Abstract
Cytidine deamination is the primary mechanism by which APOBEC3G restricts HIV-1; however, several studies have reported that APOBEC3G also inhibits virus replication via a mechanism that is independent of deamination. Using active site APOBEC3G mutants, we have re-evaluated the biological relevance of deaminase-independent APOBEC3G-mediated restriction of HIV-1. APOBEC3G proteins with Glu→Ala mutations in AS1, AS2 or AS1 and AS2 were stably expressed at physiological levels in CEM-SS T cells and 293T cells and the ability of the cells to support Δvif HIV-1 replication was then tested. The AS2 and AS1/AS2 mutants were packaged efficiently into virions but in single-cycle or multi-cycle HIV-1 replication assays, were found to lack antiviral activity. The AS1 mutant, which retained deaminase activity, maintained near wild-type antiviral function. To determine the potency of APOBEC3G antiviral activity, cell lines were established that that expressed low levels of wild-type APOBEC3G and generated virions that contained as few as 1-2 APOBEC3G molecules. Even at very low copy number, APOBEC3G caused a significant reduction in infectivity, suggesting that a single molecule of packaged APOBEC3G inactivates the virus. The high potency of APOBEC3G is consistent with a catalytic mechanism of restriction in which a single molecule can induce a string of mutations but difficult to reconcile with a deaminase-independent, non-catalytic mechanism. Analysis of the reverse transcript sequences showed that the G→A mutations were clustered, likely reflecting the action of single APOBEC3G molecules acting processively. We conclude that cytidine deamination is the mechanism by which APOBEC3G restricts HIV-1.
Introduction
APOBEC3 cytidine deaminases constitute a critical arm of the innate immune system that potently restricts retrovirus replication (Harris and Liddament, 2004). HIV-1 escapes APOBEC3G-mediated restriction by encoding Vif, an accessory protein that induces the degradation of APOBEC3G through a proteasome dependent pathway (Kao et al., 2003; Mariani et al., 2003; Marin et al., 2003; Mehle et al., 2004; Sheehy, Gaddis, and Malim, 2003; Stopak et al., 2003; Yu et al., 2003). Δvif HIV-1 cannot replicate in primary T cells and macrophages but, in laboratory cell lines such as HeLa, human embryonic kidney 293T or CEM-SS that do not express APOBEC3G, the virus replicates with wild-type kinetics. In cells infected with Δvif HIV-1, APOBEC3G molecules are packaged into virions as they assemble at the plasma membrane (Alce and Popik, 2004; Khan et al., 2005; Schafer, Bogerd, and Cullen, 2004; Zennou et al., 2004). In the next round of virus replication, the packaged APOBEC3G deaminates the minus-strand reverse transcript as it is synthesized from the plus-stranded genomic RNA template (Harris et al., 2003; Mariani et al., 2003; Zhang et al., 2003). Δvif virions produce a reduced number of full-length reverse transcripts, either as a result of degradation of the uracil-containing cDNA or effects of APOBEC3G on reverse transcriptase-mediated synthesis (Bishop, Holmes, and Malim, 2006; Guo et al., 2007; Iwatani et al., 2007; Yang et al., 2007).
The APOBEC3G protein contains two cytidine deaminase domains, each of which harbors an active site containing a conserved Cys/His-Xaa-Glu-Xaa23-28-Pro-Cys-Xaa2-4-Cys motif. In this motif, the Cys/His residues coordinate a Zn2+ ion and the glutamic acid acts as proton shuttle in catalysis (Carlow, Short, and Wolfenden, 1996). Mutation of the active site 2 (AS2) glutamic acid residue (Glu259) abolishes catalytic activity (Navarro et al., 2005; Newman et al., 2005) while mutation of this residue in active site 1 (AS1)(Glu67) has little effect. This finding demonstrated that catalysis is mediated by AS2. In some reports, Glu259-mutated APOBEC3G was found to have little or no antiviral activity (Navarro et al., 2005). This finding led to the conclusion that that cytidine deamination is required for virus restriction. Mutation of Glu67 in AS1 does not affect APOBEC3G-mediated antiviral activity, but mutation of the Cys/His Zn2+ coordination residues reduces virion packaging of the enzyme and thus prevents antiviral activity (Navarro et al., 2005; Newman et al., 2005). Further support for the requirement for deaminase activity was provided by an analysis of mouse Apobec3. The mouse genome encodes a single Apobec3 protein in which the roles of the two cytidine deaminase domains are reversed, such that AS1 mediates the catalytic activity while AS2 mediates virion packaging (Hakata and Landau, 2006). In the mouse protein, the AS1, but not the AS2 glutamic acid residue was required for antiviral activity. Taken together, these findings supported the conclusion that the principal mechanism by which APOBEC3G restricts HIV-1 is cytidine deamination.
In spite of these findings, there is evidence that APOBEC3G can also restrict HIV-1 by a deaminase-independent mechanism. Newman et al. reported that APOBEC3G with a Glu259Gln mutation of AS2 lacked cytidine deaminase activity yet maintained potent antiviral activity (Newman et al., 2005). In resting primary T cells, APOBEC3G was found to restrict HIV-1 in the target cell, and the majority of newly synthesized reverse transcripts lacked G→A hypermutation (Chiu et al., 2005). APOBEC3G was also found to restrict Hepatitis B virus in the absence of G→A hypermutation (Turelli et al., 2004). Another APOBEC3 deaminase, APOBEC3A, was found to restrict LTR and non-LTR retrotransposons and adeno-associated virus in the absence of detectable G→A hypermutation (Bogerd et al., 2006; Chen et al., 2006).
While cytidine deamination is clearly an important mechanism by which APOBEC3G restricts HIV-1, whether a cytidine deaminase-independent mechanism plays a significant role remains unresolved. The notion of deaminase-independent restriction of HIV-1 by APOBEC3G was challenged in the recent reports of Schumacher et al. and Miyagi et al. who found that deaminase-deficient APOBEC3G mutants had reduced or undetectable antiviral activity (Miyagi et al., 2007; Schumacher et al., 2008). To further address this question, we generated transformed T cell and 293T cell lines that stably expressed wild-type or active site mutants of APOBEC3G at levels comparable to that of primary T cells, and tested their ability to restrict replication-competent and single cycle Δvif NL4-3. The AS2 deaminase-deficient mutants failed to restrict Δvif NL4-3 in either assay. In addition, APOBEC3G restricted Δvif NL4-3 when present in virions that contained as few as 1-2 molecules, a finding that consistent with an enzymatic mechanism of restriction. For viruses that contained limited amounts of APOBEC3G, the G→A mutations occurred in local clusters, consistent with a processive mechanism of deamination. We conclude that cytidine deaminase independent effects of APOBEC3G are likely not to play a biologically relevant role in HIV-1 restriction.
Materials and Methods
Cell Lines and primary cells
H9 and CEM-SS cells were cultured in RPMI 1640 medium supplemented with 10% FBS. HEK 293T, HOS human osteosarcoma and GHOST-X4R5 reporter cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. pMIGR.APOBEC3G retroviral vector stocks were prepared by calcium phosphate transfection of 293T cells. The pMIGR plasmid was originally constructed by Warren Pear (University of Pennsylvania). CEM-SS and 293T cells were infected with 1.0 ml of virus-containing supernatant. After five days, the cells were sorted by FACS for EGFP expressed from the pMIGR IRES-EGFP cassette. Single cell clones were generated by limiting dilution for CEM-SS and by isolation of single colonies for 293T. Primary CD4+ T cells were obtained from anonymous healthy donor peripheral blood mononuclear cells that were positively selected on magnetic anti-CD4-coated beads and then activated with anti-CD28/CD3 coated beads. The resulting cells were >95% CD45RO+. Protein lysates were harvested at 8 days post activation.
Western Blots
Cells in a confluent 6-well plate were washed with PBS and lysed in buffer containing 150 μl of lysis buffer (10 mM TRIS, pH 7.5; 1.5 M NaCl; 2mM EDTA; 0.5 % v/v NP-40) for 30 min at 4 °C. The lysates were cleared by centrifugation for 10 min at 14,000 X g at 4 °C. The lysate was mixed with reducing sample buffer, heated for 7 min at 95 °C and an amount corresponding to 20 μg of protein was subjected to SDS-PAGE on a 4-12 % gradient gel. Baculovirus expressed recombinant human APOBEC3G (Immunodiagnostics Inc.) was run in parallel to determine copy number. To isolate virion proteins, virus-containing supernatants were clarified by centrifugation at 2,000 rpm for 5 mins, filtered through a 0.45 μM filter and then pelleted by ultracentrifugation through a 1.0 ml 20% sucrose cushion for 90 mins at 25,000 rpm in an SW41 rotor. The pellet was resuspended in PBS, dissolved in loading buffer and the proteins were separated by SDS-PAGE. The gel was transferred to a nitrocellulose membrane and probed with rabbit anti-APOBEC3G serum (provided by W. Greene, University of California, San Francisco) or mouse anti-tubulin monoclonal antibody and then hybridized with biotinylated goat anti-rabbit or goat anti-mouse secondary antibody. The filter was developed by incubation with Streptavidin DayLight 680 conjugate and quantitated on an Odyssey Infrared Imaging System at 700 nm.
Sequence analysis of reverse transcripts
HOS cells were infected with DNAse-I treated virus-containing supernatant (50 ng of p24) for 2 h. The virus was removed by three washes in PBS. After 24 h, the cells were lysed in digestion buffer (100 mM NaCl, 10 mM Tris pH 8, 25 mM EDTA, 0.5% SDS, 0.1 mg/mL proteinase K). The lysate was incubated overnight at 50° C and the cellular DNA was ethanol precipitated and resuspended in ddH20 overnight at 50° C. Viral DNA sequences were amplified from 1 μl of cellular DNA template with primers complementary to env that contained terminal EcoRI sites (forward primer: TGTGTGGAATTCTCAGCACTTGTGGAGATGGG and reverse primer: TGTGTGGAATTCGACATTTGTACATGGTCCTG). The PCR products were cloned into pCDNA3 at the EcoRI site.
Single-cycle infection
For data shown in Fig. 2, reporter virus stocks were generated by transfection of 293T cells using calcium phosphate with pNL.Luc.R-E- (Connor et al., 1995) or pNL.Luc.R-E-V- that contains a firefly luciferase gene in place of nef. 293T cells were seeded at 4.0 X 105 cells/well in a 6-well plate and the next day transfected with 5 μg reporter virus plasmid and 1.5 μg of pcVSVG. Two days later, the supernatant was harvested, centrifuged for 5 min at 400 X g, filtered through a 0.45-μm filter and frozen at -80° C in aliquots. p24 in the supernatant was quantitated by ELISA. To determine the infectivity of the virus, HOS.T4.X4 cells were seeded in a 96-well plate at 1.0 X 104 per well and the next day infected in triplicate with virus-containing supernatant (0.25 ng p24). After three days, luciferase activity was measured using Steady Lite HTS (PerkinElmer Life Sciences). For data shown in Fig. 4, 293T clones were infected with 50 ng of NL4-3 or Δvif NL4-3 that had been pseudotyped with VSV-G. At 48 h post-infection, viral supernatants were harvested, quantitated for p24 levels, and used to infect GHOST X4R5 cells (2.5 X 104) that were seeded in a 12-well plate. After two days, EGFP expression was evaluated by flow cytometry.
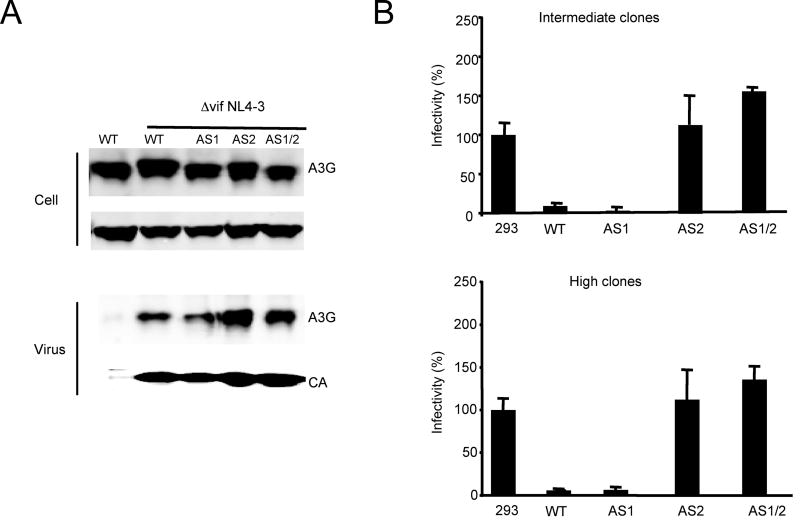
Figure 2. An AS2 cytidine deaminase mutant does not inhibit Δvif NL4-3 virion infectivity in a single-cycle infection.
A. 293T cell clones expressing intermediate levels of APOBEC3G were transfected with Δvif NL4-3 and VSV-G plasmids. After 48h, the virions were pelleted by ultracentrifugation. APOBEC3G and p24 content of the virions was visualized on an immunoblot probed with antiserum specific for these proteins. B. 293T clones expressing intermediate (upper panel) or high levels (lower panel) of APOBEC3G were transfected with wild-type or Δvif pNL-Luc and pVSV-G to generate single-cycle virus. The infectivity of the virus, normalized for p24, was determined by infection in triplicate of HOS cells. The cells were lysed after three days and luciferase activity was measured. The results are presented as the luciferase activity of the Δvif NL4-3 divided by wild-type, and is the average of triplicates.
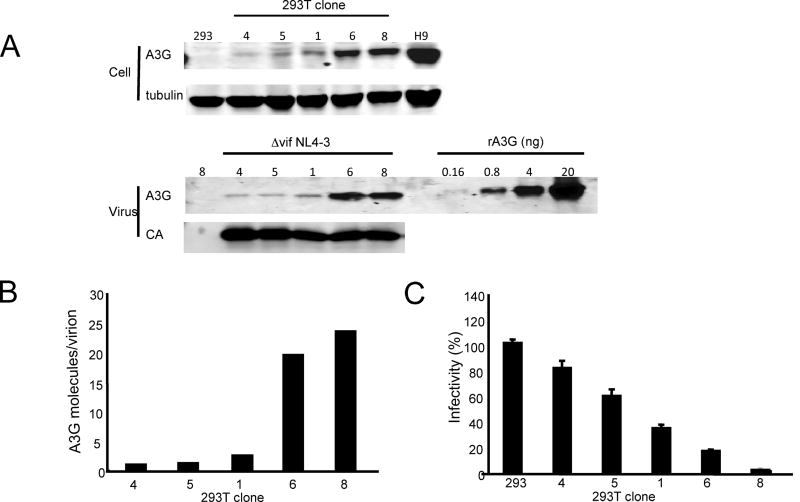
Figure 4. Quantitation of the number of APOBEC3G molecules per virion required to inhibit Δvif NL4-3 infectivity.
A. 293T clones expressing a range of wild type APOBEC3G levels were infected with NL4-3 (VSV-G) or Δvif NL4-3 (VSV-G) at an MOI of 0.5. At 72 h post-infection, culture supernatants were harvested and cell lysates were prepared. Virions were prepared from a portion of the supernatant and the remainder was used to quantitate p24 and determine virus titer. The cell lysates and virions were analyzed on an immunoblot probed with anti-APOBEC3G and anti-capsid. A serial dilution of recombinant APOBEC3G was included to determine copy number. Mock virions prepared from uninfected clone 8 cells had no detectable APOBEC3G. B. To determine the number of APOBEC3G copies per virion, the number of APOBEC3G molecules per ng of p24 was determined by standardizing the intensity of the virion APOBEC3G band against the recombinant APOBEC3G serial dilution curve for a fixed mass of p24. The number of virions corresponding to 1 ng of p24 was calculated assuming 2,000 copies/virion (Briggs et al., 2004). C. Virus infectivity was determined by single round infection of GHOST-X4R5 cells. The infectivity of virus produced by the 239T cell clones is shown as determined by the Δvif NL4-3 infectivity normalized to wild-type. The results are the average of triplicate measurements.
HIV-1 replication kinetics
Replication-competent NL4-3 and Δvif NL4-3 stocks were produced by transfection of 293T cells, quantitated and frozen in aliquots. CEM-SS cell clones (1 X 105) were infected with 40 ng of wild-type or Δvif NL4-3 corresponding to an MOI of 0.05 in a 12 well dish. To facilitate infection, the cells were centrifuged for two hrs at 400 X g. After 24 h, the input virus was removed with by washing twice with PBS. The culture medium was sampled every other day over two weeks and p24 was quantitated.
Results
Generation of stable cell lines that express physiological levels of wild-type and deaminase-deficient APOBEC3G
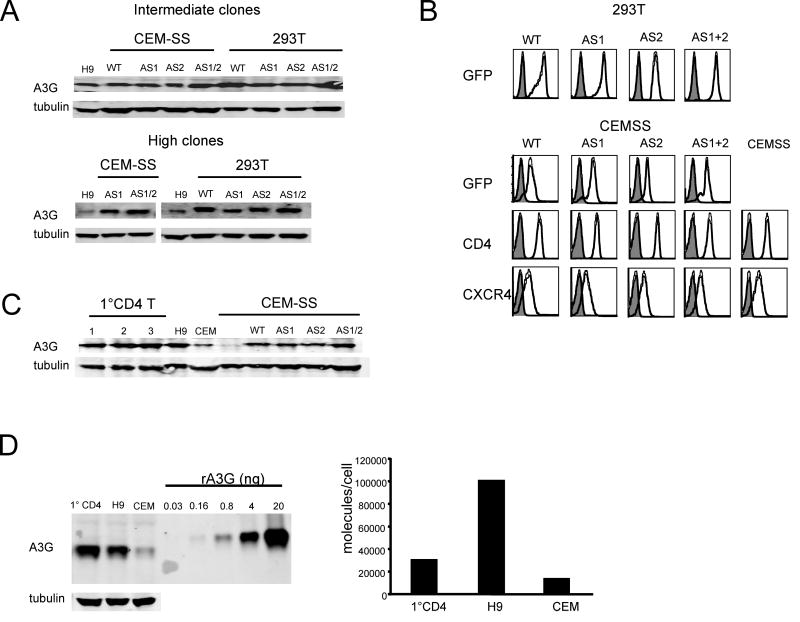
Most studies that have detected deaminase-independent antiviral activity of APOBEC3G have used virions that were generated by the transient transfection of 293T cells with Δvif NL4-3 proviral DNA and APOBEC3G expression vector. In vivo, however, APOBEC3G is expressed from a chromosomal gene and the virus is produced from an integrated provirus by infected T cells and macrophages. To study APOBEC3G restriction under conditions that are closer to those that pertain in vivo, we used the retroviral vector pMIGR, which has an IRES-EGFP, to construct expression vectors for wild-type (WT), AS1 (Glu67Ala), AS2 (Glu259Ala) or AS1 and AS2 (Glu67Ala, Glu259Ala) APOBEC3G. We transduced CEM-SS T cells and 293T cells, neither of which expresses endogenous APOBEC3G, with retroviral expression vector to generate cell lines that stably expressed the enzyme at levels similar to that of primary activated CD4+ T cells. The transduced cells were cloned by limiting dilution and their APOBEC3G expression level was determined on an immunoblot (Fig. 1A). The H9 T cell line was used for comparison as these cells express a level of APOBEC3G that is similar to that of primary activated CD4+ T cells (Fig. 1C). Two panels of CEM-SS and 293T cell clones were assembled, one termed “intermediate” and a second termed “high” based on their APOBEC3G expression relative to H9. We were not able to generate a CEM-SS high clone for the AS2 single mutant. The clonality of the cell lines was confirmed by analysis of EGFP fluorescence which showed a single sharp peak by flow cytometry (Fig. 1B). Equivalent levels of surface CD4 and CXCR4 were confirmed on the CEM-SS cell lines (Fig. 1B).
Figure 1. Wild-type APOBEC3G and active site mutant expression in CEM-SS and 293T stable cell lines.
293T cells and CEM-SS cells were transduced with pMIGR retroviral vectors that encode wild type APOBEC3G (WT), AS1 (E67A), AS2 (E259A) or AS1/AS2 (E67A/E259A) mutants and contain a downstream IRES-EGFP. A. Cell clones were isolated and their APOBEC3G expression levels were determined on an immunoblot probed with anti-APOBEC3G and anti-tubulin serum. H9 cells were used for comparison. The cell clones were grouped as “intermediate” and “high” based on APOBEC3G expression level. B. The EGFP fluorescence of the 293T and CEM-SS cell clones was measured, and the CEM-SS cell clones were analyzed for CD4 and CXCR4 expression by flow cytometry. C. APOBEC3G expression in the intermediate CEM-SS clones was compared to primary activated CD4+ T cells isolated from three healthy donors on an immunoblot probed with anti-APOBEC3G serum. D. The copy number of APOBEC3G molecules in human T cell lines was determined on an immunoblot standardized with recombinant APOBEC3G (left panel). To determine the copy number of APOBEC3G per cell, lysates containing a fixed mass of protein and corresponding to a known cell number was analyzed. The amount of APOBEC3G per cell was then calculated, standardized to the recombinant APOBEC3G control (right panel).
To determine whether the expression levels of APOBEC3G in the CEM-SS clones were similar to that of primary activated CD4+ T cells, we compared them to cell lysates from three healthy donors on an immunoblot (Fig. 1C). The level of APOBEC3G in the intermediate cell lines was somewhat lower than primary T cells but higher than that of CEM, a Δvif HIV-1 non-permissive T cell line. Using recombinant APOBEC3G as a standard and cell lysate derived from a known number of cells (Fig. 1D left panel), we calculated the APOBEC3G copy number per cell (Fig. 1D, right panel). Primary CD4+ T cells contained about 30,000 molecules per cell, while H9 cells, which are larger, contained 100,000 and CEM contained 20,000. Thus, the amount of APOBEC3G expressed by H9 and primary T cells is well above what is required to block virus replication. We conclude that the CEM-SS cell lines expressed APOBEC3G at levels comparable to H9 and primary T cells and that this amount is more than sufficient to detect antiviral activity.
Stably expressed deaminase-deficient APOBEC3G lacks detectable antiviral activity in single-cycle and multi-cycle replication
To determine whether the mutant APOBEC3G proteins could be efficiently packaged into HIV-virions, we transfected the panel of 293T cell lines with Δvif NL4-3 plasmid, and after two days, harvested the virus-containing supernatant. An analysis of the APOBEC3G content of the pelleted virions on an immunoblot showed that the wild-type APOBEC3G and active site mutants were packaged at similar levels (Fig. 2A). This result suggests that the mutants were properly folded and packaged into virions at levels sufficient to mediate deaminase-dependent and independent activities.
In a previous study, we reported that deaminase-deficient APOBEC3G mutants packaged in virions produced by transient transfection of 293T cells caused a 50% reduction in infectivity (Navarro et al., 2005), a result that could have suggested deaminase-independent restriction. To determine whether this effect was also present when the APOBEC3G mutants were stably expressed, we transfected the 293T stable cell lines with wild-type or Δvif NL4-3 luciferase reporter virus plasmid and VSV-G expression vector. The resulting virions were normalized for p24 and tested for infectivity by infection of target cells. The results showed that the AS1 mutant maintained its antiviral activity but that the AS2 and AS1/AS2 APOBEC3G mutants caused no detectable reduction of infectivity (Fig. 2B, upper panel). Similar results were found using the 293T cells that over-expressed the wild-type and mutant APOBEC3G proteins (Fig. 2B, lower panel). These results suggested that the modest effect of transiently expressed deaminase-deficient APOBEC3G disappears when it is expressed stably from an integrated cassette.
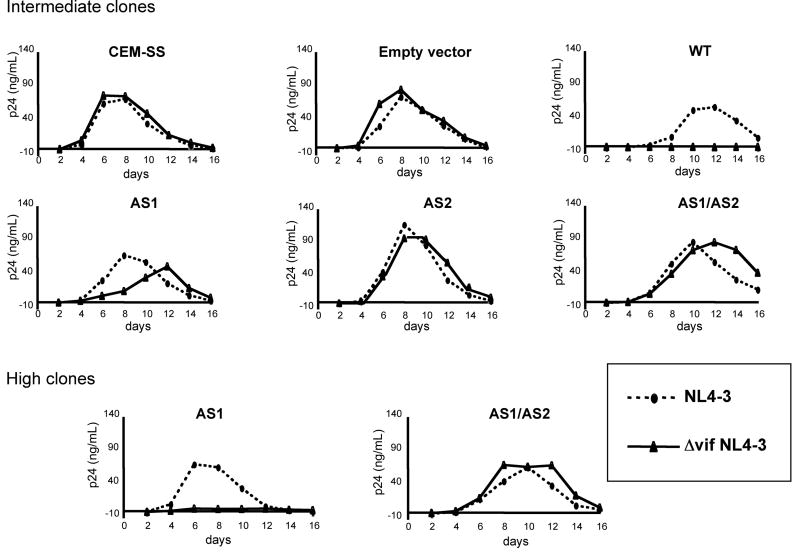
We next tested the effect of the APOBEC3G mutants on HIV-1 replication kinetics, a setting in which multiple rounds of virus replication amplifies modest antiviral effects. For the analysis, the CEM-SS clonal cell lines were infected with wild-type or Δvif NL4-3 and supernatant p24 was measured over two weeks. Comparison of wild-type and Δvif virus served to control for possible clone-to-clone differences in virus replication unrelated to APOBEC3G. The results showed that the empty vector control cell clones supported similar levels of wild-type and mutant virus replication, peaking on day eight (Fig. 3). Wild-type APOBEC3G blocked the replication of Δvif NL4-3 and had a small effect on wild-type virus replication, delaying peak replication until day 12 and reducing virus production by about 50%. This delay indicates that APOBEC3G can apply pressure to HIV-1 even when Vif is produced by the virus and suggests that its levels are a limiting factor for HIV-1 replication in cells that produce physiological levels of APOBEC3G. At an intermediate level of expression, the AS1 mutant delayed but did not completely block Δvif NL4-3 replication. When expressed at higher level, this mutant blocked Δvif NL4-3 replication. In contrast, Δvif NL4-3 replicated with wild-type kinetics in cells that expressed the AS2 or AS1/2 mutants. When expressed at higher levels the AS1/2 mutant also lacked detectable activity. Taken together, these results demonstrate that deaminase-deficient APOBEC3G lacks antiviral activity when expressed within physiological levels in T cells.
Figure 3. An APOBEC3G AS2 mutant does not inhibit Δvif NL4-3 replication in T cells.
CEM-SS cell clones that express intermediate or high levels of wild-type APOBEC3G (WT), AS1, AS2, or AS1/AS2 mutants were infected with wild-type or Δvif NL4-3. The culture medium was sampled every other day for two weeks and p24 was quantitated by ELISA.
Restriction of HIV-1 requires only a small number of packaged APOBEC3G molecules
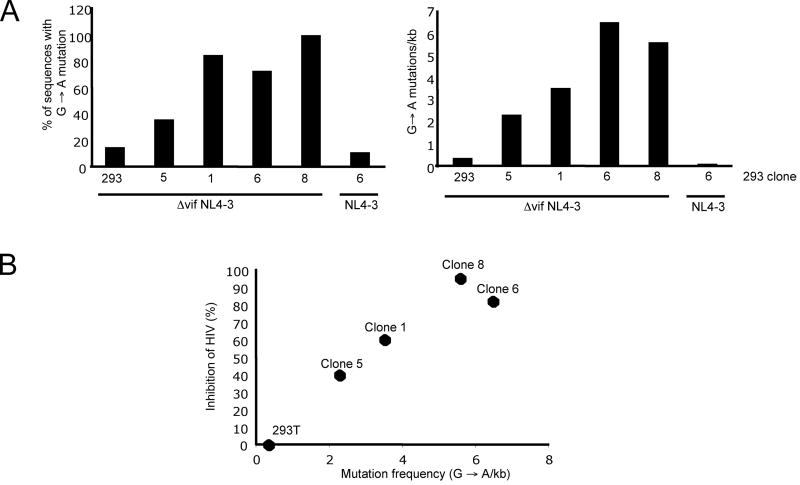
To determine the potency of the APOBEC3G restriction, we used 293T cell clones that expressed low levels of APOBEC3G. Five clones were chosen that expressed graded levels of wild-type APOBEC3G, from approximately 5% to 46% of the amount expressed by H9 (Fig. 4A). To determine the potency of APOBEC3G, we generated virions from the 293T cell lines by infection with Δvif NL4-3(VSV-G) and then measured their APOBEC3G content (Fig. 4A and 4B) and corresponding level of restriction (Fig. 4C). The two lowest expressing clones, 4 and 5, produced virions that contained 1.3 and 1.4 APOBEC3G molecules per virion while clones 6 and 8 produced virions that contained 17 and 22 molecules, respectively. The amount of APOBEC3G in the virions paralleled the amount present in the cell lysates. To determine infectivity, the virions were titered on GHOST cells. These cells contain an LTR-EGFP cassette that is transactivated by Tat upon infection. The titer of viruses produced by clones 4 and 5 were reduced by 20% and 40%, respectively (Fig. 4C). Expression of larger amounts of APOBEC3G in clone 8 reduced the titer by 95%. The results demonstrated that only a few, and possibly one packaged APOBEC3G molecule, can inhibit HIV-1 virion infectivity.
Antiviral activity correlates with G→A mutational frequency
To determine whether APOBEC3G antiviral activity was correlated with G→A mutations in the cDNA, we determined the frequency of G→A mutations in the viral cDNA produced by virions containing limiting amounts of APOBEC3G. To do this, we infected HOS cells with viral supernatants derived from Δvif NL4-3 infected 293T cell lines used in Figure 4. At 24 h post infection, we isolated cellular DNA from the HOS cells and PCR amplified a 700 base pair fragment of env. We determined the nucleotide sequence of 11-15 cloned fragments derived from each cell line. To analyze the sequences, we calculated the proportion of clones that had at least one G→A change (Fig. 5A left panel) and the average number of G→A mutations per kilobase (Fig. 5A right panel). Virions from clone 5 generated cDNAs in which 38% had at least one mutation and with an average of 2.3 mutations per kilobase, while clone 8 virions generated cDNAs that were nearly all mutant and had 5.6 mutations per kilobase. A plot of the reduction in titer against the number of mutations generated shows a linear relationship (Fig. 5B). A mutation frequency of 3.2 per kilobase reduced infectivity by 50%. Three mutations per kilobase corresponds to about 30 per genome (not accounting for variation in the mutational frequency over the genome). Infectivity was determined using GHOST indicator cells that contain a trans-activated LTR-GFP, and for a cell to score as infected in this assay, it needs only to harbor a provirus that expresses a functional Tat. Thus, viruses with G→A mutations in the structural genes will not be detected by the assay. It was unexpected that a such low mutational frequency would have have a significant impact on infectivity. This finding suggests that it is not the changes in nucleotide coding capacity that caused the reduction in infectivity but rather a direct effect on the reverse transcript caused either by its degradation or a reduction in elongation of the viral cDNA.
Figure 5. The antiviral activity of APOBEC3G correlates with frequency of G→A mutation.
The 293T cell clones were infected with wild-type or Δvif NL4-3 (VSV-G), and viral supernatants harvested at 48h post infection. The supernatants were then used to infect HOS cells and after 24 h of infection, cellular DNA was isolated. The DNA was used as a template for PCR amplification of a 700 bp fragment of env and the products were cloned into pCDNA3. The nucleotide sequence of 11-15 clones from each infection was determined. A. The proportion of inserts containing G→A mutations (left panel) and the average number of G→A mutations per kilobase are shown (right panel). B. The percent inhibition mediated by APOBEC3G in each of the 293T cell lines as shown in Fig. 4C was plotted against the frequency of G→A changes in the viral DNA as shown in the panel above on the right. The individual cell clone number is shown above each data point.
Deamination of the minus-strand reverse transcript is processive over a short distance
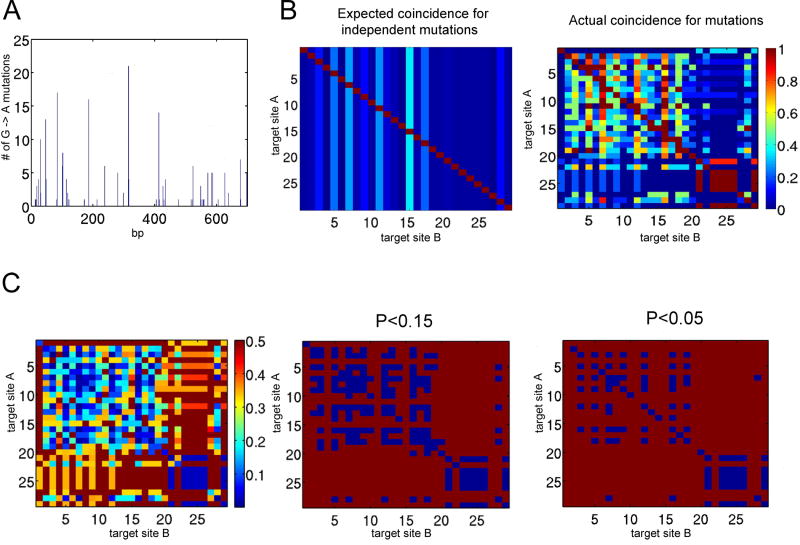
APOBEC3G could deaminate viral DNA distributively, catalyzing a single mutation and then disengaging from the DNA, or processively, by sliding along the DNA generating a string of mutations. Support for both models has been derived using purified enzyme in vitro on oligonucleotide substrate (Chelico et al., 2006; Coker and Petersen-Mahrt, 2007; Nowarski et al., 2008; Pham, Chelico, and Goodman, 2007). An analysis of the mutational patterns of the cDNA derived from the 293T clones allowed us to address this question in the context of the virion rather than model substrates. In virions that contained low levels of APOBEC3G, clustered stretches of mutated sequence are likely to have been generated by the action of a single molecule. The 700 bp region that we sequenced contained 28 APOBEC3G target sites as defined by bases that had been mutated more than once in the data set (Fig. 6A). As a measure of processivity, we calculated pair-wise values for the frequency with which a mutation at one site was accompanied by mutation at neighboring sites. The frequency of mutation at a second site will also be affected by the number of APOBEC3G molecules in the virion, but if deamination is distributive, this effect should apply equally to all other target sites of equivalent susceptibility. Processive deamination, by constrast, predicts that the frequency of mutation at a nearby second site will be affected more than a distant site with similar overall mutation frequency. The analysis showed that second site mutations at nearby target sites had a higher frequency than expected for independently occurring mutations (Fig. 6B right panel), consistent with processive deamination. The probability of two target sites being mutated in the same cDNA was lower for more distant pairs of sites. The statistical significance of this finding was demonstrated by a chi-squared test of independence for each pair wise relationship (Fig. 6 C). Interestingly, the increased frequency of paired mutations fell into two distinct clusters. A mutation at one base within a cluster was accompanied by an increased probability of a second mutation within that cluster but was not associated with an increased likelihood of a mutation in the other cluster. The clustering of mutations suggests processive deamination occurs in local domains but that there are boundary sequences over which the enzyme is less likely to cross.
Figure 6. APOBEC3G deaminates Δvif NL4-3 in local clusters.
APOBEC3G target sites in clones of the 700 bp env fragment generated in cells infected with the 293T cell line-derived virions were identified. Patterns of mutation in 55 clones were analyzed using Matlab software (Mathworks). A. The location of each target site and frequency with which it was mutated is plotted. B. The expected coincident frequency of a G→A mutation at a base (Target site B) when one site is mutated (Target site A) if all mutations occur independently is shown in the left panel. The actual coincident mutational frequency from the dataset is shown with values that range from no coincidence (dark blue) to 100% coincidence (red) in the right panel. C. The statistical significance of the difference between the actual and expected values was calculated as P values using a chi-squared test of independence and plotted (left panel). Pairs of target sites with P values of less than 0.15 and 0.05 are highlighted with blue squares (middle and right panels).
Discussion
We report here, that under conditions that resemble those that occur in vivo, APOBEC3G restriction of HIV-1 is cytidine deaminase-dependent. We based this conclusion on the finding that an APOBEC3G AS2 mutant and an AS1/AS2 double mutant that were stably expressed at levels similar to that of primary CD4+ T cells, lacked detectable antiviral activity in single-cycle and multi-cycle replication assays. Our findings are consistent with those of Schumacher et al. who reported that a CEM-SS APOBEC3G Glu259Gln stable cell line did not restrict HIV-1 replication, and with Miyagi et al. who found that an APOBEC3G AS2 mutant expressed stably in a HeLa cell line had only minimal effect on infection (Miyagi et al., 2007; Schumacher et al., 2008).
APOBEC3G has been reported to interfere with various steps in HIV-1 reverse transcription, including initiation and elongation, tRNA priming and first strand transfer (Bishop, Holmes, and Malim, 2006; Guo et al., 2007; Iwatani et al., 2007; Li et al., 2007). These findings are not inconsistent with the conclusion that restriction is caused by deamination. It is possible that deamination of the reverse transcript during synthesis could interfere with elongation, strand transfer or tRNA priming. However, these effects have generally been detected in with virions generated by transiently expressed APOBEC3G, a method that can cause an artifactual reduction in virion infectivity. Deaminase-deficient APOBEC3G expressed by transient transfection caused a two-fold reduction in virus infectivity (Navarro et al., 2005) but this antiviral activity was absent when the APOBEC3G mutants were expressed stably by an integrated retroviral vector. The reason for the difference is not clear, but could potentially be caused by the wide range of expression level per cell in transient transfection as compared to the relatively constant level in stably transduced cell clones. Further experimentation will be required to determine whether the effects of deaminase-deficient APOBEC3G on the various steps in viral genome replication are active in viruses produced by methods other than transient transfection.
Our results differ from those of Newman et al. who found that an AS2 (Glu259Gln) APOBEC3G mutant retained most of its antiviral activity (Newman et al., 2005) and those of Bishop et al. who found that antiviral activity did not correlate with the mutational frequency induced by APOBEC3G/APOBEC3F chimeras (Bishop, Holmes, and Malim, 2006). The cause of the different results is not clear. Our AS2 mutant had a Glu→Ala rather than Glu→Gln substitution. It is possible that, as shown previously, the AS2 glutamic acid to glutamine mutant retains residual catalytic activity sufficient to restrict the virus but does not induce enough G→A mutations to readily detectable by sequencing (Shindo et al., 2003). In addition, as noted above, we expressed the APOBEC3G mutants stably, rather than transiently.
Δvif virions produced by cells that expressed near physiological levels of APOBEC3G contained on average 17-22 molecules of APOBEC3G, close to the seven copies found in virions produced by activated peripheral blood mononuclear cells (Xu et al., 2007). When the virions were produced by cells that expressed low levels of APOBEC3G, they contained 1.3 molecules per virion and this resulted in a 20% reduction in infectivity. Assuming that APOBEC3G is a dimer (Miller, Presnyak, and Smith, 2007), a Poisson distribution predicts that 52% of the virions contained no APOBEC3G while 33% contain one dimer. Thus, a single dimer typically renders the virus noninfectious. This highly potent activity is consistent with an enzymatic reaction such as deamination, that can be mediated by a single molecule acting catalytically, rather than a mechanism such as occlusion of virion components from the reverse transcription complex or physical interference with progression of reverse transcriptase.
The frequency of APOBEC3G-generated mutations correlated inversely with infectivity, and a relatively small number of mutations had an unexpectedly large impact on infectivity. A mutational frequency of three/kilobase, corresponding to about 30 per viral genome, reduced virus titers by 50% on GHOST cells, that require only Tat expression from the provirus to score as infected. If inhibition of HIV-1 by APOBEC3G occurs primarily via the generation of inactivating mutations in essential coding proteins or controlling regions, only rare mutations in Tat itself or the LTR promoter will affect the titer as determined by this method. Thus, most of the effect on infectivity was likely due to a direct effect of deamination on the reverse transcript, probably caused either by interference with elongation of the reverse transcript or degradation by repair enzymes. Consistent with this conclusion, Bishop et al. recently reported that APOBEC3G causes reverse transcriptase to disengage from minus-strand synthesis with increasing distance from the initiation site (Bishop et al., 2008).
Using model oligonucleotide substrates, APOBEC3G-mediated deamination has been found to be either processive or distributive (Chelico et al., 2006; Coker and Petersen-Mahrt, 2007; Pham, Chelico, and Goodman, 2007). In our study we were able to address the question of processivity as it occurs in the reverse transcription complex of a newly infected cell. For this, we analyzed the clustering of mutations in the cDNA generated by viruses that contained limited numbers of APOBEC3G molecules. In viruses that contain one or a small number of APOBEC3G molecules, mutations were rare, but occurred in patches, presumably the result of a single APOBEC3G molecule acting processively. An analysis of the clustering of the mutations showed that the enzyme acts on discrete domains of the reverse transcript that are flanked by regions of sequence that cause the enzyme to disengage. These boundaries could be caused by unfavorable nucleotide sequences or by a tendency for RNAse-H to leave stretches of double stranded sequence that would be protected from deamination.
Our conclusion that APOBEC3G-mediated restriction is caused by cytidine deamination does not extend to other systems in which APOBEC3 family members are active. Restriction of HTLV-1 by APOBEC3G was found to occur in the absence of hypermutation (Sasada et al., 2005). Mouse mammary tumor virus and murine leukemia virus were found to be suppressed by murine Apobec3 in the absence of hypermutation (Browne and Littman, 2008; Okeoma et al., 2007; Rulli et al., 2008). In cell culture, adeno-associated virus was restricted by APOBEC3A without hypermutation and a catalytically inactive mutant APOBEC3A maintained its antiviral activity (unpublished observation). In addition, APOBEC3A was active against endogenous retroelements without causing hypermutation (Bogerd et al., 2006).
The critical role of cytidine deamination in the restriction of HIV-1 provides a rationale for the evolutionary conservation APOBEC3 catalytic activity. The catalytic mechanism provides a means by which a relatively small number of packaged molecules can inflict considerable damage to the viral genome. The mechanism by which APOBEC3 proteins restrict the replication of other viruses still needs to be addressed.
Acknowledgments
We thank Derya Unutmaz and Lina Kozhaya for primary CD4+ T cells and Cynthia Rudin for the bioinformatic analysis. Anti-APOBEC3G antiserum # 9968 and recombinant APOBEC3G # 9968 were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH from Warner Greene and Immunodiagnostics Inc., respectively. We thank Paulette Conger and Paul Vial (Universidad del Desarrollo) for support.
This work was supported by NIH grants T32 AI007647-09 and AI058864.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279(33):34083–6. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol. 2006;80(17):8450–8. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4(12):e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34(1):89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11(7):672–5. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- Browne EP, Littman DR. Species-specific restriction of apobec3-mediated hypermutation. J Virol. 2008;82(3):1305–13. doi: 10.1128/JVI.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlow DC, Short SA, Wolfenden R. Role of glutamate-104 in generating a transition state analogue inhibitor at the active site of cytidine deaminase. Biochemistry. 1996;35(3):948–54. doi: 10.1021/bi951498y. [DOI] [PubMed] [Google Scholar]
- Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3′ --> 5′ on single-stranded DNA. Nat Struct Mol Biol. 2006;13(5):392–9. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16(5):480–5. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435(7038):108–14. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Coker HA, Petersen-Mahrt SK. The nuclear DNA deaminase AID functions distributively whereas cytoplasmic APOBEC3G has a processive mode of action. DNA Repair (Amst) 2007;6(2):235–43. doi: 10.1016/j.dnarep.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206(2):935–44. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- Guo F, Cen S, Niu M, Yang Y, Gorelick RJ, Kleiman L. The interaction of APOBEC3G with human immunodeficiency virus type 1 nucleocapsid inhibits tRNA3Lys annealing to viral RNA. J Virol. 2007;81(20):11322–31. doi: 10.1128/JVI.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281(48):36624–31. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113(6):803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4(11):868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35(21):7096–108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol. 2003;77(21):11398–407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA, Kao S, Miyagi E, Takeuchi H, Goila-Gaur R, Opi S, Gipson CL, Parslow TG, Ly H, Strebel K. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J Virol. 2005;79(9):5870–4. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G inhibits DNA strand transfer during HIV-1 r. J Biol Chem. 2007;282(44):32065–74. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114(1):21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9(11):1398–403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279(9):7792–8. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Miller JH, Presnyak V, Smith HC. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol. 2007;81(24):13346–53. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, Landau NR. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333(2):374–86. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15(2):166–70. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Nowarski R, Britan-Rosich E, Shiloach T, Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat Struct Mol Biol. 2008;15(10):1059–66. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445(7130):927–30. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- Pham P, Chelico L, Goodman MF. DNA deaminases AID and APOBEC3G act processively on single-stranded DNA. DNA Repair (Amst) 2007;6(6):689–92. doi: 10.1016/j.dnarep.2007.01.001. author reply 693-4. [DOI] [PubMed] [Google Scholar]
- Rulli SJ, Jr, Mirro J, Hill SA, Lloyd P, Gorelick RJ, Coffin JM, Derse D, Rein A. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J Virol. 2008;82(13):6566–75. doi: 10.1128/JVI.01357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasada A, Takaori-Kondo A, Shirakawa K, Kobayashi M, Abudu A, Hishizawa M, Imada K, Tanaka Y, Uchiyama T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005;2:32. doi: 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328(2):163–8. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J Virol. 2008;82(6):2652–60. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9(11):1404–7. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J Biol Chem. 2003;278(45):44412–6. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12(3):591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303(5665):1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- Xu H, Chertova E, Chen J, Ott DE, Roser JD, Hu WS, Pathak VK. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360(2):247–56. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Yang Y, Guo F, Cen S, Kleiman L. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology. 2007;365(1):92–100. doi: 10.1016/j.virol.2007.03.022. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302(5647):1056–60. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78(21):12058–61. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424(6944):94–8. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]