Abstract
Mass spectrometry (MS)-based quantitative proteomics has matured into a methodology able to detect and quantitate essentially all proteins of model microorganisms, allowing for unprecedented depth in systematic protein analyses. The most accurate quantitation approaches currently require lysine auxotrophic strains, which precludes analysis of most existing mutants, strain collections, or commercially important strains (e.g. those used for brewing or for the biotechnological production of metabolites). Here, we used MS-based proteomics to determine the global response of prototrophic yeast and bacteria to exogenous lysine. Unexpectedly, down-regulation of lysine synthesis in the presence of exogenous lysine is achieved via different mechanisms in different yeast strains. In each case, however, lysine in the medium down-regulates its biosynthesis, allowing for metabolic proteome labeling with heavy-isotope-containing lysine. This strategy of native stable isotope labeling by amino acids in cell culture (nSILAC) overcomes the limitations of previous approaches and can be used for the efficient production of protein standards for absolute SILAC quantitation in model microorganisms. As proof of principle, we have used nSILAC to globally analyze yeast proteome changes during salt stress.
Breakthroughs in proteomics (1–4) open up new possibilities for biological systems analysis. Central to these approaches is the necessity to accurately quantitate protein abundances. The most accurate quantitation is achieved by means of stable isotope labeling by amino acids in cell culture (SILAC)1 using heavy-isotope-containing amino acids (5). This approach has been widely used in many different experimental systems (6–11). SILAC relies on the metabolic incorporation of isotope-labeled lysine and/or arginine into proteins. Samples from differently labeled cells are mixed and analyzed, for example, after one of them has been subjected to a different experimental condition. During sample preparation, proteins are digested to yield peptides containing one differentially labeled amino acid. As a consequence, mass spectrometry reveals “SILAC pairs” for each peptide, containing peaks corresponding to the unlabeled and the labeled peptides. The abundance ratio between the two reflects the different abundances of the protein in the starting samples. Lysine, which is commonly used in SILAC experiments, is an essential amino acid in higher eukaryotes that is obtained exclusively from food, but it can be synthesized in a tightly controlled fashion by plants, bacteria, and many fungi. Therefore, analysis of these prototroph organisms using SILAC has relied largely on mutants in lysine biosynthesis. In contrast to other biochemical pathways of amino acid metabolism, lysine biosynthesis occurs through distinct sets of reactions in different organisms that possess this biosynthetic capability. In bacteria, plants, and some fungi, lysine is produced in nine enzymatically catalyzed steps via the diaminopimelate pathway from aspartate. In contrast, most fungi synthesize lysine from the citrate cycle intermediate 2-oxoglutarate in ten steps, a pathway known as the α-aminoadipate pathway (12). Enzymes of this pathway are evolutionary conserved between such diverse species as S. pombe and S. cerevisiae, which are separated by 400 million years of evolution (13). Because of the specificity of lysine biosynthesis pathways for bacteria and fungi, the enzymes participating in these reactions have become targets for the treatment of infections (14). An understanding of lysine regulation is important for understanding the effects of such drugs, and it might also help in optimizing the industrial production of lysine and other derived metabolites in these microorganisms.
Both the diaminopimelate and the α-aminoadipate pathways of lysine biosynthesis are exquisitely controlled: In S. cerevisiae, in which the regulation of lysine metabolism has been studied in the most detail, the α-aminoadipate pathway is under combinatorial control. Product inhibition by lysine controls homocitrate synthase, the first enzyme of the lysine synthesis pathway, as well as the expression of key lysine biosynthetic enzymes. Substrate feed-forward regulation by the intermediate l-2-aminoadipate-6-semialdehyde functions as a transcriptional co-activator, acting together with Lys14 to increase the amount of lysine synthesis enzymes (15).
Similarly, bacterial lysine synthesis via the diaminopimelate pathway is regulated by a combination of transcriptional and post-transcriptional mechanisms. Specifically, dihydrodipicolinate synthase catalyzing the committing step of lysine biosynthesis in bacteria is allosterically regulated by lysine, and a number of genes in the pathways are transcriptionally regulated via lysine repression (16).
In order to determine the effects of lysine in growth medium and to gain further insights into the biochemical regulation of lysine synthesis pathways, we analyzed global proteome changes in different lysine prototrophic model microorganisms in the presence or absence of lysine. Our results reveal important differences in lysine regulation between S. cerevisiae and S. pombe.
Despite these differences, both organisms allowed us to exploit the regulation of lysine biosynthesis for the development of a novel strategy for labeling prototrophic microorganisms. This strategy not only avoids complications of studying amino acid prototrophic mutant strains (e.g. when studying cellular metabolism), but also enables analysis of the large arsenal of mutants and systematic strain collections available for these model systems of cell biology, as well as industrially or medically important yeast and bacterial strains.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
All experiments with S. cerevisiae were performed with the lysine prototroph strain W303 MAT a (TWY#139). A lysine auxotroph strain was generated by deleting the LYS2 gene via homologous recombination (17) to yield the strain W303 lys2Δ::NATR (TWY#1050). All experiments with S. pombe were performed with the lysine prototroph strain MKSP1. Experiments with E. coli were performed with the lysine prototroph strain E. coli BL21 (DE3). To express the recombinant protein, glutathione S-transferase (GST) cells were transformed with the plasmid pGex6P1 (TWP#41). To express the fusion protein GST-Pil1, cells were transformed with TWP#121 in which the yeast gene PIL1 is cloned after GST.
Cell Culture
S. cerevisiae strains were grown in synthetic medium containing 6.7 g/l yeast nitrogen base, 2 g/l drop out mix (United States Biological, Salem, MA) containing all amino acids except lysine, and 2% glucose. Lysine (Sigma) or heavy (13C6/15N2) l-lysine (Cambridge Isotope Labs, Andover, MA) was added to a final concentration of 30 mg/l as indicated. Cells were pre-cultured in 5 ml medium containing light, heavy, or no lysine overnight at 30 °C. 50 ml of medium were inoculated from the pre-cultures to A600 = 0.001, and cells were grown to a final A600 = 0.7 matching roughly ten doublings. To induce osmotic stress, NaCl was added to a final concentration of 0.4 m, and cells were grown for an additional 20 min.
S. pombe strains were grown in Edinburgh minimal medium (Sunrise Science Products, San Diego, CA) supplemented with 75 mg/l leucine, histidine, uracil, and adenine. Lysine was added to a final concentration of 30 mg/l as indicated. Cells were grown in a 5 ml pre-culture containing either heavy (13C6/15N2) l-lysine or no lysine overnight at 30 °C. Pre-cultures were used to inoculate fresh medium to A600 = 0.001, and cells were grown to a final A600 = 0.7 at 30 °C.
E. coli BL21 (DE3) cells were grown in M9 minimal medium. A 5× stock of M9 salts containing 64 g/l Na2HPO4.7H2O, 15 g/l KH2PO4, 2.5 g/l NaCl, and 5 g/l NH4Cl was prepared. The M9 minimal medium contained 1 × M9 salts, 2 mm MgSO4, 0.1 mm CaCl2, and 0.4% glucose. Lysine or lysine 8 was added to a final concentration of 30 mg/l as indicated. Cells were pre-cultured in 5 ml of medium containing lysine, heavy (13C6/15N2) l-lysine, or no lysine overnight at 37 °C. The next day, cultures were inoculated to A600 = 0.001 and grown to a final A600 = 0.7. To induce recombinant protein expression, isopropyl β-D-thiogalactopyranoside was added to a final concentration of 1 mm, and cells were grown for an additional 1 h.
Sample Preparation
For the incorporation test, 40 OD units of heavy labeled cells were harvested via centrifugation. For all other experiments, 20 OD units of light and heavy labeled cells were mixed and harvested via centrifugation. Cells were lysed in 200 μl buffer containing 50 mm Tris/HCl pH = 9.0, 5% SDS, and 100 mm DTT for 30 min at 55 °C. Lysates were cleared via centrifugation at 17,000g for 10 min. Supernatants were diluted with buffer UA (8 m urea, 0.1 m Tris/HCl pH = 8.5) to a final concentration of 0.5% SDS. Proteins were digested with the endoproteinase LysC following the protocol for filter aided sample preparation (18). Briefly, protein extracts were loaded on a 30k Amicon Ultra filter unit (Millipore, Billerica, MA) by means of centrifugation at 14,000g. Samples were washed twice by the addition of 200 μl buffer UA and alkylated for 20 min in the dark by the addition of 5.5 mm iodoacetamide in 200 μl buffer UA. Samples were washed an additional four times via the addition of 200 μl buffer UA and centrifugation. 60 μl of buffer UA containing 0.5 mg/ml LysC were added to the filter units and incubated at 37 °C overnight. Peptides were recovered via centrifugation into a fresh tube and additional elution with 200 μl of 0.5 m NaCl. Samples were acidified by the addition of trifluoroacetic acid and cleared of precipitates via centrifugation at 17,000g for 5 min. The peptide concentration was measured, and 5 μg of peptides were desalted following the protocol for StageTip purification (19). Samples were eluted with 60 μl buffer B (80% acetonitrile, 0.1% formic acid in H20) and reduced in a Vacufuge plus (Eppendorf, Hauppauge, New York) to a final volume of 3 μl. 2 μl of buffer A (0.1% formic acid in H20) were added, and the resulting 5 μl were injected into the high-performance liquid chromatograph.
Chromatography and Mass Spectrometry
Reversed-phase chromatography was performed on a Thermo Easy nLC 1000 system connected to a Q Exactive mass spectrometer (Thermo) through a nano-electrospray ion source. Peptides were separated on 50 cm or 15 cm columns (New Objective, Woburn, MA) with an inner diameter of 75 μm packed in house with 1.9 μm C18 resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Baden-Würtemberg, Germany). Peptides were eluted from 50-cm columns with a linear gradient of acetonitrile from 5%–27% in 0.1% formic acid for 240 min at a constant flow rate of 250 nl/min. For 15-cm columns, peptides were eluted with a linear gradient of acetonitrile from 5%–27% in 0.1% formic acid for 95 min at a constant flow rate of 250 nl/min. The column temperature was kept at 35 °C by an oven (Sonation GmbH, Biberach, Baden-Württemberg, Germany) with a Peltier element. Eluted peptides from the column were directly electrosprayed into the mass spectrometer. Mass spectra were acquired on the Q Exactive in a data-dependent mode to automatically switch between full scan MS and up to ten data-dependent MS/MS scans. The maximum injection time for full scans was 20 ms, with a target value of 3,000,000 at a resolution of 70,000 at m/z = 200. The ten most intense multiply charged ions (z ≥ 2) from the survey scan were selected with an isolation width of 1.6 Th and fragment with higher energy collision dissociation (20) with normalized collision energies of 25. Target values for MS/MS were set at 1,000,000 with a maximum injection time of 60 ms at a resolution of 17,500 at m/z = 200. To avoid repetitive sequencing, the dynamic exclusion of sequenced peptides was set at 45 s for 240-min runs and 20 s for 95-min runs.
Data Analysis
The resulting MS and MS/MS spectra were analyzed using MaxQuant (version 1.3.0.5), utilizing its integrated ANDROMEDA search algorithms (21, 22). Peak lists were searched against local databases for S. cerevisiae (obtained from the Saccharomyces Genome Database, Stanford University; 6641 entries, July 26, 2012), S. pombe (obtained from PomBase, University of Cambridge; 5089 entries, August 26, 2012), or E. coli BL21-DE3 (obtained from Uniprot, “Escherichia coli(strain B/BL21-DE3)[469008]”; 4726 entries, March 25, 2012), with common contaminants added. Additionally, the sequences of GST and the yeast protein Pil1 were added to the E. coli database. The search included carbamidomethlyation of cysteine as a fixed modification and methionine oxidation and N-terminal acetylation as variable modifications. The maximum allowed mass deviation was 6 ppm for MS peaks and 20 ppm for MS/MS peaks. The maximum number of missed cleavages was two. The false discovery rate was determined by searching a reverse database. The maximum false discovery rate was 0.01 on both the peptide and the protein level. The minimum required peptide length was six residues. Proteins with at least two peptides (one of them unique) were considered identified. The “match between runs” option was enabled with a time window of 2 min to match identification between replicates. The requant option of MaxQuant was disabled.
Label-free quantitation was done with the QUBIC software package as described elsewhere (23). Significance values of proteins were calculated as described previously (24). For incorporation tests, the peptide list was filtered for lysine containing peptides with a valid heavy/light ratio. For each peptide, the incorporation was calculated as 1 − (1/(ratio H/L + 1)). The maximum of a density distribution of all peptides represents the estimated incorporation level. All calculations and plots were done with the R software package.
RESULTS
The Abundance of Lysine Metabolism Enzymes Is Regulated by Feedback
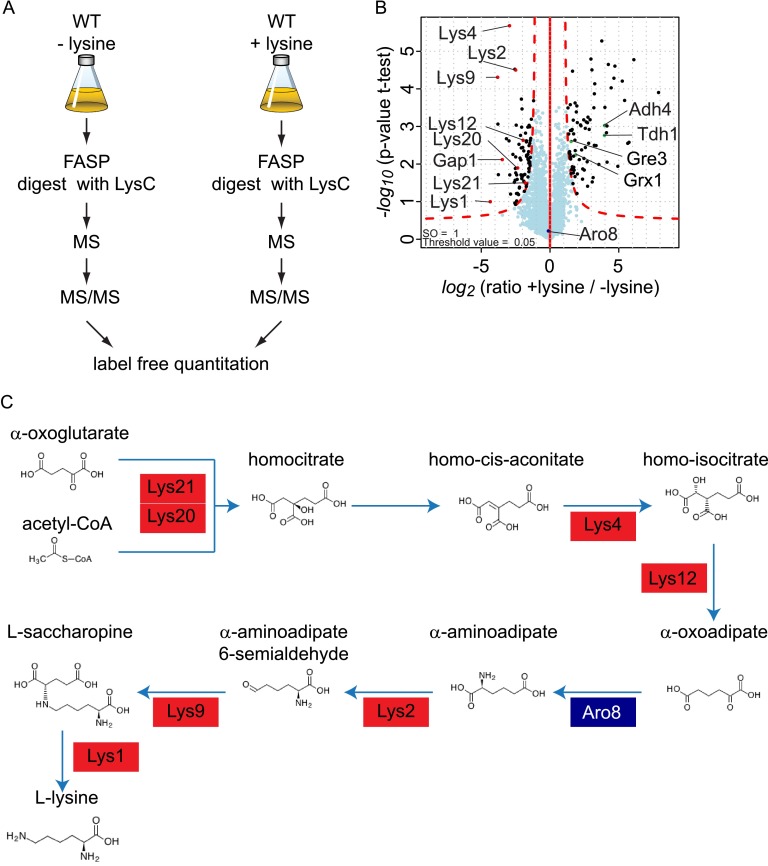
To determine changes of the yeast proteome due to the presence or absence of lysine in the culture medium, we measured proteome changes of prototrophic S. cerevisiae strains under these conditions while using label-free quantitation of triplicate liquid chromatography runs coupled online to high-resolution mass spectrometry (LC-MS) (see Fig. 1A for the experimental strategy). All measurements presented here were performed for independent biological replicates, from which we in each case obtained highly reproducible results (data not shown and supplemental Fig. S3). For presentation, we analyzed the independent runs together and filtered out the few proteins not giving reproducible results. This strategy led to the identification of 4361 proteins in total, representing almost the entire expressed proteome of S. cerevisiae (25). We identified proteins significantly changed in abundance from cells grown in the presence or absence of lysine (23) (see supplemental Table S1 for an overview of MS data from the experiments presented here). Our analysis of 2906 proteins detected in at least five of the six experiments revealed few changes in the proteome overall. Among the significantly changing proteins, enzymes acting in the lysine biosynthetic pathway were much less abundant with the presence of lysine in the medium (Fig. 1B). Particularly, Lys20, Lys21, Lys4, Lys12, Lys1, Lys2, and Lys9 were strongly down-regulated in cells grown in the presence of lysine (Fig. 1B). This represents all enzymes in the α-aminoadipate pathway of lysine synthesis in S. cerevisiae except Aro8, which might also function in other amino acid metabolism pathways (26) (Fig. 1C). This is consistent with previous findings that lysine metabolism is controlled by feedback inhibition in which lysine represses the expression of Lys20, Lys4, Lys12, Lys2, Lys9, and Lys1, potentially mediated by the Lys14 transcriptional regulator (15). We also found that the expression of amino acid membrane transporters, such as Gap1, was down-regulated in the presence of lysine. These data suggest that cells adapt to the presence of lysine by decreasing their lysine synthesis and by adjusting their import capacity for lysine. We also found that proteins of the oxidative stress response were up-regulated in response to lysine in the growth medium (e.g. Grx1, Gre3). Potentially, this is due to the decreased withdrawal of Krebs cycle intermediates for lysine biosynthesis and, conversely, increased levels of acetyl-CoA for β-oxidation under these conditions. Consistent with this notion, we found Adh4 and Tdh1, involved in gluconeogenesis, among the most up-regulated proteins in the presence of lysine.
Fig. 1.
Proteome changes of prototrophic yeast in medium containing or lacking lysine. A, experimental design for the proteome analysis of S. cerevisiae grown in the presence or absence of lysine. FASP, filter aided sample preparation; WT, wild-type. B, S. cerevisiae down-regulates the abundance of lysine biosynthetic enzymes in response to external lysine. In the volcano plot, the protein abundance ratios in the presence or absence of lysine are plotted against the negative log10 of the p value of the t test for each protein. C, the lysine biosynthesis pathway of S. cerevisiae is down-regulated (denoted by red boxes).
Prototrophic S. cerevisiae Cells Preferentially Use Lysine Available in the Medium
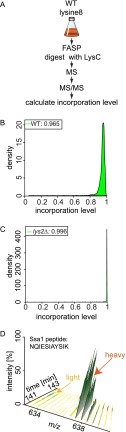
Based on these findings, we hypothesized that decreased lysine synthesis and increased lysine uptake lead to the preferential incorporation of lysine from the growth medium into the cellular proteome. To test this model, we measured the efficiency of the incorporation of heavy-isotope-containing (13C6/15N2) l-lysine (lysine 8) in the growth medium into the proteome of lysine prototrophic S. cerevisiae (see Fig. 2A for the experimental strategy). Consistent with our prediction, 96% of peptides from the proteome of lysine prototrophic yeast cells grown in minimal medium containing lysine 8 contained only the heavy-isotope-containing amino acid (Fig. 2B). Several repetitions of this experiment showed that metabolic labeling is highly reproducible (data not shown). At the level of an individual peptide mass spectrum, efficient labeling is reflected in the almost complete absence of the light-labeled peak in a SILAC pair (Fig. 2D). The level of labeling achieved in the prototrophic strain (96.5% incorporation) was almost as efficient as that of a lys2Δ lysine auxotrophic strain (99.6% incorporation), which cannot produce lysine and thus relies entirely on the exogenously provided lysine (Fig. 2C). The efficiency of metabolic labeling determines the maximal ratios—and thus the dynamic range—that can be achieved in a SILAC experiment. The maximal possible ratio is given by the fraction of labeled versus unlabeled proteins. In our experiments with prototroph yeast, one can therefore obtain maximal ratios of ∼28. Importantly, when we normalized our data for differences in labeling by factoring in small differences in labeling, we obtained qualitatively the same results as obtained by directly comparing different conditions (data not shown). Moreover, we overall found uniform labeling efficiency across the spectrum of protein abundances and detected no bias for the labeling of proteins belonging to specific gene ontology classes (F.F. and T.C.W., data not shown). However, we found lysine enzymes as notable exceptions to uniform labeling. We hypothesize that during labeling with lysine, these enzymes are not synthesized, and thus they are more resistant to metabolic labeling.
Fig. 2.
Prototrophic S. cerevisiae efficiently incorporates exogenous lysine. A, experimental strategy to determine incorporation of heavy labeled lysine into S. cerevisiae, analyzed via proteomics. FASP, filter aided sample preparation; WT, wild-type. B, density function of the rates of heavy lysine incorporation of all lysine-containing peptides for the lysine prototroph S. cerevisiae W303 strain. Maximum of the density function representing the most abundant incorporation rate is given above the graph. C, density function of the heavy lysine incorporation rate in the lysine auxotroph W303 lys2Δ strain. Maximum of the density function representing the most abundant incorporation rate is given above the graph. D, example MS spectrum plotted against chromatography runtime of Ss a1 peptide NQIESIAYSIK from the lysine prototroph strain W303.
Native SILAC Allows Accurate Proteome Quantification in Lysine Prototroph Yeast
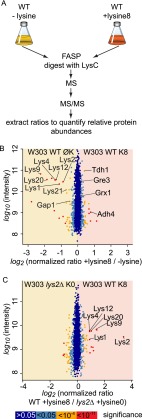
We next tested whether complete metabolic labeling of prototrophic strains allows proteome quantitation in comparison to an unlabeled control strain (see Fig. 3A for the experimental strategy). Proteome analysis of prototrophic yeast grown in the presence or absence of lysine 8 yielded the identification of 3232 proteins in two chromatographic runs, representing roughly 70% of the cellular proteome, with 2417 of them quantified at least twice (supplemental Table S1). Consistent with the results from label-free quantitation, enzymes of the lysine biosynthetic pathway were significantly down-regulated (Fig. 3B).
Fig. 3.
Native SILAC (nSILAC) detects proteome changes in cells grown with or without lysine. A, experimental design for the proteome analysis of S. cerevisiae grown in the presence or absence of heavy-isotope-containing lysine. FASP, filter aided sample preparation; WT, wild-type. B, plot of protein intensities against normalized H/L SILAC ratios of WT W303. S. cerevisiae cells grown in absence (light lysine, L) or presence of non-radioactive stable-isotope-containing lysine (heavy lysine 8, H). Significant outliers are colored in red (p < 1e−11), orange (p < 1e−4), or light blue (p < 0.05); other proteins are shown in dark blue. C, proteome comparison of lysine prototroph and lysine auxotroph S. cerevisiae strains. Protein intensities are plotted against normalized SILAC ratios of heavy (lysine prototroph strain labeled with lysine 8, H) to light (lysine auxotroph strain labeled with light lysine, L). Significant outliers are colored in red (p < 1e−11), orange (p < 1e−4). or light blue (p < 0.05); other proteins are shown in dark blue.
Because lysine auxotrophic strains are most commonly used in S. cerevisiae proteomic experiments (27), we compared the proteome of the lysine auxotrophic benchmark strain to a lysine prototrophic strain. Consistent with our previous observation from diploid cells (24), we found that a lysine auxotrophic lys2Δ strain down-regulated the expression of other enzymes of the lysine biosynthetic pathway relative to the prototrophic strain even in the presence of lysine (Fig. 3C). This finding is consistent with the total absence of L2-aminoadipate-6-semialdehyde, a transcriptional co-activator of lysine biosynthesis in lys2Δ cells (28). Together, our data show that lysine biosynthesis enzyme expression is down-regulated via negative feedback when lysine is present in the medium or when the cells do not have the capacity to synthesize lysine.
Furthermore, these data show that S. cerevisiae lysine prototrophic strains can be efficiently metabolically labeled with heavy-isotope-containing amino-acids provided in the growth medium, a labeling strategy that we call native SILAC (nSILAC).
The Proteome of Lysine Prototrophic S. pombe Can Be Efficiently Labeled by Exogenously Provided Lysine
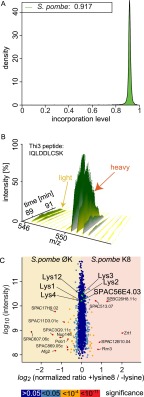
We next tested whether the application of nSILAC afforded by the regulation of lysine biosynthesis was specific to S. cerevisiae or could be applied more broadly to microorganisms. Because S. pombe is separated from S. cerevisiae by ∼400 million years of evolution (13) and is widely used as a model system for biological research, we investigated the regulation of lysine biosynthesis enzymes and the possibility of nSILAC labeling in this organism. S. pombe could be almost as efficiently labeled with lysine 8 as S. cerevisiae (ca. 92% lysine-containing peptides contained lysine 8 with uniform labeling across abundance and function of proteins; Fig. 4A and data not shown), leading to the almost complete absence of the light peak in a SILAC pair (Fig. 4B). This result shows that also in S. pombe, the presence of exogenous lysine led to the shutdown of cellular lysine production and consequently the almost complete replacement of the lysine pool available for protein synthesis with amino acids imported from the medium. In marked contrast to findings for S. cerevisiae, however, comparison of the proteome (1746 quantitated proteins) of prototrophic strains grown in the presence or absence of lysine in the culture medium revealed that enzymes of the lysine biosynthetic pathway do not change in abundance (Fig. 4C). This argues that the down-regulation of lysine biosynthesis apparent in the lysine 8 incorporation experiments is meditated primarily by post-translational mechanisms affecting enzyme activity instead of abundance.
Fig. 4.
Abundance of lysine biosynthesis of S. pombe is not regulated in response to external lysine. A, lysine prototrophic S. pombe incorporates heavy lysine 8 from the medium. A density function for lysine incorporation in S. pombe is shown. B, example MS spectrum plotted against chromatography runtime for the Thi3 peptide IQLDDLCSK from the lysine prototroph S. pombe strain grown in the presence of lysine 8. C, abundances of lysine biosynthetic enzymes are not changed in response to external lysine. Protein intensities are plotted against normalized SILAC ratios of heavy (heavy lysine 8 as external source) to light (no exogenous lysine). Significant outliers are colored in red (p < 1e−11), orange (p < 1e−4), or light blue (p < 0.05); other proteins are shown in dark blue. The enzymes of the lysine biosynthesis pathway are colored in green.
The E. coli Proteome Can Be Efficiently Labeled via nSILAC
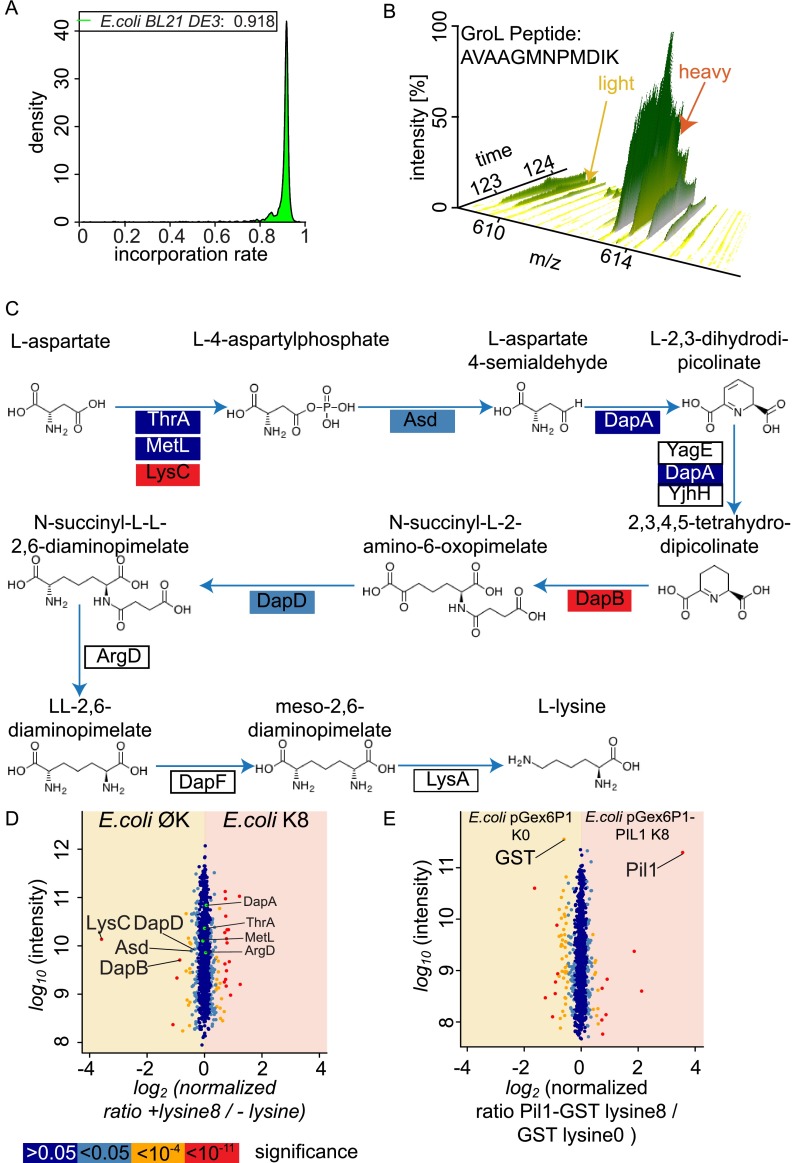
Lysine biosynthesis is controlled not only in eukaryotes, but also in bacteria. Because the model system E. coli is widely used for studying Gram-negative bacteria and provides a convenient system for the expression of heterologous proteins, we assayed whether the application of nSILAC could be extended to this organism, which tightly regulates lysine biosynthesis (29–32). When E. coli BL21 DE3 cells were grown in synthetic lysine-8-containing medium, the exogenous amino acid efficiently replaced non-labeled lysine (92% of lysine-containing peptides labeled with lysine 8 with uniform labeling across abundance and function of proteins; Fig. 5A and data not shown), leading to an almost complete replacement of light labeled peaks in SILAC pairs (Fig. 5B). Lysine biosynthesis via the diaminopimelate pathway in E. coli differs from the reactions in yeasts, as primarily aspartate is converted to lysine, rather than lysine being synthesized from the citrate cycle intermediate 2-oxoglutarate (Fig. 5C). We tested whether, despite this difference, similar mechanisms as in S. cerevisiae contribute to the labeling of E. coli. Consistent with this notion, we found that the expression of several genes encoding enzymes of the lysine synthesis pathway, including LysC, DapD, and Asd, were down-regulated in medium containing lysine relative to medium not including lysine (Fig. 5D; 1140 proteins quantitated). Together, these data show that E. coli can be labeled via nSILAC and that this is likely because of similar feedback regulation of lysine biosynthesis.
Fig. 5.
Regulation of lysine biosynthesis in E. coli allows for efficient proteome labeling. A, E. coli incorporates heavy lysine from the medium efficiently. A density function for lysine incorporation in E. coli is shown. B, example MS spectrum plotted against chromatography runtime for the GroL peptide AVAAGMNPMDIK from the lysine prototroph E. coli BL21 (DE3) strain is shown. C, enzymes of the diaminopimelate pathway are color coded according to the statistical significance of their abundance change. D, E. coli down-regulates lysine biosynthetic enzymes in response to external lysine. Protein intensities are plotted against SILAC ratios of heavy (lysine 8 as external source) to light (no external lysine). Significant outliers are colored in red (p < 1e−11), orange (p < 1e−4), or light blue (p < 0.05); other proteins are shown in dark blue. E, E. coli can be used to produce heavy-labeled recombinant proteins. An E. coli BL21 (DE3) strain carrying the plasmid pGex6P1 encoding the protein glutathione S-transferase (GST) was labeled with light lysine. A strain carrying the plasmid pGex6P1-PIL1 encoding a fusion protein of GST to the S. cerevisiae protein Pil1 was labeled with heavy lysine 8. After 1 h of recombinant protein induction, cells were analyzed via MS. Protein intensities are plotted against normalized heavy/light SILAC ratios. Significant outliers are colored in red (p < 1e−11), orange (p < 1e−4), or light blue (p < 0.05); other proteins are shown in dark blue.
E. coli is often used to produce heterologous proteins from various sources. Versions of such proteins labeled with heavy-isotope-containing amino acids can be used as standards for the absolute quantitation of proteins, an alternative to relative quantitation between conditions known as absolute SILAC (33). To test whether nSILAC could be used to produce such lysine-8-labeled heterologous proteins, we expressed either GST alone or GST fused to the S. cerevisiae protein Pil1 in E. coli grown in medium with only light- or heavy-isotope-containing lysine, respectively. When we analyzed a 1:1 protein mixture from those two samples, we found that Pil1 was efficiently labeled (supplemental Fig. S1). Because the GST moiety is present in both conditions, it has an abundance ratio between heavy- and light-containing versions of the protein of close to one, with a slightly higher abundance when expressed alone relative to the fusion protein (Fig. 5E). These results show that nSILAC provides a simple means to label heterologous proteins in E. coli, which could be used as standards in absolute SILAC applications.
nSILAC Provides a Robust Method for Global Proteome Analysis
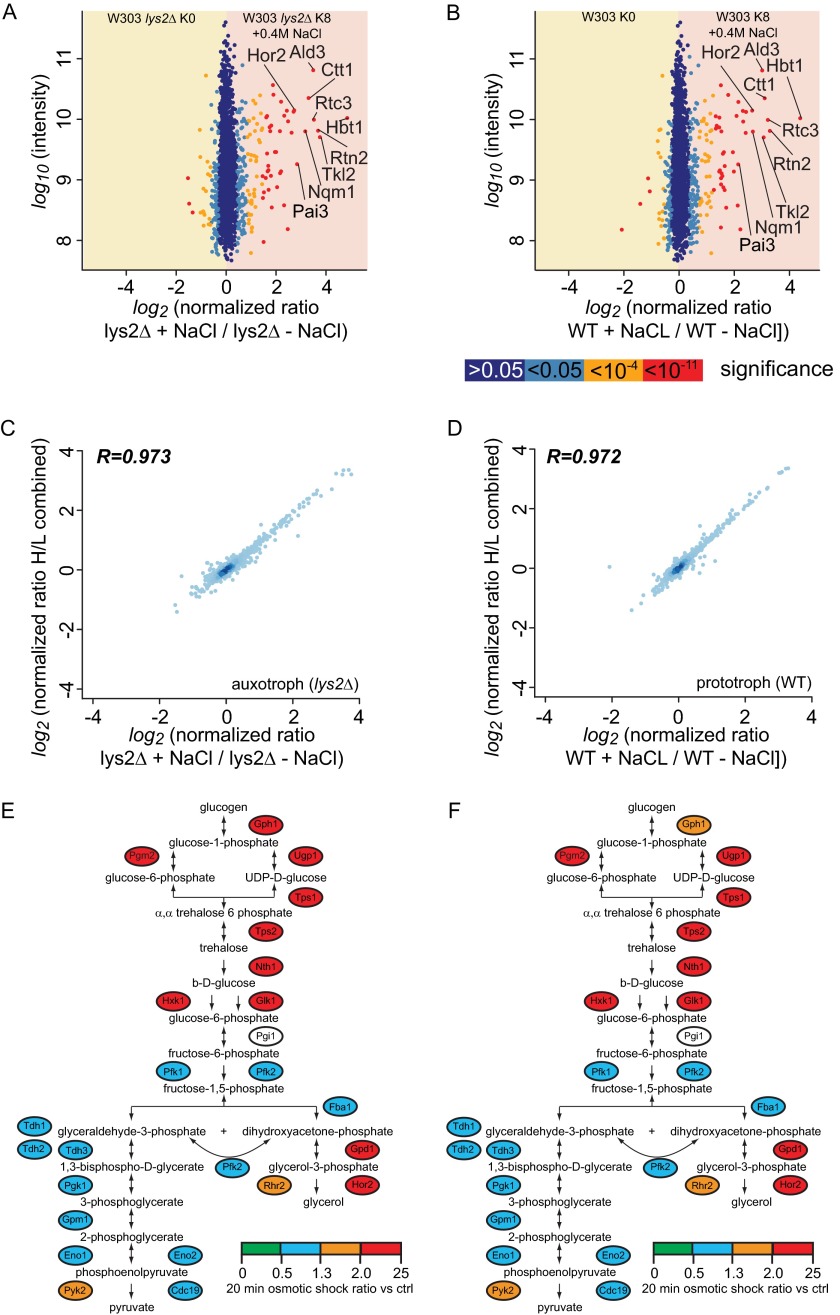
We reasoned that nSILAC could be used for the quantitation of proteome changes in different conditions. We therefore globally determined changes of the S. cerevisiae proteome in either lysine prototrophic or lys2Δ auxotrophic cells grown under conditions of salt stress (i.e. either normal medium or medium containing 0.4 m NaCl). We chose salt stress for a proof-of-principle experiment because there already are proteome data available for yeast cultured under these conditions (34). In our analyses, the few outliers with a ratio of heavy to light peptides indicative of a significant change in protein abundance during salt stress were basically identical for the experiments with either lysine prototrophic or auxotrophic strains (Figs. 6A, 6B; 2485 quantitated proteins in prototrophic and lys2Δ cells). The correspondence of the experiments in the two different strains was reflected in the very high correlation of ratios detected in each experiment to the average of ratios for the same proteins in both experiments (Figs. 6C, 6D). In both cases, and fully consistent with previous measurements of yeast proteome changes due to salt stress (supplemental Fig. S2) (34), we measured significant changes in the abundance of enzymes involved in the synthesis of the osmo-stabilizer glycerol (Figs. 6E, 6F).
Fig. 6.
Lysine-labeled prototroph yeast can be used for SILAC-based quantitative proteomics. Lysine auxotrophic (A) or prototrophic (B) S. cerevisiae strains were labeled with either light or heavy lysine. Heavy-labeled strains were exposed to a hyperosmotic shock (0.4 m NaCl for 20 min). Protein intensities are plotted against heavy/light SILAC ratios. Significant outliers are colored in red (p < 1e−11), orange (p < 1e−4), or light blue (p < 0.05); other proteins are shown in dark blue. Ratio scatter plots show protein abundance changes between 0 and 20 min of salt stress as measured in a lysine auxotroph (C) or prototroph (D) strain plotted versus a combined ratio (22). The Pearson correlation coefficient is shown in the top left corner of each plot. Osmotic shock regulates glycolysis and glycerol synthesis. Enzymes are color coded according to their fold abundance change in response to a hyperosmotic shock in a lysine auxotroph (E) or prototroph (F) strain.
DISCUSSION
Here, we show that the culturing of prototroph microorganisms with stable isotopes of lysine can be used for proteome quantitation, a novel strategy we call nSILAC. nSILAC is made possible by the regulation of lysine metabolism by different mechanisms in different prototrophic microorganisms.
After determining the global proteome changes of cells grown in medium with or without lysine, we surprisingly found that despite the high conservation of the enzymes of the α-aminoadipate pathway during 400 million years of evolution from S. cerevisiae to S. pombe, the regulation of enzyme abundance differs between the species. Efficient labeling of the proteome by exogenous isotope-marked lysine shows that nonetheless, in both cases a shutoff of the endogenous synthesis is achieved, reducing the pool of light-isotope-containing lysine available for protein synthesis. One possibility is that in S. pombe the regulation of enzyme activity dominates over the regulation of the enzymes' abundances, which might be more important in S. cerevisiae. Consistent with this notion that biosynthetic pathways evolve slower than regulatory networks, analysis of phosphorylation has led to the suggestion that differences in the regulation of processes by kinases provide phenotypic diversity during evolution (35). Future studies will show how the regulation of lysine enzymes via other mechanisms, such as allosteric regulation, participates in the evolution of pathway regulation. For the diaminopimelate pathway in bacteria, differences in the allosteric regulation of dihydrodipicolinate synthase have already emerged between E. coli and Corynebacterium glutamicum (16). Interestingly, we found that many genes of the lysine biosynthetic pathway are regulated in E. coli, whereas DapA, encoding dihydrodipicolinate synthase, remains unaltered. This suggests that at least in these bacteria, the regulation of enzyme activity and abundance contributes to regulation. By providing a global analysis of the proteome for such regulation, our data will likely help in deciphering these differences and thus might in the long term lead to microbial strains more efficiently producing lysine.
However, our analysis already has practical avail in the development of nSILAC. In order for proteomics to be useful for most cell and systems biology applications, it needs to deliver quantitative data on changes of protein abundance (1). Several different methods have been developed to obtain this information (5, 36–40). Among them, quantitation based on the ratio of ion currents resulting from peptides derived from proteins metabolically labeled with amino acids containing heavy, non-radioactive isotopes (SILAC) (5) provides a highly accurate and direct measurement of relative protein abundances, as in this case samples are mixed and processed together. Using SILAC for the analysis of global proteomes by quantitative proteomics adds an important dimension to cell and systems biology, particularly of model organisms, in that it involves direct analysis of the effects of perturbations on the levels of proteins, which provide the biologically active agents in cells.
A barrier to the adaptation of SILAC-based quantitative proteomics for the systems and cell biology of microorganisms such as yeasts and bacteria is that it requires metabolic labeling of the organisms with heavy-isotope-containing amino acids. In contrast to mammals, most microorganisms can produce all amino acids biosynthetically and thus are prototrophic for them. As a consequence, SILAC labeling of such organisms has relied on the generation of auxotrophic strains in which one or several genes encoding enzymes of amino acid biosynthetic pathways have been genetically deleted, rendering the resulting cells dependent on the availability of the corresponding amino acid in the medium (27). Although this approach using lysine and/or arginine auxotrophic microorganisms has yielded important data (e.g. 24, 27, 41, 42), it might complicate the interpretation of some experiments, for example, by creating a focus on metabolism or other biological processes linked to amino acid metabolism, which might be altered in auxotrophic mutants. Moreover, dependence on auxotrophic strains precludes the use of previously generated mutants harboring, for example, temperature-sensitive alleles, which provide major advantages in the study of biological processes but are sometimes difficult to transport into other backgrounds (i.e. auxotrophic cells). In addition, the necessity of using lysine auxotroph mutants precludes the use of most systematically generated collections for high-throughput proteomic measurements, which would allow for the integration of proteomic data in the network analysis of model organisms. The restriction of metabolic labeling to auxotrophic organisms also does not permit the proteomic analysis of commercially important microorganisms, such as yeast strains used for brewing, baking, and the industrial production of metabolites (43, 44).
nSILAC overcomes this barrier for the application of comprehensive, quantitative proteomics to model microorganisms, building on data concerning the regulation of lysine biosynthetic pathways. Our systematic development of nSILAC extends previous observations on the metabolic labeling of S. cerevisiae that support our findings (24, 45). The nSILAC approach faithfully recapitulates results obtained with the conventional lysine auxotrophic strains generated by us and others (e.g. for the analysis of the response to osmotic stress) (34). The development of this approach enables labeling and analysis of the majority of available strains generated in S. cerevisiae, S. pombe, and E. coli. As microorganisms are used as food sources for other model systems, application of this strategy also might be useful for their study. Drosophila, for example, feed on yeast, and C. elegans on E. coli. In the latter context, extensive libraries of E. coli expressing double-stranded RNA for interference with gene expression after feeding C. elegans have been established (46, 47). Combining these libraries with nSILAC of the respective strains might enable metabolic labeling and RNAi-mediated depletion of target proteins at the same time. The nSILAC approach will also facilitate the production of metabolically labeled heterologous proteins in these organisms, which could be used as spike-in standards for the absolute quantification of protein abundance via MS.
Supplementary Material
Acknowledgments
We thank Drs. Robert V. Farese, Jr., and Boumediene Soufi for critical comments on our manuscript, and Dr. Megan King for supplying S. pombe strains. We also particularly thank Drs. Nagaraj Nagarjuna and Matthias Mann for invaluable advice on proteomics.
The data associated with this manuscript may be downloaded from the Proteome Commons Tranche using the following hash:
/YDnJo3Ku5OpCAZbcmCMF1×8u4aTgyCc7LOZWKeageDhCZ7DvKnctXr+tCcSDG0jFlaEqA7xA8aq0cDi2cNlD343P/YAAAAAAAAXaw==
During submission, these data are protected by the passphrase FFnSILAC.
Footnotes
* This work was supported by National Institutes of Health grants R01GM097194 and R01GM095982 (T.C.W.).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- E. coli
- Escherichia coli
- S. cerevisiae
- Saccharomyces cerevisiae
- SILAC
- stable isotope labeling by amino acids in cell culture
- S. pombe
- Schizosaccharomyces pombe.
REFERENCES
- 1. Walther T. C., Mann M. (2010) Mass spectrometry-based proteomics in cell biology. J. Cell Biol. 190, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bensimon A., Heck A. J., Aebersold R. (2012) Mass spectrometry-based proteomics and network biology. Ann. Rev. Biochem. 81, 379–405 [DOI] [PubMed] [Google Scholar]
- 3. Yates J. R., Ruse C. I., Nakorchevsky A. (2009) Proteomics by mass spectrometry: approaches, advances, and applications. Ann. Rev. Biomed. Eng. 11, 49–79 [DOI] [PubMed] [Google Scholar]
- 4. Bantscheff M., Lemeer S., Savitski M. M., Kuster B. (2012) Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 404, 939–65 [DOI] [PubMed] [Google Scholar]
- 5. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–86 [DOI] [PubMed] [Google Scholar]
- 6. Martinovic S., Veenstra T. D., Anderson G. A., Pasa-Tolic L., Smith R. D. (2002) Selective incorporation of isotopically labeled amino acids for identification of intact proteins on a proteome-wide level. J. Mass Spectrom. 37, 99–107 [DOI] [PubMed] [Google Scholar]
- 7. Dreisbach A., Otto A., Becher D., Hammer E., Teumer A., Gouw J. W., Hecker M., Volker U. (2008) Monitoring of changes in the membrane proteome during stationary phase adaptation of Bacillus subtilis using in vivo labeling techniques. Proteomics 8, 2062–76 [DOI] [PubMed] [Google Scholar]
- 8. Nirmalan N., Sims P. F., Hyde J. E. (2004) Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol. Microbiol. 52, 1187–99 [DOI] [PubMed] [Google Scholar]
- 9. Huttlin E. L., Hegeman A. D., Harms A. C., Sussman M. R. (2007) Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol. Cell. Proteomics 6, 860–81 [DOI] [PubMed] [Google Scholar]
- 10. Kolkman A., Daran-Lapujade P., Fullaondo A., Olsthoorn M. M., Pronk J. T., Slijper M., Heck A. J. (2006) Proteome analysis of yeast response to various nutrient limitations. Mol. Syst. Biol. 2, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sury M. D., Chen J. X., Selbach M. (2010) The SILAC fly allows for accurate protein quantification in vivo. Mol. Cell. Proteomics 9, 2173–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolfner M., Yep D., Messenguy F., Fink G. R. (1975) Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J. Mol. Biol. 96, 273–90 [DOI] [PubMed] [Google Scholar]
- 13. Sipiczki M. (2000) Where does fission yeast sit on the tree of life? Genome Biol. 1, 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berges D. A., DeWolf W. E., Jr., Dunn G. L., Grappel S. F., Newman D. J., Taggart J. J., Gilvarg C. (1986) Peptides of 2-aminopimelic acid: antibacterial agents that inhibit diaminopimelic acid biosynthesis. J. Med. Chem. 29, 89–95 [DOI] [PubMed] [Google Scholar]
- 15. Feller A., Dubois E., Ramos F., Pierard A. (1994) Repression of the genes for lysine biosynthesis in Saccharomyces cerevisiae is caused by limitation of Lys14-dependent transcriptional activation. Mol. Cell. Biol. 14, 6411–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geng F., Chen Z., Zheng P., Sun J., Zeng A. P. (2013) Exploring the allosteric mechanism of dihydrodipicolinate synthase by reverse engineering of the allosteric inhibitor binding sites and its application for lysine production. Appl. Microbiol. Biotechnol. 97, 1963–71 [DOI] [PubMed] [Google Scholar]
- 17. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–62 [DOI] [PubMed] [Google Scholar]
- 18. Wisniewski J. R., Zielinska D. F., Mann M. (2011) Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal. Biochem. 410, 307–9 [DOI] [PubMed] [Google Scholar]
- 19. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–70 [DOI] [PubMed] [Google Scholar]
- 20. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–12 [DOI] [PubMed] [Google Scholar]
- 21. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–805 [DOI] [PubMed] [Google Scholar]
- 22. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–72 [DOI] [PubMed] [Google Scholar]
- 23. Hubner N. C., Bird A. W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., Mann M. (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189, 739–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Frohlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–4 [DOI] [PubMed] [Google Scholar]
- 25. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–41 [DOI] [PubMed] [Google Scholar]
- 26. Pirkov I., Norbeck J., Gustafsson L., Albers E. (2008) A complete inventory of all enzymes in the eukaryotic methionine salvage pathway. FEBS J. 275, 4111–20 [DOI] [PubMed] [Google Scholar]
- 27. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–27 [DOI] [PubMed] [Google Scholar]
- 28. Ramos F., Dubois E., Pierard A. (1988) Control of enzyme synthesis in the lysine biosynthetic pathway of Saccharomyces cerevisiae. Evidence for a regulatory role of gene LYS14. Eur. J. Biochem. 171, 171–6 [DOI] [PubMed] [Google Scholar]
- 29. Dobson R. C., Devenish S. R., Turner L. A., Clifford V. R., Pearce F. G., Jameson G. B., Gerrard J. A. (2005) Role of arginine 138 in the catalysis and regulation of Escherichia coli dihydrodipicolinate synthase. Biochemistry 44, 13007–13 [DOI] [PubMed] [Google Scholar]
- 30. Ou J., Yamada T., Nagahisa K., Hirasawa T., Furusawa C., Yomo T., Shimizu H. (2008) Dynamic change in promoter activation during lysine biosynthesis in Escherichia coli cells. Mol. Biosyst. 4, 128–34 [DOI] [PubMed] [Google Scholar]
- 31. Chenais J., Richaud C., Ronceray J., Cherest H., Surdin-Kerjan Y., Patte J. C. (1981) Construction of hybrid plasmids containing the lysA gene of Escherichia coli: studies of expression in Escherichia coli and Saccharomyces cerevisiae. Mol. Gen. Genet. 182, 456–61 [DOI] [PubMed] [Google Scholar]
- 32. Haziza C., Stragier P., Patte J. C. (1982) Nucleotide sequence of the asd gene of Escherichia coli: absence of a typical attenuation signal. EMBO J. 1, 379–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hanke S., Besir H., Oesterhelt D., Mann M. (2008) Absolute SILAC for accurate quantitation of proteins in complex mixtures down to the attomole level. J. Proteome Res. 7, 1118–30 [DOI] [PubMed] [Google Scholar]
- 34. Soufi B., Kelstrup C. D., Stoehr G., Frohlich F., Walther T. C., Olsen J. V. (2009) Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst. 5, 1337–46 [DOI] [PubMed] [Google Scholar]
- 35. Beltrao P., Trinidad J. C., Fiedler D., Roguev A., Lim W. A., Shokat K. M., Burlingame A. L., Krogan N. J. (2009) Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 7, e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–69 [DOI] [PubMed] [Google Scholar]
- 37. Wong J. W., Cagney G. (2010) An overview of label-free quantitation methods in proteomics by mass spectrometry. Methods Mol. Biol. 604, 273–83 [DOI] [PubMed] [Google Scholar]
- 38. Schmidt A., Claassen M., Aebersold R. (2009) Directed mass spectrometry: towards hypothesis-driven proteomics. Curr. Opin. Chem. Biol. 13, 510–7 [DOI] [PubMed] [Google Scholar]
- 39. Dephoure N., Gygi S. P. (2012) Hyperplexing: a method for higher-order multiplexed quantitative proteomics provides a map of the dynamic response to rapamycin in yeast. Sci. Signal. 5, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raijmakers R., Berkers C. R., de Jong A., Ovaa H., Heck A. J., Mohammed S. (2008) Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell. Proteomics 7, 1755–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aguilar P. S., Frohlich F., Rehman M., Shales M., Ulitsky I., Olivera-Couto A., Braberg H., Shamir R., Walter P., Mann M., Ejsing C. S., Krogan N. J., Walther T. C. (2010) A plasma-membrane E-MAP reveals links of the eisosome with sphingolipid metabolism and endosomal trafficking. Nat. Struct. Mol. Biol. 17, 901–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henriksen P., Wagner S. A., Weinert B. T., Sharma S., Bacinskaja G., Rehman M., Juffer A. H., Walther T. C., Lisby M., Choudhary C. (2012) Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol. Cell. Proteomics 11, 1510–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peralta-Yahya P. P., Zhang F., del Cardayre S. B., Keasling J. D. (2012) Microbial engineering for the production of advanced biofuels. Nature 488, 320–8 [DOI] [PubMed] [Google Scholar]
- 44. Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S. B., Keasling J. D. (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–62 [DOI] [PubMed] [Google Scholar]
- 45. Dilworth D. J., Saleem R. A., Rogers R. S., Mirzaei H., Boyle J., Aitchison J. D. (2010) QTIPS: a novel method of unsupervised determination of isotopic amino acid distribution in SILAC experiments. J. Am. Soc. Mass Spectrom. 21, 1417–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kamath R. S., Ahringer J. (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–21 [DOI] [PubMed] [Google Scholar]
- 47. Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., Welchman D. P., Zipperlen P., Ahringer J. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.