Abstract
The Tibetan and Andean Plateaus and Ethiopian highlands are the largest regions to have long-term high-altitude residents. Such populations are exposed to lower barometric pressures and hence atmospheric partial pressures of oxygen. Such “hypobaric hypoxia” may limit physical functional capacity, reproductive health, and even survival. As such, selection of genetic variants advantageous to hypoxic adaptation is likely to have occurred. Identifying signatures of such selection is likely to help understanding of hypoxic adaptive processes. Here, we seek evidence of such positive selection using five Ethiopian populations, three of which are from high-altitude areas in Ethiopia. As these populations may have been recipients of Eurasian gene flow, we correct for this admixture. Using single-nucleotide polymorphism genotype data from multiple populations, we find the strongest signal of selection in BHLHE41 (also known as DEC2 or SHARP1). Remarkably, a major role of this gene is regulation of the same hypoxia response pathway on which selection has most strikingly been observed in both Tibetan and Andean populations. Because it is also an important player in the circadian rhythm pathway, BHLHE41 might also provide insights into the mechanisms underlying the recognized impacts of hypoxia on the circadian clock. These results support the view that Ethiopian, Andean, and Tibetan populations living at high altitude have adapted to hypoxia differently, with convergent evolution affecting different genes from the same pathway.
Keywords: adaptation to high altitude, natural selection
Introduction
Barometric pressure falls with ascent to altitude and with it the partial pressure of inspired oxygen. The resulting reduction in systemic oxygen availability (hypobaric hypoxia) impairs physical performance and can be detrimental to health and survival (Monge et al. 1992; Leon-Velarde et al. 2005). It may also impair placental function and neonatal survival (Niermeyer et al. 2009). Over generations, such effects are likely to have exerted pressure for the selection of beneficial genetic variants in humans who have settled the three major high-altitude regions of the world: the Tibetan Plateau, the Andean Plateau, and the Ethiopian highlands. However, the genetic targets of selection may, in part, have differed between these populations: for example, hemoglobin concentration and oxygen saturation vary between them (Beall 2006). Indeed, hemoglobin concentrations are largely independent of altitude (up to 4,000 m) in Tibetans but rise with altitude in the high-altitude Andean population (Beall 2006). Tibetan individuals also have lower arterial oxygen saturations than Andeans at the same altitude (Beall 2006). Both traits are highly heritable (Moore et al. 2001; Niermeyer et al. 2009). In contrast, among male Ethiopians of mostly Amharic ethnicity living at altitudes of 3,500 m, both hemoglobin concentration and arterial oxygen saturation remain almost the same as those of US sea level males (Beall et al. 2002) and show small changes compared with lowland Amhara individuals (Alkorta-Aranburu et al. 2012). In contrast, Scheinfeldt et al. (2012) observes higher hemoglobin concentration in the high-altitude Amhara compared with Hamer individuals from altitudes of approximately 1,000 m. These observations suggest that the genetic adaptation to high altitude in Ethiopians may differ from that in Tibetans and Andeans.
At the cellular level, activation of the HIF hypoxia-sensing pathway is the key response to a reduced oxygen environment, primarily through the activity of the HIF-1α and HIF-2α transcription factors. In the case of Tibetans, two genes within the HIF hypoxia-sensing pathway exhibiting strong signatures of positive selection in response to high altitude have been identified (e.g., EPAS1 and EGLN1; Beall et al. 2010; Bigham et al. 2010; Simonson et al. 2010; Yi et al. 2010; Peng et al. 2011; Wang et al. 2011; Xu et al. 2011). In Andeans, EGLN1 is the only gene so far identified, which also has strong signatures of positive selection in Tibetans (Bigham et al. 2010). In Tibetans, variants in these genes are associated with a minimal increase in hemoglobin concentration at high altitude.
Recently, Scheinfeldt et al. (2012) searched for signatures of positive selection across the genome in one high-altitude Ethiopian population—the Amhara. Using a number of statistical methods for detecting selection (including per single-nucleotide polymorphism [SNP] FST between pairs of populations, locus-specific branch length [LSBL], integrated haplotype score [iHS], cross population composite likelihood ratio [XP-CLR], and SNP-phenotype association), they proposed several genes as candidate targets of positive selection. More recently, Alkorta-Aranburu et al. (2012) analyzed both the Amhara and the Oromo, two populations that inhabit high-altitude regions in Ethiopia. In that study, they conducted many population comparisons designed to detect selection in high-altitude Amhara and Oromo and look for enrichment of hypoxia genes in their results. They also employ genotype–phenotype associations to propose some candidate genes.
However, studying high-altitude adaptation in Ethiopia is challenging for at least two reasons. First, because of its geographic location, it is likely that there has been gene flow from sub-Saharan Africa, northern Africa, and the Middle East into Ethiopia. Indeed, Ethiopian populations share a non-negligible proportion of their genetic material with other non-African populations (Semino et al. 2002)—perhaps as high as 40–50% (Pagani et al. 2012). Also, there has likely been substantial gene flow between low- and high-altitude populations within Ethiopia and nearby regions. Second the Ethiopian highlands are in a lower altitude than the Andean and Tibetan Plateaus.
Here, we undertake an analysis of the Amhara, Tigray, and Oromo genotype data published in Pagani et al. (2012), which are individuals sampled at intermediate altitudes of approximately 1,800 m. Archeological studies in Ethiopia show that humans have inhabited regions of more than 2,000 m for thousands of years (Brandt 1986; Pleurdeau 2006). Both the Amhara and the Oromo populations have inhabited regions of more than 2,500 m for many generations. Alkorta-Aranburu et al. (2012) assume 5,000 years as a reasonable estimate for the Amhara high-altitude settlement in Ethiopia. In contrast, the estimates of when the Oromo settled in regions of high altitude are far more recent at approximately 500 years (Lewis 1966; Hassen 1990). Part of the analysis from Alkorta-Aranburu et al. (2012) shows that genome-wide genetic differentiation between the high- and low-altitude Amhara or between the high- and low-altitude Oromo is almost negligible as measured by principle components analysis (PCA) and FST analyses (see fig. S3 and text S2 in Alkorta-Aranburu et al. [2012]). We also show that when comparing the high-altitude Amhara from Scheinfeldt et al. (2012) with the Amhara considered in this study, we do not observe evidence for population subdivision between these two populations (supplementary fig. S1A and S2B, Supplementary Material online), which suggests continual gene flow between the high- and intermediate-altitude populations or a small divergence time. Therefore, we expect the signal of positive selection to remain detectable in the populations considered here, even if their current environment does not expose them to such extreme selection pressure.
Both the Amhara and Tigray populations share the same Semitic language group, cluster in PCA (Pagani et al. 2012), and live at similar elevations. We, therefore, scanned both a separate and a pooled sample of Amhara–Tigray individuals for signals of positive selection and compared our findings with those from previous studies on high altitude adaptation in Ethiopians. For completeness, we carried out the same analysis with Oromo samples, both separately and when pooled with the Amhara–Tigray groups (see Materials and Methods for exact altitude of the sampled populations). The low altitude populations employed in the study consist of the Afar and the Anuak from Pagani et al. (2012). Finally, we conducted a simulation study to assess the effects of admixture on our ability to detect true signatures of positive selection in admixed populations.
To detect selection, we used a statistical method that, for each gene, ranks signals based on a previously developed score, termed the population branch statistic (PBS; Yi et al. 2010). The PBS method has been proven effective in detecting selected loci among high-altitude Tibetan populations. It employs three populations, such that a population’s PBS value corresponds to the magnitude of the allele frequency change at a given locus relative to the divergence from the other two populations (see Materials and Methods). In this study, we sought to identify signals of positive selection specifically in Ethiopians hypothesized to be high altitude adapted. Therefore, we applied the test on the Amhara, Tigray, and Oromo separately as well as the Amhara–Tigray or Amhara–Tigray–Oromo pooled data. Briefly, we computed the PBS for each gene region after correcting for non-African admixture (see Materials and Methods). When allele frequencies were corrected for admixture from European or Middle Eastern populations, the top candidate is BHLHE41, a gene of functional relevance to both hypoxic-response and circadian rhythm pathways.
Results
Non-African Admixture in Ethiopians
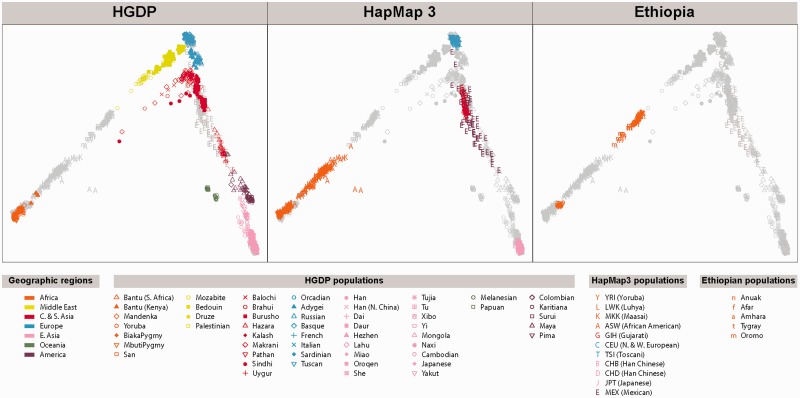
The Ethiopian populations considered in this study share a moderate degree of genetic similarity with some non-African populations. Figure 1 shows Ethiopian populations clustering between sub-Saharan Africans and non-Africans. Notably, African Americans also lie between sub-Saharan Africans and non-Africans, and they are known to have about 20% of European ancestry on average (Tang et al. 2006). This clustering pattern is consistent with previous results reported for the same samples, where it was shown that it is likely due to European admixture (between 40% and 50%) into Ethiopians at approximately 3,000 years ago (Pagani et al. 2012). In addition, by fitting a model of population splits with three migration events using the software TreeMix (Pickrell and Pritchard 2012), we observe that the most likely model has migration events stemming from an ancestral (or unsampled) population from the non-African populations (supplementary fig. S2, Supplementary Material online). Though the current understanding of the demographic history for Ethiopians remains incomplete, such admixture could result in spurious signals of positive selection if the admixture proportion differs among Ethiopian populations. In fact, when the selection scan was performed without correcting for admixture, the first and second most significant genes for the Tigray–Amhara scan were MYEF2 and SLC24A5 (supplementary table S1, Supplementary Material online), which both display strong signatures of positive selection in Europeans, and are suggested to relate to lighter skin pigmentation (Lamason et al. 2005). Therefore, we propose that the higher European admixture proportion observed for these genes in the high altitude compared with the low-altitude Ethiopians led to the potentially spurious signal of altitude adaptation. Indeed, if we calculate the average European admixture in the MYEF2-SLC24A5 region (see Materials and Methods), then we find it to be about 22% in the low-altitude Afar, but 48% and 44% in the high-altitude Amhara and Tigray, respectively. We cannot rule out, however, that the European alleles were indeed differentially selected in the high-altitude population due to an unknown selective pressure. However, we will in the following use an admixture-corrected version of the PBS by Yi et al. (2010) to detect selection (see Materials and Methods for details on this method).
Fig. 1.
Multidimensional scaling for the HGDP, HapMap 3, and Ethiopian populations. Note that the Anuak individuals lie between the Yoruban and African American individuals, and the Afar, Amhara, Tigray, and Oromo individuals lie between the African American and Middle Eastern individuals.
Simulation Results: Correcting for Admixture Leads to Fewer False Positives
To assess whether correcting for admixture leads to fewer false positives, we performed two types of simulations (see Materials and Methods for details). In the first scenario, we simulated two admixed populations: one with selection at a given locus and with a higher admixture proportion, representing the non-African admixed highland Ethiopians, and the other without selection and a lower admixture proportion, representing non-African admixture in the lowland Ethiopians (see supplementary fig. S3A, Supplementary Material online). In the second scenario, the non-African population itself experiences positive selection at a locus before the admixture event (e.g., SLC24A5 locus in Europeans), but the admixed populations experience no selective event (see supplementary fig. S3B, Supplementary Material online). This scenario evaluates the false positive rate for our PBS with or without the admixture correction.
In supplementary figure S4, Supplementary Material online, we plot receiver operator characteristic (ROC) curves under the first simulation scenario (supplementary fig. S3A, Supplementary Material online) and identify signals of selection with the PBS. The null distribution of the PBS is derived from the same demographic models in supplementary figure S3A and B, Supplementary Material online, without any selective event (i.e., neutrality). The ROC curves reveal that correction for admixture improves the sensitivity (i.e., the true positive rate as defined by the proportion of true selection signals correctly identified as selection signals by the PBS) and lowers the false positive rate for detecting selection in the admixed population. However, in practice, one is often only concerned with a method’s performance at reasonably low false positive rates. For two of the parameter values displayed in supplementary figure S4, supplementary figure S5 (Supplementary Material online), focuses on more realistic false positive rates, varying from 0.0 to 0.05. It shows that within this range correcting for admixture affords approximately a 20% increase in sensitivity when compared with not correcting for admixture. In addition, supplementary figure S6, Supplementary Material online, shows that correcting for admixture correctly down weighs the false signal of selection in the admixed population that arises from an adaptive event in the non-African group (i.e., under the setting displayed in supplementary fig. S3B, Supplementary Material online). It is worth noting, however, that even without correction, the PBS is mostly robust to admixture at the level simulated here.
Selection Scans
Selection Scan in Amhara–Tigray After Correcting for Admixture
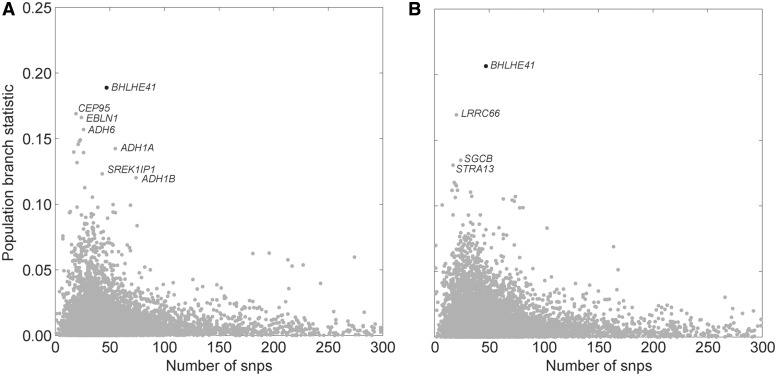
Figure 2A plots the PBS values for each gene and groups them with PBS values from other genes containing identical numbers of SNPs. Supplementary table S2, Supplementary Material online, lists the top 25 genes after correcting for admixture when the Amhara and the Tigray populations are combined.
Fig. 2.
The empirical distribution of the population branch statistic (PBS) values per gene region as a function of the number of SNPs in the gene. The y-axis is the corresponding PBS value of the gene region with a given number of SNPs (the x-axis). The x-axis has been truncated at 300 SNPs. (A) Results for the Tygray-Amhara comparison. (B) Results for the Oromo comparison.
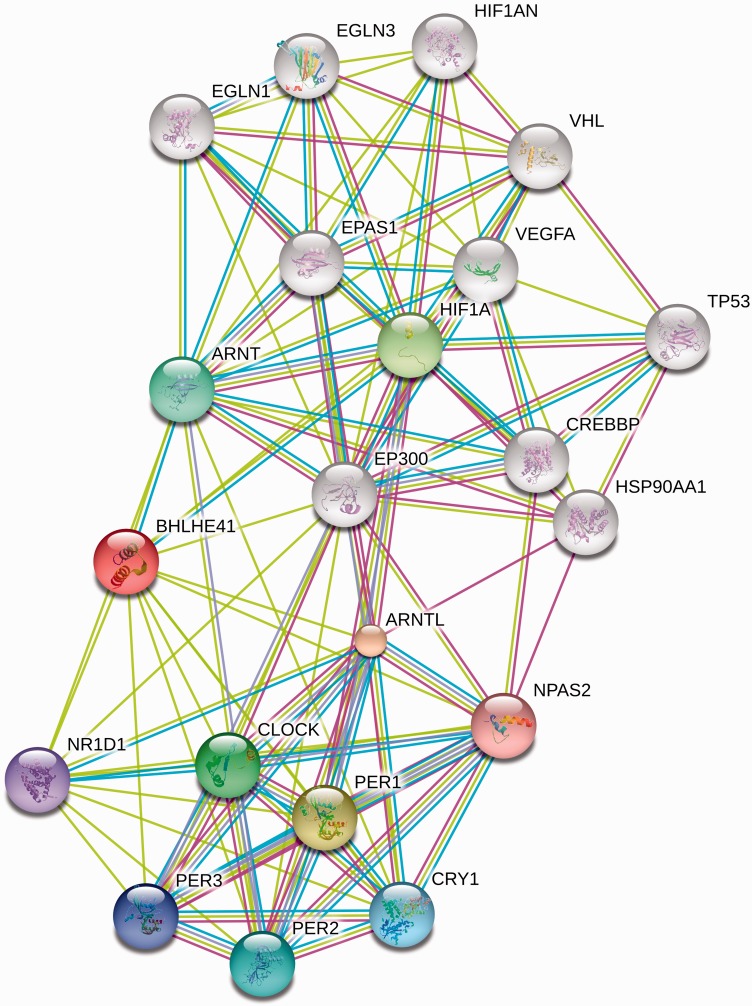
At the top of the list is BHLHE41 (also known as DEC2 or SHARP1), which is also an extreme outlier with respect to the empirical distribution of PBS values for genes with comparable numbers of SNPs (supplementary fig. S7A, Supplementary Material online). This gene is a biologically plausible candidate for selection, being involved in hypoxic-response pathways and having a physical interaction with HIF-1α. Figure 3 illustrates the known and predicted interactions for BHLHE41 from the STRING database (Jensen et al. 2009), including several components of the hypoxia pathway. The HIF-1α/ARNT1 protein heterodimer plays a critical role in the hypoxia-induced transcription of vascular endothelial growth factor (VEGF) (Forsythe et al. 1996), and BHLHE41 negatively regulates VEGF expression by its interaction with HIF-1α/ARNT1 activation (Sato et al. 2008). In addition, the promoter region of BHLHE41 contains a hypoxia response element that is bound and transcriptionally regulated by HIF-1α, generating an apparent negative feedback loop (Miyazaki et al. 2002). More recently, experiments in breast cancer cell lines have confirmed the direct interaction of BHLHE41 with HIF-1α and demonstrated that BHLHE41 is a global inhibitor of HIF-1α and HIF-2α’s transcriptional activity via downregulation of HIF-1α and HIF-2α protein expression (Montagner et al. 2012). BHLHE41 is proposed to facilitate the delivery of the HIF proteins to the proteasome (Montagner et al. 2012). BHLHE41 also ranked highly when the analysis is performed using only the Tigray or only the Amhara population (supplementary tables S3 and S4, Supplementary Material online, respectively).
Fig. 3.
STRING 9.0 (Jensen et al. 2009) database of interactions with BHLHE41, including up to 10 direct links with other genes and 10 genes separated by two links from BHLHE41. Colored links connected to BHLHE41 are from PubMed co-occurrence (yellow) and comembership in pathways from the NCI-Nature Pathways Interaction Database (light blue).
BHLHE41 is also a component of the circadian clock pathway (Honma et al. 2002; Kato et al. 2010), and a mutation in BHLHE41 is associated with a short-sleep phenotype in humans (He et al. 2009). Interestingly, the genes at the 18th and 19th position (DKFZp779M0652 and SLC35C1, respectively) in supplementary table S2, Supplementary Material online, are within 100 kb of CRY2, a gene that is also a member of the circadian clock pathway. CRY2 may indirectly suppress HIF-1α/ARNT1 activity through the transcriptional regulation of the circadian PER1 gene, which is known to interact with HIF-1α via the PAS domain (Koyanagi et al. 2003). Extensive crosstalk between circadian clock and hypoxia pathways has been previously elucidated (Chilov et al. 2001). Another gene, SMURF2 (SMAD-specific E3 ubiquitin-protein ligase 2), plays a role in the vascular inflammatory response in the presence of hypoxia in endothelial cells through an upregulation of TGF-β signaling (Akman et al. 2001). In addition, CASP1 (caspase 1) is in the hypoxia response pathway and has been implicated in the pathogenesis of many disorders including cardiovascular disease. Interestingly, the alcohol dehydrogenase genes ADH6, ADH1A, ADH1B, and ADH1C are differentiated between the low- (Afar and Anuak) and the high-altitude populations (Tigray and Amhara). They have previously been observed to display strong signals of positive selection, concurrent with the introduction of agriculture and fermentation in human societies (Peng et al. 2008), and were also identified in the recent study of Amhara high-altitude populations (Alkorta-Aranburu et al. 2012).
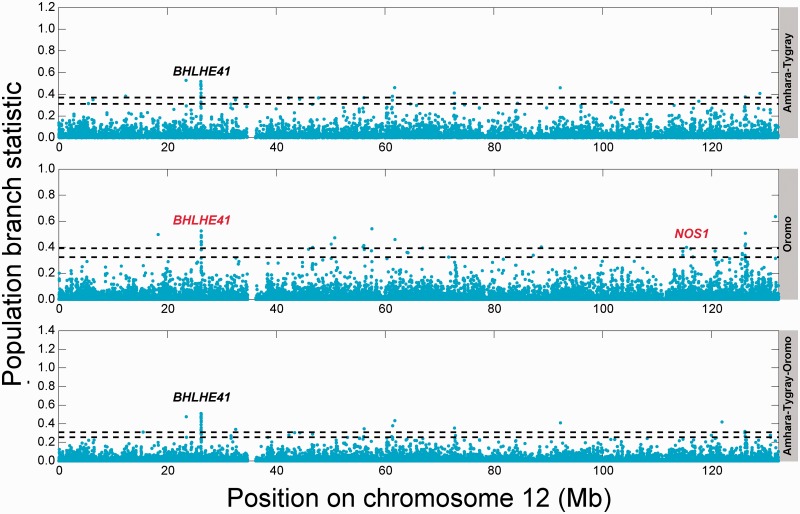
The ranking in supplementary table S2, Supplementary Material online, discussed in the previous section is based on PBS calculated from the aggregation of all the SNPs in the immediate region (i.e., within 50 kb upstream or downstream) of the gene. If we instead calculate PBS for each SNP separately, and then rank genes that are within 50 kb of each SNP, we again retrieve BHLHE41, SMURF2, and CASP1 (fig. 4 [Amhara–Tigray comparison], and supplementary fig. S8, Supplementary Material online) as having a group of SNPs with PBS values above the 0.10% cutoff of the empirical distribution of all PBS values. These three genes thus rank highly when either “per SNP" or “per genic-region" analyses are performed.
Fig. 4.
Per-SNP PBS results. Gaps represent regions of the genome that were not covered. Names of genes in black contain at least one SNP in the top 0.10% of SNPs that is located inside the gene. Genes in red contain at least one SNP in the top 0.10% that is located within 50 kb of the gene but not within the gene. The top and bottom horizontal dotted lines are the 0.05% and 0.10% empirical cutoffs, respectively. The names in the shaded gray area are the population(s) considered (Amhara–Tigray, Oromo, and Amhara–Tigray–Oromo). Supplementary figures S8–S10, Supplementary Material online, display all 22 autosomes for each population comparison shown here.
Selection Scan in the Oromo
Applying the same methodology to the Oromo population, and employing the same low-altitude (the Afar) and outgroup (the Anuak) populations as controls, BHLHE41 again emerges as the most significant locus, with or without admixture correction (see fig. 2B and supplementary table S5, Supplementary Material online). In the Oromo, BHLHE41 is also an extreme outlier with respect to the empirical distribution of PBS values for genes with comparable numbers of SNPs (supplementary fig. S7B, Supplementary Material online). If we instead calculate PBS for each SNP separately, and rank genes that are within 50 kb of each SNP, then we still retrieve BHLHE41 (see fig. 4 [Oromo comparison], supplementary fig. S9, Supplementary Material online). Interestingly, unlike the Amhara and Tigray, neither the alcohol genes nor the pigmentation genes show strong differentiation between the Oromo and the low-altitude Afar. If we pool the Oromo with the Amhara and the Tigray, BHLHE41 remains the top candidate (fig. 4 [Amhara–Tigray–Oromo comparison], supplementary figs. S6C and S10 and table S6, Supplementary Material online).
The identification of the BHLHE41 gene in the Oromo population supports the hypothesis that it arose by a single early selective event affecting an ancestral population, rather than by two independent selective events. The genetic differentiation between the Oromo and the Amhara (FST = 0.01, Alkorta-Aranburu et al. [2012]; FST = 0.02, Pagani et al. [2012]) is sufficiently small to support a scenario in which selection occurred in an ancestral Amhara–Tigray–Oromo population. Alternatively, it is possible that we observe selection on BHLHE41 in the Oromo due to recent gene flow followed by selection for the variant, causing its frequency to increase. Unlike the Amhara, records point to a recent (∼500 years) settlement of the Ethiopian highlands by the Oromo population (Lewis 1966; Hassen 1990). This recent estimate would support the scenario of selection aided by gene flow. In fact, in the genic region of BHLHE41, the FST between the Oromo and the Amhara and between the Oromo and the Tigray is smaller than between the Oromo and the Afar despite the latter pair belonging to the same language group (Cushitic). Furthermore, the FST between Amhara and Oromo in that gene region (FST = 0.01) is smaller than the median across all gene regions (median FST = 0.03), and the same observation holds true between the Oromo and the Tigray populations. The FST values, therefore, are on the lower end of the distribution of FST values across all gene regions, suggesting that selection has acted to reduce genetic differentiation between high-altitude adapted populations at this locus. If this scenario is true, then this is the first example in humans of natural selection acting to reduce FST between populations in a genomic region. Populations of Ethiopia, however, have a complex demographic history, and more studies are needed to reconcile these observations and attest their statistical significance.
Comparison with Other Studies
Scheinfeldt et al. (2012) performed a scan for positive selection in the Amhara, one of the high-altitude Ethiopian populations analyzed here, albeit sampled from a different location in Ethiopia. Overall, they found no enrichment for HIF pathway genes but did propose a number of candidate genes: VAV3, CBARA1, THRB, ARNT2, PIK3CB, ARHGAP15, and RNF216. All these genes have SNPs with LSBL values in the top 0.10% of their analysis. If we intersect their full gene list from the top 0.10% of SNPs (listed in table S2 in Scheinfeldt et al. [2012]) with our hypoxia-related genes (see Materials and Methods for definition of this hypoxia set), we obtain DDIT4, NARFL, RYR2, RYR1, ARNT2, and GATA6. For an equivalent analysis, we examined our Amhara sample without correcting for admixture and retrieved the SNPs with PBS values in the top 0.10%, revealing a collection of hypoxia-related genes: PPARA, ANGPT2, RYR2, SFRP1, ITPR2, TP53, CHRNB2, FLT1, and PYGM. From our top-ranking list, only RYR2 overlapped with their LSBL list. They do, however, identify PPARA from the XP-CLR test of selection, and if we extend the interval to include genes within 100 kb of the top 0.10% of SNPs, then VAV3 and THRB also appear in common. In our data set, we could not identify any SNPs in or near CBARA1, ARNT2, or PIK3CB with PBS values in the top 0.10%.
The more recent study by Alkorta-Aranburu et al. (2012) made multiple Ethiopian high- and low-altitude population comparisons (table S22 in Alkorta-Aranburu et al. [2012]), and they included results with the same PBS we apply here. They concluded that with the PBS metric, the most significant enrichment of hypoxia-related genes emerged when comparing a mixed high- and low-altitude Amhara population to the Masai (Luhya as outgroup). Their results are marginally significant when comparing high-altitude Amhara to low-altitude Amhara, which suggests, analogous to what we find between Amhara and Tigray, that the population groups are so closely related that they both still harbor the selected loci at similar frequencies. However, their candidate gene list with extreme PBS values does not include BHLHE41, and none of their PBS candidates are significantly associated with hemoglobin concentration or oxygen saturation phenotypes. Interestingly, one SNP within a hypoxia-related gene, RORA, does associate with hemoglobin concentration in the Amhara despite lacking a signature of positive selection.
Expected Number of Hypoxia-Related Genes
We performed a permutation test (see Materials and Methods) to assess the number of hypoxia-related genes that would be expected by chance to be found in a random selection of the same number of genes that we observe in the top 0.10% of SNPs and found that the expectation varies from 6 to 12 genes depending on the high-altitude population considered (supplementary table S7, Supplementary Material online). Our analysis identified between 7 and 16 genes in the hypoxia-related set, which is not a statistically significant enrichment. For a comparison of gene lists that include our admixture correction and all the high-altitude populations, we calculated a PBS score for each SNP in 14 cases: before and after correcting for admixture, for the Amhara, Tigray, and Oromo separately as well as to their pooled data. We then identified genes within 50 kb of the top SNPs. Supplementary table S8, Supplementary Material online, displays the results of intersecting the genes within the top-ranked 0.10% of SNPs with the hypoxia-related gene set, with the exception of BHLHE41 (which was not included in the gene set definition but retrospectively appears to be highly relevant for hypoxic response). Notably, BHLHE41 and RYR2 are the only genes that appear under all the 14 scenarios considered.
Discussion
In this study, we have compared Ethiopian populations to identify genes that are likely involved in the Ethiopians’ adaptation to high altitude. Although the current sampled populations live at intermediate elevations, it is likely that they are descendants of high-altitude adapted populations who lived at elevations greater than 2,500 m, and therefore, we expect the signals of selection to remain detectable in the modern populations. In addition, the current altitude of residence (∼1,800 m) is not free of selective pressure. Standard barometric pressure at 1,800 m is 604 mmHg at 2,000 m, a reduction of nearly 20% when compared with sea level, which has clear biological effects. Indeed, low birth weight is three times more common at elevations greater than 2,000 m even in the United States, with a threshold effect of 1,500 m (Yip 1987). Furthermore, using a candidate gene approach, Pagani et al. (2011) identified highly differentiated regions of strong relevance to the hypoxia response (in both EGLN1 and HIF1A) between low-altitude populations and Daghestani populations living at moderate altitudes of approximately 2,000 m.
One challenging aspect of our analysis is that Ethiopians have a complex demographic history, involving, among other events, admixture with non-Africans (Pagani et al. 2012). If admixture is not corrected for, then genes such as SLC24A5, which are involved in lighter skin pigmentation in Europeans, show strong signals of positive selection in the high-altitude populations. Accounting for admixture in the pooled Amhara–Tigray samples results in a decrease in the strength of these potentially spurious signals of high-altitude adaptation, and, instead, yields the strongest signal from the BHLHE41 gene. Furthermore, in the Oromo, the strongest signal is also in the BHLHE41 gene. This is a functionally relevant candidate gene with a major regulatory role in the same hypoxia-sensing pathway that has undergone selection in Tibetan and Andean populations. It is transcriptionally regulated by HIF-1α, binds HIF-1α, and represses many of the hypoxia-induced transcriptional targets including VEGF, likely due to the increased degradation of HIF-1α and HIF-2α proteins by BHLHE41 (Miyazaki et al. 2002; Sato et al. 2008; Montagner et al. 2012). In addition, it is a component of the circadian clock pathway (Honma et al. 2002; Kato et al. 2010), and a mutation in BHLHE41 is associated with a short-sleep phenotype in humans (He et al. 2009).
A role in hypoxic responses offers a clear target for selection in response to altitude, but a role in the regulation of circadian cycles is perhaps less clear. However, extensive circadian-hypoxia pathway crosstalk occurs (Chilov et al. 2001). Indeed, hypoxia-mediated changes in circadian rhythms have been suggested to be a key driver of the sleep fragmentation and poor sleep quality seen in lowlanders at high altitude (Mortola 2007). In agreement, sleep quality is better in the native high-altitude populations of Tibet and the Andes (Coote et al. 1992, 1993; Plywaczewski et al. 2003).
The genes highlighted in our analyses were not detected, however, in previous studies of Ethiopian highland populations (Alkorta-Aranburu et al. 2012; Scheinfeldt et al. 2012), and a number of differences between our analyses and theirs could account for this. The first relates to the choice of low-altitude reference group. We compared the high-altitude Amhara, Tigray, and Oromo populations against the Afar, a low-altitude population with a similar genetic and linguistic (Afro-Asiatic language group) background. Scheinfeldt et al. (2012) used the low-altitude Omotic group, which is less closely related to the Amhara, as shown using comparable samples in Pagani et al. (2012). In contrast, Alkorta-Aranburu et al. (2012) used multiple low-altitude groups (table S22 in Alkorta-Aranburu et al. [2012]). In one of their PBS three-population combinations, they compare the low-altitude Amhara and high-altitude Amhara and find marginal enrichment for hypoxia-related genes, but it may be that continued gene flow between the two groups have kept the putative selected mutations at similar frequencies. Accordingly, when they pooled the low- and high-altitude Amhara, they find stronger enrichment in hypoxia-related genes. The second difference is that we chose the Anuak as our outgroup; in Scheinfeldt et al. (2012), two outgroup populations (the Yorubans and the Europeans) were employed, whereas Alkorta-Aranburu et al. (2012) used various outgroup populations not including the Anuak. The third difference is that we found it important to correct for population admixture (supplementary figs. S4–S6, Supplementary Material online). If SNP frequencies are left uncorrected, then much of the selection signal could derive from differences in admixture proportions. Scheinfeldt et al. (2012) and Alkorta-Aranburu et al. (2012) utilized a European population as an outgroup, and this potentially indirectly removed the non-African admixture effect in their analyses. The fourth difference is that the populations of this study are living at intermediate altitudes. However, as we mentioned in the Results section, when we compare high-altitude Amhara populations from Scheinfeldt et al. (2012) with the intermediate-altitude Amhara population from this study, we do not observe any significant population structure (supplementary fig. S1, Supplementary Material online). This genetic evidence suggests that the high- and low-altitude Amhara are likely derived from the same ancestral population. Thorough meta-analyses, sequencing of larger samples, and the collection of other relevant phenotypes should help resolve the inconsistent results among the Ethiopian studies so far.
We agree with the conclusions of Scheinfeldt et al. (2012) and Alkorta-Aranburu et al. (2012) that high-altitude adaptation can take place by distinct genetic alterations, as there is no overlap with the candidate genes from this study and those of previous Tibetan and Andean studies. However, Tibetan and Andean environments are considerably more extreme at elevations greater than 4,000 m, and phenotypic differences exist in hemoglobin concentrations and oxygen saturation. Thus, the underlying genetic differences may reflect different biological adaptation mechanisms. Furthermore, the high-altitude Ethiopian populations are the least isolated of the three global high-altitude populations, increasing the difficulty of uncovering signatures of adaptation. Nevertheless, at the pathway level, we demonstrated broadly shared biological processes targeted by selection in each of the adapted high-altitude populations, indicative of convergent evolution. The top gene revealed by our analyses, BHLHE41, is an excellent candidate for further studies as it has an important function in the hypoxia response pathway. Given its role in the circadian clock, it also provides justification to explore the relationship between hypoxic conditions and the circadian cycles in future studies.
Materials and Methods
Data
We analyzed genetic data of individuals from Ethiopia available from Pagani et al. (2012): namely, three high-altitude populations (26 Amhara, 21 Tigray, and 21 Oromo individuals) and two low-altitude populations (12 Afar and 23 Anuak individuals). The Amhara and Tigray are members of the Semitic and the Afar and the Oromo of the Cushitic linguistic groups, both belonging to the Afro-Asiatic linguistic family. The Anuak are members of the Nilotic language group, a member of the Nilo-Saharan linguistic family. The Amhara, Tigray, Oromo, Afar, and Anuak samples were collected at altitudes of 1,829 m, 1,695 m, 1,758 m, 400 m, and 500 m, respectively. Though the samples were not collected at extremely high altitudess, the Amhara and Oromo populations have been residing in regions of Ethiopia higher than 2,500 m for many generations (Lewis 1966; Hassen 1990; Alkorta-Aranburu et al. 2012). The Tigray individuals were chosen, so that they had both parents and all grandparents living at greater than 2,000 m. Using these five populations, we performed five selection scans without correcting for admixture and another five selection scans after correcting for admixture. In one scan, we combined the two high-altitude populations (Amhara and Tigray), and in another scan, we combined the three high-altitude populations (Amhara, Tigray, and Oromo). In the other three scans, we considered the Amhara, Tigray, and Oromo separately. All consent information can be found in Pagani et al. (2012).
Multidimensional Scaling
Between each pair of individuals 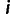 and
and 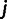 in our data set, we computed the allele sharing distance. For a particular site k in the genome, the pair of individuals
in our data set, we computed the allele sharing distance. For a particular site k in the genome, the pair of individuals 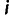 and
and 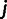 have a distance, denoted as
have a distance, denoted as 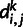 , of 0.0 if they both have the same genotypes, they have a distance of 0.5 if one has a homozygous and the other has a heterozygous genotype, and they have a distance of 1.0 if they are both homozygous but for different alleles. Assume that there are
, of 0.0 if they both have the same genotypes, they have a distance of 0.5 if one has a homozygous and the other has a heterozygous genotype, and they have a distance of 1.0 if they are both homozygous but for different alleles. Assume that there are 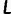 sites in the genome for which neither individual
sites in the genome for which neither individual  nor
nor 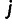 is missing any genotype and that these sites are indexed as
is missing any genotype and that these sites are indexed as 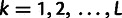 . Then, we compute the allele sharing distance between individuals i and j as
. Then, we compute the allele sharing distance between individuals i and j as
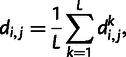 |
such that 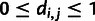 for
for 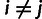 and that
and that 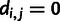 for
for 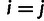 . We then construct a matrix of allele sharing distances between all pairs of individuals and apply classical multidimensional scaling to obtain components displayed in figure 1.
. We then construct a matrix of allele sharing distances between all pairs of individuals and apply classical multidimensional scaling to obtain components displayed in figure 1.
Principal Components Analysis
For principal components analysis (PCA) in this study, we focused on one data set that contained the Anuak, Afar, Oromo, Tygray, and Amhara from Pagani et al. (2012) and the Amhara from Scheinfeldt et al. (2012) and a second data set with only the Amhara from Pagani et al. (2012) and the Amhara from Scheinfeldt et al. (2012). For a given data set, we considered site 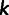 in the genome only if no individual had a missing genotype at that site. Further, for a given data set, we considered site
in the genome only if no individual had a missing genotype at that site. Further, for a given data set, we considered site 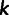 in the genome only if it was polymorphic within the sample of individuals in that data set. That is, if there are
in the genome only if it was polymorphic within the sample of individuals in that data set. That is, if there are 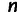 individuals in the data set, and
individuals in the data set, and 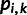 ,
, 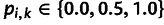 , is the frequency of the reference allele at site
, is the frequency of the reference allele at site 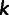 in individual
in individual  , then site
, then site 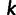 is considered only if
is considered only if 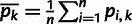 is neither 0.0 nor 1.0. Each individual
is neither 0.0 nor 1.0. Each individual 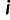 at site
at site 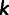 was represented by their reference allele frequency
was represented by their reference allele frequency 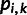 , and we created an
, and we created an 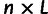 matrix
matrix 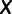 , with
, with 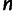 individuals representing the rows,
individuals representing the rows,  sites used for the data set representing the columns, and the entry in row
sites used for the data set representing the columns, and the entry in row  and column
and column 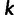 being
being 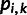 . We then centered
. We then centered 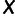 by subtracting all entries in column
by subtracting all entries in column 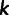 by
by 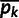 . We performed singular value decomposition on this centered matrix and extracted the first two eigenvectors to represent the first two principal components displayed in supplementary figure S1, Supplementary Material online.
. We performed singular value decomposition on this centered matrix and extracted the first two eigenvectors to represent the first two principal components displayed in supplementary figure S1, Supplementary Material online.
TreeMix Analysis
The data set that we use for TreeMix analysis contains the eight African populations from the HGDP data set (San, Mbuti Pygmy, Biaka Pygmy, Bantu from South Africa, Bantu from Kenya, Yoruban, and Mandenka), two African population from the HapMap3 data set (YRI and LWK), five Ethiopian populations from Pagani et al. (2012) data set (Anuak, Afar, Amhara, Tigray, and Oromo), four Middle Eastern population from the HGDP data set (Mozabite, Bedouin, Palestinian, and Druze), eight European populations from the HGDP data set (Adygei, Italian, Basque, French, Orcadian, Russian, Tuscan, and Sardinian), and two European populations from the HapMap3 data set (TSI and CEU). We considered only sites in a data set for which there was no population with all individuals missing their genotypes at that site. We ran TreeMix using the San as an outgroup, the sample size correction option, and with exactly three migrations to produce supplementary figure S2, Supplementary Material online.
Admixture Analyses
We compared unrelated individuals from the Ethiopian (Pagani et al. 2012) populations to unrelated individuals from the HapMap phase 3 populations (International HapMap 3 Consortium 2010; Pemberton et al. 2010) as well as to unrelated individuals from the HGDP populations (Rosenberg 2006; Li et al. 2008). From these comparisons, and from previous results in Pagani et al. (2012), we observe that the individuals in our Ethiopian data set are probably admixed (fig. 1), and it was, therefore, necessary to control for admixture within our analyses because admixture can mimic signals of positive selection. To correct for admixture, we employ the European population as a proxy to represent the non-African population that contributed genetic material to the Ethiopian populations. For each population (i.e., Amhara, Tigray, Oromo, Afar, Anuak, and European), we calculated allele frequencies at each locus. To control for admixture, we followed Bhatia et al. (2011). We assumed that the low- and high-altitude Ethiopian populations were a mixture between the Anuak (the outgroup population) and the European population (the nine unrelated CEU individuals from the Complete Genomics data set; Drmanac et al. 2009). This assumption is reasonable given that the Afar, the Amhara, and the Tigray cluster between Europeans and the Anuak (see fig. 1). Under this assumption, at a given locus k, we can calculate the pseudo unadmixed allele frequency for each population by
 |
where α is the proportion of European admixture (Bhatia et al. 2011). We employed the value of α that minimizes 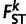 between the Anuak and the corresponding population (e.g., Afar, Tigray, Amhara, Oromo, or the combined Amhara–Tigray or Amhara–Tigray–Oromo populations). To compute
between the Anuak and the corresponding population (e.g., Afar, Tigray, Amhara, Oromo, or the combined Amhara–Tigray or Amhara–Tigray–Oromo populations). To compute 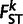 , we used the formula derived in Reynolds et al. (1983).
, we used the formula derived in Reynolds et al. (1983).
To calculate the mean European admixture in the MYEF2-SLC24A5 region for the Afar, Amhara, and Tigray, we averaged the values of α across all SNPs within the region ranging from 50 kb upstream of SLC24A5 to 50 kb downstream of MYEF2.
We did not find the same MYEF2-SLC24A5 region to be under selection in the Oromo population. However, results from Pagani et al. (2012) show that the Oromo also have received some non-African admixture, and thus, we also applied the correction to the Oromo population.
Population Branch Statistic
To detect regions under selection, we calculated the PBS (Yi et al. 2010). This test of selection takes three populations that have an evolutionary relationship illustrated in supplementary figure S11, Supplementary Material online. Population branch statistic (PBS) is similar to the “locus-specific branch length” statistic by Shriver et al. (2004), except that we use a log-transformation. We assume that the Anuak are an outgroup population to the Afar and the high-altitude populations—the Amhara, the Tigray, and the Oromo. Under a scenario of only genetic drift, we expect the high-altitude populations and the Afar to be more genetically similar than the high altitude–Anuak or the Afar–Anuak. If, however, there has been local adaptation in the high-altitude populations, then the regions targeted by positive selection would be highly diverged between the Afar and the high altitude, and the Afar–Anuak would be more similar than the high altitude–Anuak comparison (supplementary fig. S11, Supplementary Material online). Therefore, we should only detect genes that have been targeted by selection in the high-altitude populations. Because we know that one of the selective pressures is lower oxygen level, we expect to observe genes involved in the response to hypoxia.
We obtained RefSeq gene annotations from http://genome.ucsc.edu/, and we used the longest RefSeq identifier for the analysis. For a given gene, FST between a pair of populations, which is a measure of genetic differentiation between a pair of populations, was calculated using the formula from Reynolds et al. (1983) across all SNPs within the genic region, such that each SNP is located within 50 kb of the transcription start and end of the gene. For each gene (±50 kb), we computed FST between population pairs Amhara–Afar, Amhara–Anuak, Tigray–Afar, Tigray–Anuak, (Amhara–Tigray)-Afar, (Amhara–Tigray)–Anuak, Oromo–Afar, Oromo–Anuak, (Amhara–Tigray–Oromo)–Afar, (Amhara–Tigray–Oromo)–Anuak, and Afar–Anuak. Using Anuak as our outgroup, Afar as our lowland population, and Amhara, Tigray, Oromo, Amhara–Tigray, or Amhara–Tigray–Oromo as our highland population. We calculated the PBS of the high-altitude population using the following formula:
where 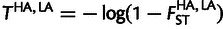 is an estimate of the divergence time between the high altitude (HA) and low altitude (LA) populations. Similarly,
is an estimate of the divergence time between the high altitude (HA) and low altitude (LA) populations. Similarly,  is an estimate of the divergence time between the high altitude and outgroup populations and
is an estimate of the divergence time between the high altitude and outgroup populations and  is an estimate of the divergence time between the low-altitude and outgroup populations. We calculated PBS (Yi et al. 2010) for each (highland, lowland, and outgroup) triple at each gene. Additionally, for each SNP used in the FST calculations, we required that at least 10 alleles (i.e., five individuals) were observed in each population within a (highland, lowland, and outgroup) triple. We computed PBS before and after correction of admixture, and the results are shown in supplementary tables S1 and S2, Supplementary Material online, respectively. These two tables correspond to the case in which the two high-altitude populations (the Amhara and the Tigray) were combined. In both cases, the Anuak population was used as the outgroup population. We also performed the analysis requiring at least 20 alleles (10 individuals), and BHLHE41 dropped from first place to second place. In the Oromo population, BHLHE41 remained at the top of the list before and after correcting for admixture (supplementary table S5, Supplementary Material online, lists results after correcting for admixture).
is an estimate of the divergence time between the low-altitude and outgroup populations. We calculated PBS (Yi et al. 2010) for each (highland, lowland, and outgroup) triple at each gene. Additionally, for each SNP used in the FST calculations, we required that at least 10 alleles (i.e., five individuals) were observed in each population within a (highland, lowland, and outgroup) triple. We computed PBS before and after correction of admixture, and the results are shown in supplementary tables S1 and S2, Supplementary Material online, respectively. These two tables correspond to the case in which the two high-altitude populations (the Amhara and the Tigray) were combined. In both cases, the Anuak population was used as the outgroup population. We also performed the analysis requiring at least 20 alleles (10 individuals), and BHLHE41 dropped from first place to second place. In the Oromo population, BHLHE41 remained at the top of the list before and after correcting for admixture (supplementary table S5, Supplementary Material online, lists results after correcting for admixture).
Simulations with Selection and Admixture
Based on our observed results and those from Pagani et al. (2012), it is likely that the low- and high-altitude Ethiopian populations are admixed with non-African populations. Therefore, we wanted to investigate the effect that admixture has on our ability to identify selection signals, as it may mimic the effect of positive selection in the high-altitude population. Moreover, it is possible that the non-African population has itself experienced recent selection; therefore, it was also important to understand the influence of admixture in this case. We performed simulations to determine the impact of admixture on the PBS, to assess whether correcting for admixture improves inferences of positive selection, and to quantify how often we identify selection when it is in fact admixture from a population that has recently experienced positive selection. We considered two selection scenarios on the same demographic model. First, we considered the case in which the high-altitude population is targeted by positive selection and has a genetic contribution from a non-African population that has experienced a bottleneck in its demographic history. Second, we considered the case in which the high-altitude population receives a genetic contribution from a non-African population that has experienced both a bottleneck and positive selection. For an illustration of the models, see supplementary figure S3, Supplementary Material online. We employed the software SFS_CODE (Hernandez 2008) to simulate under the two selection scenarios. We simulated a region of 10 kb with per-generation per-site mutation and recombination rates of 10−3. We investigated a population-scaled selection coefficient S of 150 and 250, admixture proportions into the low- (α1) and high-altitude (α2) populations of (α1, α2) = (0.1, 0.2), (0.1, 0.3), (0.1, 0.4), and (0.2, 0.4), a time at which admixture occurs TADM of 1.5 and 3 thousand years ago (kya), with all other demographic parameters remaining fixed (see supplementary fig. S3, Supplementary Material online). For each simulation, we sampled 25 individuals from each population. The motivation for these recent admixture times stems from Pagani et al. (2012), who found that admixture into these populations was recent (∼2.5–3 kya). In addition, under the scenario in which selection occurs in the high-altitude population (supplementary fig. S3A, Supplementary Material online), we used a time of selection TSEL of 1.5 kya and 3 kya, and under the scenario in which selection occurs in the non-African population (supplementary fig. S3B, Supplementary Material online), we used a time of selection of 5 kya. These time estimates are comparable to those estimated in Pagani et al. (2012).
We obtained 103 simulated data sets under each parameter combination. For the scenarios in which selection occurs in the high-altitude population, we calculated the proportion of true positives under a specified false positive rate that was based on the corresponding neutral scenario (see supplementary fig. S3, Supplementary Material online). For two parameter combinations, we ran 104 simulations and plotted the true positive rate as a function of false positive rate in the range from 0.0 to 0.05 (see supplementary fig. S5, Supplementary Material online). Supplementary figure S3A, Supplementary Material online, corresponds to TADM = 1.5 kya, S = 150, TSEL = 1.5 kya, α1 = 0.1, and α2 = 0.4. Supplementary figure S3B, Supplementary Material online, corresponds to TADM = 1.5 kya, S = 150, TSEL = 1.5 kya, α1 = 0.2, and α2 = 0.4. For the scenario with selection in the non-African population, we calculated the proportion of times (out of 103) that a simulation is falsely called a positive for a specified false positive rate (see supplementary fig. S6, Supplementary Material online).
Expected Number of Hypoxia Genes
Because of multiple testing, the probability of finding a SNP in or near a hypoxia-related gene increases as the number of tests increase. We restricted our set of genes to those that are within 50 kb of any SNP to estimate the total number of all possible genes that could be captured with the given data. Then, focusing on the top 0.10% of SNPs in our study, we identified the number of genes that were within 50 kb of these SNPs. To approximate the number of hits for hypoxia genes that we should expect by chance, we sampled the same number of unique genes that we identified using the top 0.10% of SNPs from the set of all possible genes. The reason for sampling the same number of unique genes that we identified using the top 0.10% of SNPs is because we wanted to maintain the SNP structure found in the real data. Finally, we counted the number of genes in the intersection of that random set and the hypoxia-related gene set (see Definitions of Hypoxia Gene Set). We repeated this experiment 103 times to derive an empirical null distribution and compute the expected number of hits by chance. Supplementary table S7, Supplementary Material online, contains the median number of hypoxia-related genes we would expect to see based on this empirical null distribution under each of the different population scenarios.
Definitions of Hypoxia Gene Set
The AmiGO tool (http://amigo.geneontology.org, last accessed December 9, 2010) was used to list all genes within the Gene Ontology biological process term “response to hypoxia” plus all descendent terms (GO:0001666 “response to hypoxia," GO:0071456 “cellular response to hypoxia” and GO:0070483 “detection of hypoxia”). This resulted in a set of 152 unique human genes (see supplementary table S9, Supplementary Material online).
Supplementary Material
Supplementary figures S1–S11 and tables S1–S9 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by research grants from the US NSF to E.H.-S. (DBI-0906065) and to M.D. (DBI-1103639) and the US NIH (R01HG003229) to R.N. and to E.H.S (R01HG003229-08S2). We are grateful to two anonymous reviewers for their comments on an earlier version of the manuscript.
References
- Akman HO, Zhang H, Siddiqui MAQ, Solomon W, Smith ELP, Batuman OA. Response to hypoxia involves transforming growth factor-β2 and Smad proteins in human endothelial cells. Blood. 2001;98:3324–3331. doi: 10.1182/blood.v98.12.3324. [DOI] [PubMed] [Google Scholar]
- Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, Di Rienzo A. The genetic architecture of adaptations to high altitude in Ethiopia. PLoS Genet. 2012;8:e1003110. doi: 10.1371/journal.pgen.1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM. Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr Comp Biol. 2006;46:8–24. doi: 10.1093/icb/icj004. [DOI] [PubMed] [Google Scholar]
- Beall CM, Cavalleri LG, Deng L, et al. (29 co-authors) Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, Decker MJ, Brittenham GM, Kushner I, Gebremedhin A, Strohl KP. An Ethiopian pattern of human adaptation to high-altitude hypoxia. Proc Natl Acad Sci U S A. 2002;99:17215–17218. doi: 10.1073/pnas.252649199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia G, Patterson N, Pasaniuc B, et al. (40 co-authors) Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am J Hum Genet. 2011;89:368–381. doi: 10.1016/j.ajhg.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigham A, Bauchet M, Pinto D, et al. (14 co-authors) Identifying signature of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6:e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt SA. The upper pleistocene and early holocene prehistory of the horn of Africa. African Archaeol Rev. 1986;4:41–82. [Google Scholar]
- Chilov D, Hofer T, Bauer C, Wenger RH, Gassmann M. Hypoxia affects expression of circadian genes PER1 and CLOCK in mouse brain. FASEB J. 2001;15:2613–2622. doi: 10.1096/fj.01-0092com. [DOI] [PubMed] [Google Scholar]
- Coote JH, Stone B, Tsang G. Sleep of Andean high altitude natives. Eur J Appl Physiol. 1992;64:178–181. doi: 10.1007/BF00717957. [DOI] [PubMed] [Google Scholar]
- Coote JH, Tsang G, Baker A, Stone B. Respiratory changes and structure of sleep in young high-altitude dwellers in the Andes of Peru. Eur J Appl Physiol. 1993;66:249–253. doi: 10.1007/BF00235102. [DOI] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, et al. (65 co-authors) Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2009;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassen M. The Oromo of Ethiopia: a history, 1570–1860. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- He Y, Jones CR, Fijiki N, Xu Y, Guo B, Holder JL, Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD. A flexible forward simulator for populations subject to selection and demography. Bionformatics. 2008;24:2786–2787. doi: 10.1093/bioinformatics/btn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takag Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, et al. (12 co-authors) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Noshiro M, Fujimoto K, Kawamoto T. Roles of Dec1 and Dec2 in the core loop of the circadian clock, and clock outputs metabolism. Hirosaki Med. J. 2010;61:s34–s42. [Google Scholar]
- Koyanagi S, Kuramoto Y, Nakagawa H, Aramaki H, Ohdo S, Soeda S, Shimeno H. A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 2003;63:7277–7283. [PubMed] [Google Scholar]
- Lamason RL, Mohideen M-APK, Mest JR, et al. (25 co-authors) SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 2005;310:1782–1786. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- Leon-Velarde F, Maggiorini M, Reeves JT, et al. (17 co-authors) Consensus statement on chronic subacute high altitude diseases. High Alt Med Biol. 2005;6:147–157. doi: 10.1089/ham.2005.6.147. [DOI] [PubMed] [Google Scholar]
- Lewis H. The origins of the Galla and Somali. J Afr Hist. 1966;7:27–46. [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. (11 co-authors) Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem. 2002;277:47014–47021. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- Monge CC, Arregui A, Leon-Velarde F. Pathophysiology and epidemiology of chronic mountain sickness. Int J Sports Med. 1992;13:S79–S81. doi: 10.1055/s-2007-1024603. [DOI] [PubMed] [Google Scholar]
- Montagner M, Enzo E, Forcato M, et al. (12 co-authors) SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487:380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- Moore LG, Young D, McCullough RE, Droma T, Zamudio S. Tibetan protection from intrauterine growth restriction (IUGR) and reproductive loss at high altitude. Am J Hum Biol. 2001;13:635–644. doi: 10.1002/ajhb.1102. [DOI] [PubMed] [Google Scholar]
- Mortola JP. Hypoxia and circadian patterns. Respir Physiol Neurobiol. 2007;158:274–279. doi: 10.1016/j.resp.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Niermeyer S, Andrade Mollinedo P, Huicho L. Child health and living at high altitude. Arch Dis Child. 2009;94:806–811. doi: 10.1136/adc.2008.141838. [DOI] [PubMed] [Google Scholar]
- Pagani L, Ayub Q, MacArthur DG, et al. (13 co-authors) High altitude adaptation in Daghestani populations from the Caucasus. Hum Genet. 2011;131:423–433. doi: 10.1007/s00439-011-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Kivisild T, Tarekegn A, et al. (14 co-authors) Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Wang C, Li JZ, Rosenberg NA. Inference of unexpected genetic relatedness among individuals in HapMap phase III. Am J Hum Genet. 2010;87:457–464. doi: 10.1016/j.ajhg.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Shi H, Qi X, Xia C, Zhong H, Ma RZ, Su B. The ADH1B Arg47His polymorphism in East Asian populations and expansion of rice domestication in history. BMC Evol Biol. 2008;20:1–15. doi: 10.1186/1471-2148-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, et al. (15 co-authors) Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28:1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleurdeau D. Human technical behavior in the African middle stone age: the Lithic Assemblange of Porc-Epic Cave (Dire Dawa, Ethiopia) Afr Archaeol Rev. 2006;22:177–197. [Google Scholar]
- Plywaczewski R, Wu T-Y, Wang X-Q, Cheng H-W, Sliwinski P, Zielinski J. Sleep structure and periodic breathing in Tibetans and Han at simulated altitude of 5000 m. Respir Physiol Neurobiol. 2003;136:187–197. doi: 10.1016/s1569-9048(03)00081-8. [DOI] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Sato F, Bhawal UK, Kawamoto T, et al. (13 co-authors) Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates vascular endothelial growth factor expression. Genes Cells. 2008;13:131–144. doi: 10.1111/j.1365-2443.2007.01153.x. [DOI] [PubMed] [Google Scholar]
- Scheinfeldt LB, Soi S, Thompson S, et al. (11 co-authors) Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Kennedy GC, Parra EJ, Lawson HA, Sonpar V, Huang J, Akey JM, Jones KW. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, et al. (12 co-authors) Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- Tang H, Jogersen E, Gadde M, Kardia SL, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N. Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Human Genet. 2006;119:624–633. doi: 10.1007/s00439-006-0175-4. [DOI] [PubMed] [Google Scholar]
- Wang B, Zhang Y-B, Zhang F, et al. (18 co-authors) On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS One. 2011;6:e17002. doi: 10.1371/journal.pone.0017002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Li S, Yang Y, et al. (13 co-authors) A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 2011;28:1003–1011. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- Yi X, Liang Y, Huerta-Sanchez E, et al. (70 co-authors) Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip R. Altitude and birth weight. J Pediatr. 1987;111:869–876. doi: 10.1016/s0022-3476(87)80209-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.