Abstract
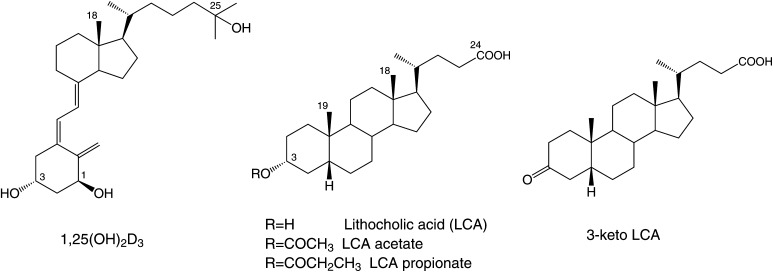
The secondary bile acid lithocholic acid (LCA) and its derivatives act as selective modulators of the vitamin D receptor (VDR), although their structures fundamentally differ from that of the natural hormone 1α,25-dihydroxyvitamin D3 [1,25(OH)2D3)]. Here, we have determined the crystal structures of the ligand-binding domain of rat VDR (VDR-LBD) in ternary complexes with a synthetic partial peptide of the coactivator MED1 (mediator of RNA polymerase II transcription subunit 1) and four ligands, LCA, 3-keto LCA, LCA acetate, and LCA propionate, with the goal of elucidating their agonistic mechanism. LCA and its derivatives bind to the same ligand-binding pocket (LBP) of VDR-LBD that 1,25(OH)2D3 binds to, but in the opposite orientation; their A-ring is positioned at the top of the LBP, whereas their acyclic tail is located at the bottom of the LBP. However, most of the hydrophobic and hydrophilic interactions observed in the complex with 1,25(OH)2D3 are reproduced in the complexes with LCA and its derivatives. Additional interactions between VDR-LBD and the C-3 substituents of the A-ring are also observed in the complexes with LCA and its derivatives. These may result in the observed difference in the potency among the LCA-type ligands.
Keywords: nuclear receptor, structure-function relationship, bile acid, hypercalcemia
The active metabolite of vitamin D3, 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], regulates calcium homeostasis (1). It also promotes cellular differentiation, inhibits cellular proliferation, and suppresses the immune system (2–7). It has been used clinically to treat renal osteodystrophy, vitamin D-dependent rickets type I, and X-linked hypophosphatemic rickets, among other conditions (8–14). Most of its effects are mediated by its specific binding to the vitamin D receptor (VDR), which is a member of nuclear receptor (NR) super family (15). When 1,25(OH)2D3 is bound to VDR, it activates it by inducing conformational changes. The activated complex, VDR/1,25(OH)2D3, binds as a heterodimer with the retinoid X receptor (RXR) to vitamin D response elements located in the promoter region of the target genes. Recruitment of coactivator proteins to this heterodimer is also essential to the transactivation. However, clinical use of 1,25(OH)2D3 is limited because therapeutic doses can give rise to significant hypercalciuria and hypercalcemia (16). A number of synthetic ligands to VDR have been developed for medical use; however, most of them can also cause similar problems because they are derived from 1,25(OH)2D3.
Several synthetic compounds without the vitamin D3 scaffold have been reported to bind to VDR and have VDR-modulating activities, including growth inhibition of cancer cells and keratinocytes and induction of leukemic cell differentiation, with less calcium mobilization side effects than 1,25(OH)2D3 (17). Therefore, these synthetic compounds are expected to be therapeutics for cancer, leukemia, and psoriasis. Subsequently, Makishima et al. discovered that secondary bile acids, including lithocholic acid (LCA) and its derivatives, also behaved as VDR agonists (18–20). LCA acts as a detergent to stabilize fats for absorption, and it has been implicated in human and experimental animal carcinogenesis. However, the agonistic behavior of LCA as a ligand recognized by VDR was not common knowledge because the structure of LCA is completely different from that of vitamin D3. Additional studies showed that VDR had dual functions as a metabolic sensor of bile acids and as an endocrine receptor for 1,25(OH)2D3. Both functions are closely related to colon cancer suppression (21). Although theoretical studies were performed to elucidate how LCA binds to 1,25(OH)2D3, the agonistic mechanism of VDR by LCA was still unclear (18, 22).
Here we determined the crystal structures of the ligand binding domain (LBD) of rat VDR in ternary complexes with a synthetic peptide containing the target sequence of the coactivator MED1 (mediator of RNA polymerase II transcription subunit 1, also known as ARC205 and DRIP205) and the ligands LCA, 3-keto LCA, LCA acetate, and LCA propionate (Fig. 1) to investigate how LCA and its derivatives bind to VDR and how they act as the agonists. The structures reveal that LCA and its derivatives bind to the same ligand-binding pocket (LBP) of VDR that 1,25(OH)2D3 binds to (23–31), but in the opposite orientation. Comparison of these structures also show how LCA and its derivatives mimic 1,25(OH)2D3 and give insight into how the C-3 substituents on the A-ring affect the activity of each ligand. The structures also provide a sound basis for designing new compounds using the scaffold of LCA.
Fig. 1.
Chemical structures of 1,25(OH)2D3, LCA, and LCA derivatives.
MATERIALS AND METHODS
Preparation of LCA ligands
LCA, 3-keto LCA, and LCA acetate were commercially obtained. LCA propionate was synthesized as follows. Boron trifruoride diethyl ether complex (44 μl, 0.33 mmol) was added to a stirred solution of LCA (504.5 mg, 1.34 mmol) in a mixture of propionic anhydride (1.67 ml) and tetrahydrofuran (5 ml) at 0°C, and the resulting solution was stirred at room temperature for 14.5 h. Aqueous sodium hydrogen carbonate was added to this solution, and the reaction mixture was stirred at room temperature for 1 h. Crude LCA propionate was extracted from the reaction mixture with ethyl acetate (AcOEt). The organic layer containing LCA propionate was washed with brine and dried over anhydrous magnesium sulfate, and the solvents were evaporated under house vacuum. The residue was purified by chromatography on silica gel (15 g) with 10% AcOEt/hexane to yield 460 mg (79%) of LCA propionate. The product was recrystallized on AcOEt/hexane.
Protein expression, purification, and crystallization
Rat VDR-LBD (residues 116–423, Δ165–211) was expressed, purified, and crystallized as described by Vanhooke et al. and Nakabayashi et al. (31, 32). The purity and homogeneity of the protein was assessed by SDS-PAGE. The concentration of the protein was determined by UV absorption at 280 nm with molar extinction coefficients estimated using the method developed by Pace et al. (33). The 13mer synthetic oligopeptide (KNHPMLMNLLKDN), which corresponds to residues 625–637 of rat MED1, was purchased from World Gene Co., Ltd. (Tokyo, Japan). Each of the four ligands, LCA, 3-keto LCA, LCA acetate, and LCA propionate, was used to prepare a ternary complex of VDR-LBD/peptide/ligand in 10 mM Tris·HCl (pH 7.0), 10 mM dithiothreitol, and 0.02% sodium azide. All the ternary complexes were crystallized at 20°C in a series of precipitant solutions containing 0.1–0.4 M sodium formate, 12–22% (w/v) polyethylene glycol 4000, and 0–10% (v/v) ethylene glycol.
X-ray diffraction data collection and structural analysis
The crystals were flash-frozen using mother liquor supplemented with 10% ethylene glycol. Diffraction data for the ternary complexes were collected at 95 K at beamline BL-6A at the Photon Factory of the High Energy Accelerator Research Organization and were integrated and scaled with HKL2000 (HKL Research, Inc.). The space group for each complex is C2; the unit cell dimensions are listed in Table 1 with one complex per asymmetrical unit. The structures were solved by molecular replacement by using the crystal structure of the ternary complex reported by Vanhooke et al. (PDB code: 1RK3) as the search model in CNS (31, 34). Refinement was performed with CNS and XtalView (35).
TABLE 1.
Data collection and refinement statistics
| Ligands | LCA | LCA Acetate | LCA Propionate | 3-keto LCA |
| Data collection | ||||
| Unit cell dimensions | ||||
| a (Å) | 154.5 | 154.4 | 154.5 | 153.8 |
| b (Å) | 42.4 | 42.0 | 42.8 | 42.3 |
| c (Å) | 41.6 | 41.5 | 41.6 | 41.5 |
| β (degree) | 96.8 | 96.2 | 96.3 | 96.4 |
| Resolution (Å) | 50–1.9 (1.96–1.90) | 50–2.2 (2.28–2.20) | 50–2.2 (2.28–2.20) | 50–1.9 (1.96–1.90) |
| Completeness (%) | 99.2 (99.9) | 99.3 (95.9) | 99.6 (96.8) | 99.2 (99.8) |
| Redundancy | 3.6 (3.6) | 4.3 (4.0) | 3.6 (3.4) | 3.6 (3.5) |
| I/σ(I) | 43.1 (6.2) | 26.2 (4.4) | 30.3 (3.9) | 41.3 (4.7) |
| Rsym (%) | 3.2 (26.4) | 5.8 (29.5) | 4.5 (35.7) | 4.4 (32.9) |
| Refinement | ||||
| R (%) | 23.3 | 22.0 | 21.1 | 23.4 |
| R-free (%) | 26.3 | 27.6 | 26.5 | 27.9 |
| RMSD bond lengths (Å) | 0.0058 | 0.0068 | 0.0069 | 0.0065 |
| RMSD bond angles (degree) | 1.25 | 1.19 | 1.18 | 1.22 |
| Number of atoms | ||||
| Protein | 1925 | 1942 | 1942 | 1926 |
| Peptide | 92 | 92 | 92 | 92 |
| Ligand | 27 | 30 | 31 | 27 |
| Water | 108 | 49 | 56 | 110 |
| Average B factor (Å2) | 41.0 | 50.1 | 49.9 | 43.8 |
RESULTS
Overall structures of the ternary complexes
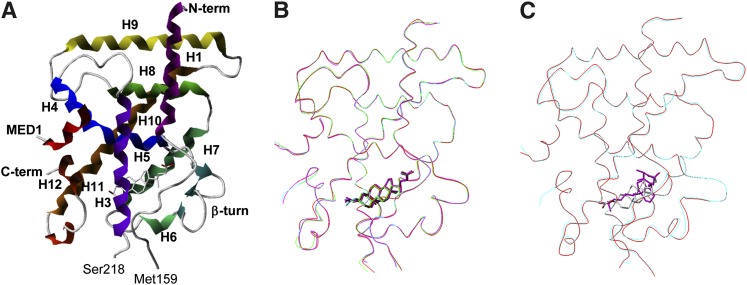
The crystal structures of the ternary complexes of VDR-LBD with LCA and its three derivatives were determined at 1.9–2.2 Å resolution by X-ray crystallography (Table 1 and Fig. 2A). Most of the residues in the complexes were unambiguously determined; however, the N-terminal region (Ala116–Gln122), the middle region (Asp160–Gly164 and Ser212–Leu217), and the C-terminal end (Ser423) of VDR-LBD, and Asp636 and Asn637 of the MED1 peptide were not detected, probably due to fluctuation in these regions. Two more residues at the C-terminal end (Glu421 and Ile422) were also undetectable in the complexes with LCA and 3-keto LCA. Most of these missing residues were previously reported as invisible in studies of other ternary complexes of VDR and are likely a characteristic common to crystals of VDR complexes (23–31).
Fig. 2.
Overall structures of ternary complexes of VDR with LCA derivatives. A: Complex structure of VDR with LCA. VDR and MED1 are represented by ribbons, and LCA is represented by a stick. The helices of VDR are numbered after that of the human RXR. B: Superposition of Cα traces of the four ternary complexes of VDR with LCA derivatives. Complexes with LCA, 3-keto LCA, LCA acetate, and LCA propionate are represented in red, green, cyan, and magenta, respectively. C: Superposition of Cα trace of ternary complexes of VDR with LCA (cyan) and 1,25(OH)2D3 (red) complex.
The overall structures of VDR-LBD in the four complexes are nearly identical (Fig. 2B). The root-mean-square deviations (RMSD) between the proteins in the LCA complex and each of the 3-keto LCA, LCA acetate, and LCA propionate complexes are 0.34, 0.52, and 0.52 Å, respectively, using the Cα atoms of Lys123–Met159 and Ser218–Asn420. Furthermore, the RMSD between the proteins in the LCA and 1,25(OH)2D3 complexes is 0.49 Å for the overall structure (Fig. 2C). Therefore, no significant structural differences were found among the proteins in the ternary complexes with LCA, its derivatives, or 1,25(OH)2D3 (28, 31).
Structures of the ligands and their interactions with VDR-LDB
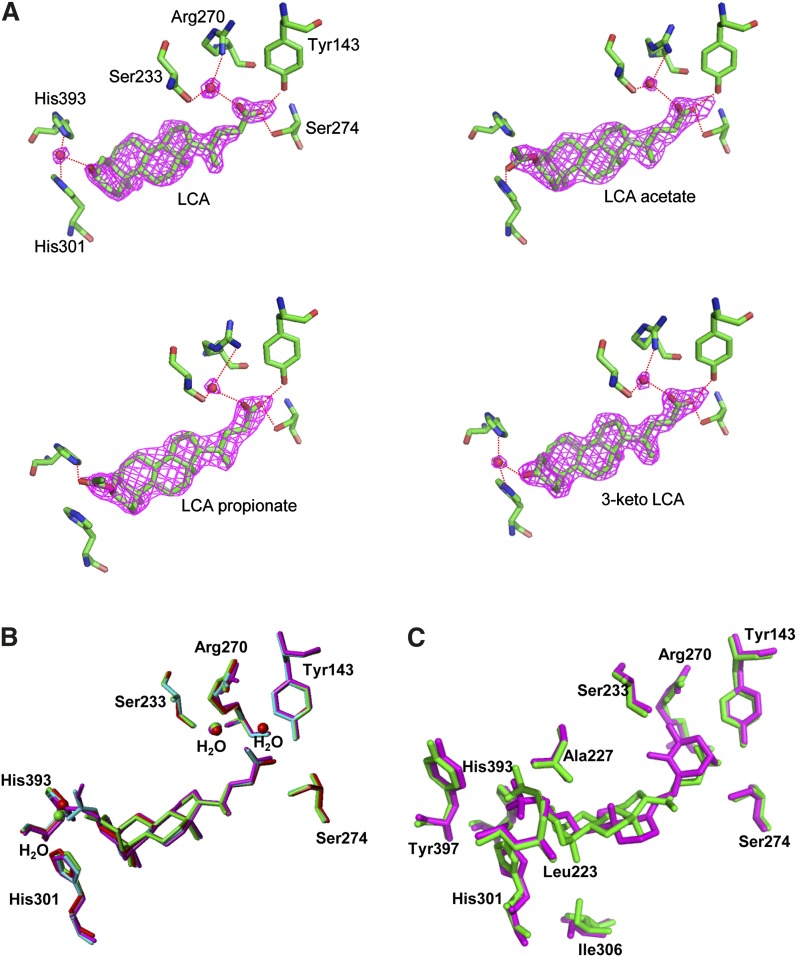
Proteins in the NR super family have a common ligand-binding pocket (LBP). Residues in helices 1, 3, 5, 11, and 12, all β-turns, and loops 6–7 and 11–12 form the framework for the LBP of VDR. The natural hormone 1,25(OH)2D3 is accommodated in the LBP. In the present study, we observed clear electron density in the LBP, as was previously reported in the complex with 1,25(OH)2D3 (Fig. 3A) (28), and crystallographic refinement allowed us unambiguous determination of the structure of LCA and its derivatives in the complex (Figs. 2A and 3A).
Fig. 3.
A: Hydrogen-bonding network between VDR and LCA derivatives, LCA, 3-keto LCA, LCA acetate, and LCA propionate. The purple fishnet is the Fo-Fc omit-annealed difference Fourier electron-density map contoured at 3σ level from the average of the map. The oxygen atoms of water are represented by red spheres. B: Superposition of the four LCA derivatives in VDR complexes. LCA, 3-keto LCA, LCA acetate, and LCA propionate are represented in red, green, cyan, and magenta, respectively. C: Superposition of LCA acetate (green) and 1,25(OH)2D3 (magenta) in VDR complexes.
Except for their respective substituents, LCA and its three derivatives are accommodated in the LBP of VDR-LBD with almost identical structures. However, their orientation is opposite to that of 1,25(OH)2D3 in both the horizontal and vertical planes (Fig. 3C). The 24-carboxyl group is directed toward the β-turns, and the β-face of the steroid is directed toward helix 7 in the bottom of the LBP, while the A-ring faces helix 12 (Fig. 2A). LCA forms three hydrogen bonds at the carboxyl group. One oxygen atom of the carboxyl group directly forms hydrogen bonds with the hydroxyl groups in the side chains of Tyr143 in helix 1 (the distance between the oxygen atoms of the carboxyl group and the hydroxyl group is 2.46 Å) and Ser274 in helices 4/5 (the distance between the oxygen atoms of the carboxyl group and the hydroxyl group is 2.76 Å). The other oxygen atom of the same carboxyl group interacts via a water molecule (the distance between the oxygen atoms of the water and the carboxyl group is 2.69 Å), with the hydroxyl group in the side chain of Ser233 in helix 3 and the guanidinium group in the side chain of Arg270 in helix 4/5 (the distance between the water molecule and these residues are 2.92 Å and 3.01 Å, respectively) (Fig. 3A). These hydrogen bonds are also observed in the other three complexes. The four rings of the steroid in each of the complexes interact with hydrophobic residues in the LBP through hydrophobic interactions. There are 12 residues (Leu226, Leu229, Val230, Ile264, Ile276, Met268, Trp282, Val296, Ala299, Leu305, Ile306, and Leu309) distributed within 4.3 Å from the rings. Such hydrophobic interactions are also conserved in the other complexes.
Hydrogen bonds between VDR and the ligands are also observed at the other end of the ligands, the C-3 position of the A-ring. The four ligands differ in their substituents at this position. The hydroxyl group of LCA, the carbonyl group of 3-keto LCA, the propionyl group of LCA propionate, and the acetyl group of LCA acetate interact with residues in helix 6, loop 6–7, and helix 11 (Figs. 2A and 3A). In the complexes with LCA and 3-keto LCA, the oxygen atoms of the respective hydroxyl and carbonyl groups of the substituents interact via a water-mediated hydrogen bond with the nitrogen atoms of imidazole rings of His301 in helix 6 and His393 in helix 11. In contrast, in the complex with LCA acetate, the oxygen atom of the acetyl group directly forms a hydrogen bond with the nitrogen atom of the imidazole ring of His301. In the complex with LCA propionate, the oxygen atom of the propionyl group also directly forms a hydrogen bond with the nitrogen atom of the imidazole ring of His393. Furthermore, the alkyl parts of the two substituents interact with the aromatic rings of Tyr397 in helix 11 and Phe418 in helix 12 and with the side chains of Leu410 and Val414 in helix 12, stabilizing the binding of the two derivatives to the LBP of VDR-LBD (Fig. 3A). From this viewpoint, LCA propionate may be the most effective of the four ligands because it has the longest alkyl part in the substituent.
Structure of the MED1 peptide
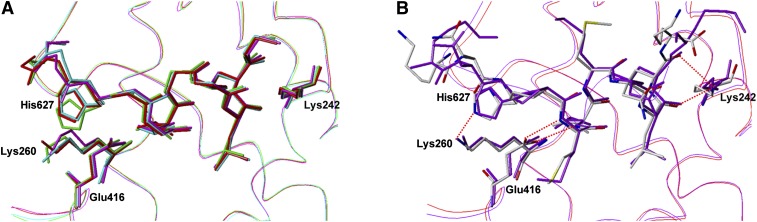
NR super family proteins generally include two domains for two types of transactivations, a constitutive activation function (AF-1) and a ligand-dependent activation function (AF-2). The AF-2 domain of the VDR-LBD consists of helices 3, 4, and 12, and loop 3–4, and it interacts with the LXXLL motif known as the NR-box. In the present study, we synthesized a peptide containing the target sequence of the MED1 coactivator and determined the structure of the peptide in the four ternary complexes to investigate whether or not LCA and its derivatives affect coactivator binding. The peptide binds to the AF-2 domain in each of the four complexes similar to the complex with 1,25(OH)2D3. Although two residues of the peptide, Asp636 and Asn637, have no detectable structure as described above, the rest of the peptide forms an α-helix with a kink at Pro628 in each complex (Fig. 4A). The structure of the peptide in the four complexes is nearly identical. The RMSDs between the peptides in the LCA complex and the 3-keto LCA, LCA acetate, and LCA propionate complexes are 0.22, 0.58, and 0.69 Å, respectively, using the Cα atoms of Lys625–Lys635. The structure of the peptide in the LCA complex was also compared with that in the 1,25(OH)2D3 complex. The RMSD between the two peptides was calculated at 0.76 Å, indicating no significant structural differences among the peptides in the ternary complexes with LCA, its derivatives, and 1,25(OH)2D3 (Fig. 4B) (28, 31).
Fig. 4.
Structures of MED1 peptides in VDR complexes. MED1 peptide and Lys242 and Glu416 of each VDR complex are represented by sticks; the other parts are represented by ribbons. A: Superposition of MED1 peptides in VDR complexes with LCA (red), 3-keto LCA (green), LCA acetate (cyan), and LCA propionate (magenta). B: Superposition of MED1 peptides in VDR complexes with LCA (magenta) and 1,25(OH)2D3 (white carbons).
The AF-2 domain forms a shallow pit consisting of five hydrophobic residues: Ile238 in helix 3, Ile256 in helix 4/5, Leu259 in helix 4/5, Leu413 in helix 12, and Val417 in helix 12. The peptide binds to this pit through the hydrophobic interactions between the LXXLL motif of the peptide and the complementary pit of the protein. The polar side chains of Lys242 in helix 3 and Glu416 in helix 12 also facilitate the binding of the peptide by clamping it on the both edges of the AF-2 domain (a charge clamping). Two hydrogen bonds are formed between the oxygen atom of the side chain carboxyl group of Glu416 and the amide nitrogen of Met629, and the nitrogen atom of the side chain amino group of Lys242 and the carbonyl oxygen of Leu633. All these interactions observed in the present study are the same as those seen in the complex with 1,25(OH)2D3. Therefore, these results indicate that the interactions between the coactivator MED1 and the AF-2 domain are well conserved in the ternary complexes with LCA and its derivatives.
DISCUSSION
Since the discovery of its function as an agonist of VDR, LCA has been expected to be used a vitamin D alternative, especially because LCA appears to activate VDR without causing hypercalcemia. However, because the functional mechanism of LCA was still unclear, LCA derivatives with higher activities have been found mainly by trial and error. In the present study, we determined the structures of ternary complexes of VDR-LBD with LCA and its derivatives and elucidated how they bind to VRD-LDB.
LCA and its derivatives bind to the same LBP that 1,25(OH)2D3 binds to. However, their orientation is opposite to that of 1,25(OH)2D3 (Fig. 3C). Its A-ring was set on the inlet of the LBP, while its 24-carboxyl group wedged itself into the LBP. Interestingly this orientation is the same as that of 6α-ethyl-chenodeoxycholic acid (6α-ethyl-CDCA) in the farnesoid X receptor (FXR) (36). LCA and its derivatives are similar in size (14–16 Å × 5 Å × 3 Å length × width × thickness) to natural 1,25(OH)2D3 (approximately 15 Å × 5 Å × 2 Å), and although their shapes are somewhat different, the nonplanar cis A/B rings of the steroid scaffold of LCA mimic the curvature of the 9,10-secosteriod portion of 1,25(OH)2D3 (Fig. 3C). Often, the size seems to be critical to the agonistic activity of a ligand. Agonistic ligands induce a conformational change of loop 11–12 to bring helix 12 into the active conformation, which is stabilized by hydrophobic interactions between the ligand and Val418 and Phe422 (Val414 and Phe418 of rat VDR, respectively). This conformation creates the AF-2 surface and allows the coactivator to bind, which is required for the VDR activity. Large ligands could collide with helix 12 and destabilize the active conformation. Indeed, several larger ligands have been reported to show antagonistic activity instead of agonistic activity (32, 37).
Despite their opposite orientation, LCA and its derivatives can reproduce the interactions between VDR and 1,25(OH)2D3, which consists of a hydrophobic secosteroid framework and three polar groups. The polar groups are located at the both ends of the ligand and stabilize ligand binding through several hydrogen bonds, while the hydrophobic secosteroid frame just fits the hydrophobic tunnel of the LBP. In the complex with 1,25(OH)2D3, the A-ring of 1,25(OH)2D3 deeply wedges itself into the LBP and its 25-hydroxyl group is set on the inlet of the LBP. There are two polar groups at the C-1 and C-3 positions of the A-ring of 1,25(OH)2D3, whose oxygen atoms form two pairs of bifurcated hydrogen bonds with the side chains of Tyr143 and Ser274, and Ser233 and Arg270, respectively. Furthermore, the 25-hydroxyl group of 1,25(OH)2D3 also forms a bifurcated hydrogen bond with the nitrogen atoms of the imidazole rings of His301 and His393. In contrast, LCA and its derivatives consist of the hydrophobic steroid framework and two polar groups at the both ends (Fig. 1). However, they maintain most of the hydrogen bonds between the ligand and the protein by exchanging the hydrogen-bonding partners between the A-ring and the carboxyl group. Two oxygen atoms of the 24-carboxyl group form, with the help of a water molecule, two pairs of bifurcated hydrogen bonds directly with the side chains of Tyr143 and Ser274, and indirectly with Ser233 and Arg270, respectively. The hydrogen bonds with the nitrogen atoms of the imidazole rings of His301 and His393 are partially formed, depending on the substituents at the C-3 position of the A-ring. Therefore, the hydrogen-bonding network between the ligand and the protein is essentially the same for both LCA and 1,25(OH)2D3, even though their orientations are opposite. Adachi et al. also indicated the importance of the hydrogen-bonding network for agonistic activity by showing that esterification of the carboxyl group of LCA weakened its agonistic activity (18).
Although LCA mimicked the dimensions and chemical properties of 1,25(OH)2D3 as discussed above, its affinity is much lower than 1,25(OH)2D3. In a competitive binding assay using VDR fused to glutathione S-transferase, IC50 values were estimated as 0.08 nM and 300 μM for 1,25(OH)2D3 and LCA, respectively (19). As the accessible surface areas of the hydrophobic surface of LCA and 1,25(OH)2D3 are 371 Å2 and 442 Å2, respectively, the total hydrophobic interaction of LCA with VDR would be expected to be smaller than that of 1,25(OH)2D3. Also the simple difference of the size between LCA (C24H40O3) and 1,25(OH)2D3 (C27H44O3) suggests that the LBP accommodates LCA somewhat loosely. Such a feature allows several water molecules to penetrate the LBP of VDR. One of the water molecules is involved in the hydrogen bond network around the 24-carboxyl group. Another water molecule is incorporated in the hydrogen bond network around the C-3 position of the A-ring in the complex with LCA. However, given the affinity analyses mentioned above, the interactions mediated by hydrogen bonding seem to be less important for stabilizing the complex than the hydrophobic interactions.
The substituents at the C-3 position of the A-ring of LCA also affect the activation of human VDR, and LCA acetate and LCA propionate are more potent agonists than LCA and 3-keto LCA (18, 19, 22, 38). The biding assay mentioned above showed that IC50 values for LCA acetate and LCA propionate are 30 μM, about 10 times more potent than LCA (19). Another cotransfection assay showed LCA acetate (EC50 = 0.40 μM) is about 30 times more effective than LCA (EC50 = 12.1 μM), whereas 3-keto LCA (EC50 = 6.8 μM) is comparable to LCA (18). Our crystal structures show that the C-3 substituents of the former derivatives interact directly with VDR via a hydrogen bond, while the latter ones require a water molecule to form indirect hydrogen bonds with VDR. In addition, the alkyl part of these acyl groups interacts with the hydrophobic residues of VDR, including Val418 and Phe422 (Val414 and Phe418 of rat VDR, respectively), which are key residues in the VDR activation mechanism described above. These interactions are probably the reason for the higher potency of these two ligands because this valine is not directly involved in the interaction with the ligands in the case of LCA and 3-keto LCA. These additional interactions at the C-3 position may also explain some of the mutation analyses; the human VDR-S275A and S278A mutations (corresponding to S271A and S274A of rat VDR, respectively) almost completely abolished the activity of LCA, whereas they were still activated by LCA acetate (18). The extra interactions at the C-3 position compensate for the loss of the hydrogen bond due to the mutation to some extent.
Some of the other results from the mutation analyses could also be explained from our structure (18, 19, 22, 38). The VDR-S237M mutant (S233M of rat VDR) can respond to LCA but not to 1,25(OH)2D3. The serine residue is directly interacts with the A-ring of 1,25(OH)2D3 through a hydrogen bond, and the mutation to methionine would not only lose the capability of the interaction but also hinder the ligand binding. On the other hand, the bulky A-ring is replaced by a linear alkyl group in LCA (Fig. 3C), creating enough room to accommodate in the methionine. In contrast, the VDR-S278V mutant (S274V of rat VDR) is activated by 1,25(OH)2D3 but not by LCA. The side chain of the serine also directly interacts with the hydroxyl group of the A-ring of 1,25(OH)2D3 and the carboxyl group of LCA. The mutation to valine would lose the hydrogen bond, but there is enough room to accommodate the replaced side chain in both cases. LCA might disfavor the mutant that would bring the hydrophobic valine close to the negative charge of the carboxyl group.
The role of His305 (His301 of rat VDR) in the interactions with the LCA-related ligands seems more complicated. Although LCA and 3-keto LCA interact with His305 in a very similar manner (Fig. 3A), the VDR-H305A (H301A of rat VDR) mutant significantly diminishes the LCA activity but has little effect on activation by LCA acetate or 3-keto LCA. The indole ring lies almost parallel to the A-ring of LCA, making van der Waals contacts. It is also involved in the hydrogen bond directly (LCA acetate) or indirectly (LCA and 3-keto LCA). These interactions are almost the only interactions between the ligand and loop 6–7, and the mutation to alanine may perhaps trigger a large conformational change of the loop, which could even expose the ligand to the solvent.
Although LCA activates VDR, it functions differently from 1,25(OH)2D3; for example, it does not induce hypercalcemia. The most prominent difference in the ligand binding between 1,25(OH)2D3 and the LCA-related ligands reported here are the pattern of the hydrogen bonds with Ser233 in helix 3 and Arg270 in helix 4/5. While 1α-hydroxyl group of 1,25(OH)2D3 directly forms the hydrogen bonds with these two residues, all the LCA-related ligands studied here require a water molecule to form indirect hydrogen bonds with them, which is likely to weaken the interaction with VDR. Also, replacing the A-ring of 1,25(OH)2D3 with a linear alkyl group seems to loosen the hydrophobic interactions around it, as highlighted by the VDR-S237M mutant discussed above. These differences may affect the structure of the AF-2 surface and, therefore, the interactions between VDR and the coactivators statically and/or dynamically.
However, we did not detect significant differences in the interactions between VDR and the coactivator peptide. One possible reason is the crystal packing. The coactivator peptide not only interacts with VDR but also plays an important role in the crystal packing, and even antagonists could be trapped in its active form at high concentrations as in the crystallization conditions (32). Thus the structural differences that may be caused by LCA could have been suppressed by the crystal packing. The possibility that LCA behaves differently from 1,25(OH)2D3 through a nonstructural mechanism also cannot be excluded. Their metabolic behavior and/or cellular distribution would be different in vivo, causing the functional differences. To rationally elucidate the underlying reason for the difference in the agonistic activity between the ligands, it will be necessary to analyze the higher-ordered complexes, perhaps using full-length VDR. The crystal structures reported here nonetheless have shown the similarities and differences between LCA-related ligands and 1,25(OH)2D3 in their interactions with VDR and should provide a sound basis for the design of new ligands based on LCA, hopefully with better pharmaceutical features.
Data deposition
The coordinates of the determined structures have been deposited in the Protein Data Bank with accession numbers 3W5P, 3W5Q, 3W5R, and 3W5T for the LCA, 3keto-LCA, LCA acetate, and LCA propionate complexes, respectively.
Acknowledgments
The authors thank Professor H. Kagechika and Professor H. Tamamura of Tokyo Medical and Dental University for supporting this research project. The authors also thank the technical staff at the Photon Factory of the High Energy Accelerator Research Organization for maintenance of the beam line.
Footnotes
Abbreviations:
- AcOEt
- ethyl acetate
- AF2
- activation function 2
- LBD
- ligand-binding domain
- LBP
- ligand-binding pocket
- LCA
- lithocholic acid
- MED1
- mediator of RNA polymerase II transcription subunit 1
- NR
- nuclear receptor
- 1
- 25(OH)2D3, 1α,25-dihydroxyvitamin D3
- RMSD
- root-mean-square deviation
- RXR
- retinoid X receptor
- VDR
- vitamin D receptor
REFERENCES
- 1.Haussler M. R., Whitfield G. K., Haussler C. A., Hsieh J. C., Thompson P. D., Selznick S. H., Dominguez C. E., Jurutka P. W. 1998. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J. Bone Miner. Res. 13: 325–349 [DOI] [PubMed] [Google Scholar]
- 2.Abe E., Miyaura C., Sakagami H., Takeda M., Konno K., Yamazaki T., Yoshiki S., Suda T. 1981. Differentiation of mouse myeloid leukemia cells induced by 1 alpha,25-dihydroxyvitamin D3. Proc. Natl. Acad. Sci. USA. 78: 4990–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLuca H. F. 2004. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 80(Suppl.): 1689–1696 [DOI] [PubMed] [Google Scholar]
- 4.Hosomi J., Hosoi J., Abe E., Suda T., Kuroki T. 1983. Regulation of terminal differentiation of cultured mouse epidermal cells by 1 alpha,25-dihydroxyvitamin D3. Endocrinology. 113: 1950–1957 [DOI] [PubMed] [Google Scholar]
- 5.Lemire J. M. 1992. Immunomodulatory role of 1,25-dihydroxyvitamin D3. J. Cell. Biochem. 49: 26–31 [DOI] [PubMed] [Google Scholar]
- 6.Smith E. L., Walworth N. C., Holick M. F. 1986. Effect of 1 alpha,25-dihydroxyvitamin D3 on the morphologic and biochemical differentiation of cultured human epidermal keratinocytes grown in serum-free conditions. J. Invest. Dermatol. 86: 709–714 [DOI] [PubMed] [Google Scholar]
- 7.Tanaka H., Abe E., Miyaura C., Kuribayashi T., Konno K., Nishii Y., Suda T. 1982. 1 alpha,25-Dihydroxycholecalciferol and a human myeloid leukaemia cell line (HL-60). Biochem. J. 204: 713–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bortman P., Folgueira M. A., Katayama M. L., Snitcovsky I. M., Brentani M. M. 2002. Antiproliferative effects of 1,25-dihydroxyvitamin D3 on breast cells: a mini review. Braz. J. Med. Biol. Res. 35: 1–9 [DOI] [PubMed] [Google Scholar]
- 9.Fraser D., Kooh S. W., Kind H. P., Holick M. F., Tanaka Y., DeLuca H. F. 1973. Pathogenesis of hereditary vitamin-D-dependent rickets. An inborn error of vitamin D metabolism involving defective conversion of 25-hydroxyvitamin D to 1 alpha,25-dihydroxyvitamin D. N. Engl. J. Med. 289: 817–822 [DOI] [PubMed] [Google Scholar]
- 10.Glorieux F. H., Marie P. J., Pettifor J. M., Delvin E. E. 1980. Bone response to phosphate salts, ergocalciferol, and calcitriol in hypophosphatemic vitamin D-resistant rickets. N. Engl. J. Med. 303: 1023–1031 [DOI] [PubMed] [Google Scholar]
- 11.Hayes C. E. 2000. Vitamin D: a natural inhibitor of multiple sclerosis. Proc. Nutr. Soc. 59: 531–535 [DOI] [PubMed] [Google Scholar]
- 12.Konety B. R., Getzenberg R. H. 2002. Vitamin D and prostate cancer. Urol. Clin. North Am. 29: 95–106, ix [DOI] [PubMed] [Google Scholar]
- 13.Lamberg-Allardt C. 1991. Is there a role for vitamin D in osteoporosis? Calcif. Tissue Int. 49(Suppl.): S46–S49 [DOI] [PubMed] [Google Scholar]
- 14.Langner A., Verjans H., Stapor V., Mol M., Fraczykowska M. 1993. Topical calcitriol in the treatment of chronic plaque psoriasis: a double-blind study. Br. J. Dermatol. 128: 566–571 [DOI] [PubMed] [Google Scholar]
- 15.Yamada S., Shimizu M., Yamamoto K. 2003. Vitamin D receptor. Endocr. Dev. 6: 50–68 [DOI] [PubMed] [Google Scholar]
- 16.Bouillon R., Okamura W. H., Norman A. W. 1995. Structure-function relationships in the vitamin D endocrine system. Endocr. Rev. 16: 200–257 [DOI] [PubMed] [Google Scholar]
- 17.Boehm M. F., Fitzgerald P., Zou A., Elgort M. G., Bischoff E. D., Mere L., Mais D. E., Bissonnette R. P., Heyman R. A., Nadzan A. M., et al. 1999. Novel nonsecosteroidal vitamin D mimics exert VDR-modulating activities with less calcium mobilization than 1,25-dihydroxyvitamin D3. Chem. Biol. 6: 265–275 [DOI] [PubMed] [Google Scholar]
- 18.Adachi R., Honma Y., Masuno H., Kawana K., Shimomura I., Yamada S., Makishima M. 2005. Selective activation of vitamin D receptor by lithocholic acid acetate, a bile acid derivative. J. Lipid Res. 46: 46–57 [DOI] [PubMed] [Google Scholar]
- 19.Ishizawa M., Matsunawa M., Adachi R., Uno S., Ikeda K., Masuno H., Shimizu M., Iwasaki K., Yamada S., Makishima M. 2008. Lithocholic acid derivatives act as selective vitamin D receptor modulators without inducing hypercalcemia. J. Lipid Res. 49: 763–772 [DOI] [PubMed] [Google Scholar]
- 20.Makishima M., Lu T. T., Xie W., Whitfield G. K., Domoto H., Evans R. M., Haussler M. R., Mangelsdorf D. J. 2002. Vitamin D receptor as an intestinal bile acid sensor. Science. 296: 1313–1316 [DOI] [PubMed] [Google Scholar]
- 21.Degirolamo C., Modica S., Palasciano G., Moschetta A. 2011. Bile acids and colon cancer: solving the puzzle with nuclear receptors. Trends Mol. Med. 17: 564–572 [DOI] [PubMed] [Google Scholar]
- 22.Choi M., Yamamoto K., Itoh T., Makishima M., Mangelsdorf D. J., Moras D., DeLuca H. F., Yamada S. 2003. Interaction between vitamin D receptor and vitamin D ligands: two-dimensional alanine scanning mutational analysis. Chem. Biol. 10: 261–270 [DOI] [PubMed] [Google Scholar]
- 23.Ciesielski F., Rochel N., Mitschler A., Kouzmenko A., Moras D. 2004. Structural investigation of the ligand binding domain of the zebrafish VDR in complexes with 1alpha,25(OH)2D3 and Gemini: purification, crystallization and preliminary X-ray diffraction analysis. J. Steroid Biochem. Mol. Biol. 89–90: 55–59 [DOI] [PubMed] [Google Scholar]
- 24.Eelen G., Verlinden L., Rochel N., Claessens F., De Clercq P., Vandewalle M., Tocchini-Valentini G., Moras D., Bouillon R., Verstuyf A. 2005. Superagonistic action of 14-epi-analogs of 1,25-dihydroxyvitamin D explained by vitamin D receptor-coactivator interaction. Mol. Pharmacol. 67: 1566–1573 [DOI] [PubMed] [Google Scholar]
- 25.Hourai S., Fujishima T., Kittaka A., Suhara Y., Takayama H., Rochel N., Moras D. 2006. Probing a water channel near the A-ring of receptor-bound 1 alpha,25-dihydroxyvitamin D3 with selected 2 alpha-substituted analogues. J. Med. Chem. 49: 5199–5205 [DOI] [PubMed] [Google Scholar]
- 26.Rochel N., Hourai S., Perez-Garcia X., Rumbo A., Mourino A., Moras D. 2007. Crystal structure of the vitamin D nuclear receptor ligand binding domain in complex with a locked side chain analog of calcitriol. Arch. Biochem. Biophys. 460: 172–176 [DOI] [PubMed] [Google Scholar]
- 27.Rochel N., Wurtz J. M., Mitschler A., Klaholz B., Moras D. 2000. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell. 5: 173–179 [DOI] [PubMed] [Google Scholar]
- 28.Shimizu M., Miyamoto Y., Takaku H., Matsuo M., Nakabayashi M., Masuno H., Udagawa N., DeLuca H. F., Ikura T., Ito N. 2008. 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvita min D3 analogs: synthesis, biological evaluation, and crystal structure. Bioorg. Med. Chem. 16: 6949–6964 [DOI] [PubMed] [Google Scholar]
- 29.Tocchini-Valentini G., Rochel N., Wurtz J. M., Mitschler A., Moras D. 2001. Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands. Proc. Natl. Acad. Sci. USA. 98: 5491–5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tocchini-Valentini G., Rochel N., Wurtz J. M., Moras D. 2004. Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification. J. Med. Chem. 47: 1956–1961 [DOI] [PubMed] [Google Scholar]
- 31.Vanhooke J. L., Benning M. M., Bauer C. B., Pike J. W., DeLuca H. F. 2004. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 43: 4101–4110 [DOI] [PubMed] [Google Scholar]
- 32.Nakabayashi M., Yamada S., Yoshimoto N., Tanaka T., Igarashi M., Ikura T., Ito N., Makishima M., Tokiwa H., DeLuca H. F., et al. 2008. Crystal structures of rat vitamin D receptor bound to adamantyl vitamin D analogs: structural basis for vitamin D receptor antagonism and partial agonism. J. Med. Chem. 51: 5320–5329 [DOI] [PubMed] [Google Scholar]
- 33.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4: 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921 [DOI] [PubMed] [Google Scholar]
- 35.McRee D. E. 1999. XtalView/Xfit--a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125: 156–165 [DOI] [PubMed] [Google Scholar]
- 36.Mi L. Z., Devarakonda S., Harp J. M., Han Q., Pellicciari R., Willson T. M., Khorasanizadeh S., Rastinejad F. 2003. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol. Cell. 11: 1093–1100 [DOI] [PubMed] [Google Scholar]
- 37.Inaba Y., Nakabayashi M., Itoh T., Yoshimoto N., Ikura T., Ito N., Shimizu M., Yamamoto K. 2010. 22S-butyl-1alpha,24R-dihydroxyvitamin D3: recovery of vitamin D receptor agonistic activity. J. Steroid Biochem. Mol. Biol. 121: 146–150 [DOI] [PubMed] [Google Scholar]
- 38.Sato H., Macchiarulo A., Thomas C., Gioiello A., Une M., Hofmann A. F., Saladin R., Schoonjans K., Pellicciari R., Auwerx J. 2008. Novel potent and selective bile acid derivatives as TGR5 agonists: biological screening, structure-activity relationships, and molecular modeling studies. J. Med. Chem. 51: 1831–1841 [DOI] [PubMed] [Google Scholar]