Abstract
Click chemistry is evolving as a powerful tool in biological applications because it allows the sensitive and specific detection of compounds with alkyne or azido groups. Here we describe the use of alkyne lipids as substrates for in vitro enzymatic assays of lipid modifying enzymes. The small alkyne moiety is introduced synthetically at the terminus of the hydrocarbon chain of various substrate lipids. After the assay, the label is click-reacted with the azide-bearing fluorogenic dye 3-azido-7-hydroxycoumarin, followed by the separation of the lipid mix by thin-layer chromatography and fluorescence detection, resulting in high sensitivity and wide-range linearity. Kinetic analyses using alkyne-labeled substrates for lysophosphatidic acid acyltransferases, lysophosphatidylcholine acyltransferases, and ceramide synthases resulted in Michaelis-Menten constants similar to those for radiolabeled or natural substrates. We tested additional alkyne substrates for several hydrolases and acyltransferases in lipid metabolism. In this pilot study we establish alkyne lipids as a new class of convenient substrates for in vitro enzymatic assays.
Keywords: ceramide synthase, acyltransferase, fatty acid amide hydrolase, click lipid, click-labeling, lipid substrate, enzyme kinetics, in vitro assay
In vitro enzymatic assays are a major source of information about the characteristics of enzymes. In particular the determination of functional parameters relies on data in well-defined in vitro systems. Enzymes of lipid metabolism have been investigated with radiolabeled substrates for decades (1). They represent ideal tracers that display all the characteristics of the unlabeled compound, but their synthesis or use can be laborious, requires special equipment, is often subject to special regulations, and the quantification of signal can be tedious. In recent times, fluorescently tagged lipids have been developed as an alternative to radioactive labeling. They are convenient and sensitive reporters, but in general contain spacious tags of high impact on the structure, likely to influence specificity and kinetic parameters of enzymatic reactions. Although there are reports of assays with unchanged affinity of the enzyme to fluorescently labeled substrates, notably nitrobenzoxadiazole (NBD)-sphingolipids [e.g., NBD-sphinganine (2), NBD-sphingosine-1-phosphate (3)] the applicability of a substrate has to be validated thoroughly. For instance, when the boron dipyrromethene (BDP) analog, BDP-sphingosine-1-phosphate, was developed as a more photostable and hydrophobic alternative to NBD-sphingosine-1-phosphate, the BDP-, but not the NBD-derivate displayed a higher Km value than the natural substrate (4). A smaller nonradioactive universally applicable tag with less impact on the substrate structure would hence be beneficial.
Click chemistry (5), and especially the copper-catalyzed Huisgen 1,3-dipolar cycloaddition between a terminal alkyne group and an azide, provide new possibilities for the bioorthogonal labeling of molecules like amino acids (6), sugars (7), and nucleotides (8), which have already triggered many applications in life sciences (9, 10). In lipid biology, alkyne and azide analogs and precursors are already applied broadly, e.g., in protein lipidation analyses [reviewed in (11)], receptor binding studies (12), tracing of fatty acid metabolism (13), and the labeling of lipids in vivo followed by fluorescence imaging (14, 15). In addition to these applications, it is especially worthwhile to test the feasibility of click-based in vitro assays in lipid metabolism, for which other labeled substrates often are not readily at hand.
As the alkyne moiety is a small, neither overly hydrophobic nor hydrophilic label, a good affinity of enzymes to alkyne-modified substrates can be expected. For the first time this study addresses the usage of alkyne lipids in enzymatic assays through the determination of Michaelis-Menten kinetics for these substrates. Here we describe a click-based in vitro enzymatic assay to study enzymes of lipid metabolism, notably from the families of lysophosphatidic acid acyltransferases (LPAATs), lysophosphatidylcholine acyltransferases (LPCATs), and ceramide synthases (CerSs).
MATERIALS AND METHODS
Alkyne lipids
The chemical syntheses of the alkyne lipids are available in the supplementary material to this study, or were described previously (13).
Escherichia coli culture and preparation of microsomal fractions
E. coli (strain RosettaTM 2 pLysS, Merck Millipore) cultured in Luria Broth at 37°C to an OD600 of 1.6 were harvested by centrifugation at 3,000 g for 10 min. Microsomal fractions were prepared according to Lewin, Wang, and Coleman (16), frozen in liquid nitrogen and stored at −80°C. The protein concentration in the microsomes was determined with the BioRad Bradford assay kit using BSA as a standard.
Cell culture and preparation of cell lysate
HuH7 cells were grown in RPMI 1640 (PAN Biotech P04-17500) supplemented with 10 mM HEPES, 0.1 mM nonessential amino acids, 2 mM l-glutamine, 10% FBS, at 5% CO2. Cells were washed, scraped into ice-cold buffer (20 mM HEPES/NaOH, pH 7.0, 200 mM sucrose) and homogenized in a cooled EMBL cell cracker (HGM, Heidelberg, Germany) with five double strokes and a maximum clearance of 18 μm. The lysate was centrifuged at 500 g for 5 min at 4°C, the supernatant frozen in liquid nitrogen and stored at −80°C. The protein content of the lysate was determined with the bicinchoninic acid assay kit (Pierce) using BSA as a standard.
Animals and preparation of tissue microsomal fractions
Membrane fraction samples were prepared of the brain, liver, and kidneys of C57BL/6 wild-type (+/+) and CerS2-deficient mice (−/−) (one individual each, littermates, 13–14 weeks old) according to the protocol described by Imgrund et al. (17), frozen in liquid nitrogen and stored at −80°C. A total lysate of wild-type liver was also prepared. The protein content of the microsomal fractions, and of liver lysate was determined with the bicinchoninic assay kit (Pierce) using BSA as a standard.
Enzymatic assays
All assays were performed in 1.5 ml reaction tubes in a total volume of 100 μl. The reaction was started by addition of the prewarmed reaction mix to the enzyme preparation and the tubes were incubated in a heating block (Eppendorf Thermomixer comfort) under shaking (1,100 rpm). Incubation times and the amount of enzyme preparation were optimized for each assay (data not shown) to ensure substrate saturation and linearity of the reaction rate with regard to time and enzyme amount.
LPAAT assay
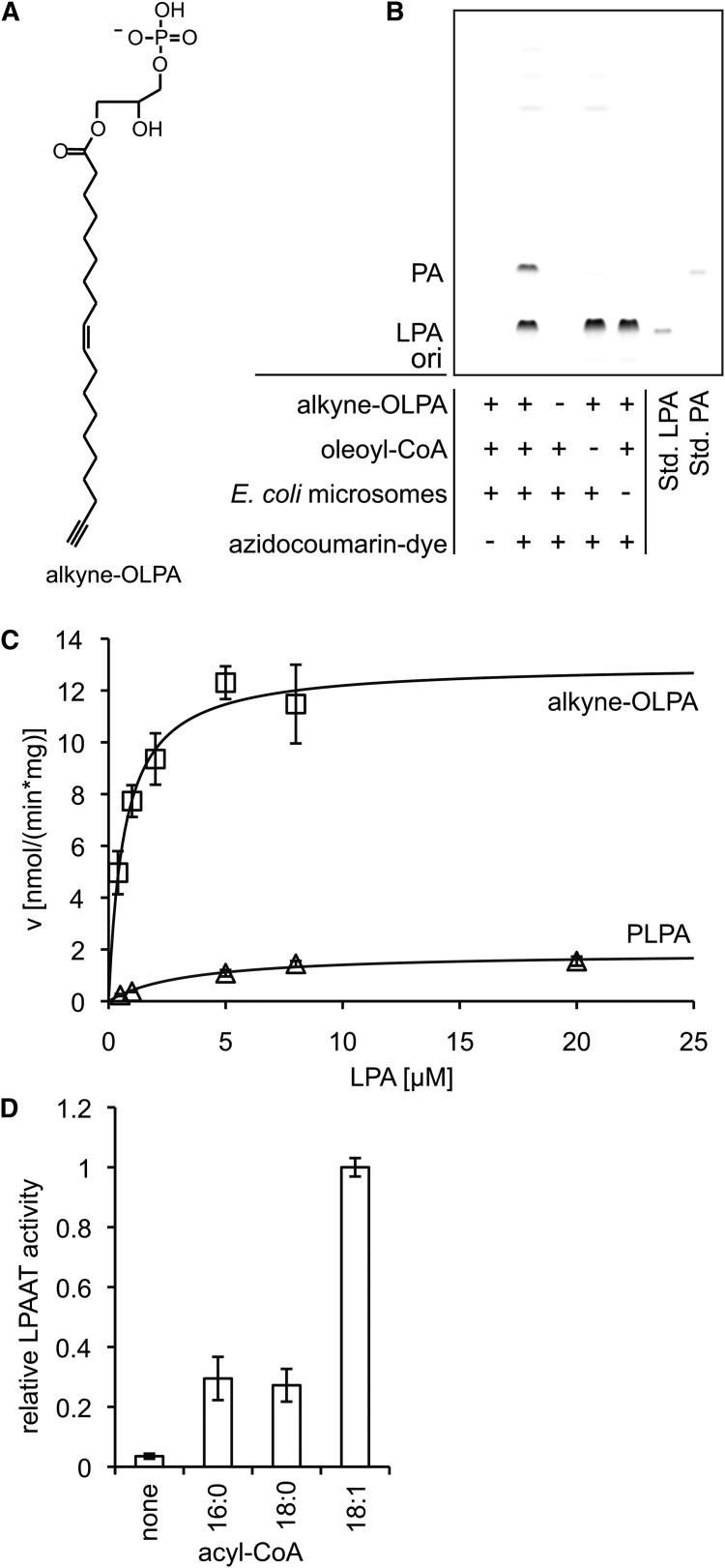
For the kinetic studies, different concentrations of 1-(nonadec-9-cis-en-18-ynoyl)-sn-glycero-3-phosphate (alkyne-OLPA) and 50 μM oleoyl-CoA, or various concentrations of 1-palmitoyl-lysophosphatidic acid (PLPA) and 50 μM nonadec-9-cis-en-18-ynoyl CoA (alkyne-oleoyl-CoA) were incubated with 2.06 μg (protein) E. coli microsomes in LPAAT buffer (18) [60 mM Tris/HCl, pH 7.5, 3 mM MgCl2, 0.6 mg/ml lipid-free BSA (Applichem A0848)] for 5 min at 30°C. For the measurement of the acyl-CoA specificity (Fig. 2D, 5 min incubation) and the TLC displayed in the results section (Fig. 2C, 10 min incubation), 4 μM alkyne-OLPA and 100 μM acyl-CoA were used.
Fig. 2.
LPAAT assay using alkyne-OLPA or alkyne-oleoyl-CoA. Alkyne-OLPA [structure in panel (A)] or alkyne-oleoyl-CoA were used as labeled substrates in a LPAAT assay with E. coli microsomes. B: Fluorescent TLC image of the LPAAT assay with alkyne-OLPA. Product PA was identified using comigrating synthetic alkyne-OOPA. For assay details, see Materials and Methods. “ori” depicts the origin of the TLC. C: Michaelis-Menten kinetics measured for alkyne-OLPA (squares) and PLPA (triangles). Oleoyl-CoA was the acyl chain donor for alkyne-OLPA, whereas alkyne-oleoyl-CoA was used to detect enzymatic activity toward PLPA. Line graphs show the reaction rate (v) calculated with the Michaelis-Menten equation using the values for Vmax and Km obtained by nonlinear regression fitting. Data are mean ± SD of triplicate determinations. D: Acyl-CoA specificity of the E. coli LPAAT using alkyne-OLPA. The monounsaturated 18:1-CoA is favored over the saturated acyl-CoAs (16:0 and 18:0).
LPCAT assay
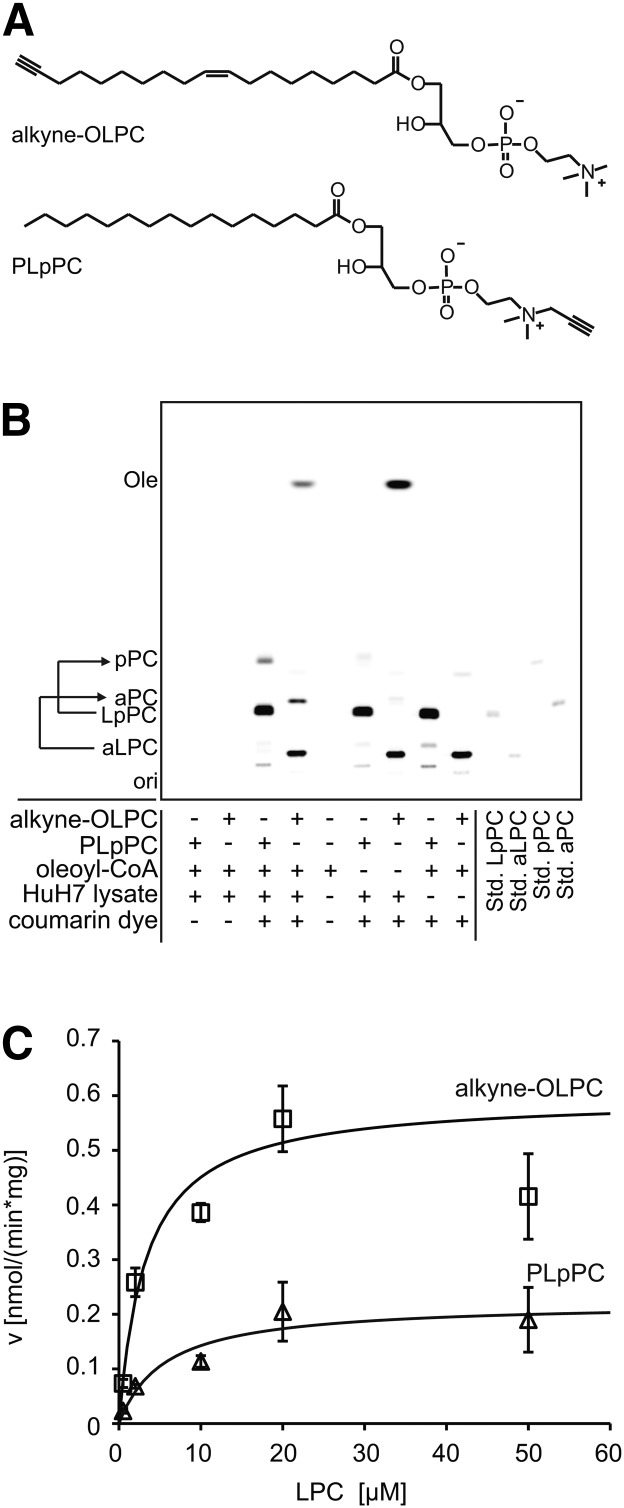
For the kinetic studies, different concentrations of 1-(nonadec-9-cis-en-18-ynoyl)-sn-glycero-3-phosphocholine (alkyne-OLPC) or palmitoyl-lyso-propargyl-phosphatidylcholine (PLpPC) and 150 μM oleoyl-CoA were incubated with 5 μg (protein) HuH7 lysate for 15 min at 30°C in LPCAT buffer (60 mM Tris/HCl, pH 7.4, 3 mM MgCl2, 0.6 mg/ml lipid-free BSA). For the TLC that shows the setup of the assay (Fig. 3B), 20 μM lysophosphatidylcholine, 50 μM oleoyl-CoA, and 5 μg (protein) HuH7 lysate were incubated for 30 min.
Fig. 3.
LPCAT assay using alkyne-OLPC or PLpPC. A: Structures of the substrates alkyne-OLPC (acyl chain-labeled) and PLpPC (headgroup-labeled). B: Fluorescent image of a TLC that shows a LPCAT assay using either alkyne-OLPC or PLpPC, oleoyl-CoA, and HuH7 cell lysate. For simplicity of the labeling of the TLC plate, we used aPC and aLPC for fatty acid-labeled PC and LPC, respectively, and pPC and LpPC for the headgroup-labeled analogs. Products were identified using comigrating synthetic standards (Std.). “ori” depicts the origin of the TLC. For assay details, see Materials and Methods. C: Michaelis-Menten kinetics measured for alkyne-OLPC (squares) and PLpPC (triangles). Line graphs show the reaction rate (v) calculated with the Michaelis-Menten equation using the values for Vmax and Km obtained by nonlinear regression fitting. Data are mean ± SD of triplicate determinations.
CerS assay
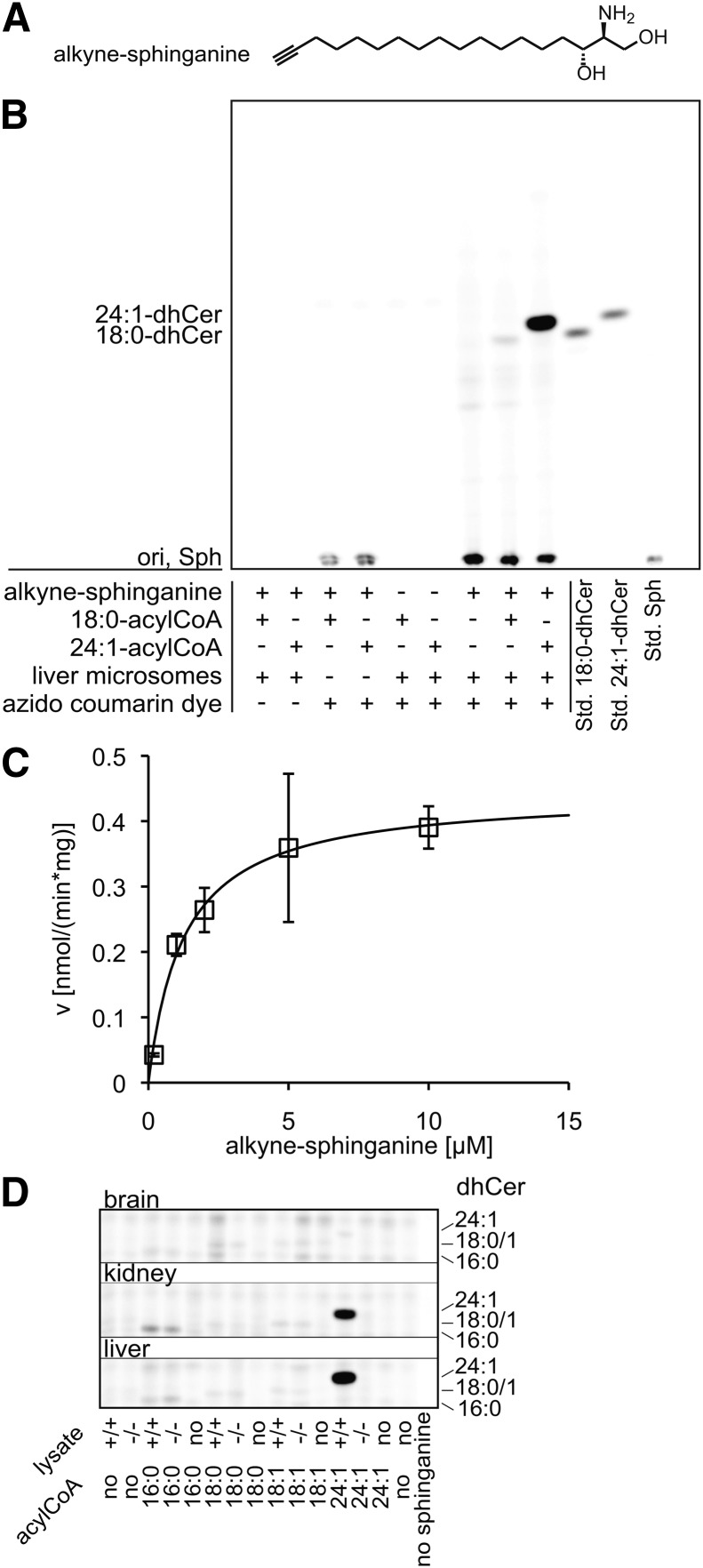
The kinetic measurements were performed with different concentrations of (2S,3R)-2-aminooctadec-17-yn-1,3-diol (alkyne-sphinganine), 50 μM nervonoyl(24:1)-CoA, and 5 μg (protein) mouse wild-type liver microsomes. The assay was incubated for 20 min at 37°C in CerS buffer (20 mM HEPES/KOH, pH 7.4, 25 mM KCl, 250 mM sucrose, 2 mM MgCl2, 0.34 mg/ml lipid-free BSA) (17). For the TLC that shows the setup of the assay (Fig. 4B), 20 μM alkyne-sphinganine, 50 μM nervonoyl- or stearoyl-CoA, and 20 μg (protein) liver wild-type microsomes were used. For screening various tissues for CerS activity (Fig. 4D), 10 μg (protein) brain, kidney, or liver microsomes of wild-type or CerS2−/− mice were incubated with 20 μM alkyne-sphinganine and 50 μM acyl-CoA.
Fig. 4.
CerS assay using alkyne-sphinganine. Alkyne-sphinganine [structure in panel (A)] was applied in a CerS assay using microsomes from mouse tissues. B: Fluorescent TLC image of the assay performed with liver microsomes from a C57BL/6 wild-type mouse. Products were identified using comigrating synthetic standards (Std.). “ori” depicts he origin of the TLC. For assay details, see Materials and Methods. C: Michaelis-Menten kinetics measured for alkyne-sphinganine (squares). The line graph shows the reaction rate (v) calculated with the Michaelis-Menten equation using the values for Vmax and Km obtained by nonlinear regression fitting. Data are mean ± SD of triplicate determinations. D: CerS activity toward the acyl-CoAs 16:0, 18:0, 18:1, and 24:1 was measured in microsomes from brain, kidney, and liver from wild-type (+/+) or CerS2 knockout (−/−) mice. The image shown here is slightly overexposed for the two strongest signals to enhance the visibility of weaker signals. The setup allows the fast detection of the CerS activity profile for various acyl-CoAs in multiple tissues.
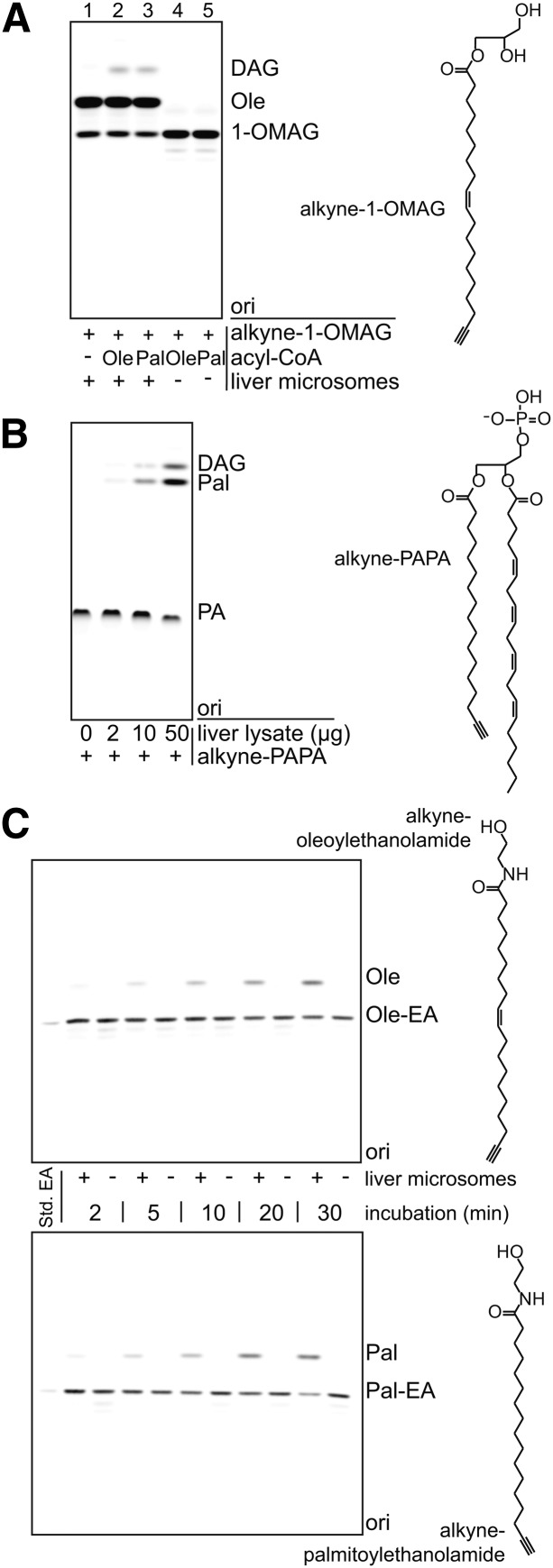
Incubation of additional substrates with mouse liver microsomes or lysate
Incubation of 1-(nonadec-9-cis-en-18-ynoyl)-monoacylglycerol (alkyne-1-OMAG) (50 μM) was performed in LPCAT buffer for 30 min at 30°C with oleoyl-CoA or palmitoyl-CoA (100 μM) and mouse liver microsomes (50 μg protein). Incubation of 1-(16-heptadecanoyl)-2-((5Z,8Z,11Z,14Z)-5,8,11,14-eicosatetraenoyl)-sn-glycero-3-phosphate (alkyne-PAPA) (50 μM) was performed with different amounts of mouse total liver lysate (0, 2, 10, and 50 μg protein) in ammonium acetate buffer (60 mM ammonium acetate, pH 7.0, 0.6 mg/ml lipid-free BSA) for 30 min at 30°C. Alkyne-palmitoylethanolamide or alkyne-oleoylethanolamide (50 μM) were incubated in LPCAT buffer at 30°C with or without the addition of mouse liver microsomes (10 μg protein) for different timespans (2–30 min).
Lipid extraction, click reaction, and TLC
All assay reactions were stopped by the addition of 500 μl chloroform/methanol 1/3 (v/v) to the assay mixture. Samples were then incubated for 5 min in an ultrasonic bath and 500 μl 1% acetic acid were added, followed by vortexing and centrifugation at 14,000 g for 2 min. One hundred microliters of the chloroform phase were transferred to a new 1.5 ml reaction vessel. The aqueous phase was again extracted with 200 μl chloroform. The combined organic phases were dried in a speed-vac and the click reaction performed as described previously (13): To the dried extracts, 7 μl chloroform were added to redissolve lipids. Then 30 μl click reaction mix (10 μl of 2 mg/ml 3-azido-7-hydroxycoumarin, 250 ul of 10 mM [acetonitrile]4CuBF4 in acetonitrile, 850 μl ethanol) were added and the tubes incubated in a heating block (Eppendorf Thermomixer comfort) at 43°C without shaking until all solvent was condensed under the lid (3 h).
After brief centrifugation, addition of 30 μl chloroform, and 5 min incubation in an ultrasonic bath, the samples were applied onto TLC silica plates (Merck 1.05721.0001). Plates were developed in the respective solvent system: a) for all assays except CerS: first chloroform/methanol/water/acetic acid 65/25/4/1 for 13 cm, then hexane/ethyl acetate 1/1 for 18 cm, with gentle drying between the two solvents; or b) for the CerS assay: chloroform/methanol/water 80/10/1 (2).
Extraction efficiency experiments
Diluted solutions (0.5 μM) of 1,2-di-(nonadec-9-cis-en-18-ynoyl)-sn-glycero-3-phosphate (alkyne-OOPA), alkyne-18:0-dihydroceramide (dhCer), alkyne-PC, and propargyl-phosphatidylcholine (pPC) in ethanol were prepared freshly, 10, 20, or 40 μl of this solution added to a reaction vessel, the solvent evaporated, and the lipids dissolved in the appropriate assay buffer (see above). After the addition of heat-inactivated enzyme preparation, the reaction mix was incubated according to the respective assay protocol and subjected to extraction, click reaction, and TLC. Quantification against alkyne-oleate was performed as given below, using the same molar amounts of alkyne-oleate as for the extracted lipids.
Detection and quantification
Shortly before fluorescence detection, the dry TLC was soaked for 5 s in 4% (v/v) N,N-diisopropylethylamine in hexane and excess solvent was allowed to evaporate in a hood. The system used for the imaging of the TLC plates and image quantification was described earlier (13). Briefly, standard LEDs (10 × 1 W 420 nm LEDs; Roithner Lasertechnik, Vienna, Austria) filtered through a colored glass filter (HEBO V01, Hebo Spezialglas) were used for excitation. Images were acquired with a Rolera MGI plus electron multiplying charge-coupled device (EMCCD) camera (Decon Science Tec), equipped with a 494/20 (channel for detection of the coumarin fluorescence) and 572/28 (channel for detection and correction of background fluorescence) bandpass emission filter wheel, all under control of GelPro analyzer software (Media Cybernetics). Note that the high sensitivity of the EMCCD camera (which is part of our electrochemiluminescence Western blot detection system) is not necessary to record the images, which typically can be seen already by visual inspection, but its large dynamic range is advantageous for image quantification. We also use a system with a much simpler camera with good results. Fluorescent signals were correlated to the lipid amount detected by drying two different defined amounts of alkyne-oleate solution in the speed-vac, subjecting them to the click reaction, and applying them to separate lanes in the TLC. All signals of a TLC were quantified against the weighted mean signal of the alkyne-oleate signals.
Statistical analysis and nonlinear regression
Unless stated otherwise (extraction efficiency measurements), all data are presented as mean values ± standard deviations (n = 3). Michaelis-Menten kinetics were assessed in three independent measurements, a representative graph is shown. For each measurement, the kinetic constants Km and Vmax were calculated from the data using nonlinear regression in Microsoft Excel (19), and the mean value and standard deviation of these three calculations are given.
RESULTS AND DISCUSSION
Assay setup and method of quantification
The general workflow, as outlined in Fig. 1, consists of the enzymatic reaction using alkyne substrate, lipid extraction, and reaction of the alkyne moieties with the fluorogenic dye 3-azido-7-hydroxycoumarin in a quantitative copper(I)-catalyzed cycloaddition. After separation by TLC, lipids are analyzed by fluorescence detection.
Fig. 1.
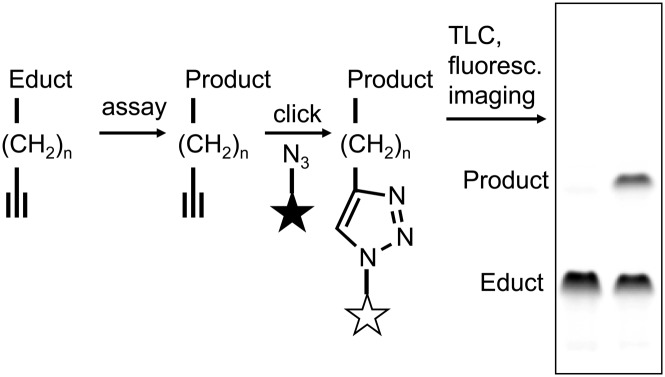
Typical workflow of the click chemistry-based enzymatic assay. Alkyne lipids were used as substrates in enzymatic assays. After lipid extraction the alkyne moiety was quantitatively click-reacted with the fluorogenic dye 3-azido-7-hydroxycoumarin (symbolized as a star) to yield fluorescently labeled lipids. These were separated by TLC and analyzed by fluorescence detection.
The high sensitivity and wide linear dynamic range of this procedure regarding the click reaction, TLC, and imaging were established earlier (13) and are comparable to methods using [3H]-labeled lipids. For an accurate quantification of the assay products, a reliable and near complete recovery is necessary. To determine the efficiency of the lipid extraction protocol, defined amounts of alkyne lipids that represent products of the LPAAT, LPCAT, or CerS assay were subjected to the standard procedures (in vitro enzymatic reaction, extraction, click reaction, detection), but using heat-inactivated enzyme preparations. The extraction procedure was evaluated by quantification of fluorescent signal against that of defined amounts of alkyne-oleate that had not been subjected to assay incubation and extraction. High recovery rates were measured for all tested lipids, alkyne-OOPA (97%), alkyne-dhCer (85%), alkyne-PC (88%), and pPC (82%), with good to very good reproducibility. This demonstrates the applicability of the extraction protocol (which had been optimized for polar lipids) for the assays performed in this study, and of the quantification method. Thus, an exact correlation of fluorescent signal to the amount of lipid produced in the assay is achieved.
LPAAT assay with alkyne-OLPA or alkyne-oleoyl-CoA
In E. coli, the conversion of lysophosphatidic acid (LPA) to phosphatidic acid (PA) is catalyzed by the gene product of plsC, 1-acyl-sn-glycero-3-phosphate acyltransferase (20). Its activity has been assessed with [32P]LPA (21). To the best of our knowledge, kinetic constants have not been determined for this enzyme.
We established a LPAAT assay with alkyne-OLPA (Fig. 2A), the labeled analog of sn-1-oleoyl-lysophosphatidic acid using E. coli microsomes (Fig. 2B), and determined the constants Vmax and Km for this substrate (Fig. 2C, Table 1). Conversely, by applying alkyne-oleoyl-CoA as an acyl chain donor, we measured Vmax and Km for PLPA. The apparent Km for alkyne-OLPA (0.53 ± 0.18 μM) was significantly lower than for PLPA (3.64 ± 0.24 μM). Also, Vmax was apparently increased for alkyne-OLPA compared with PLPA, but the differences did not reach statistical significance due to large variations between different enzyme preparations. In other organisms, Km had previously been determined for LPAAT enzymes using radiolabeled substrates. A similar acyl chain specificity in the acceptor was found in Spinacia oleracea (22); with oleoyl-CoA as the donor, a Km of 5.3 μM was measured for [14C]1-oleoyl-lysophosphatidic acid (OLPA), and of 3.0 μM for [14C]PLPA. For the human LPAAT-α, a Km of 6.49 μM toward OLPA with [14C]palmitoyl-CoA as the acyl chain donor (23), and of 6.0 μM with [3H]oleoyl-CoA (24), respectively was reported. For human LPAAT-β, Km values in the same range were measured toward OLPA with the acyl chain donor oleoyl-CoA [2.0 μM (25) and 8.29 μM (24)]. For both human isoforms, a preference of OLPA over PLPA as the acceptor was detected as well (24, 25). Compared with palmitate and stearate, oleate is preferred by the E. coli enzyme also as the donor acyl-CoA (Fig. 2D) for the incorporation at the sn-2 position, in accordance with previous findings (26).
TABLE 1.
Kinetic constants obtained by nonlinear regression analysis
| Assay | Substrate | Km (μM)a | Vmax [nmol/(min·mg)]a |
| LPAAT | Alkyne-OLPA | 0.53 ± 0.18b | 6.83 ± 4.39c |
| LPAAT | PLPA | 3.64 ± 0.24b | 2.05 ± 0.21c |
| LPCAT | Alkyne-OLPC | 2.46 ± 1.62d | 0.51 ± 0.08e |
| LPCAT | PLpPC | 5.46 ± 2.23d | 0.30 ± 0.07e |
| CerS | Alkyne-sphinganine | 2.29 ± 2.08 | 0.71 ± 0.38 |
Mean values and standard deviations from three independent triplicate determinations. Enzyme preparations, acyl chain donors, and further assay conditions used are given in the text.
P< 0.0001, cP = 0.1327, dP = 0.1325, eP = 0.0267 (these P values refer to the comparison of different substrates in the same enzymatic assay; two-tailed P values from Student's unpaired t-test).
Our results demonstrate that alkyne-OLPA and alkyne-oleoyl-CoA are good substrates for the E. coli LPAAT. They indicate that the bacterial enzyme may have a lower Km for lysophosphatidic acids (especially OLPA) than the plant or mammalian enzymes investigated so far.
The two alkyne substrates described above represent the two strategies that are available for an acyltransferase assay with alkyne lipids, i.e., labeling of the acyl chain acceptor or the acyl chain donor. While the latter is the more general approach, enabling the investigation of all acyltransferase reactions with a relatively small set of labeled alkyne-acyl-CoAs, it suffers from the limited storage stability of acyl-CoAs and the frequent observation of significant amounts of labeled side products arising from both enzymatic and nonenzymatic conversion of the acyl-CoAs. In our experiments, the use of alkyne-oleoyl-CoA led to a few fluorescent signals derived from side reactions of the reactive compound including the hydrolysis to oleate. The labeled acceptors on the other hand expressed high specificity for the enzymatic reactions of interest. They essentially gave one or two strong product signals besides the fluorescent educt (Figs. 2B, 3B, 4B), which might be beneficial for automated image analysis. In addition, alkyne-labeled acyl acceptors show better stability for long-term storage.
LPCAT assay with alkyne-OLPC or PLpPC
Lysophosphatidylcholine (LPC) is converted to phosphatidylcholine (PC) by lysophosphatidylcholine acyltransferases. In mammals, four isoforms (LPCAT1–4) are known. HuH7 cells express LPCAT1 and LPCAT3, but not LPCAT2 (18, 27). We synthesized two alkyne-labeled LPC species, i.e., side chain-labeled alkyne-OLPC and headgroup-labeled PLpPC (Fig. 3A). Note that because PLpPC is derived by enzymatic headgroup exchange from egg yolk PC, it contains a mixture of fatty acids at sn-1, dominated by palmitate (28). Figure 3B shows the application of the two substrates in a LPCAT assay on HuH7 lysate using oleoyl-CoA as the acyl chain donor. The Km value we measured (Fig. 3C, Table 1) for alkyne-OLPC (2.46 ± 1.62 μM) was very similar to that determined for palmitoyl-LPC with [14C]palmitoyl-CoA for murine recombinant LPCAT1 (2.3 μM) (29). PLpPC had a higher apparent Km (5.46 ± 2.23 μM), which might reflect a somewhat stronger effect of the alkyne label when incorporated in the headgroup of a phospholipid rather than at the terminus of an acyl chain. However, the differences between the Km values for alkyne-OLPC and PLpPC were not statistically significant and the differential acyl chains at the sn-1 positions have to be taken into account. In addition, we observed a significant difference (P < 0.05) in activity (Vmax) for the two substrates, 0.51 ± 0.08 nmol/(mg·min) for alkyne-OLPC compared with 0.30 ± 0.07 nmol/(mg·min) for PLpPC. On the basis of these results, alkyne-OLPC may be considered as the substrate of first choice for enzymatic assays compared with PLpPC. Nevertheless, the latter also displayed near to natural kinetic constants and allows for the direct combination of enzyme kinetics with microscopic imaging after labeling with propargyl-choline (14).
Ceramide synthase assay with alkyne-sphinganine
Ceramide synthases catalyze the acyl-CoA-dependent synthesis of dihydroceramide from sphinganine. The six mammalian isoforms display different tissue distributions and acyl chain specificities (30). CerS activity has been measured with radioactive and NBD-labeled sphinganine analogs (2, 31). We synthesized the new substrate alkyne-sphinganine (Fig. 4A) and employed it in a CerS assay on mouse liver microsomes using nervonoyl-CoA (Fig. 4B). In kinetic measurements in the same system, we determined an apparent Km of 2.29 ± 2.08 μM and a Vmax of 0.71 ± 0.38 nmol/(mg·min) for alkyne-sphinganine (Fig. 4C, Table 1). In the liver, CerS2 is the dominant isoform with activity toward long-chain acyl-CoAs; hence the activity we measured in this assay may almost solely be attributed to CerS2 (32). The Km value is in agreement with recent studies: using tritiated sphinganine, Lahiri et al. (31) reported Km values in the range of 2–5 μM for all six (murine or human) CerS isoforms, even for those which use multiple acyl-CoAs. That study reported a Km of 4.8 ± 0.4 μM for human Cers2 with lignoceroyl (24:0)-CoA. Kim et al. (2) developed a fluorescent assay with NBD-sphinganine and nervonoyl-CoA and found Km to be comparable to natural sphinganine (3.61 ± 1.86 μM vs. 3.05 ± 0.81 μM, respectively). However, Vmax was elevated for the NBD derivative.
CerS2 is expressed in various tissues besides the liver, with high mRNA levels in the kidney and moderate levels in the brain (30). Imgrund et al. (17) created a CerS2-deficient mouse and measured the CerS activity toward tritiated sphinganine and various acyl-CoAs (16:0, 18:0, 20:0, 22:0, and 24:1) in brain and liver microsomes. To demonstrate that alkyne-sphinganine can be used as a convenient alternative substrate for such applications, we performed a similar small screen with brain, liver, and kidney microsomes and several acyl-CoAs (16:0, 18:0, 18:1, and 24:1). Figure 4D shows fluorescent images of the TLC plates of this pilot study. Like Imgrund et al. (17), we observed a strong activity toward 24:1-CoA in the wild-type liver and a weak one in the brain. We also measured a strong 24:1 activity in kidney microsomes. For all tissues, 24:1-CoA was not converted to dhCer in the CerS2-deficient samples. For the acyl-CoAs of shorter chain length, the CerS activity was similar for wild-type and −/− mice, reflecting that it was not derived from CerS2, but other isoforms. We detected activity toward 16:0-CoA in kidney (moderate) and liver (weak), but not in brain. Weak activity was also found for 18:0-CoA in brain and liver, both in wild-type and −/− mice. 18:1-CoA was converted to dhCer in kidney and very little in liver. Because we show here that alkyne-sphinganine is a useful substrate to screen for CerS activity in tissue samples, more detailed quantitative investigations can be carried out in the future.
Application of the method in assays for other lipid-modifying enzymes
We utilized mouse liver microsomes or mouse liver lysate to demonstrate the principal feasibility of assays with four additional examples of alkyne-labeled substrates (Fig. 5). All substrates were converted by one or more enzymes in the liver fractions to alkyne-labeled products.
Fig. 5.
Application of alkyne lipids in assays for other lipid-modifying enzymes. Four different alkyne lipids were incubated with mouse liver microsomes or lysate (C57BL/6 wild-type) to test their applicability as substrates in enzymatic assays. A: Alkyne-1-OMAG deacylation and acylation. Alkyne-1-OMAG was incubated with liver microsomes in the absence (−) or presence (+) of acyl-CoAs as indicated. The data indicate both hydrolysis to oleate (Ole) in the presence of liver microsomes and acylation to DAG if an acyl-CoA is present. B: Alkyne-PAPA deacylation and dephosphorylation. Alkyne-PAPA was incubated with different amounts of liver lysate as indicated. We observed both release of alkyne-palmitate (Pal) and dephosphorylation to DAG. C: Time course of the hydrolysis of fatty acid ethanolamides by liver microsomes. Alkyne-oleoylethanolamide (Ole-EA) and alkyne-palmitoylethanolamide (Pal-EA) were hydrolyzed upon incubation with the microsomes, but not in their absence, to give the alkyne-labeled fatty acids. A–C: Products were identified using comigrating synthetic standards (Std. EA). “ori” depicts the origin of the TLC. For assay details, see Materials and Methods.
Deacylation of alkyne-1-OMAG (structure depicted in Fig. 5A) by monoacylglycerol lipases (MAGLs) yielded alkyne-oleate (Fig. 5A, lane 1). Upon addition of oleoyl-CoA or palmitoyl-CoA, alkyne-1-OMAG was acylated at the sn-2 position to form diacylglycerol (DAG) (Fig. 5A, lane 2+3) by monoacylglycerol acyltransferase (MGAT) activities, presumably the murine MGAT1 and lysophosphatidylglycerol acyltransferase 1 (33).
Alkyne-PAPA (structure depicted in Fig. 5B) was converted to DAG in the presence of liver lysate (Fig. 5B), presumably by phosphatidate phosphatases (34). In parallel, we observed the release of alkyne-palmitate from the sn-1 position, either by a phospholipase 1 activity acting on the labeled PA, or by a DAG lipase activity acting on the released DAG.
N-oleoylethanolamide and N-palmitoylethanolamide are bioactive lipids that are cleaved by the enzymes fatty acid amide hydrolase (FAAH) (35) and N-acylethanolamine-hydrolyzing acid amidase (36) yielding the corresponding fatty acids. Both the lipids and the enzymes have gained increasing interest as therapeutic targets for the control of pain, inflammation, or food intake (37, 38). We were able to survey the hydrolysis of the fatty acid amides alkyne-oleoylethanolamide and alkyne-palmitoylethanolamide in a time-course experiment (Fig. 5C) upon incubation with mouse liver microsomes. Several enzymatic assays are reported to follow this reaction [see (35) for a review and (39)]. We will conduct further studies concerning the kinetic properties of these alkyne-labeled substrates, which would expand the existing techniques by a very convenient and direct assay procedure.
The various substrates discussed above therefore demonstrate how the scope of the assay can be extended to more acyltransferases (MGAT), lipases (MAGL, phospholipases A1 and A2) and other hydrolases (PA phosphatase, FAAH). This general applicability will prove useful for the kinetic characterization of many enzymes in lipid metabolism.
Scope and limitations of the method
All alkyne lipids that we tested in this pilot study were used as substrates by lipid-modifying enzymes. No substantial shift in affinity (Km) of the enzymes compared with the natural substrates was detected in our kinetic studies. We thus introduce enzymatic assays with alkyne lipids as a reliable and convenient alternative to fluorescent or radioactive assays.
As with the latter two methods, the TLC separation in our assay does not achieve species resolution of the lipids. This can however be overcome by a subsequent mass-spectrometric analysis [compare (13)]. Depending on the location of the alkyne label in the molecule, not all reactions can be followed with every alkyne lipid. β-oxidation of fatty acids leads to the loss of the label (13), as does the headgroup cleavage of pPC to PA by phospholipase D.
Our studies suggest that the alkyne label does not generally interfere with the affinity of enzymes to the substrates, especially if the alkyne label is attached to the terminus of the alkyl chain. Such fatty acids of different length and unsaturation are synthetically available (13, 40) and can serve as a basis of a versatile toolbox of tailor-made lipid substrates with the acyl chain(s) of choice at defined sn-positions, including phospholipids, lysophospholipids, acyl-CoAs, and acylglycerols, using standard chemical and enzymatic procedures. Similar to this strategy, alkyne-sphingolipids can be synthesized from alkyne-sphinganine. Hence, we expect alkyne lipids to become more and more commercially available in the coming years, both as substrates for enzymatic assays as well as standards for identification after TLC separation. The migration behavior of lipids coupled to the coumarin dye is shifted, but, in general, the separation is possible with the same solvent mixes as for radiolabeled lipids or slightly adapted versions thereof and the retention factors of the substances usually are in the regular order. The adaptation of radiolabeled assays to the use of alkyne lipid substrates should therefore be neither costly nor overly time-consuming.
Alkyne lipids are promising substrates for use in in vitro enzymatic assays, as they are inexpensive, versatile, convenient and, as we show here for the first time, display the kinetic characteristics of the natural substrates.
Supplementary Material
Acknowledgments
The authors would like to thank Christiane Kremser and Prof. Dr. K. Willecke for providing the mice used in the CerS assay.
Footnotes
Abbreviations:
- alkyne-1-OMAG
- 1-(nonadec-9-cis-en-18-ynoyl)-monoacylglycerol
- alkyne-oleoyl-CoA
- nonadec-9-cis-en-18-ynoyl coenzyme A
- alkyne-OLPA
- 1-(nonadec-9-cis-en-18-ynoyl)-sn-glycero-3-phosphate
- alkyne-OLPC
- 1-(nonadec-9-cis-en-18-ynoyl)-sn-glycero-3-phosphocholine
- alkyne-OOPA
- 1,2-di-(nonadec-9-cis-en-18-ynoyl)-sn-glycero-3-phosphate
- alkyne-PAPA
- 1-(16-heptadecanoyl)-2-((5Z,8Z,11Z,14Z)-5,8,11,14-eicosatetraenoyl)-sn-glycero-3-phosphate
- alkyne-sphinganine
- (2S,3R)-2-aminooctadec-17-yn-1,3-diol
- BDP
- boron dipyrromethene
- CerS
- ceramide synthase
- DAG
- diacylglycerol
- dhCer
- dihydroceramide
- EMCCD
- electron multiplying charge-coupled device
- FAAH
- fatty acid amide hydrolase
- LPA
- lysophosphatidic acid
- LPAAT
- lysophosphatidic acid acyltransferase
- LPC
- lysophosphatidylcholine
- LPCAT
- lysophosphatidylcholine acyltransferase
- MAGL
- monoacylglycerol lipase
- MGAT
- monoacylglycerol acyltransferase
- NBD
- nitrobenzoxadiazole
- OLPA
- 1-oleoyl-lysophosphatidic acid
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PLPA
- 1-palmitoyl-lysophosphatidic acid
- PLpPC
- palmitoyl-lyso-propargyl-phosphatidylcholine
- pPC
- propargyl-phosphatidylcholine
The authors thankfully acknowledge financial support by the German Federal Ministry of Science and Education (Virtual Liver Network) and the Deutsche Forschungsgemeinschaft (Transregio TRR83 TP12).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of text.
REFERENCES
- 1.Weiss S. B., Kennedy E. P., Kiyasu J. Y. 1960. The enzymatic synthesis of triglycerides. J. Biol. Chem. 235: 40–44 [PubMed] [Google Scholar]
- 2.Kim H. J., Qiao Q., Toop H. D., Morris J. C., Don A. S. 2012. A fluorescent assay for ceramide synthase activity. J. Lipid Res. 53: 1701–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandhuvula P., Fyrst H., Saba J. D. 2007. A rapid fluorescence assay for sphingosine-1-phosphate lyase enzyme activity. J. Lipid Res. 48: 2769–2778 [DOI] [PubMed] [Google Scholar]
- 4.Bandhuvula P., Li Z., Bittman R., Saba J. D. 2009. Sphingosine 1-phosphate lyase enzyme assay using a BODIPY-labeled substrate. Biochem. Biophys. Res. Commun. 380: 366–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolb H. C., Finn M. G., Sharpless K. B. 2001. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 40: 2004–2021 [DOI] [PubMed] [Google Scholar]
- 6.Dieterich D. C., Link A. J., Graumann J., Tirrell D. A., Schuman E. M. 2006. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. USA. 103: 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxon E., Bertozzi C. R. 2000. Cell surface engineering by a modified Staudinger reaction. Science. 287: 2007–2010 [DOI] [PubMed] [Google Scholar]
- 8.Salic A., Mitchison T. J. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA. 105: 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baskin J. M., Bertozzi C. R. 2007. Bioorthogonal click chemistry: covalent labeling in living systems. QSAR Comb. Sci. 26: 1211–1219 [Google Scholar]
- 10.Best M. D. 2009. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 48: 6571–6584 [DOI] [PubMed] [Google Scholar]
- 11.Charron G., Wilson J., Hang H. C. 2009. Chemical tools for understanding protein lipidation in eukaryotes. Curr. Opin. Chem. Biol. 13: 382–391 [DOI] [PubMed] [Google Scholar]
- 12.Smith M. D., Sudhahar C. G., Gong D., Stahelin R. V., Best M. D. 2009. Modular synthesis of biologically active phosphatidic acid probes using click chemistry. Mol. Biosyst. 5: 962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiele C., Papan C., Hoelper D., Kusserow K., Gaebler A., Schoene M., Piotrowitz K., Lohmann D., Spandl J., Stevanovic A., et al. 2012. Tracing fatty acid metabolism by click chemistry. ACS Chem. Biol. 7: 2004–2011 [DOI] [PubMed] [Google Scholar]
- 14.Jao C. Y., Roth M., Welti R., Salic A. 2009. Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc. Natl. Acad. Sci. USA. 106: 15332–15337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neef A. B., Schultz C. 2009. Selective fluorescence labeling of lipids in living cells. Angew. Chem. Int. Ed. Engl. 48: 1498–1500 [DOI] [PubMed] [Google Scholar]
- 16.Lewin T. M., Wang P., Coleman R. A. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 38: 5764–5771 [DOI] [PubMed] [Google Scholar]
- 17.Imgrund S., Hartmann D., Farwanah H., Eckhardt M., Sandhoff R., Degen J., Gieselmann V., Sandhoff K., Willecke K. 2009. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 284: 33549–33560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moessinger C., Kuerschner L., Spandl J., Shevchenko A., Thiele C. 2011. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 286: 21330–21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemmer G., Keller S. 2010. Nonlinear least-squares data fitting in Excel spreadsheets. Nat. Protoc. 5: 267–281 [DOI] [PubMed] [Google Scholar]
- 20.Coleman J. 1992. Characterization of the Escherichia coli gene for 1-acyl-sn-glycerol-3-phosphate acyltransferase (plsC). Mol. Gen. Genet. 232: 295–303 [DOI] [PubMed] [Google Scholar]
- 21.Coleman J. 1990. Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J. Biol. Chem. 265: 17215–17221 [PubMed] [Google Scholar]
- 22.Hares W., Frentzen M. 1987. Properties of the microsomal acyl-CoA: sn-1-acyl-glycerol-3-phosphate acyltransferase from spinach (Spinacia oleracea L.) leaves. J. Plant Physiol. 131: 49–59 [Google Scholar]
- 23.Yamashita A., Nakanishi H., Suzuki H., Kamata R., Tanaka K., Waku K., Sugiura T. 2007. Topology of acyltransferase motifs and substrate specificity and accessibility in 1-acyl-sn-glycero-3-phosphate acyltransferase 1. Biochim. Biophys. Acta. 1771: 1202–1215 [DOI] [PubMed] [Google Scholar]
- 24.Agarwal A. K., Sukumaran S., Cortes V. A., Tunison K., Mizrachi D., Sankella S., Gerard R. D., Horton J. D., Garg A. 2011. Human 1-acylglycerol-3-phosphate O-acyltransferase isoforms 1 and 2: biochemical characterization and inability to rescue hepatic steatosis in Agpat2(-/-) gene lipodystrophic mice. J. Biol. Chem. 286: 37676–37691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollenback D., Bonham L., Law L., Rossnagle E., Romero L., Carew H., Tompkins C. K., Leung D. W., Singer J. W., White T. 2006. Substrate specificity of lysophosphatidic acid acyltransferase beta–evidence from membrane and whole cell assays. J. Lipid Res. 47: 593–604 [DOI] [PubMed] [Google Scholar]
- 26.Weier D., Lühs W., Dettendorfer J., Frentzen M. 1998. sn-1-Acylglycerol-3-phosphate acyltransferase of Escherichia coli causes insertion of cis-11 eicosenoic acid into the sn-2 position of transgenic rapeseed oil. Mol. Breed. 4: 39–46 [Google Scholar]
- 27.Zhao Y., Chen Y-Q., Bonacci T. M., Bredt D. S., Li S., Bensch W. R., Moller D. E., Kowala M., Konrad R. J., Cao G. 2008. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J. Biol. Chem. 283: 8258–8265 [DOI] [PubMed] [Google Scholar]
- 28.Schreiner M., Moreira R. G., Hulan H. W. 2006. Positional distribution of fatty acids in egg yolk lipids. J. Food Lipids. 13: 36–56 [Google Scholar]
- 29.Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. 2006. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J. Biol. Chem. 281: 20140–20147 [DOI] [PubMed] [Google Scholar]
- 30.Levy M., Futerman A. H. 2010. Mammalian ceramide synthases. IUBMB Life. 62: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R. N., Futerman A. H. 2007. Kinetic characterization of mammalian ceramide synthases: determination of K(m) values towards sphinganine. FEBS Lett. 581: 5289–5294 [DOI] [PubMed] [Google Scholar]
- 32.Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H. J., Futerman A. H. 2008. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283: 5677–5684 [DOI] [PubMed] [Google Scholar]
- 33.Hiramine Y., Emoto H., Takasuga S., Hiramatsu R. 2010. Novel acyl-coenzyme A:monoacylglycerol acyltransferase plays an important role in hepatic triacylglycerol secretion. J. Lipid Res. 51: 1424–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282: 3450–3457 [DOI] [PubMed] [Google Scholar]
- 35.Ueda N., Puffenbarger R. A., Yamamoto S., Deutsch D. G. 2000. The fatty acid amide hydrolase (FAAH). Chem. Phys. Lipids. 108: 107–121 [DOI] [PubMed] [Google Scholar]
- 36.Tsuboi K., Takezaki N., Ueda N. 2007. The N-acylethanolamine-hydrolyzing acid amidase (NAAA). Chem. Biodivers. 4: 1914–1925 [DOI] [PubMed] [Google Scholar]
- 37.Petrosino S., Iuvone T., Di Marzo V. 2010. N-palmitoyl-ethanolamine: biochemistry and new therapeutic opportunities. Biochimie. 92: 724–727 [DOI] [PubMed] [Google Scholar]
- 38.Thabuis C., Tissot-Favre D., Bezelgues J. B., Martin J. C., Cruz-Hernandez C., Dionisi F., Destaillats F. 2008. Biological functions and metabolism of oleoylethanolamide. Lipids. 43: 887–894 [DOI] [PubMed] [Google Scholar]
- 39.Huang H., Nishi K., Tsai H. J., Hammock B. D. 2007. Development of highly sensitive fluorescent assays for fatty acid amide hydrolase. Anal. Biochem. 363: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milne S. B., Tallman K. A., Serwa R., Rouzer C. A., Armstrong M. D., Marnett L. J., Lukehart C. M., Porter N. A., Brown H. A. 2010. Capture and release of alkyne-derivatized glycerophospholipids using cobalt chemistry. Nat. Chem. Biol. 6: 205–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.