Abstract
HDL subclasses detection, in cardiovascular risk, has been limited due to the time-consuming nature of current techniques. We have developed a time-saving and reliable separation of the principal HDL subclasses employing iodixanol density gradient ultracentrifugation (IxDGUC) combined with digital photography. HDL subclasses were separated in 2.5 h from prestained plasma on a three-step iodixanol gradient. HDL subclass profiles were generated by digital photography and gel scan software. Plasma samples (n = 46) were used to optimize the gradient for the resolution of HDL heterogeneity and to compare profiles generated by IxDGUC with gradient gel electrophoresis (GGE); further characterization from participants (n = 548) with a range of lipid profiles was also performed. HDL subclass profiles generated by IxDGUC were comparable to those separated by GGE as indicated by a significant association between areas under the curve for both HDL2 and HDL3 (HDL2, r = 0.896, P < 0.01; HDL3, r = 0.894, P < 0.01). The method was highly reproducible, with intra- and interassay coefficient of variation percentage < 5 for percentage area under the curve HDL2 and HDL3, and < 1% for peak Rf and peak density. The method provides time-saving and cost-effective detection and preparation of the principal HDL subclasses.
Keywords: high density lipoprotein, HDL subfractions, self-generating gradients, LDL subclasses, atherogenic lipoprotein phenotype, plasma triacylglycerol, HDL2, HDL3, plasma lipoproteins, cardiovascular disease
Several epidemiological and prospective studies have identified low high density lipoprotein cholesterol (HDL-C) as an independent risk factor for cardiovascular disease (CVD) (1, 2). While pharmacological interventions elevating HDL-C have, to some extent, shown a reduction in CVD risk (3, 4), there is also evidence that the quality of HDL may also be important (5, 6). It has been suggested that HDL subclasses may show a variable relationship with CVD risk. In patients undergoing coronary angiography, Drexel et al. have shown HDL2 to be the strongest predictor of the extent of coronary artery disease (7); however, the number of studies investigating associations of HDL subclasses with CVD risk is relatively small and with mixed results, with some demonstrating an inverse association of HDL2 with CVD risk or individual risk factors (8–11) and others either showing no association with subclass distribution or an inverse association with HDL3 (12–14). The interpretation of such studies is made increasingly difficult by the range of methods used to identify HDL subclasses and the paucity of large studies, possibly due to the arduous nature of the separation techniques available.
Currently there are no standardized reference methods for the separation of lipoprotein subclasses; however, ultracentrifugation is a well-established research method for both preparative and quantitative analysis of LDL and HDL subclasses (15, 16). The majority of ultracentrifuge methods to separate HDL subfractions involve the use of salt (KBr or NaBr) gradients (17, 18). However, Graham et al. (1996) revolutionized ultracentrifugation methods by utilizing iodixanol, a derivative of triiodobenzoic acid, in conjunction with vertical and near vertical rotors, to separate plasma lipoproteins in a run time of 3 h (19). Iodixanol is nontoxic to cells, noninhibitory to enzymes, and iso-osmotic at all densities (20), iodixanol reduces the potential for the dissociation or disruption of apolipoprotein that has been associated with salt gradient techniques (21), and it allows the use of gradient fractions for supplementary analysis without prior dialysis. The use of iodixanol has been adapted to further separate LDL into its principal subclasses by the method of Davies et al. (22). This procedure used self-generated gradients of iodixanol coupled with digital photography to generate LDL subclass profiles, comparable with those generated by salt-density gradient ultracentrifugation and gradient gel electrophoresis (GGE), in a run time of 2.5 h.
We have developed a three-step gradient using iodixanol which allows identification of the principal HDL subclasses (HDL2 and HDL3) using the same rotor and conditions as previously described for LDL subclasses (22). Furthermore, the technique also utilizes digital photography coupled with gel scan software to generate HDL profiles.
MATERIALS AND METHODS
Materials
Iodixanol was supplied as a 60% w/v solution (Optiprep™) by Axis-Shield (UK). Optiseal tubes (4.9 ml) and 12 ml ultraclear tubes were supplied by Beckman Coulter (UK). Cholesterol, triacylglycerol (TAG), and apolipoprotein calibrators; assays; and serum lipid controls level I, level II, and level III were supplied by Randox UK (Co Antrim). Precast polyacrylamide gels (4–30%) and electrophoresis equipment were supplied by CBS Scientific (CA). Phosphate buffered saline (PBS), potassium bromide (KBr), methanol, glacial acetic acid, boric acid, sulphosalicylic acid, Sudan Black B, and ethylene glycol were supplied by Sigma-Aldrich (UK).
Blood samples
Blood samples (10 ml) were taken from 46 normal, healthy male and female volunteers who, on the basis of their blood lipid profile, were expected to show a range of HDL subclass patterns. These samples were used to establish optimal gradient conditions and to compare subclass patterns generated by IxDGUC and GGE. HDL subclasses were separated from a total of 548 plasma samples from healthy volunteers to examine associations between HDL subclass profiles and other plasma lipids and lipoproteins. Volunteers were participants recruited for dietary intervention studies. All studied individuals had been informed in writing of the intended use of their sample and provided written consent. The intervention studies were approved by the Multi-centred Research Ethics Committee and the University of Surrey Ethics Committee. All blood samples were taken by venepuncture from volunteers who had fasted for 12 h and were collected into vacutainers containing K2EDTA (1 g/l). Plasma was harvested by low-speed centrifugation (2,000 g for 20 min at 4°C) in a Heraeus Labofuge 400R centrifuge. All plasma was stored at −80°C before analysis.
Preparation of the iodixanol gradient
The iodixanol gradient and visualization of separated HDL subclasses were based on a procedure for the separation of LDL subclasses described by Davies et al. (22). Davies and coworkers identified the distribution of LDL subclasses by prestaining plasma with Coomassie Blue, similar to the method of Swinkels et al. (23). The presence of other plasma proteins in the same density region as HDL precluded the use of Coomassie Blue as a protein stain. The technique of Terpstra et al. (24), which uses Sudan Black B in a solution of ethylene glycol to stain lipids, was modified and used as an alternative stain for HDL subclasses. Briefly, although Terpstra et al. (24) stained plasma with 200 µl of Sudan Black B at a concentration of 0.1 g/100 ml, our method used higher concentrations with a lower volume (described below), as the larger volume resulted in overstaining. The iodixanol gradient was generated by initially creating three density layers: the bottom layer consisted of 23% w/v iodixanol/PBS solution; the middle layer, a 17.6% w/v solution of iodixanol mixed with plasma and stain; and the top layer, a 15% w/v solution of iodixanol/PBS.
Plasma was mixed with Optiprep™ (iodixanol at 60% w/v) and prestained with Sudan Black B (80 μl of 2 g/100 ml in ethylene glycol) to give a final concentration of 17.6% (w/v) with respect to iodixanol. For example, plasma (1.4 ml) was added to Optiprep™ (0.6 ml) and 80 μl of Sudan Black B or PBS to provide a working sample of 2 ml. The lower layer was prepared by mixing Optiprep™ with PBS to provide a 23% iodixanol solution (final density 1.118 kg/l). The upper layer was also prepared by mixing Optiprep with PBS to give a 15% w/v iodixanol solution (final density 1.108 kg/l).
To prepare the gradient, 1.7 ml aliquots of the 15% iodixanol solution were dispensed into 4.9 ml Optiseal centrifuge tubes. Then 1.5 ml of the prestained working sample was carefully underlayered using a Luer fitting steel cannula and syringe. Finally, 1.7 ml of the 23% iodixanol solution was underlayered, again using a cannula and syringe. Centrifuge tubes were housed in a Beckman NVT 65.2 rotor and centrifuged at 65,000 rpm (371,000 g(av)), 16°C, acceleration program 5, deceleration program 5, for 2.5 h in Beckman Optima L-100 ultracentrifuge.
Measurement of the gradient density
To determine the density profile of the gradient, unstained blank samples were prepared with 1.4 ml PBS in place of plasma and 80 μl of PBS in place of the Sudan Black B stain. Samples were then centrifuged under the conditions described. Following centrifugation, samples were fractionated into 200 μl fractions using a Labonco Autodensiflow and Gilson FC203P fraction collector. The refractive index of each fraction was determined using a Bellingham Stanley DR-103 digital refractometer and converted to density using the following formula:
where a = 3.4193, b = 3.56, η = refractive index, and ρ = density. A blank tube was marked at 200 μl intervals corresponding to the fractions collected; each fraction was then assigned a density value. Nonstained plasma fractions were used to determine the apolipoprotein profile of the lipoprotein separation and to confirm the nature and position of the HDL classes using GGE.
Generation of HDL subclass profiles by digital photography
HDL subclass profiles were generated using digital photography coupled with Total Lab 1D gel scan software (Pharmacia, UK). Immediately after centrifugation, Optiseal tubes containing lipid-stained HDL bands were photographed against a vertical light box using a Nikon D1× digital camera set at the highest resolution. All photographs were taken with the camera set at a fixed distance from the rack housing the Optiseal tubes. Photographs were downloaded to a PC and evaluated using Total Lab 1D gel scan software. The gel scan software converted the photographs of the stained HDL bands into HDL profiles with an x axis of distance (mm) against a y axis of pixel intensity. The gel scan software was then used to assign relative electrophoretic migration distance (Rf) values to the HDL peaks. The peak Rf values were converted to density by means of cross-reference to a photograph of the blank tube with calibrated density intervals. The gel scan software also calculated areas under the curve for each HDL peak.
Gradient gel electrophoresis
HDL subclasses were coisolated by GGE using precast, nondenaturing acrylamide gradient gels (4–30%) using a pore gradient gel lipoprotein electrophoresis system (GGE, C.B.S. Scientific). Whole plasma- and relevant IxDGUC gradient fractions (50 μl) were prestained 1:1 with a 2% w/v solution of Sudan Black B in ethylene glycol. The samples (20 µl) were loaded onto precast 4–30% acrylamide gels that had been equilibrated at 70 V, 65 mA for 30 min. Gels were then run for 20 min at 20 V, 50 mA, followed by 30 min at 70 V, 65 mA, and finally for 24 h at 120 V, 100 mA. Following electrophoresis, gels were directly photographed and analyzed using TotaLab 1D software, with no further staining required.
Additionally, HDL subclasses were identified using a protein stain following the initial separation of lipoproteins by flotation to remove interfering proteins present in whole plasma. Whole plasma was adjusted to a density of 1.21 kg/l by the addition of KBr(s); 0.326 g of KBr was added to 1 ml of plasma and transferred to a 12 ml Beckman Ultraclear centrifuge tube. Density-adjusted plasma was mixed with 11 ml of a 1.21 kg/l KBr, 1% EDTA density solution. Tubes were housed in a Beckman 70.1 Ti rotor and centrifuged for 24 h at 117,734 g(av) at 15°C. Following centrifugation, the upper yellow supernatant (lipoprotein top) was aspirated. Bromophenol Blue was added as a color marker to the lipoprotein fraction (100 µl) and to relevant IxDGUC gradient fractions to give a final concentration of 5% w/v. Ten microliters of sample was then loaded onto a precast 4–30% acrylamide gel that had been equilibrated and then run under the same conditions as previously described. Following electrophoresis, gels were removed and fixed with a solution of 10% w/v sulphosalicylic acid for 30 min. The fixative was poured off, and the gels were immersed in protein stain (0.1% w/v Coomassie Blue R250, methanol, glacial acetic acid, RO water in a 5:1:4 ratio) for 1 h. Gels were destained with a solution of methanol:glacial acetic acid:RO water (50:75:875 ml) for 24 h. Gels were then photographed using a Nikon D1× digital camera and analyzed using TotaLab software.
Analysis of HDL fractions and plasma lipids
Whole plasma was analyzed for cholesterol, TAG, LDL-C, HDL-C, and apoA-I using commercially available assays on a SpACE automated analyzer (Schipparelli Biosystems, NJ). Additionally, apo A-I was measured in each 200 μl unstained fraction.
Analytical performance
Blood samples (60 ml) were taken from two fasted volunteers to examine the precision of the HDL separation both within and between rotors. To examine intra- (within) rotor variability, 10 replicate profiles were prepared for each participant, and an additional 10 replicates were prepared from the same individuals in a separate run to examine inter- (between) rotor variability. Within rotor variability was also examined using 4 replicate HDL profiles for eight participants (four male, four female) with a range of lipid profiles.
Statistics
All data were analyzed using SPSS version 14 (SPSS Inc., IL). All data were checked for normality using the Kolmogorov-Smirnov test; data that were not normally distributed were log- or square root-transformed. Differences between groups were assessed by one-way ANOVA. Associations of plasma lipids with HDL subclasses were assessed using Pearson's correlation.
RESULTS
Gradient characteristics
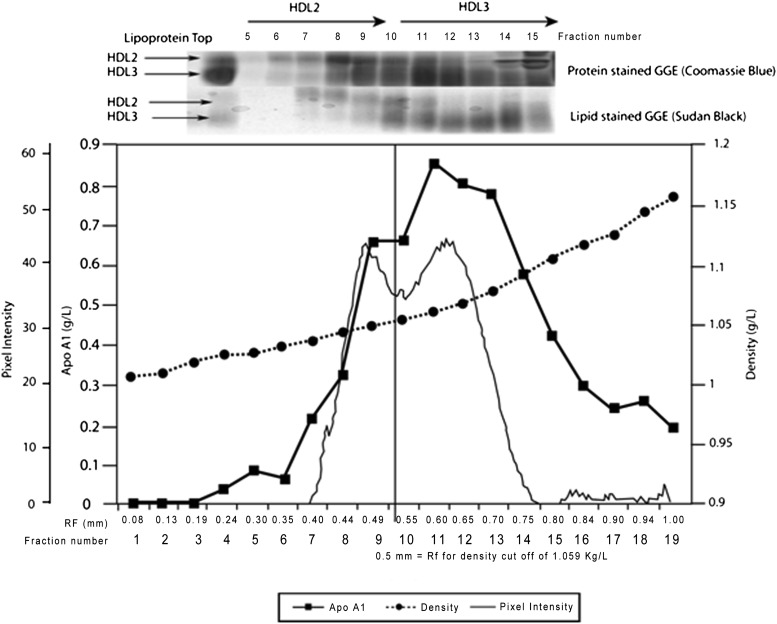
The iodixanol gradient was curvilinear with an extended linear region between fractions 1 and 14 (Fig. 1). The apoA-I profile showed a bimodal distribution that corresponded to the HDL2 and HDL3 subclasses, as indicated by both IxDGUC and GGE. Iodixanol profiles showed a peak corresponding to LDL (fractions 3–6, density 1.022–1.034 kg/l) and peaks corresponding to HDL subclasses (fractions 8–13, density 1.046–1.089 kg/l). The cutoff density and corresponding Rf for HDL2 was determined by the separation of the sample using IxDGUC and the separation of both lipid- and protein-stained fractions on 4–30% GGE. The results from the unstained fractionated tubes showed the density cutoff between HDL2 and HDL3 to be ∼1.059 kg/l. This value was then used in the analysis of HDL subclass distribution of 46 participants, displaying a variety of profiles.
Fig. 1.
Density gradient and distribution of plasma apolipoproteins. An unstained plasma sample was fractionated, in triplicate, into equal fractions of 200 μl. ApoA-I and density profiles are shown together with GGE (4–30%) for fractions 5–15. The HDL profile generated by IxDGUC and TotaLab analysis is shown as a solid line. The apoA-I profile was bimodal and corresponded to the HDL subclass profile generated by IxDGUC and with the fractions run using GGE. Delineation occurred at a density of ∼1.059 kg/l, corresponding to an Rf of 0.52 mm.
HDL heterogeneity in iodixanol
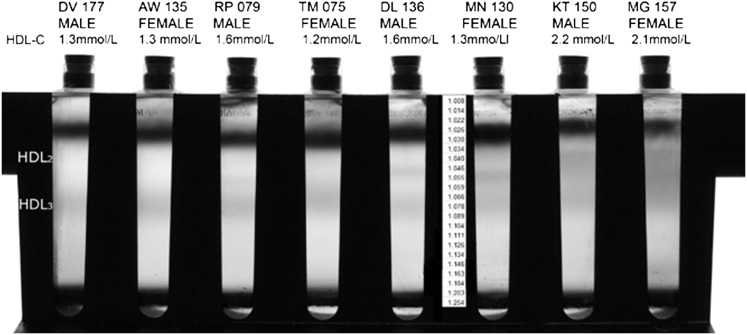
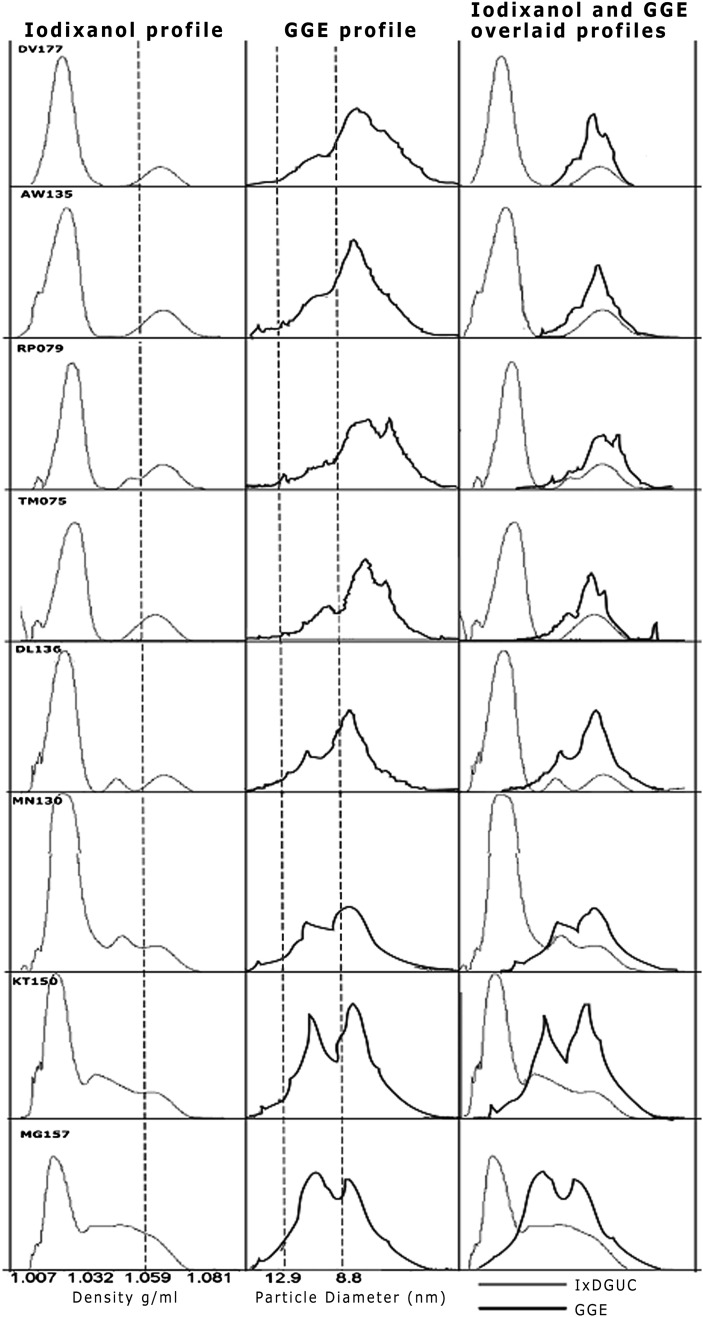
HDL subclasses separated from prestained plasma displayed variability in banding patterns both within and between participants. Shown in Figs. 2 and 3 are the HDL subclasses separated from the prestained plasma of eight individuals by IxDGUC and by GGE. The clear banding patterns within individuals indicated that IxDGUC resolves structural heterogeneity in HDL that corresponds to that achieved with electrophoresis. HDL profiles obtained by GGE and IxDGUC for these eight individuals were generally comparable, but there were some subtle differences. Profiles generated by GGE showed pronounced points of inflection or “shoulders” on the HDL2 subclass in participants with low HDL-C; these shoulders were not observed using IxDGUC.
Fig. 2.
HDL subclass profiles for eight individuals with varying HDL subclass distributions. The central panel denotes the density associated with Rf. There was clear evidence of variability in HDL subclass profile with HDL2, visible in both males and females.
Fig. 3.
IxDGUC and GGE profiles for eight participants with varying total HDL-C. Density profiles from IxDGUC are shown on the left, and the center column shows GGE lipid-stained profiles for the same participants. Particle size was determined using a set of protein standards of known molecular mass and diameter to calibrate the gel. The column on the right depicts overlaid IxDGUC and GGE profiles.
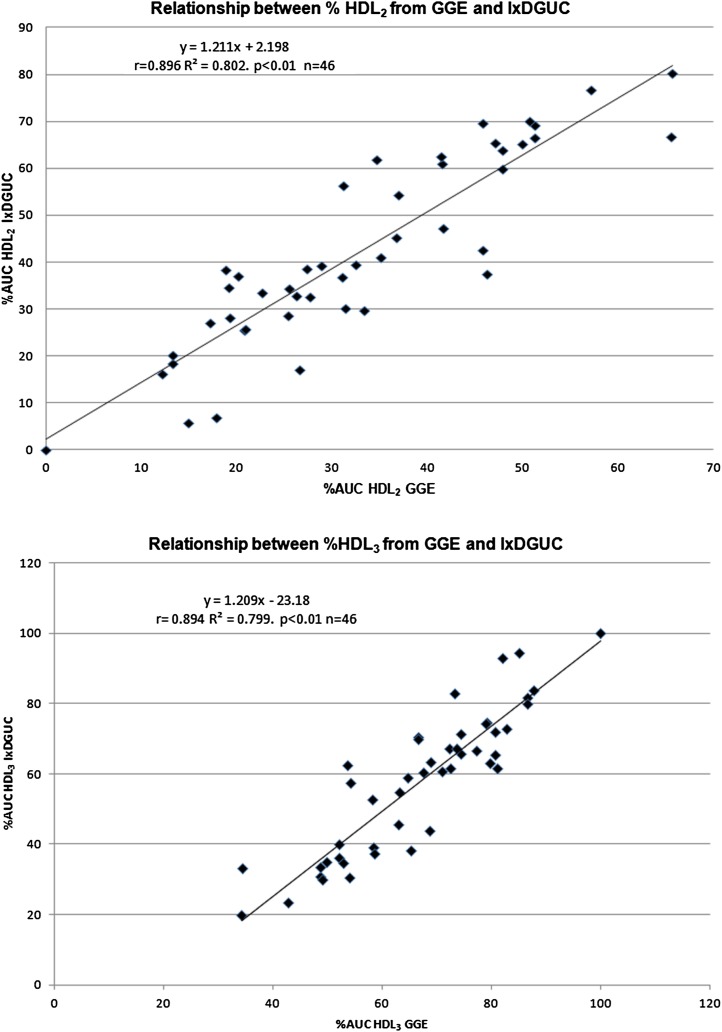
The percentage area under the curve (%AUC) for both HDL2 and HDL3 was determined for 46 participants using a density cutoff of 1.059 kg/l. The %AUC for each subclass was compared with those of GGE for the same samples. There was a significant correlation between HDL subclasses for both GGE (lipid-stained) and IxDGUC methods as measured by AUC (HDL2, r = 0.896, P < 0.01; HDL3, r = 0.894, P < 0.01) (Fig. 4).
Fig. 4.
Relationship of %AUC generated by GGE and IxDGUC. % HDL2 and % HDL3 generated by IxDGUC was significantly correlated with the %AUC as determined by GGE. [HDL2, r = 0.896 (r2 = 80.2%) P < 0.01; HDL3, r = 0.894 (r2 = 79.9%) P < 0.01].
Reproducibility of HDL separation on iodixanol gradient
Within-rotor variability.
The within-rotor coefficient of variation (CV) for %AUC HDL2 for two different individuals (10 samples/rotor) were 2.5% (HDL2 = 46%, density = 1.046 kg/l) and 3.93% (HDL2 = 14%, density = 1.049 kg/l). Within-rotor variability calculated from replicate (×4) HDL profiles taken from four males and four females was less than 4% and less than 1% for HDL2 (% AUC) and HDL2 peak density, respectively (Table 1).
TABLE 1.
Within rotor variation
| Participant | Gender | Total HDL-C (mmol/l) | %HDL2 (AUC) | HDL2 Peak Density (kg/l) | CV %HDL2 (AUC) | CV HDL2 %Peak Density |
| DV 177 | Male | 1.3 | 5.59 | 1.055 | 3.77 | 0.06 |
| AW 135 | Female | 1.3 | 25.86 | 1.052 | 3.43 | 0.25 |
| RP 079 | Male | 1.6 | 28.14 | 1.050 | 1.55 | 0.03 |
| TM 075 | Female | 1.2 | 32.87 | 1.050 | 2.13 | 0.08 |
| DL 136 | Male | 1.6 | 38.62 | 1.043 | 3.98 | 0.03 |
| MN 130 | Female | 1.3 | 62.70 | 1.045 | 1.21 | 0.10 |
| KT 150 | Male | 2.2 | 69.59 | 1.035 | 1.73 | 0.14 |
| MG 157 | Female | 2.1 | 70.12 | 1.032 | 1.10 | 0.10 |
Within rotor variation for eight participants (four males and four females) with a range of HDL patterns and total HDL concentrations.
Between-rotor variability.
There was a nonsignificant variation in %HDL2 and peak density values from two individuals (peak densities 1.046 kg/l and 1.064 kg/l) between rotors [2 × 10 samples mean (SD) %HDL2 = 46.88 (1.17) versus 45.96 (1.15), CV = 2.64%; and 14.36 (0.44) versus 14.52 (0.54), CV = 3.73%). The CV for HDL peak density was 0.09% (density = 1.046 kg/l) and 0.07% (density = 1.064 kg/l).
Relationship of HDL subclasses with other lipid risk markers
The relationship between the %AUC and peak density for HDL2 were compared with other markers of cardiovascular risk, including plasma total cholesterol, plasma TAG, LDL-C, HDL-C, and apoA-I in 548 participants. Percentage AUC of HDL2 was positively correlated with total HDL-C and apoA-I, and it was negatively correlated with plasma TAG and LDL-C (Table 2). There was no association with plasma total cholesterol.
TABLE 2.
Correlation of %AUC HDL2 with plasma lipids and apoA-I
| Plasma Lipid/Apolipoprotein | Pearson Correlation (r) with %AUC HDL2 | Pearson Correlation (P) |
| T-CHOL (mmol/l) | −0.41 | NS |
| TAG (mmol/l) | −0.348 | <0.01 |
| HDL-C (mmol/l) | 0.571 | <0.01 |
| LDL-C (mmol/l) | −0.149 | <0.01 |
| ApoA-I (g/l) | 0.328 | <0.01 |
Plasma lipids and lipoproteins for 548 participants were correlated with % HDL2 measured by IxDGUC. Significant correlations were observed for TAG, HDL-C, LDL-C, and apoA-I.
Unlike LDL, there is no currently accepted phenotypic pattern for HDL subclasses. K-means cluster analysis of %HDL2 was used to identify a predefined number of clusters in the data set (Table 3). In an attempt to determine HDL subclass patterns associated with low, medium, and high %HDL2, three clusters were chosen. Cluster analysis of 548 samples defined the three cluster centers at 18.4, 38.5, and 62.4 %HDL2. For ease of use, these clusters were grouped according to low (I), medium (II), and high (III) patterns according to >60, 40–60, and <40 %HDL2, respectively. Plasma lipid profiles were completed for all participants, and when grouped according to subclass pattern, significant differences were observed between groups for plasma TAG, HDL-C, LDL-C, and apoA-I. Participants grouped into the low %HDL2 pattern (I) had significantly lower TAG and LDL-C (P < 0.001) and significantly higher HDL-C and apoA-I (P < 0.01) compared with medium and high %HDL2 groups. There was no difference between groups for total cholesterol.
TABLE 3.
Mean plasma lipid/lipoprotein concentration according to HDL subclass pattern (IxDGUC %HDL2)
| Plasma Lipid/Apolipoprotein | Pattern I | Pattern II | Pattern III | One-way ANOVA between Groups (P) |
| >60% | 40–60% | <40% | ||
| N = 105 | N = 161 | N = 282 | ||
| T-CHOL (mmol/l) | 5.63 | 5.60 | 5.62 | NS |
| TAG (mmol/l) | 1.18 | 1.37 | 1.68 | <0.0001 |
| HDL-C (mmol/l) | 1.70 | 1.43 | 1.23 | <0.0001 |
| LDL-C (mmol/l) | 3.40 | 3.54 | 3.63 | 0.034 |
| ApoA-I (g/l) | 1.38 | 1.22 | 1.13 | <0.0001 |
Samples were grouped into three clusters according to %HDL2. Significant differences were observed between groups for TG, total HDL-C, LDL-C, and apoA-I. The mean values for T-CHOL, TAG, HDL-C, LDL-C, and apoA-I are presented for each of the HDL subclass patterns.
DISCUSSION
While methods for the measurement for HDL subclasses are labor intensive, time consuming, and expensive, some of the latest techniques, including VAP-II auto-profiling and proton NMR, have overcome these problems, but they are not without disadvantages. The VAP-II autoprofiling system has reduced ultracentrifugation time to 47 min, although the method requires analysis of the cholesterol content of the gradient followed by the application of complex equations to quantify HDL subclasses (25). Proton NMR was described by two different groups in the early 1990s (26, 27). NMR has the advantage of requiring a small sample volume and being capable of high sample throughput; however, the equipment is expensive and the method cannot be used preparatively. Iodixanol is a density media that is iso-osmotic and nontoxic to cells; the use of iodixanol in two-step gradients was employed by Graham et al. (19) to separate lipoprotein classes. Sawle et al. (28) also used IxDGUC coupled with fractionation to identify the major lipoprotein classes as well as the principal LDL subclasses. In 2003, Davies et al. (22) developed this method further by designing a gradient that would specifically identify LDL subclasses without the need for fractionation by using digital photography to generate LDL profiles. Most recently, Yee at al. (29) have described a method for the separation of the major lipoproteins and subclasses of LDL; however, this technique did not discriminate between the principal HDL subclasses and, furthermore, required fractionation of the sample tube for the identification of lipoproteins. Therefore, the aim of the present study was to develop a rapid technique for both the identification and preparation of the principal HDL subclasses using iodixanol as a density media. The gradient described is a three-step gradient, in which prestained plasma is “sandwiched” between two solutions of iodixanol with differing densities; this permits the separation and identification of the principal HDL subclasses in a run time of 2.5 h. Previous methods have used ultracentrifugal separations followed by an elution of the gradient and continuous spectrophotometric detection of the separated fractions to generate lipid profiles. Davies et al. (22) used digital photography followed by gel scan software to generate LDL subclass profiles. The use of digital photography eliminates the need for time-consuming fractionation and improves the reproducibility of the method; therefore, it was adopted in our study for the identification of HDL subclasses. To allow identification of the HDL subclasses, plasma was prestained with Sudan Black B at physiological pH. This technique produced profiles displaying HDL heterogeneity that was visible within both males and females, across a range of total plasma HDL-C concentrations (Fig. 2).
The percentage of the principal HDL subclasses as measured by the AUC correlated well with the established GGE methods (Fig. 4). Profiles were on the whole very similar; however, IxDGUC detected less HDL2 in participants with low HDL-C and predominantly small HDL3 compared with lipid-stained GGE. This may be, in part, due to the molecular sieving effect on electrophoretic gels that produces a greater resolution of HDL subclasses by GGE. Nevertheless, a spectrum of samples was analyzed using IxDGUC and results were found to correlate significantly with those of GGE.
To date, no classification of HDL subclasses has been defined in terms of CVD risk. LDL subclasses, on the other hand, can be categorized into LDL phenotypes. Three major patterns exist and are distinguished based upon the percentage of each LDL subclasses present (30). sdlDL becomes the predominant subclass when plasma TAG concentrations exceed 1.5 mmol/l (31); the classification of LDL subclasses into phenotypes has been shown to have clinical utility as a risk marker, with LDL subclass pattern B being associated with a 3-fold increase in CVD risk (32). A similar classification for HDL has been proposed by Rosenson et al. (33) from a range of diverse methods; the proposal suggests classifying HDL heterogeneity into five categories, ranging from very small HDL to very large HDL. We agree that a standardized nomenclature would improve the ability to predict cardiovascular risk within these categories, and our study adds to the literature by assessing three distinct HDL subclass patterns in relation to other lipid risk markers. Participants were separated according to the %HDL2 into the three patterns using K-means cluster analysis. When separated according to these patterns, participants exhibited significant differences between total HDL-C, apoA-I, and TAG (Table 3). The elevated TAG in pattern III (mean 1.68 mmol/l) supports previous observations of an inverse relationship between TAG and the abundance of HDL2 (34–36), and it suggests that elevated TAG not only increases the predominance of sdlDL but also that of smaller, dense HDL3. Although the three patterns of HDL subclasses displayed differences in some lipids and apolipoproteins, there were no significant differences between groups for total cholesterol, supporting previous evidence that shows an increase in the prevalence of sdlDL at TAG concentrations greater than 1.5 mmol/l with no change in total cholesterol concentration (37). Austin et al. (38) defined an atherogenic lipoprotein phenotype (ALP) as a cluster of elevated plasma TAG, low HDL-C, and predominance of sdlDL. The correlation between HDL2 plasma TAG and sdlDL suggests that a predominance of HDL3 should also be included in the definition. Without a definitive classification of HDL phenotypes, the validation of IxDGUC in terms of its ability to predict increased CVD risk is limited.
In conclusion, IxDGUC is a reproducible method with a considerable saving in ultracentrifugation time. The centrifugation parameters used are the same as those described for the NVT 65.2 and separation of LDL subclasses, thus giving the added advantage of being able to determine both LDL and HDL subclass profiles simultaneously. This method should be advantageous in determining CVD risk, particularly in groups with “normal” concentrations of total plasma and LDL cholesterol whose increased CVD risk may be overlooked. This method provides a time-saving and cost-effective analytical procedure for the detection of the principal HDL subclasses, and it can also be used preparatively to isolate HDL2 and HDL3.
Footnotes
Abbreviations:
- AUC
- area under the curve
- CV
- coefficient of variation
- CVD
- cardiovascular disease
- GGE
- gradient gel electrophoresis
- IxDGUC
- iodixanol density gradient ultracentrifugation
- TAG
- triacylglycerol
Funding for this study was provided by the Food Standards Agency, UK.
REFERENCES
- 1.Gordon T., Castelli W. P., Hjortland M. C., Kannel W. B., Dawber T. R. 1977. High-density lipoprotein as a protective factor against coronary heart-disease. The Framingham Study. Am. J. Med. 62: 707–714 [DOI] [PubMed] [Google Scholar]
- 2.Castelli W. P., Garrison R. J., Wilson P. W., Abbott R. D., Kalousdian S., Kannel W. B. 1986. Incidence of coronary heart-disease and lipoprotein cholesterol levels. The Framingham-Study. JAMA. 256: 2835–2838 [PubMed] [Google Scholar]
- 3.Brown B. G., Zhao X. Q., Chait A., Fisher L. D., Cheung M. C., Morse J. S., Dowdy A. A., Marino E. K., Bolson E. L., Alaupovic P., et al. 2001. Simvastatin and niacin, antioxidant vitamins, or the combination for the prevention of coronary disease. N. Engl. J. Med. 345: 1583–1592 [DOI] [PubMed] [Google Scholar]
- 4.Frick M. H., Elo O., Haapa K., Heinonen O. P., Heinsalmi P., Helo P., Huttunen J. K., Kaitaniemi P., Koskinen P., Manninen V., et al. 1987. Helsinki Heart-Study: primary-prevention trial with gemfibrozil in middle-aged men with dyslipidemia. Safety of treatment, changes in risk-factors, and incidence of coronary heart disease. N. Engl. J. Med. 317: 1237–1245 [DOI] [PubMed] [Google Scholar]
- 5.Barter P. J., Caulfield M., Eriksson M., Grundy S. M., Kastelein J. J., Komajda M., Lopez-Sendon J., Mosca L., Tardif J. C., Waters D. D., et al. 2007. Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med. 357: 2109–2122 [DOI] [PubMed] [Google Scholar]
- 6.Otocka-Kmiecik A., Mikhailidis D. P., Nicholls S. J., Davidson M., Rysz J., Banach M. 2012. Dysfunctional HDL: a novel important diagnostic and therapeutic target in cardiovascular disease? Prog. Lipid Res. 51: 314–324 [DOI] [PubMed] [Google Scholar]
- 7.Drexel H., Amann F. W., Rentsch K., Neuenschwander C., Luethy A., Khan S. I., Follath F. 1992. Relation of the level of high-density-lipoprotein subfractions to the presence and extent of coronary-artery disease. Am. J. Cardiol. 70: 436–440 [DOI] [PubMed] [Google Scholar]
- 8.Asztalos B. F., Collins D., Cupples L. A., Demissie S., Horvath K. V., Bloomfield H. E., Robins S. J., Schaefer E. J. 2005. Value of high-density lipoprotein (HDL) subpopulations in predicting recurrent cardiovascular events in the Veterans Affairs HDL Intervention Trial. Arterioscler. Thromb. Vasc. Biol. 25: 2185–2191 [DOI] [PubMed] [Google Scholar]
- 9.Salonen J. T., Salonen R., Seppänen K., Rauramaa R., Tuomilehto J. 1991. HDL, HDL2, and HDL3 subfractions, and the risk of acute myocardial-infarction. A prospective population study in eastern Finnish men. Circulation. 84: 129–139 [DOI] [PubMed] [Google Scholar]
- 10.Williams P. T., Haskell W. L., Vranizan K. M., Krauss R. M. 1995. The associations of high-density-lipoprotein subclasses with insulin-levels and glucose-levels, physical-activity, resting heart-rate, and regional adiposity in men with coronary-artery-disease: the Stanford Coronary Risk Intervention Project baseline survey. Metabolism. 44: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Superko H. R., Pendyala L., Williams P. T., Momary K. M., King S. B., 3rd, Garrett B. C. 2012. High-density lipoprotein subclasses and their relationship to cardiovascular disease. J. Clin. Lipidol. 6: 496–523 [DOI] [PubMed] [Google Scholar]
- 12.Buring J. E., O'Connor G. T., Goldhaber S. Z., Rosner B., Herbert P. N., Blum C. B., Breslow J. L., Hennekens C. H. 1992. Decreased HDL2 and HDL3 cholesterol, Apo A-I and Apo A-II, and increased risk of myocardial infarction. Circulation. 85: 22–29 [DOI] [PubMed] [Google Scholar]
- 13.Sweetnam P. M., Bolton C. H., Yarnell J. W., Bainton D., Baker I. A., Elwood P. C., Miller N. E. 1994. Associations of the HDL2 and HDL3 cholesterol subfractions with the development of ischemic heart disease in British men. The Caerphilly and Speedwell Collaborative Heart Disease Studies. Circulation. 90: 769–774 [DOI] [PubMed] [Google Scholar]
- 14.Yu S., Yarnell J. W., Sweetnam P., Bolton C. H. 2003. High density lipoprotein subfractions and the risk of coronary heart disease: 9-years follow-up in the Caerphilly Study. Atherosclerosis. 166: 331–338 [DOI] [PubMed] [Google Scholar]
- 15.Chung M., Lichtenstein A. H., Ip S., Lau J., Balk E. M. 2009. Comparability of methods for LDL subfraction determination: a systematic review. Atherosclerosis. 205: 342–348 [DOI] [PubMed] [Google Scholar]
- 16.Kontush A., Chantepie S., Chapman M. J. 2003. Small, dense HDL particles exert potent protection of atherogenic LDL against oxidative stress. Arterioscler. Thromb. Vasc. Biol. 23: 1881–1888 [DOI] [PubMed] [Google Scholar]
- 17.Groot P. H., Scheek L. M., Havekes L., van Noort W. L., van't Hooft F. M. 1982. A one-step separation of human serum high density lipoproteins 2 and 3 by rate-zonal density gradient ultracentrifugation in a swinging bucket rotor. J. Lipid Res. 23: 1342–1353 [PubMed] [Google Scholar]
- 18.Chung B. H., Segrest J. P., Cone J. T., Pfau J., Geer J. C., Duncan L. A. 1981. High resolution plasma lipoprotein cholesterol profiles by a rapid, high volume semi-automated method. J. Lipid Res. 22: 1003–1014 [PubMed] [Google Scholar]
- 19.Graham J. M., Higgins J. A., Gillott T., Taylor T., Wilkinson J., Ford T., Billington D. 1996. A novel method for the rapid separation of plasma lipoproteins using self-generating gradients of iodixanol. Atherosclerosis. 124: 125–135 [DOI] [PubMed] [Google Scholar]
- 20.Ford T., Graham J., Rickwood D. 1994. Iodixanol: a nonionic iso-osmotic centrifugation medium for the formation of self-generated gradients. Anal. Biochem. 220: 360–366 [DOI] [PubMed] [Google Scholar]
- 21.Murdoch S. J., Breckenridge W. C. 1994. Development of a density gradient ultracentrifugation technique for the resolution of plasma lipoproteins which avoids apo E dissociation. Anal. Biochem. 222: 427–434 [DOI] [PubMed] [Google Scholar]
- 22.Davies I. G., Graham J. M., Griffin B. A. 2003. Rapid separation of LDL subclasses by iodixanol gradient ultracentrifugation. Clin. Chem. 49: 1865–1872 [DOI] [PubMed] [Google Scholar]
- 23.Swinkels D. W., Haklemmers H. L. M., Demacker P. N. M. 1987. Single spin-density gradient ultracentrifugation method for the detection and isolation of light and heavy low-density-lipoprotein subfractions. J. Lipid Res. 28: 1233–1239 [PubMed] [Google Scholar]
- 24.Terpstra A. H. M., Woodward C. J. H., Sanchez-Muniz F. J. 1981. Improved techniques for the separation of serum lipoproteins by density gradient ultracentrifugation: visualization by prestaining and rapid separation of serum lipoproteins from small volumes of serum. Anal. Biochem. 111: 149–157 [DOI] [PubMed] [Google Scholar]
- 25.Kulkarni K. R., Marcovina S. M., Krauss R. M., Garber D. W., Glasscock A. M., Segrest J. P. 1997. Quantification of HDL2 and HDL3 cholesterol by the Vertical Auto Profile-II (VAP-II) methodology. J. Lipid Res. 38: 2353–2364 [PubMed] [Google Scholar]
- 26.Otvos J. D., Jeyarajah E. J., Bennett D. W., Krauss R. M. 1992. Development of a proton nuclear-magnetic-resonance spectroscopic method for determining plasma-lipoprotein concentrations and subspecies distributions from a single, rapid measurement. Clin. Chem. 38: 1632–1638 [PubMed] [Google Scholar]
- 27.Ala-Korpela M., Korhonen A., Keisala J., Hörkkö S., Korpi P., Ingman L. P., Jokisaari J., Savolainen M. J., Kesäniemi Y. A. 1994. 1H NMR-based absolute quantitation of human lipoproteins and their lipid contents directly from plasma. J. Lipid Res. 35: 2292–2304 [PubMed] [Google Scholar]
- 28.Sawle A., Higgins M. K., Olivant M. P., Higgins J. A. 2002. A rapid single-step centrifugation method for determination of HDL, LDL, and VLDL cholesterol, and TG, and identification of predominant LDL subclass. J. Lipid Res. 43: 335–343 [PubMed] [Google Scholar]
- 29.Yee M. S., Pavitt D. V., Tan T., Venkatesan S., Godsland I. F., Richmond W., Johnston D. G. 2008. Lipoprotein separation in a novel iodixanol density gradient, for composition, density, and phenotype analysis. J. Lipid Res. 49: 1364–1371 [DOI] [PubMed] [Google Scholar]
- 30.Griffin B. A., Caslake M. J., Yip B., Tait G. W., Packard C. J., Shepherd J. 1990. Rapid isolation of low density lipoprotein (LDL) subfractions from plasma by density gradient ultracentrifugation. Atherosclerosis. 83: 59–67 [DOI] [PubMed] [Google Scholar]
- 31.Griffin B. A., Freeman D. J., Tait G. W., Thomson J., Caslake M. J., Packard C. J., Shepherd J. 1994. Role of plasma triglyceride in the regulation of plasma low-density-lipoprotein (LDL) subfractions: relative contribution of small, dense ldl to coronary heart-disease risk. Atherosclerosis. 106: 241–253 [DOI] [PubMed] [Google Scholar]
- 32.Austin M. A., Breslow J. L., Hennekens C. H., Buring J. E., Willet W. C., Krauss R. M. 1988. Low density lipoprotein subclass patterns and risk of myocardial infarction. JAMA. 260: 1917–1921. [PubMed] [Google Scholar]
- 33.Rosenson R. S., Brewer H. B., Chapman M. J., Fazio S., Hussain M. M., Kontush A., Krauss R. M., Otvos J. D., Remaley A. T., Schaefer E. J. 2011. HDL measures, particle heterogeneity, proposed nomenclature, and relation to atherosclerotic cardiovascular events. Clin. Chem. 57: 392–410 [DOI] [PubMed] [Google Scholar]
- 34.Syvänne M., Ahola M., Lahdenperä S., Kahri J., Kuusi T., Virtanen K. S., Taskinen M. R. 1995. High density-lipoprotein subfractions in non-insulin-dependent diabetes mellitus and coronary artery disease. J. Lipid Res. 36: 573–582 [PubMed] [Google Scholar]
- 35.Xu Y., Fu M. 2003. Alterations of HDL subclasses in hyperlipidemia. Clin. Chim. Acta. 332: 95–102 [DOI] [PubMed] [Google Scholar]
- 36.Yang Y., Yan B., Fu M., Xu Y., Tian Y. 2005. Relationship between plasma lipid concentrations and HDL subclasses. Clin. Chim. Acta. 354: 49–58 [DOI] [PubMed] [Google Scholar]
- 37.Griffin B. A., Minihane A. M., Furlonger N., Chapman C., Murphy M., Williams D., Wright J. J., Williams C. M. 1999. Inter-relationships between small, dense low-density lipoprotein (LDL), plasma triacylglycerol and LDL apoprotein B in an atherogenic lipoprotein phenotype in free-living subjects. Clin. Sci. (Lond.). 97: 269–276 [PubMed] [Google Scholar]
- 38.Austin M. A., King M. C., Vranizan K. M., Krauss R. M. 1990. Atherogenic lipoprotein phenotype. A proposed genetic marker for coronary heart disease risk. Circulation. 82: 495–506 [DOI] [PubMed] [Google Scholar]