Abstract
In an effort to understand molecular mechanisms of human disease and to determine genes responsible, we systematically examine relationships between 3,949 genes, 62,663 mutations and 3,453 associated disorders within the framework of a three-dimensional structurally resolved human interactome, consisting of 4,222 high-quality binary protein-protein interactions with their atomic-resolution interfaces. We find that in-frame mutations (missense point mutations and in-frame insertions and deletions) are enriched on the interaction interfaces of proteins associated with the corresponding disorders, indicating that alteration of specific interactions by in-frame disease mutations is critical in understanding the pathogenesis of many genes. Furthermore, locations of mutations on proteins with regard to interaction interfaces are significantly associated with underlying pathogenic processes and the disease specificity for different mutations of the same gene. Based on these findings, we generate 292 new gene candidates for 694 unknown disease-to-gene associations with proposed molecular mechanism hypotheses, readily expanding our understanding of human genetic diseases and corresponding therapeutic possibilities.
Over the past few decades, a tremendous amount of resources and effort have been invested in mapping human disease loci genetically and later physically1. Since the completion of the human genome sequence, especially with advances in genome-wide association studies and on-going cancer genome sequencing projects, an impressive list of disease-associated genes and their mutations have been produced2. However, it has rarely been possible to translate this wealth of information on individual mutations and their association with disease into biological or therapeutic insights3. Most of the US Food and Drug Administration approved drugs today are palliative4 – they merely treat symptoms, rather than targeting specific genes or pathways responsible, even if associated genes are known. One main reason for this lack of success is the complex genotype-to-phenotype relationships among diseases and their associated genes and mutations. In particular, (a) the same gene can be associated with multiple disorders (gene pleiotropy); and (b) mutations in any one of many genes can cause the same clinical disorder (locus heterogeneity). For example, mutations in TP53 are linked to 32 clinically distinguishable forms of cancer and cancer-related disorders, whereas mutations in any of at least 12 different genes can lead to long QT syndrome.
With the publication of several large-scale protein-protein interaction networks in human5-8, researchers have recently begun to use complex cellular networks to explore these genotype-to-phenotype relationships2,9, on the basis that many proteins function by interacting with other proteins. However, most analyses model proteins as graph-theoretical nodes, ignoring the structural details of individual proteins and the spatial constraints of their interactions. Here, we investigate on a large-scale the underlying molecular mechanisms for the complex genotype-to-phenotype relationships by integrating three-dimensional (3D) atomic-level protein structure information with high-quality large-scale protein-protein interaction data. Within the framework of this structurally resolved protein interactome, we examine the relationships among human diseases and their associated genes and mutations.
Results
Construction of structurally resolved protein interactome for human disease
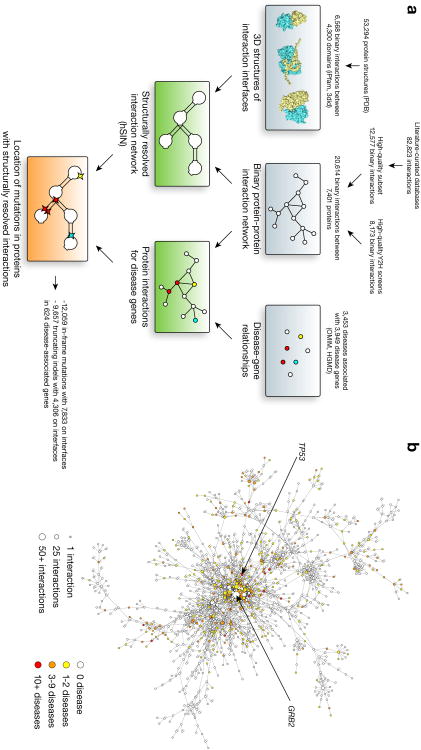
We first combined 12,577 reliable literature-curated binary interactions filtered from six widely used databases10-15 (Online Methods) and 8,173 well-verified high-throughput yeast two-hybrid (Y2H) interactions5-8 to produce the high-quality human protein interaction network (hPIN) with 20,614 interactions between 7,401 proteins (Fig. 1a).
Figure 1.
Disease-associated proteins in the human structural interaction network (hSIN). (a) The procedure used to create the structural interaction network and to relate disease genes and mutations to this network. (b) Network representation of the main connected component of hSIN. Nodes represent proteins and edges correspond to structurally resolved interactions. Colored nodes indicate disease-associated proteins. The arrows point to the two main hubs: disease hub TP53 with 32 diseases and interaction hub GRB2 with 56 structurally resolved interactions. (c) Co-expression correlation of interacting proteins in the unfiltered interaction network, hPIN and hSIN. (d) Enrichment of functionally similar pairs in the unfiltered interaction network, hPIN and hSIN.
Next, we structurally resolved the interfaces of these interactions using a homology modeling approach16. We used both iPfam17 and 3did18 to identify the interfaces of two interacting proteins by mapping them to known atomic-resolution 3D structures of interactions in the Protein Data Bank (PDB)19 (Fig. 1a). Only those interactions in which the interacting domains of both partners (or their homologues) can be found in a 3D structure of an interaction are kept, resulting in the human structural interaction network (hSIN) of 4,222 structurally resolved interactions between 2,816 proteins (Fig. 1a). Here, we carefully selected high-quality direct physical interactions between human proteins because interaction databases often contain low quality and/or non-binary interactions20-22, for which interaction interfaces do not exist.
Finally, to compile a comprehensive list of disease-associated genes and their mutations, we combined information from both Online Mendelian Inheritance in Man (OMIM)23 and the Human Gene Mutation Database (HGMD)24 (Fig. 1a). In total, we were able to collect 62,663 Mendelian mutations in 3,949 protein-coding genes associated with 3,453 clinically distinct disorders (Supplementary Note 1), of which 21,716 mutations in 624 disease-associated genes were mapped to corresponding proteins in hSIN (Fig. 1a,b).
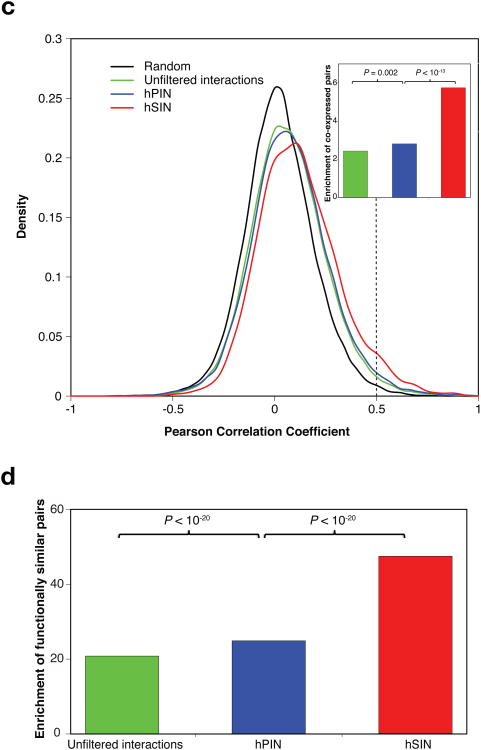
To evaluate the reliability of our homology modeling approach, we cross-validated domain-domain interactions in 1,456 interactions with co-crystal structures and found that over 94% can be correctly inferred from their homologous domains of other interacting pairs in the dataset (Supplementary Note 2). To further verify the quality of hPIN and hSIN, we investigated enrichment of highly co-expressed and functionally similar25 interacting pairs in these networks as well as unfiltered interactions relative to random pairs (Supplementary Note 3). We found that hPIN has a significantly higher enrichment for co-expressed and functionally similar pairs than unfiltered interactions (P = 0.002 and P < 10−20 by cumulative binomial tests, respectively; Fig. 1c,d), verifying the high quality of hPIN and our filtering process. More importantly, hSIN exhibits an even higher enrichment (P < 10−13 and P < 10−20 by cumulative binomial tests, respectively; Fig. 1c,d), illustrating the importance of structural resolution.
Enrichment of in-frame disease mutations on interaction interfaces
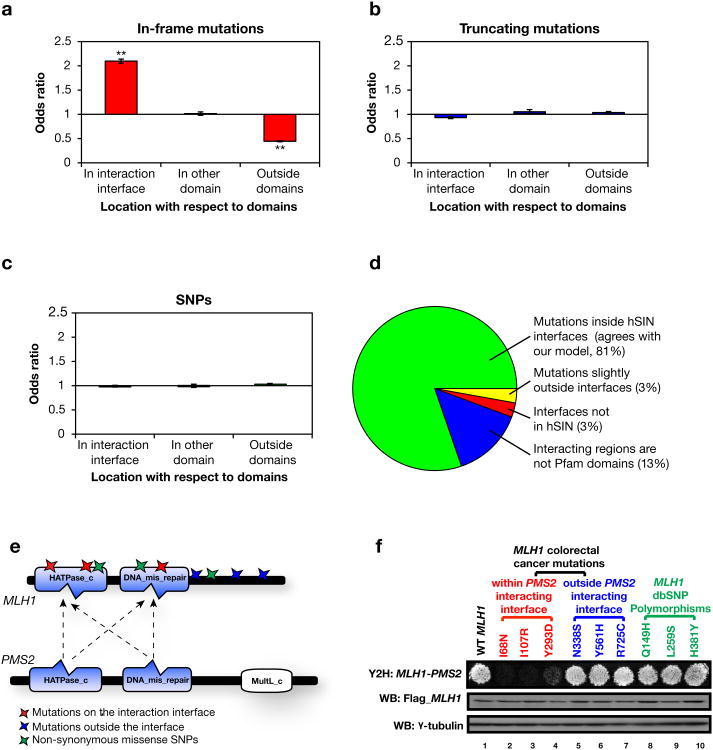
Disease mutations can be classified into two broad categories - in-frame mutations (including missense point mutations and in-frame insertions or deletions) and truncating mutations (including nonsense point mutations and frameshift insertions or deletions). Disease alleles with in-frame mutations are likely to produce full-length proteins with local defects, whereas those with truncating mutations will only give rise to incomplete fragments. Our list comprises 12,059 in-frame mutations and 9,657 truncating mutations from 624 genes in hSIN.
Although individual experiments have shown that in-frame mutations can lead to loss of interactions26, previous studies have concluded that only a small fraction of disease-associated mutations are expected to specifically affect protein-protein interactions27,28. To explore the relationships between mutations and their associated disorders, we investigated positions of the disease-associated mutations with regard to interaction interfaces on the corresponding proteins. Among the 12,059 in-frame mutations, we found that 7,833 are located on interaction interfaces, which is significantly enriched with respect to the relative length of interfaces to whole proteins (Odds ratio = 2.1, P < 10−20 with a Z-test; Fig. 2a). In contrast, an enrichment of in-frame mutations was not detected in other non-interacting domains (Odds ratio = 1.0, P = 0.70 with a Z-test; Fig. 2a). This indicates that specific alteration (disruption or enhancement; Supplementary Note 4) of protein-protein interactions plays an important role in the pathogenesis of many disease genes, more than previously expected27 (Supplementary Note 5). On the other hand, truncating mutations seem to be distributed randomly throughout the protein (Fig. 2b). We also examined the distribution of 13,783 non-synonymous single nucleotide polymorphisms (SNPs)29 in 806 disease genes in hSIN and found that they too are randomly distributed (Fig. 2c and Supplementary Note 6). These results further confirm our conclusion because alleles with truncating mutations are more likely to produce non-functional products26 and most SNPs in dbSNP are considered to be non-disease-related30.
Figure 2.
Analysis of disease-associated mutations with respect to interaction interfaces. (a) Odds ratios for the distribution of in-frame mutations in different locations on proteins in hSIN. **P < 10−20. P-values calculated using Z-tests for the log odds ratios. Error bars indicate ± SE. (b) Odds ratios for the distribution of truncating mutations in different locations on proteins in hSIN. (c) Odds ratios for the distribution of non-synonymous SNPs in different locations on proteins in hSIN. (d) Comparison of hSIN with mutations known to modify protein-protein interactions. (e) Illustration of MLH1 and PMS2 interaction interfaces. Colored stars indicate locations of experimentally tested in-frame mutations and SNPs. (f) Effects of in-frame mutations and SNPs on the MLH1-PMS2 interaction tested by Y2H. Flag tagged wild-type and mutant MLH1 were expressed in HEK293T cells, western blot analysis showed similar levels of MLH1 proteins. γ -tubulin was used as a loading control.
To verify that the in-frame mutations on the interfaces in hSIN can interfere with protein interactions, we manually compared them with an independent list of known interaction-altering missense mutations that could be mapped to genes in hSIN27. The majority (81%) of these mutations (72 mutations in total) are indeed localized on the interaction interfaces according to hSIN (Fig. 2d), confirming the coverage and quality of hSIN (Supplementary Note 7).
We also experimentally evaluated the effects of disease-associated mutations and non-disease-related SNPs found in MLH1, a well characterized human DNA mismatch repair gene frequently mutated in hereditary nonpolyposis colorectal cancer (HNPCC)31. MLH1 is known to interact with many proteins, including its heterodimeric partner PMS2, but the structural basis of most interactions, including with PMS2, still remains unknown. Our hSIN predicts that the HATPase_c domain and the DNA_mis_repair domain on MLH1 are potentially responsible for MLH1's interaction with PMS2 (Fig. 2e). Therefore we hypothesized that mutations within these two domains are likely to alter this interaction. To test our hypothesis, six different in-frame colorectal-cancer-associated mutations and three non-synonymous SNPs found in MLH1 were tested by Y2H for their abilities to alter the MLH1-PMS2 interaction (Supplementary Note 8 and Supplementary Fig. 1). Compared to the wild-type MLH1, only missense mutations (I68N, I107R, Y293D) within the predicted PMS2 interacting interface greatly reduce the MLH1-PMS2 interaction (Fig. 2f). These experimental results further confirm the validity of our predicted interaction interfaces in hSIN. Moreover, they show that in-frame mutations enriched on interfaces could indeed alter corresponding interactions.
Pleiotropy of disease genes - effects of mutations on different interaction interfaces of the same protein
Disease genes are often associated with multiple clinically distinct disorders2. To investigate how mutations in the same gene can cause different phenotypes, we examined the relationships between potentially interaction-altering in-frame disease-associated mutations within our atomic-resolution structural interaction network, hSIN.
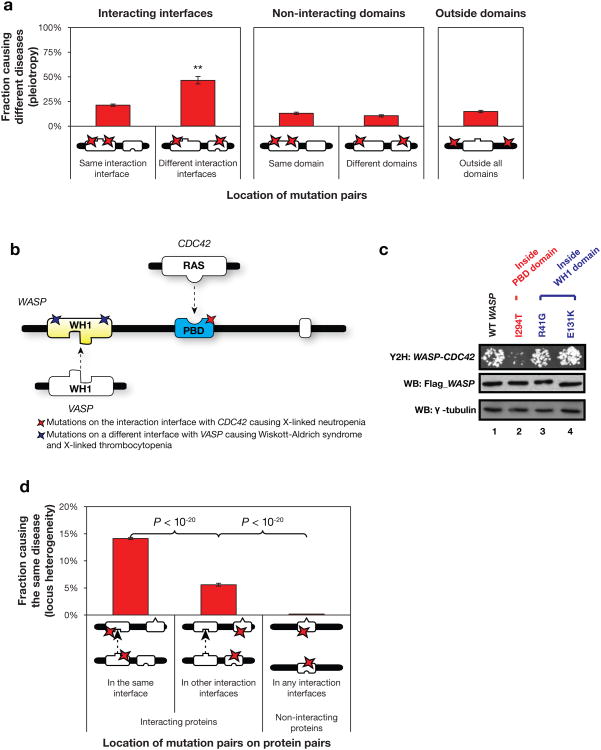
By analyzing the distribution of in-frame mutation pairs on the same gene (Supplementary Note 9), we found that in-frame mutation pairs on different interaction interfaces are more than twice as likely to cause different disorders as those on the same interface (46% and 21% respectively, P < 10−20 by a cumulative binomial test; Fig. 3a). This suggests that the number of interactions and interfaces are key in understanding the pleiotropic effects of disease genes. Mutations on interaction interfaces of the same protein mediating different interactions are more likely to cause distinct interruptions in the overall interactome and can therefore result in different biological consequences and lead to pleiotropic effects. Interestingly, there is no such difference between mutations in different non-interacting domains, further underscoring the importance of protein-protein interactions and their role in understanding disease.
Figure 3.
Analysis of pleiotropy and locus heterogeneity. (a) Fraction of mutation pairs on the same protein causing different diseases. **P < 10−20. P-values calculated using binomial tests. (b) Illustration of WASP and its interaction interfaces with CDC42 and VASP. Colored stars indicate locations of experimentally tested mutations. (c) Effects on the WASP-CDC42 interaction by mutations on different interaction interfaces tested by Y2H. Flag tagged wild-type and mutant WASP were expressed in HEK293T cells, western blot analysis showed similar levels of WASP proteins. γ -tubulin was used as a loading control. (d) Fraction of mutation pairs on two proteins causing the same disease.
One well-studied example of pleiotropy is the Wiskott-Aldrich syndrome protein (WASP)32 (Fig. 3b). Mutations in this protein can give rise to three diseases: Wiskott-Aldrich syndrome (WAS), X-linked thrombocytopenia (XLT) or X-linked neutropenia (XLN). WAS and XLT are related diseases with XLT being a milder form of WAS, both of which are clinically distinct from XLN (Supplementary Note 10). Based on our 3D structural analysis using hSIN, mutations associated with WAS and XLT are in or around the WH1 domain, which is responsible for interaction with VASP; mutations for XLN on the other hand are all inside the PBD domain, which performs an entirely different function by interacting with CDC42 and regulating the auto-inhibition and potentially localization of WASP33-35(Fig. 3b). More interestingly, our experimental results confirm that mutations on different interfaces of WASP function differently in terms of altering protein interactions. Specifically, we compared interactions of CDC42 with the wild-type WASP and three disease-associated variants using Y2H. Neither mutation (R41G and E131K; associated with WAS/XLT) located within WH1 domain affects WASP's interaction with CDC42 (Fig. 3c, Lanes 3 and 4). However, for the first time we provided experimental evidence that one amino acid change within the PBD domain (I294T; associated with XLN) greatly reduces the WASP-CDC42 interaction (Fig. 3c, Lane 2). Previous in vitro analysis has shown that I294T increases WASP activity36, our result suggests that I294T might function by disrupting the WASP-CDC42 interaction, therefore affecting WASP's regulation by CDC42.
Locus heterogeneity – effects of mutations on the corresponding interfaces of two interacting proteins
Uncovering the mechanisms through which mutations in different genes can lead to the same disease is critical in finding novel disease-associated genes and ultimately understanding and treating the corresponding disease. Based on the widely accepted “guilt-by-association” principle, interacting proteins have been shown to have a tendency of sharing similar functions and causing the same disorders37. Earlier implementations of this idea had a significant impact and led to the determination of important disease associations for genes38. However, the fraction of successful predictions is still relatively small39. One main reason is that most interacting protein pairs only share a subset of their associated disorders.
To understand the underlying molecular mechanism for this phenomenon, we calculated the distribution of in-frame mutation pairs on two different proteins that cause the same disorder (Supplementary Note 9). We found, in agreement with previous studies2, that in-frame mutations on interacting proteins are generally much more likely to cause the same disorder (12%) than random expectation (0.17%, P < 10−20 by a cumulative binomial test; Fig. 3d). More importantly, our results show that the likelihood for two in-frame mutations on the corresponding interfaces of the interacting proteins to cause the same disorder (14%) is significantly higher than that for two in-frame mutations on two interfaces not mediating their interaction (5.6%, P < 10−20 by a cumulative binomial test; Fig. 3d). These results further indicate that alteration of specific interactions, caused by mutations on corresponding interfaces of two interacting proteins, plays an important role in the pathogenesis of the same disorder. An interesting example is the hemolytic uremic syndrome, which is associated with mutations on the corresponding interaction interfaces of both CFH and C3 that mediate the interaction between the two proteins40 (Supplementary Note 11 and Supplementary Fig. 2).
Modeling potential molecular mechanisms of disease genes
Our 3D structural analysis provides potential atomic-level understanding for some of the complex genotype-to-phenotype relationships. More importantly, these results enable us to generate a concrete molecular mechanism hypothesis for mutations of a certain disorder enriched on a specific interaction interface – they may cause their associated disorders via alteration of the interactions mediated by the corresponding interfaces (Fig. 4a, Supplementary Fig. 3 and Supplementary Note 4). Based on this proposed model, we can further predict new disease-associated genes (those that interact with known disease genes through the interfaces enriched with mutations associated with a certain disease; Supplementary Note 12 and Supplementary Fig. 4). Therefore, our analysis provides a much higher resolution application of the “guilt-by-association” principle. We then applied this principle to uncover unknown disease-associated genes using hSIN. For each disease, we selected proteins in hSIN that have at least 3 mutations associated with a certain disease and at least 1.5-fold enrichment on interaction interfaces (Online Methods and Supplementary Note 13). Other proteins interacting through the interfaces with enriched disease-specific mutations are predicted to be associated with the corresponding disease. In total, we predicted 292 new disease genes for 182 different diseases, representing 694 novel disease-to-gene associations. Using three-fold cross-validation, we confirmed that our structurally resolved interactome greatly improves the performance of predicting disease-associated genes, compared with existing interaction networks where proteins are modeled as simple graph-theoretical nodes (Supplementary Note 13 and Supplementary Figs. 5 and 6).
Figure 4.
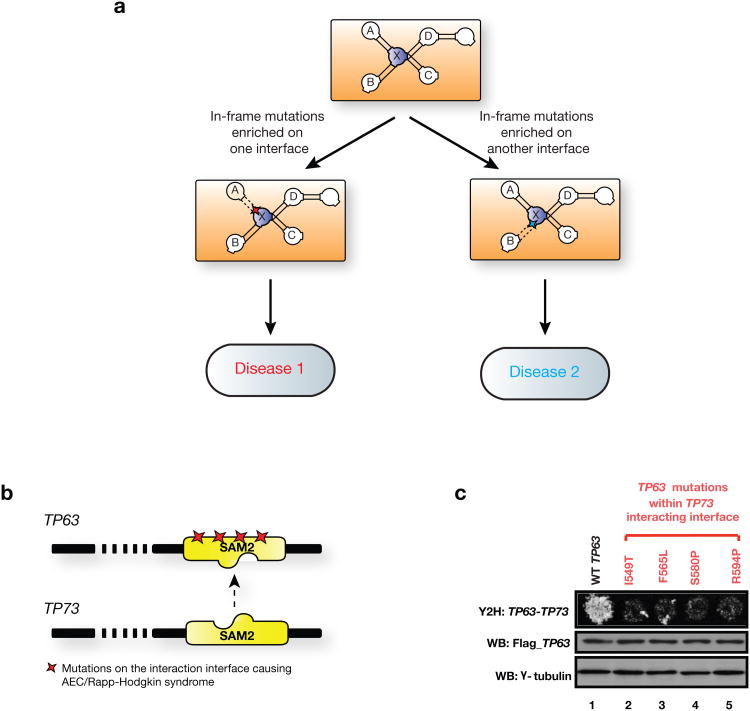
Modeling molecular mechanisms of disease genes and mutations through our structurally resolved interaction network. (a) Schematic illustration of using hSIN to understand complex genotype-to-phenotype relationships. In-frame mutations enriched on an interaction interface of protein X likely alter the interaction between protein X and A, leading to one disease, while mutations enriched on a different interface likely to alter the interaction between X and B, leading to another disease. Interactions between protein X and C, as well as X and D are likely to be intact under both scenarios. (b) Illustration of TP63 and its predicted interaction interface with TP73. Colored stars indicate locations of experimentally tested mutations. (c) Effects on the TP63-TP73 interaction by mutations on the predicted interacting interface tested by Y2H. Flag tagged wild-type and mutant TP63 were expressed in HEK293T cells, western blot analysis showed similar levels of TP63 proteins. γ -tubulin was used as a loading control.
To further experimentally validate our predictions, we examined the TP63-TP73 interaction. Unlike its paralog, the well-known tumor suppressor gene TP53, TP63 has an important role in epithelial development41. Sequence analysis suggested TP63 mutations are responsible for Ankyloblepharon-ectodermal defect-cleft lip/palate (AEC) and Rapp-Hodgkin syndrome, two clinically similar disorders (Supplementary Note 14)42. Interestingly, most of mutations cluster in the SAM2 domain of TP63. Based on the known co-crystal structure of DGKD homodimer43, we predict that the SAM2 domain is potentially part of the interface for the TP63-TP73 interaction (Fig. 4b). Therefore, we hypothesized that mutations in the SAM2 domain could affect this interaction. We examined four mutations associated with AEC/Rapp-Hodgkin syndrome in the SAM2 domain (I549T, F565L, S580P, R594P) using Y2H. The protein expression levels of the mutants are comparable to the wild-type TP63 (Fig. 4c, middle panel). Our Y2H results indicate that all four mutations cause great reduction of the TP63-TP73 interaction. This suggests that the disruption of proper binding between TP63 and TP73 might contribute to the observed phenotypes, and thus TP73 might also be involved in AEC/Rapp-Hodgkin syndrome.
Discussion
From our 3D analysis of disease-associated mutations and their corresponding genes within the atomic-level structurally resolved human protein interactome, we find that specific alteration of protein interactions by in-frame mutations plays an important role in the pathogenesis of many disease genes. More importantly, our results show that the locations of the mutations with respect to the interaction interfaces are crucial in understanding the complex genotype-to-phenotype relationships, including pleiotropy and locus heterogeneity. All observations are demonstrated to be robust to the removal of random interactions and proteins as well as interaction, disease and domain hubs, potential biases that might be present in our datasets (Supplementary Note 15 and Supplementary Figs. 7-22). Furthermore, all observations remain the same when the calculations are repeated using only known domain-domain interactions from existing co-crystal structures (Supplementary Note 16 and Supplementary Fig. 22). Our findings are directly applicable to understanding molecular mechanisms of human genetic diseases and discovering new disease-associated genes and mutations both experimentally and computationally, which is of significant interest to both pharmaceutical and medical industries and especially important for treating diseases currently with undruggable target genes. To this end, we provide a list of novel disease-to-gene associations and generate many new hypotheses. Moreover, with the development of exome sequencing, many mutations are being discovered in every study44. It is difficult to determine their functional relevance experimentally all at once. Our analysis could potentially provide a novel approach to prioritize mutations discovered in large-scale sequencing projects, especially for protein pairs without known co-crystal structures.
The construction of our structurally resolved protein interactome largely relies on the availability of 3D co-crystal structures, which limits the coverage of our network. However with the rapid growth of PDB45, more co-crystal information will become available and the same principles that we developed here can be readily applied to uncover potential molecular mechanisms of many more disease genes whose structural information is currently missing. Another limiting factor is that some interaction interfaces fall outside of the known domain structures, including the disordered regions46. Incorporating this type of information will further improve the coverage of hSIN. Moreover, other parts of the protein, especially regions immediately outside of the interacting domains we predicted, might also contribute to the interaction directly or contribute to the correct folding of the corresponding domains. For example, a previous study indicated that the SAM2 domain alone might not be sufficient for the TP63-TP73 interaction and suggested that residues upstream and downstream of the SAM2 domain and the P53_tetramer domain could also be involved in the interaction47. Accordingly, based on the known co-crystal structure of TP53 homodimer48, we also predicted in hSIN that the P53_tetramer domain of TP63 could also be part of the interface for this interaction.
Although we have shown that the interaction pairs in hSIN have significantly higher co-expression correlation and functional similarity in general, further studies can be carried out by considering gene expression under disease-specific conditions and/or within corresponding tissues for specific disorders. Moreover, study of changes in the protein-protein interaction network during disease progression can also assist the identification of disease biomarkers and modules49. In addition to genetic mutations, many other factors including environmental stress, epigenetic modifications and invasion of pathogens might also contribute to human clinical disorders50. Integrating these factors in the follow-up studies of the hypotheses generated by our analysis will likely expand our understanding of many human genetic disorders in the near future.
Supplementary Material
References
- 1.Pasternak J. An Introduction to Human Molecular Genetics. 2nd. Wiley; Hoboken, NJ: 2005. [Google Scholar]
- 2.Goh KI, et al. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dermitzakis ET, Clark AG. Genetics. Life after GWA studies. Science. 2009;326:239–240. doi: 10.1126/science.1182009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 5.Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 6.Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 7.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, et al. Next-generation sequencing to generate interactome datasets. Nat Methods. 2011;8:478–480. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proc Natl Acad Sci U S A. 2008;105:4323–4328. doi: 10.1073/pnas.0701722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aranda B, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceol A, et al. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Z, et al. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner B, et al. iRefWeb: interactive analysis of consolidated protein interaction data and their supporting evidence. Database (Oxford) 2010;2010:baq023. doi: 10.1093/database/baq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim PM, Lu LJ, Xia Y, Gerstein MB. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 17.Finn RD, Marshall M, Bateman A. iPfam: visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 18.Stein A, Panjkovich A, Aloy P. 3did Update: domain-domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37:D300–304. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cusick ME, et al. Literature-curated protein interaction datasets. Nat Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turinsky AL, Razick S, Turner B, Donaldson IM, Wodak SJ. Literature curation of protein interactions: measuring agreement across major public databases. Database (Oxford) 2010;2010:baq026. doi: 10.1093/database/baq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Jansen R, Stolovitzky G, Gerstein M. Total ancestry measure: quantifying the similarity in tree-like classification, with genomic applications. Bioinformatics. 2007;23:2163–2173. doi: 10.1093/bioinformatics/btm291. [DOI] [PubMed] [Google Scholar]
- 26.Zhong Q, et al. Edgetic perturbation models of human inherited disorders. Mol Syst Biol. 2009;5:321. doi: 10.1038/msb.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster-Bockler B, Bateman A. Protein interactions in human genetic diseases. Genome Biol. 2008;9:R9. doi: 10.1186/gb-2008-9-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrer-Costa C, Orozco M, de la Cruz X. Characterization of disease-associated single amino acid polymorphisms in terms of sequence and structure properties. J Mol Biol. 2002;315:771–786. doi: 10.1006/jmbi.2001.5255. [DOI] [PubMed] [Google Scholar]
- 29.Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium TGP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peltomaki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 32.Thrasher AJ, Burns SO. WASP: a key immunological multitasker. Nat Rev Immunol. 2010;10:182–192. doi: 10.1038/nri2724. [DOI] [PubMed] [Google Scholar]
- 33.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 34.Higgs HN, Pollard TD. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J Cell Biol. 2000;150:1311–1320. doi: 10.1083/jcb.150.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moulding DA, et al. Unregulated actin polymerization by WASp causes defects of mitosis and cytokinesis in X-linked neutropenia. J Exp Med. 2007;204:2213–2224. doi: 10.1084/jem.20062324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ancliff PJ, et al. Two novel activating mutations in the Wiskott-Aldrich syndrome protein result in congenital neutropenia. Blood. 2006;108:2182–2189. doi: 10.1182/blood-2006-01-010249. [DOI] [PubMed] [Google Scholar]
- 37.Oliver S. Guilt-by-association goes global. Nature. 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Gulbahce N, Yu H. Network-based methods for human disease gene prediction. Brief Funct Genomics. 2011 doi: 10.1093/bfgp/elr024. [DOI] [PubMed] [Google Scholar]
- 39.Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein-protein interactions. J Med Genet. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noris M, Remuzzi G. Atypical hemolyticuremic syndrome. N Engl J Med. 2009;361:1676–1687. doi: 10.1056/NEJMra0902814. [DOI] [PubMed] [Google Scholar]
- 41.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 42.Bougeard G, Hadj-Rabia S, Faivre L, Sarafan-Vasseur N, Frebourg T. The Rapp-Hodgkin syndrome results from mutations of the TP63 gene. Eur J Hum Genet. 2003;11:700–704. doi: 10.1038/sj.ejhg.5201004. [DOI] [PubMed] [Google Scholar]
- 43.Harada BT, et al. Regulation of enzyme localization by polymerization: polymer formation by the SAM domain of diacylglycerol kinase delta1. Structure. 2008;16:380–387. doi: 10.1016/j.str.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 44.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011 doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 45.Chandonia JM, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–351. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 46.Neduva V, et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi SW, Ayed A, Arrowsmith CH. Solution structure of a conserved C-terminal domain of p73 with structural homology to the SAM domain. EMBO J. 1999;18:4438–4445. doi: 10.1093/emboj/18.16.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clore GM, et al. Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat Struct Biol. 1995;2:321–333. doi: 10.1038/nsb0495-321. [DOI] [PubMed] [Google Scholar]
- 49.Hwang D, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.