SUMMARY
Autotransporters, the largest family of secreted proteins in Gram negative bacteria, perform a variety of functions, including adherence, cytotoxicity, and immune evasion. In Yersinia pestis the autotransporter YapE has adhesive properties and contributes to bubonic infection of the mouse model. Here, we demonstrate that omptin cleavage of Y. pestis YapE is required to mediate bacterial aggregation and adherence to eukaryotic cells. We demonstrate that omptin cleavage is specific for the Y. pestis and Y. pseudotuberculosis YapE orthologs but is not conserved in the Y. enterocolitica protein. We also show that cleavage of YapE occurs in Y. pestis but not in the enteric Yersinia species, and requires the omptin Pla (plasminogen activator protease), which is encoded on the Y. pestis-specific plasmid pPCP1. Together, these data show that post-translation modification of YapE appears to be specific to Y. pestis, was acquired along with the acquisition of pPCP1 during the divergence of Y. pestis from Y. pseudotuberculosis, and are the first evidence of a novel mechanism to regulate bacterial adherence.
INTRODUCTION
The genus Yersinia currently comprises 14 species, of which three are considered human pathogens. Y. enterocolitica and Y. pseudotuberculosis are enteric pathogens that infect humans through contaminated food or water (Naktin & Beavis, 1999, Bottone, 1997). Y. pestis recently diverged from Y. pseudotuberculosis to cause an acute systemic infection known as plague (Achtman et al., 1999, Wren, 2003). During the course of its evolution, Y. pestis has adapted to exploit an insect vector as its primary means for transmission (Hinnebusch, 2005). After deposition of bacteria in the dermis by the flea, Y. pestis rapidly colonizes the draining lymph node (Perry & Fetherston, 1997, Butler, 1983). Without proper treatment, the bacteria can disseminate to the blood to cause septicemic plague. From the blood, other organs can be colonized by the bacteria, including the lungs, which can lead to the development of secondary pneumonic plague. Pneumonic plague patients can spread Y. pestis directly to naive individuals via aerosol droplets. Aerosol transmission of Y. pestis is also a major factor contributing to the concerns of the potential use of plague as a biological weapon (Inglesby et al., 2000).
Despite differences in lifestyle, Y. pestis retained important virulence factors that contribute to enteric Yersinia infections (Chain et al., 2004). An important example is the ~70 kb plasmid, conserved in all three species, that encodes a type three secretion system (T3SS) and the effector proteins secreted by this system. In Y. pestis, this plasmid is required for mammalian colonization and the T3SS has been linked to cytotoxicity, inhibition of phagocytosis, and modulation of the mammalian immune response (for review see (Viboud & Bliska, 2005)). In addition to conserved virulence factors, the acquisition of new virulence factors by Y. pestis also has contributed to the adaptation to a vector-borne disease (Wren, 2003). These acquisitions included two plasmids that are not found in the other Yersinia species (Ferber & Brubaker, 1981, Chain et al., 2004, Thomson et al., 2006). Genes on these plasmids contribute to both flea transmission (e.g., murine toxin) and virulence in the mammalian host (e.g., pla and the caf operon) (Weening et al., 2011, Sodeinde et al., 1992, Hinnebusch et al., 2002). Changes to an acute vector-borne disease also have led to mutations in Y. pestis genes that are important for enteric infection but appear to be dispensable for plague. Two significant examples are the adhesins Invasin and YadA (Chain et al., 2004). These adhesins are important virulence factors for Y. enterocolitica and Y. pseudotuberculosis, contributing to host colonization and promoting interactions with host cells required for efficient type three secretion, but are inactive pseudogenes in Y. pestis (Bliska et al., 1993, Miller & Falkow, 1988, Isberg & Falkow, 1985, Paerregaard et al., 1991). The inactivation of inv and yadA suggests either that the functions of Invasin and YadA are not required by Y. pestis or their loss is compensated by other Y. pestis factors. Ongoing genetic divergence of Y. pestis from the enteric Yersinia is likely to be accompanied by the further differentiation of conserved and species specific virulence factors.
Autotransporters are a large family of proteins found in Gram negative bacteria (Dautin & Bernstein, 2007, Benz & Schmidt, 2011, Henderson et al., 2004). They are composed of outer membrane and secreted proteins that use the type V secretion system for export. The family can be further segregated into conventional and trimeric autotransporters, but both autotransporter types have three functional domains. A variable domain, referred to as the passenger domain, is flanked by two conserved domains, an N-terminal signal sequence and C-terminal β-domain. The signal sequence and β-domain are required for translocation of the passenger domain to the outer surface of the bacteria. Furthermore, recent data indicates important roles for periplasmic chaperones and the Bam (Omp85) complex in autotransporter secretion (Bodelon et al., 2009, Jong et al., 2010, Ruiz-Perez et al., 2009, Ruiz-Perez et al., 2010, Sauri et al., 2009). Upon export across the outer membrane, the passenger domain can remain anchored to the β-domain or be proteolytically cleaved. Passenger domains that are cleaved can remain associated with the membrane by non-covalent interactions or diffuse away from the bacteria. Autotransporter cleavage can occur through an autocatalytic mechanism or be mediated in trans by another protein. The variable nature of the passenger domain allows this system to export peptides capable of encoding a variety of functions, including, but not limited to, adhesion, proteolysis, and cytotoxicity (Nishimura et al., 2010, Henderson & Nataro, 2001). Interestingly, many autotransporters described are pathogenesis factors contributing to disease.
In Y. pestis, 10 conventional autotransporters have been identified (Yen et al., 2007, Lenz et al., 2011). We previously demonstrated that seven of these proteins are exported to the outer membrane and three (YapA, YapG and YapE) are secreted into the culture supernatant (Lawrenz et al., 2009, Lenz et al., 2011). The secretion of all three is dependent on the outer surface protease Pla. Genome comparisons indicate that all 10 autotransporters were acquired by Y. pseudotuberculosis prior to the divergence of Y. pestis, but only YapE appears also to be conserved in Y. enterocolitica. In addition, we previously showed that YapE is required by Y. pestis for efficient colonization of the draining lymph node during bubonic infection of mice (Lawrenz et al., 2009). Furthermore, YapE has adhesive properties and can mediate aggregation of bacteria and adherence to eukaryotic cells. In this study, we expand the analysis of YapE to the other Yersinia species and specifically compare the functional properties of Y. pestis and Y. enterocolitica YapE proteins. We demonstrate differences in YapE processing by different Yersinia species, define the effect of processing on function, and use these differences to identify the cleavage sites in Y. pestis YapE.
RESULTS
YapE is conserved in the Yersiniae
YapE was originally identified as a pathogenicity factor of Y. pestis that contributes to lymph node colonization during bubonic infection (Lawrenz et al., 2009). Our previous analyses revealed that YapE is the only conventional autotransporter conserved in the three species of Yersinia that cause human disease. With the availability of additional genome data, we extended our in silico analysis to the other Yersinia species and identified YapE orthologs in Y. aldovae, Y. bercovieri, Y. mollaretii, and Y. rohdei. Phylogenetic analysis indicated that YapE proteins fall into two major groups within the genus (Supporting Information, Fig. 1). Group A proteins are conserved in Y. pestis and Y. pseudotuberculosis. These proteins share >97% amino acid identity over the entire sequence. The second group had a broader distribution in multiple species (Group B). Consequently, variability is higher among Group B proteins than among Group A proteins. However, Group B proteins differ more significantly from Group A proteins than from other members of Group B (65% amino acid sequence identity to Group A). While Y. rohdei appears to encode a YapE ortholog, this ortholog is in a different location within the genome as seen for the other species and has diverged significantly from both Group A and Group B proteins. In Y. frederiksenii yapE appears to be a pseudogene and Y. ruckeri lacks not only yapE but also several of the flanking genes within the yapE region. Interestingly, the genes downstream of yapE in Y. pestis (YPO3983-YPO3978) are only conserved in Y. pestis and Y. pseudotuberculosis and are not found in the other species. Finally, our analysis indicated that YapE is a Yersinia specific protein, as it had little similarity to proteins in other bacteria. Proteins from other genera with any similarity, including the Rahnella protein serving as an outgroup for the phylogenetic analysis (Supplementary Material), tend to share higher degrees of similarity within the autotransporter-specific conserved domains but have limited similarity with the YapE passenger domain (≤38% amino acid identity).
Concentrating on the Y. enterocolitica YapE as a representative of Group B proteins, we identified a putative signal peptide and autotransporter β-domain within the protein (data not shown). The protein shares about 65% identity to YapE from Y. pestis and this identity is over the entire length of the protein, with both conserved domains (e.g. β- and pertactin domains) and the passenger domains having undergone similar degrees of divergence. The Y. enterocolitica ortholog is nine amino acids longer than Y. pestis YapE and has two small insertions (or perhaps deletions in the Y. pestis protein) at amino acids 547–553 (ASPLNII) and 751–760 (DPGTGIGVDP).
YapE from Y. enterocolitica does not promote bacterial aggregation
Previous studies demonstrated that expression of Y. pestis YapE in E. coli results in aggregation of the bacteria (Lawrenz et al., 2009). We hypothesized that divergence of YapE in Y. enterocolitica could impact these functions. To determine if the Y. enterocolitica ortholog promotes aggregation, the protein was expressed in E. coli and aggregation was determined by monitoring settling of the cultures. As previously reported, expression of Y. pestis YapE resulted in aggregation of bacteria that settled in the culture tube (Fig. 1A), forming a rippled pellet morphology. The Y. pseudotuberculosis ortholog, which shares 97% identity to the Y. pestis protein, also promoted aggregation of E. coli at a similar rate. In contrast, E. coli expressing Y. enterocolitica YapE did not settle during the entire course of the assay, even when the amount of inducer was doubled (data not shown). These data indicate that Y. enterocolitica YapE does not promote bacterial aggregation.
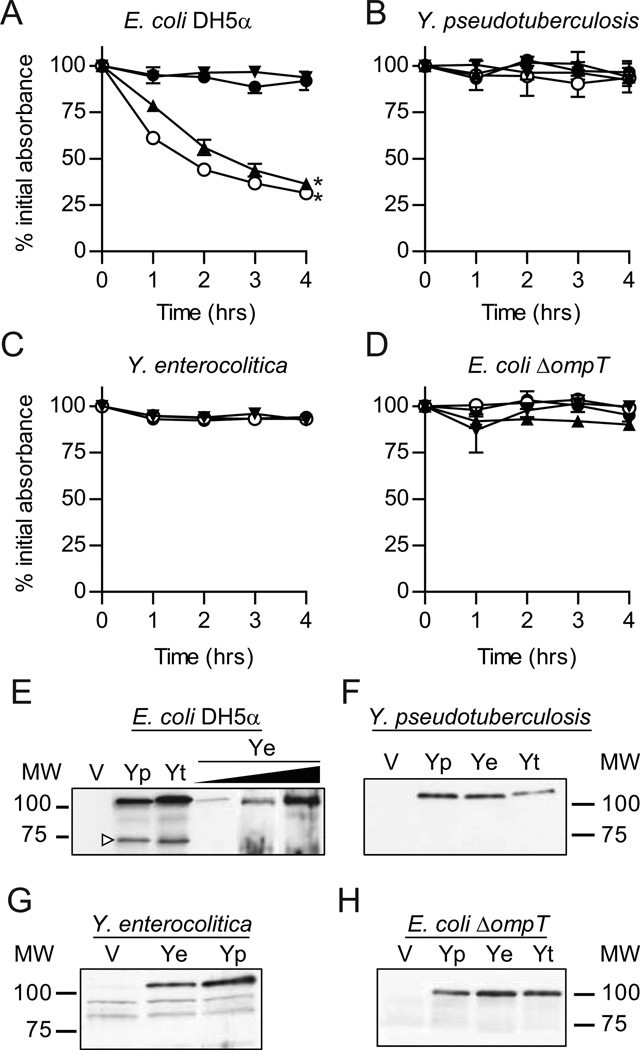
Figure 1. Analysis of YapE-mediated aggregation.
For settling assays, YapE otholog expression was induced for 2 hrs and then samples were harvest from static cultures at a specific depth at 1 hr intervals. The absorbance (OD600) of these samples were determined and compared to the OD600 at T=0. (A) E. coli, (B) Y. pseudotuberculosis, (C) Y. enterocolitica, and (D) E. coli ΔompT. ● = Vector only (pLP-PROTet- 6xHN); ○ = Y. pestis YapE (pMBL295); ▲ = Y. pseudotuberculosis YapE (pMBL299); ▼ = Y. enterocolitica YapE (pMBL291). Three independent biological replicates were performed for each bacterial strain and each assay was repeated three times (n=9). Each symbol represents the mean percent initial absorbance ± the standard deviation; *=P≤0.0005. At 4 hrs, 1 OD600 equivalents of total bacterial proteins were isolated from one representative sample and YapE processing was determined by Western blot with anti-YapE serum. (E) E. coli, (F) Y. pseudotuberculosis, (G) Y. enterocolitica, (H) E. coli ΔompT. V = vector; Yp = Y. pestis YapE; Yt = Y. pseudotuberculosis YapE; Ye = Y. enterocolitica YapE. White arrowhead indicates processed YapE. Black triangle represents addition of increasing concentrations of anhydrous tetracycline to demonstrate that increasing the levels of expression does not result in cleavage of Ye.
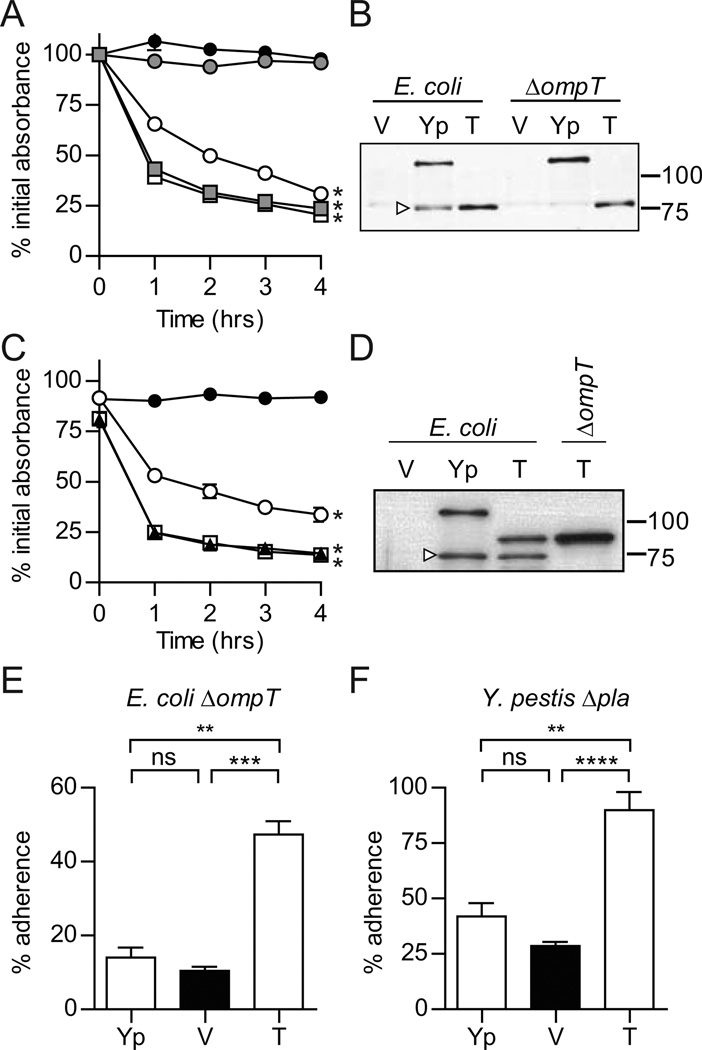
To determine whether or not the Y. enterocolitica YapE phenotype was specific to E. coli, we performed similar settling assays in Y. enterocolitica and Y. pseudotuberculosis. Unexpectedly, none of the three YapE orthologs promoted aggregation in these Yersinia backgrounds (Fig. 1B and 1C). To eliminate the possibility that YapE was not expressed in the Yersinia strains, Western blot analysis of the bacterial cell pellets with anti-YapE serum was performed (Fig. 1E–G). Each YapE ortholog was expressed in the Yersinia strains at similar levels as seen in E. coli; thus, lack of protein expression was not the cause of the non-aggregating phenotype. However, this analysis revealed that the Y. pestis and Y. pseudotuberculosis YapE orthologs were processed in E. coli but not in the Yersinia strains. In contrast, the Y. enterocolitica YapE was not processed in any of the strains, including E. coli. This difference in processing suggested that YapE processing is required for aggregation.
YapE is processed by the omptin proteases OmpT (in E. coli) and Pla (in Y. pestis) (Lawrenz et al., 2009). Therefore, to determine the impact of omptin cleavage on YapE-mediated aggregation, we generated an E. coli strain with a deletion in ompT. Upon induction of YapE expression in this background, we did not observe aggregation by any of the three orthologs (Fig. 1D). Western blot analysis confirmed that none of the three proteins were processed in the ΔompT E. coli strain (Fig. 1H). Together these data suggest that (a) omptin cleavage of Y. pestis/Y. pseudotuberculosis YapE proteins is necessary to promote aggregation, and (b) the Y. enterocolitica ortholog differs from Group A proteins by the absence of an OmpT cleavage site and an inability to promote aggregation.
Y. pestis YapE requires omptin cleavage for adherence to eukaryotic cells
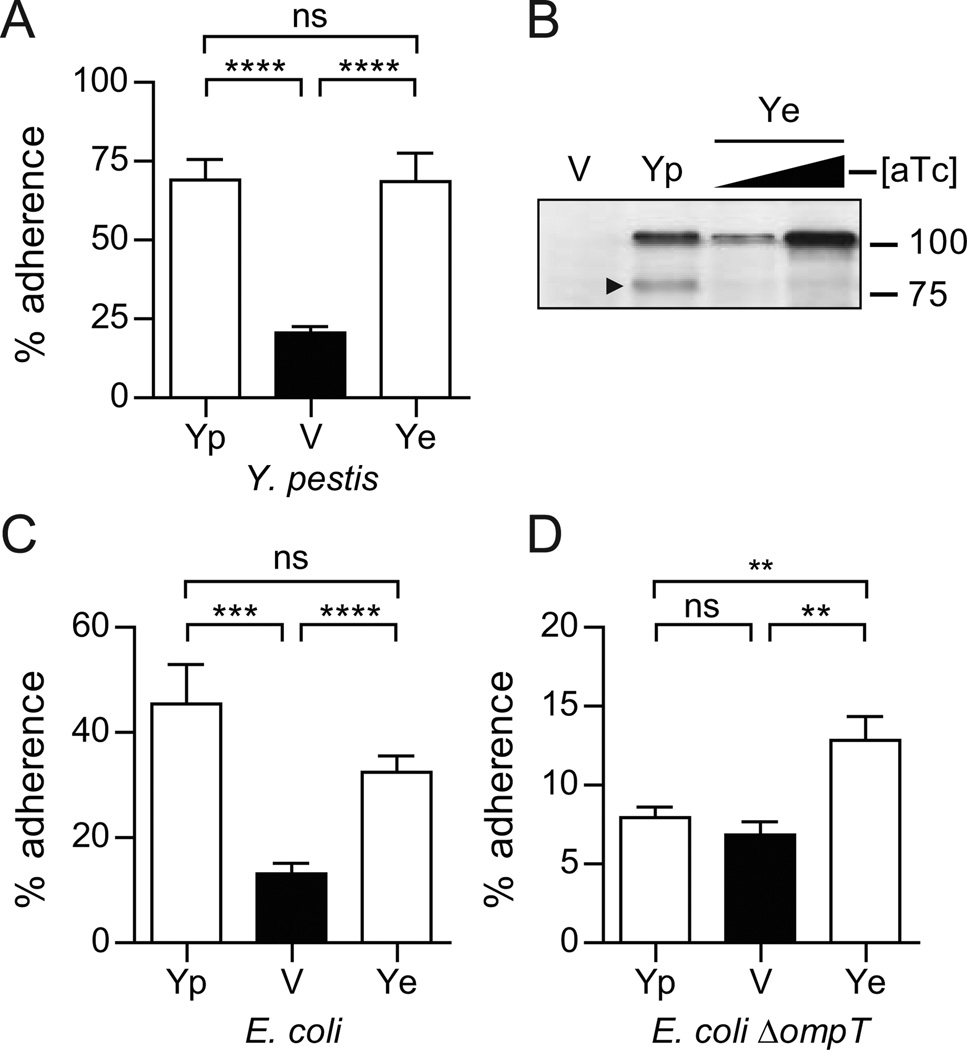
In addition to promoting bacterial aggregation, we previously demonstrated that Y. pestis YapE is an adhesin for eukaryotic cells (Lawrenz et al., 2009). The sequence divergence and differences in the aggregation phenotype raised the question of whether or not the Y. enterocolitica YapE has the other adherence functions. To determine if Y. enterocolitica YapE promotes adherence to eukaryotic cells, we incubated Y. pestis strains expressing either the Y. pestis or Y. enterocolitica YapE protein with RAW264.7 macrophage-like cells. After 30 minutes of incubation, unbound bacteria were removed by washing, and adherent bacteria were enumerated. The percent of adherent bacteria was determined by comparing to unwashed wells (total bacteria). When compared to Y. pestis with the vector only, expression of native Y. pestis YapE significantly increased the number of adherent bacteria by 3.5-fold (Fig. 2A, P<0.0001). Interestingly, expression of the Y. enterocolitica ortholog in Y. pestis also promoted a significant increase in adherence (Fig. 2A, P<0.0001). We also performed adherence assays using E. coli to determine if YapE alone (without other Y. pestis proteins) could confer adherence to RAW264.7 cells. As seen in Y. pestis, expression of both orthologs in E. coli promoted adherence to the cells (Fig. 2C, P<0.001 and P<0.0001, for Y. pestis and Y. enterocolitica YapE orthologs, respectively). Together these data demonstrate that while the Y. enterocolitica YapE does not promote autoaggregation, it still promotes adherence to eukaryotic cells.
Figure 2. YapE adherence to RAW264.7 macrophages.
The ability of YapE orthologs to mediate adherence to eukaryotic cells was determined with RAW264.7 cells. (A) Adherence assays were performed with Y. pestis Δcaf1, ΔpsaA expressing the YapE orthologs. Yp = Y. pestis YapE (pMBL313); Ye = Y. enterocolitica YapE (pMBL311); V = vector control (pMWO-034). (B) Western blot analysis of YapE expression in Y. pestis strains. Black arrowhead indicates processed YapE. Triangle represents addition of increasing concentrations of anhydrous tetracycline in Ye samples to demonstrate that higher levels of expression does not result in cleavage of Ye. (C) Adherence assays with E. coli or (D) E. coli ΔompT expressing the YapE orthologs. YP = Y. pestis YapE (pMBL313); Ye = Y. enterocolitica YapE (pMBL311); V=vector control (pMWO-034). Percent adherence was calculated as the number of cell-associated cfu divided by total cfu from a separate tissue culture well. Three independent biological replicates were performed for each adherence assay and each assay was repeated three times (n=9). Each bar represents the mean percent adherence ± the standard deviation; ** = P≤0.01; *** = P≤0.001; **** = P≤0.0001.
Omptin processing of Y. pestis YapE is required for aggregation, suggesting that omptin cleavage may also be required for YapE mediated adherence to eukaryotic cells. To determine the impact of omptin cleavage of YapE on adhesion, we performed adherence assays with the ΔompT strain of E. coli expressing the different YapE orthologs (Fig. 2D). Deletion of ompT did not affect adherence mediated by the Y. enterocolitica YapE protein (still observed ~2-fold increase in adherence; P>0.001). However, deletion of the protease, and thus proteolytic cleavage of the Y. pestis YapE protein, resulted in loss of adherence to the cells. These data suggest that both Y. pestis and Y. enterocolitica YapE proteins are eukaryotic adhesins, but only the Y. pestis YapE requires omptin cleavage for adherence to cells.
YapE cleavage differs between E. coli and Y. pestis
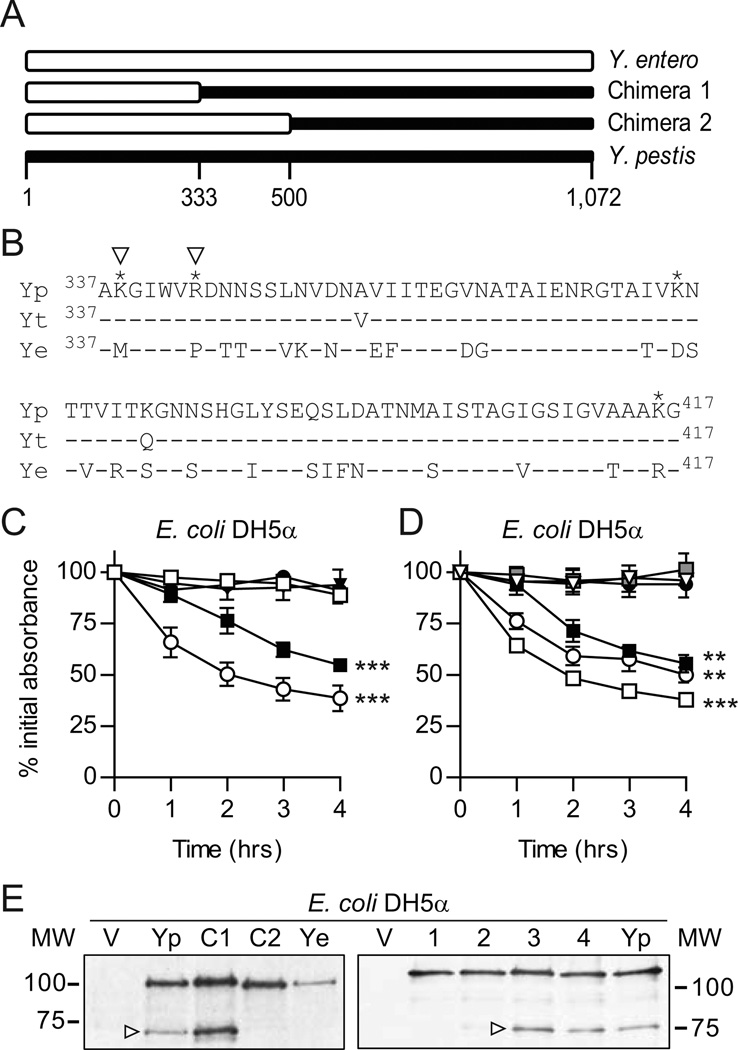
Our data from the omptin mutants demonstrated that Y. pestis YapE cleavage is necessary for the protein to mediate adherence. Current data suggest that omptins preferentially cleave between two basic amino acids (arginine or lysine), but a consensus recognition sequence has not been well characterized (−/−/a/RK*Rk/agv/ar/g with * representing the cleavage site; http://merops.sanger.ac.uk). The Y. pestis YapE protein contains 54 arginine or lysine residues and based on estimated mass of the cleavage product, several of these residues could be part of the cleavage site. However, because Y. enterocolitica YapE is not cleaved by omptins, we devised a strategy to narrow the possible region/candidates for the omptin cleavage site. Toward this end, we generated two chimeric proteins by replacing the N-terminal portion of the Y. pestis YapE protein with the equivalent regions from Y. enterocolitica (Fig. 3A). Expression of these chimeric proteins in E. coli demonstrated that while Chimera 1 retained its ability to mediate aggregation, E. coli expressing Chimera 2 did not aggregate (Fig. 3C). Western analysis confirmed that Chimera 2 was not processed in E. coli (Fig. 3E). These results suggested that the OmpT cleavage site for Y. pestis YapE is between amino acids 333 and 500.
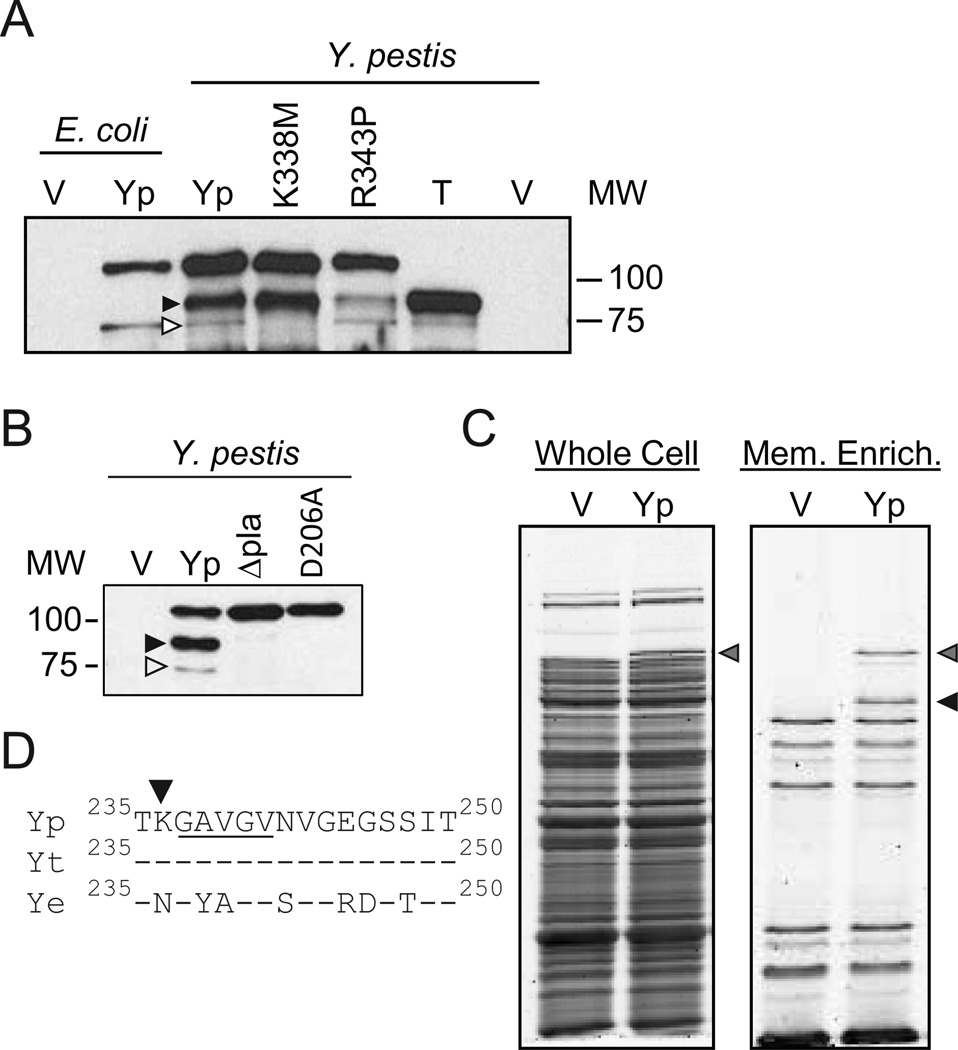
Figure 3. Identification of OmpT cleavage site in YapE.
To identify the OmpT cleavage site in Y. pestis YapE, we constructed chimeric proteins consisting of the N-terminal portion of the Y. enterocolitica protein and the C-terminal portion of the Y. pestis protein. These chimeric proteins allowed for the identification of regions where the OmpT cleavage was located. Once the region was identified, K and R residues were identified in this region that differed between the two Yersinia orthologs and site directed mutagenesis was used to identify residues required for cleavage. (A) Schematic of the chimeric proteins. White and black boxes represent Y. enterocolitica and Y. pestis sequences, respectively. (B) Alignment of YapE proteins highlighting K and R residues (*) that differ between Y. pestis and Y. enterocolitica. Yp = Y. pestis YapE, Yt = Y. pseudotuberculosis YapE; Ye = Y. enterocolitica YapE; * = residues mutated in Y. pestis YapE pt mutants. (C) Settling assay with E. coli expressing YapE chimeric proteins. ● = Vector only (pMWO-034); ○ = Y. pestis YapE (pMBL313); ▼ = Y. enterocolitica YapE (pMBL311); ■ = Chimera 1 (pMBL326); □ = Chimera 2 (pLOU013). (D) Settling assay with E. coli expressing Y. pestis YapE point mutations. ● = Vector only (pMWO-034); ○ = Y. pestis YapE (pMWO-034); ∇ = K338M (pLOU021);  = R343P (pLOU022); ■ = K375D (pLOU023); □ = K416R (pLOU024). For settling assays, YapE otholog expression was induced for 2 hrs and then samples were harvest from static cultures at a specific depth at 1 hr intervals. The absorbance (OD600) of these samples were determined and compared to the OD600 at T=0. Three independent biological replicates were performed for each bacterial strain and each assay was repeated three times (n=9). Each symbol represents the mean percent initial absorbance ± the standard deviation; ** = P≤0.002; *** = P≤0.0002. (E) Western blot analysis of YapE cleavage from strains in C and D, respectively. V = Vector only; Yp = WT Y. pestis YapE; C1 = Chimera 1; C2 = Chimera 2; Ye = Y. enterocolitica YapE; 1 = K338M; 2 = R343P; 3 = K375D; 4 = K416R. White arrowheads represent YapE OmpT cleavage site/product.
= R343P (pLOU022); ■ = K375D (pLOU023); □ = K416R (pLOU024). For settling assays, YapE otholog expression was induced for 2 hrs and then samples were harvest from static cultures at a specific depth at 1 hr intervals. The absorbance (OD600) of these samples were determined and compared to the OD600 at T=0. Three independent biological replicates were performed for each bacterial strain and each assay was repeated three times (n=9). Each symbol represents the mean percent initial absorbance ± the standard deviation; ** = P≤0.002; *** = P≤0.0002. (E) Western blot analysis of YapE cleavage from strains in C and D, respectively. V = Vector only; Yp = WT Y. pestis YapE; C1 = Chimera 1; C2 = Chimera 2; Ye = Y. enterocolitica YapE; 1 = K338M; 2 = R343P; 3 = K375D; 4 = K416R. White arrowheads represent YapE OmpT cleavage site/product.
To further refine the region cleaved by OmpT, we identified lysine and arginine residues that differed between Y. pestis and Y. enterocolitica in this region of YapE. Four candidates were identified: K338, R343, K375, and K416 (K382 is not conserved in Y. pseudotuberculosis YapE and was thus excluded as a candidate) (Fig. 3B). Using site directed mutagenesis, we mutated each candidate amino acid individually in Y. pestis YapE to the Y. enterocolitica equivalent. Expression of the point mutant constructs in E. coli revealed that changes to K338 or R343 inhibited aggregation and cleavage (Fig. 3D and E). Because both K338 and R343 affected cleavage, we were unable to define the exact residue cleaved by OmpT. However, because these amino acids are only 5 residues apart it is possible that one amino acid represents the cleavage site while disruption of the other interferes with OmpT recognition of or binding to the peptide.
Initially we suspected that OmpT and Pla cleaved YapE at the same location. However, when we analyzed E. coli and Y. pestis samples on the same Western blot, we realized that the truncated YapE from Y. pestis migrates at a different size than the truncated form in E. coli (Fig. 4A). Furthermore, expression of the K338M and R343P point mutations in Y. pestis did not appear to alter cleavage of Y. pestis YapE (Fig. 4A, lanes K338M and R343P, black arrowhead). However, increasing the film exposure time revealed a second Pla cleavage product in the control sample that migrated at the same size as truncated YapE in E. coli (Fig. 4A, lane YP, white arrowhead). This product was absent in the strain expressing the K338M site directed YapE mutant but not the R343P site directed YapE mutant (Fig. 4A), indicating that Pla also cleaves Y. pestis YapE at the K338M site which results in a minor product that is similar in size as the OmpT cleavage product. Furthermore, these results suggest that in addition to the OmpT cleavage site, Pla also recognizes a second site that OmpT does not, and this is the primary cleavage site in Y. pestis. Finally, we also showed that both cleavage products are dependent on Pla as both are absent in Δpla and catalytically inactive Pla mutants of Y. pestis (Fig. 4B).
Figure 4. Identification of Pla cleavage sites in YapE.
(A)To identify the Pla cleavage sites in Y. pestis YapE, YapE point mutants that altered OmpT cleavage of YapE were expressed in Y. pestis and Western blots were performed with anti-YapE serum. V = Vector only (pMWO-034); Yp = WT Y. pestis YapE (pMBL313); K338M = K338M pt. mutant (pLOU021); R343P = R343P pt. mutant (pLOU022); T = YapEΔ34-236 (pLOU109). As a reference, V and Yp samples expressed in E. coli are also included. (B) Anti-YapE Western blots of Y. pestis YapE expressed in Y. pestis (Yp), Y. pestis Δpla, or Y. pestis with catalytically inactive Pla (D206A). V = vector control. (C)Whole cell proteins and outer membrane enriched samples from Y. pestis stained with Flamingo fluorescent stain. Yp = Y. pestis YapE (pMBL313); V = Vector control (pMWO-034). (D) Alignment of YapE proteins between amino acids 235 and 250. Yp = Y. pestis YapE, Yt = Y. pseudotuberculosis YapE; Ye = Y. enterocolitica YapE; underlined region represent the N-terminal amino acids of the Pla cleaved YapE peptide. Gray arrowheads represent full length YapE, white arrowheads represent YapE OmpT cleavage site/product, and black arrowheads represent primary YapE Pla cleavage site/product.
These data indicated that the Pla primary cleavage site is closer to the N-terminal end of YapE than the OmpT cleavage site. To identify this cleavage site, we used sarkosyl enrichment to isolate outer membrane proteins from Y. pestis expressing YapE. This approach allowed us to isolate the major truncated peptide from other Y. pestis proteins and use N-terminal sequencing to identify the Pla cleavage site (Fig. 4C). N-terminal sequencing of the isolated fragment revealed that the N-terminal amino acids of the primary truncated YapE peptide were GAVGV. These residues are directly downstream of K236, supporting the consensus data that omptins cleave after basic amino acids (Fig. 4D). We further generated a truncated form of YapE lacking the region between the signal sequence and K236 and observed that this protein migrates at the same size as the major Pla truncated peptide (Fig. 4A, Lane T).
The secreted N-terminal portion of YapE is not required for adherence
While YapE cleavage is necessary for adherence, it is unclear if the secreted portion of the passenger domain directly contributes to this function. To determine if the membrane anchored region is sufficient for adherence, we generated truncated forms of Y. pestis YapE that lacked the secreted regions of the passenger domain between the signal sequence and omptin cleavage sites (Δ34-337 and Δ34-236, OmpT and Pla cleavage sites respectively). Both constructs were subsequently expressed in E. coli and settling assays were performed to determine if the truncated proteins could mediate aggregation. As expected, aggregation mediated by full length YapE was dependent on the presence of OmpT (Fig. 5A). However, expression of either of the truncated constructs resulted in settling of both the E. coli and ΔompT strains (Fig 5A and C). Western blot analysis confirmed protein expression in all strains (Fig. 5B and D).
Figure 5. Analysis of adherence by truncated YapE.
Y. pestis YapE proteins lacking the N-terminal portions of the passenger domain were expressed in E. coli and Y. pestis omptin mutants to determine if the truncated proteins could restore adherence. (A) Settling assays with WT and ΔompT E. coli expressing YapEΔ34-337. ● = WT E. coli, vector only (pMWO-034); ○ = WT E. coli, Y. pestis YapE (pMBL313);  = ΔompT, Y. pestis YapE (pMBL313); □ = WT E. coli, YapEΔ34-337 (pLOU059);
= ΔompT, Y. pestis YapE (pMBL313); □ = WT E. coli, YapEΔ34-337 (pLOU059);  = ΔompT, YapEΔ34-337 (pLOU059). YapE expression was induced for 2 hrs and then samples were harvest from static cultures at a specific depth at 1 hr intervals. The absorbance (OD600) of these samples were determined and compared to the OD600 at T=0. Three independent biological replicates were performed for each bacterial strain and each assay was repeated three times (n=9). Each symbol represents the mean percent initial absorbance ± the standard deviation; * = P≤0.0001 (B) Western blot of YapE proteins from A. V = vector control; Yp = Y. pestis YapE; T = YapEΔ34-337. White arrowhead indicates OmpT cleavage site. (C) Settling assays (performed as described in A) with WT and ΔompT E. coli expressing YapEΔ34-236. ● = WT E. coli, vector only (pMWO-034); ○ = WT E. coli, Y. pestis YapE (pMBL313); □ = WT E. coli, YapEΔ34-236 (pLOU109); ▲ = ΔompT, YapEΔ34-236 (pLOU109). * = P≤0.0001. (D) Western blot of YapE proteins from C. V = vector control; Yp = Y. pestis YapE; T = YapEΔ34-236. White arrowhead indicates OmpT cleavage site. (E and F) Adherence assays with E. coli ΔompT (E) or Y. pestis Δpla (F) expressing truncated YapE. Yp = Y. pestis YapE (pMBL313); V = vector control (pMWO-034); T = truncated YapE (YapEΔ34-337 [pLOU059] or YapEΔ34-236 [pLOU109] for E and F, respectively). Fold adherence was calculated as the number of cell-associated cfu divided by total cfu from a separate tissue culture well. Three independent biological replicates were performed for each adherence assay and each assay was repeated three times (n=9). Each bar represents the mean percent adherence ± the standard deviation; ** = P≤0.002; *** = P≤0.0006; **** = P≤0.0001.
= ΔompT, YapEΔ34-337 (pLOU059). YapE expression was induced for 2 hrs and then samples were harvest from static cultures at a specific depth at 1 hr intervals. The absorbance (OD600) of these samples were determined and compared to the OD600 at T=0. Three independent biological replicates were performed for each bacterial strain and each assay was repeated three times (n=9). Each symbol represents the mean percent initial absorbance ± the standard deviation; * = P≤0.0001 (B) Western blot of YapE proteins from A. V = vector control; Yp = Y. pestis YapE; T = YapEΔ34-337. White arrowhead indicates OmpT cleavage site. (C) Settling assays (performed as described in A) with WT and ΔompT E. coli expressing YapEΔ34-236. ● = WT E. coli, vector only (pMWO-034); ○ = WT E. coli, Y. pestis YapE (pMBL313); □ = WT E. coli, YapEΔ34-236 (pLOU109); ▲ = ΔompT, YapEΔ34-236 (pLOU109). * = P≤0.0001. (D) Western blot of YapE proteins from C. V = vector control; Yp = Y. pestis YapE; T = YapEΔ34-236. White arrowhead indicates OmpT cleavage site. (E and F) Adherence assays with E. coli ΔompT (E) or Y. pestis Δpla (F) expressing truncated YapE. Yp = Y. pestis YapE (pMBL313); V = vector control (pMWO-034); T = truncated YapE (YapEΔ34-337 [pLOU059] or YapEΔ34-236 [pLOU109] for E and F, respectively). Fold adherence was calculated as the number of cell-associated cfu divided by total cfu from a separate tissue culture well. Three independent biological replicates were performed for each adherence assay and each assay was repeated three times (n=9). Each bar represents the mean percent adherence ± the standard deviation; ** = P≤0.002; *** = P≤0.0006; **** = P≤0.0001.
Next we examined whether the truncated proteins could mediate adherence to eukaryotic cells. While the full length Y. pestis YapE protein did not increase adherence to eukaryotic cells in the omptin mutants, expression of the Δ34-337 construct in the E. coli ΔompT strain or the Δ34-236 construct in Y. pestis Δpla significantly increased the number of bacteria adhering to eukaryotic cells (Fig. 5E and F; P≤0.001 and P≤0.0001, respectively). These data demonstrate that the N-terminal portion of the Y. pestis YapE passenger domain is not required for adherence and suggest that this region inhibits the adhesive domain of YapE in the unprocessed, full length protein.
DISCUSSION
Our previous work demonstrated that three conventional autotransporters identified in Y. pestis are processed after translocation to the cell surface and secreted into the culture supernatant (Lawrenz et al., 2009, Lenz et al., 2011). While it appears that the majority of the passenger domains of YapA and YapG are secreted, YapE is unique in that processing results in only a small portion of the passenger domain being released. Therefore, a substantial portion of the YapE passenger domain remains associated with the bacteria. Several examples of post-translational modification of autotransporters have been reported, including cleavage of the passenger domain after translocation. The AIDA-1 adhesin from E. coli is cleaved through an autoproteolytic mechanism, but the passenger domain remains non-covalently attached to the bacteria to mediate adherence. However, cleavage is not necessary for adherence, and protease mutants demonstrate wild type levels of adherence (Charbonneau et al., 2006). The adhesin Hap from H. influenzae undergoes an intermolecular autoproteolysis, but the passenger domain is released into the supernatant after cleavage (Fink et al., 2001). Hap can only mediate autoaggregation and adherence to epithelial cells when the protein is at low densities on the bacterial surface or when autoproteolysis is inhibited (Fink & St Geme, 2003). Therefore, the full length protein mediates adherence, and cleavage appears to be a release mechanism to remove the adhesin. More recently, the autotransporter EatA was shown to mediate adherence by targeting another adhesin, EtpA (Roy et al., 2011). EtpA is an adhesin secreted via the two partner secretion system (related to the autotransporter secretion system) and promotes adherence to intestinal epithelial cells (Fleckenstein et al., 2006). Similar to AIDA-1 and Hap, cleavage of EtpA by EatA results in decreased adherence. Unlike these proteins, Y. pestis YapE requires proteolytic processing by an omptin to become an active adhesin. Based on our data with truncated YapE, our working model indicates that the N-terminal region of the YapE passenger domain hides or inhibits an adhesive domain in the unprocessed, full length protein. When cleaved by OmpT or Pla, the N-terminal region is released, either exposing the adhesive domain or resulting in a conformational change in the remaining portion of the protein that activates the adhesive domain. To our knowledge, this is the first demonstration of a bacterial adhesin requiring protease cleavage to become active, representing a novel post-translational mechanism to regulate bacterial adherence.
Several studies suggest that optimal Pla activity coincides with increased YapE expression, further supporting Pla activity as a post-transcriptional regulatory mechanism of YapE. Pla expression and activity are thermoregulated and induced under in vitro conditions mimicking mammalian infection (Chromy et al., 2005, McDonough & Falkow, 1989). Furthermore, changes in the LPS composition of Y. pestis at 37°C result in increased Pla activity at mammalian temperatures (Kukkonen et al., 2004). We demonstrated that YapE, while not regulated by temperature, is induced during mammalian infection, especially within the lymph nodes (Lenz et al., 2011). These data support a hypothesis that maximal expression of functional forms of YapE on the surface of Y. pestis most likely occurs during mammalian infection when yapE transcription and Pla activity are likely highest. Future analysis of the role of YapE adherence during early stages of bubonic infection will lead to a better understanding of the protein’s role in colonization and in identifying targets of YapE-mediated adherence.
The mechanism of Y. pestis dissemination from the inoculation site to the draining lymph node is not well understood, but Pla has an important role during bubonic plague and a pla mutant is significantly defective in dissemination from the site of infection (Sodeinde et al., 1992). Pla actively cleaves several host substrates and has been extensively studied for its role in host plasminogen activation (reviewed in (Suomalainen et al., 2007) and (Haiko et al., 2009)). The current hypothesis to explain defective dissemination by the pla mutant is that plasmin production by Pla degrades fibrin deposits, allowing Y. pestis to disseminate from the inoculation site. However, a yapE mutant is also defective in dissemination, and specifically in colonization of the draining lymph node (Lawrenz et al., 2009). While host plasminogen activation is clearly important for Y. pestis pathogenesis (Degen et al., 2007), the absence of YapE cleavage also may contribute to the dissemination phenotype observed in the pla mutant. YapE adherence may promote interactions with host cells at the site of infection or within the lymph nodes that are beneficial to colonization, and these interactions would be absent in the pla mutant. Future work to compare the interactions with host cells by wild type Y. pestis and the yapE mutant at the site of infection and the lymph nodes will provide a better understanding of the role of YapE adherence in pathogenesis.
Previous work has established that OmpT and Pla have different affinities for substrates. For example, Pla is much more efficient at cleaving host plasminogen and α2-antiplasmin than OmpT (Kukkonen et al., 2001), suggesting that these two omptins may recognize different cleavage sites. Furthermore, we have observed that Pla can be more promiscuous than OmpT, especially when the primary cleavage site is mutated ((Lenz et al., 2011) and M.C. Lane and V.L. Miller, unpublished data). Our attempts to identify the omptin cleavage sites in YapE confirm these observations. OmpT cleaves YapE at only one site, but Pla cleaves the protein at two sites. Furthermore, while Pla and OmpT recognized the same site, Pla appears to preferentially cleave at a site not recognized by OmpT. However, cleavage at either site results in activation of the adhesive domain of YapE, indicating that the adherence domain(s) is C-terminal to the OmpT cleavage site.
The regulation of YapE adherence by omptin cleavage also raises interesting questions about the evolution of YapE. Y. pseudotuberculosis YapE contains an omptin cleavage site and requires omptin processing to become an active adhesin, but we have been unable to demonstrate YapE processing in Y. pseudotuberculosis. Y. pseudotuberculosis does not encode pla, a Y. pestis specific gene found on the pPCP1 plasmid which is absent in the other Yersiniae. The acquisition of this plasmid by the ancestral bacterium that eventually became Y. pestis is thought to be an important step in the evolution from an enteric pathogen to a vector borne pathogen. However, the question remains: why did Y. pseudotuberculosis YapE evolve and acquire an omptin cleavage site in the absence of Pla? One possible explanation is that Y. pseudotuberculosis has another omptin that recognizes YapE. The genome sequence of Y. pseudotuberculosis indicates the presence of an open reading frame (YPTB1266; also referred to as pla2 or ycoB) on the chromosome that appears to encode an omptin. However, protease activity for YcoB has yet to be shown (Haiko et al., 2010). Sequence analysis of this open reading frame indicates it has a mutation in one of the predicted omptin active site Asp residues (D84). Perhaps, this omptin was once active and capable of cleaving YapE, providing a mechanism of selection for the evolution of YapE processing. Interestingly, homologs of YPTB1266 exist in Y. pestis and Y. enterocolitica, but these also appear to encode inactive proteases (the same point mutation exists in Y. pestis, and the Y. enterocolitica 8081 homolog contains a frame shift mutation). The conservation of the active site mutation in Y. pestis YcoB suggests the mutation occurred prior to the divergence of Y. pseudotuberculosis and Y. pestis. It appears that through the acquisition of pPCP1 and pla, Y. pestis also acquired a cryptic function in YapE that is currently hidden in Y. pseudotuberculosis. Furthermore, two other autotransporters, YapA and YapG, also are cleaved in a Pla dependent manner (Lenz et al., 2011). These findings suggest that the acquisition of Pla not only provided a mechanism to cleave host proteins but may also have resulted in the processing of Y. pseudotuberculosis proteins that contributed to the ability of Y. pestis to alter its lifestyle. Further identification and analysis of other Y. pestis proteins processed by Pla may identify additional cryptic virulence factors in the Y. pseudotuberculosis genome.
Unlike the Group A YapE orthologs, the Y. enterocolitica protein does not appear to contain an omptin cleavage site, at least one recognized by Pla or OmpT. The Y. enterocolitica YapE protein has diverged considerably from the Group A proteins. Our cell adherence data suggest that unlike the Y. pestis protein, Y. enterocolitica YapE does not require processing to adhere to host cells. However, we did not observe Y. enterocolitica YapE-mediated bacterial aggregation. In addition, bacteria expressing processed Y. pestis YapE had consistantly higher adherence rates than those expressing the Y. enterocolitica protein. Selection for increased adhesive properties in YapE by Y. pestis may compensate for the lack of the two major adhesins, Invasin and YadA, in the plague pathogen. We also cannot rule out the possibility that YapE-mediated aggregation in Y. pestis contributes to biofilm formation necessary for flea colonization and efficient transmission.
In conclusion, we describe here the first bacterial adhesin that requires proteolytic cleavage to become active. This represents a novel post-translational mechanism to regulate adherence in bacteria. This mechanism relies on a protease from a common bacterial protease family, the omptins, found in many species of Gram negative bacteria and whose members are capable of processing a large variety of proteins. Therefore, it is possible that YapE may be the first example of a larger group of proteins that use proteolysis to activate adhesive properties. Additional studies to define the impact of omtpin cleavage of other bacterial outer membrane or secreted proteins may identify a larger role for these proteins in bacterial virulence.
EXPERIMENTAL PROCEDURES
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1 in the Supporting Information. E. coli, Y. enterocolitica, and Y. pseudotuberculosis were grown in Luria-Bertani (LB) broth at 37°C or 26°C. Y. pestis was grown in Brain Heart Infusion (BHI) broth at 26°C. When appropriate, antibiotics were used at the following concentrations: kanamycin, µg ml−1 (E. coli), 25 µg ml−1 (Yersinia) ; carbenicillin, 50 µg ml−1; spectinomycin, 100 µg ml−1; nalidixic acid, 20 µg ml−1; and irgasan, 2 µg ml−1.
All DNA manipulations were performed using standard molecular techniques. The primers used in this study are listed in Table 2 in the Supporting Information. To generate inducible YapE constructs, the yapE genes from Y. pestis CO92, Y. pseudotuberculosis IP32953, and Y. enterocolitica 8081 were amplified using PCR, treated with indicated restriction enzymes (Supporting Information, Table 2), and ligated into pLP-PROTet-6xHN or pMWO-034 downstream of the tet operator (Obrist & Miller, 2012). Chimeric yapE genes were constructed using the splicing by overlap extension (SOE) method (Horton et al., 1989), treated with indicated restriction enzymes (Supporting Information, Table 2) and ligated into pMWO-034. yapE site directed mutations were generated by inverse PCR as described previously (Price et al., 2010) using pMBL313 DNA as template and primers listed in Table 2 in the Supporting Information. All yapE constructs were validated by DNA sequencing not to contain PCR errors prior to protein studies.
Phylogenetic analysis
YapE orthologs were identified by BLAST analysis of GenBank using YapE from Y. pestis CO92 as the query sequence. Phylogenetic analysis of the orthologs was performed using Phylogeny.fr (http://www.phylogeny.fr/) (Dereeper et al., 2008). Protein alignments were performed with MUSCLE and phylogeny with PhyML (bootstrapped 500 times). Due to sequence conservation and the large number of Y. pestis sequences available, phylogenetic analysis was limited to include only one representative of YapE from each of the Y. pestis biovars (orientalis, mediaevalis, and antiqua) and the subspecies Pestoides F. To root the tree, an autotransporter (YP_004212153) from Rahnella sp. Y9602 that shares 48% amino acid similarity to YapE was included as an outgroup in the analysis.
Aggregation and adherence assays
For settling assays to monitor bacterial aggregation, E. coli, Y. enterocolitica, and Y. pseudotuberculosis containing the pTETR plasmid (encodes the tetracycline repressor protein) were transformed with pLP-PROTet-6xHN, pMBL295, pMBL299, or pMBL291. These Y. enterocolitica and Y. pseudotuberculosis strains retain the native yapE genes, but expression of the native genes is low during in vitro growth and below our level of detection, as evident by absence of YapE in the vector only controls of the Western blots. E. coli and Yersinia overnight cultures were diluted 1:50 and 1:12.5, respectively, and grown for 3 hrs at 37°C and 26°C, respectively. YapE expression was induced with 40 ng of anhydrous tetracycline ml−1 for an additional 2 hrs prior to settling assay. Settling assays were performed as described previously (Lawrenz et al., 2009). For adherence assays, wells of a 24-well plate were seeded with 4×105 RAW264.7 cells and grown overnight in DMEM, 10% FBS at 37°C with 5% CO2 to obtain ca. 80% confluent monolayers. yapE expression was induced as described above for settling assays, and 0.03 OD600 equivalents of bacteria (MOI 20–40 cfu/cell) were added to RAW264.7 cells. Plates were centrifuged for 5 min at 200 × g to initiate contact between bacteria and cells and incubated for 30 (Y. pestis) or 60 (E. coli) min. To remove nonadherent bacteria, the culture medium was removed, and wells were washed four times with PBS. Adherent bacteria were recovered by treatment with 1% Triton for 5 min to lyse the eukaryotic cells. Serial dilutions of recovered bacteria were plated on agar. To calculate the percentage of adherent bacteria, the number of adherent bacteria was divided by the number of total bacteria as determined from an independent well which was not washed (represents total bacteria added to the cells). These studies do not differentiate between intracellular and extracellular bacteria; therefore, adherence accounts for total number of bacteria associated with the host cells. All assays were repeated independently three times and consisted of three biological replicates each time.
Western blots
For Western blots, samples were normalized by determining the absorbance of the cultures at 600 nm (OD600) and 1 OD600 equivalent was harvested from each culture. 0.1 OD600 equivalents of total bacterial proteins were then separated by SDS-PAGE, transferred to PVDF, and Western blot analysis of YapE expression and processing was performed as described previously with anti-YapE serum (Lawrenz et al., 2009).
Outer membrane protein enrichment and N-terminal sequencing
To isolate the Pla-cleaved YapE peptide, Y. pestis overnight cultures with pMWO-034 (vector control) or pMBL313 (Y. pestis YapE) were diluted 1:12.5 and grown for 3 hrs at 26°C. YapE expression was induced with 80 ng of anhydrous tetracycline ml−1 (Sigma) for an additional 3 hrs and 32 OD600 equivalents of bacteria were harvested. Outer membrane proteins were isolated using sarkosyl enrichment as described by Myers-Morales et al. (Myers-Morales et al., 2007) and separated by SDS-PAGE. The gel was stained with Flamingo fluorescent strain (BioRad) and two peptides present in the YapE sample, but absent in the vector control sample, were confirmed as being YapE by mass spectrometry (performed by the University of Louisville Protein Core). A second batch of outer membrane proteins were prepared, separated by SDS-PAGE, and transferred to PVDF (Millipore). The smaller YapE peptide was cut out from the membrane and sent for N-terminal sequencing (performed by the Iowa State University Protein Facility).
Statistical analysis
Statistical significance was calculated using a two-tailed, unpaired Student's t test. All data is represented as mean ± standard deviations from three independent experiments.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Markus Obrist for pMWO-034, Timo Korhonen and Liisa Laakkonen for helpful discussions on Yersinia omptins, and Ralph Isberg for Y. pseudotuberculosis IP32953. We are also grateful to Sue Straley for advice on Y. pestis outer membrane preparations and Michael Merchant for help with mass spectrometry identification of YapE peptides. This work was supported by funding from the University of Louisville, including support from the School of Medicine Research Committee, to M.B.L. and from National Institutes of Health grants R56AI078930, R21AI64313, and U54AI057157 (Southeast Regional Center for Biodefense and Emerging Infectious Diseases, project 006) to V.L.M.
REFERENCES
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol. 2011;301:461–468. doi: 10.1016/j.ijmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Bliska JB, Copass MC, Falkow S. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into Hep-2 cells. Infect. Immun. 1993;61:3914–3921. doi: 10.1128/iai.61.9.3914-3921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelon G, Marin E, Fernandez LA. Role of periplasmic chaperones and BamA (YaeT/Omp85) in folding and secretion of intimin from enteropathogenic Escherichia coli strains. J Bacteriol. 2009;191:5169–5179. doi: 10.1128/JB.00458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone EJ. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T. Plague and Other Yersinia Infections. New York: Plenum Medical Book Company; 1983. pp. 1–220. [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U S A. 2004;101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau ME, Berthiaume F, Mourez M. Proteolytic processing is not essential for multiple functions of the Escherichia coli autotransporter adhesin involved in diffuse adherence (AIDA-I) J Bacteriol. 2006;188:8504–8512. doi: 10.1128/JB.00864-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chromy BA, Choi MW, Murphy GA, Gonzales AD, Corzett CH, Chang BC, Fitch JP, McCutchen-Maloney SL. Proteomic characterization of Yersinia pestis virulence. J Bacteriol. 2005;187:8172–8180. doi: 10.1128/JB.187.23.8172-8180.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- Degen JL, Bugge TH, Goguen JD. Fibrin and fibrinolysis in infection and host defense. Journal of thrombosis and haemostasis : JTH. 2007;5(Suppl 1):24–31. doi: 10.1111/j.1538-7836.2007.02519.x. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber DM, Brubaker RR. Plasmids in Yersinia pestis. Infect Immun. 1981;31:839–841. doi: 10.1128/iai.31.2.839-841.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink DL, Cope LD, Hansen EJ, Geme JW., 3rd The Hemophilus influenzae Hap autotransporter is a chymotrypsin clan serine protease and undergoes autoproteolysis via an intermolecular mechanism. J Biol Chem. 2001;276:39492–39500. doi: 10.1074/jbc.M106913200. [DOI] [PubMed] [Google Scholar]
- Fink DL, St Geme JW., 3rd Chromosomal expression of the Haemophilus influenzae Hap autotransporter allows fine-tuned regulation of adhesive potential via inhibition of intermolecular autoproteolysis. J Bacteriol. 2003;185:1608–1615. doi: 10.1128/JB.185.5.1608-1615.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein JM, Roy K, Fischer JF, Burkitt M. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect Immun. 2006;74:2245–2258. doi: 10.1128/IAI.74.4.2245-2258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiko J, Laakkonen L, Juuti K, Kalkkinen N, Korhonen TK. The omptins of Yersinia pestis and Salmonella enterica cleave the reactive center loop of plasminogen activator inhibitor 1. J Bacteriol. 2010;192:4553–4561. doi: 10.1128/JB.00458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiko J, Suomalainen M, Ojala T, Lahteenmaki K, Korhonen TK. Invited review: Breaking barriers--attack on innate immune defences by omptin surface proteases of enterobacterial pathogens. Innate Immun. 2009;15:67–80. doi: 10.1177/1753425909102559. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Nataro JP. Virulence functions of autotransporter proteins. Infect. Immun. 2001;69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ. The evolution of flea-borne transmission in Yersinia pestis. Curr. Issues Mol. Biol. 2005;7:197–212. [PubMed] [Google Scholar]
- Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Inglesby TV, Dennis DT, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Koerner JF, Layton M, McDade J, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Schoch-Spana M, Tonat K. Plague as a biological weapon: medical and public health management. Working Group on Civilian Biodefense. JAMA. 2000;283:2281–2290. doi: 10.1001/jama.283.17.2281. [DOI] [PubMed] [Google Scholar]
- Isberg RR, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- Jong WS, ten Hagen-Jongman CM, Ruijter E, Orru RV, Genevaux P, Luirink J. YidC is involved in the biogenesis of the secreted autotransporter hemoglobin protease. J Biol Chem. 2010;285:39682–39690. doi: 10.1074/jbc.M110.167650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SA, Badger JL, Bryant GO, Pepe JC, Miller VL. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- Kukkonen M, Lahteenmaki K, Suomalainen M, Kalkkinen N, Emody L, Lang H, Korhonen TK. Protein regions important for plasminogen activation and inactivation of alpha2-antiplasmin in the surface protease Pla of Yersinia pestis. Mol Microbiol. 2001;40:1097–1111. doi: 10.1046/j.1365-2958.2001.02451.x. [DOI] [PubMed] [Google Scholar]
- Kukkonen M, Suomalainen M, Kyllonen P, Lahteenmaki K, Lang H, Virkola R, Helander IM, Holst O, Korhonen TK. Lack of O-antigen is essential for plasminogen activation by Yersinia pestis and Salmonella enterica. Mol Microbiol. 2004;51:215–225. doi: 10.1046/j.1365-2958.2003.03817.x. [DOI] [PubMed] [Google Scholar]
- Lathem WW, Price PA, Miller VL, Goldman WE. A plasminogen-activating protease specifically controls the development of primary pneumonic plague. Science. 2007;315:509–513. doi: 10.1126/science.1137195. [DOI] [PubMed] [Google Scholar]
- Lawrenz MB, Lenz JD, Miller VL. A novel autotransporter adhesin is required for efficient colonization during bubonic plague. Infect. Immun. 2009;77:317–326. doi: 10.1128/IAI.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JD, Lawrenz MB, Cotter DG, Lane MC, Gonzalez RJ, Palacios M, Miller VL. Expression during host infection and localization of Yersinia pestis autotransporter proteins. J Bacteriol. 2011;193:5936–5949. doi: 10.1128/JB.05877-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough KA, Falkow S. A Yersinia pestis-specific DNA fragment encodes temperature-dependent coagulase and fibrinolysin-associated phenotypes. Mol Microbiol. 1989;3:767–775. doi: 10.1111/j.1365-2958.1989.tb00225.x. [DOI] [PubMed] [Google Scholar]
- Merriam JJ, Mathur R, Maxfield-Boumil R, Isberg RR. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 1997;65:2497–2501. doi: 10.1128/iai.65.6.2497-2501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Falkow S. Evidence for two genetic loci from Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers-Morales T, Cowan C, Gray ME, Wulff CR, Parker CE, Borchers CH, Straley SC. A surface-focused biotinylation procedure identifies the Yersinia pestis catalase KatY as a membrane-associated but non-surface-located protein. Appl. Environ. Microbiol. 2007;73:5750–5759. doi: 10.1128/AEM.02968-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naktin J, Beavis KG. Yersinia enterocolitica and Yersinia pseudotuberculosis. Clin. Lab. Med. 1999;19:523–536. [PubMed] [Google Scholar]
- Nishimura K, Tajima N, Yoon YH, Park SY, Tame JR. Autotransporter passenger proteins: virulence factors with common structural themes. J Mol Med (Berl) 2010;88:451–458. doi: 10.1007/s00109-010-0600-y. [DOI] [PubMed] [Google Scholar]
- Obrist MW, Miller VL. Low copy expression vectors for use in Yersinia sp. and related organisms. Plasmid. 2012 doi: 10.1016/j.plasmid.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerregaard A, Espersen F, Skurnik M. Role of the Yersinia outer membrane protein YadA in adhesion to rabbit intestinal tissue and rabbit intestinal brush border membrane vesicles. APMIS. 1991;99:226–232. doi: 10.1111/j.1699-0463.1991.tb05143.x. [DOI] [PubMed] [Google Scholar]
- Perry RD, Fetherston JD. Yersinia pestis: etiologic agent of plague. Clin. Microbiol. Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CT, Al-Khodor S, Al-Quadan T, Abu Kwaik Y. Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect Immun. 2010;78:2079–2088. doi: 10.1128/IAI.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J Biol Chem. 2011;286:29771–29779. doi: 10.1074/jbc.M111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Perez F, Henderson IR, Leyton DL, Rossiter AE, Zhang Y, Nataro JP. Roles of periplasmic chaperone proteins in the biogenesis of serine protease autotransporters of Enterobacteriaceae. J Bacteriol. 2009;191:6571–6583. doi: 10.1128/JB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Perez F, Henderson IR, Nataro JP. Interaction of FkpA, a peptidyl-prolyl cis/trans isomerase with EspP autotransporter protein. Gut microbes. 2010;1:339–344. doi: 10.4161/gmic.1.5.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauri A, Soprova Z, Wickstrom D, de Gier JW, Van der Schors RC, Smit AB, Jong WS, Luirink J. The Bam (Omp85) complex is involved in secretion of the autotransporter haemoglobin protease. Microbiology. 2009;155:3982–3991. doi: 10.1099/mic.0.034991-0. [DOI] [PubMed] [Google Scholar]
- Sodeinde OA, Subrahmanyam YV, Stark K, Quan T, Bao Y, Goguen JD. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- Suomalainen M, Haiko J, Ramu P, Lobo L, Kukkonen M, Westerlund-Wikstrom B, Virkola R, Lahteenmaki K, Korhonen TK. Using every trick in the book: the Pla surface protease of Yersinia pestis. Adv Exp Med Biol. 2007;603:268–278. doi: 10.1007/978-0-387-72124-8_24. [DOI] [PubMed] [Google Scholar]
- Thomson NR, Howard S, Wren BW, Holden MT, Crossman L, Challis GL, Churcher C, Mungall K, Brooks K, Chillingworth T, Feltwell T, Abdellah Z, Hauser H, Jagels K, Maddison M, Moule S, Sanders M, Whitehead S, Quail MA, Dougan G, Parkhill J, Prentice MB. The Complete Genome Sequence and Comparative Genome Analysis of the High Pathogenicity Yersinia enterocolitica Strain 8081. PLoS Genet. 2006;2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud GI, Bliska JB. Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 2005;59:69–89. doi: 10.1146/annurev.micro.59.030804.121320. [DOI] [PubMed] [Google Scholar]
- Weening EH, Cathelyn JS, Kaufman G, Lawrenz MB, Price P, Goldman WE, Miller VL. The dependence of the Yersinia pestis capsule on pathogenesis is influenced by the mouse background. Infect Immun. 2011;79:644–652. doi: 10.1128/IAI.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren BW. The yersiniae--a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol. 2003;1:55–64. doi: 10.1038/nrmicro730. [DOI] [PubMed] [Google Scholar]
- Yen YT, Karkal A, Bhattacharya M, Fernandez RC, Stathopoulos C. Identification and characterization of autotransporter proteins of Yersinia pestis KIM. Mol Membr Biol. 2007;24:28–40. doi: 10.1080/09687860600927626. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.