Abstract
Diacylglycerol kinase (DGK)θ is a lipid kinase that phosphorylates diacylglycerol to form phosphatidic acid (PA). We have previously shown that PA is a ligand for the nuclear receptor steroidogenic factor 1 (SF1) and that cAMP-stimulated expression of SF1 target genes requires DGKθ. In this study, we sought to investigate the role of cAMP signaling in regulating DGKθ gene expression. Real time RT-PCR and Western blot analysis revealed that dibutyryl cAMP (Bt2cAMP) increased the mRNA and protein expression, respectively, of DGKθ in H295R human adrenocortical cells. SF1 and sterol regulatory element binding protein 1 (SREBP1) increased the transcriptional activity of a reporter plasmid containing 1.5 kb of the DGKθ promoter fused to the luciferase gene. Mutation of putative cAMP responsive sequences abolished SF1- and SREBP-dependent DGKθ reporter gene activation. Consistent with this finding, chromatin immunoprecipitation assay demonstrated that Bt2cAMP signaling increased the recruitment of SF1 and SREBP1 to the DGKθ promoter. Coimmunoprecipitation assay revealed that SF1 and SREBP1 interact, suggesting that the two transcription factors form a complex on the DGKθ promoter. Finally, silencing SF1 and SREBP1 abolished cAMP-stimulated DGKθ expression. Taken together, we demonstrate that SF1 and SREBP1 activate DGKθ transcription in a cAMP-dependent manner in human adrenocortical cells.
Keywords: diacylglycerol kinase θ, adrenal cortex, cAMP
Diacylglycerol kinases (DGKs) are intracellular lipid kinases that phosphorylate diacylglycerol (DAG) to form phosphatidic acid (PA), which is linked to lipid metabolism and signaling (1–3). For example, targeted disruption of DGKδ in mice impairs epidermal growth factor receptor expression and increases protein kinase C (PKC) activity (4). DGKϵ-null mice exhibit several neural abnormalities, including a higher resistance of electroconvulsive shock (5) and increased cyclooxygenase 2 and tyrosine hydroxylase expression (6), suggesting a role for DGKϵ in regulating synaptic activity. Mice lacking DGKα (7) or DGKζ (8) exhibit enhanced T cell function and demonstrate a role for these kinases in controlling DAG metabolism during the immune response. DGK isoforms have been implicated in various other cellular processes including inhibition of Rap1 signaling (9) and retinoblastoma-mediated cell cycle control (10). DGKθ is activated by nerve growth factor in PC12 cells (11) and thrombin in IIC9 fibroblasts (12, 13), whereas DGKζ promotes myogenesis in C2C12 cells (14) and DGKγ plays a role in regulating the cell cycle in CHO-K cells (15).
To date, 10 mammalian DGKs have been identified that are divided into five groups based on functional domains (16, 17). However, all isoforms contain cysteine-rich zinc finger-like structures, a conserved catalytic region (18–21). DGKθ, the sole member of group V, is comprised of three cysteine-rich domains (CRDs), a proline/glycine-rich domain at its N terminus, and a pleckstrin homology (PH) with an overlapping Ras-binding domain (22). While the functions of many of the other domains in DGKθ are unclear, the catalytic activity requires all domains of the enzyme (23). It has been postulated that the CRDs of the enzyme are required both for correct folding of the protein and for substrate presentation (23). Mutation of the CRD of DGKθ diminishes DAG-induced translocation of the enzyme to the plasma membrane (24); whereas the interaction between DGKθ and the nuclear receptor steroidogenic factor 1 (SF1) requires the PH domain (25).
The ability of distinct isoforms to exert regulatory control occurs through unique interactions with protein partners, and differential subcellular localization of DGK isoforms is thought to enable local regulation of DAG and PA concentrations for spatial and temporally separated cellular processes. Many studies have demonstrated roles for compartmentalized DGK activity in nuclear processes. Both DGKθ (26) and DGKζ (14) are localized in punctate structures that are enriched in pre-mRNA splicing factors called nuclear speckles. DGKθ is colocalized with hyperphosphorylated RNA polymerase II and the splicing factor SC-35 in the nuclear speckles of various cell types, including PC12, HeLa, and MCF-7 (26). Interestingly, nuclear speckles have been shown to sequester posttranslationally modified SF1 (27, 28).
SF1 induces the transcription of genes involved in steroid hormone biosynthesis and endocrine development and function (29–31). We have previously shown that cAMP signaling increases the transcription of CYP17A1 by stimulating the binding of SF1 to the CYP17A1 promoter (32, 33). We have also shown that DGKθ regulates the production of PA, a ligand for SF1 that is produced in response to cAMP signaling (25). DGKθ acts as a coregulatory protein by binding to SF1 when the receptor is bound to chromatin. The PA produced in response to DGKθ activation stimulates SF1-dependent gene transcription by promoting coactivator recruitment to SF1 target genes, thereby inducing the mRNA expression of CYP17A1 and several other steroidogenic genes. In contrast, inhibition of DGK activity attenuates the binding of SF1-dependent gene expression, and silencing the expression of DGKθ expression inhibits cAMP-dependent CYP17A1 transcription. Finally, we have also shown that LXXLL motifs in DGKθ mediate a direct interaction of SF1 with the kinase and may facilitate ligand delivery (25). To date, studies have demonstrated that DGKθ is regulated by intracellular targeting (24), membrane lipids (12), protein-protein interactions (34), and intrinsic activity (12). However, the factors that control DGKθ gene expression in the adrenal cortex are poorly understood. In this study, we defined the role of cAMP signaling in regulating the expression of DGKθ.
MATERIALS AND METHODS
Materials
Dibutyryl cAMP (Bt2cAMP) and tetracycline were obtained from Sigma (St. Louis, MO) and H89 from EMD Biosciences (La Jolla, CA).
Cell culture
H295R adrenocortical cells (35, 36) were generously donated by Dr. William E. Rainey (University of Michigan, Ann Arbor, MI) and cultured in Dulbecco's modified Eagle's medium/F-12 (DMEM/F-12) (Invitrogen, Carlsbad, CA) supplemented with 10% Nu-Serum I (BD Bioscience, Palo Alto, CA), 1% ITS Plus (BD Bioscience), antibiotics, and antimycotics. CV-1 monkey kidney cells were obtained from American Type Culture Collection (ATCC) (Manassas, VA) and cultured in Eagle's minimum essential medium (MEM) (Mediatech, Inc., Manassas, VA) supplemented with 10% fetal bovine serum (Mediatech, Inc.), antibiotics, and antimycotics. SF1 and sterol regulatory element binding protein 1 (SREBP1) knockdown cell lines were generated by transfecting H295R cells with short hairpin RNA (shRNA) plasmids (in the pGFP-V-RS HuSH vector; Origene, Rockville, MD) containing the following oligonucleotides: SF1 5′-TCC TGG CCG TGC CAT CAA GTC TGA GTA CC and SREBP1 5′-ATC TAT GTG GCG GCT GCA TTG AGA GTG AA. Stable clones were selected using 10 μg/ml puromycin (Mediatech, Inc.). H295R cells expressing tetracycline-inducible DGKθ shRNA were generated using the BLOCK-iT Inducible H1 RNAi Entry Vector Kit (Invitrogen) as previously described (37). To construct an inducible vector for DGKθ shRNA, the following sequences were cloned into pENTR/H1/TO: 5′-ACC GCC CAG TAT TGA AGG CCT CAT CTT CAC GAA TGA AGA TGA GGC CTT CAA TAC TGG G-3′ and 5′-AAA CCC AGT ATT GAA GGC CTC ATC TTC ATT CGT GAA GAT GAG GCC TTC AAT ACT GGG C-3′. H295R-TetR cells were stably transfected with the constructed pENTR/H1/TO-DGKθ shRNA expression vector or the control vector using GeneJuice (EMD Biosciences), and cell clones were selected using 50 μg/ml zeocin. Clones were treated with 5 μg/ml tetracycline for 96 h and suppression of DGKθ protein levels in each clone was confirmed by Western blotting using an anti-DGKθ antibody (Sigma).
Cloning and mutagenesis
The human DGKQ promoter was cloned using LA Taq DNA polymerase (Takara, Madison, WI), 500 ng of human genomic DNA (Promega, Madison, WI) and 300 nM of the following primers: forward 5′-CGA GCT CTT ACG CGT CTA GCT CTC CCA GGG CCC and reverse 5′-CTT AGA TCG CAG ATC TCT CGG CCG CCG CCG C. PCR fragments were cloned into the MluI (5′) and BglII (3′) sites of the pGL3 Firefly luciferase vector (Promega). Putative SF1 and SREBP1 response elements were identified by in silico analysis using MatInspector (Genomatix Software, Ann Arbor, MI) and site-directed mutagenesis performed using the following primers: M1 forward 5′-CCT TCC CTC CAG AGT AAA CAG CCC CCA GCC and reverse 5′-GGC TGG GGG CTG TTT ACT CTG GAG GGA AGG, M2 forward 5′-CCC CCA GCC CCT TTC AAA CGT CTC CCC ACA GGC and reverse 5′-GCC TGT GGG GAG ACG TTT GAA AGG GGC TGG GGG, M3 forward 5′-TGC TGC GAT GGC CCT AAA GCC CTG CCC TCT GC and reverse 5′-GCA GAG GGC AGG GCT TTA GGG CCA TCG CAG CA, M4 forward 5′-GGC CCA CGG GGG CAA AAA CCC AGA CTG CTG CC and reverse 5′-GGC AGC AGT CTG GGT TTT TGC CCC CGT GGG CC, M5 forward 5′-GGG GTG ACC CGC GTA AAC GCG GCT CTC AAA GG and reverse 5′-CCT TTG AGA GCC GCG TTT ACG CGG GTC ACC CC, M6 forward 5′-GGA CGC GGC TCT CAA AAA ACA CCA GCG CCA CC and reverse 5′-GGT GGC GCT GGT GTT TTT TGA GAG CCG CGT CC. Wild-type and mutant pGL3-DGKQ constructs were confirmed by sequencing.
Transient transfection and reporter gene analysis
H295R cells were sub-cultured onto 24-well plates and transfected with 20 ng of pGL3-DGKθ, 1 ng pRL-CMV (Promega), and/or 25 ng of pCMV6-GFP-SF1 (RC207577; Origene), pCDNA3.1-SREBP1a, pCDNA3.1-SREBP1c [Addgene, Cambridge, MA; generated by Dr. Timothy Osborne, Sanford-Burnham Institute, FL (38)] using Genejuice (Novagen, Madison, WI). Twenty-four hours after transfection, the cells were treated with 0.4 mM Bt2cAMP for 24 h and the transcriptional activity of DGKθ reporter gene measured using a dual luciferase assay kit (Promega). Firefly (pLG3-DGKθ) luciferase activity was normalized to Renilla luciferase activity (pRL-CMV, Promega) and expressed as fold change over the mean of the untreated control group.
RNA isolation and quantitative RT-PCR
Cells were sub-cultured onto 12-well plates and 24 h later treated with 0.4 mM Bt2cAMP for 1–24 h. Total RNA was extracted using Iso-RNA Lysis Reagent (5 Prime Inc., Gaithersburg, MD) and amplified using a One-Step SYBR Green RT-PCR Kit (Thermo Fisher Scientific Inc., Waltham, MA) and the primer pairs described in Li et al. (25). DGK expression was normalized to β-actin content and calculated using delta-delta cycle threshold (ΔΔCT) method.
Western blotting
H295R cells were sub-cultured onto 6-well plates and treated with 0.4 mM Bt2cAMP for 24 h, 48 h, or 72 h and harvested into radioimmunoprecipitation assay (RIPA) buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease inhibitors [150 nM aprotinin, 1 mM leupeptin, 1 mM E-64, 500 mM 4-(2-aminoethyl)benzenesulfonylfluoride); EMD Biosciences]. Cells were then lysed by sonication (one 5 s burst) followed by incubation on ice for 30 min. Lysates were centrifuged for 10 min at 4°C and the supernatant collected for analysis by SDS-PAGE. Protein concentrations were determined by bicinchoninic acid (BCA) protein assay (Pierce). Aliquots of each sample (25 μg of protein) were run on 8% SDS-PAGE gels and transferred to Immobilon-FL polyvinylidene difluoride (PVDF) membranes (IPFL00010; Millipore, Billerica, MA). Blots were probed with an anti-DGKθ (1:1000, HPA026797; Sigma-Aldrich, St. Louis, MO), anti-SREBP1 (1:1000, sc-8984; Santa Cruz Biotechnology, Santa Cruz, CA), SF1 (1:4000; Millipore, Temecula, CA), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:10,000, sc-25778; Santa Cruz Biotechnology) and an anti-rabbit or mouse secondary antibody (1:5000, ECF Western blotting reagent; GE Healthcare, Piscataway, NJ). Blots were scanned on a VersaDoc 4000 Imager (Bio-Rad, Hercules, CA) and densitometric analysis carried out using Quantity One software (Bio-Rad).
Nuclear and cytoplasmic extract isolation
H295R cells were cultured in 100 mm dishes and treated with Bt2cAMP for 48 h. Cytoplasmic and nuclear extracts were harvested from H295R cells and separated using Thermo NE-PER® nuclear and cytoplasmic extraction reagents (Pierce Biotechnology, Rockford, IL). Western blotting analysis was carried out as described above. Blots were probed with anti-DGKθ (1:1000, HPA026797; Sigma) and anti-lamin A/C (1:5000, sc-376248; Santa Cruz Biotechnology) or anti-β-tubulin (1:2000, sc-23949; Santa Cruz Biotechnology).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described previously in (32, 39, 40). Briefly, H295R cells were sub-cultured onto 150 mm dishes and treated with Bt2cAMP for 60 min. Cells were treated with 1% formaldehyde for 10 min at room temperature and then incubated for 5 min with 0.125 M glycine. After twice washing with PBS, cells were harvested into RIPA buffer. The purified chromatin solutions were immunoprecipitated using 5 μg of anti-acetyl-K5, K8, K12, K16 histone H4 (06-866; Millipore, Temecula, CA), anti-SF1 (07-618; Millipore), SREBP1 (sc-8984; Santa Cruz Biotechnology, Santa Cruz, CA), and anti-IgG protein A/G plus (sc-2003; Santa Cruz Biotechnology). Real-time PCR was carried out using the following primer sets: forward 5′-CAG AGT CCA CAG CCC CCA GCC CCT TTC AGG and reverse 5′-CTG CCT CGT GCG CGC CAC GGG TCT TGT TCA. Output DNA (immunoprecipitated promoter region) was normalized to input DNA. PCR products were separated on 2% agarose gels and the EtBr-stained bands imaged using a VersaDoc 4000 (Bio-Rad).
Coimmunoprecipitation
CV1 cells were plated onto 100 mm dishes and transfected with pCMV6-GFP-SF1, pcDNA 3.1-FLAG SREBP1c for 48 h. Five percent of lysates were retained as input and the remaining cell lysates were incubated with an anti-FLAG M2 mouse monoclonal antibody (5 μg; F1804, Sigma) and protein A/G agarose beads (Santa Cruz Biotechnology) overnight at 4°C with rotation. Beads were washed three times with RIPA buffer and twice with PBS and the immobilized proteins separated by SDS-PAGE. Output blots were probed with anti-SF1 (1:5000, 07-618; Millipore) and input blots with anti-FLAG (1:2500, F1804, Sigma). Expression was detected using an ECF Western blotting kit (GE Biosciences) and visualized using a VersaDoc 4000 imager (Bio-Rad).
PA assay
H295R cells were grown on 100 mm dishes and then treated with Bt2cAMP from 24 h to 48 h and total lipid extract was harvested. Nuclei were purified using a Nuclei Pure kit (Sigma) and PA content was quantified using a Total PA kit (Cayman, Ann Arbor, MI) on a SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices, LLC, Sunnyvale, CA) at an excitation wavelength of 530–540 nM and an emission wavelength of 585–595 nm. Data was quantified using through SoftMax Pro software (Molecular Devices).
DAG assay
H295R cells were cultured onto 6-well plates and treated with Bt2cAMP from 72 h and cells were harvested with PBS. PBS was aspirated and the content of DAG in each sample was determined using a Human DAG ELISA kit (MyBioSource, Inc., San Diego, CA).
Statistical analysis
One-way ANOVA and Tukey-Kramer multiple comparisons were performed using Prism 5.0 (GraphPad Software, San Diego, CA). Significant difference value was set as P < 0.05.
RESULTS
cAMP induces DGKθ mRNA expression
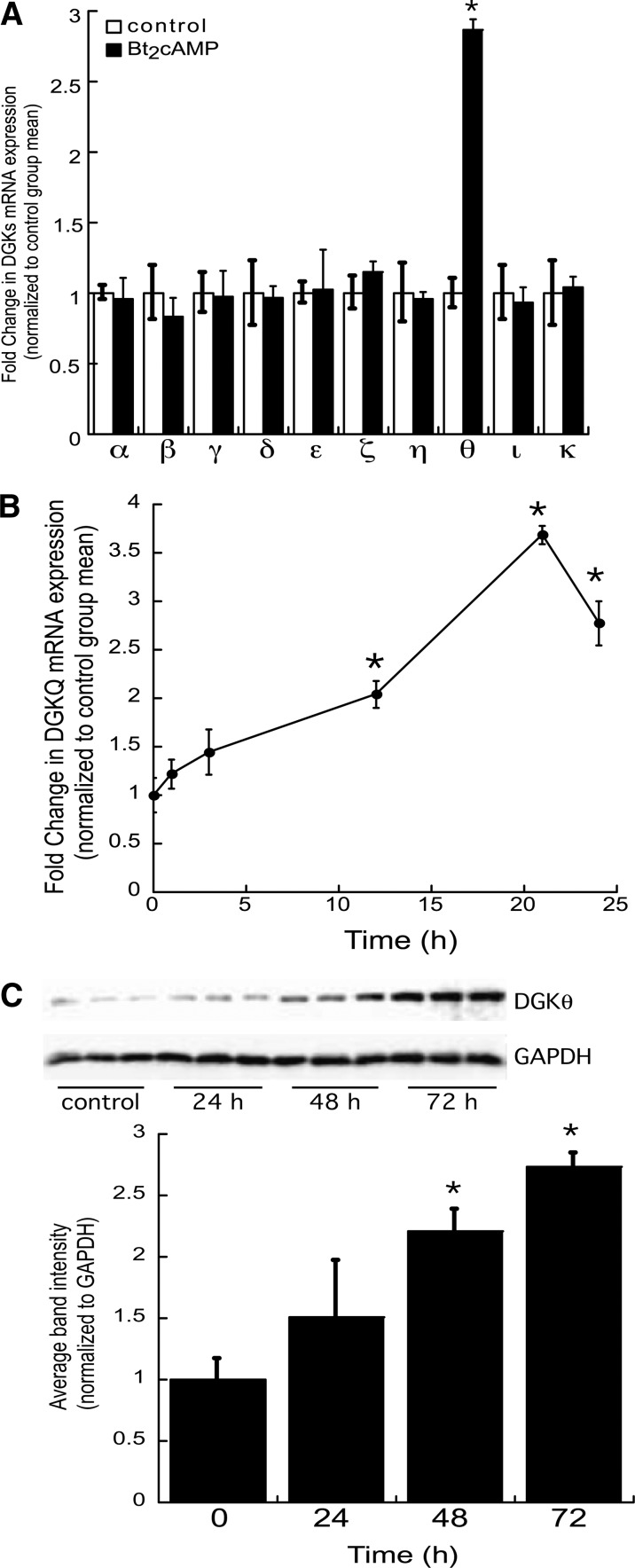
We have previously shown that adrenocorticotropic hormone (ACTH) signaling rapidly increases DGKθ activity (25). Therefore, in this study we sought to determine the effect of increased intracellular cAMP on DGKθ gene expression. H295R human adrenocortical cells were treated with Bt2cAMP for 24 h and RNA isolated for qRT-PCR. DGKθ mRNA expression was increased by 2.9-fold after 24 h treatment with Bt2cAMP (Fig. 1A), but had no effect on the mRNA expression of other DGK isoforms (Fig. 1A). Next, we assessed the kinetics of the DGKθ response to Bt2cAMP by treating H295R cells for 1–24 h. The results revealed that Bt2cAMP activation rapidly increased DGKθ mRNA expression by 1.5-fold at the 3 h time point with a maximal 3.7-fold increase in DGKθ mRNA expression occurring at the 21 h time point (Fig. 1B). Consistent with an increase in mRNA expression, Bt2cAMP treatment led to an increase in DGKθ protein expression by 2.2- and 2.7-fold after 48 h and 72 h treatment, respectively (Fig. 1C).
Fig. 1.
Bt2cAMP increases DGKθ mRNA and protein expression. A: H295R cells were cultured onto 12-well plates and treated for 24 h with 0.4 mM Bt2cAMP. Total RNA was isolated for analysis of DGK and β-actin mRNA expression by qRT-PCR. Data are graphed as fold change in DGK mRNA expression normalized to the mRNA expression of β-actin and represent the mean ± SEM of three separate experiments, each performed in triplicate. *Statistically different from untreated control group, P < 0.05. B: H285R cells were treated for 1–24 h with 0.4 mM Bt2cAMP and DGKθ mRNA expression quantified by real time RT-PCR. Data are graphed as fold change in DGKθ mRNA content and is normalized to the mRNA expression of β-actin. Shown is the mean ± SEM of three individual experiments, each performed in triplicate. Asterisk denotes statistically different from untreated control group, P < 0.05. C: H295R cells were cultured onto 6-well plates and incubated for 24–72 h with 0.4 mM Bt2cAMP. Whole cell lysates were harvested and analyzed by SDS-PAGE and Western blotting using anti-DGKθ antibody. Data graphed are densitometric analysis of Western blots of DGKθ protein expression in cells treated for 24–72 h with 0.4 mM Bt2cAMP. DGKθ protein expression normalized to GAPDH expression is graphed and represents the mean ± SEM of four separate experiments, each carried out in triplicate. Asterisks indicate a statistically significant difference compared with the untreated 0 h control group (P < 0.05).
Effect of kinase on cAMP-stimulated DGKθ mRNA expression
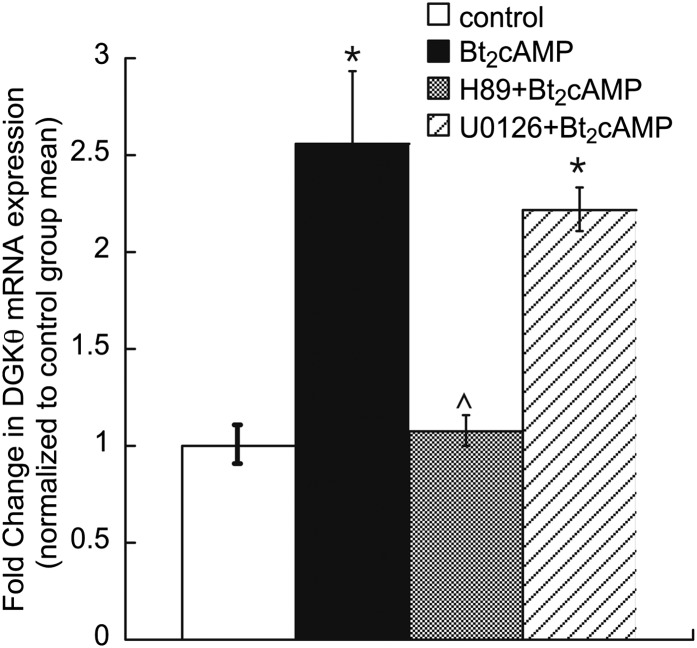
In the human adrenal cortex, the action of cAMP is mediated by the cAMP-dependent protein kinase A (PKA) (41). To determine if Bt2cAMP stimulated DGKθ expression required PKA, H295R cells were treated with H89 or the mitogen-activated protein kinase inhibitor U0126. As shown in Fig. 2, H89 treatment attenuated the cAMP activation on DGKθ mRNA expression. Conversely, no significant effect was observed with U0126.
Fig. 2.
PKA inhibitor decreases Bt2cAMP activation on DGKθ mRNA expression. H295R cells were cultured onto 12-well plates and incubated with kinase inhibitors for 1 h, followed by treatment with 0.4 mM Bt2cAMP for 24 h. DGKθ mRNA expression was assessed by qRT-PCR and normalized to β-actin content. Data graphed represent the mean ± SEM of three separate experiments (performed in triplicate). Asterisks (*) and carats (^) indicate a statistically significant difference (P < 0.05) from control group and Bt2cAMP-treated groups, respectively.
cAMP increases DGKθ reporter gene activity
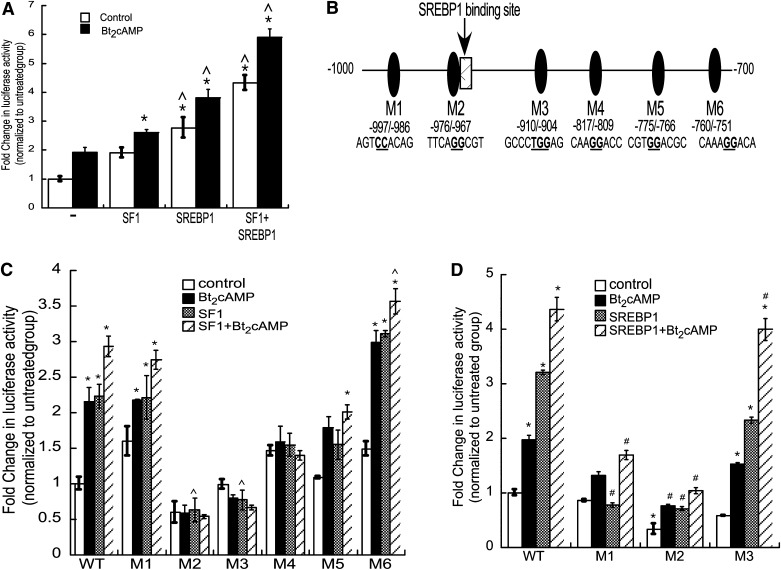
Next we sought to define the mechanism by which cAMP stimulation induces DGKθ expression and cloned 1.5 kb of the DGKθ promoter into a reporter gene plasmid fused to the Firefly luciferase gene and transfected the construct into H295R cells. As shown in Fig. 3A, Bt2cAMP treatment significantly increased the transcriptional activity of the 1.5 kb reporter gene by 1.9-fold. In silico analysis of the DGKθ promoter revealed several putative SF1 binding sites (Fig. 3B). Notably, one of these putative SF1 binding sites overlapped with response elements for the SREBP family. SREBPs are a family of basic helix-loop-helix leucine zipper (bHLHLZ) transcription factors that regulate fatty acid, triglyceride, and cholesterol metabolism (42–44). In contrast to other bHLHLZ transcription factors, SREBPs bind to both E-boxes (5′-CANNTG-3′) and sterol regulatory element (SRE) sequences (5′-TCACNCCAC-3′) (45). There are three isoforms of SREBPs in mammals: SREBP1a, SREBP1c, and SREBP2. However, because SREBP1s are more specific for activation of fatty acid synthesis and SREBP1c is the predominant isoform in murine and human tissues such as liver, adrenal gland, and brain (46), whereas SREBP2 is more selective for regulating cholesterol production (42), we focused on SREBP1c. Further, our previous work has shown that sphingosine-1-phosphate-stimulated CYP17A1 transcription requires SREBP1c (47).
Fig. 3.
SF1 and SREBP1 confer cAMP-stimulated increased DGKθ reporter gene activity. A: H295R cells were transiently transfected with pGL3-DGKθ, pCMV6-GFP-SF-1, pCDNA3.1-SREBP1c, and pRL-CMV and then treated 24 h later with Bt2cAMP (0.4 mM) for 24 h. Luciferase activity in lysates isolated from control (−) and Bt2cAMP-treated cells (+) was quantified by luminometry. Data are expressed as the fold change in pGL3-DGKθ (Firefly luciferase) reporter gene activity over the untreated control group mean, are normalized to pRL-CMV (Renilla luciferase) activity and represent the mean ± SEM of three separate experiments, each performed in triplicate. Asterisks (*) and carats (^) indicate a statistically significant difference (P < 0.05) from the pGL3-DGKθ control and Bt2cAMP-treated group, respectively. B: Depiction of −1,000 to −700 bp of the DGKθ promoter. Putative SF1/SREBP binding sites are denoted by ovals and labeled M1 to M6. C: H295R cells were transiently transfected with wild-type or mutant (M1 to M6) pGL3-DGKθ, pCMV6-GFP-SF-1, and pRL-CMV and luciferase activity quantified by luminometry. Data are expressed as the fold change in pGL3-DGKθ reporter gene activity over the untreated control group mean and represent the mean ± SEM of three separate experiments, each performed in triplicate. Asterisks (*) and carats (^) indicate a statistically significant difference (P < 0.05) from the untreated control group and untreated SF1-transfected group, respectively. D: Luciferase activity was quantified in lysates that were isolated from H295R cells that were transfected with wild-type or mutant (M1, M2, and M3) pGL3-DGKθ, pRL-CMV, and pcDNA3.1-SREBP1c expression plasmids. Changes in DGKθ promoter activity are normalized to Renilla luciferase activity and graphed as fold change over wild-type untreated control group. Asterisks (*) and hash (#) indicate a statistically significant difference (P < 0.05) from the untreated control group and the untreated SREBP1c-transfected group, respectively.
To determine the effect of SF1 and SREBP1c on DGKθ reporter gene transcription, we transfected expression plasmids for these transcription factors in H295R cells and quantified luciferase activity. Consistent with the Bt2cAMP effect, cotransfection with an SF1 expression and SREBP1c plasmids resulted in a 1.8- and 2.7-fold increase in DGKθ reporter gene activity, respectively. Moreover, overexpression of both transcription factors resulted in a 4.2-fold increase in DGKθ luciferase activity, with Bt2cAMP further stimulating DGKθ reporter gene transcription. Transfection of DGKθ reporter gene plasmids harboring mutations at putative SF1/SREBP1c sites (Fig. 3B) revealed that mutation of region M5 (−775/−776) had no significant effect on basal DGKθ promoter reporter gene activity, whereas mutation of regions M1 (997/−986) and M6 (760/−751) increased basal luciferase activity (Fig. 3C). Compared with the wild-type promoter, mutation of M2 (−976/−967), M3 (−910/−904), and M4 (−817/−809) significantly attenuated the SF1 response. As shown in Fig. 3D, mutation of regions M1 and M2 significantly reduced SREBP1c-stimulated DGKθ reporter gene activity. In contrast to the requirement of region M3 for SF1-dependent transcription, mutation of M3 had no effect on SREBP1-stimulated transcriptional activity of the DGKθ reporter gene. Further, mutation of M4, M5, and M6 were unable to reduce SREBP1-stimulated DGKθ luciferase activity. Collectively, these studies indicate that M2 (−976/−967), M3 (−910/−904), M4 (−817/−809) contribute to SF1-dependent transactivation, whereas SREBP1 requires region M2.
cAMP promotes the recruitment of SF1 and SREBP1 to the DGKθ promoter
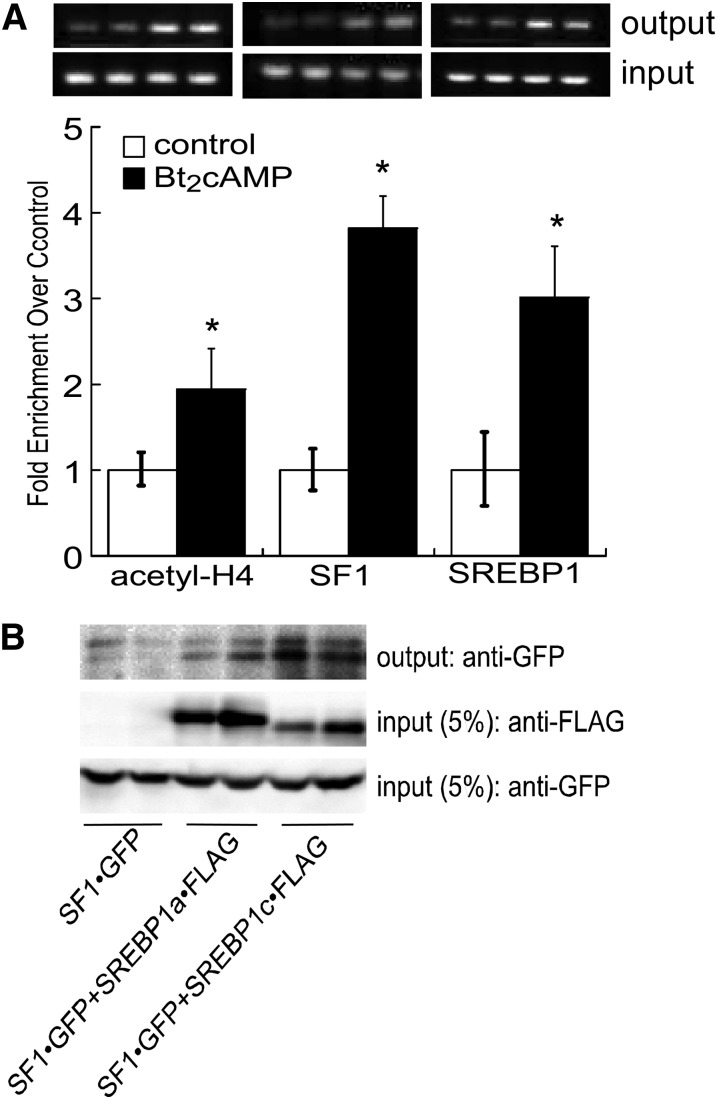
We next determined the effect of cAMP stimulation on the recruitment of SF1 and SREBP1 to the endogenous DGKθ promoter by performing ChIP assays using chromatin isolated from H295R cells that were treated with 0.4 mM Bt2cAMP for 1 h and found that Bt2cAMP increased the acetylation of histone H4 (Fig. 4A). cAMP stimulation promoted the enrichment of SF1 and SREBP1 at the DGKθ promoter by 3.8- and 3-fold, respectively. The proximity of the SF1 and SREBP1 binding sites on the DGKθ promoter (Fig. 3B) and the effect of mutating the M2 region on the ability of both SF1 and SREBP1 to increase DGKθ reporter gene activity (Fig. 3C, D) promoted us to determine if the two proteins interact. As shown in Fig. 4B, FLAG-tagged SREBP1c coimmunoprecipitates with GFP-tagged SF1.
Fig. 4.
cAMP stimulates the recruitment of SF1 and SREBP1 to the DGKθ promoter. A: H295R cells were incubated with 0.4 mM Bt2cAMP, cross-linked with formaldehyde, and the sheared chromatin immunoprecipitated with antibodies against anti-SF1, anti-acetyl histone H4, or anti-SREBP1 and recruitment to the DGKθ promoter (−1,000/−700) assessed by qPCR and normalized to the ΔCt values of input DNA. Data are expressed as fold change over untreated control and represent the mean ± SD of four separate experiments, each performed in duplicate. A representative gel of PCR reaction is shown where reactions were subjected to agarose (2%) gel electrophoresis and the EtBr-stained PCR products (top bands are output and lower bands input) imaged using a VersaDoc scanner (Bio-Rad). B: CV1 cells were transfected with expression plasmids for GFP-tagged SF1 and FLAG-tagged SREBP1a or SREBP1c and harvested 48 h after transfection. Lysates were subjected to IP using an anti-FLAG antibody and protein A/G agarose. Immobilized proteins were washed, separated by SDS-PAGE, and analyzed by Western blotting. Blots were hybridized to anti-SF1 (upper and lower panel) or anti-FLAG (5% of input lysates; middle panel) antibodies. Shown are representative blots of coimmunoprecipitation experiments that were performed on five separate occasions, each time in duplicate.
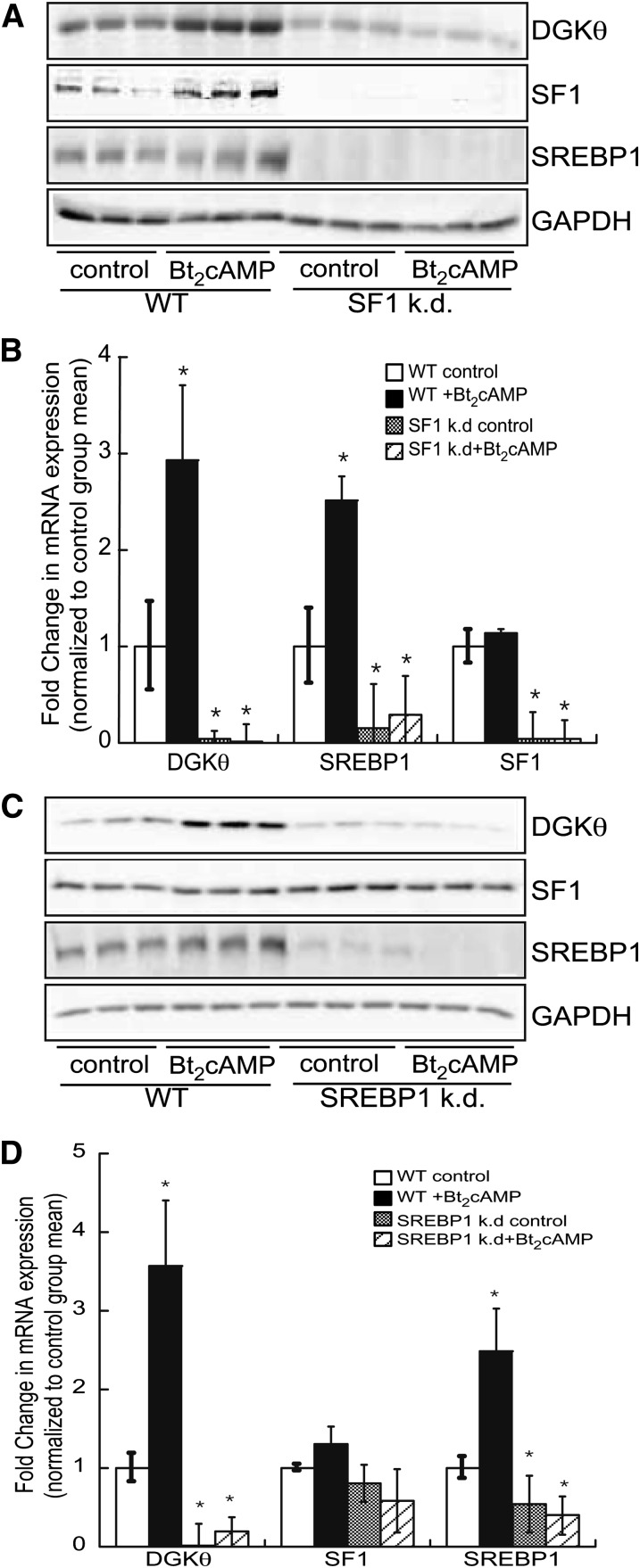
Silencing SF1 or SREBP1 represses DGKθ mRNA
Next we stably knocked down the expression of SF1 in the H295R cell line and determined the effect on DGKθ gene expression. Suppressing SF1 abolished both basal and Bt2cAMP-stimulated DGKθ protein (Fig. 5A) and mRNA (Fig. 5B) expression. Interestingly, silencing SF1 also attenuated the expression of SREBP1 (Fig. 5A, B), suggesting that the receptor regulates the expression of SREBP1 in the human adrenal cortex. Consistent with the effect of suppressing SF1 on DGKθ expression, silencing SREBP1 attenuated DGKθ protein (Fig. 5C) and mRNA (Fig. 5D), demonstrating that SREBP1 is required for both basal and cAMP-stimulated DGKθ transcription. Finally, silencing SREBP1 had no effect on the expression of SF1 (Fig. 5C).
Fig. 5.
Silencing SF1 and SREBP1 suppresses DGKθ gene expression. A: H295R wild-type and SF1 knockdown (k.d.) cells were treated with 0.4 mM Bt2cAMP for 48 h and cell lysates were harvested and analyzed by SDS-PAGE (8%), followed by Western blotting for DGKθ, SF1, SREBP1, and GAPDH. B: Real time RT-PCR was used to assess the mRNA expression of DGKθ, SREBP1, and SF1 using total RNA that was isolated from wild-type and SF1 knockdown H295R. Data are graphed as fold change in DGKθ, SREBP1, or SF1 expression mRNA expression normalized to the mRNA expression of β-actin and represent the mean ± SEM of three separate experiments, each performed in triplicate. *Statistically different from untreated control group, P < 0.05. C: Wild-type and SREBP1 knockdown cells were treated with 0.4 mM Bt2cAMP for 48 h and cell lysates were harvested and analyzed by SDS-PAGE and Western blotting for DGKθ, SF1, SREBP1, and GAPDH. D: RNA isolated from wild-type and SREBP1 knockdown H295R cells was subjected to qRT-CPR analysis. Data are graphed as fold change in DGKθ, SREBP1, or SF1 expression mRNA expression normalized to the mRNA expression of β-actin and represent the mean ± SEM of three separate experiments, each performed in triplicate. *Statistically different from untreated control group, P < 0.05.
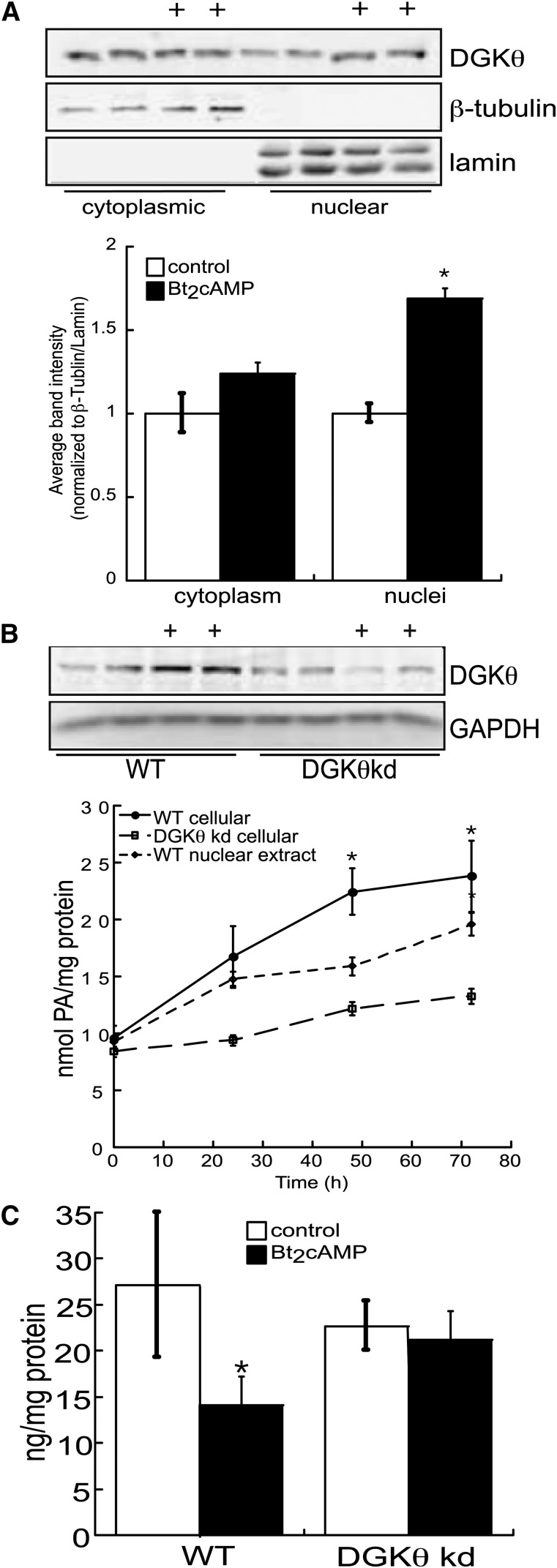
cAMP increases DGKθ nuclear expression and PA concentration
In agreement with our previous findings (25), and as shown in Fig. 6A, DGKθ is located in both the cytoplasmic and nuclear compartments of H295R cells. However, although Bt2cAMP increased the amount of DGKθ in the cytoplasm by 1.2-fold, there was a 1.8-fold increase in nuclear DGKθ protein expression (Fig. 6A). To determine if the effect of cAMP on DGKθ expression led to an increase in activity, we quantified PA concentrations in cells treated for 24–72 h with Bt2cAMP. As shown in Fig. 6B, the total PA concentration was increased by 3-fold at the 72 h time point, with a concomitant 48% decrease in the cellular amount of DAG (Fig. 6C). Consistent with the increase in total cellular PA, nuclear PA concentrations were also increased with Bt2cAMP treatment in a time-dependent manner. Finally, to assess the relative contribution of DGKθ to the cAMP-stimulated increase in PA, we quantified the concentrations of PA in wild-type and DGKθ knockdown (Fig. 6B) H295R cell lines. Knockdown of DGKθ reduced both basal and cAMP-stimulated cellular PA concentrations (Fig. 6B) and resulted in prevention of the cAMP-stimulated reduction in DAG (Fig. 6C). Taken together, our data suggested that DGKθ plays a major role in the cAMP-dependent increase in PA production.
Fig. 6.
cAMP increases PA production. A: H295R cells were grown on 10 cm dishes and treated with 0.4 mM Bt2cAMP and the cytoplasmic and nuclear fractions isolated for SDS-PAGE and Western blotting for DGKθ, β-tubulin, and lamin. Data graphed represent densitometric analysis of DGKθ cytoplasmic and nuclear expression, normalized to β-tubulin and lamin expression, respectively. B: Wild-type or DGKθ knockdown (kd) cells were treated with Bt2cAMP for 24–72 h and the cellular or nuclear lipids isolated for quantification of PA. The cellular or nuclear amount of PA was normalized to the protein concentration. Data graphed represent the mean ± SEM of three separate experiments, each performed in triplicate. *Statistically different from untreated control group, P < 0.05. Inset: Representative Western blot of controls and tetracycline treated H295R cells demonstrating decreased DGKθ protein levels. C: Wild-type or DGKθ knockdown H295R cells were grown on 6-well plates and treated with 0.4 mM Bt2cAMP for 72 h and the cellular content of DAG quantified by ELISA. The graphed data represent the mean ± SEM of three independent experiments, each performed in triplicate.
DISCUSSION
DGKs modulate the concentration of DAG and PA, key second messengers in numerous signaling pathways (48–52). Recent studies have revealed that DGKs regulate immunity, inflammation, and the nervous system (53–57), and aberrant DGK activity is implicated in the etiology of type 2 diabetes, cardiovascular disease, and cancer (49, 58, 59). We have previously identified a role for DGKθ in glucocorticoid production. By virtue of its ability to produce PA, a ligand for the nuclear receptor SF1, DGKθ regulates the transcription of multiple genes required for cortisol biosynthesis, including CYP17A1 (25). Our present studies provide a further support that phospholipid metabolism plays a key role in cAMP-dependent steroidogenesis. We demonstrate that the expression of DGKθ is induced by cAMP (Fig. 1A). Although DGKα, DGKγ, DGKδ, DGKϵ, DGKη, DGKθ, and DGKζ are expressed in H295R cells (25), the mRNA expression of these isoforms is not affected by Bt2cAMP. In agreement with our previous findings (25), our data suggest that DGKθ is the main PA source in cAMP-stimulated human adrenocortical cells (Fig. 6B). We previously demonstrated that cAMP rapidly induced nuclear DGKθ catalytic activity within 5 min (25). Herein, we showed that cAMP, in addition to an acute effect on DGK enzymatic activity and activation of the cAMP signaling pathway, also chronically increased the expression (Fig. 1) and activity (Fig. 6A) of DGKθ.
Luciferase reporter assays revealed that SF1 and SREBP1c increased the transcriptional activity of a DGKθ reporter gene (Fig. 3). The activation of DGKθ luciferase activity and the recruitment of the receptor to the endogenous DGKθ promoter (Fig. 4A) suggest that cAMP signaling may activate a feed-forward mechanism that enables the sustained activation of SF1 target genes that are required for glucocorticoid production. We envision that optimal steroid hormone production requires not only a rapid increase in nuclear PA production in response to ACTH/cAMP (25), but also a mechanism to facilitate the continued ability of SF1 to activate target gene transcription. One mechanism to achieve SF1 activation is to allow for an increase in DGKθ expression, and subsequently PA production. SF1 plays an essential role in inducing the transcription of multiple steroidogenic genes, including cytochrome CYP17A1 in the adrenal cortex and gonads. The ability of SF1 to activate target genes is regulated by mechanisms including coregulatory proteins (60–63), posttranslational modification (27, 28, 64–68), and ligand binding (25, 69–72).
SREBPs are considered as master regulators of lipid metabolism. In general, SREBP target genes include cholesterol biosynthetic (e.g., HMG-CoA synthase, LDLR receptor) and lipogenic genes (e.g., acetyl-CoA carboxylase, fatty acid synthase). However, we have also previously shown that SREBP1 is recruited to the CYP17A1 promoter in response to stimulation by sphingosine-1-phosphate (47). Our current studies demonstrate that SREBP1 is recruited to the DGKθ promoter (Fig. 4A) and is required for both basal and cAMP-stimulated DGKθ expression (Fig. 5C, D).
Interestingly, we also found that SREBP1 and SF1 interact (Fig. 4B), suggesting coordinated action between these two transcription factors. The ability of SF1 to act cooperatively with other transcription factors is well documented. SF1 synergizes with several transcription factors, including GATA transcription factors (73–76), cAMP regulatory element binding proteins (77–79), AP1 family members (80), and β-catenin (81). Significantly, the likelihood of a physical interaction between SF1 and SREBP1 is supported by studies demonstrating that both SREBP1 and SREBP2 interact with hepatic nuclear factor 4 (82) and with the liver receptor homolog (LRH)1 (83). LRH1 and SF1 belong to the NR5A subfamily of nuclear receptors and share greater than 90% conservation in the DNA binding domain and are >50% conserved in the ligand binding domain (71, 84), so it is not surprising that SF1 also interacts with SREBP1. However, despite similarities in the ability of the two NR5A family members to interact with SREBP1, the functional consequences on target gene expression differ. Whereas we show herein that SREBP1 and SF1 cooperate in the activation of DGKθ reporter gene activity (Fig. 3A), SREBPs inhibit the ability of LRH1 to activate target genes in HepG2 and Huh7 hepatoma cells by preventing the interaction of LRH1 with the coactivator PGC1α (peroxisome proliferator-activated receptor γ coactivator 1α) (83).
We also observed that silencing SF1 in the H295R cell line suppresses the expression of SREBP1 (Fig. 5C, D). Microarray analysis (K. Cai et al., unpublished observations) revealed that silencing SF1 reduced the expression of several genes in the SREBP regulatory pathway, including SREBP2, insulin induced gene 1 (INSIG1), and SREBP cleavage-activating protein, suggesting a role for the nuclear receptor in regulating cholesterol homeostasis in the adrenal cortex. These findings are inconsistent with studies performed in Huh7 human hepatoma cells demonstrating that silencing LRH1 led to an increase in the expression of SREBP target genes when the cells were cultured in cholesterol-free media (83). However, further studies are required to delineate the role of SF1 in regulating the expression of SREBP1.
Our data demonstrate that the cellular content of PA increases in response to Bt2cAMP treatment, concomitant with a decrease in DAG (Fig. 6). The time course of this increase supports a role for cAMP-stimulated DGKθ transcription in mediating PA production. However, given our previous studies demonstrating that Bt2cAMP rapidly increases nuclear PA (25), it is likely that activation of the cAMP signaling pathway acutely regulates DGKθ activity and chronically regulates DGKθ expression. Indeed, we have preliminary mass spectrometric evidence that DGKθ is phosphorylated at multiple sites (D. Li et al., unpublished observations). Published findings from other laboratories have demonstrated that phosphorylation plays a key role in regulating DGK activity, thus it is plausible that posttranslational modification also modulates DGKθ function. PKA and PKC have been shown to phosphorylate DGK in COS7 cells (85). Phosphorylation of DGKα by the tyrosine kinase Src confers hepatocyte growth factor-induced cell motility (86, 87), whereas PKC-catalyzed phosphorylation of DGKζ promotes the dissociation of the lipid kinase from PKC (88, 89). Given that DAG stimulates PKC activity, the association with DGKζ provides a mechanism to limit the ability of PKC to phosphorylate target proteins. Studies are ongoing to investigate the role of phosphorylation in regulating DGKθ function in response to activation of the cAMP signaling cascade.
Consistent with our previous studies (25), and the work of others, DGKθ is expressed in the nucleus of H295R cells. As shown in Fig. 6B, most of the increase in PA production in response to cAMP at the 24 h time point is due to an increase in nuclear PA biosynthesis, demonstrating the importance of spatially regulated phospholipid metabolism in cell signaling. Because other DGK isoforms also exhibit nuclear localization (90, 91), DGKζ for example (25, 92–95), it was important to determine the relative contribution of DGKθ to the increased PA production observed in response to Bt2cAMP. Though DGKθ was the sole isoform whose mRNA expression increased after cAMP stimulation (Fig. 1A), it is possible that other isoforms may be positively regulated by cAMP at the posttranscriptional level. H295R cells that were stably expressing a shRNA targeted against DGKθ exhibited a 50% decrease in basal and Bt2cAMP-stimulated concentrations of PA, indicating that DGKθ plays a prominent role in the capacity of adrenocortical cells to produce PA in response to cAMP signaling. However, these findings also suggest that other DGK isoforms or members of the phospholipase D family may also contribute to the increased biosynthesis of PA. In an elegant study recently reported by Mitra et al. (96), targeted disruption of the PA phosphatase lipin1 in adipocytes revealed a novel role for the transcriptional coactivator and lipid phosphatase in the regulation of cAMP/PKA signaling. These findings provide support for the role of lipid-metabolizing enzymes as key regulators, not only of lipid homeostasis, but also of signal transduction and cellular processes.
In summary, we found that the expression of DGKθ is induced by cAMP. Both SF1 and SREBP1 are required for constitutive and cAMP-stimulated DGKθ expression. Additionally, SF1 is a novel regulator of SREBP1 expression. Given the role of DGKθ in synthesizing the agonist for SF1 (25), our studies identify a feed-forward mechanism by which the capacity of adrenocortical cells to produce PA in response to cAMP is regulated by SF1.
Footnotes
Abbreviations:
- ACTH
- adrenocorticotropic hormone
- bHLHLZ
- basic helix-loop-helix leucine zipper
- Bt2cAMP
- dibutyryl cAMP
- ChIP
- chromatin immunoprecipitation
- CRD
- cysteine-rich domain
- DAG
- diacylglycerol
- DGK
- diacylglycerol kinase
- LRH
- liver receptor homolog
- PA
- phosphatidic acid
- PH
- pleckstrin homology
- PK
- protein kinase
- RIPA
- radioimmunoprecipitation
- SF1
- steroidogenic factor 1
- shRNA
- short hairpin RNA
- SREBP
- sterol regulatory element binding protein
This work is supported by the National Institutes of Health grant DK084178 (to M.B.S.).
REFERENCES
- 1.Cai J., Abramovici H., Gee S. H., Topham M. K. 2009. Diacylglycerol kinases as sources of phosphatidic acid. Biochim. Biophys. Acta. 1791: 942–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Topham M. K. 2006. Signaling roles of diacylglycerol kinases. J. Cell. Biochem. 97: 474–484 [DOI] [PubMed] [Google Scholar]
- 3.Wattenberg B. W., Pitson S. M., Raben D. M. 2006. The sphingosine and diacylglycerol kinase superfamily of signaling kinases: localization as a key to signaling function. J. Lipid Res. 47: 1128–1139 [DOI] [PubMed] [Google Scholar]
- 4.Crotty T., Cai J., Sakane F., Taketomi A., Prescott S. M., Topham M. K. 2006. Diacylglycerol kinase delta regulates protein kinase C and epidermal growth factor receptor signaling. Proc. Natl. Acad. Sci. USA. 103: 15485–15490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez de Turco E. B., Tang W., Topham M. K., Sakane F., Marcheselli V. L., Chen C., Taketomi A., Prescott S. M., Bazan N. G. 2001. Diacylglyerol kinse epsilon regulates seizure susceptibility and long-term potentiation through arachidonoyl-inositol lipid signaling. Proc. Natl. Acad. Sci. USA. 98: 4740–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukiw W. J., Cui J. G., Musto A. E., Musto B. C., Bazan N. G. 2005. Epileptogenesis in diacylglycerol kinase epsilon deficiency up-regulates COX-2 and tyrosine hydroxylase in hippocampus. Biochem. Biophys. Res. Commun. 338: 77–81 [DOI] [PubMed] [Google Scholar]
- 7.Olenchock B. A., Guo R., Carpenter J. H., Jordan M., Topham M. K., Koretzky G. A., Zhong X. P. 2006. Disruption of diacylglycerol metabolism impairs the induction of T cell anergy. Nat. Immunol. 7: 1174–1181 [DOI] [PubMed] [Google Scholar]
- 8.Zhong X. P., Hainey E. A., Olenchock B. A., Jordan M. S., Maltzman J. S., Nichols K. E., Shen H., Koretzky G. A. 2003. Enhanced T cell responses due to diacylglycerol kinase zeta deficiency. Nat. Immunol. 4: 882–890 [DOI] [PubMed] [Google Scholar]
- 9.Regier D. S., Higbee J., Lund K. M., Sakane F., Prescott S. M., Topham M. K. 2005. Diacylglycerol kinase iota regulates Ras guanyl-releasing protein 3 and inhibits Rap1 signaling. Proc. Natl. Acad. Sci. USA. 102: 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Los A. P., Vinke F. P., de Widt J., Topham M. K., van Blitterswijk W. J., Divecha N. 2006. The retinoblastoma family proteins bind to and activate diacylglycerol kinase zeta. J. Biol. Chem. 281: 858–866 [DOI] [PubMed] [Google Scholar]
- 11.Tabellini G., Billi A. M., Fala F., Cappellini A., Evagelisti C., Manzoli L., Cocco L., Martelli A. M. 2004. Nuclear diacylglycerol kinase-theta is activated in response to nerve growth factor stimulation of PC12 cells. Cell. Signal. 16: 1263–1271 [DOI] [PubMed] [Google Scholar]
- 12.Tu-Sekine B., Ostroski M., Raben D. M. 2007. Modulation of diacylglycerol kinase theta activity by alpha-thrombin and phospholipids. Biochemistry. 46: 924–932 [DOI] [PubMed] [Google Scholar]
- 13.Bregoli L., Baldassare J. J., Raben D. M. 2001. Nuclear diacylglycerol kinase-theta is activated in response to alpha-thrombin. J. Biol. Chem. 276: 23288–23295 [DOI] [PubMed] [Google Scholar]
- 14.Evangelisti C., Riccio M., Faenza I., Zini N., Hozumi Y., Goto K., Cocco L., Martelli A. M. 2006. Subnuclear localization and differentiation-dependent increased expression of DGK-zeta in C2C12 mouse myoblasts. J. Cell. Physiol. 209: 370–378 [DOI] [PubMed] [Google Scholar]
- 15.Matsubara T., Shirai Y., Miyasaka K., Murakami T., Yamaguchi Y., Ueyama T., Kai M., Sakane F., Kanoh H., Hashimoto T., et al. 2006. Nuclear transportation of diacylglycerol kinase gamma and its possible function in the nucleus. J. Biol. Chem. 281: 6152–6164 [DOI] [PubMed] [Google Scholar]
- 16.Mérida I., Avila-Flores A., Merino E. 2008. Diacylglycerol kinases: at the hub of cell signalling. Biochem. J. 409: 1–18 [DOI] [PubMed] [Google Scholar]
- 17.Sakane F., Imai S., Kai M., Yasuda S., Kanoh H. 2007. Diacylglycerol kinases: why so many of them? Biochim. Biophys. Acta. 1771: 793–806 [DOI] [PubMed] [Google Scholar]
- 18.Kanoh H., Yamada K., Sakane F. 2002. Diacylglycerol kinases: emerging downstream regulators in cell signaling systems. J. Biochem. 131: 629–633 [DOI] [PubMed] [Google Scholar]
- 19.van Blitterswijk W. J., Houssa B. 2000. Properties and functions of diacylglycerol kinases. Cell. Signal. 12: 595–605 [DOI] [PubMed] [Google Scholar]
- 20.Topham M. K., Prescott S. M. 1999. Mammalian diacylglycerol kinases, a family of lipid kinases with signaling functions. J. Biol. Chem. 274: 11447–11450 [DOI] [PubMed] [Google Scholar]
- 21.Sakane F., Kanoh H. 1997. Molecules in focus: diacylglycerol kinase. Int. J. Biochem. Cell Biol. 29: 1139–1143 [DOI] [PubMed] [Google Scholar]
- 22.Houssa B., Schaap D., van der Wal J., Goto K., Kondo H., Yamakawa A., Shibata M., Takenawa T., van Blitterswijk W. J. 1997. Cloning of a novel human diacylglycerol kinase (DGKtheta) containing three cysteine-rich domains, a proline-rich region, and a pleckstrin homology domain with an overlapping Ras-associating domain. J. Biol. Chem. 272: 10422–10428 [DOI] [PubMed] [Google Scholar]
- 23.Los A. P., van Baal J., de Widt J., Divecha N., van Blitterswijk W. J. 2004. Structure-activity relationship of diacylglycerol kinase theta. Biochim. Biophys. Acta. 1636: 169–174 [DOI] [PubMed] [Google Scholar]
- 24.van Baal J., de Widt J., Divecha N., van Blitterswijk W. J. 2005. Translocation of diacylglycerol kinase theta from cytosol to plasma membrane in response to activation of G protein-coupled receptors and protein kinase C. J. Biol. Chem. 280: 9870–9878 [DOI] [PubMed] [Google Scholar]
- 25.Li D., Urs A. N., Allegood J., Leon A., Merrill A. H., Jr, Sewer M. B. 2007. Cyclic AMP-stimulated interaction between steroidogenic factor 1 and diacylglycerol kinase theta facilitates induction of CYP17. Mol. Cell. Biol. 27: 6669–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabellini G., Bortul R., Santi S., Riccio M., Baldini G., Cappellini A., Billi A. M., Berezney R., Ruggeri A., Cocco L., et al. 2003. Diacylglycerol kinase-theta is localized in the speckle domains of the nucleus. Exp. Cell Res. 287: 143–154 [DOI] [PubMed] [Google Scholar]
- 27.Chen W. Y., Lee W. C., Hsu N. S., Huang F., Chung B. C. 2004. SUMO modification of repression domains modulates function of the nuclear receptor 5A1 (steroidogenic factor-1). J. Biol. Chem. 279: 38730–38735 [DOI] [PubMed] [Google Scholar]
- 28.Chen W. Y., Juan L. J., Chung B. C. 2005. SF-1 (nuclear receptor 5A1) activity is activated by cyclic AMP via p300-mediated recruitment to active foci, acetylation, and increased DNA binding. Mol. Cell. Biol. 25: 10442–10453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo X., Ikeda Y., Parker K. L. 1994. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 77: 481–490 [DOI] [PubMed] [Google Scholar]
- 30.Parker K. L., Rice D. A., Lala D. S., Ikeda Y., Luo X., Wong M., Bakke M., Zhao L., Frigeri C., Hanley N. A., et al. 2002. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog. Horm. Res. 57: 19–36 [DOI] [PubMed] [Google Scholar]
- 31.Schimmer B. P., White P. C. 2010. Minireview: steroidogenic factor 1: its roles in differentiation, development, and disease. Mol. Endocrinol. 24: 1322–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dammer E. B., Leon A., Sewer M. B. 2007. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3′,5′-monophosphate-dependent cytochrome P450c17 transcription rate. Mol. Endocrinol. 21: 415–438 [DOI] [PubMed] [Google Scholar]
- 33.Sewer M. B., Nguyen V., Huang C-J., Tucker P. W., Kagawa N., Waterman M. R. 2002. Transcriptional activation of human CYP17 in H295R adrenocortical cells depends on complex formation between p54nrb/NonO, PSF and SF-1, a complex which also participates in repression of transcription. Endocrinology. 143: 1280–1290 [DOI] [PubMed] [Google Scholar]
- 34.McMullan R., Hiley E., Morrison P., Nurrish S. J. 2006. Rho is a presynaptic activator of neurotransmitter release at pre-existing synapses in C. elegans. Genes Dev. 20: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staels B., Hum D. W., Miller W. L. 1993. Regulation of steroidogenesis in NCI-H295 cells: a cellular model of the human fetal adrenal. Mol. Endocrinol. 7: 423–433 [DOI] [PubMed] [Google Scholar]
- 36.Rainey W. E., Bird I. M., Mason J. I. 1994. The NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Mol. Cell. Endocrinol. 100: 45–50 [DOI] [PubMed] [Google Scholar]
- 37.Lucki N. C., Bandyopadhyay S., Wang E., Merrill A. H., Sewer M. B. 2012. Acid ceramidase (ASAH1) is a global regulator of steroidogenic capacity and adrenocortical gene expression. Mol. Endocrinol. 26: 228–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toth J. I., Datta S., Athanikar J. N., Freedman L. P., Osborne T. F. 2004. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol. Cell. Biol. 24: 8288–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucki N., Sewer M. B. 2009. The cAMP-responsive element binding protein (CREB) regulates the expression of acid ceramidase (ASAH1) in H295R human adrenocortical cells. Biochim. Biophys. Acta. 1791: 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucki N. C., Li D., Bandyopadhyay S., Wang E., Merrill A. H., Sewer M. B. 2012. Acid ceramidase (ASAH1) represses steroidogenic factor 1-dependent gene transcription in H295R human adrenocortical cells by binding to the receptor. Mol. Cell Biol. 32: 4419–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de la Iglesia N., Konopka G., Puram S. V., Chan J. A., Bachoo R. M., You M. J., Levy D. E., Depinho R. A., Bonni A. 2008. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 22: 449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raghow R., Yellaturu C., Deng X., Park E. A., Elam M. B. 2008. SREBPs: the crossroads of physiological and pathophysiological lipid homeostasis. Trends Endocrinol. Metab. 19: 65–73 [DOI] [PubMed] [Google Scholar]
- 44.Shimano H. 2009. SREBPs: physiology and pathophysiology of the SREBP family. FEBS J. 276: 616–621 [DOI] [PubMed] [Google Scholar]
- 45.Kim J. B., Spotts G. D., Halvorsen Y. D., Shih H. M., Ellenberger T., Towle H. C., Spiegelman B. M. 1995. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol. Cell. Biol. 15: 2582–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimano H., Shimomura I., Hammer R. E., Herz J., Goldstein J. L., Brown M. S., Horton J. D. 1997. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J. Clin. Invest. 100: 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozbay T., Rowan A., Leon A., Patel P., Sewer M. B. 2006. Cyclic adenosine 5′-monophosphate-dependent sphingosine-1-phosphate biosynthesis induces human CYP17 gene transcription by activating cleavage of sterol regulatory element binding protein 1. Endocrinology. 147: 1427–1437 [DOI] [PubMed] [Google Scholar]
- 48.Brose N., Betz A., Wegmeyer H. 2004. Divergent and convergent signaling by the diacylglycerol second messenger pathway in mammals. Curr. Opin. Neurobiol. 14: 328–340 [DOI] [PubMed] [Google Scholar]
- 49.Griner E. M., Kazanietz M. G. 2007. Protein kinase C and other diacylglycerol effectors in cancer. Nat. Rev. Cancer. 7: 281–294 [DOI] [PubMed] [Google Scholar]
- 50.Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 294: 1942–1945 [DOI] [PubMed] [Google Scholar]
- 51.Baillie G. S., Huston E., Scotland G., Hodgkin M., Gall I., Peden A. H., MacKenzie C., Houslay E. S., Currie R., Pettitt T. R., et al. 2002. TAPAS-1, a novel microdomain within the unique N-terminal region of the PDE4A1 cAMP-specific phosphodiesterase that allows rapid, Ca2+-triggered membrane association with selectivity for interaction with phosphatidic acid. J. Biol. Chem. 277: 28298–28309 [DOI] [PubMed] [Google Scholar]
- 52.Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 304: 1644–1647 [DOI] [PubMed] [Google Scholar]
- 53.Sanjuán M. A., Pradet-Balade B., Jones D. R., Martinez A. C., Stone J. C., Garcia-Sanz J. A., Mérida I. 2003. T cell activation in vivo targets diacylglycerol kinase alpha to the membrane: a novel mechanism for Ras attenuation. J. Immunol. 170: 2877–2883 [DOI] [PubMed] [Google Scholar]
- 54.Liu C. H., Machado F. S., Guo R., Nichols K. E., Burks A. W., Aliberti J. C., Zhong X. P. 2007. Diacylglycerol kinase zeta regulates microbial recognition and host resistance to Toxoplasma gondii. J. Exp. Med. 204: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams J. M., Pettitt T. R., Powell W., Grove J., Savage C. O., Wakelam M. J. 2007. Antineutrophil cytoplasm antibody-stimulated neutrophil adhesion depends on diacylglycerol kinase-catalyzed phosphatidic acid formation. J. Am. Soc. Nephrol. 18: 1112–1120 [DOI] [PubMed] [Google Scholar]
- 56.Goto K., Kondo H. 1999. Diacylglycerol kinase in the central nervous system–molecular heterogeneity and gene expression. Chem. Phys. Lipids. 98: 109–117 [DOI] [PubMed] [Google Scholar]
- 57.Clarke C. J., Ohanian V., Ohanian J. 2007. Norepinephrine and endothelin activate diacylglycerol kinases in caveolae/rafts of rat mesenteric arteries: agonist-specific role of PI3-kinase. Am. J. Physiol. Heart Circ. Physiol. 292: H2248–H2256 [DOI] [PubMed] [Google Scholar]
- 58.Verrier E., Wang L., Wadham C., Albanese N., Hahn C., Gamble J. R., Chatterjee V. K., Vadas M. A., Xia P. 2004. PPARgamma agonists ameliorate endothelial cell activation via inhibition of diacylglycerol-protein kinase C signaling pathway: role of diacylglycerol kinase. Circ. Res. 94: 1515–1522 [DOI] [PubMed] [Google Scholar]
- 59.Takahashi H., Takeishi Y., Seidler T., Arimoto T., Akiyama H., Hozumi Y., Koyama Y., Shishido T., Tsunoda Y., Niizeki T., et al. 2005. Adenovirus-mediated overexpression of diacylglycerol kinase-zeta inhibits endothelin-1-induced cardiomyocyte hypertrophy. Circulation. 111: 1510–1516 [DOI] [PubMed] [Google Scholar]
- 60.Li L-A., Chiang E. F-L., Chen J-C., Hsu N-C., Chen Y-J., Chung B. 1999. Function of steroidogenic factor 1 domains in nuclear localization, transactivation, and interaction with transcription factor TFIIB and c-Jun. Mol. Endocrinol. 13: 1588–1598 [DOI] [PubMed] [Google Scholar]
- 61.Li L-A., Lala D. S., Chung B. 1998. Function of steroidogenic factor 1 (SF1) ligand-binding domain in gene activation and interaction with AP1. Biochem. Biophys. Res. Commun. 250: 318–320 [DOI] [PubMed] [Google Scholar]
- 62.Monté D., DeWitte F., Hum D. W. 1998. Regulation of the human P450scc gene by steroidogenic factor 1 is mediated by CBP/p300. J. Biol. Chem. 273: 4585–4591 [DOI] [PubMed] [Google Scholar]
- 63.Ou Q., Mouillet J-F., Yan X., Dorn C., Crawford P. A., Sadovsky Y. 2001. The DEAD box protein DP103 is a regulator of steroidogenic factor-1. Mol. Endocrinol. 15: 69–79 [DOI] [PubMed] [Google Scholar]
- 64.Hammer G. D., Krylova I., Zhang Y., Darimont B. D., Simpson K., Weigel N. L., Ingraham H. A. 1999. Phosphorylation of the nuclear receptor SF-1 modulates cofactor recruitment: integration of hormone signaling in reproduction and stress. Mol. Cell. 3: 521–526 [DOI] [PubMed] [Google Scholar]
- 65.Jacob A. L., Lund J., Martinez P., Hedin L. 2001. Acetylation of steroidogenic factor 1 protein regulates its transcriptional activity and recruits the coactivator GCN5. J. Biol. Chem. 276: 37659–37664 [DOI] [PubMed] [Google Scholar]
- 66.Komatsu T., Mizusaki H., Mukai T., Ogawa H., Baba D., Shirakawa M., Hatakeyama S., Nakayama K. I., Yamamoto H., Kikuchi A., et al. 2004. Small ubiquitin-like modifier 1 (SUMO-1) modification of the synergy control motif of Ad4 binding protein/steroidogenic factor 1 (Ad4BP/SF-1) regulates synergistic transcription between Ad4BP/SF-1 and Sox9. Mol. Endocrinol. 18: 2451–2462 [DOI] [PubMed] [Google Scholar]
- 67.Lee M. B., Lebedeva L. A., Suzawa M., Wadekar S. A., Desclozeaux M., Ingraham H. A. 2005. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 25: 1879–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis A. E., Rusten M., Hoivik E. A., Vikse E. L., Hansson M. L., Wallberg A. E., Bakke M. 2008. Phosphorylation of steroidogenic factor 1 is mediated by cyclin-dependent kinase 7. Mol. Endocrinol. 22: 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ingraham H. A., Redinbo M. R. 2005. Orphan nuclear receptors adopted by crystallography. Curr. Opin. Struct. Biol. 15: 708–715 [DOI] [PubMed] [Google Scholar]
- 70.Krylova I. N., Sablin E. P., Moore J., Xu R. X., Waitt G. M., MacKay J. A., Juzumiene D., Bynum J. M., Madauss K., Montana V., et al. 2005. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 120: 343–355 [DOI] [PubMed] [Google Scholar]
- 71.Li Y., Choi M., Cavey G., Daugherty J., Suino K., Kovach A., Bingham N. C., Kliewer S. A., Xu H. E. 2005. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell. 17: 491–502 [DOI] [PubMed] [Google Scholar]
- 72.Urs A. N., Dammer E., Sewer M. B. 2006. Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology. 147: 5249–5258 [DOI] [PubMed] [Google Scholar]
- 73.Tremblay J. J., Viger R. S. 1999. Transcription factor GATA-4 enhances Müllerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol. Endocrinol. 13: 1388–1401 [DOI] [PubMed] [Google Scholar]
- 74.Tremblay J. J., Viger R. S. 2001. Nuclear receptor Dax-1 represses the transcriptional cooperation between GATA-4 and SF-1 in Sertoli cells. Biol. Reprod. 64: 1191–1199 [DOI] [PubMed] [Google Scholar]
- 75.Lo A., Zheng W., Gong Y., Crochet J. R., Halvorson L. M. 2011. GATA transcription factors regulate LHbeta gene expression. J. Mol. Endocrinol. 47: 45–58 [DOI] [PubMed] [Google Scholar]
- 76.Jimenez P., Saner K., Mayhew B., Rainey W. E. 2003. GATA-6 is expressed in the human adrenal and regulates transcription of genes required for adrenal androgen biosynthesis. Endocrinology. 144: 4285–4288 [DOI] [PubMed] [Google Scholar]
- 77.Ito M., Park Y., Weck J., Mayo K. E., Jameson J. L. 2000. Synergistic activation of the inhibin alpha-promoter by steroidogenic factor-1 and cyclic adenosine 3′,5′-monophosphate. Mol. Endocrinol. 14: 66–81 [DOI] [PubMed] [Google Scholar]
- 78.Carlone D. L., Richards J. S. 1997. Evidence that functional interactions of CREB and SF-1 mediate hormone regulated expression of the aromatase gene in granulosa cells and constitutive expression in R2C cells. J. Steroid Biochem. Mol. Biol. 61: 223–231 [PubMed] [Google Scholar]
- 79.Zheng W., Jefcoate C. R. 2005. Steroidogenic factor-1 interacts with cAMP response element-binding protein to mediate cAMP stimulation of CYP1B1 via a far upstream enhancer. Mol. Pharmacol. 67: 499–512 [DOI] [PubMed] [Google Scholar]
- 80.Martin L. J., Tremblay J. J. 2009. The nuclear receptors NUR77 and SF1 play additive roles with c-JUN through distinct elements on the mouse Star promoter. J. Mol. Endocrinol. 42: 119–129 [DOI] [PubMed] [Google Scholar]
- 81.Gummow B. M., Winnay J. N., Hammer G. D. 2003. Convergence of Wnt signaling and steroidogenic factor-1 (SF-1) on transcription of the rat inhibin alpha gene. J. Biol. Chem. 278: 26572–26579 [DOI] [PubMed] [Google Scholar]
- 82.Misawa K., Horiba T., Arimura N., Hirano Y., Inoue J., Emoto N., Shimano H., Shimizu M., Sato R. 2003. Sterol regulatory element-binding protein-2 interacts with hepatocyte nuclear factor-4 to enhance sterol isomerase gene expression in hepatocytes. J. Biol. Chem. 278: 36176–36182 [DOI] [PubMed] [Google Scholar]
- 83.Kanayama T., Arito M., So K., Hachimura S., Inoue J., Sato R. 2007. Interaction between sterol regulatory element-binding proteins and liver receptor homolog-1 reciprocally suppresses their transcriptional activities. J. Biol. Chem. 282: 10290–10298 [DOI] [PubMed] [Google Scholar]
- 84.Sablin E. P., Krylova I. N., Fletterick R. J., Ingraham H. A. 2003. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol. Cell. 11: 1575–1585 [DOI] [PubMed] [Google Scholar]
- 85.Schaap D., van der Wal J., van Blitterswijk W. J., van der Bend R. L., Ploegh H. L. 1993. Diacylglycerol kinase is phosphorylated in vivo upon stimulation of the epidermal growth factor receptor and serine/threonine kinases, including protein kinase C-epsilon. Biochem. J. 289: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baldanzi G., Cutrupi S., Chianale F., Gnocci V., Rainero E., Porporato P., Filigheddu N., van Blitterswijk W. J., Parolini O., Bussolino F., et al. 2008. Diacylglycerol kinase-alpha phosphorylation by Src on Y335 is required for activation, membrane recruitment and Hgf-induced cell motility. Oncogene. 27: 942–956 [DOI] [PubMed] [Google Scholar]
- 87.Cutrupi S., Baldanzi G., Gramaglia D., Maffe A., Schaap D., Giraudo E., van Blitterswijk W. J., Bussolino F., Comoglio P. M., Graziani A. 2000. Src-mediated activation of -diacylglycerol kinase is required for hepatocyte growth factor-induced cell motility. EMBO J. 19: 4614–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo B., Prescott S. M., Topham M. K. 2003. Protein kinase C alpha phosphorylates and negatively regulates diacylglycerol kinase zeta. J. Biol. Chem. 278: 39542–39547 [DOI] [PubMed] [Google Scholar]
- 89.Luo B., Prescott S. M., Topham M. K. 2003. Association of diacylglycerol zeta with protein kinase C alpha: spatial regulation of diacylglycerol signaling. J. Cell Biol. 160: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Evangelisti C., Bortul R., Fala F., Tabellini G., Goto K., Martelli A. M. 2007. Nuclear diacylglycerol kinases: emerging downstream regulators in cell signaling networks. Histol. Histopathol. 22: 573–579 [DOI] [PubMed] [Google Scholar]
- 91.Raben D. M., Tu-Sekine B. 2008. Nuclear diacylglycerol kinases: regulation and roles. Front. Biosci. 13: 590–597 [DOI] [PubMed] [Google Scholar]
- 92.Evangelisti C., Bortul R., Tabellini G., Papa V., Cocco L., Martelli A. M. 2006. Nuclear expression of diacylglycerol kinases: possible involvement in DNA replication. Eur. J. Histochem. 50: 9–13 [PubMed] [Google Scholar]
- 93.Evangelisti C., Tazzari P. L., Riccio M., Fiume R., Hozumi Y., Falà F., Goto K., Manzoli L., Cocco L., Martelli A. M. 2007. Nuclear diacylglycerol kinase-zeta is a negative regulator of cell cycle progression in C2C12 mouse myoblasts. FASEB J. 21: 3297–3307 [DOI] [PubMed] [Google Scholar]
- 94.Hasegawa H., Nakano T., Hozumi Y., Takagi M., Ogino T., Okada M., Iseki K., Kondo H., Watanabe M., Martelli A. M., et al. 2008. Diacylglycerol kinase zeta is associated with chromatin, but dissociates from condensed chromatin during mitotic phase in NIH3T3 cells. J. Cell. Biochem. 105: 756–765 [DOI] [PubMed] [Google Scholar]
- 95.Topham M. K., Bunting M., Zimmerman G. A., McIntyre T. M., Blackshear P. J., Prescott S. M. 1998. Protein kinase C regulates the nuclear localization of diacylglycerol kinase-zeta. Nature. 394: 697–700 [DOI] [PubMed] [Google Scholar]
- 96.Mitra M. S., Chen Z., Ren H., Harris T. E., Chambers K. T., Hall A. M., Nadra K., Klein S., Chrast R., Su X., et al. 2013. Mice with an adipocyte-specific lipin 1 separation-of-function allele reveal unexpected roles for phosphatidic acid in metabolic regulation. Proc. Natl. Acad. Sci. USA. 110: 642–647 [DOI] [PMC free article] [PubMed] [Google Scholar]