mean ± S.E. (n = 6–7 each group). *, p ≤ 0.05 versus HFD. C, Effect of HFD feeding and treatment on plasma alanine transaminase (ALT) levels. D, Effect of HFD feeding and treatment on liver TNFα protein concentration (n = 6–7 each group). E, Effect of HFD feeding and treatment on liver glycogen (n = 7–12). Data are expressed as mean ± S.E. * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001 versus HFD.
Liver Phosphoproteome
To further study inflammatory processes in the liver, we analyzed the liver phosphoproteomes of LFD, HFD, and HFD+A1prev mice, and identified 8732 unique phosphorylation sites in phospho-enriched samples, belonging to 6956 proteins. We observed mostly Ser phosphorylation (7691, 88.1%), followed by Thr (977, 11.2%) and Tyr (64, 0.7%). In the case of HFD versus LFD amino acid motifs surrounding each phosphorylation site were 46.5% proline-directed (.SP.) and 20.5% (R..S) motif-directed.
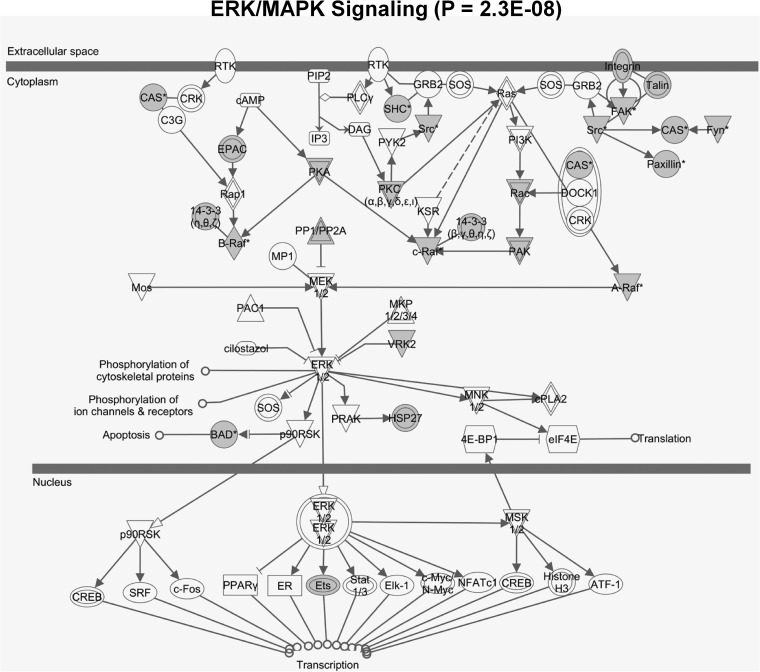
HFD-triggered phosphorylation of liver proteins was partly suppressed by preventive treatment with A1 (Fig. 6B, supplemental Table S4). Differential enrichment analysis revealed characteristically increased phosphorylation of kinase substrates that are known to be involved in insulin-resistant states, most importantly substrates of glycogen synthase kinase β (GSKβ) (Figs. 6C and supplemental Fig. S7). GSK is normally inactivated by phosphorylation via nutrient signaling pathways (for example via mTOR and in particular AKT kinase signaling). A1 prevention suppressed this protein signaling axis concomitant with significantly increasing storage of liver glycogen compared with HFD livers (Fig. 7E), suggesting a beneficial switch from fat to glycogen storage in the liver. HFD led to further differential regulation of phosphorylation pathways (as is exemplarily shown for the ERK/MAPK protein signaling network in Fig. 8), and specifically for decreased phosphorylation of the apoptosis factor BAD at serine 155, which was efficiently reconstituted by preventive A1 treatment (Fig. 6D). Phosphorylation of BAD at serine 155 inhibits association with Bcl-2 and thus promotes cell survival (41, 42). These data suggest liver-protective effects of preventive A1 treatment by modulating phosphorylation pathways and rescuing BAD-mediated cell death.
Fig. 8.
Enrichment of hyper- and hypophosphorylated peptides in the ERK/MAPK signaling pathway in the phosphoproteome of murine liver on treatment with high-fat versus low-fat diet. Proteins with phosphopeptide ratios ≤0.75 or ≥1.33 were marked in gray. The ERK/MAPK pathway includes the apoptosis initiating BAD protein (Fig. 6D). Enrichment analysis and visualization were performed with Ingenuity Pathway Analysis (IPA).
In summary, in liver RSG and A1 feeding to obese mice had no significant impact, but strikingly preventive A1 substitution saved the liver from developing HFD-induced steatosis.
DISCUSSION
Protein Set Analyses
Gene set enrichment analysis is based on the concept that changes in gene expression manifest at the level of coregulated or interacting genes, rather than individually. This functional genomics concept proved to be very powerful, as it is based on a fundamental principle of biological organization. Single-gene or as shown here single-protein events are rather important when the individual gene effect is strong and the variance is small across individuals, which is rarely the case in robust homeostatic or physiological systems, or in many common disease states. In these cases, complex disorders typically result from slight variation in the expression of activities of multiple genes or proteins. PSEA as applied in this study, provides an adequate framework to investigate protein state changes operating at a higher level of organization. PSEA in combination with quantitative mass spectrometry is an adequate tool to describe functional links or causality of complex physiological crosstalk in an in vivo context. Moreover, the method provides unbiased insights to pinpoint to pathways underlying physiological changes. The integration of multiple proteins in coregulated sets further provides diagnostic robustness for preclinical evaluation of drug candidates. In contrast to GSEA, PSEA has the advantage to detect expression changes on the protein level, which in general provides (more) relevant information with regard to functional outcomes. Second, as shown in our study, protein expression change analyses can be complemented by analyses of post-translational modification to extract regulated signaling pathways.
The observed subtle expression patterns of proteins, and additionally the consistence of RNA and protein expression on the pathway, but not necessarily on the individual gene protein level, support the hypothesis that physiological effects emerge as a new, partly unpredictable property of the context-specific interaction of various biomolecules in vivo (43). Moreover, the detected characteristic stability of regulation on the level of functionally defined protein sets renders the methodology to predict physiological changes unbiased and robust against nondetected proteins or still missing information on the function of particular proteins or genes.
Conclusions and Outlook
Quantitative protein set analysis including comprehensive protein expression and post-translational modification data revealed disease-relevant physiological pathways in DIO mice and demonstrated significant differences in the outcomes of treatment with two different PPARγ activators, the full agonist RSG and the partial agonist A1. We treated our mice with RSG for 3 weeks only, which was already predictive for potential systemic cardiovascular complications. In humans, these side effects occurred after several years of treatment. Further increase of sample throughput and detection sensitivity of mass spectrometry analysis using for example new benchtop orbitrap mass spectrometers for combining single-shot proteome and metabolome analyses (44), and/or applying alternatively targeted proteomics approaches (45), will enable even more detailed insight into the actual proteomic state of, in the best case, all organs of a diseased organism (46). Using isotopically labelled human reference cells or label-free quantitative mass spectrometry approaches, protein set analysis can be extended for clinical applications including disease and treatment monitoring in human patients.
Physiology relies on robust and redundant systems to react to environmental changes to sustain homeostasis. Although the various underlying molecular processes may interact deterministically, these systems are by no means linear. Physiological outcomes of differential drug treatments as shown here depend on interconnected protein pathway regulation in various tissues, which can, only in the case of highly connected “hub” genes or proteins, be adequately reduced to simple molecular mechanisms of action, for example by applying knock-down or over-expression experiments. Our approach is in concordance with genome-wide association meta-analyses, which correlated and grouped a number of genetic variants to various genes with subtle effects to pathways that were implicated in complex diseases (47). We thus argue for differential functional protein network analyses to capture the underlying biology of complex disease processes and their treatments to complement reductionist experimental approaches.
Protein sets can be used to functionally describe complex in vivo effects such as drug treatment or multifactorial disorders. Such phenomena emerge on a higher level of biological organization, and may for example be efficiently captured by applying information or pattern recognition theory. In other words, protein sets can generate causative links to explain physiological phenomena based on the properties of molecular networks.
Supplementary Material
Acknowledgments
We thank K. Hansen, C. Franke, and U. Schroeder for technical assistance during the animal studies. This work is part of the Ph.D. thesis of C.W.
Footnotes
* This work was supported by the German Ministry for Education and Research (BMBF, grant number 0315082), the European Union (FP7/2007–2013), under grant agreement n° 262055 (ESGI)), and the Max Planck Society.
 This article contains supplemental Figs. S1 to S7 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S7 and Tables S1 to S4.
1 The abbreviations used are:
- A1
- Amorfrutin A1
- DIO
- Diet-Induced Obesity
- HFD
- High-Fat Diet
- LFD
- Low-Fat Diet
- PDM
- Protein Distance Matrix
- PSEA
- Protein Set Enrichment Analysis
- RSG
- Rosiglitazone
- SILAC
- Stable isotope labeling by amino acids in cell culture.
REFERENCES
- 1. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 2. Sauer S., Kliem M. (2010) Mass spectrometry tools for the classification and identification of bacteria. Nat. Rev. Microbiol. 8, 74–82 [DOI] [PubMed] [Google Scholar]
- 3. Ong S. E., Mann M. (2005) Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 [DOI] [PubMed] [Google Scholar]
- 4. Ong S. E., Mann M. (2006) A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1, 2650–2660 [DOI] [PubMed] [Google Scholar]
- 5. Millioni R., Puricelli L., Iori E., Arrigoni G., Tessari P. (2010) The effects of rosiglitazone and high glucose on protein expression in endothelial cells. J. Proteome Res. 9, 578–584 [DOI] [PubMed] [Google Scholar]
- 6. Ahmed M., Neville M. J., Edelmann M. J., Kessler B. M., Karpe F. (2010) Proteomic Analysis of Human Adipose Tissue After Rosiglitazone Treatment Shows Coordinated Changes to Promote Glucose Uptake. Obesity 18, 27–34 [DOI] [PubMed] [Google Scholar]
- 7. Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krüger M., Moser M., Ussar S., Thievessen I., Luber C. A., Forner F., Schmidt S., Zanivan S., Fässler R., Mann M. (2008) SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 [DOI] [PubMed] [Google Scholar]
- 10. Kahn B. B., McGraw T. E. (2010) Rosiglitazone, PPARgamma, and type 2 diabetes. N. Engl. J. Med. 363, 2667–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weidner C., de Groot J. C., Prasad A., Freiwald A., Quedenau C., Kliem M., Witzke A., Kodelja V., Han C. T., Giegold S., Baumann M., Klebl B., Siems K., Müller-Kuhrt L., Schürmann A., Schüler R., Pfeiffer A. F., Schroeder F. C., Büssow K., Sauer S. (2012) Amorfrutins are potent antidiabetic dietary natural products. Proc. Natl. Acad. Sci. U.S.A. 109, 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monetti M., Nagaraj N., Sharma K., Mann M. (2011) Large-scale phosphosite quantification in tissues by a spike-in SILAC method. Nat. Methods 8, 655–658 [DOI] [PubMed] [Google Scholar]
- 13. Kaiser P., Meierhofer D., Wang X., Huang L. (2008) Tandem affinity purification combined with mass spectrometry to identify components of protein complexes. Methods Mol. Biol. 439, 309–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Villén J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiśniewski J. R., Zougman A., Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 16. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 17. Oppermann F. S., Gnad F., Olsen J. V., Hornberger R., Greff Z., Kéri G., Mann M., Daub H. (2009) Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics 8, 1751–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 20. Battke F., Symons S., Nieselt K. (2010) Mayday–integrative analytics for expression data. BMC Bioinformatics 11, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- 22. Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R. (2006) The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 313, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 23. Kuhn M., Campillos M., Letunic I., Jensen L. J., Bork P. (2010) A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 6, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lachmann A., Ma'ayan A. (2009) KEA: kinase enrichment analysis. Bioinformatics 25, 684–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Srere P. A., Brooks G. C. (1969) The circular dichroism of glucagon solutions. Arch. Biochem. Biophys. 129, 708–710 [DOI] [PubMed] [Google Scholar]
- 26. Rustin P., Chretien D., Bourgeron T., Gérard B., Rötig A., Saudubray J. M., Munnich A. (1994) Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta 228, 35–51 [DOI] [PubMed] [Google Scholar]
- 27. Mayr J. A., Paul J., Pecina P., Kurnik P., Förster H., Fötschl U., Sperl W., Houstek J. (2004) Reduced respiratory control with ADP and changed pattern of respiratory chain enzymes as a result of selective deficiency of the mitochondrial ATP synthase. Pediatr. Res. 55, 988–994 [DOI] [PubMed] [Google Scholar]
- 28. Deng W. J., Nie S., Dai J., Wu J. R., Zeng R. (2010) Proteome, phosphoproteome, and hydroxyproteome of liver mitochondria in diabetic rats at early pathogenic stages. Mol. Cell. Proteomics 9, 100–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Isserlin R., Merico D., Alikhani-Koupaei R., Gramolini A., Bader G. D., Emili A. (2010) Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps. Proteomics 10, 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cha S., Imielinski M. B., Rejtar T., Richardson E. A., Thakur D., Sgroi D. C., Karger B. L. (2010) In situ proteomic analysis of human breast cancer epithelial cells using laser capture microdissection: annotation by protein set enrichment analysis and gene ontology. Mol. Cell. Proteomics 9, 2529–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taubes G. (2009) Insulin resistance. Prosperity's plague. Science 325, 256–260 [DOI] [PubMed] [Google Scholar]
- 32. Patti M. E., Brambilla E., Luzi L., Landaker E. J., Kahn C. R. (1998) Bidirectional modulation of insulin action by amino acids. J. Clin. Invest. 101, 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ruggero D., Pandolfi P. P. (2003) Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 34. Peters J. M., Shah Y. M., Gonzalez F. J. (2012) The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat. Rev. Cancer 12, 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosen C. J. (2010) Revisiting the rosiglitazone story–lessons learned. N. Engl. J. Med. 363, 803–806 [DOI] [PubMed] [Google Scholar]
- 36. Shimoyama M., Ogino K., Tanaka Y., Ikeda T., Hisatome I. (1999) Hemodynamic basis for the acute cardiac effects of troglitazone in isolated perfused rat hearts. Diabetes 48, 609–615 [DOI] [PubMed] [Google Scholar]
- 37. Cosselman K. E., Krishnan R. M., Oron A. P., Jansen K., Peretz A., Sullivan J. H., Larson T. V., Kaufman J. D. (2012) Blood pressure response to controlled diesel exhaust exposure in human subjects. Hypertension 59, 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marchesini G., Brizi M., Bianchi G., Tomassetti S., Bugianesi E., Lenzi M., McCullough A. J., Natale S., Forlani G., Melchionda N. (2001) Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes 50, 1844–1850 [DOI] [PubMed] [Google Scholar]
- 39. Sertznig P., Seifert M., Tilgen W., Reichrath J. (2008) Peroxisome proliferator-activated receptors (PPARs) and the human skin: importance of PPARs in skin physiology and dermatologic diseases. Am. J. Clin. Dermatol. 9, 15–31 [DOI] [PubMed] [Google Scholar]
- 40. Piro R. M., Ala U., Molineris I., Grassi E., Bracco C., Perego G. P., Provero P., Di Cunto F. (2011) An atlas of tissue-specific conserved coexpression for functional annotation and disease gene prediction. Eur. J. Hum. Genet. 19, 1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang E., Zha J., Jockel J., Boise L. H., Thompson C. B., Korsmeyer S. J. (1995) Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell 80, 285–291 [DOI] [PubMed] [Google Scholar]
- 42. Datta S. R., Katsov A., Hu L., Petros A., Fesik S. W., Yaffe M. B., Greenberg M. E. (2000) 14–3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6, 41–51 [PubMed] [Google Scholar]
- 43. Feldmann R., Fischer C., Kodelja V., Behrens S., Haas S., Vingron M., Timmermann B., Geikowski A., Sauer S. (2013) Genome-wide analysis of LXRalpha activation reveals new transcriptional networks in human atherosclerotic foam cells. Nucleic Acids Res. 41, 3518–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, 1239–1247 M111.011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reiter L., Rinner O., Picotti P., Huttenhain R., Beck M., Brusniak M. Y., Hengartner M. O., Aebersold R. (2011) mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 [DOI] [PubMed] [Google Scholar]
- 46. Sauer S., Lange B. M., Gobom J., Nyarsik L., Seitz H., Lehrach H. (2005) Miniaturization in functional genomics and proteomics. Nat. Rev. Genet. 6, 465–476 [DOI] [PubMed] [Google Scholar]
- 47. Morris A. P., Voight B. F., Teslovich T. M., Ferreira T., Segre A. V., Steinthorsdottir V., Strawbridge R. J., Khan H., Grallert H., Mahajan A., Prokopenko I., Kang H. M., Dina C., Esko T., Fraser R. M., Kanoni S., Kumar A., Lagou V., Langenberg C., Luan J., Lindgren C. M., Muller-Nurasyid M., Pechlivanis S., Rayner N. W., Scott L. J., Wiltshire S., Yengo L., Kinnunen L., Rossin E. J., Raychaudhuri S., Johnson A. D., Dimas A. S., Loos R. J., Vedantam S., Chen H., Florez J. C., Fox C., Liu C. T., Rybin D., Couper D. J., Kao W. H., Li M., Cornelis M. C., Kraft P., Sun Q., van Dam R. M., Stringham H. M., Chines P. S., Fischer K., Fontanillas P., Holmen O. L., Hunt S. E., Jackson A. U., Kong A., Lawrence R., Meyer J., Perry J. R., Platou C. G., Potter S., Rehnberg E., Robertson N., Sivapalaratnam S., Stancakova A., Stirrups K., Thorleifsson G., Tikkanen E., Wood A. R., Almgren P., Atalay M., Benediktsson R., Bonnycastle L. L., Burtt N., Carey J., Charpentier G., Crenshaw A. T., Doney A. S., Dorkhan M., Edkins S., Emilsson V., Eury E., Forsen T., Gertow K., Gigante B., Grant G. B., Groves C. J., Guiducci C., Herder C., Hreidarsson A. B., Hui J., James A., Jonsson A., Rathmann W., Klopp N., Kravic J., Krjutskov K., Langford C., Leander K., Lindholm E., Lobbens S., Mannisto S., Mirza G., Muhleisen T. W., Musk B., Parkin M., Rallidis L., Saramies J., Sennblad B., Shah S., Sigurethsson G., Silveira A., Steinbach G., Thorand B., Trakalo J., Veglia F., Wennauer R., Winckler W., Zabaneh D., Campbell H., van Duijn C., Uitterlinden A. G., Hofman A., Sijbrands E., Abecasis G. R., Owen K. R., Zeggini E., Trip M. D., Forouhi N. G., Syvanen A. C., Eriksson J. G., Peltonen L., Nothen M. M., Balkau B., Palmer C. N., Lyssenko V., Tuomi T., Isomaa B., Hunter D. J., Qi L., Shuldiner A. R., Roden M., Barroso I., Wilsgaard T., Beilby J., Hovingh K., Price J. F., Wilson J. F., Rauramaa R., Lakka T. A., Lind L., Dedoussis G., Njolstad I., Pedersen N. L., Khaw K. T., Wareham N. J., Keinanen-Kiukaanniemi S. M., Saaristo T. E., Korpi-Hyovalti E., Saltevo J., Laakso M., Kuusisto J., Metspalu A., Collins F. S., Mohlke K. L., Bergman R. N., Tuomilehto J., Boehm B. O., Gieger C., Hveem K., Cauchi S., Froguel P., Baldassarre D., Tremoli E., Humphries S. E., Saleheen D., Danesh J., Ingelsson E., Ripatti S., Salomaa V., Erbel R., Jockel K. H., Moebus S., Peters A., Illig T., de Faire U., Hamsten A., Morris A. D., Donnelly P. J., Frayling T. M., Hattersley A. T., Boerwinkle E., Melander O., Kathiresan S., Nilsson P. M., Deloukas P., Thorsteinsdottir U., Groop L. C., Stefansson K., Hu F., Pankow J. S., Dupuis J., Meigs J. B., Altshuler D., Boehnke M., McCarthy M. I. (2012) Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.