Abstract
Apolipoprotein (apo)A-I is the principal protein component of HDL, and because of its conformational adaptability, it can stabilize all HDL subclasses. The amphipathic α-helix is the structural motif that enables apoA-I to achieve this functionality. In the lipid-free state, the helical segments unfold and refold in seconds and are located in the N-terminal two thirds of the molecule where they are loosely packed as a dynamic, four-helix bundle. The C-terminal third of the protein forms an intrinsically disordered domain that mediates initial binding to phospholipid surfaces, which occurs with coupled α-helix formation. The lipid affinity of apoA-I confers detergent-like properties; it can solubilize vesicular phospholipids to create discoidal HDL particles with diameters of approximately 10 nm. Such particles contain a segment of phospholipid bilayer and are stabilized by two apoA-I molecules that are arranged in an anti-parallel, double-belt conformation around the edge of the disc, shielding the hydrophobic phospholipid acyl chains from exposure to water. The apoA-I molecules are in a highly dynamic state, and they stabilize discoidal particles of different sizes by certain segments forming loops that detach reversibly from the particle surface. The flexible apoA-I molecule adapts to the surface of spherical HDL particles by bending and forming a stabilizing trefoil scaffold structure. The above characteristics of apoA-I enable it to partner with ABCA1 in mediating efflux of cellular phospholipid and cholesterol and formation of a heterogeneous population of nascent HDL particles. Novel insights into the structure-function relationships of apoA-I should help reveal mechanisms by which HDL subclass distribution can be manipulated.
Keywords: ATP binding cassette transporter A1, amphipathic α-helix, apoA-I, apoE, cholesterol, helix bundle, lipoprotein, membrane solubilization, phospholipid
The purpose of this review is to summarize current understanding of how apolipoproteins (apoA-I in particular) determine the structures of HDL particles. The focus is on recent advances in the field, so the coverage of the literature on HDL structure is not comprehensive. Knowledge of HDL structure at the molecular level is critical for understanding how this lipoprotein achieves the multiple functions elaborated in other reviews in this thematic series.
In human plasma, HDL is a heterogeneous collection of particles ranging 7–12 nm in diameter and 1.063–1.21 g/ml in density (1–4). The predominant species of HDL are spherical microemulsion particles, in which a core of neutral cholesteryl ester (CE) and triacylglycerol (TG) is encapsulated by a monolayer of phospholipid (PL), unesterified (free) cholesterol (FC), and protein (5). The protein and PL constituents comprise approximately 50 and 25%, respectively, of the mass of such particles, with the CE, FC, and TG components making up the remainder. Larger, less dense HDL particles have a higher lipid-to-protein mass ratio. Approximately 70% of total plasma HDL protein is apoA-I (which is present in normolipidemic human plasma at ∼130 mg/dl), and it is located in essentially every HDL particle. The second most abundant protein is apoA-II, which comprises 15–20% of total plasma HDL protein, but this component is not present in all HDL particles. In human plasma, about 25% of apoA-I is present in HDL particles containing only apoA-I (LpA-I); the remaining HDL particles contain both apoA-I and apoA-II (LpA-I+A-II), typically in a molar ratio of 1-2/1 (6). ApoA-I and apoA-II are the “scaffold” proteins that primarily determine HDL particle structure. Other members of the exchangeable apolipoprotein gene family that are associated with HDL include apoA-IV, apoC, and apoE; these proteins comprise ≤ 10% of HDL protein and do not significantly affect overall particle structure. Small populations of HDL particles containing mainly either apoA-IV or apoE exist in normal human plasma and also in plasma from apoA-I-deficient subjects (7). The many other types of protein molecules identified by proteomic analysis of HDL (8, 9) (also see review on HDL proteomics in this thematic series) are sequestered on compositionally distinct particles (9) (present at levels < 1 molecule/particle) and serve a metabolic rather than structural role. Besides being present in spherical HDL particles, apoA-I is a critical component of nascent HDL formed by the action of ATP binding cassette transporter A1 (ABCA1). The structures of these discoidal HDL particles, which contain a segment of PL bilayer, are largely determined by the properties of the apoA-I molecule. In addition to the above lipid-bound forms, some 5–10% of the apoA-I in human plasma is present in a lipid-free/poor state, designated pre-β1-HDL (10, 11).
In this review, the focus is to understand how the structure of the apoA-I molecule allows it to exist in the various physical states mentioned above and to determine the structure of HDL particles. The fact that the functional motif in exchangeable apolipoproteins is the amphipathic α-helix has been established for some time (for a review, see Ref. 12). After considering how the amphipathic α-helix determines the structure of lipid-free apoA-I, this review addresses the mechanisms by which apoA-I interacts with lipids to form HDL particles and discusses current understanding of the organization of apoA-I on discoidal and spherical HDL particles. This structural information forms a basis for understanding why HDL exists as a heterogeneous population of particles.
STRUCTURES OF HDL EXCHANGEABLE APOLIPOPROTEINS IN THE LIPID-FREE STATE
Primary and secondary structures
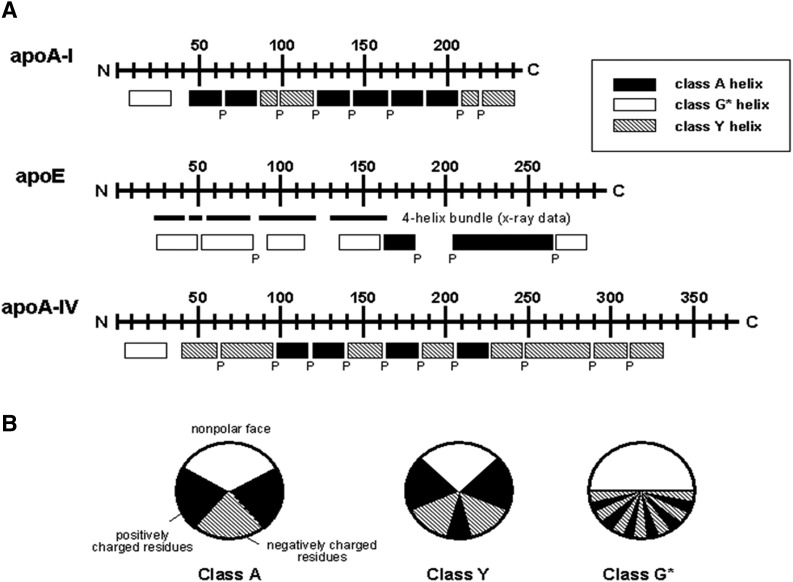
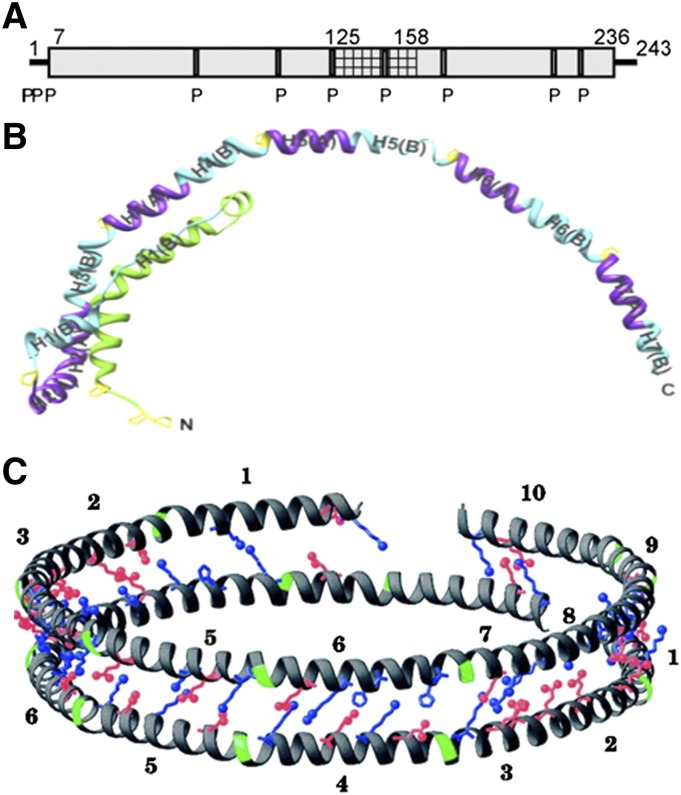
Human exchangeable apolipoproteins (apoA, apoC, apoE) have the same genomic structure and are members of a multigene family that probably evolved from a common ancestor (13). The last exon codes for primary structures of 11- and 22-amino acid tandem repeats that span amino acid residues 44–243, 40–77, 40–397 and 62–299 in apoA-I, apoA-II, apoA-IV, and apoE, respectively. Each of these repeats has the periodicity of an amphipathic α-helix, and these helices are often separated by a proline residue (12, 13). The distributions of all the potential α-helices along the amino sequences of apoA-I, apoA-IV, and apoE are shown in Fig. 1A. The predicted α-helices for apoA-I include ∼80% of the amino acids and represent essentially the maximal helix content. Amphipathic α-helices have been classified into several distinct classes according to the distribution of charged residues around the axis of the helix (12). The class A helix is a major lipid-binding motif of exchangeable apolipoproteins and is characterized by the location of positively charged residues near the hydrophilic/hydrophobic interface and negatively charged residues clustered at the center of the polar face (Fig. 1B). Class G* and class Y helices have also been identified in the exchangeable apolipoproteins, and these types of amphipathic α-helix are proposed to have reduced lipid affinity. The class G* helix is distinguished by a random radial arrangement of positively charged and negatively charged amino acid residues in the polar face, whereas the class Y helix is characterized by the presence of three clusters of positively charged amino acids in the polar face forming a Y pattern. The repeating amphipathic α-helical segments are critical for the ability of exchangeable apolipoproteins to interact with PL and stabilize HDL particles. In the case of apoA-I, the importance is reflected in the conservation of this structural motif across multiple species (14, 15).
Fig. 1.
(A) Distribution of amphipathic α-helices in the human exchangeable apolipoproteins apoA-I, apoA-IV, and apoE. The letter “P” below the rectangles indicates positions of all proline residues. (B) Amphipathic helix classes found in the exchangeable apolipoproteins. Classification is based on the distribution of charged residues. Adapted from Segrest et al. (12).
A longstanding question has been the relationship of the helix locations and overall helix content derived from analysis of the amino acid sequence (Fig. 1A) to the secondary structure of the monomeric lipid-free apoA-I molecule in aqueous solution. It is difficult to study this issue because, at concentrations above about 0.1 mg/ml, apoA-I self-associates (16) with a concomitant increase in helix content (16–18). Circular dichroism (CD) measurements made at <0.1 mg/ml in which the apoA-I is monomeric indicate that the α-helix content is ∼50% (19–21), which is much lower than the content depicted in Fig. 1A. Site-directed spin labeling followed by electron paramagnetic resonance (EPR) spectroscopy has been applied to define the locations of the elements of secondary structure in lipid-free apoA-I in the oligomeric state (2–5 mg apoA-I/ml) (22–24). The results indicated that the total α-helix content was 53% with the helices being distributed along the length of the apoA-I molecule. Interestingly, about 12% of the amino acids were found to be located in β-strand. These findings do not agree very well with the helix assignments for the native apoA-I molecule in the monomeric state (0.07 mg apoA-I/ml) derived by hydrogen-deuterium exchange and mass spectrometry (HX-MS) analysis (see below). The discrepancies are probably a consequence of the use in the EPR experiments of mutated (cysteine residues substituted at each position with a spin-label attached) apoA-I molecules in an oligomeric state. The resultant intermolecular interactions may have promoted the observed β-strand formation.
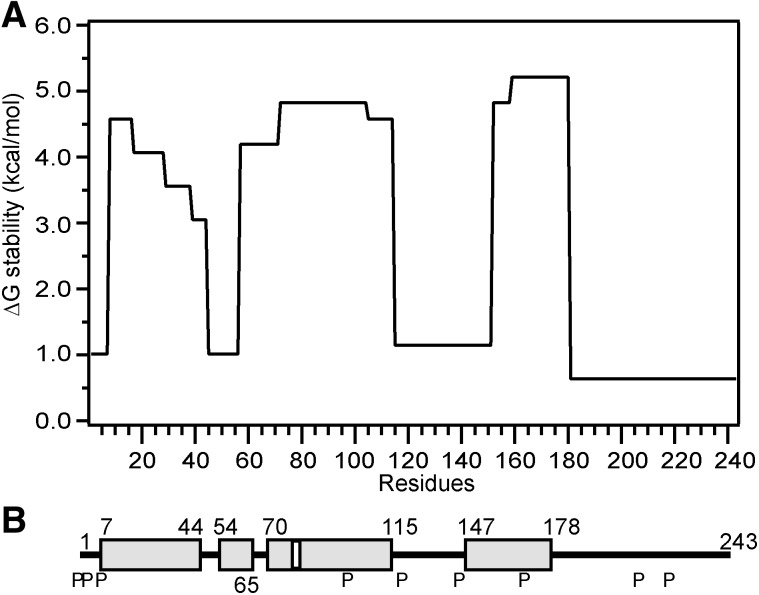
HX-MS methods can be used to probe at close to amino acid resolution the secondary structure of the native apoA-I molecule (unmutated and unlabeled) as lipid-free monomer in dilute solution. The process involves simply monitoring the naturally occurring exchange of amide hydrogens with water (25, 26). Such exchange requires transient H-bonding of the amide hydrogen with a solvent hydroxyl ion, so α-helix has to unfold for HX to occur. As a result, relative to random coil structure, helices are protected against HX and the degree of protection (slowing of amide HX) gives a measure of helix stability (free energy of unfolding). This approach has yielded the locations and stabilities of helices in the apoA-I molecule. As shown in Fig. 2, the C-terminal region formed by residues 179–243 is unstructured. There are five helical segments that span residues 7–44, 54–65, 70–78, 81–115, and 147–178, giving a total helix content of about 50%, which is consistent with CD measurements for monomeric apoA-I in dilute solution (25). The structure in Fig. 2 agrees with the results of other biophysical studies showing that the helices are located in the terminal half of the apoA-I molecule (19, 20, 27–29). The helices exhibit free energies of stabilization in the range of 3–5 kcal/mol which is significantly lower than the 5–10 kcal/mol typically observed for globular proteins. The stabilization free energy of 3–5 kcal/mol corresponds to an unfolding equilibrium constant of approximately 10−3, meaning that the apoA-I α-helices spend about 0.1% of the time unfolded. Because α-helix folding can occur on a submillisecond timescale, the α-helices in lipid-free apoA-I probably unfold (and refold) on a timescale of seconds or faster. The dynamic nature of apoA-I structure and its relative instability presumably explain the facile ability of its helical segments to unfold and refold during HDL formation and maturation.
Fig. 2.
Summary of the HX-derived secondary structure assignments and α-helix stabilities for lipid-free human apoA-I. (A) Site-resolved stability of apoA-I in the monomeric state. (B) The gray cylinders represent α-helices, and the lines indicate disordered secondary structure. The positions of proline residues (P), whose presence leads to some perturbation of α-helix organization, are marked. The helical structure is dynamic, unfolding and refolding in seconds. Adapted from Ref. 25.
Tertiary structure
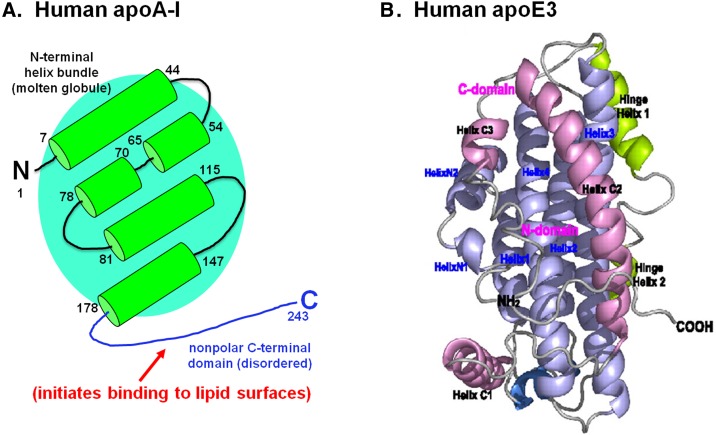
The stabilities of 3–5 kcal/mol for the apoA-I helices described in Fig. 2 are far greater than can be attained by isolated helices (30). This strongly suggests that the helices are organized into a mutually stabilizing helix bundle, as has been observed by fluorescence and other measurements (27–29). Stability is promoted by juxtaposition of the nonpolar faces of the amphipathic α-helices in the interior of the bundle and favorable cross-helix ion pairs (31). The helices are folded into an antiparallel bundle as depicted in Fig. 3A. Helix cross-linking (21) and fluorescence resonance energy transfer (29) data for lipid-free apoA-I support this conclusion. The precise locations of the helices with respect to one another are not known because the apoA-I molecule in the monomeric state in dilute solution is not amenable to study by either X-ray crystallography or NMR. A high-resolution structure for a native human apolipoprotein in the monomeric state has not been reported; the closest available structure that may provide additional insight into the organization of the apoA-I helix bundle domain (Fig. 3A) is that shown in Fig. 3B for a monomeric apoE3 variant (32). In this structure, the N-terminal domain (residues 1–167) comprises an antiparallel four-helix bundle that is similar to the isolated apoE3 N-terminal domain structure determined by both X-ray crystallography (33) and NMR (34). The helix bundle is stabilized mainly by hydrophobic interactions, while the interactions between the helix bundle and the helical C-terminal domain are stabilized by buried salt bridges and hydrogen bonds (32). X-ray crystallography and NMR experiments have demonstrated the existence of helix bundles in lipid-free human apoA-IV (35), as well as in insect apolipoproteins (36–38). It should be noted that a crystal structure for full-length apoA-I in the lipid-free state (39) has been reported to be fraudulent (40). Crystallization of lipid-free human apoA-I molecules with truncations at either the N-terminal (41) or C-terminal (42) end leads to the protein adopting an extended conformation that is similar to the structure of lipid-associated apoA-I (discussed below).
Fig. 3.
Structures of human apoA-I and apoE3 in the lipid-free monomeric state. Both molecules adopt a two-domain tertiary structure in which the N-terminal two thirds is folded into an anti-parallel helix bundle, and the C-terminal region forms a distinct domain. (A) The helix locations along the amino acid sequence of the apoA-I molecule are taken from Fig. 2. (B) NMR structure of an apoE3 monomeric variant containing the mutations F257A/W264R/V269A/L279Q/V287E to prevent self-association. The average structure is shown as a ribbon representation in which the N-terminal helices in the anti-parallel bundle are colored blue. The C-terminal helices are shown in pink, and the hinge region that links the N- and C-terminal domains is colored green. ApoE3 figure reproduced with permission from Ref. 32.
It is well established that human apoA-I and apoE3 adopt similar overall two-domain structures comprising an N-terminal helix bundle and separately folded C-terminal domain (Fig. 3). However, the detailed characteristics of these domains in the two proteins are different (20, 43). The helix bundle in apoE3 is relatively stable (midpoint of denaturation D1/2 = 2.5 M GdnHCl and free energy of stabilization = 6.6 kcal/mol, as monitored by CD) (44) with specific interhelix interactions and helix positions so that there is a defined tertiary structure (Fig. 3B), as determined by NMR (32, 34). The helix bundle in apoA-I is significantly less stable (D1/2 = 1.0 M GdnHCl and free energy of stabilization = 4.5 kcal/mol) (45). and the exposure of hydrophobic surface, as reflected by binding of 8-anilino-1-napthalenesulfonic acid, is significantly greater than in the apoE3 helix bundle (43). Furthermore, as mentioned earlier with regard to Fig. 2, HX experiments indicate that the individual helices in apoA-I exist transiently, opening and closing on a timescale of seconds. The above characteristics are consistent with the helix bundle existing in a molten globular state, as has been inferred from studies of the thermal denaturation of apoA-I (46). Thus, the apoA-I N-terminal domain (Fig. 3A) may be best viewed as having a characteristic antiparallel four-helix bundle fold but not possessing fixed and specific tertiary (interhelix) interactions. The helix bundle domain probably exists as an ensemble of similar conformations that exhibit a degree of global cooperativity. As such, the apoA-I molecule is highly flexible and can readily adopt different conformations; this low stability structure facilitates functional remodeling of HDL particles (47). The helix bundle structure can be very sensitive to perturbation by point mutations. For example, the presence of a G26R mutation in apoA-IIowa leads to unfolding of the helix spanning residues 7–44 (Fig. 3A) which, in turn, destabilizes the helix bundle (48); this destabilization promotes proteolysis at residue 83 and formation of amyloid (49).
The C-terminal domains are organized differently in the monomeric apoA-I and apoE3 molecules. This domain contains α-helices in the latter case (Fig. 3B), and the segment spanning residues 260–270 forms an exposed hydrophobic surface that can initiate lipid binding (32). In contrast, the entire C-terminal domain (residues 180–243) in apoA-I is disordered (Figs. 2 and 3A). This domain lacks a well-structured three-dimensional fold and has the characteristics of an intrinsically unstructured protein (50, 51). Such proteins or protein domains require stabilizing interactions to fold, and they exhibit coupled folding and binding. The apoA-I C-terminal domain behaves in this manner and forms α-helix when it either self-associates or interacts with a PL-water interface. Site-specific fluorescence labeling indicates that the apoA-I C-terminal domain can also participate in intramolecular interactions that stabilize the N-terminal helix bundle domain (52). As expected, the stability of the helix bundle is sensitive to the primary structure. Thus, mouse apoA-I, which also adopts a two-domain tertiary structure, has 70% sequence identity to human apoA-I in the helix bundle domain, but it forms a much less stable structure (45). Human and mouse apoA-I have very different tertiary structure domain contributions for achieving functionality with regard to interacting with PL and forming HDL particles.
INTERACTION OF APOA-I WITH LIPIDS
The amphipathic α-helices in apoA-I (Fig. 1) are well adapted to bind to a polar/nonpolar interface, such as the PL-water interface. The ability of the nonpolar face of such a helix to insert into hydrophobic regions of a PL monolayer is the basis for the stabilization of spherical microemulsion HDL particles by apoA-I. ApoA-I also binds effectively to the surface of PL vesicles leading, under appropriate conditions, to solubilization of the PL bilayer and formation of discoidal HDL particles. The above behavior is consistent with apoA-I possessing detergent-like properties.
Molecular mechanism of ApoA-I binding to lipids
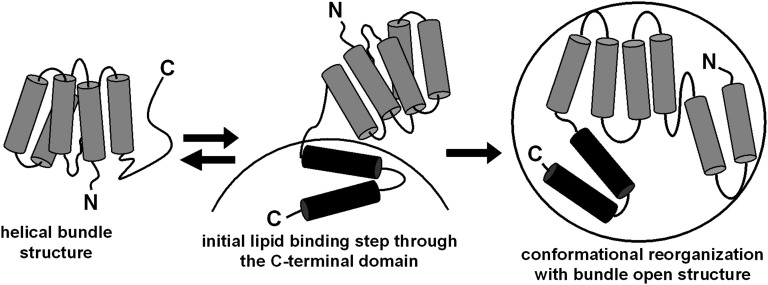
It is well established that the C-terminal domain of the human apoA-I molecule (Fig. 3A) plays a critical role in lipid-binding because deletion of this domain drastically reduces the level of binding (reviewed in Refs. 14, 43, 53, 54). This fact, coupled with the two-domain structure adopted by the human apoA-I molecule in the lipid-free state, led us to propose a two-step model for binding to lipid surfaces (20). This model is depicted in Fig. 4. The sequential interactions of the C- and N-terminal domains of the apoA-I molecule with the lipid surface have been demonstrated directly by comparing the kinetics of binding to PL vesicles of apoA-I fluorescently labeled in a domain-specific manner (55). Similar behavior has been confirmed with apoE, which also adopts a two-domain tertiary structure (Fig. 3B) (56).
Fig. 4.
Model of the two-step mechanism of human apoA-I binding to a spherical lipid particle. In the lipid-free state, apoA-I is organized into two structural domains as shown in Fig. 3A. Lipid binding is initiated by the C-terminal domain and is coupled to an increase in α-helicity. Subsequently, the helix bundle undergoes a conformational opening, converting hydrophobic helix-helix interactions to helix-lipid interactions; this second step is only slowly reversible. Reproduced with permission from Ref. 20.
As shown in Fig. 4, lipid binding is initiated by contact of the disordered C-terminal domain with the surface of the target lipid particle. This interaction is coupled with a conformational change from random coil to α-helix in the C-terminal domain of the apoA-I molecule. As noted above, such coupling of folding with binding is typical of intrinsically unstructured proteins (50, 51). An advantage of the conformational flexibility exhibited by this class of proteins and protein domains is that molding to fit diverse binding surfaces is possible. In the case of the apoA-I C-terminal domain, these surfaces are different sizes of HDL particles and various lipid membranes. The facility of the human apoA-I C-terminal domain to form α-helix upon interaction with lipid (19, 20, 22, 24) is critical for step 1 in the binding model depicted in Fig. 4. The process is enthalpically driven, with the enthalpy of binding being linearly correlated with the number of amino acids forming α-helix upon lipid binding (57). The increase in α-helix content from about 50 to 80% that occurs when apoA-I binds PL increases the favorable free energy of binding by 2.5 kcal/mol (43, 57). It should be noted that the C-terminal domain does not play such a critical role in the lipid binding of all species of apoA-I. Thus, mouse apoA-I possesses a relatively polar C-terminal domain, and deleting it enhances lipid binding (45).
Step 2 in the lipid-binding mechanism involves opening of the N-terminal helix bundle (Fig. 4). ApoA-I associated with the surface of a PL vesicle can adopt conformations in which the N-terminal helix bundle is either open or closed (55). The helix bundle tends to be closed and out of contact with the lipid surface at higher apoA-I surface concentrations. Such conformational rearrangement is also seen with apoE (53, 58); in this case, the helix bundle-open conformation is required for recognition by the LDL receptor. The highly dynamic, molten globule structure adopted by the human apoA-I helix bundle domain (Fig. 3A) is well adapted to open readily and expose hydrophobic surfaces for interaction with PL-water surface. Reductions in helix bundle stability promote opening and enhance lipid binding (20, 59, 60). The precise pathway by which the helix bundle opens in apoA-I and other helix bundle-forming apolipoproteins has been the subject of extensive investigation. The topic has been comprehensively reviewed recently (61); there is no single pathway by which such bundles open to expose the nonpolar faces of the amphipathic α-helices. Opening involves initial separation around a hinge of two pairs of helices, but at this stage, there is not a clear understanding of which helices are paired.
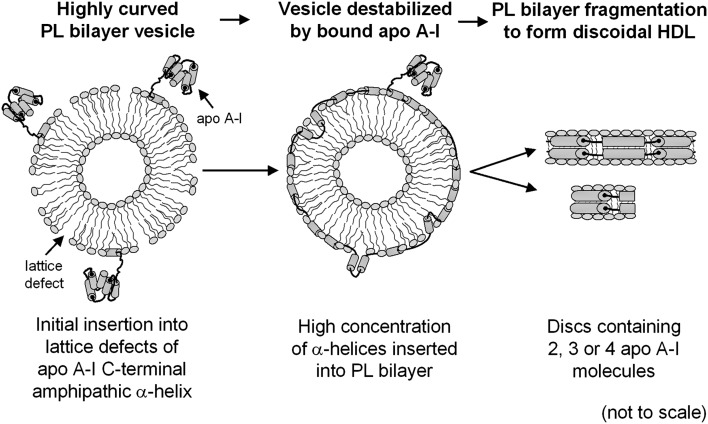
Lipid solubilization by apoA-I
As mentioned earlier, apoA-I binding to a PL-coated emulsion particle containing a core of TG and CE molecules does not lead to particle destabilization, whereas binding to a PL vesicle can be followed by particle rearrangement and solubilization of the bilayer. A proposed mechanism of this solubilization process is summarized in Fig. 5. The ability of apoA-I to mediate a spontaneous solubilization reaction was first established with dimyristoyl phosphatidylcholine (DMPC) multilamellar vesicles, which are converted into discoidal HDL particles when incubated with apoA-I at 24°C (for a review, see Ref. 54). Detailed kinetic studies of this process by Pownall and colleagues established that apoA-I interacts with lattice defects in the DMPC bilayer, and once a critical concentration of α-helices in such defects is attained, the bilayer becomes destabilized and rearranges into discoidal HDL particles (62) (cf. Fig. 5). The spontaneous solubilization process is under kinetic control, and the reaction is sensitive to factors such as PL bilayer physical state and cholesterol content (63, 64). The size distribution of the HDL particles formed is also influenced by these factors. The discoidal HDL particles created by interaction of human apoA-I with DMPC multilamellar vesicles typically contain two or three apoA-I molecules. Larger discs contain more protein molecules and have a higher lipid/protein ratio. All of the human exchangeable apolipoproteins can participate in this reaction because they contain amphipathic α-helices; the rate of solubilization is sensitive to the protein molecular weight, hydrophobicity, and state of self-association (62, 65). ApoE reacts similarly with DMPC multilamellar vesicles; the rate is sensitive to the stability of the N-terminal helix bundle domain (60). The solubilization reaction is initiated by binding via their C-terminal domains of monomeric apoA-I or apoE molecules to the DMPC surface (Fig. 5). In the case of apoE, which exists as oligomers (primarily tetramers) in dilute solution, the rate-limiting step for the solubilization reaction is the dissociation of the apoE tetramers to monomers (56, 66).
Fig. 5.
Suggested molecular mechanism for the solubilization of PL bilayers by apoA-I to create discoidal HDL particles. Binding of apoA-I to the PL bilayer occurs in two steps as summarized in Fig. 4; formation of the C-terminal amphipathic α-helix by the apoA-I molecule is coupled with interaction of the protein with the PL surface. The discoidal HDL products of the solubilization process have structures like those depicted in Figs. 6C and 7. This process is envisaged to underlie the formation of nascent HDL particles in the apoA-I/ABCA1 reaction depicted in Fig. 9.
Both the rate of vesicle solubilization and the size distribution of the HDL products are sensitive to apoA-I domain structure (45). Essentially all of the lipid-solubilizing capacity of human apoA-I resides in the C-terminal domain because this domain in isolation functions similarly to the intact protein (45). The relatively high hydrophobicity of the C-terminal domain in human apoA-I compared with mouse apoA-I promotes formation of smaller discoidal HDL particles (67, 68). This effect is probably a consequence of the enhanced affinity of this domain for the surface of a PL bilayer vesicle, giving rise to a higher surface concentration of human apoA-I molecules. The resultant enhanced penetration of amphipathic helices into the PL bilayer (69) and reduction in the PL/apoA-I ratio leads to fragmentation of the PL bilayer into smaller segments (cf. Fig. 5). Studies of the effects of deleting the C-terminal domain indicate that the isolated helix bundle domain in mouse apoA-I is more efficient in solubilizing DMPC than is its more stable human counterpart (45, 67). Overall, it is clear that the stability of the N-terminal helix bundle domain and the hydrophobicity of the C-terminal domain are critical factors in determining the lipid-binding and lipid-solubilizing properties of the apoA-I molecule.
APOA-I ORGANIZATION IN DISCOIDAL HDL
The most detailed structural information about apoA-I organization in HDL particles has been obtained with homogeneous preparations of reconstituted discoidal HDL particles that contain a single type of PL molecule and have a fixed lipid/protein stoichiometry. Such particles can be formed by either direct apoA-I solubilization of PL vesicles (cf. Fig. 5) or formation of ternary PL/apoA-I/cholate micelles and removal of the cholate by dialysis (70). The structure of a discoidal HDL particle (∼10 nm hydrodynamic diameter) containing a 160 molecule segment of POPC bilayer stabilized by two apoA-I molecules has been characterized thoroughly by a range of methods (for reviews, see Refs. 71–74). As summarized above, it is well established that the α-helix content of the apoA-I molecule increases upon interaction with PL. However, exactly which amino acids along the length of the protein molecule are involved in this conformational change has not been known until the question was answered recently by application of HX-MS methods (26). The locations of the α-helices in the apoA-I molecule when present in a 9.6 nm diameter discoidal HDL particle are shown in Fig. 6A. Comparison of this structure to that presented in Fig. 2B for lipid-free apoA-I reveals that residues 45–53, 66–69, 116–146, and 179–236 change conformation from random coil to α-helix upon incorporation into the HDL particle. The conformational change of these 102 residues corresponds to an increase in helix content of ∼40% which is similar to the value indicated by CD. The major contribution to helix formation comes from the C-terminal domain, in agreement with earlier EPR measurements (22). Except for the terminal amino acids 1–6 and 237–243, nearly the entire apoA-I molecule in the lipid-bound state forms a continuous helical structure, punctuated by kinks at proline residues.
Fig. 6.
Structure of apoA-I in a discoidal HDL particle. (A) Summary of the HX-derived apoA-I helix locations in a 9.6 nm diameter particle that contains 140 molecules of POPC and 2 molecules of apoA-I. Residues 125–158 (cross-hatched) exist in two populations (helix and loop). The equivalent secondary structure information for lipid-free apoA-I is shown in the same representation in Fig. 2B. Reproduced with permission from Ref. 26. (B) Structure of apoA-I (Δ185–243) monomer obtained from a crystal structure at 2.2 Å resolution. This truncated apoA-I molecule adopts an extended conformation in the crystal and forms a half circle dimer. Reproduced with permission from Ref. 42. (C) Double-belt model for apoA-I structure at the edge of a discoidal HDL particle. Two ring-shaped molecules of apoA-I are stacked on top of each other with both molecules in an anti-parallel orientation, allowing the helix registry to maximize intermolecular salt-bridge interactions. Only the charged residues at selected positions are explicitly displayed; positively charged residues are represented in blue, negatively charged residues in red, and prolines in green. The numbers 1–10 represent the ten helical domains identified by computer analysis of the human apoA-I amino acid sequence between residues 44 and 243 (cf. Fig. 1A). Reproduced with permission from Ref. 76.
The first direct experimental determination of the organization of these helical apoA-I molecules around the edge of a discoidal HDL particle in solution involved the use of polarized internal reflection infrared spectroscopy (75). The results verified that the disc contains PL bilayer structure with the apoA-I helices oriented parallel to the bilayer surface and arranged in a belt-like fashion around the edge of the disc. This extended apoA-I organization is consistent with a similar conformation observed in the 4 Å resolution crystal structure of a lipid-free, N-terminally truncated variant of human apoA-I in which the molecule forms a horseshoe-shaped, pseudo-continuous, amphipathic α-helix (41). A more recent 2.2 Å resolution crystal structure of a C-terminally truncated variant of human apoA-I in the lipid-free state confirms that the protein can form elongated helix with kinks at proline residues (Fig. 6B) (42). In the crystal, the protein forms a dimer with the two elongated helices packed in an antiparallel fashion to form a half-circle with hydrophobic surface on the inside. Intrahelical salt bridges provide helix stabilization while salt bridge networks between monomers help stabilize the dimer. This high-resolution structural information lends support to the generally accepted “double-belt” model for apoA-I organization in a discoidal HDL particle (75), a detailed model for which (Fig. 6C) has been proposed by Segrest and colleagues (76). Chemical cross-linking and mass spectrometry studies are consistent with this concept and also show that a central helical segment (residues 121–142) in one apoA-I molecule is packed adjacent to the same segment in its partner (reviewed in Refs. 73, 74). This helix registry between the two apoA-I molecules may be preferred (77) but is probably not unique because crosslinking experiments have shown that variable registry can occur (73, 78), presumably reflecting independent circular motion of the two protein monomers (79). Because available crystal structures are for truncated lipid-free apoA-I variants, models of the organization of the terminal regions of the protein in discoidal HDL particles are less well developed. However, there is evidence that the N-terminal region can fold back on itself (cf. Fig. 6B), with the degree to which this occurs being a function of particle size (42, 74, 80, 81).
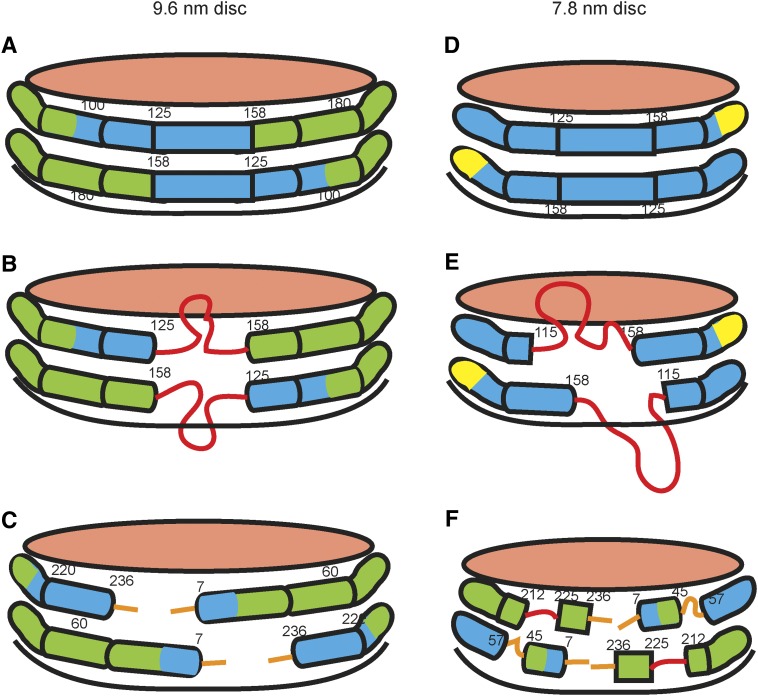
Since apoA-I forms discoidal HDL particles of different sizes, there has been great interest in understanding how it accommodates such variations. We employed HX-MS to compare the locations, stabilities, and dynamics of apoA-I helical segments in large (9.6 nm) and small (7.8 nm) discoidal HDL particles (26). As summarized in Fig. 7, the two apoA-I molecules in the larger 9.6 nm disc assume the secondary structure shown in Fig. 6A. This structure is not significantly affected by either incorporation of 10 mol% cholesterol into the disc or alteration of the PL acyl chain composition. The segment spanning amino acids 125–158 can form either a helix with residues in direct contact with the PL molecules at the edge of the disc or a disordered loop that protrudes into the aqueous phase. Exchange between these two coexisting states occurs on the timescale of minutes and the relative occupancy of the loop structure is about 20% at 5°C. The finding that residues 125–158 readily desorb from the disc edge is consistent with spectroscopic results showing that residues 130–174 (82) and residues 133–146 (83) form flexible loops. It should be noted that, contrary to a prior claim (84), residues 159–180, which are in the LCAT-activating region of apoA-I, do not form such a loop but rather remain helical (26).
Fig. 7.
Diagram comparing the secondary structures of apoA-I molecules in 9.6 nm (A–C) and 7.8 nm (D–F) discoidal HDL particles, as determined by HX-MS. The amphipathic α-helices are represented by cylinders colored according to their stability. The values of the free energy of stabilization (kcal/mol) are as follows: red, ≤1; orange, 1–2; yellow, 2–3; green, 3–4; blue >4. Importantly, the structure is not static but highly dynamic; the α-helices unfold and refold in seconds or less. The pink surfaces of the discs are covered by PL polar groups. The apoA-I molecules are organized according to the double-belt model (Fig. 6C). (A) and (B) show the states in which residues 125–158 adopt α-helical and disordered loop conformations, respectively. The diagrams show the two apoA-I molecules on each disc acting in concert and adopting the same conformation, but the segment spanning residues 125–158 also could be α-helical in one molecule and loop in the other. The orientation in (C) is obtained by a 180° clockwise rotation of the disc in (A) and depicts the nonhelical structures formed by the 6 or 7 amino acids at the ends of the apoA-I molecules. (D–F) show the equivalent information for a 7.8 nm discoidal HDL particle. The approximately 20% smaller area available at the edge of the 7.8 nm disc leads to displacement of about 20% more apoA-I amino acid residues from contact with PL molecules and enhances formation of protruding disordered loops [red in (E) and (F)]. The surface-dissociated and surface-associated states coexist on the time scale of minutes or longer. Reproduced with permission from Ref. 26.
When incorporated around the edge of the smaller 7.8 nm disc, the increase in packing density of the two apoA-I molecules causes more residues to be displaced from the particle surface and form disordered loops (Fig. 7). Residues 45–57, 115–158, and 190–243 are involved in this helix unfolding, and these are mostly the segments that are nonhelical in lipid-free apoA-I (Fig. 2). Apparently, the propensity of these regions to maintain disordered structure in lipid-free apoA-I similarly favors disordered loop structures in the lipid-bound state. This behavior is likely to be the basis for the minimum in free energy associated with selective formation of the 7.8 nm HDL disc. The selective formation of particles with 9.6 nm diameter probably reflects the optimized coverage of the hydrophobic surface formed by PL acyl chains achieved by the two maximally α-helical apoA-I molecules. These structural characteristics explain why apoA-I interacts with PL in a quantized fashion to stabilize discoidal HDL particles of a defined size rather than to form a continuum of particle sizes.
Increases in discoidal HDL partial diameter beyond 10 nm are associated with incorporation of more apoA-I molecules. In particles containing three apoA-I molecules, at least one of them must adopt a helical hairpin conformation (71, 85). Cross-linking and mass spectrometry studies have shown that apoA-II in discoidal HDL particles also forms such a hairpin structure (86). The presence of apoA-II in discoidal LpA-I+A-II HDL particles alters the conformation of apoA-I in a site-specific manner (87). It has been argued that this interaction could restrict the conformational space available to apoA-I, thereby hindering the remodeling of LpA-I+A-II HDL to larger particles (88).
The apoA-I amphipathic α-helices in discoidal HDL particles have relatively low stability, in the range 3–5 kcal/mol (Fig. 7), indicating high flexibility and dynamic unfolding and refolding in seconds or less (26). These free energies of helix stabilization are the same as for apoA-I in the lipid-free state (cf. Fig. 2A). This finding from HX-MS experiments is consistent with the results of GdnHCl denaturation experiments showing that the free energies of apoA-I helix unfolding are similar in the lipid-free (3.0 kcal/mol) (89) and lipid-bound (3.5 kcal/mol) (45) states. It follows that the dynamic nature of the apoA-I molecule is maintained in the presence of PL molecules. Such conformational plasticity facilitates remodeling of HDL particles during maturation and metabolism. The fact that the apoA-I molecule samples an ensemble of conformations on a biologically significant timescale contrasts to the depiction in Fig. 6C of a single conformation in the original double-belt model that is derived from a crystal structure. Crystal lattice effects induce highly ordered structure, but this situation does not apply to apoA-I molecules wrapped around the edge of a fluid discoidal HDL particle. In this case, the PL molecules are in a liquid-crystalline state in which the carbon-carbon bonds in their acyl chains undergo gauche-trans isomerizations, allowing the apoA-I structure to be relatively labile. It follows that the apoA-I molecules located in discs exist in an energy landscape that is not sharply predetermined and that they can accommodate readily to alternative free energy minima.
In light of the highly dynamic and flexible structure adopted by apoA-I in discoidal HDL particles, it follows that a detailed, all-atom structure is, at best, a representation of one of many coexisting, rapidly interchanging conformations. Furthermore, such models are necessarily speculative because structural investigations of apoA-I and HDL discs in solution have been limited to low-resolution methods. These points need to be borne in mind when considering results of small-angle neutron scattering experiments (90, 91) and models derived from molecular dynamics simulations (92, 93) in which the choice of starting structure has a major influence on the outcome. Given these uncertainties, the validity of models of discoidal HDL particle structure showing apoA-I organizations that differ significantly from the double-belt arrangement (Figs. 6C and 7) is questionable (94).
APOA-I ORGANIZATION IN SPHERICAL HDL
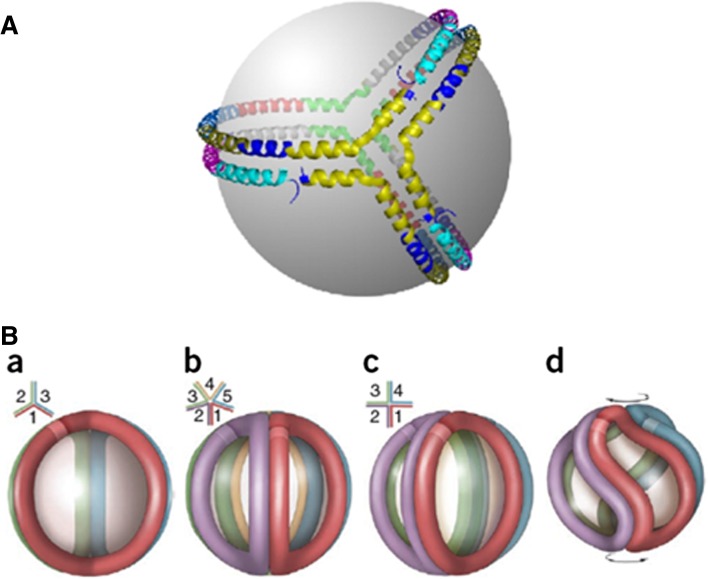
The spherical CE-containing particles that are the predominant form of HDL in human plasma arise by the action of LCAT on FC-containing discoidal HDL particles whose structures are discussed above. The double-belt model (Figs. 6C and 7) explains how apoA-I determines the structure of discoidal HDL, and it is important to know how the protein reorganizes to allow the transition to spherical HDL. Davidson and colleagues have performed elegant chemical cross-linking and mass spectrometry studies to address this question (95). Strikingly, they found that the intermolecular cross-linking pattern that characterizes the double-belt organization of apoA-I in discoidal HDL particles is maintained in spherical HDL particles, regardless of diameter. It follows that the double-belt model represents a common structural framework for apoA-I in both discs and spheres, irrespective of the size and number of apoA-I molecules present. The trefoil model depicted in Fig. 8A explains this effect; half of each apoA-I molecule in the double-belt arrangement is bent 60° out of the plane of the particle and the spherical geometry is achieved by addition of a third apoA-I molecule bent in the same fashion. The trefoil model in Fig. 8A was generated for apoA-I lacking residues 1–43, and the hinging of the apoA-I molecule is supposed to occur near residues 133 and 233 (95). More recently, an alternative trefoil arrangement for the full-length protein has been suggested; in this case, the hinging is proposed to occur near residues 65 and 185 (81). The trefoil organization of three apoA-I molecules is highly symmetric, but the observed intermolecular cross-linking patterns are also consistent with other apoA-I arrangements. Such an alternative arrangement has been suggested to involve two of the apoA-I molecules forming a twisted helical dimer and the third molecule forming a helical hairpin (96). Given the highly dynamic nature of the apoA-I molecule, it is possible that both the trefoil and helical dimer/hairpin structures coexist on the surface of spherical HDL particles. In either case, it is apparent that the apoA-I molecules form a quasi-scaffold to maintain the highly curved surface of ∼10 nm diameter spherical HDL particles.
Fig. 8.
Models of apoA-I organization on spherical HDL particles. (A) Trefoil organization of three apoA-I molecules on the surface of a 9.6 nm diameter spherical particle. Each putative helical domain shown in Fig. 6C is represented as a separate color: helix 1, teal; helix 2, purple; helix 3, dark blue; helix 4, gray; helix 5, green; helix 6, red; helix 7, light blue; helix 8, dark yellow; helix 9, navy blue; and helix 10, yellow. All helix-to-helix interactions present in the double belt between two molecules of apoA-I in a disc (Fig. 6C) are also present among three apoA-I molecules in the trefoil. Reproduced with permission from Ref. 95. (B) Incorporation of additional apoA-I molecules to the trefoil model and apoA-I adaptation to smaller particle diameters. (a) Schematic representation of the three-molecule trefoil model with each molecule of apoA-I shown in a different color; See (A) for more detail. The lighter colored band on each molecule represents the N-terminus (residue 44, as the model was built in the absence of residues 1–43). The inset is a schematic top view showing the bend angles of each apoA-I. (b) Pentameric complex proposed for the structure of larger HDL. (c) An idealized, fully extended tetrameric complex. (d) Twisted tetrameric complex with a reduced particle diameter as proposed for smaller HDL. Reproduced with permission from Ref. (97).
The structure of apoA-I in LpA-I HDL fractions isolated from human plasma has also been investigated by the combined chemical cross-linking and mass spectrometry approach (97). The particles ranged in diameter from 8.8 to 11.2 nm and contained 3–7 apoA-I molecules. The cross-linking pattern was identical for all particles, indicating that the intermolecular apoA-I contacts are substantially the same. The trefoil-based models depicted in Fig. 8B explain this finding and show that changes in spherical HDL particle size and lipid content are accommodated by twisting of the apoA-I molecules. Small-angle neutron scattering studies of apoA-I organization in spherical HDL particles are consistent in showing that the protein envelope is twisted (96). HX-MS measurements (96a) demonstrate that the secondary structure of apoA-I in a 10 nm spherical LpA-I particle is very similar to that in a discoidal HDL particle of the same size (Fig. 6A). The only significant difference is that the length of the loop formed by the central segment of the apoA-I molecule is increased, as observed when the HDL disc size is decreased (cf. Fig. 7). This effect occurs because increased apoA-I packing density in both HDL discs and spheres forces more residues out of direct contact with the particle surface. In agreement with this concept, studies of the binding of epitope-defined monoclonal antibodies showed that the conformation of the region between residues 121 and 165 of the apoA-I molecule is particularly sensitive to changes in diameter of spherical HDL particles (98). Importantly, HX kinetics are the same for apoA-I located on discoidal and spherical HDL, indicating that helix stabilities and dynamics are similar in both situations. As discussed in detail above for discoidal HDL, it follows that the diagrams of the trefoil apoA-I organization in spherical HDL particles (Fig. 8) represent static snapshots of a highly dynamic protein organization in which the helices unfold and refold on a seconds timescale. It remains to be discovered how the presence of apoA-II molecules in LpA-I+A-II HDL particles affects apoA-I organization, but it seems that the trefoil structure is perturbed. Thus, in contrast to LpA-I particles which apolipoprotein cross-linking shows contain up to four proximal apoA-I molecules, LpA-I+A-II particles contain only two proximal apoA-I molecules and up to three proximal dimeric apoA-II molecules (6).
The apoA-I scaffold on the surface of spherical HDL particles occupies ∼80% of the total surface area with the remainder being covered with PL molecules (97, 99, 100); the proportion of total surface area accounted for by PL progressively decreases as the particle diameter decreases (97). Consequently, proteins that associate with HDL can potentially participate in protein-protein as well as protein-lipid interactions, with the former being favored in smaller particles. Surface plasmon resonance and fluorescence measurements have shown that both types of interaction are involved in the binding of apoA-I (99) and apoE (56, 101) to HDL particles. ApoA-I binds reversibly to HDL particles (99), consistent with the existence of a more labile, easily dissociable pool (102) in addition to the trefoil apoA-I scaffold (Fig. 8). The coexistence of several apoA-I conformations on the surface of fluid spherical HDL particles is consistent with the highly dynamic and flexible nature of the protein. A conformation that would promote ready dissociation of apoA-I molecules is depicted in Fig. 4 in which the N-terminal helix bundle is closed and only the C-terminal domain is interacting with the HDL particle surface. ApoA-I molecules in the readily exchangeable pool of apoA-I on the surface of spherical HDL particles are precursors of circulating lipid-free (poor) apoA-I (pre-β1-HDL) (11, 103, 104). This partitioning of apoA-I molecules from the surface into the aqueous phase occurs more readily with small HDL than with larger particles (105).
INFLUENCE OF APOA-I STRUCTURE ON NASCENT HDL BIOGENESIS
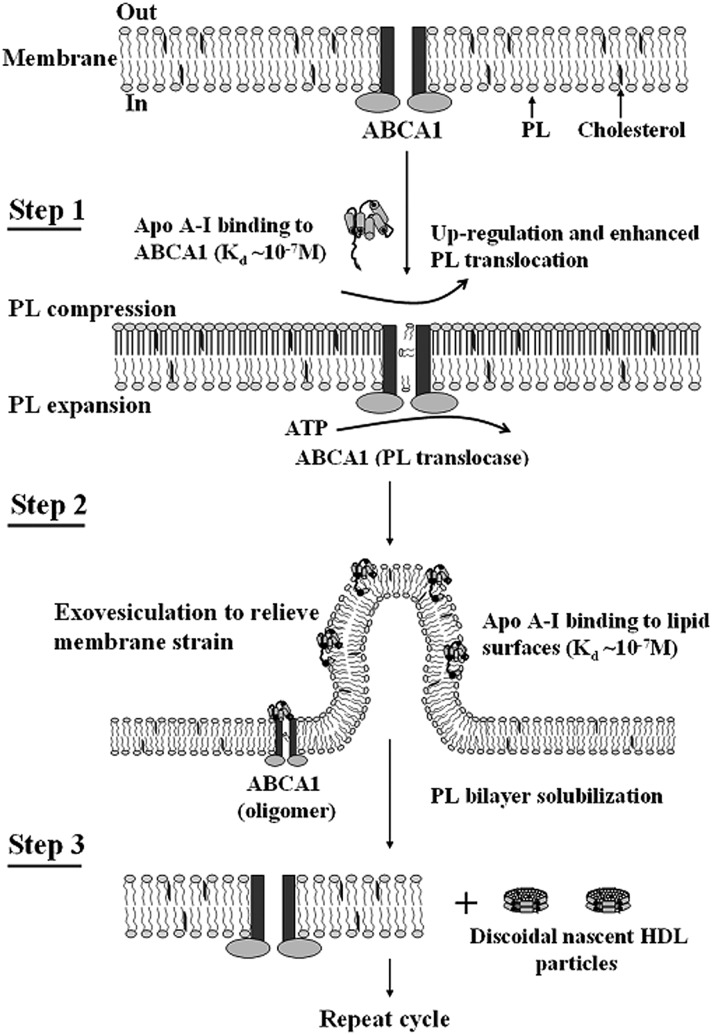
ABCA1 located in the plasma membrane of cells plays an essential role in HDL biogenesis (for reviews, see Refs. 54, 106). The lipid-binding and lipid-solubilizing properties of apoA-I summarized above are critical for the ability of apoA-I to partner with ABCA1 in creating nascent HDL particles (107). As summarized in Fig. 9, the mechanism involves ABCA1-mediated vesiculation of membrane PL/FC domains, binding of apoA-I to them (cf. Fig. 4), followed by apoA-I-mediated solubilization of these domains (cf. Fig. 5) to create discoidal nascent HDL particles (cf. Figs. 6 and 7). The latter membrane microsolubilization step is rate-limiting for the overall process of PL/FC efflux from the cell and HDL particle formation; the Km is ∼0.1 µM (3 µg/ml) human apoA-I (67, 107, 108). As the amphipathic α-helices in apoA-I are responsible for the above functions, it is to be expected that other exchangeable apolipoproteins can participate in the reaction to create nascent HDL particles. In particular, human apoA-II and apoE behave similarly to apoA-I (109–111).
Fig. 9.
Mechanism of interaction of apoA-I with ABCA1 and efflux of cellular phospholipids and cholesterol. The reaction in which apoA-I binds to ABCA1 and membrane lipids to create discoidal nascent HDL particles comprised three steps. Step 1 involves the high affinity binding of a small amount of apoA-I to ABCA1 located in the plasma membrane PL bilayer; this regulatory pool of apoA-I upregulates ABCA1 activity, thereby enhancing the active translocation of membrane PL from the cytoplasmic to exofacial leaflet. This translocase activity leads to lateral compression of the PL molecules in the exofacial leaflet and expansion of those in the cytoplasmic leaflet. Step 2 involves the bending of the membrane to relieve the strain induced by the unequal molecular packing density across the membrane and the formation of an exovesiculated domain to which apoA-I can bind with high affinity (cf. Fig. 4). This interaction with the highly curved membrane surface involves apoA-I/membrane lipid interactions and creates a relatively large pool of bound apoA-I. Step 3 involves the spontaneous solubilization (cf. Fig. 5) by the bound apoA-I of membrane PL and cholesterol in the exovesiculated domains to create discoidal HDL particles (cf. Figs. 6 and 7) containing two, three, or four apoA-I molecules/particle. In the diagram, the two transmembrane six-helix domains of ABCA1 are represented as rectangles, whereas the two ATPase domains are shown as ovals. The space between the two rectangles represents the chamber in which translocation of PL molecules occurs. Reproduced with permission from Ref. 107.
The PL/FC-containing nascent HDL products of the apoA-I/ABCA1 reaction are primarily discoidal particles (see discussion above). These particles contain two, three, or four apoA-I molecules and range 7–17 nm in diameter (112–116). There are variations in the distributions of classes of PL molecules across the HDL species, and larger particles are relatively FC-enriched. In addition, there is production of ∼7 nm diameter lipid-poor apoA-I (pre-β1-HDL) that comprises an apoA-I monomer associated with three to nine PL molecules and one or two FC molecules (114, 117). The population of nascent HDL particles is heterogeneous regardless of the cell type in which the ABCA1 is located, and the particles of different sizes are created simultaneously because there are not precursor-product relationships between them (114, 115). The essential characteristics of the lipid solubilization reaction that occurs at the surface of ABCA1-expressing cells (Fig. 9) are the same as for the process in which apoA-I solubilizes PL vesicles in a cell-free system to create discoidal HDL particles (Fig. 5). Thus, when the ratio of cell lipid made available through ABCA1 activity to extracellular apoA-I concentration is high, formation of larger HDL is promoted (117a). The influence of lipid-protein ratio on HDL disc size distribution is the same when DMPC vesicles react with apoA-I (see discussion above).
The properties of the tertiary structure domains of the apoA-I molecule influence how the protein functions in the pathway for nascent HDL particle formation in Fig. 9. For example, the C-terminal domain of human apoA-I is critical for effective lipid binding (cf. Fig. 4), so deleting it greatly inhibits cellular PL/FC efflux and nascent HDL production (108, 118, 119). Indeed, removal or disruption of the C-terminal α-helix alone is sufficient to achieve this effect (108, 112, 120). The C-terminal α-helix is the most hydrophobic segment of the human apoA-I molecule, and this feature enables human apoA-I to efflux cellular FC efficiently and form smaller nascent HDL particles compared with mouse apoA-I, which possess a relatively polar C-terminal helix (67). Manipulation of the hydrophobicity of the human apoA-I C-terminal α-helix by altering the balance of aromatic and aliphatic amino acids confirms that raising the hydrophobicity enhances the rate of lipid solubilization and formation of smaller HDL particles, in both cell and cell-free systems (68). The properties of the N-terminal helix bundle domain also influence the efficacy of apoA-I with respect to cellular PL/FC efflux and nascent HDL production. Thus, a hybrid apoA-I molecule containing the relatively unstable mouse helix bundle domain coupled to the human C-terminal domain gives greater PL/FC efflux from ABCA1-expressing cells than wild-type human apoA-I does under the same conditions (67). Furthermore, compared with the consequences of expressing wild-type human apoA-I, expression of the mouse-human hybrid apoA-I molecule in apoA-I-null mice gives rise to a gain of function with respect to macrophage reverse cholesterol transport (121). These findings suggest that engineering either some destabilization into the N-terminal helix bundle domain or increasing the hydrophobicity of the C-terminal domain of human apoA-I would enhance the antiatherogenic properties of the protein.
SUMMARY AND CONCLUSIONS
Significant progress is being made in understanding the structures of human exchangeable apolipoproteins, especially apoA-I and apoE. In the case of apoA-I, which stabilizes HDL particles, the highly dynamic and flexible conformation adopted by the molecule underlies its ability to adapt readily to particles of different sizes and shapes. The key structural motif in apoA-I is the amphipathic α-helix; these segments are relatively unstable and unfold and refold on a timescale of seconds. The lipid-free human apoA-I molecule has a two-domain tertiary structure comprising an N-terminal helix bundle and an intrinsically disordered C-terminal domain. The protein binds readily to PL surfaces by a two-step mechanism involving initial interaction of the C-terminal domain followed by opening of the helix bundle domain. The binding of apoA-I to lipid involves a coupled change in conformation of segments, particularly in the C-terminal domain, from random coil to α-helix. The structure of apoA-I in reconstituted homogeneous discoidal HDL particles is understood in the most detail. Such particles are stabilized by two apoA-I molecules wrapped around the edge of the disc in an antiparallel, double-belt arrangement so that the hydrophobic PL acyl chains are protected from exposure to water. These apoA-I molecules are in a highly dynamic state and adapt to discs of different sizes by certain segments forming loops that detach reversibly from the particle surface. The protein maintains similar protein-protein contacts when an HDL disc changes to a sphere as LCAT converts FC to CE because it is sufficiently flexible to bend and form a trefoil scaffold structure that stabilizes the highly curved surface of a mature spherical HDL particle. ApoA-I possesses detergent-like properties and can solubilize vesicular PL because its amphipathic α-helices can insert among PL molecules in a bilayer, causing destabilization and fragmentation into small discoidal HDL particles. This mechanism is the basis for nascent HDL biogenesis in which ABCA1 located in the plasma membrane of cells creates vesiculated domains which apoA-I binds to and subsequently solubilizes. The essentials of the vesicle solubilization reaction are similar at the surface of ABCA1-expressing cells and in cell-free HDL reconstitution systems. Overall, recognition of the highly dynamic nature of the apoA-I molecule should enhance understanding of the molecular details of how apoA-I determines HDL subclass structures. Such knowledge should aid in the design of ways to control HDL heterogeneity and select for desired functionalities, such as those described in other articles in this thematic review series. Furthermore, knowledge at the molecular level of the properties of apoA-I should facilitate understanding of changes caused by mutations and chemical modifications such as oxidation, as well as benefit the design of superior apoA-I mimetics for therapeutic applications.
Acknowledgments
The author is indebted to all colleagues for their valuable contributions to the studies from this laboratory described here.
Footnotes
Abbreviations:
- CD
- circular dichroism
- CE
- cholesteryl ester
- DMPC
- dimyristoyl phosphatidylcholine
- EPR
- electron paramagnetic resonance
- FC
- free (unesterified) cholesterol
- GdnHCl
- guanidinium hydrochloride
- HX-MS
- hydrogen exchange-mass spectrometry
- PL
- phospholipid
- TG
- triacylglycerol
This work was supported by National Institutes of Health Grants HL-22633 and HL-56083. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Lund-Katz S., Liu L., Thuahnai S. T., Phillips M. C. 2003. High density lipoprotein structure. Front. Biosci. 8: d1044–d1054 [DOI] [PubMed] [Google Scholar]
- 2.Kontush A., Chapman M. J. 2006. Functionally defective high-density lipoprotein: a new therapeutic target at the crossroads of dyslipidemia, inflammation, and atherosclerosis. Pharmacol. Rev. 58: 342–374 [DOI] [PubMed] [Google Scholar]
- 3.Jonas A., Phillips M. C. 2008. Lipoprotein. In Biochemistry structure of Lipids, Lipoproteins and Membranes. 5th edition. D. E. Vance and J. E. Vance, editors. Elsevier, Oxford, UK. 485–506. [Google Scholar]
- 4.Rothblat G. H., Phillips M. C. 2010. High-density lipoprotein heterogeneity and function in reverse cholesterol transport. Curr. Opin. Lipidol. 21: 229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen B. W., Scanu A. M., Kezdy F. J. 1977. Structure of human serum lipoproteins inferred from compositional analysis. Proc. Natl. Acad. Sci. USA. 74: 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gauthamadasa K., Rosales C., Pownall H. J., Macha S., Jerome W. G., Huang R., Silva R. A. 2010. Speciated human high-density lipoprotein protein proximity profiles. Biochemistry. 49: 10656–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos R. D., Schaefer E. J., Asztalos B. F., Polisecki E., Wang J., Hegele R. A., Martinez L. R., Miname M. H., Rochitte C. E., Da Luz P. L., et al. 2008. Characterization of high density lipoprotein particles in familial apolipoprotein A-I deficiency. J. Lipid Res. 49: 349–357 [DOI] [PubMed] [Google Scholar]
- 8.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson W. S., Silva R. A., Chantepie S., Lagor W. R., Chapman M. J., Kontush A. 2009. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler. Thromb. Vasc. Biol. 29: 870–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Connor P. M., Zysow B. R., Schoenhaus S. A., Ishida B. Y., Kunitake S. T., Naya-Vigne J. M., Duchateau P. N., Redberg R. F., Spencer S. J., Mark S., et al. 1998. Prebeta-1 HDL in plasma of normolipidemic individuals: influences of plasma lipoproteins, age, and gender. J. Lipid Res. 39: 670–678 [PubMed] [Google Scholar]
- 11.Rye K. A., Barter P. J. 2004. Formation and metabolism of prebeta-migrating, lipid-poor apolipoprotein A-I. Arterioscler. Thromb. Vasc. Biol. 24: 421–428 [DOI] [PubMed] [Google Scholar]
- 12.Segrest J. P., Jones M. K., De Loof H., Brouillette C. G., Venkatachalapathi Y. V., Anantharamaiah G. M. 1992. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J. Lipid Res. 33: 141–166 [PubMed] [Google Scholar]
- 13.Li W. H., Tanimura M., Luo C. C., Datta S., Chan L. 1988. The apolipoprotein multigene family: biosynthesis, structure, structure-function relationships, and evolution. J. Lipid Res. 29: 245–271 [PubMed] [Google Scholar]
- 14.Brouillette C. G., Anantharamaiah G. M., Engler J. A., Borhani D. W. 2001. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim. Biophys. Acta. 1531: 4–46 [DOI] [PubMed] [Google Scholar]
- 15.Bashtovyy D., Jones M. K., Anantharamaiah G. M., Segrest J. P. 2011. Sequence conservation of apolipoprotein A-I affords novel insights into HDL structure-function. J. Lipid Res. 52: 435–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborne J. C., Jr, Brewer H. B., Jr 1977. The plasma lipoproteins. Adv. Protein Chem. 31: 253–337 [DOI] [PubMed] [Google Scholar]
- 17.Stone W. L., Reynolds J. A. 1975. The self-association of the apo-Gln-I and apo-Gln-II polypeptides of human high density serum lipoproteins. J. Biol. Chem. 250: 8045–8048 [PubMed] [Google Scholar]
- 18.Swaney J. B., O'Brien K. 1978. Cross-linking studies of the self-association properties of apo-A-I and apo-A-II from human high density lipoprotein. J. Biol. Chem. 253: 7069–7077 [PubMed] [Google Scholar]
- 19.Davidson W. S., Hazlett T., Mantulin W. M., Jonas A. 1996. The role of apolipoprotein AI domains in lipid binding. Proc. Natl. Acad. Sci. USA. 93: 13605–13610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito H., Dhanasekaran P., Nguyen D., Holvoet P., Lund-Katz S., Phillips M. C. 2003. Domain structure and lipid interaction in human apolipoproteins A-I and E: a general model. J. Biol. Chem. 278: 23227–23232 [DOI] [PubMed] [Google Scholar]
- 21.Silva R. A. G., Hilliard G. M., Fang J., Macha S., Davidson W. S. 2005. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence tracking. Biochemistry. 44: 2759–2769 [DOI] [PubMed] [Google Scholar]
- 22.Oda M. N., Forte T. M., Ryan R. O., Voss J. C. 2003. The C-terminal domain of apolipoprotein A-I contains a lipid-sensitive conformational trigger. Nat. Struct. Biol. 10: 455–460 [DOI] [PubMed] [Google Scholar]
- 23.Lagerstedt J. O., Budamagunta M. S., Oda M. N., Voss J. C. 2007. EPR spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apo AI in solution. J. Biol. Chem. 282: 9143–9149 [DOI] [PubMed] [Google Scholar]
- 24.Lagerstedt J. O., Budamagunta M. S., Liu G. S., Devalle N. C., Voss J. C., Oda M. N. 2012. The “beta-clasp” model of apolipoprotein A-I - A lipid-free solution structure determined by electron paramagnetic resonance spectroscopy. Biochim. Biophys. Acta. 1821: 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chetty P. S., Mayne L., Lund-Katz S., Stranz D., Englander S. W., Phillips M. C. 2009. Helical structure and stability in human apolipoprotein A-I by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevugan Chetty P., Mayne L., Kan Z-Y., Lund-Katz S., Englander S. W., Phillips M. C. 2012. Apolipoprotein A-I helical structure and stability in discoidal high density lipoprotein (HDL) particles by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. USA. 109: 11687–11692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davidson W. S., Arnvig-McGuire K., Kennedy A., Kosman J., Hazlett T., Jonas A. 1999. Structural organization of the N-terminal domains of apolipoprotein A-I: studies of tryptophan mutants. Biochemistry. 38: 14387–14395 [DOI] [PubMed] [Google Scholar]
- 28.Roberts L. M., Ray M. J., Shih T. W., Hayden E., Reader M. M., Brouillette C. G. 1997. Structural analysis of apolipoprotein A-I: limited proteolysis of methionine-reduced and -oxidized lipid-free and lipid-bound human apoA-I. Biochemistry. 36: 7615–7624 [DOI] [PubMed] [Google Scholar]
- 29.Brouillette C. G., Dong W. J., Yang Z. W., Ray M. J., Protasevich I. I., Cheung H. C., Engler J. A. 2005. Forster resonance energy transfer measurements are consistent with a helical bundle model for lipid-free apolipoprotein A-I. Biochemistry. 44: 16413–16425 [DOI] [PubMed] [Google Scholar]
- 30.Muñoz V., Serrano L. 1994. Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Biol. 1: 399–409 [DOI] [PubMed] [Google Scholar]
- 31.Gorshkova I. N., Liu T., Horng-Yuan K., Chroni A., Zannis V. I., Atkinson D. 2006. Structure and stability of apolipoprotein A-I in solution and in discoidal high-density lipoprotein probed by double charge ablation and deletion mutation. Biochemistry. 45: 1242–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Li Q., Wang J. 2011. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc. Natl. Acad. Sci. USA. 108: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson C., Wardell M. R., Weisgraber K. H., Mahley R. W., Agard D. A. 1991. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 252: 1817–1822 [DOI] [PubMed] [Google Scholar]
- 34.Sivashanmugam A., Wang J. 2009. A unified scheme for initiation and conformational adaptation of human apolipoprotein E N-terminal domain upon lipoprotein-binding and for receptor-binding activity. J. Biol. Chem. 284: 14657–14666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng X., Morris J., Dressmen J., Tubb M. R., Tso P., Jerome W. G., Davidson W. S., Thompson T. B. 2012. The structure of dimeric apolipoprotein A-IV and its mechanism of self-association. Structure. 20: 767–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breiter D. R., Kanost M. R., Benning M. M., Wesenberg G., Law J. H., Wells M. A., Rayment I., Holden H. M. 1991. Molecular structure of an apolipoprotein determined at 2.5-A resolution. Biochemistry. 30: 603–608 [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Gagne S. M., Sykes B. D., Ryan R. O. 1997. Insight into lipid surface recognition and reversible conformational adaptations of an exchangeable apolipoprotein by multidimensional heteronuclear NMR techniques. J. Biol. Chem. 272: 17912–17920 [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Sykes B. D., Ryan R. O. 2002. Structural basis for the conformational adaptability of apolipophorin III, a helix-bundle exchangeable apolipoprotein. Proc. Natl. Acad. Sci. USA. 99: 1188–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ajees A. A., Anantharamaiah G. M., Mishra V. K., Hussain M. M., Murthy S. 2006. Crystal structure of human apolipoprotein A-I: Insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. USA. 103: 2126–2131 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Borrell B. 2009. Fraud rocks protein community. Nature. 462: 970. [DOI] [PubMed] [Google Scholar]
- 41.Borhani D. W., Rogers D. P., Engler J. A., Brouillette C. G. 1997. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA. 94: 12291–12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mei X., Atkinson D. 2011. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. J. Biol. Chem. 286: 38570–38582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito H., Lund-Katz S., Phillips M. C. 2004. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog. Lipid Res. 43: 350–380 [DOI] [PubMed] [Google Scholar]
- 44.Morrow J. A., Segall M. L., Lund-Katz S., Phillips M. C., Knapp M., Rupp B., Weisgraber K. H. 2000. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 39: 11657–11666 [DOI] [PubMed] [Google Scholar]
- 45.Tanaka M., Koyama M., Dhanasekaran P., Nguyen D., Nickel M., Lund-Katz S., Saito H., Phillips M. C. 2008. Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry. 47: 2172–2180 [DOI] [PubMed] [Google Scholar]
- 46.Gursky O., Atkinson D. 1996. Thermal unfolding of human high-density apolipoprotein A-I: implications for a lipid-free molten globular state. Proc. Natl. Acad. Sci. USA. 93: 2991–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guha M., Gao X., Jayaraman S., Gursky O. 2008. Correlation of structural stability with functional remodeling of high-density lipoproteins: the importance of being disordered. Biochemistry. 47: 11393–11397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chetty P. S., Ohshiro M., Saito H., Dhanasekaran P., Lund-Katz S., Mayne L., Englander S. W., Phillips M. C. 2012. Effects of the Iowa and Milano mutations on apolipoprotein A-I structure and dynamics determined by hydrogen exchange and mass spectrometry. Biochemistry. 51: 8993–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagerstedt J. O., Cavigiolio G., Roberts L. M., Hong H. S., Jin L. W., Fitzgerald P. G., Oda M. N., Voss J. C. 2007. Mapping the structural transition in an amyloidogenic apolipoprotein A-I. Biochemistry. 46: 9693–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyson H. J., Wright P. E. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6: 197–208 [DOI] [PubMed] [Google Scholar]
- 51.Gsponer J., Babu M. M. 2009. The rules of disorder or why disorder rules. Prog. Biophys. Mol. Biol. 99: 94–103 [DOI] [PubMed] [Google Scholar]
- 52.Koyama M., Tanaka M., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2009. Interaction between the N- and C-terminal domains modulates the stability and lipid binding of apolipoprotein A-I. Biochemistry. 48: 2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narayanaswami V., Ryan R. O. 2000. Molecular basis of exchangeable apolipoprotein function. Biochim. Biophys. Acta. 1483: 15–36 [DOI] [PubMed] [Google Scholar]
- 54.Lund-Katz S., Phillips M. C. 2010. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell. Biochem. 51: 183–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kono M., Okumura Y., Tanaka M., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2008. Conformational flexibility of the N-terminal domain of apolipoprotein A-I bound to spherical lipid particles. Biochemistry. 47: 11340–11347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizuguchi C., Hata M., Dhanasekaran P., Nickel M., Phillips M. C., Lund-Katz S., Saito H. 2012. Fluorescence analysis of the lipid binding-induced conformational change of apolipoprotein E4. Biochemistry. 51: 5580–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saito H., Dhanasekaran P., Nguyen D., Deridder E., Holvoet P., Lund-Katz S., Phillips M. C. 2004. Alpha-helix formation is required for high affinity binding of human apolipoprotein A-I to lipids. J. Biol. Chem. 279: 20974–20981 [DOI] [PubMed] [Google Scholar]
- 58.Saito H., Dhanasekaran P., Baldwin F., Weisgraber K., Lund-Katz S., Phillips M. C. 2001. Lipid binding-induced conformational change in human apolipoprotein E. J. Biol. Chem. 276: 40949–40954 [DOI] [PubMed] [Google Scholar]
- 59.Tanaka M., Dhanasekaran P., Nguyen D., Nickel M., Takechi Y., Lund-Katz S., Phillips M. C., Saito H. 2011. Influence of N-terminal helix bundle stability on the lipid-binding properties of human apolipoprotein A-I. Biochim. Biophys. Acta. 1811: 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segall M. L., Dhanasekaran P., Baldwin F., Anantharamaiah G. M., Weisgraber K., Phillips M. C., Lund-Katz S. 2002. Influence of apoE domain structure and polymorphism on the kinetics of phospholipid vesicle solubilization. J. Lipid Res. 43: 1688–1700 [DOI] [PubMed] [Google Scholar]
- 61.Narayanaswami V., Kiss R. S., Weers P. M. 2010. The helix bundle: a reversible lipid binding motif. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 155: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pownall H. J., Massey J. B., Sparrow J. T., Gotto A. M. 1987. Lipid-protein interactions and lipoprotein reassembly. In Plasma Lipoproteins. A. M. Gotto, editor. Elsevier Science Publishers B.V., Amsterdam. 95–127. [Google Scholar]
- 63.Fukuda M., Nakano M., Sriwongsitanont S., Ueno M., Kuroda Y., Handa T. 2007. Spontaneous reconstitution of discoidal HDL from sphingomyelin-containing model membranes by apolipoprotein A-I. J. Lipid Res. 48: 882–889 [DOI] [PubMed] [Google Scholar]
- 64.Massey J. B., Pownall H. J. 2008. Cholesterol is a determinant of the structures of discoidal high density lipoproteins formed by the solubilization of phospholipid membranes by apolipoprotein A-I. Biochim. Biophys. Acta. 1781: 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonas A. 1992. Lipid-binding properties of apolipoproteins. In Structure and Function of Apolipoproteins. M. Rosseneu, editor. CRC Press, Boca Raton. 217–250. [Google Scholar]
- 66.Garai K., Baban B., Frieden C. 2011. Dissociation of apolipoprotein E oligomers to monomer is required for high-affinity binding to phospholipid vesicles. Biochemistry. 50: 2550–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vedhachalam C., Chetty P. S., Nickel M., Dhanasekaran P., Lund-Katz S., Rothblat G. H., Phillips M. C. 2010. Influence of apolipoprotein (Apo) A-I structure on nascent high density lipoprotein (HDL) particle size distribution. J. Biol. Chem. 285: 31965–31973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyssenko N. N., Hata M., Dhanasekaran P., Nickel M., Nguyen D., Chetty P. S., Saito H., Lund-Katz S., Phillips M. C. 2012. Influence of C-terminal alpha-helix hydrophobicity and aromatic amino acid content on apolipoprotein A-I functionality. Biochim. Biophys. Acta. 1821: 456–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gillotte K. L., Zaiou M., Lund-Katz S., Anantharamaiah G. M., Holvoet P., Dhoest A., Palgunachari M. N., Segrest J. P., Weisgraber K. H., Rothblat G. H., et al. 1999. Apolipoprotein-mediated plasma membrane microsolubilization. J. Biol. Chem. 274: 2021–2028 [DOI] [PubMed] [Google Scholar]
- 70.Jonas A. 1986. Reconstitution of high-density lipoproteins. Methods Enzymol. 128: 553–582 [DOI] [PubMed] [Google Scholar]
- 71.Davidson W. S., Silva R. A. G. 2005. Apolipoprotein structural organization in high density lipoproteins: belts, bundles, hinges and hairpins. Curr. Opin. Lipidol. 16: 295–300 [DOI] [PubMed] [Google Scholar]
- 72.Thomas M. J., Bhat S., Sorci-Thomas M. G. 2006. The use of chemical cross-linking and mass spectrometry to elucidate the tertiary conformation of lipid-bound apolipoprotein A-I. Curr. Opin. Lipidol. 17: 214–220 [DOI] [PubMed] [Google Scholar]
- 73.Davidson W. S., Thompson T. B. 2007. The structure of apolipoprotein A-I in high density lipoproteins. J. Biol. Chem. 282: 22249–22253 [DOI] [PubMed] [Google Scholar]
- 74.Thomas M. J., Bhat S., Sorci-Thomas M. G. 2008. Three-dimensional models of HDL apoA-I: implications for its assembly and function. J. Lipid Res. 49: 1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koppaka V., Silvestro L., Engler J. A., Brouillette C. G., Axelsen P. H. 1999. The structure of human lipoprotein A-I: evidence for the “belt” model. J. Biol. Chem. 274: 14541–14544 [DOI] [PubMed] [Google Scholar]
- 76.Segrest J. P., Jones M. K., Klon A. E., Sheldahl C. J., Hellinger M., DeLoof H., Harvey S. C. 1999. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J. Biol. Chem. 274: 31755–31758 [DOI] [PubMed] [Google Scholar]
- 77.Li L., Li S., Jones M. K., Segrest J. P. 2012. Rotational and hinge dynamics of discoidal high density lipoproteins probed by interchain disulfide bond formation. Biochim. Biophys. Acta. 1821: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H. H., Lyles D. S., Pan W., Alexander E., Thomas M. J., Sorci-Thomas M. G. 2002. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J. Biol. Chem. 277: 39093–39101 [DOI] [PubMed] [Google Scholar]
- 79.Caulfield T. R. 2011. Inter-ring rotation of apolipoprotein A-I protein monomers for the double-belt model using biased molecular dynamics. J. Mol. Graph. Model. 29: 1006–1014 [DOI] [PubMed] [Google Scholar]
- 80.Bhat S., Sorci-Thomas M. G., Tuladhar R., Samuel M. P., Thomas M. J. 2007. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 46: 7811–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gursky O. 2013. Crystal structure of delta(185–243)ApoA-I suggests a mechanistic framework for the protein adaptation to the changing lipid load in good cholesterol: from flatland to sphereland via double belt, belt buckle, and double hairpin trefoil/tetrafoil. J. Mol. Biol. 425: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maiorano J. N., Jandacek R. J., Horace E. M., Davidson W. S. 2004. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 43: 11717–11726 [DOI] [PubMed] [Google Scholar]
- 83.Martin D. D., Budamagunta M. S., Ryan R. O., Voss J. C., Oda M. N. 2006. Apolipoprotein A-I assumes a “looped belt” conformation on reconstituted high density lipoprotein. J. Biol. Chem. 281: 20418–20426 [DOI] [PubMed] [Google Scholar]
- 84.Wu Z., Wagner M. A., Zheng L., Parks J. S., Shy J. M., III, Smith J. D., Gogonea V., Hazen S. L. 2007. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14: 861–868 [DOI] [PubMed] [Google Scholar]
- 85.Li L., Chen J., Mishra V. K., Kurtz J. A., Cao D., Klon A. E., Harvey S. C., Anantharamaiah G. M., Segrest J. P. 2004. Double belt structure of discoidal high density lipoproteins: molecular basis for size heterogeneity. J. Mol. Biol. 343: 1293–1311 [DOI] [PubMed] [Google Scholar]
- 86.Silva R. A., Schneeweis L. A., Krishnan S. C., Zhang X., Axelsen P. H., Davidson W. S. 2007. The structure of apolipoprotein A-II in discoidal high density lipoproteins. J. Biol. Chem. 282: 9713–9721 [DOI] [PubMed] [Google Scholar]
- 87.Gauthamadasa K., Vaitinadin N. S., Dressman J. L., Macha S., Homan R., Greis K. D., Silva R. A. 2012. Apolipoprotein A-II-mediated conformational changes of apolipoprotein A-I in discoidal high density lipoproteins. J. Biol. Chem. 287: 7615–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao X., Yuan S., Jayaraman S., Gursky O. 2012. Role of apolipoprotein A-II in the structure and remodeling of human high-density lipoprotein (HDL): protein conformational ensemble on HDL. Biochemistry. 51: 4633–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kono M., Tanaka T., Tanaka M., Vedhachalam C., Chetty P. S., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2010. Disruption of the C-terminal helix by single amino acid deletion is directly responsible for impaired cholesterol efflux ability of apolipoprotein A-I Nichinan. J. Lipid Res. 51: 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu Z., Gogonea V., Lee X., Wagner M. A., Li X. M., Huang Y., Undurti A., May R. P., Haertlein M., Moulin M., et al. 2009. Double superhelix model of high density lipoprotein. J. Biol. Chem. 284: 36605–36619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gogonea V., Wu Z., Lee X., Pipich V., Li X. M., Ioffe A. I., Didonato J. A., Hazen S. L. 2010. Congruency between biophysical data from multiple platforms and molecular dynamics simulation of the double-super helix model of nascent high-density lipoprotein. Biochemistry. 49: 7323–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Segrest J. P., Jones M. K., Catte A., Thirumuruganandham S. P. 2012. Validation of previous computer models and MD simulations of discoidal HDL by a recent crystal structure of apoA-I. J. Lipid Res. 53: 1851–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gu F., Jones M. K., Chen J., Patterson J. C., Catte A., Jerome W. G., Li L., Segrest J. P. 2010. Structures of discoidal high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285: 4652–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jones M. K., Zhang L., Catte A., Li L., Oda M. N., Ren G., Segrest J. P. 2010. Assessment of the validity of the double superhelix model for reconstituted high density lipoproteins: a combined computational-experimental approach. J. Biol. Chem. 285: 41161–41171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Silva R. A. G., Huang R., Morris J., Fang J., Gracheva E. O., Ren G., Kontush A., Jerome W. G., Rye K. A., Davidson W. S. 2008. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA. 105: 12176–12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu Z., Gogonea V., Lee X., May R. P., Pipich V., Wagner M. A., Undurti A., Tallant T. C., Baleanu-Gogonea C., Charlton F., et al. 2011. The low resolution structure of ApoA1 in spherical high density lipoprotein revealed by small angle neutron scattering. J. Biol. Chem. 286: 12495–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96a.Chetty P. S., Nguyen D., Nickel M., Lund-Katz S., Mayne L., Englander S. W., Phillips M. C. 2013. Comparison of apoA-I helical structure in discoidal and spherical HDL particles by HX and mass spectrometry. J. Lipid Res. 54: 1589–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., Davidson W. S. 2011. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18: 416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Curtiss L. K., Bonnett D. J., Rye K. A. 2000. The conformation of apolipoprotein A-I in high-density lipoproteins is influenced by core lipid composition and particle size: a surface plasmon resonance study. Biochemistry. 39: 5712–5721 [DOI] [PubMed] [Google Scholar]
- 99.Lund-Katz S., Nguyen D., Dhanasekaran P., Kono M., Nickel M., Saito H., Phillips M. C. 2010. Surface plasmon resonance analysis of the mechanism of binding of apoA-I to high density lipoprotein particles. J. Lipid Res. 51: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguyen D., Dhanasekaran P., Nickel M., Nakatani R., Saito H., Phillips M. C., Lund-Katz S. 2010. Molecular basis for the differences in lipid and lipoprotein binding properties of human apolipoproteins E3 and E4. Biochemistry. 49: 10881–10889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nguyen D., Dhanasekaran P., Phillips M. C., Lund-Katz S. 2009. Molecular mechanism of apolipoprotein E binding to lipoprotein particles. Biochemistry. 48: 3025–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cavigiolio G., Geier E. G., Shao B., Heinecke J. W., Oda M. N. 2010. Exchange of apolipoprotein A-I between lipid-associated and lipid-free states: a potential target for oxidative generation of dysfunctional high density lipoproteins. J. Biol. Chem. 285: 18847–18857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Asztalos B. F., Roheim P. S. 1995. Presence and promotion of ‘free apolipoprotein A-I-like’ particles in human plasma. Arterioscler. Thromb. Vasc. Biol. 15: 1419–1423 [DOI] [PubMed] [Google Scholar]
- 104.Kane J. P., Malloy M. J. 2012. Prebeta-1 HDL and coronary heart disease. Curr. Opin. Lipidol. 23: 367–371 [DOI] [PubMed] [Google Scholar]
- 105.Pownall H. J., Hosken B. D., Gillard B. K., Higgins C. L., Lin H. Y., Massey J. B. 2007. Speciation of human plasma high-density lipoprotein (HDL): HDL stability and apolipoprotein A-I partitioning. Biochemistry. 46: 7449–7459 [DOI] [PubMed] [Google Scholar]
- 106.Oram J. F., Heinecke J. W. 2005. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 85: 1343–1372 [DOI] [PubMed] [Google Scholar]
- 107.Vedhachalam C., Duong P. T., Nickel M., Nguyen D., Dhanasekaran P., Saito H., Rothblat G. H., Lund-Katz S., Phillips M. C. 2007. Mechanism of ATP-binding cassette transporter A1-mediated cellular lipid efflux to apolipoprotein A-I and formation of high density lipoprotein particles. J. Biol. Chem. 282: 25123–25130 [DOI] [PubMed] [Google Scholar]
- 108.Vedhachalam C., Liu L., Nickel M., Dhanasekaran P., Anantharamaiah G. M., Lund-Katz S., Rothblat G., Phillips M. C. 2004. Influence of apo A-I structure on the ABCA1-mediated efflux of cellular lipids. J. Biol. Chem. 279: 49931–49939 [DOI] [PubMed] [Google Scholar]
- 109.Remaley A. T., Stonik J. A., Demosky S. J., Neufeld E. B., Bocharov A. V., Vishnyakova T. G., Eggerman T. L., Patterson A. P., Duverger N., Santamarina-Fojo S., et al. 2001. Apolipoprotein specificity for lipid efflux by the human ABCA1 transporter. Biochem. Biophys. Res. Commun. 280: 818–823 [DOI] [PubMed] [Google Scholar]
- 110.Krimbou L., Denis M., Haidar B., Carrier M., Marcil M., Genest J., Jr 2004. Molecular interactions between apoE and ABCA1: impact on apoE lipidation. J. Lipid Res. 45: 839–848 [DOI] [PubMed] [Google Scholar]
- 111.Vedhachalam C., Narayanaswami V., Neto N., Forte T. M., Phillips M. C., Lund-Katz S., Bielicki J. K. 2007. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 46: 2583–2593 [DOI] [PubMed] [Google Scholar]
- 112.Liu L., Bortnick A. E., Nickel M., Dhanasekaran P., Subbaiah P. V., Lund-Katz S., Rothblat G. H., Phillips M. C. 2003. Effects of apolipoprotein A-I on ATP-binding cassette transporter A1-mediated efflux of macrophage phospholipid and cholesterol - formation of nascent high density lipoprotein particles. J. Biol. Chem. 278: 42976–42984 [DOI] [PubMed] [Google Scholar]
- 113.Krimbou L., Hassan H. H., Blain S., Rashid S., Denis M., Marcil M., Genest J. 2005. Biogenesis and speciation of nascent apoA-I-containing particles in various cell lines. J. Lipid Res. 46: 1668–1677 [DOI] [PubMed] [Google Scholar]
- 114.Mulya A., Lee J. Y., Gebre A. K., Thomas M. J., Colvin P. L., Parks J. S. 2007. Minimal lipidation of pre-beta HDL by ABCA1 results in reduced ability to interact with ABCA1. Arterioscler. Thromb. Vasc. Biol. 27: 1828–1836 [DOI] [PubMed] [Google Scholar]
- 115.Duong P. T., Collins H. L., Nickel M., Lund-Katz S., Rothblat G. H., Phillips M. C. 2006. Characterization of nascent HDL particles and microparticles formed by ABCA1-mediated efflux of cellular lipids to apoA-I. J. Lipid Res. 47: 832–843 [DOI] [PubMed] [Google Scholar]
- 116.Sorci-Thomas M. G., Owen J. S., Fulp B., Bhat S., Zhu X., Parks J. S., Shah D., Jerome W. G., Gerelus M., Zabalawi M., et al. 2012. Nascent high density lipoproteins formed by ABCA1 resemble lipid rafts and are structurally organized by three apoA-I monomers. J. Lipid Res. 53: 1890–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Duong P. T., Weibel G. L., Lund-Katz S., Rothblat G. H., Phillips M. C. 2008. Characterization and properties of pre beta-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 49: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117a.Lyssenko N. N., Nickel M., Tang C., Phillips M. C. 2013. Factors controlling nascent high-density lipoprotein particle heterogeneity: ATP-binding cassette transporter A1 activity and cell lipid and apolipoprotein AI availability. FASEB J. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Favari E., Bernini F., Tarugi P., Franceschini G., Calabresi L. 2002. The C-terminal domain of apolipoprotein A-I is involved in ABCA1-driven phospholipid and cholesterol efflux. Biochem. Biophys. Res. Commun. 299: 801–805 [DOI] [PubMed] [Google Scholar]
- 119.Chroni A., Liu T., Gorshkova I., Kan H. Y., Uehara Y., von Eckardstein A., Zannis V. I. 2003. The central helices of apoA-I can promote ATP-binding cassette transporter AI (ABCA1)-mediated lipid efflux. J. Biol. Chem. 278: 6719–6730 [DOI] [PubMed] [Google Scholar]
- 120.Panagotopulos S. E., Witting S. R., Horace E. M., Hui D. Y., Maiorano J. N., Davidson W. S. 2002. The role of apolipoprotein A-I helix 10 in apolipoprotein-mediated cholesterol efflux via the ATP-binding cassette transporter ABCA1. J. Biol. Chem. 277: 39477–39484 [DOI] [PubMed] [Google Scholar]
- 121.Alexander E. T., Vedhachalam C., Sankaranarayanan S., de la Llera-Moya M., Rothblat G. H., Rader D. J., Phillips M. C. 2011. Influence of apolipoprotein A-I domain structure on macrophage reverse cholesterol transport in mice. Arterioscler. Thromb. Vasc. Biol. 31: 320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]