Abstract
Low density lipoprotein (LDL) cholesterol is taken up into cells via clathrin-mediated endocytosis of the LDL receptor (LDLR). Following dissociation of the LDLR-LDL complex, LDL is directed to lysosomes whereas the LDLR recycles to the plasma membrane. Activation of the sterol-sensing nuclear receptors liver X receptors (LXRs) enhances degradation of the LDLR. This depends on the LXR target gene inducible degrader of the LDLR (IDOL), an E3-ubiquitin ligase that promotes ubiquitylation and lysosomal degradation of the LDLR. How ubiquitylation of the LDLR by IDOL controls its endocytic trafficking is currently unknown. Using genetic- and pharmacological-based approaches coupled to functional assessment of LDL uptake, we show that the LXR-IDOL axis targets a LDLR pool present in lipid rafts. IDOL-dependent internalization of the LDLR is independent of clathrin, caveolin, macroautophagy, and dynamin. Rather, it depends on the endocytic protein epsin. Consistent with LDLR ubiquitylation acting as a sorting signal, degradation of the receptor can be blocked by perturbing the endosomal sorting complex required for transport (ESCRT) or by USP8, a deubiquitylase implicated in sorting ubiquitylated cargo to multivesicular bodies. In summary, we provide evidence for the existence of an LXR-IDOL-mediated internalization pathway for the LDLR that is distinct from that used for lipoprotein uptake.
Keywords: liver X receptor, low density lipoprotein receptor, inducible degrader of low density lipoprotein receptor, E3-ubiquitin ligase, endocytosis, lipoprotein receptors, epsins
The endocytosis of LDL via the LDL receptor (LDLR) has served as a paradigm for receptor-mediated endocytosis since its initial description by the Brown and Goldstein laboratory (1). At the plasma membrane, LDLR binds extracellular LDL and is endocytosed by clathrin-dependent mechanisms in clathrin-coated pits. Acidification of early endocytic vesicles liberates LDL from the receptor and allows the cargo to be delivered to lysosomes where the lipoprotein particle is degraded and cholesterol is salvaged for cellular use. In contrast, the LDLR recycles back to the plasma membrane and can be reused in a new round of endocytosis (2). Clathrin is not sufficient to support LDLR internalization. Efficient internalization of the LDLR requires adaptor proteins that tether the receptor to the endocytic machinery. The best-described adaptor proteins are the autosomal recessive hypercholesterolemia (ARH) and disabled 2 (DAB2) (3, 4). These adaptors interact with AP2 and clathrin and in parallel via their phospho-tyrosine binding domain with the NPxY endocytosis motif present in the intracellular tail of the LDLR (5–7). Michaely et al. (8) recently reported that a HIC sequence-motif present downstream of the canonical NPxY motif can also be used for clathrin-dependent uptake of β-VLDL, thus representing a second clathrin-dependent route for LDLR internalization. The clathrin-mediated endocytic strategy is evolutionarily conserved, and is used by different receptors employing distinct adaptor proteins (9). This route can be used as a means to control turnover of membrane proteins, uptake of extracellular cargo as is the case for the LDLR and the transferrin receptor (10), or alternatively to regulate downstream signaling as is exemplified by the epidermal growth factor receptor (EGFR) (11). Other clathrin-independent entry routes, such as caveolin-dependent pathways, also exist and are defined by the use of alternative adaptor proteins, dynamin, and membrane sub-domains (12). Together, these pathways control proper spatial and temporal signaling and sorting of receptors and cargo (13). Ubiquitylation forms an added layer of regulation in these processes and is involved in controlling signaling, endocytosis, and targeting of receptors toward lysosomal degradation (14).
Endocytosis of LDL via the “classical” LDLR pathway is particularly important when cells are faced with declining intracellular sterol levels. Under this condition, cholesterol synthesis and LDL-cholesterol uptake via the LDLR pathway are increased (15). Elegantly, the sterol regulatory element binding protein (SREBP) transcription factors regulate both processes by increasing expression of the complete set of enzymes required to produce cholesterol as well as expression of the LDLR (16). The LDLR pathway has important clinical implications. For example, LDLR mutants that display defective endocytosis, e.g., the Y828C mutation that prevents binding of the LDLR to ARH, manifest clinically as hypercholesterolemia due to decreased hepatic clearance of LDL (17). Similarly, homozygous carriers of loss-of-function ARH alleles suffer from a severe form of recessive hypercholesterolemia (3). The existence of several endocytosis/recycling-defective LDLR mutants emphasizes the importance of mapping the cellular trafficking of the LDLR (18).
When faced with increasing cellular sterol levels, cells turn down the LDLR pathway by several mechanisms. Increased cholesterol content in the ER membrane prevents maturation of the ER-resident precursor SREBPs to their nuclear active form resulting in decreased LDLR mRNA (15). Additionally, elevated levels of cellular cholesterol lead to activation of the sterol-sensing nuclear receptors, liver X receptor (LXR)α and LXRβ (19, 20), and transcriptional induction of their target gene, inducible degrader of the LDL receptor (IDOL) (21). We have previously shown that the E3-ubiquitin ligase IDOL binds to the LDLR, promotes its poly-K63-linked ubiquitylation, and subsequently leads to lysosomal degradation of the receptor (21, 22). Thus, the LXR-IDOL-LDLR axis offers cells an efficient mechanism, which circumvents the SREBP transcriptional pathway and the long half-life of the LDLR, to limit cholesterol uptake (23). However, how ubiquitylation of the LDLR by IDOL is coupled to endocytosis and lysosomal degradation of the receptor is not well understood. We show here that IDOL-stimulated internalization of the LDLR defines a clathrin-independent route that is distinct from the one used for LDL uptake via the LDLR pathway.
MATERIALS AND METHODS
Reagents
Bafilomycin A1, GW3965, methyl-β-cyclodextrin (MβCD), and filipin were from Sigma. Dynasore was purchased from Ascent and simvastatin salt from Calbiochem. All other reagents were purchased from Sigma.
Cell culture and transfections
HEK293T, HeLa, and HepG2 cells were obtained from the American Type Culture Collection. Cells were maintained in DMEM supplemented with 10% FBS at 37°C and 5% CO2. A431 cells were a kind gift from Drs. Vilja Pietiainen and Elina Ikonen (University of Helsinki). Atg5(−/−) and corresponding wild-type (WT) cells were a gift from Dr. Eric Reits (University of Amsterdam). Cav1(−/−) and corresponding WT cells were from Dr. Miguel Angel del Pozo (Spanish National Cardiovascular Research Centre) (24). Human fibroblasts (with no detectable LDLR) and their derivatives in which WT LDLR or Y828C (JD) LDLR were stably introduced were a kind gift from Dr. Peter Michaely (UTSW) (25). Doxycycline-inducible clathrin heavy chain (CHC) knockdown HeLa cells were a generous gift from Pier Paolo Di Fiore. Six days of doxycycline treatment (0.5 μg/ml) were required to achieve a >90% decrease of CHC levels and this condition was used in the experiments shown in Fig. 2. Where indicated, cells were depleted of sterols by culture in sterol-depletion medium (DMEM supplemented by 10% LPDS, 5 μg/ml simvastatin, and 100 μM mevalonate) and with 1 μM GW3965 to induce LXR signaling. HEK293T, HeLa, A431, and HepG2 cells were transfected with the indicated amounts of plasmids using the JetPrime reagent. Transfection efficiency was monitored by cotransfecting an expression plasmid for GFP and was consistently >85% in HEK293T and HeLa cells.
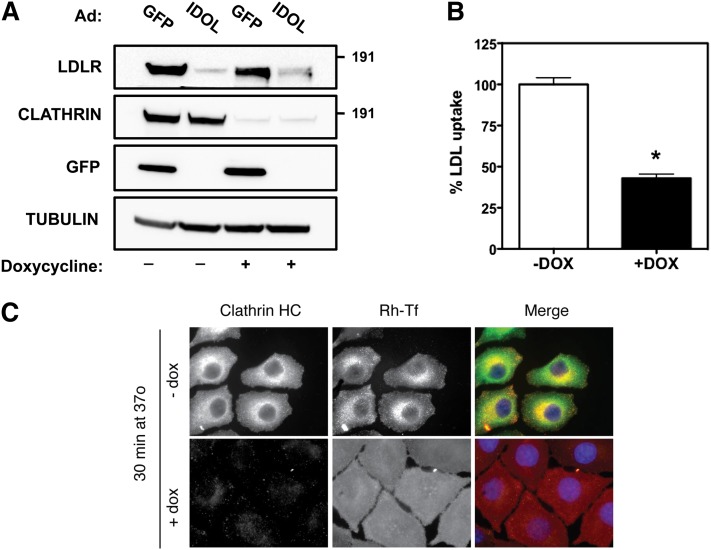
Fig. 2.
IDOL-mediated degradation of the LDLR is clathrin independent. A: Inducible CHC knockdown HeLa cells were cultured in the presence (+) or absence (−) of doxycycline for 6 days to induce silencing of the CHC. Subsequently, cells were infected with a GFP- or IDOL-expressing adenovirus for 10 h at an MOI of 5. Total cell lysates were immunoblotted as indicated. B: Inducible CHC knockdown cells were cultured as in (A). Subsequently, cells were shifted to sterol-depletion medium for 16 h and then incubated for 30 min with 5 μg/ml DyLight 488-labeled LDL. After extensive washing, internalized LDL was quantified by measuring fluorescence in total cell lysates. LDL uptake in −Dox cells was set to 100%. Each bar and error represent the average ± SD (n = 8; *P < 0.01; DOX/dox, doxycycline). C: Inducible CHC knockdown cells were cultured as in (A) and then treated with rhodamine-transferrin (red) for 30 min. After fixation, cells were stained for clathrin (green), and counterstained with DAPI (blue). Note that in clathrin knockdown cells transferrin internalization is blocked, while in control cells transferrin is internalized and colocalized with clathrin. Immunoblots are representative of three independent experiments.
Plasmids and expression constructs
Expression plasmids for IDOL, LDLR, and GFP were previously reported (21). WT and mutant (E228Q) human VPS4A were a kind gift of Dr. Sylvie Urbè (University of Liverpool) (26). WT and mutant rat Epsin1 expression plasmids were a gift from Dr. Inger Madshus (University of Oslo) (27). All plasmids used in transfection experiments were isolated by CsCl2 gradient centrifugation. DNA sequencing was used to verify the correctness of all the constructs used in this study.
Antibodies and immunoblot analysis
Cell lysates were prepared in RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.1% sodium deoxycholate, 0.1% SDS, 100 mM Tris-HCl, pH 7.4) supplemented with protease inhibitors. Lysates were cleared by centrifugation at 4°C for 10 min at 10,000 g. Protein concentration was determined using the Bradford assay with BSA as reference. Samples (10–40 ug) were separated on NuPAGE BisTris gels and transferred to nitrocellulose. Membranes were probed with the following antibodies: LDLR (Epitomics, clone EP1553Y, 1:4,000), tubulin (Sigma, clone DM1A, ascites fluid, 1:5,000), ABCA1 (Novus Biologicals, NB400-105, 1:1,000), FLAG-HRP (Sigma, clone M2, 1:1,000), GFP (affinity purified rabbit polyclonal anti-GFP was a gift from Dr. Mireille Riedinger, University of California Los Angeles, 1:5,000), Myc (Cell Signaling, clone 9B11, 1:4,000), CHC (BD Transduction Laboratories, clone 23, 1:1,000), HA (Covance, clone 16B12, ascites fluid, 1:6,000), Flotillin1 (BD Transduction Laboratories, clone 18, 1:1,000), Caveolin1 (BD Transduction Laboratories, polyclonal, 1:1,000), ATG5 (Novus Biologicals, NB110-53818, 1:500), TSG101 (GeneTex, clone 4A10, 1:500), dynamin (Santa Cruz, clone D5, 1:500), ubiquitin (ENZO, clone FK2), Epsin1 (rabbit polyclonal serum raised against amino acids 249–401 of rat Epsin1). Secondary HRP-conjugated antibodies (Zymed Laboratories Inc.) were used and visualized with chemiluminescence on a Fuji LAS4000 (GE Healthcare). Unless otherwise indicated, all immunoblots shown are representative of at least three independent experiments with similar results. Where indicated, numbers represent the average normalized intensity of the LDLR band from these experiments.
LDL uptake assays
DyLight 488-labeled LDL was produced as previously described (28). Briefly, Hepg2, HeLa, or A431 cells were incubated in sterol-depletion medium for 16 h prior to adding LDL. To measure LDL uptake, cells were washed twice with PBS and incubated with 5 μg/ml DyLight488-labeled LDL in DMEM supplemented with 0.5% BSA for the indicated time at 37°C. Subsequently, cells were washed twice with PBS supplemented with 0.5% BSA and lysed in RIPA buffer. LDL uptake was determined by quantification of the fluorescence signal on a Typhoon imager and presented as mean ± SD.
Measurement of cell-surface LDLR
Cells were cultured as indicated above for LDL uptake assays and subsequently in the presence or absence of 2 μM of the synthetic LXR agonist GW3965 for 4 h and 100 nM bafilomycin A1. Cells were dissociated with TrypLE Express (Invitrogen) and incubated in FACS blocking buffer (PBS supplemented with 2 mM EDTA, 0.5% BSA, 2% goat serum) for 30 min on ice. Subsequently, 100,000 cells were stained in 50 μl FACS buffer with a PE-conjugated mouse anti-human LDLR antibody (R and D Systems, catalog #FAB2148P) for 1 h on ice. Cells were then washed three times with FACS buffer and directly analyzed on a FACSCalibur. Viable cells were gated and 10,000 events per condition acquired. Data were analyzed using the CellQuest software package (BD Biosciences).
Immunofluorescence and internalization studies
Inducible CHC knockdown HeLa cells were either induced or not induced with doxycycline treatment (0.5 μg/ml) for six days, split on coverslips, and then serum-starved for 4 h in binding buffer (DMEM, 0.1% BSA, 20 mM HEPES). After starvation, cells were incubated in the presence of 50 μg/ml of rhodamine-Tf (Molecular Probes) for 30 min, followed by fixation (4% paraformaldehyde) and treatment with anti-clathrin antibody (X22, Affinity BioReagents), Alexa 488-conjugated secondary (Molecular Probes), and DAPI staining.
siRNA interference experiments
To silence ARH, TSG101, DNM1, and DNM2 (Dynamin1 and Dynamin2) we used On-Target smart pools (Dharmacon). A431 and HeLa cells were transfected with control and specific siRNA pool (final concentration 20 nM) using the JetPrime reagent (PolyPlus). HepG2 cells were similarly transfected, but using the RNAiMAX reagent (Invitrogen). Silencing of target genes was assessed by quantitative PCR or Western blotting. Unless otherwise indicated, siRNAs were transfected and analyzed 48 h later.
RNA isolation and quantitative PCR
Total RNA was isolated from cells using TRIzol (Invitrogen). One microgram of total RNA was reverse-transcribed with random hexamers using iScript reverse transcription reagents kit (Bio-Rad). SYBR Green real-time quantitative PCR assays were performed on a Lightcycler 480 II apparatus (Roche Applied Science) and are shown as mean ± SE. Primer sequences are available upon request.
Membrane fractionation
Cellular membranes were isolated and fractionated following the methodology of Song et al. (29). Briefly, two confluent 150 mm plates of cells were washed with ice-cold buffered saline and scraped into 1.5 ml of 500 mM sodium carbonate (pH 11.0) containing protease inhibitors. Cells were homogenized by 15 passages through a 23 gauge needle and sonicated three times for 20 s (60% intensity) in a MSE Soniprep 150. The homogenate was mixed with an equal volume of 90% sucrose prepared in MES-buffered saline (25 mM MES, pH 6.5, 0.15 M NaCl) and placed at the bottom of an ultracentrifuge tube. A 5% to 35% sucrose discontinuous gradient was formed above (4 ml of 5% sucrose/4 ml of 35% sucrose; both in MES-buffered saline with 250 mM sodium carbonate). Gradients were centrifuged for 18 h at 39,000 rpm at 4°C in a SW41Ti rotor. Ten fractions were collected from the top of each gradient. For SDS-PAGE analysis an equal amount of protein was loaded from each fraction.
Statistical analysis
Data were analyzed using the Prism software package and differences in surface LDLR and LDL uptake were determined with one-way ANOVA. A probability value of P < 0.05 was considered statistically significant.
RESULTS
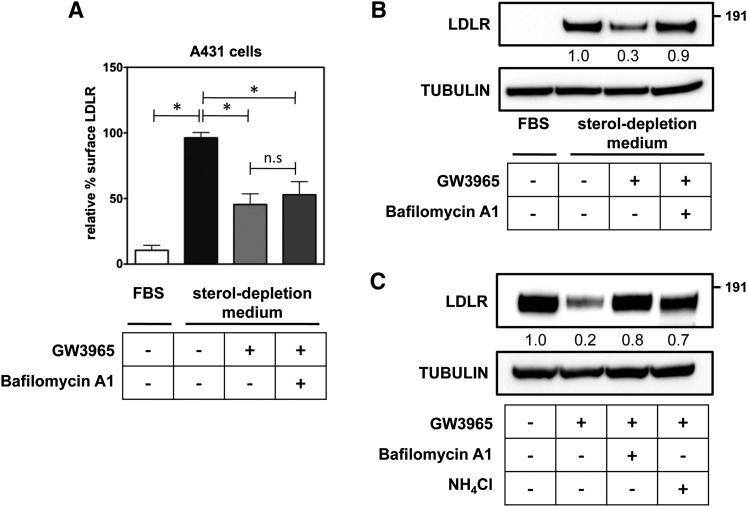
The LDLR is a recycling receptor that constitutively traffics between the plasma membrane and endosomal compartments (2). Therefore, at least two dynamic LDLR pools, on the cell surface and in intracellular recycling vesicles, can be defined. We have previously reported that in HepG2 cells the LDLR is able to recruit IDOL to the plasma membrane, suggesting that IDOL targets the plasma membrane-resident LDLR (22). To address this experimentally we followed surface levels of LDLR by FACS analysis in the epidermoid carcinoma cell line A431 and in HeLa cells. The former has particularly high levels of endogenous LDLR and is very suitable for mechanistic studies. In both cell lines, shifting the cells from a FBS-containing medium to sterol-depletion medium increases levels of endogenous LDLR protein, which is accompanied by a marked increase of LDLR at the cell surface (Fig. 1A, B, supplementary Fig. IA). In line with our previous report (21), treating the cells for a short (4 h) period with a synthetic LXR ligand to induce IDOL expression reduces the level of total LDLR. This also leads to a sharp decrease in the levels of LDLR in the plasma membrane (Fig. 1A). Consistent with lysosomal degradation of the LDLR (19, 30), cotreating the cells with the lysosomotropic agents, bafilomycin A1, or ammonium chloride blocks LXR-stimulated degradation of the LDLR (Fig. 1C, supplementary Fig. IB). However, in contrast to its effect on total cellular levels of the LDLR, bafilomycin A1 is unable to rescue the level of the receptor on the cell surface (Fig. 1A, B, supplementary Fig. I). Therefore, our observations suggest that by promoting LDLR ubiquitylation, the LXR-IDOL pathway targets the plasma membrane LDLR for lysosomal degradation and impedes receptor recycling.
Fig. 1.
The LXR-IDOL axis targets the LDLR membrane pool for lysosomal degradation. A: A431 cells were cultured as indicated in the presence (+) or absence (−) of 2 μM of the synthetic LXR agonist GW3965 for 4 h and 100 nM bafilomycin A1 or 10 mM NH4Cl. LDLR at the cell surface was determined by FACS analysis. Cell surface LDLR in sterol-depletion medium was set to 100% (n = 6; *P < 0.01; n.s, not significant). B, C: Total cell lysates were immunoblotted as indicated. Immunoblots are representative of three independent experiments.
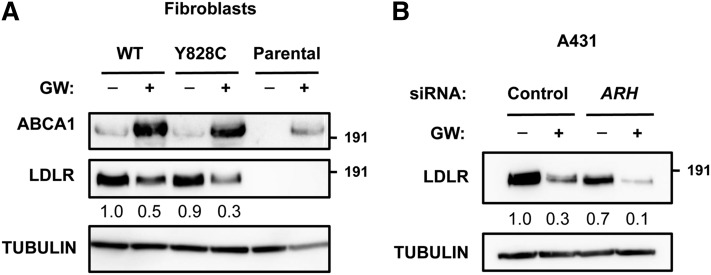
It is well established that endocytosis of LDL via the LDLR pathway is a clathrin-dependent process (2). Clathrin is required for forming coated pits that later pinch from the plasma membrane as coated vesicles that carry the LDLR-LDL complex. As IDOL activity limits LDL uptake into cells (21), we hypothesized that efficient targeting of the LDLR by IDOL will also proceed via the same clathrin-dependent route. Silencing of the CHC limits entry of several well-studied cargos, including transferrin, epidermal growth factor, and LDL (4, 10, 11). We therefore took advantage of a stable and inducible HeLa cell line in which we obtained >90% reduction in the level of CHC protein upon doxycycline treatment (Fig. 2A). In line with the essential role of clathrin for cargo entry, uptake of both LDL (Fig. 2B) and transferrin (Fig. 2C) were severely diminished in CHC-silenced cells. However, even in the face of this impairment IDOL was still able to promote degradation of the LDLR pointing toward a clathrin-independent process (Fig. 2A). In hepatocytes and lymphocytes tethering of the LDLR to the clathrin machinery requires the adaptor ARH (3, 31). These cells lack functional DAB2 and accordingly, mutations in the LDLR that prevent association with ARH (e.g., Y828C) lead to impaired hepatic clearance of LDL and hypercholesterolemia (3). To test whether IDOL requires ARH for LDLR internalization, we used fibroblasts previously reported by Michaely et al. (25). These cells have very low basal levels of LDLR protein and LDL uptake. Reconstitution of WT LDLR in these cells promotes cellular uptake of LDL, which remains severely impaired when LDLRY828C is introduced instead [supplementary Fig. IIA and (25)]. This also indicates that DAB2, an alternative LDLR adaptor protein found in fibroblasts (32, 33), is unable to compensate for impaired ARH binding in these cells. Activation of LXRs led to an increased protein level of the canonical LXR target ABCA1 (Fig. 3A), and to degradation of both WT and LDLRY828C by IDOL; the latter despite absence of functional ARH binding. Similarly, silencing ARH with siRNAs in A431 cells, which have low amounts of DAB2 [(34) and not shown], resulted in a 90 ± 4% decrease in ARH mRNA levels (n = 3, P < 0.01) and LDL uptake (57 ± 8% of control, n = 3, P < 0.01), yet did not prevent degradation of the LDLR by activated LXRs (Fig. 3B). Similar results were obtained in HepG2 (supplementary Fig. IIB), although unlike A431 cells, LDL uptake remained unchanged following effective knockdown of ARH, likely due to moderate levels of DAB2 in these cells (4). It is important to point out that ubiquitylation of the LDLR is not critically required for LDL uptake; a LDLR mutant lacking the IDOL-sensitive ubiquitylation sites displays only a minimal reduction in endocytosis of LDL as compared with WT LDLR (73 ± 5%, n = 3, P < 0.05). Collectively, this series of experiments indicates that degradation of the LDLR by the LXR-IDOL axis does not require clathrin-mediated internalization and the adaptor protein ARH.
Fig. 3.
IDOL-mediated degradation of the LDLR does not require ARH. A: Human fibroblasts (parental) and their engineered derivatives in which WT LDLR or Y828C LDLR were stably introduced and cultured in the absence (−) or presence (+) of 1 μM GW3965 for 24 h. Total cell lysates were immunoblotted as indicated. B: A431 cells were transfected with 20 nM control or ARH siRNA. Subsequently, cells were shifted to sterol-depletion medium for 16 h and then treated for an additional 6 h with 2 μM GW3965. Total cell lysates were immunoblotted as indicated. Immunoblots are representative of three independent experiments.
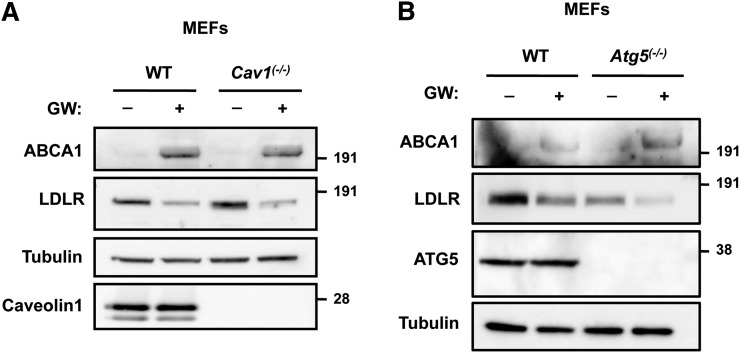
Having ruled out clathrin and ARH involvement in IDOL-mediated degradation of the LDLR, we considered the potential involvement of alternative entry portals. First, we tested whether IDOL uses caveolin-mediated endocytosis to promote LDLR internalization by studying Cav1(−/−) and corresponding WT fibroblasts. Loss of Cav1 in these cells eliminates morphological caveolae (24). Nevertheless, LXR activation increased ABCA1 and promoted LDLR degradation in these cells (Fig. 4A). Similarly, IDOL-mediated degradation of the LDLR did not require macroautophagy-related pathways, a general clearance mechanism for bulk cargo degradation (35), as demonstrated by normal LDLR degradation in Atg5(−/−) fibroblasts (Fig. 4B).
Fig. 4.
Degradation of the LDLR by IDOL is independent of caveolin and macroautophagy. Cav1(−/−) (A) and Atg5(−/−) (B) mouse embryonic fibroblasts and their corresponding WT cells were incubated for 16 h in sterol-deficient medium with vehicle (−) or 1 μM GW3965 (+). Total cell lysates were immunoblotted as shown. Immunoblots are representative of three independent experiments.
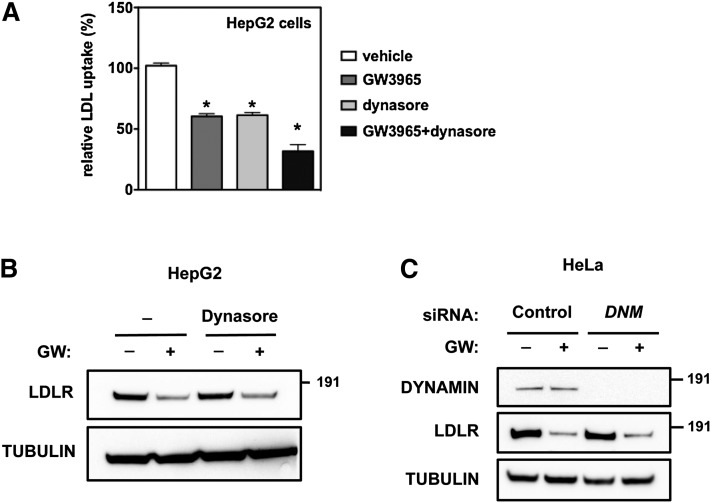
Several additional entry routes, distinct from the ones studied above, have been recently reported (12). While their molecular detail is still not well understood, many require activity of dynamin GTPases for pinching of the membrane invagination from the plasma membrane. The chemical dynasore is an inhibitor of dynamin-dependent processes, and consistent with the requirement of dynamin for clathrin-mediated processes, dynasore substantially inhibited LDL uptake in HepG2, A431, and HeLa cells (Fig. 5A, supplementary Fig. IIIB, D). However, dynasore did not prevent degradation of the LDLR by LXR activation (Fig. 5B, supplementary Fig. IIIA, C). Additionally, silencing the two major dynamins, DNM1 and DNM2, in HeLa cells did not prevent degradation of the LDLR by IDOL (Fig. 5C). Collectively, our genetic and pharmacological experiments indicate that the LXR-IDOL axis defines an uncharacterized pathway for LDLR internalization that is clathrin-, caveolin-, and dynamin-independent.
Fig. 5.
IDOL-mediated LDLR degradation is dynamin independent. A, B: HepG2 cells were cultured for 16 h in sterol-depletion medium to stimulate LDLR expression. Subsequently, cells were treated with 1 μM GW3965 or vehicle in the presence (+) or absence (−) of 80 μM dynasore for 4 h. A: Cells were incubated for 30 min with 5 μg/ml DyLight 488-labeled LDL. LDL uptake in vehicle-treated cells was set to 100% (n = 4, *P < 0.01 from vehicle treated cells). B: Total cell lysates were immunoblotted as indicated. C: HeLa cells were transfected with 20 nM control or DNM1/2 siRNA and subsequently incubated for 16 h with sterol-depletion medium to induce LDLR expression. Cells were then treated for 6 h with vehicle or 1 μM GW3965. Immunoblots are representative of three independent experiments.
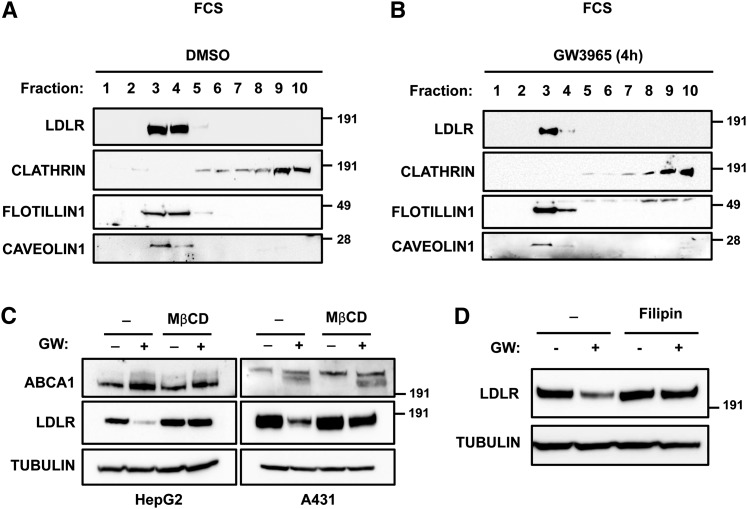
In order to characterize the environmental requirements for IDOL's activity toward the LDLR, we employed a sucrose gradient-based cell fractionation method to isolate cell membrane fractions from HepG2 cells. We collected membrane fractions from HepG2 cells cultured in FBS-containing or sterol-depletion medium. Under both culture conditions the majority of LDLR in HepG2 cells was detected in low-density membrane fractions (Fig. 6A, supplementary Fig. IV). These fractions, which were enriched in Flotillin1 and Caveolin1 and were largely devoid of clathrin, are generally considered to represent lipid rafts. Notably, LXR activation and subsequent IDOL activity led to a marked decrease in the lipid raft-associated LDLR, without changing the distribution of the receptor along the gradient (Fig. 6B, supplementary Fig. IV). We point out that this distribution differs from that previously reported for the LDLR in fibroblasts, in which ∼25% of the LDLR could be localized to coated-pit structures by electron microscopy (8). The discrepancy between HepG2 cells and fibroblasts in this respect may reflect intrinsic cell-type-specific differences or the different methodologies used.
Fig. 6.
The LXR-IDOL degradation pathway targets a lipid-raft resident LDLR pool. HepG2 cells were cultured in FBS-containing medium and treated with vehicle (A) or 1 μM GW3965 for 4 h (B). Membrane fractions were isolated and an equal amount of protein per fraction was separated by SDS-PAGE and immunoblotted as indicated. Immunoblots are representative of two independent experiments with similar results. C: HepG2 and A431 cells were incubated for 16 h with sterol-depletion medium and subsequently pretreated with 1 μM GW3965 (GW; +) or vehicle (−) for 30 min followed by addition of 5 mg/ml MβCD for an additional 4 h. D: HepG2 cells were cultured as in (C) except that 10 μg/ml filipin was added for an additional 3 h. Total cell lysates were immunoblotted as indicated.
Lipid rafts are specialized plasma membrane microdomains that form ordered but dynamic structures within the membrane environment and have an important role in signaling, endocytosis, and ubiquitin-mediated receptor degradation (36). Therefore, we decided to investigate whether the integrity of lipid rafts is required for IDOL-mediated degradation of the LDLR. We treated HepG2 and A431 cells with MβCD, a cholesterol-depleting agent able to perturb lipid rafts and induce delocalization of associated proteins (36). This treatment largely abrogated LDLR degradation upon LXR activation in both cell lines (Fig. 6C). However, consistent with an earlier report (37) MβCD also moderately inhibited LDL uptake by 27 ± 5% (n = 3, P < 0.05), indicating that under our experimental conditions clathrin-dependent internalization of the LDLR was partially inhibited. We therefore also tested the effect of filipin, a specific lipid-raft disrupting agent that acts by sequestering cholesterol, and found that in HepG2 cells it blocked degradation of the LDLR by IDOL (Fig. 6D). Collectively, these results support the idea that IDOL targets a LDLR pool located in lipid rafts.
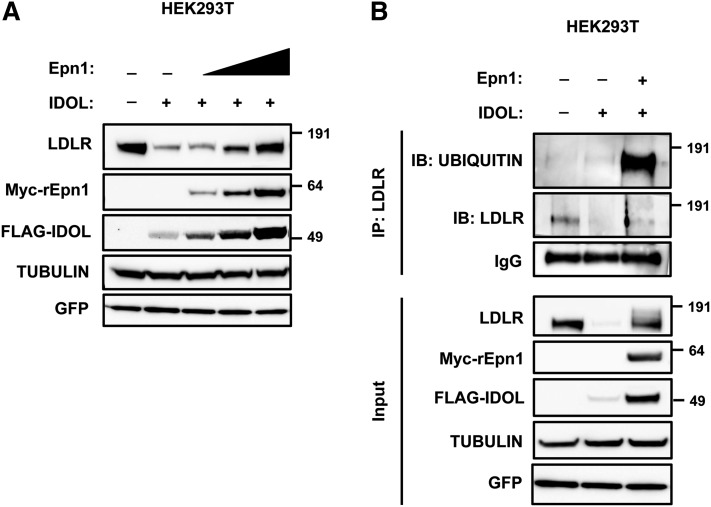
How ubiquitylated cargo is internalized and sorted from lipid rafts is not fully elucidated. Recent work implicates epsins or related ubiquitin-binding proteins as important adaptors for internalization of ubiquitylated cargo that are used in different entry routes, including those initiating at lipid rafts (38–40). Consistent with its use by multiple endocytic pathways Epsin1 was distributed along the membrane gradient in HepG2 cells (supplementary. Fig. IVA, B). Because Epsin1 was also detected in lipid rafts, we investigated the possibility that Epsin1 might be part of the IDOL-LDLR degradation network. Initially, we tested the consequence of Epsin1 silencing on IDOL-dependent degradation of the LDLR. Despite achieving effective silencing of Epsin1 in HeLa and A431 cells, IDOL was still able to promote degradation of the LDLR (not shown), most likely pointing to functional redundancy with other related ubiquitin adaptors like Eps15 and Eps15R, as previously reported for the EGFR (38). Indeed, our preliminary experiments show that IDOL is unable to promote LDLR degradation in stable EPS15/EPS15R knockdown HeLa cells [(38) and not shown]. As an alternative approach, we used over-expression of Epsin1, which is reported to act in a dominant negative fashion and inhibit pan-epsin-dependent processes (41). Coexpression of Epsin1 together with LDLR and IDOL inhibited degradation of the receptor in a dose-dependent manner (Fig. 7A), which paradoxically, was also accompanied by stabilization of IDOL protein. Similarly, Epsin1 over-expression also blocked degradation of another IDOL target, the VLDL receptor (VLDLR) (supplementary Fig. V) (30). Inhibition of LDLR and VLDLR degradation by IDOL was also accompanied by accumulation of lower mobility receptors, clearly visible with the VLDLR and requiring longer exposures with LDLR, which we identified as ubiquitylated receptors (Fig. 7B, supplementary Fig. V). Thus, our results indicate that epsin-dependent processes act downstream of IDOL-mediated ubiquitylation of the LDLR to sort the receptor toward lysosomal degradation.
Fig. 7.
Epsin1 over-expression blocks IDOL-mediated degradation of the LDLR and leads to accumulation of ubiquitylated receptor. A: HEK293T cells were transfected with expression plasmids for LDLR, FLAG-IDOL, increasing amounts of Myc-rEpn1, and GFP to monitor transfection efficiency. Total cell lysates were immunoblotted as indicated. B: HEK293T cells were transfected with expression plasmids for LDLR, FLAG-IDOL, and Myc-rEpn1. Samples were immunoprecipitated and analyzed by immunoblotting as indicated. Immunoblots are representative of three independent experiments.
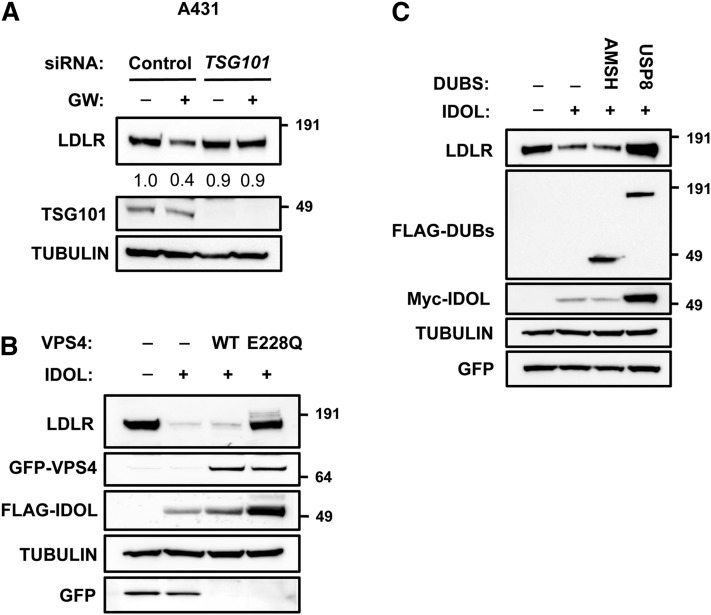
Following its ubiquitylation by IDOL and ubiquitin-dependent internalization, the LDLR needs to be sorted toward maturing multi-vesicular bodies (MVBs). The ESCRT system is implicated in these sorting processes, yet whether it acts on the LDLR is unknown (42). To determine the role of the ESCRT system in IDOL-mediated degradation of the LDLR, we used several approaches. Silencing of TSG101 in A431 cells attenuated LDLR degradation upon LXR activation without affecting LDLR and IDOL mRNA levels (Fig. 8A and not shown), in line with the critical role of TSG101 in sorting ubiquitinated cargo in endosomes. Another hallmark of ESCRT-dependent processes is their dependence on VPS4 activity. VPS4 is an ATPase required for disassembling the ESCRT machinery and promoting MVB maturation and subsequent fusion with lysosomes (26, 42). In line with its important role in ESCRT function, dominant negative mutants of VPS4 hamper lysosomal degradation of ubiquitylated cargo (43, 44). Accordingly, expression of a mutant VPS4E228Q form in HEK293T cells abolished degradation of the LDLR by IDOL (Fig. 8B), while WT VPS4 had no effect. Furthermore, because sorting into MVBs ultimately requires removal of the conjugated ubiquitin chain from the cargo protein, we tested the involvement of the ESCRT-associated deubiquitylases (DUBs) AMSH and USP8 (known also as UBPY) in LDLR degradation (45). Coexpression of AMSH was unable to prevent LDLR degradation by IDOL. However, in this setting USP8 was a potent inhibitor of IDOL-dependent degradation of the LDLR, further supporting the role of the ESCRT system in LDLR degradation (Fig. 8C). Overall, our experiments propose that IDOL-dependent degradation of the LDLR emanates from lipid rafts and requires ubiquitylation and subsequent initial sorting by epsins that is followed by ESCRT-dependent sorting into MVBs. This cellular itinerary is clathrin-independent and distinct from that used for LDL internalization.
Fig. 8.
The ESCRT system is required for sorting of ubiquitylated LDLR. A: A431 cells were transfected with 20 nM of control or TSG101 siRNA and subsequently incubated for 16 h with sterol-depletion medium followed by treatment with vehicle (−) or 1 μM GW3965 (+) for 5 h. B: HEK293T cells were transfected with expression plasmids for LDLR, FLAG-IDOL, GFP-VPS4WT, GFP-VPS4E228Q, or GFP. C: HEK293T cells were transfected with expression plasmids for LDLR, Myc-IDOL, FLAG-AMSH, FLAG-USP8, and GFP to monitor for transfection efficiency. In (A–C) total cell lysates were analyzed by immunoblotting as indicated. Immunoblots are representative of three independent experiments.
DISCUSSION
The LDLR pathway for lipoprotein uptake has, since its discovery, served as a paradigm for receptor-mediated endocytosis (1). A key feature of this pathway is that receptor internalization involves clathrin-associated adaptor proteins that couple the LDLR to the clathrin machinery (2). In this respect, the main finding of our study is that IDOL-mediated internalization and degradation of the LDLR is clathrin-independent, thus distinguishing it from the route that is used for lipoprotein uptake via the LDLR pathway. In fact, this pathway is not only clathrin-independent, but does not require caveolin or dynamin activity either. Instead, we propose that IDOL-mediated internalization of the LDLR requires epsins and subsequent sorting by the ESCRT system. Thus, our study establishes an intracellular trafficking sequence for the LDLR that emanates from lipid rafts at the plasma membrane and culminates in sorting to MVBs and receptor degradation.
Our results illuminate two important facets of IDOL-mediated degradation of the LDLR: 1) targeting of lipid raft LDLR and the involvement of epsins in the initial steps of internalization, and 2) involvement of the ESCRT system. Lipid rafts are dynamic microdomains consisting of cholesterol and sphingolipids in the plasma membrane (36). Partitioning of signaling molecules into these rafts establishes them as central nodes of signal transduction and receptor trafficking. Signaling and internalization of receptors largely depends on recruitment of specific adaptor proteins, including ubiquitin-ligases and epsins (13, 14, 46). Moreover, disrupting lipid raft integrity, e.g., with MβCD or filipin, perturbs cell-signaling and protein-interaction networks (36). Our experiments demonstrate that in HepG2 cells the bulk of the LDLR is located in lipid rafts and that this pool is particularly sensitive to IDOL-mediated degradation. Thus, we propose that lipid rafts can serve as a platform for recruiting IDOL for LDLR downregulation. Unfortunately, current IDOL antibodies, both ones we developed and those available commercially, are unable to detect endogenous levels of IDOL protein, preventing us from critically testing this. Receptor ubiquitylation must be followed by internalization and sorting of the receptor and we provide here evidence for the involvement of an epsin-dependent event in this process. Epsins are a class of ubiquitin adaptors that are implicated in internalization and sorting of plasma membrane receptors (39, 40, 47), and are implicated in tuning, among other, EGFR (27, 38), Notch (48), and VEFGR2 signaling (49). Over-expression of Epsin1, which is known to act in a dominant fashion and pan-inhibit epsin-dependent processes (41), effectively blocked IDOL-mediated degradation of the LDLR and the VLDLR. This was also accompanied by accumulation of ubiquitylated receptor, positioning epsins downstream of receptor ubiquitylation by IDOL. Because epsins control cargo internalization at the plasma membrane, this result further strengthens our conclusion that IDOL targets a plasma membrane-resident LDLR pool. The role of Epsin1 in mammalian cells is complex and seems to be cargo specific. For example, while silencing of EPN1 (EPSIN1) in monkey BSC-1 cells prevents entry of the influenza virus via clathrin-dependent endocytosis, internalization of LDL or transferrin in these cells via the same pathway remains unchanged (50). Similarly, knockdown of EPN1 in HeLa cells did not block internalization of LDL (4), supporting the idea that Epsin1 is not required for LDL or LDLR internalization. In contrast to these studies, Kang et al. (51) recently reported that in HeLa cells silencing Epsin1 attenuates internalization of LDL and of an over-expressed CD8extracellular-LDLRintracellular chimeric protein. The reason for this discrepancy is unknown. Our study was not specifically geared toward addressing the role of Epsin1 in LDL uptake, yet our results demonstrate that effective silencing of EPN1 in both HeLa and A431 cells does not abrogate the ubiquitylation-coupled internalization of the LDLR by IDOL. Our preliminary results rather suggest that EPS15 and EPS15R are involved in this process and studies to elucidate the critical role of these ubiquitin-adaptors as downstream effectors of ubiquitylated LDLR sorting are ongoing.
Our study also illustrates the role of the ESCRT system in sorting ubiquitylated LDLR toward MVBs. Functional inhibition of the ESCRT system at the level of VPS4, TSG101, or USP8 attenuated IDOL-stimulated degradation of the LDLR. This finding is consistent with our earlier report that IDOL stimulates poly-K63 ubiquitylation of the LDLR, a known lysosomal targeting tag for membrane receptors (22). The involvement of the DUB USP8 is particularly intriguing. USP8 can break down both K48- and K63-linked ubiquitin chains and can remove attached ubiquitin from cargo as well as modify the ubiquitylation state of ESCRT components (45). Likely, USP8 acts on ubiquitylated LDLR directly and shunts it away from degradation, as has been shown in the case of the EGFR (52). Intriguingly, over-expressed USP8 also stabilizes transfected IDOL, similar to what we observe by over-expression of Epsin1 or mutant VPS4E228Q, which have no deubiquitylase activity. A common feature of these perturbations is their ability to inhibit degradation of the LDLR by IDOL. A simple explanation for IDOL stabilization could be that IDOL is codegraded with the LDLR in lysosomes, and blocking the latter results in accumulation of IDOL protein. We think that this is unlikely, as we previously reported that whereas the LDLR is degraded in the lysosome, IDOL is subject to proteasomal breakdown (21). Furthermore, the lysosomotropic agent bafilomycin A1, which attenuates IDOL-mediated degradation of the LDLR, does not lead to parallel stabilization of IDOL (not shown). Functional coupling of target ubiquitylation and degradation to that of the cognate E3-ubiquitin ligase is an emerging concept in the ubiquitin field (53). Therefore, stabilization of IDOL in these scenarios may reflect part of its mechanism of action and we speculate that degradation of IDOL is coupled to degradation of the LDLR, by a mechanism that remains to be defined.
IDOL and PCSK9 represent the major posttranscriptional pathways that promote LDLR degradation, and as previously suggested these pathways represent distinct routes for LDLR degradation (23, 54). Our current study further emphasizes this point. In contrast to IDOL-stimulated degradation of the LDLR, the PCSK9 route does not require ESCRT-mediated sorting and is a clathrin-dependent process that capitalizes on the same cellular machinery used for LDL endocytosis (55). This raises the possibility that combining IDOL and PCSK9 inhibitors may act additively in a LDL-lowering regimen. While completing our study, Scotti et al. (56) reported findings similar to those reported here. Both studies, using different methodologies, demonstrate that IDOL-stimulated internalization of the LDLR is clathrin-, caveolae-, and dynamin-independent, and identify the ESCRT system and associated DUB USP8 as important determinants for sorting ubiquitylated LDLR to MVBs. A discrepancy between our studies is the possible involvement of lipid rafts. Unlike Scotti et al. (56) who observed no effect of filipin (1 μg/ml), we found that perturbing lipid raft integrity with filipin (10 μg/ml) or MβCD attenuated degradation of the LDLR by IDOL, and that IDOL preferentially targeted LDLR in membrane fractions typically associated with lipid rafts. Additionally, our study implicates epsins or related ubiquitin-interacting proteins in the initial steps of IDOL-mediated internalization of ubiquitylated LDLR.
In summary, IDOL establishes a previously unrecognized pathway for LDLR internalization that is distinct from that used for lipoprotein uptake. Defining the molecular determinants that control entry of the LDLR into these distinct internalization routes may open up new avenues to increase abundance of the receptor in therapeutic strategies to treat hypercholesterolemia.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Zelcer group, Irith Koster, and Boris Bleijlevens for critically reading the manuscript and Gilda Nappo for generating the inducible CHC knockdown HeLa cells.
Footnotes
Abbreviations:
- ARH
- autosomal recessive hypercholesterolemia
- CHC
- clathrin heavy chain
- DAB2
- disabled 2
- DUB
- deubiquitylase enzyme
- EGFR
- epidermal growth factor receptor
- IDOL
- inducible degrader of the LDL receptor
- LDLR
- LDL receptor
- LXR
- liver X receptor
- MβCD
- methyl-β-cyclodextrin
- MVB
- multi-vesicular body
- SREBP
- sterol regulatory element binding protein
- VLDLR
- VLDL receptor
- WT
- wild type
S.P. was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC, IG11627) and the EMBO Young Investigator Program. N.Z. was supported by a career development award from the Human Frontier Science Program Organization (HFSP) and by a VIDI grant from the Netherlands Organization of Scientific Research (NWO).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures.
REFERENCES
- 1.Goldstein J. L., Brown M. S. 2009. The LDL receptor. Arterioscler. Thromb. Vasc. Biol. 29: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown M. S., Goldstein J. L. 1986. A receptor-mediated pathway for cholesterol homeostasis. Science. 232: 34–47 [DOI] [PubMed] [Google Scholar]
- 3.Garcia C. K. 2001. Autosomal recessive hypercholesterolemia caused by mutations in a putative LDL receptor adaptor protein. Science. 292: 1394–1398 [DOI] [PubMed] [Google Scholar]
- 4.Keyel P. A., Mishra S. K., Roth R., Heuser J. E., Watkins S. C., Traub L. M. 2006. A single common portal for clathrin-mediated endocytosis of distinct cargo governed by cargo-selective adaptors. Mol. Biol. Cell. 17: 4300–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W. J., Goldstein J. L., Brown M. S. 1990. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J. Biol. Chem. 265: 3116–3123 [PubMed] [Google Scholar]
- 6.He G., Gupta S., Yi M., Michaely P., Hobbs H. H., Cohen J. C. 2002. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J. Biol. Chem. 277: 44044–44049 [DOI] [PubMed] [Google Scholar]
- 7.Morris S. M., Cooper J. A. 2001. Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic. 2: 111–123 [DOI] [PubMed] [Google Scholar]
- 8.Michaely P., Zhao Z., Li W-P., Garuti R., Huang L. J., Hobbs H. H., Cohen J. C. 2007. Identification of a VLDL-induced, FDNPVY-independent internalization mechanism for the LDLR. EMBO J. 26: 3273–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon H. T., Boucrot E. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12: 517–533 [DOI] [PubMed] [Google Scholar]
- 10.Motley A., Bright N. A., Seaman M. N. J., Robinson M. S. 2003. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., Di Fiore P. P. 2008. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell. 15: 209–219 [DOI] [PubMed] [Google Scholar]
- 12.Howes M. T., Mayor S., Parton R. G. 2010. Molecules, mechanisms, and cellular roles of clathrin-independent endocytosis. Curr. Opin. Cell Biol. 22: 519–527 [DOI] [PubMed] [Google Scholar]
- 13.Sigismund S., Confalonieri S., Ciliberto A., Polo S., Scita G., Di Fiore P. P. 2012. Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol. Rev. 92: 273–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacGurn J. A., Hsu P-C., Emr S. D. 2012. Ubiquitin and membrane protein turnover: from cradle to grave. Annu. Rev. Biochem. 81: 231–259 [DOI] [PubMed] [Google Scholar]
- 15.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46 [DOI] [PubMed] [Google Scholar]
- 16.Horton J. D., Goldstein J. L., Brown M. S. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis C. G., Lehrman M. A., Russell D. W., Anderson R. G., Brown M. S., Goldstein J. L. 1986. The J.D. mutation in familial hypercholesterolemia: amino acid substitution in cytoplasmic domain impedes internalization of LDL receptors. Cell. 45: 15–24 [DOI] [PubMed] [Google Scholar]
- 18.Hobbs H. H., Russell D. W., Brown M. S., Goldstein J. L. 1990. The LDL receptor locus in familial hypercholesterolemia: mutational analysis of a membrane protein. Annu. Rev. Genet. 24: 133–170 [DOI] [PubMed] [Google Scholar]
- 19.Zelcer N., Tontonoz P. 2006. Liver X receptors as integrators of metabolic and inflammatory signaling. J. Clin. Invest. 116: 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janowski B. A., Willy P. J., Devi T. R., Falck J. R., Mangelsdorf D. J. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 383: 728–731 [DOI] [PubMed] [Google Scholar]
- 21.Zelcer N., Hong C., Boyadjian R., Tontonoz P. 2009. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science. 325: 100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sorrentino V., Scheer L., Santos A., Reits E., Bleijlevens B., Zelcer N. 2011. Distinct functional domains contribute to degradation of the low density lipoprotein receptor (LDLR) by the E3 ubiquitin ligase inducible degrader of the LDLR (IDOL). J. Biol. Chem. 286: 30190–30199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorrentino V., Zelcer N. 2012. Post-transcriptional regulation of lipoprotein receptors by the E3-ubiquitin ligase inducible degrader of the low-density lipoprotein receptor. Curr. Opin. Lipidol. 23: 213–219 [DOI] [PubMed] [Google Scholar]
- 24.Razani B., Engelman J. A., Wang X. B., Schubert W., Zhang X. L., Marks C. B., Macaluso F., Russell R. G., Li M., Pestell R. G., et al. 2001. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 276: 38121–38138 [DOI] [PubMed] [Google Scholar]
- 25.Zhao Z., Michaely P. 2008. The epidermal growth factor homology domain of the LDL receptor drives lipoprotein release through an allosteric mechanism involving H190, H562, and H586. J. Biol. Chem. 283: 26528–26537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop N., Woodman P. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 11: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazazic M., Bertelsen V., Pedersen K. W., Vuong T. T., Grandal M. V., Rødland M. S., Traub L. M., Stang E., Madshus I. H. 2009. Epsin 1 is involved in recruitment of ubiquitinated EGF receptors into clathrin-coated pits. Traffic. 10: 235–245 [DOI] [PubMed] [Google Scholar]
- 28.Motazacker M. M., Pirruccello J., Huijgen R., Do R., Gabriel S., Peter J., Kuivenhoven J. A., Defesche J. C., Kastelein J. J. P., Hovingh G. K., et al. 2012. Advances in genetics show the need for extending screening strategies for autosomal dominant hypercholesterolaemia. Eur. Heart J. 33: 1360–1366. [DOI] [PubMed] [Google Scholar]
- 29.Song K. S., Li S., Okamoto T., Quilliam L. A., Sargiacomo M., Lisanti M. P. 1996. Co-purification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent-free purification of caveolae microdomains. J. Biol. Chem. 271: 9690–9697 [DOI] [PubMed] [Google Scholar]
- 30.Hong C., Duit S., Jalonen P., Out R., Scheer L., Sorrentino V., Boyadjian R., Rodenburg K. W., Foley E., Korhonen L., et al. 2010. The E3 ubiquitin ligase IDOL induces the degradation of the low density lipoprotein receptor family members VLDLR and ApoER2. J. Biol. Chem. 285: 19720–19726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilund K. R., Yi M., Campagna F., Arca M., Zuliani G., Fellin R., Ho Y-K., Garcia J. V., Hobbs H. H., Cohen J. C. 2002. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum. Mol. Genet. 11: 3019–3030 [DOI] [PubMed] [Google Scholar]
- 32.Maurer M. E., Cooper J. A. 2006. The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J. Cell Sci. 119: 4235–4246 [DOI] [PubMed] [Google Scholar]
- 33.Eden E. R., Sun X-M., Patel D. D., Soutar A. K. 2007. Adaptor protein disabled-2 modulates low density lipoprotein receptor synthesis in fibroblasts from patients with autosomal recessive hypercholesterolaemia. Hum. Mol. Genet. 16: 2751–2759 [DOI] [PubMed] [Google Scholar]
- 34.Hannigan A., Smith P., Kalna G., Lo Nigro C., Orange C., O'Brien D. I., Shah R., Syed N., Spender L. C., Herrera B., et al. 2010. Epigenetic downregulation of human disabled homolog 2 switches TGF-beta from a tumor suppressor to a tumor promoter. J. Clin. Invest. 120: 2842–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simons K., Toomre D. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1: 31–39 [DOI] [PubMed] [Google Scholar]
- 37.Subtil A., Gaidarov I., Kobylarz K., Lampson M. A., Keen J. H., McGraw T. E. 1999. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc. Natl. Acad. Sci. USA. 96: 6775–6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigismund S., Woelk T., Puri C., Maspero E., Tacchetti C., Transidico P., Di Fiore P. P., Polo S. 2005. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA. 102: 2760–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 416: 451–455 [DOI] [PubMed] [Google Scholar]
- 40.Wendland B. 2002. Epsins: adaptors in endocytosis? Nat. Rev. Mol. Cell Biol. 3: 971–977 [DOI] [PubMed] [Google Scholar]
- 41.Ford M. G. J., Mills I. G., Peter B. J., Vallis Y., Praefcke G. J. K., Evans P. R., McMahon H. T. 2002. Curvature of clathrin-coated pits driven by epsin. Nature. 419: 361–366 [DOI] [PubMed] [Google Scholar]
- 42.Raiborg C., Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 458: 445–452 [DOI] [PubMed] [Google Scholar]
- 43.Fujita H., Yamanaka M., Imamura K., Tanaka Y., Nara A., Yoshimori T., Yokota S., Himeno M. 2003. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E vps phenotype in mammalian cells. J. Cell Sci. 116: 401–414 [DOI] [PubMed] [Google Scholar]
- 44.Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 5: 709–722 [DOI] [PubMed] [Google Scholar]
- 45.Clague M. J., Urbé S. 2006. Endocytosis: the DUB version. Trends Cell Biol. 16: 551–559 [DOI] [PubMed] [Google Scholar]
- 46.Lafont F., Simons K. 2001. Raft-partitioning of the ubiquitin ligases Cbl and Nedd4 upon IgE-triggered cell signaling. Proc. Natl. Acad. Sci. USA. 98: 3180–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horvath C. A. J., Vanden Broeck D., Boulet G. A. V., Bogers J., De Wolf M. J. S. 2007. Epsin: inducing membrane curvature. Int. J. Biochem. Cell Biol. 39: 1765–1770 [DOI] [PubMed] [Google Scholar]
- 48.Chen H., Ko G., Zatti A., Di Giacomo G., Liu L., Raiteri E., Perucco E., Collesi C., Min W., Zeiss C., et al. 2009. Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both epsin 1 and epsin 2 in mice. Proc. Natl. Acad. Sci. USA. 106: 13838–13843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasula S., Cai X., Dong Y., Messa M., McManus J., Chang B., Liu X., Zhu H., Mansat R. S., Yoon S-J., et al. 2012. Endothelial epsin deficiency decreases tumor growth by enhancing VEGF signaling. J. Clin. Invest. 122: 4424–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C., Zhuang X. 2008. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc. Natl. Acad. Sci. USA. 105: 11790–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang Y. L., Yochem J., Bell L., Sorensen E. B., Chen L., Conner S. D. 2013. Caenorhabditis elegans reveals a FxNPxY-independent low-density lipoprotein receptor internalization mechanism mediated by epsin1. Mol. Biol. Cell 24: 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., Komada M. 2005. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell. 16: 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Bie P., Ciechanover A. 2011. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18: 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scotti E., Hong C., Yoshinaga Y., Tu Y., Hu Y., Zelcer N., Boyadjian R., de Jong P. J., Young S. G., Fong L. G., et al. 2011. Targeted disruption of the idol gene alters cellular regulation of the low-density lipoprotein receptor by sterols and liver x receptor agonists. Mol. Cell. Biol. 31: 1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Huang Y., Hobbs H. H., Cohen J. C. 2012. Molecular characterization of proprotein convertase subtilisin/kexin type 9-mediated degradation of the LDLR. J. Lipid Res. 53: 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scotti E., Calamai M., Goulbourne C. N., Zhang L., Hong C., Lin R. R., Choi J., Pilch P. F., Fong L. G., Zou P., et al. 2013. IDOL stimulates clathrin-independent endocytosis and multivesicular body-mediated lysosomal degradation of the low-density lipoprotein receptor. Mol. Cell. Biol. 33: 1503–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.