Abstract
The sea slug Elysia chlorotica offers a unique opportunity to study the evolution of a novel function (photosynthesis) in a complex multicellular host. Elysia chlorotica harvests plastids (absent of nuclei) from its heterokont algal prey, Vaucheria litorea. The “stolen” plastids are maintained for several months in cells of the digestive tract and are essential for animal development. The basis of long-term maintenance of photosynthesis in this sea slug was thought to be explained by extensive horizontal gene transfer (HGT) from the nucleus of the alga to the animal nucleus, followed by expression of algal genes in the gut to provide essential plastid-destined proteins. Early studies of target genes and proteins supported the HGT hypothesis, but more recent genome-wide data provide conflicting results. Here, we generated significant genome data from the E. chlorotica germ line (egg DNA) and from V. litorea to test the HGT hypothesis. Our comprehensive analyses fail to provide evidence for alga-derived HGT into the germ line of the sea slug. Polymerase chain reaction analyses of genomic DNA and cDNA from different individual E. chlorotica suggest, however, that algal nuclear genes (or gene fragments) are present in the adult slug. We suggest that these nucleic acids may derive from and/or reside in extrachromosomal DNAs that are made available to the animal through contact with the alga. These data resolve a long-standing issue and suggest that HGT is not the primary reason underlying long-term maintenance of photosynthesis in E. chlorotica. Therefore, sea slug photosynthesis is sustained in as yet unexplained ways that do not appear to endanger the animal germ line through the introduction of dozens of foreign genes.
Keywords: symbiosis, horizontal gene transfer, Elysia chlorotica, Vaucheria litorea, genomics, kleptoplasty
Introduction
Many species of herbivorous sacoglossan molluscs (sea slugs) exhibit kleptoplasty, which is the uptake and retention of plastids as a result of feeding on algal prey (see reviews by Trench 1975; Jensen 1997; Yamamoto et al. 2009; Rumpho et al. 2011; Wägele et al. 2011; Pierce and Curtis 2012). Individual kleptoplastic associations vary widely in terms of specificity of the animal toward its algal prey and in retention time and functionality of the captured plastids (see reviews by Evertsen et al. 2007; Evertsen and Johnsen 2009; Händeler et al. 2009; Wägele et al. 2011; Rumpho et al. 2011; Maeda et al. 2012). Elysia chlorotica is at one extreme because its known algal prey is limited to two marine species of the coenocytic heterokont genus Vaucheria (i.e., Vaucheria litorea and V. compacta; West 1979; unpublished observations), and because it retains functional, nondividing plastids in the cells lining the digestive tract for several months (Pierce et al. 1996; Green et al. 2000). The long-term functioning of V. litorea plastids in E. chlorotica is perplexing because no evidence exists for the presence of algal nuclei in the sea slug, and the majority of plastid proteins are encoded in the nuclear genome, in addition to all the nuclear regulatory factors that are targeted to the plastid (Rumpho et al. 2008; Woodson and Chory 2008; Puthiyaveetil et al. 2010).
Several hypotheses have been proposed to explain how the plastids continue to function within the animal cells, including horizontal gene transfer (HGT) of essential photosynthesis genes from the nuclear genome of the algal prey to the sea slug nuclear genome. The animal digestive cells presumably come into contact with algal nuclear DNA during suctorial feeding on Vaucheria, although mechanisms to explain transfer of HGT-derived genes to the germ line of the animal have not yet been proposed. The HGT hypothesis in E. chlorotica has been driven by biochemical data (Pierce et al. 1996, 2010; Rumpho et al. 2008, 2009; Soule and Rumpho 2012), polymerase chain reaction (PCR), and real time quantitative reverse transcription-PCR (qRT-PCR) data (Pierce et al. 2007; Rumpho et al. 2008, 2009; Schwartz et al. 2010; Soule and Rumpho 2012; present study), and most recently by the analysis of E. chlorotica transcriptome data (Pierce et al. 2012).
Sequence analysis of transcript data from multiple adult photosynthetic E. chlorotica has provided conflicting evidence for Vaucheria- or other algal-derived transcripts in this animal (Pelletreau et al. 2011; Pierce et al. 2012). In the initial transcriptome studies carried out by our laboratory, no evidence was found in the E. chlorotica raw or assembled transcripts for horizontally transferred algal genes (Pelletreau et al. 2011). This was based on Basic Local Alignment Search Tool (BLAST) analysis against the nonredundant National Center for Biotechnology Information (NCBI) database and also by using Phylosort, a phylogenomic pipeline designed to identify candidates of gene transfer (Moustafa et al. 2009). Although a handful of putatively transferred genes (mostly of bacterial or viral origin) emerged from these analyses, they were neither algal derived nor involved in photosynthetic processes. This study complemented the results obtained by Wägele et al. (2011) whose partial transcriptome data from two, albeit very different kleptoplastic sea slugs (E. timida and Plakobranchus ocellatus), also revealed no evidence for HGT. In contrast, Pierce et al. (2012) reported the presence of 111 algal transcripts, 52 of which are the products of nuclear genes, in the adult nuclear genome of E. chlorotica. Although each of these transcripts was only found as a singleton read in the animal transcriptome library, and none were greater than 90 bp in length, these authors presumed the presence of these transcripts indicated HGT from the algal nucleus and integration into sea slug genomic DNA (Pierce et al. 2012). More recent data clearly indicate the presence of algal transcripts in the form of singletons in the animal transcriptome; however, no data to date provide strong evidence for integration of intact algal nuclear genes into the sea slug genome or, more specifically, into the germ line (also see discussion with supplementary fig. S1, Supplementary Material online).
Given these conflicting data, we took the direct approach of generating significant genome data from naturally aposymbiotic E. chlorotica egg DNA that had never come in contact with algal prey. We analyzed individual genome reads using BLAST methods to search for evidence of HGT. To strengthen our analysis, we also generated transcriptome data from the algal prey V. litorea and used these sequences to query the egg DNA to look for instances of DNA transfer. Finally, we used all the putative HGT transcript data from Pierce et al. (2012) as a query against egg DNA. Taken together, the results of our analyses indicate the absence of algal-derived genes in the germ line of E. chlorotica. Therefore, all these results point to yet-to-be-explained mechanisms for the long-term maintenance of photosynthesis in the gut of this animal. Here, we present initial results from the PCR analysis of individual animals that suggest the existence of a novel mechanism of transient gene expression in adult sea slugs that may help explain the conflicting data.
Results and Discussion
Looking for Hits to V. litorea Transcripts in the E. chlorotica Genome
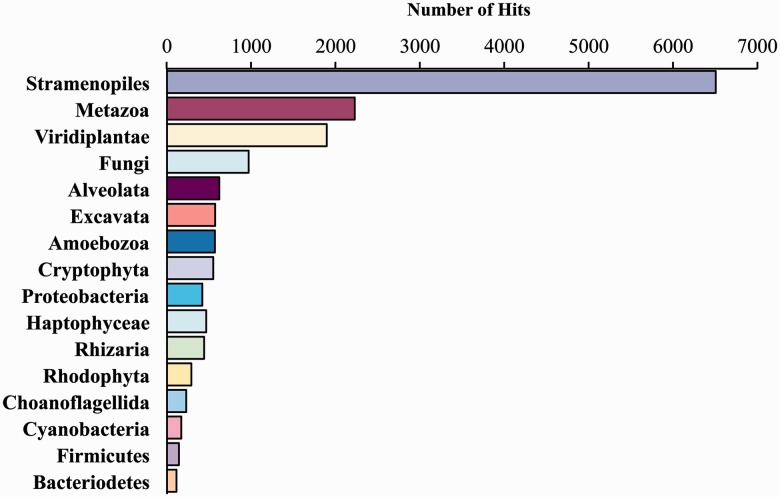
Sequencing of the V. litorea transcriptome on an Illumina MiSeq instrument generated 21.3 million paired-end reads (average size = 151 bp) totaling 3.2 Gbp. These data were assembled into 65,468 contigs (N50 = 824 bp, median coverage = 2.3×, total assembly size = 25.5 Mbp). The contigs were analyzed using BLASTx (e-value cutoff ≤ 10−5) against our comprehensive local database (e.g., Chan et al. 2011; Price et al. 2012). This database consists of 16.9 million protein sequences derived from RefSeq v.51 with the addition of sequenced eukaryote (e.g., fungal, metazoan, viridiplant, and stramenopile) genomes from the Joint Genome Institute (http://www.jgi.doe.gov/, last accessed May 16, 2013) and six-frame translated eukaryote expressed sequence tag (EST) sequences obtained from the NCBI dbEST (http://www.ncbi.nlm.nih.gov/dbEST/, last accessed May 16, 2013). The data showed that 17,311 EST contigs encode proteins with hits to the database with many showing the expected “chromalveolate” (e.g., diatoms and oomycetes [stramenopiles]) affiliation (fig. 1; supplementary table S1, Supplementary Material online). We also generated 153.2 million paired-end Illumina reads of average size 146 bp from E. chlorotica total egg genomic DNA, totaling 22.5 Gbp of data. BLASTn analysis using the assembled V. litorea cDNA sequences as a query against the sea slug egg genome database at an e-value cutoff of ≤10−5 yielded 1,298 top hits to 241 nonredundant reads from the sea slug (supplementary table S2, Supplementary Material online). Three sea slug genomic sequences that were inserted into the algal query data set as positive controls were returned in the search, indicating that our BLAST parameters were appropriate for a large database of short-read data (results not shown).
Fig. 1.
BLASTx results using assembled Vaucheria litorea cDNA contigs to query a comprehensive local genome database. This database consists of 16.9 million protein sequences derived from RefSeq v.51 plus sequenced eukaryote (e.g., fungal, metazoan, viridiplant, and stramenopile) genomes from the Joint Genome Institute (http://www.jgi.doe.gov/, last accessed May 16, 2013) and six-frame translated eukaryote EST sequences obtained from the NCBI dbEST (http://www.ncbi.nlm.nih.gov/dbEST/, last accessed May 16, 2013).
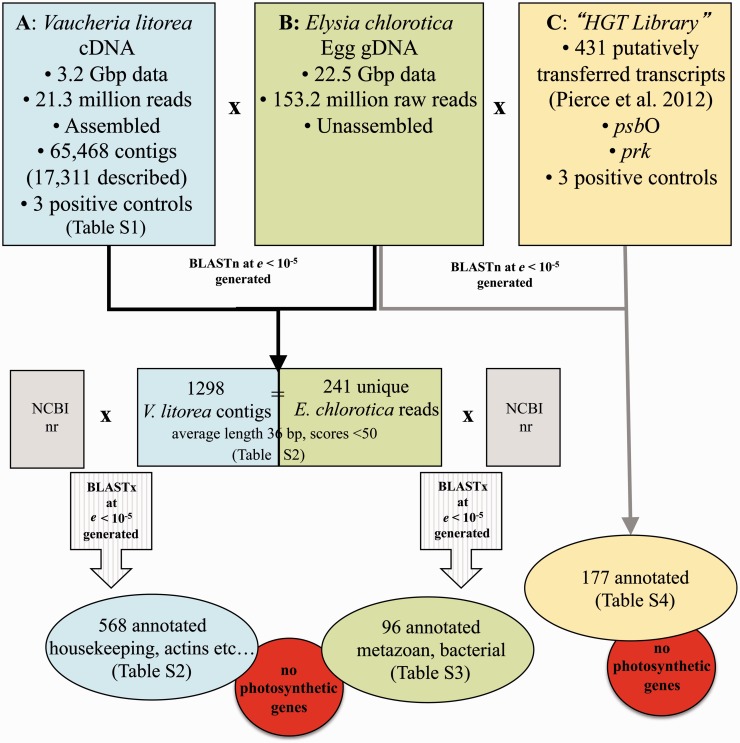
Although a large number of hits were returned between the algal EST contigs and animal singletons, the average length of these hits was 36 bp (four were >100 bp), and only 76 had an e-value less than 10−20. Nonetheless, to investigate the function of these algal hits, each sequence was annotated using the NCBI nr database. This analysis showed that 730 had no significant hit to the NCBI database, and the remaining 568 contigs were comprised predominantly of housekeeping and/or conserved genes (e.g., actins, tubulins, and some hypothetical proteins; see supplementary table S2, Supplementary Material online; fig. 2). Although this comparison generated numerous top hits to stramenopiles, this result reflects the phylogenetic affiliation of the V. litorea query sequence and does not indicate that the homolog in E. chlorotica is of algal origin. Indeed, when the corresponding E. chlorotica singletons (241 were unique of the 1,298 matches) were blasted against the NCBI nr database, 96 were annotated and many of the top hits were to metazoan or bacterial sequences (supplementary table S3, Supplementary Material online; fig. 2). A handful of genes appeared to be of plant origin; however, on closer inspection, each of these genes aligned to the same singleton (see supplementary table S3, Supplementary Material online; HWUSI-EAS1588:2:10:13875:15105#0/3) derived from E. chlorotica, which, alone, had no significant similarity to any gene in NCBI. Therefore, in spite of the finding of regions of limited similarity between the algal transcriptome and the animal genome (as might be expected for such complex data), we conclude that genomic DNA from E. chlorotica eggs is devoid of any full-length (or identifiable, partial) genic regions from V. litorea. Most importantly, our analysis does not return any genes or DNA regions shared by V. litorea and the sea slug that are involved in photosynthesis.
Fig. 2.
Comparative gernomics approach used to test for the presence of alga-derived genes (i.e., HGT candidates) in the germ line of Elysia chlorotica. (A) Algal transcriptome generated in this study. (B) Sea slug egg genomic data generated in this study. (C) Cummulative library of HGT candidates from past studies. Results of the various comparisons are illustrated and listed in detail in the indicated supplementary tables, Supplementary Material online. No algal or photosynthetic genes were detected in any of the queries performed with the E. chlorotica genomic reads.
Examining Possible Instances of HGT in the E. chlorotica Genome
The results described earlier, combined with our previous work (Pelletreau et al. 2011), stand in stark contrast to the results of Pierce et al. (2012). These latter authors compared a transcriptome from pooled adult, starved E. chlorotica against a draft assembly of the genome of V. litorea and with EST contigs (independently derived from our data) from this alga, and identified 111 HGT candidates (derived from 431 ESTs). Of these 111 animal-derived individual transcripts (out of a total of 98,238,204 EST reads [i.e., 0.00011% of the total read count]), 52 were nuclear-encoded plastid proteins with 27 involved directly in photosynthesis. Given these results, we searched the 22.5 Gbp of raw sea slug egg genome reads that we generated for hits to any of the existing photosynthesis genes that have been identified in previous transcriptome analyses of adult E. chlorotica (“HGT Library” fig. 2C). These included the 431 transcripts identified by Pierce et al. (2012) and the psbO and prk transcripts identified using targeted PCR (Rumpho et al. 2008, 2009; Schwartz et al. 2010). We found BLASTn hits (e-value cut off of ≤10−5) for 177 of the 431 transcript sequences from Pierce et al. (2012) in our E. chlorotica egg genome library (supplementary table S4, Supplementary Material online); however, none of the hits, aside from an HSP70 molecular chaperone, had a blast score more than 59 or an alignment length of greater than 46 nucleotides (see example in supplementary fig. S2, Supplementary Material online). All three positive control sequences were returned in the BLAST output. Thus, we were unable to identify any of the putative algal-derived examples of HGT in individual reads from the egg genome.
Testing the Robustness of Our Analysis
One commonly cited reason for the lack of evidence of HGT in transcriptomic and genomic analysis is insufficient coverage resulting in the failure to detect rare transcripts or low copy number genes (e.g., Pierce et al. 2012). To test the completeness of our egg genome sequence data, we mapped the reads onto the E. chlorotica mitochondrial genome (Rumpho et al. 2008) using the CLC Genomics Workbench (similarity = 0.90, length fraction = 0.90). This analysis showed that the egg data spanned 14,132 bp (100%) of the organelle genome with an average coverage of 1,976×. We repeated the analysis with the nuclear-encoded histone H3 and actin sequences available in GenBank (accession nos. JN819183 and AF448493) and found our data span 100% of each coding region at average coverage of 10,319× and 126×, respectively. Finally, we mapped the E. chlorotica egg genome reads against a subset of the assembled EST contigs from Pelletreau et al. (2011) that had a length ≥300 bp (i.e., a total of 16,012 contigs). This analysis showed that 16.01 million reads were aligned (at a stringency of 92.5% similarity over 55% coverage; the latter to allow for introns in the egg genome) to 15,771 (98.5%) of the EST contigs. The median coverage of the mapping was 19.23×, and the average coverage was 198×. These results suggest that the majority of the animal gene inventory is present in our egg genome data and, therefore, is present in the germ line. The coding regions we have identified occurred in gene-rich regions that have more than 100× average genome read coverage. Nevertheless, we stress that our results do not prove the absence of HGT candidates in the nuclear genome but do, however, argue strongly against their presence. To unambiguously prove this point will require a completed sea slug genome sequence.
Previous studies have demonstrated the presence of plastid-encoded gene transcripts in adult sea slugs, even though polyadenylated transcripts were targeted to exclude much of the plastid-derived RNA during library construction (Müjer et al. 1996; Pelletreau et al. 2011; Pierce et al. 2012). The presence of plastid transcripts is nevertheless expected because sea slugs maintain photosynthesis for protracted periods of time (at least 6 months). Given this observation, it was also crucial to show that our egg DNA assembly did not contain plastid genes. To address this issue, we used BLASTn and the completed V. litorea plastid genome (Rumpho et al. 2008) as a query against the egg DNA data. This analysis did not turn up a single significant hit (e-value cut off ≤10−5), indicating the absence of algal plastid DNA (i.e., lack of algal contamination) in our sea slug egg genomic DNA preparation.
Explaining the Inconsistency in Evidence for HGT in Sea Slugs
As discussed earlier, previous work has suggested in E. chlorotica the presence of genomic DNA or cDNA encoding Vaucheria-derived genes involved in photosynthesis, not only in adult animals but also in veliger and egg DNA (for discussion, see Pierce et al. 2012). Given that our analysis of genomic and EST data do not provide any support for the presence of algal genes of any kind in the germ line of the sea slug, it becomes important to explain the findings of algal genes in previous studies. The absence of algal genes found in our analysis of the egg genome and in a comprehensive analysis of the V. litorea cDNAs leaves at least two possible explanations for the inconsistency in current data. The first is that contamination of algal DNA or cDNAs in the PCR reactions may have provided artifactual evidence for Vaucheria sequences in the sea slug. This explanation seems on the surface to be unlikely because the PCR experiments have been done by different investigators over the course of several years and have relied on different DNA/cDNA sources and PCR reagents. The second explanation is that the trace amounts of algal DNA recovered using PCR may indicate true HGT events that have simply gone undetected in our draft assembly. If, however, our egg genome data are an accurate representation of this compartment, then it is also possible that the algal DNAs are not integrated into the egg genome but rather may reside in extrachromosomal or other forms of DNA that were not sequenced using our approach (see Discussion later). Therefore, regardless of whether evidence is ultimately found for algal DNAs in the sea slug genome, the overwhelming evidence until now suggests that, consistent with a growing list of other studies, no evidence exists for massive HGT from partner symbionts into chromosomal DNA of the host (e.g., Wisecaver and Hackett 2010; Pelletreau et al. 2011; Wägele et al. 2011; Pillet and Pawlowski 2013). An interesting exception is the recent finding of gene acquisition “from the environment” by bdelloid rotifers that provides this lineage with novel physiological adaptations (Boschetti et al. 2012).
A Final Note on Study Design and PCR Data
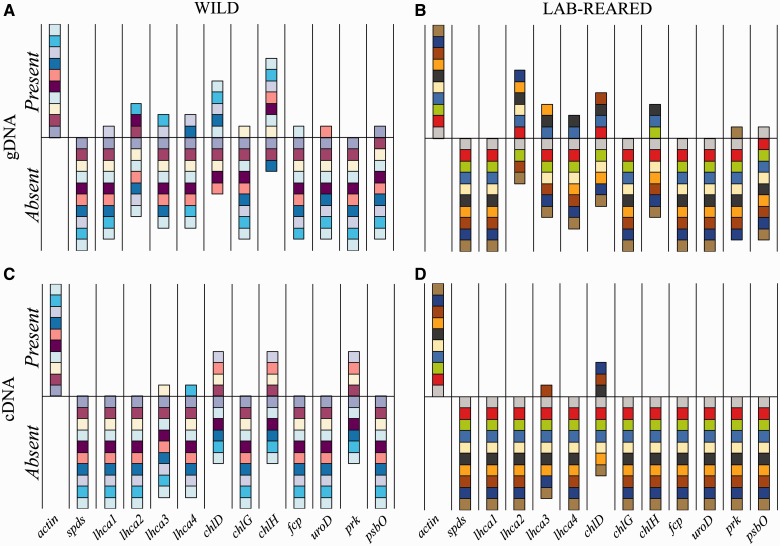
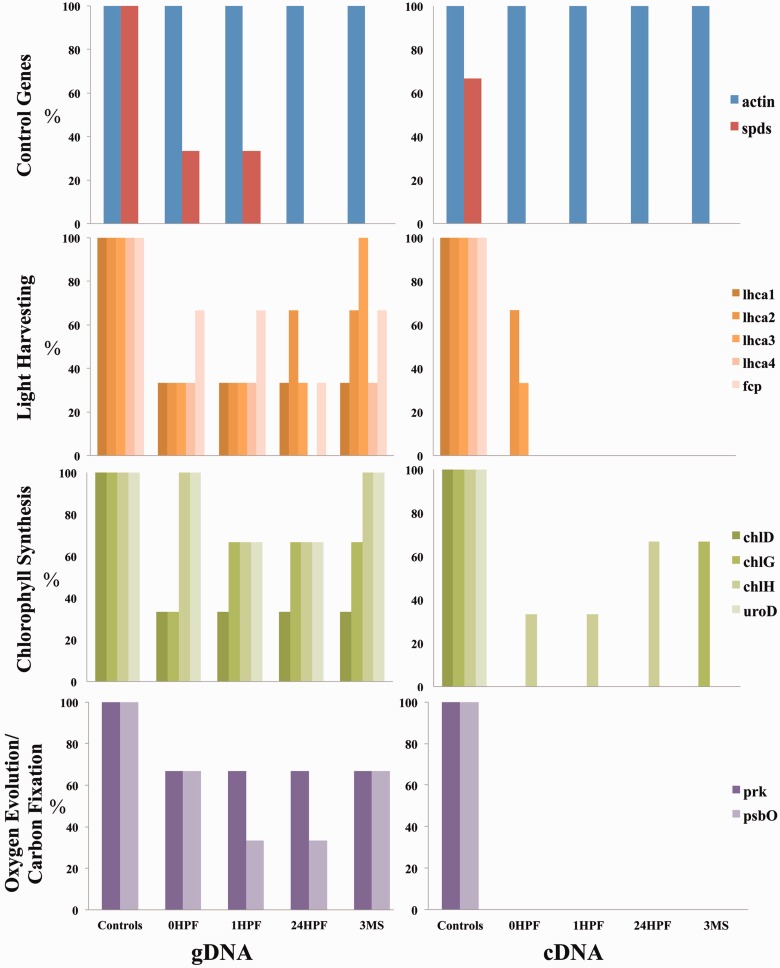
We suggest that the design of existing studies may help to explain the conflicting data regarding alga-derived HGT in the sea slug. In particular, previous studies have relied on the use of multiple pooled animals rather than individuals when characterizing HGT. Here, using PCR primers that target different HGT candidates (table 1), we found an inconsistent pattern with respect to the presence or absence of candidate genes between different individuals (fig. 3) and also over time (fig. 4). Using these preliminary data, we identified three major trends: 1) there is high variability in the presence/absence of HGT-implicated genes among individual animals, regardless of treatment (i.e., time spent feeding or breeding history), 2) the presence of a gene is not always consistent with the presence of transcripts from the same individual, supporting very low copy number of any particular transcript, and 3) traditional PCR continues to provide evidence that these genes are present in photosynthesizing adults, suggesting their role in individual animal growth and development. By using individuals and looking at exposure to algal DNA as variables, we identified a dynamic process of gene presence and expression existing in individual adult animals. For example, if one were to pool the three biological replicates used in figure 4, then all 11 of these genes would be “present” in the DNA leaving the false impression that HGT is present in all members of the sea slug population. In earlier work with this system, PCR-based approaches provided positive results for only 15–20% of the samples (Pierce et al. 2007), which may be explained by this large amount of interindividual variability.
Table 1.
Eleven Candidate Nuclear-Encoded Algal Genes That Have Been Implicated in HGT in Elysia chlorotica and Two Positive Control Genes (one algal and one animal) Used to Assess Interindividual Variability and Change over Time.
| Gene Target | Primer Name | Primer Sequence Source | Function |
|---|---|---|---|
| Spermadine synthase | Spds | Pierce et al. (2009) | Algal nuclear control |
| Beta actin | β-actin | Present study | Animal nuclear control |
| Light harvesting protein 1 | Lhc1 | Pierce et al. (2010) | Light harvesting |
| Light harvesting protein 2 | Lhc2 | Pierce et al. (2010) | Light harvesting |
| Light harvesting protein 3 | Lhc3 | Pierce et al. (2010) | Light harvesting |
| Light harvesting protein 4 | Lhc4 | Pierce et al. (2010) | Light harvesting |
| Fucoxanthin binding protein | fcp | Pierce et al. (2007) | Light harvesting |
| Uroporphyrinogen decarboxylase | uroD | Pierce et al. (2010) | Chlorophyll synthesis |
| Chlorophyll synthetase D | chlD | Pierce et al. (2010) | Chlorophyll synthesis |
| Chlorophyll synthetase G | chlG | Pierce et al. (2010) | Chlorophyll synthesis |
| Chlorophyll synthetase H | chlH | Pierce et al. (2010) | Chlorophyll synthesis |
| Manganese stabilizing protein | psbO | Rumpho et al. (2008) | Oxygen evolution |
| Phosphoribulokinase | prk.S2 | Soule (2009) | Carbon fixation cycle |
Note.—All primers were targeted to exon regions to successfully amplify both genomic DNA and cDNA of the corresponding gene.
Fig. 3.
PCR analysis showing the presence or absence of genes (A and B) or transcripts (C and D) of commonly identified HGT candidates in individual Elysia chlorotica collected from the wild (A and C) or 6-month-old laboratory-reared animals (B and D). Identical colored boxes represent the same individual being tested in panels A and C, and in panels B and D. Targeted genes are listed in table 1.
Fig. 4.
Percentage of animals (n = 3) showing the presence of genes (left) or transcripts (right) encoding 11 putative HGT candidates (divided into functional categories) and two control genes (actin and spermadine synthase). Tested animals were starved for 3 months (3MS) and then allowed to feed on Vaucheria litorea for 1 h (0HPF) and subsequently starved for 1 h (1HPF) or 24 h (24HPF). Controls indicate samples that had combined algal and animal nucleic acids as templates. Targeted genes are listed in table 1.
A possible explanation for the unstable pattern of “HGT” detected here is, as suggested earlier, the presence of extrachromosomal DNA in E. chlorotica that may encode foreign genes and/or gene fragments. Extrachromosomal DNA has been reported in a number of organisms ranging from protozoa to vertebrates (Cohen et al. 2007; Cohen and Segal 2009), and in several cases, its formation and/or disappearance is linked to specific developmental changes (Noto et al. 2003), and more recently, to distinct gene regions (Shibata et al. 2012). It is, therefore, possible that the acquisition of DNA from algal food in the sea slug differs among individuals each generation and is correlated with feeding. The gut of adult feeding E. chlorotica may function as an in vivo transfection system, whereby gene uptake is variable and random, often transient and extrachromasomal. In Caenorhabditis elegans, the model holocentric organism, extrachromasomal DNA can be heritable once integrated (albeit into the germ line; Stinchcomb et al. 1985), often in a mosaic pattern and propagated to daughter cells (Frøkjær-Jensen et al. 2008). Recent data suggest that holocentrism may be more widespread than previously thought (Melters et al. 2012).
Our work with the egg genome data suggests that foreign genes, either integrated or as extrachromosomal DNA, were not present in these data. In the latter case, the total DNA preparation we used for the egg genome would be expected to contain extrachromosomal material, including mini-circles (i.e., gradient fractionation was not included during DNA isolation). This idea is supported by the finding of many copies of mitochondrial and bacterial DNA in the egg genome data. However, the copy number of extrachromosomal DNA could be below detection limits compared with the genomic (or mitochondrial and bacterial) DNA. The DNA used to generate the E. chlorotica genome data was isolated from multiple egg ribbons produced by multiple pairs of animals, resulting in thousands of individuals contributing their nucleic acids, and yet we did not observe any evidence of HGT. Although we cannot rule out that all these egg ribbons were unique in not possessing any HGT candidates, we can conclude that HGT is not universal throughout the population of E. chlorotica collected from Martha’s Vineyard, MA.
In summary, despite the apparent complexity of the sea slug system, our data answer the fundamental question in this field: Is the ability of sea slugs to maintain algal plastids explained by massive HGT to its germ line nucleus? Our work convincingly demonstrates that this is not the case. Elysia chlorotica does not appear to “endanger” its germ line by introducing dozens of foreign algal genes to support its photosynthetic lifestyle. This leaves the far more interesting possibility that the animal is able to harvest plastids and carry out photosynthesis using nonchromosomally integrated algal-derived genes, modified animal genes, or some combination of the two. Exploring these mechanisms using a combination of developmentally targeted mRNA-seq approaches, proteomics, and deeper genome sequencing offer potential avenues to elucidate the (nearly abominable) mystery of long-term sea slug photosynthesis.
Materials and Methods
Collection of Sea Slug Tissue and Preparation of Egg Genomic DNA
Specimens of E. chlorotica were collected from Martha’s Vineyard, MA, in November 2009. Animal pairs were isolated and maintained at room temperature in the absence of algal prey in 0.2 µm filtered, autoclaved 32 practical salinity units artificial sea water (ASW; 925 mosmol kg−1 Instant Ocean, Aquarium Systems, OH) and observed for mating. The subsequent egg ribbons were removed immediately upon laying, dipped in a 5% (v/v) sodium hypochlorite solution and rinsed in fresh water. The water was aspirated, and the eggs were flash frozen in liquid nitrogen. DNA was extracted using the Qiagen Plant DNeasy Kit (Qiagen Inc., Valencia, CA) and concentrated by precipitation with ammonium acetate. Illumina paired-end sequencing libraries were prepared using the Nextera sample prep kit (Illumina Inc., San Diego, CA) before sequencing on an Illumina Genome Analyzer IIx instrument using TruSeq SBS v5-GA reagents (Illumina Inc., San Diego, CA) following the manufacturer’s protocols. Two flowcell lanes were sequenced in a paired-end (150 bp × 150 bp) manner.
Algal Culturing and Preparation of RNA
RNA was extracted from V. litorea maintained at room temperature at +5 h into the light cycle (∼30 µE/m2/s) using the Qiagen Plant RNeasy Kit. Illumina paired-end sequencing libraries were prepared using the TruSeq RNA Sample Preparation Kit v2 (Illumina, Inc., San Diego, CA) before sequencing on an Illumina MiSeq instrument using the MiSeq Reagent Kit (Illumina, Inc., San Diego, CA) in a paired-end (150 bp × 150 bp) approach.
Bioinformatic Analysis of Genomic and Transcriptomic Data
The E. chlorotica egg DNA and V. litorea RNAseq reads were quality and adapter trimmed using the CLC Genomics Workbench (CLC Bio, Aarhus, Denmark). Elysia chlorotica read data were then exported in fasta format and converted to a BLAST database (Altschul et al. 1990). The V. litorea data were assembled into cDNA contigs using the Genomics Workbench and then “seeded” with three single read sequences from the sea slug genome sequencing to act as positive controls: one 146-bp sequence of 100% similarity, one truncated 75-bp sequence of 100% similarity, and one 75-bp sequence of 90% similarity. The algal contigs were then divided into 640 subsets and sent to a 640-core Beowulf style parallel computing cluster operated by the Rutgers University School of Environmental and Biological Sciences where each was used as a BLASTn (Altschul et al. 1990) query to the sea slug read database (e-value cutoff ≤ 10−1). An additional analysis was performed using the 431 individual plastid and nuclear-encoded transcript sequences of Pierce et al. (2012), some of which were hypothesized to be genes horizontally transferred from V. litorea to the E. chlorotica nuclear genome. These sequences were used as a BLASTn query (e-value cutoff ≤ 10−1) to the E. chlorotica egg genome database created above after insertion of three positive control sequences taken from the Pierce et al. (2012) data set: one 90-bp sequence of 100% similarity, one truncated 50-bp sequence of 100% similarity, and one 90-bp sequence of 80% similarity.
PCR Analysis of Individual E. chlorotica Specimens
Interindividual Variability Experiments
Wild animals were collected from Martha’s Vineyard and maintained in the laboratory for more than 3 months without feeding. Laboratory-reared animals were raised as described by Pelletreau et al. (2012) and maintained at 10 °C on a 12:12 L:D cycle being fed V. litorea weekly.
Feeding Experiment
Wild-caught animals maintained unfed in the laboratory for 3 months were placed in individual Petri dishes filled with ASW at room temperature on a 12:12 L:D cycle and allowed to acclimate for 48 h. The experiment was then initiated 45 min after illumination by placing each animal on 0.15 g wet weight of V. litorea. Individual animals observed to feed continuously for 1 h were removed from V. litorea, rinsed in fresh water, blotted dry, and frozen in liquid nitrogen immediately after feeding (0HPF) or moved to new ASW and washed and frozen 1 h or 24 h after feeding ceased (1HPF and 24HPF, respectively). One set of animals was frozen before exposure to food and represented the state of animals after 3 months of starvation. Samples were stored at −80 °C until further processing.
DNA Extraction
Total DNA was extracted from frozen V. litorea samples using DNAzol reagent (Invitrogen Co., Carlsbad, CA). Elysia chlorotica DNA was recovered during TRIzol RNA isolation (below) by following the published TRIzol DNA precipitation protocol to enable isolation of DNA and RNA from the same individual.
RNA Isolation
Total RNA was extracted from frozen algal samples using the Qiagen RNeasy Plant Mini Kit (Qiagen Sciences, Germantown, MD) following the manufacturer’s protocol with all centrifugation steps performed at 4 °C and 8,000 × g. Total RNA was isolated from frozen sea slug samples using a modified TRIzol (Sigma, St. Louis, MO) procedure (Soule 2009).
cDNA Generation
High-quality RNA (0.5 µg) was treated with DNase I (Amplification Grade; Invitrogen), analyzed in an agarose gel to confirm the absence of genomic DNA, and first-strand cDNA was generated using Invitrogen’s oligo-dT primed Superscript III First Stand Synthesis System.
PCR Amplification
PCR reactions were set up for 11 different algal nuclear-encoded plastid targeted genes and two control genes (table 1). All PCR reactions (except for spermadine synthase) were performed using 10 ng of template and 1.25 U Promega GoTaq enzyme, 1× GoTaq Flexi buffer, 0.4 μM primers, 1.5 mM MgCl2, and 0.2 mM each dNTP, per reaction. Spermidine synthase required a final MgCl2 concentration of 3 mM. PCR conditions were optimized as follows: initial denaturing (95 °C, 2 min), 40 cycles of touchdown (95 °C, 30 s, 60–50 °C for 1 min with a decrease of 0.5 °C per cycle, and 72 °C for 45 s), and a final elongation step (72 °C, 10 min). PCR conditions for E. chlorotica β-actin and lhca1 were only run on a 30-cycle touchdown program due to highly robust amplification. For every gene tested, biological replicates (n = 3) were used in each PCR along with no reverse transcriptase (no RT) controls (cDNA) and negative controls that lacked template DNA.
Validation of PCR Results
Products were analyzed with gel electrophoresis and sequenced by the University of Maine DNA Sequencing Facility to validate observed products. Sequences were compared against the NCBI nucleotide and protein databases using a combination of BLASTn and BLASTx, respectively (Altschul et al. 1990).
Enhanced PCR to Verify Negative Trends
For each gene targeted, cDNA reactions and their corresponding no RT controls for one replicate were reamplified for an additional 20 cycles to determine whether seemingly absent transcripts were present but at levels undetectable by standard PCR and gel electrophoresis/EtBr staining. The previously described touchdown PCR was run on these templates for each time point as described earlier. Subsequently, 1 μl of this PCR reaction was used as a template for an additional 20 cycles of reamplification at 50 °C annealing temperature. PCR products were not observed in any of the reamplifications.
Supplementary Material
Supplementary tables S1–S4 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by National Science Foundation grants awarded to M.E.R. (IOS-0726178) and to D.B. (0936884), by the University of Connecticut to M.E.R., and by Rutgers University to D.B. This is Maine Agricultural and Forest Experiment Station Publication Number 3316, Hatch Project no. ME08361-08MRF (NC 1168). D.B. acknowledges generous support from members of the Genome Cooperative at Rutgers for their help in genome data generation and analysis. The Illumina sequence data generated from E. chlorotica egg DNA and from the transcriptome of V. litorea are archived at the NCBI Sequence Read Archive (SRA) under accession SRX247169. The assembled V. litorea cDNA contigs are available at http://dblab.rutgers.edu/sea_slug (last accessed May 16, 2013).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215: 403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Boschetti C, Carr A, Crisp A, Eyres I, Wang-Koh Y, Lubzens E, Barraclough TG, Micklem G, Tunnacliffe A. Biochemical diversification through foreign gene expression in bdelloid rotifers. PloS Genet. 2012;8:e1003035. doi: 10.1371/journal.pgen.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CX, Yang EC, Banerjee T, Yoon HS, Martone PT, Estevez JM, Bhattacharya D. Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr Biol. 2011;21:328–333. doi: 10.1016/j.cub.2011.01.037. [DOI] [PubMed] [Google Scholar]
- Cohen S, Houben A, Segal D. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J. 2007;53:1027–1034. doi: 10.1111/j.1365-313X.2007.03394.x. [DOI] [PubMed] [Google Scholar]
- Cohen S, Segal D. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res. 2009;124:327–338. doi: 10.1159/000218136. [DOI] [PubMed] [Google Scholar]
- Evertsen J, Burghardt I, Johnsen G, Wägele H. Retention of functional chloroplasts in some sacoglossans from the Indo-Pacific and Mediterranean. Mar Biol. 2007;151:2159–2166. [Google Scholar]
- Evertsen J, Johnsen G. In vivo and in vitro differences in chloroplast functionality in the two north Atlantic sacoglossans (Gastropoda, Opisthobranchia) Placida dendritica and Elysia viridis. Mar Biol. 2009;156:847–859. [Google Scholar]
- Frøkjær-Jensen C, Wayne Davis M, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BJ, Li WY, Manhart JR, Fox TC, Summer EJ, Kennedy RA, Pierce SK, Rumpho ME. Mollusc-algal chloroplast endosymbiosis: photosynthesis, thylakoid protein maintenance, and chloroplast gene expression continue for many months in the absence of the algal nucleus. Plant Phys. 2000;124:331–342. doi: 10.1104/pp.124.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händeler K, Grzymbowski Y, Krug PJ, Wägele H. Functional chloroplasts in metazoan cells—a unique evolutionary strategy in animal life. Front Zool. 2009;6:28. doi: 10.1186/1742-9994-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. Evolution of the Sacoglossa (Mollusca, Opisthobranchia) and the ecological association with their food plants. Evol Ecol. 1997;11:301–335. [Google Scholar]
- Maeda T, Hirose E, Chikaraishi Y, et al. (13 co-authors) Algivore or phototroph? Plakobranchus ocellatus (Gastropoda) continuously acquires kleptoplasts and nutrition from multiple algal species in nature. PLoS One. 2012;7:e42024. doi: 10.1371/journal.pone.0042024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SWL. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20:579–593. doi: 10.1007/s10577-012-9292-1. [DOI] [PubMed] [Google Scholar]
- Moustafa A, Beszteri B, Maier UG, Bowler C, Valentin K, Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Müjer CV, Andrews DL, Manhart JR, Pierce SK, Rumpho ME. Chloroplast genes are expressed during intracellular symbiotic association of Vaucheria litorea plastids with the sea slug Elysia chlorotica. Proc Natl Acad Sci U S A. 1996;93:12333–12338. doi: 10.1073/pnas.93.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto T, Yazaki K, Endoh H. Developmentally regulated extrachromosomal circular DNA formation in the mesozoan Dicyema japonicum. Chromosoma. 2003;111:359–368. doi: 10.1007/s00412-002-0216-2. [DOI] [PubMed] [Google Scholar]
- Pelletreau KN, Bhattacharya D, Price DC, Worful JM, Moustafa A, Rumpho ME. Sea slug kleptoplasty and plastid maintenance in a metazoan. Plant Phys. 2011;155:1561–1565. doi: 10.1104/pp.111.174078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletreau KN, Worful J, Sarver KE, Rumpho ME. Laboratory culturing of Elysia chlorotica reveals a shift from transient to permanent kleptoplasty. Symbiosis. 2012;58:221–232. [Google Scholar]
- Pierce SK, Biron R, Rumpho ME. Endosymbiotic chloroplasts in molluscan cells contain proteins synthesized after plastid capture. J Exp Biol. 1996;199:2323–2330. doi: 10.1242/jeb.199.10.2323. [DOI] [PubMed] [Google Scholar]
- Pierce SK, Curtis NE. Cell biology of the chloroplast symbiosis in sacoglossan sea slugs. In: Jeon WK, editor. International review of cell and molecular biology. Vol. 293. Burlington (VT): Academic Press; 2012. pp. 123–149. [DOI] [PubMed] [Google Scholar]
- Pierce SK, Curtis NE, Hanten JJ, Boerner SL, Schwartz JA. Transfer, integration and expression of functional nuclear genes between multicellular species. Symbiosis. 2007;43:57–64. [Google Scholar]
- Pierce SK, Curtis NE, Schwartz JA. Chlorophyll a synthesis by an animal using transferred algal nuclear genes. Symbiosis. 2009;49:121–131. [Google Scholar]
- Pierce SK, Fang X, Schwartz JA, Jiang X, Zhao W, Curtis NE, Kocot KM, Yang B, Wang J. Transcriptomic evidence for the expression of horizontally transferred algal nuclear genes in the photosynthetic sea slug, Elysia chlorotica. Mol Biol Evol. 2012;29:1545–1556. doi: 10.1093/molbev/msr316. [DOI] [PubMed] [Google Scholar]
- Pillet L, Pawlowski J. Transcriptome analysis of foraminiferan Elphidium margaritaceum questions the role of gene transfer in kleptoplasty. Mol Biol Evol. 2013;30:66–69. doi: 10.1093/molbev/mss226. [DOI] [PubMed] [Google Scholar]
- Price DC, Chan CX, Yoon HS, et al. (27 co-authors) Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S, Ibrahim IM, Jelicic B, Tomasic A, Fulgosi H, Allen JF. Transcriptional control of photosynthesis genes: the evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biol Evol. 2010;2:888–896. doi: 10.1093/gbe/evq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Pelletreau KN, Moustafa A, Bhattacharya D. The making of a photosynthetic animal. J Exp Biol. 2011;214:303–311. doi: 10.1242/jeb.046540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Pochareddy S, Worful JM, Summer EJ, Bhattacharya D, Pelletreau KN, Tyler MS, Lee J, Manhart JR, Soule KM. Molecular characterization of the Calvin cycle enzyme phosphoribulokinase in the stramenopile alga Vaucheria litorea and the plastid hosting mollusc Elysia chlorotica. Mol Plant. 2009;2:1384–1396. doi: 10.1093/mp/ssp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc Natl Acad Sci U S A. 2008;105:17867–17871. doi: 10.1073/pnas.0804968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JA, Curtis NE, Pierce SK. Using algal transcriptome sequences to identify transferred genes in the sea slug, Elysia chlorotica. Evol Biol. 2010;37:29–37. [Google Scholar]
- Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule K. [master’s thesis] [Orono (ME)]: University of Maine; 2009. Light regulated photosynthetic gene expression and enzyme activity in the heterokont alga Vaucheria litorea and its symbiotic partner the sacoglossan mollusc Elysia chlorotica. [DOI] [PubMed] [Google Scholar]
- Soule KM, Rumpho ME. Light-regulated photosynthetic gene expression and phosphoribulokinase enzyme activity in the heterokont alga Vaucheria litorea (Xanthophyceae) and its symbiotic molluscan partner Elysia chlorotica. J Phycol. 2012;48:373–383. doi: 10.1111/j.1529-8817.2012.01111.x. [DOI] [PubMed] [Google Scholar]
- Stinchcomb DT, Mello C, Hirsh D. Caenorhabditis elegans DNA that directs segregation in yeast cells. Proc Natl Acad Sci U S A. 1985;82:4167–4171. doi: 10.1073/pnas.82.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trench RK. Of “leaves that crawl”: functional chloroplasts in animal cells. Symp Soc Exp Biol. 1975;29:229–265. [PubMed] [Google Scholar]
- Wägele H, Deusch O, Händeler K, et al. (11 co-authors) Transcriptomic evidence that longevity of acquired plastids in the photosynthetic slugs Elysia timida and Plakobranchus ocellatus does not entail lateral transfer of algal nuclear genes. Mol Biol Evol. 2011;28:699–706. doi: 10.1093/molbev/msq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West HH. Chloroplast symbiosis and development of the ascoglossan opisthobranch Elysia chlorotica [dissertation] [Boston (MA)]: Northeastern University; 1979. pp. 1–176. [Google Scholar]
- Wisecaver JH, Hackett JD. Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC Genomics. 2010;11:366. doi: 10.1186/1471-2164-11-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Yusa Y, Yamamoto S, Hirano Y, Hirano Y, Motomura T, Tanemura T, Obokata J. Identification of photosynthetic sacoglossans from Japan. Encocytobiosis Cell Res. 2009;19:112–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.