The transcriptome during acute otitis media due to NTHi shows relatively weak immune activation
Keywords: acute otitis media, immune response signatures, non-typeable Haemophilus influenzae, transcriptome profile
Abstract
Non-typeable Haemophilus influenzae (NTHi) causes acute otitis media (AOM) in young children. In our recent paper in Microbes and Infection we described the transcriptome signature elicited from PBMCs at onset of AOM caused by Streptococcus pneumoniae. In the current study we found very different results with NTHi AOM infections; 5.1% of 29 187 genes were differentially regulated by more than 2-fold at the onset of AOM compared with the pre-infection healthy state in the same children. Among the 1487 transcripts, 100 genes associated with the immune defense response were specifically analyzed. About half of the differentially regulated genes associated with antibacterial activity and the cell-mediated immune response were activated and half were suppressed. The important signatures for NTHi in children suggested that the balance of the immune response was toward suppression. Moreover, 90% of the genes associated with a pro-inflammatory cytokine response were down-regulated. The genes associated with the classic complement pathway were down-regulated, although the alternative complement pathway genes were up-regulated. These results provide the first human transcriptome data identifying gene expression in the immune response to be predominantly down-regulated at the onset of AOM due to NTHi.
Introduction
Acute otitis media (AOM) is defined by the presence of an inflammatory infiltrate in the middle ear usually accompanied by fever and other acute signs of illness. The most frequently isolated bacteria in AOM, in countries that have adopted widespread use of pneumococcal conjugate vaccines, is non-typeable Haemophilus influenzae (NTHi) (1). We recently described the transcriptome signature elicited in PBMCs at the onset of AOM caused by Streptococcus pneumoniae (Spn) (2). For S. pneumoniae, we found that 1903 (6.5%) of 29 187 genes were differentially regulated by >2-fold at the onset of AOM compared with the pre-infection stage of the same children. The ontology of differentially regulated genes was dominated by genes involved in the immune response. At the onset of AOM caused by S. pneumoniae infection, genes associated with bacterial defenses were significantly up-regulated, including β-defensin123, S100 protein A12, Toll-like receptor 5 (TLR5), IL-10 and those involved in the classical and alternative complement pathways. Genes associated with inhibition of bacterial entry through clathrin-dependent endocytosis were also up-regulated. In contrast, genes associated with cell-mediated immune responses were broadly down-regulated.
Here we describe, for the first time in human children, an evaluation of the gene expression profile of PBMCs associated with onset of AOM caused by NTHi. This analysis was made possible during a prospective study on the immune responses of children to NTHi during AOM with comparable PBMC samples taken within 4 weeks before the onset of AOM. Double sampling from the same children reduced the complexities of analysis due to genetic heterogeneity among individuals. Closely timed sampling avoided the influence of changing age on responses in pediatric subjects. We found significant changes in regulation of immune response genes at onset of AOM caused by NTHi, differing from those observed following AOM caused by S. pneumoniae (2).
Methods
Subjects
In this study, similar to our previous work (2), we selected samples from four previously healthy children of the same age, race and severity of AOM and compared the gene change in PBMCs before and at onset of NTHi-induced AOM infection using microarray analysis. The pathogen-specific genes specifically involved in different functional immunity and defense-related mechanisms were the focus of the analysis.
The samples were collected from the four children (age: 10–15 months) as approved by the Institutional Review Board at the Rochester General Hospital. The diagnosis of AOM was made based on symptoms of fever, irritability or ear ache and physical signs of inflammation of the tympanic membrane (bulging) with the presence of middle ear fluid (pus-laden fluid) documented by tympanocentesis. The inclusion criteria were identical to those in our previous report (2): Caucasian children, pre-infection PBMC/serum sample within 4 weeks before the onset of AOM and the collection of an acute stage of illness PBMC/serum sample within 24h of onset of AOM, AOM caused by NTHi as proven by tympanocentesis, and moderate to severe AOM as judged by the physician. None of the children had an immunodeficiency, a history of chronic or recurrent AOM, any chronic disease or any infectious disease other than a concurrent common cold; none was receiving steroids or other immunomodulatory agents.
Identification of pathogens
Identification of NTHi was accomplished as described previously (3). The culture results were further verified by multiplex PCR as described previously (4).
RNA isolation
Heparinized peripheral venous blood (4–10ml) was collected for isolation of PBMCs and sera. PBMCs were isolated using a Ficoll gradient and total RNA was extracted from PBMCs as previously described (2).
Microarray analysis
DNA microarray analysis was performed as previously described (2). Gene expression was assessed in three replicates of each PBMC sample as previously described (2).
Quantitative real-time reverse transcriptase–PCR analysis
For real-time reverse transcriptase (RT)–PCR, an aliquot of the RNA preparation that was used in the microarray experiments was analyzed as previously described (2).
Proteome assays
The protein levels of CCL2 (also known as MCP-1), CXCL10 (also known as IP-10) IL-6, IL1-β and INF-γ in the serum samples were determined in the same children for whom microarray was performed, using a Bio-Plex Pro Assay (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions and read with a Luminex instrument. Five additional pairs of serum samples were identified for the study of cytokine levels in NTHi-induced AOM children; the subjects met the sample inclusion criteria as stated in sample selection, but PBMCs were not available. Cytokine levels were also determined in mucosal samples from the site of NTHi colonization (nasal wash secretions or NW and AOM infection samples (middle ear fluid or MEF) from the same nine children (four children for whom transcriptome analysis was performed and the same five additional pairs that were tested for serum cytokine levels)).
Data analysis
DNA microarray analysis was performed as previously described (2) in a blinded manner by Phalanx Biotech using the Human Whole Genome OneArray™ (Phalanx Biotech, Palo Alto, CA, USA), which included 29 187 human genome probes and 1088 experimental control probes. The data with a consistent and high-quality read-out, verified by quality control at Phalanx Biotech, of microarray were analyzed at the Rochester Institute of Technology (by L.C.) for biological function groupings. The data from 12 hybridizations of 4 children were pooled and the fold change was calculated by comparing the read-out at onset of AOM compared with the pre-infection healthy state. Genes with >2-fold increase or decrease in all four children were considered significant and included for further analysis. The biological themes associated with these genes were analyzed using DAVID software (http://david.abcc.ncifcrf.gov). To identify lexical enrichments, the terminology defined by the Gene Ontology Consortium (http://www.geneontology.org) was used.
Results
Transcriptome responses in children with AOM due to NTHi
Characteristics of the AOM patients are reported in Table 1. The healthy controls are the same children at their pre-infection healthy state. We identified 1487 (5.1%) of 29 187 genes with transcripts differentially expressed between the pre-infection healthy state and the NTHi-infected AOM state. Of this subset, 962 transcripts (65% of the total) were up-regulated, whereas 525 transcripts (35% of the total) were down-regulated at the onset of AOM relative to the healthy pre-infection state. The complete list of differentially expressed genes with the corresponding human RefSeq (Entrez gene ID) is shown in Supplementary Table 1, available at International Immunology Online, and can be tracked from the website of http://www.ncbi.nlm.nih.gov/geo/ with the accession number GSE27990. The functional annotation clustering analysis of the 1487 gene transcripts showed that 100 (6.72%) of the genes were grouped in the host immune defense category and were analyzed further.
Table 1.
Clinical and demographic characteristics of the AOM children (n = 4)
| Clinical features | AOM children |
|---|---|
| Age (months) | 10–15 |
| Male (%) | 100 |
| Caucasian (%) | 100 |
| NTHi positive in MEF (%) | 100 |
| Other otopathogens (%) | 0 |
| Duration between cold and AOM onset (days) | 5–7 |
| Fever (%) | 100 |
| Ear pulling (%) | 100 |
| Middle ear fluid (%) | 100 |
Genes associated with antibacterial activity
Of the 100 genes in the host immune defense category, 20 were classified in the antibacterial gene cluster, including 11 down-regulated and 9 up-regulated genes (Table 2). The highest down-regulated gene was CCL2 (−31-fold change), which displays chemotactic activity for monocytes and basophils. IL1-β, a pro-inflammatory cytokine that stimulates thymocyte proliferation, B-cell maturation and proliferation and that is involved in the inflammatory response, was down-regulated −6.2-fold. Other antibacterial genes that were down-regulated included β-defensin 1 (DEFB1) and DEFB112. The highest up-regulated gene was azurocidin (AZU1, +31-fold change), which encodes a neutrophil granule-derived antibacterial and monocyte- and fibroblast-specific chemotactic glycoprotein and possesses a specific cytotoxic action to species of Gram-negative bacteria. In the regulated cluster, there were nine genes with functions in response to LPS stimulation, including ADM, THBD, SLC11A1, VLDLR, STAT1, PTGS2, IL1B, IDO1 and CCL2 (Table 2).
Table 2.
The differentially expressed genes with the function of antimicrobial activity
| Entrez gene ID | Gene symbol | Gene description | Fold change |
|---|---|---|---|
| 566 | AZU1 | Azurocidin 1 | 31.38 |
| 5553 | PRG2 | Proteoglycan 2 | 3.78 |
| 8349 | HIST2H2B | Histone cluster 2 | 2.95 |
| 1670 | DEFA5 | Defensin, α 5, | 2.59 |
| 133 | ADM a | Adrenomedullin | 2.52 |
| 245929 | DEFB115 | Defensin, β 115 | 2.3 |
| 7056 | THBD | Thrombomodulin | 2.28 |
| 6556 | SLC11A1 | Solute carrier family 11 | 2.18 |
| 7436 | VLDLR | Very low-density lipoprotein receptor | 2.03 |
| 245915 | DEFB112 | Defensin, β 112 | –2.03 |
| 1672 | DEFB1 | Defensin, β 1 | –2.04 |
| 6772 | STAT1 | Signal transducer and activator of transcription 1 | –2.08 |
| 8915 | BCL10 | B-cell CLL/lymphoma 10 | –2.29 |
| 5743 | PTGS2 | Prostaglandin-endoperoxide synthase 2 | –2.45 |
| 7124 | TNFA | Tumor necrosis factor | -3.42 |
| 3569 | IL6 | Interleukin 6 | –3.71 |
| 3553 | IL1B | Interleukin 1, β | –6.22 |
| 6364 | CCL20 | Chemokine (C–C motif) ligand 20 | –7.2 |
| 3620 | IDO1 | Indoleamine 2,3-dioxygenase 1 | –21.18 |
| 6347 | CCL2 | Chemokine (C–C motif) ligand 2 | –31.39 |
aThe bold italic letters indicate the genes that have the function of LPS response as well.
Genes associated with cytokines
Nineteen genes relating to cytokine production were differentially regulated (Table 3). Among these, 89.5% (17/19) of the cytokine genes were down-regulated. The strongest down-regulated gene was CXCL10 with a −92-fold change; it has a role in stimulation of monocytes, natural-killer- and T-cell migration and modulation of adhesion molecule expression. CCL2 was down-regulated −31-fold; CCL2 recruits monocytes, memory T cells and dendritic cells to sites of tissue injury, infection and inflammation. IL-6 was down-regulated −3.7-fold; it is a cytokine produced by a variety of cell types including monocytes/macrophages, neutrophils and endothelial cells in response to LPS stimulation. There were two up-regulated genes controlling cytokines: CCL7 (+26-fold, a cytokine that specifically attracts monocytes and regulates macrophage function) and GREM2 (gremlin 2; 2-fold), whose function in bacterial immunity is not clear (Table 3).
Table 3.
Cytokines differentially expressed in children with AOM due to NTHi
| Entrez gene ID | Gene symbol | Gene description | Fold change |
|---|---|---|---|
| 6354 | CCL7 | Chemokine (C–C motif) ligand 7 | 25.63 |
| 64388 | GREM2 | Gremlin 2, cysteine knot superfamily | 2.06 |
| 2920 | CXCL2 | Chemokine (C–X–C motif) ligand 2 | –2.1 |
| 8744 | TNFSF9 | Tumor necrosis factor (ligand) superfamily, member 9 | –2.17 |
| 6366 | CCL21 | Chemokine (C–C motif) ligand 21 | –2.31 |
| 8743 | TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | –2.37 |
| 8200 | GDF5 | Growth differentiation factor 5 | –2.41 |
| 6348 | CCL3 | Chemokine (C–C motif) ligand 3 | –3.09 |
| 3448 | IFNA14 | Interferon, α 14 | –3.26 |
| 7124 | TNFA | Tumor necrosis factor (TNF superfamily, member 2) | –3.42 |
| 3569 | IL6 | Interleukin 6 (interferon, β 2) | –3.71 |
| 6355 | CCL8 | Chemokine (C–C motif) ligand 8 | –4.27 |
| 57152 | SLURP1 | Secreted LY6/PLAUR domain containing 1 | –5.22 |
| 655 | BMP7 | Bone morphogenetic protein 7 | –5.24 |
| 3553 | IL1B | Interleukin 1, β | –6.22 |
| 6364 | CCL20 | Chemokine (C–C motif) ligand 20 | –7.2 |
| 3552 | IL1A | Interleukin 1, α | –9.65 |
| 6347 | CCL2 | Chemokine (C–C motif) ligand 2 | –31.39 |
| 3627 | CXCL10 | Chemokine (C–X–C motif) ligand 10 | –92.01 |
Genes associated with T-cell activation, migration and proliferation
Seventeen differentially regulated genes involved T-cell activation, migration and proliferation. Among these, eight genes were down-regulated (Table 4). Four cytokine genes involved in this group were all significantly down-regulated, including CCL2 and IL-6. TLR7, which participates in T-cell activation, was also significantly down-regulated (−2.8-fold). Nine genes were up-regulated, with the highest one being PODXL (+3.9-fold); PODXL is involved in the regulation of adhesion and cell morphology and functions as an anti-adhesive molecule. One T-cell surface marker, CD276, was up-regulated +2.06-fold.
Table 4.
Genes involved in T-cell activation, migration and proliferation
| Entrez gene ID | Gene symbol | Gene description | Fold change |
|---|---|---|---|
| 5420 | PODXL | Podocalyxin-like | 3.88 |
| 1910 | EDNRB | Endothelin receptor type B | 3.61 |
| 3684 | ITGAM | Integrin, α M | 2.43 |
| 80329 | ULBP1 | UL16-binding protein 1 | 2.36 |
| 10014 | HDAC5 | Histone deacetylase 5 | 2.32 |
| 5896 | RAG1 | Recombination-activating gene 1 | 2.26 |
| 6556 | SLC11A1 | Solute carrier family 11, member 1 | 2.18 |
| 80381 | CD276 | CD276 molecule | 2.06 |
| 7535 | ZAP70 | ζ-chain (TCR)-associated protein kinase, 70 kDa | 2.01 |
| 684 | BST2 | NPC-A-7; bone marrow stromal cell antigen 2 | –2.02 |
| 4478 | MSN | Moesin | –2.13 |
| 6441 | SFTPD | Surfactant protein D | –2.43 |
| 51284 | TLR7 | Toll-like receptor 7 | –2.76 |
| 7124 | TNFA | Tumor necrosis factor | –3.42 |
| 3569 | IL6 | Interleukin 6 | –3.71 |
| 3553 | IL1B | Interleukin 1, β | –6.22 |
| 6347 | CCL2 | Chemokine (C–C motif) ligand 2 | –31.39 |
Genes associated with the complement system
Eight differentially regulated genes were involved in the complement pathway. Important components in the classical pathway, such as C1QB and C2, were down-regulated (both −2.2-fold), although C4BPA, encoding the protein needed to control the classical pathway of complement activation, was up-regulated +4.2-fold. The highest up-regulated gene was complement factor B (CFB, +5.3-fold), an important component of the alternative complement pathway. Taken together, the data suggest that the classical pathway of the complement system was not a predominant defense mechanism in children with AOM due to NTHi (Table 5).
Table 5.
The differentially regulated genes involved in the complement cascade
| Entrez gene ID | Gene symbol | Gene description | Fold change |
|---|---|---|---|
| 629 | CFB | Complement factor B | 5.34 |
| 716 | C1S | Complement component 1, s subcomponent | 4.57 |
| 722 | C4BPA | Complement component 4 binding protein, α | 4.22 |
| 1191 | CLU | Clusterin | 2.71 |
| 735 | C9 | Complement component 9 | 2.4 |
| 10747 | MASP2 | Mannan-binding lectin serine peptidase 2 | 2.26 |
| 717 | C2 | Complement component 2 | –2.17 |
| 713 | C1QB | Complement component 1, q subcomponent, B chain | –2.23 |
Comparison of differentially expressed genes in AOM due to NTHi and S. pneumoniae
We examined the 100 differentially expressed genes in children with AOM due to NTHi infection with the results of children at onset of AOM caused by S. pneumoniae as previously described (2). Of the 100 transcripts differentially expressed at onset of NTHi AOM, only 22 (22%) were differentially expressed in S. pneumoniae-induced AOM. Among the 22 genes, 17 were regulated in the same direction, but 8, such as CXCL10, showed a level different by >2-fold (Fig. 1). Five genes (23%), such as C1QB and C4BPA, were regulated conversely. Among the nine differentially expressed genes that were related to an LPS response in NTHi patients, two genes (IL1-β and CCL2) showed differential regulation in S. pneumoniae-induced AOM, but only CCL2 showed a similar level change (Fig. 1).
Fig. 1.
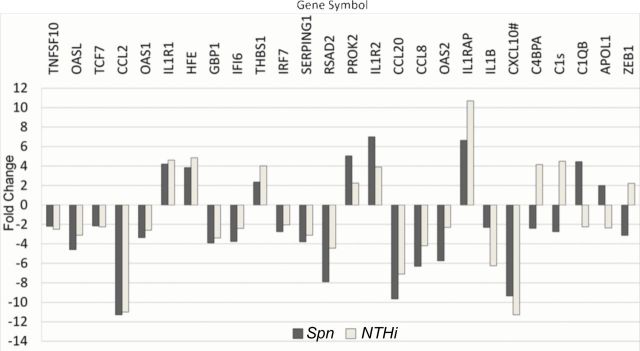
The common differentially regulated genes involved in immune defense responses in children with AOM caused by S. pneumoniae (Spn)/NTHi. The fold change derived from the comparison between AOM stage and their pre-infection healthy stage is demonstrated. White indicates NTHi-induced AOM versus their pre-infection healthy stage from four children. Grey indicates Spn-induced AOM versus their pre-infection healthy stage from four children. # The values for CXCL10 are shown after being reduced 10 times to fit the figure.
Gene verification by real time RT–PCR
To validate the microarray data, qRT–PCR was performed with the RNA used in the microarray. Because the expression of IL-6 was down-regulated in our study, which conflicted with the in vitro observations reported by other scientists, we selected IL-6 as a target for further verification. Real-time RT–PCR showed that IL-6 was down-regulated −4.8-fold at onset of AOM compared with the levels in the healthy state, which was similar to the result of −3.7-fold obtained in the microarray.
Protein analysis
To evaluate whether the transcriptome results were reflected in the proteome, we evaluated the concentrations of CCL2 (MCP-1), CXCL10 (IP-10), IL-6, IL1-β and IFN-γ proteins in the sera, nasal secretions and MEF of the study children. Similar to the transcriptome results, the levels of CCL2 in the serum samples of children at onset of NTHi-induced AOM (229ng ml–1) were significantly lower (P = 0.04) compared with the levels in the children when they were healthy (505ng ml–1; Fig. 2). However, the levels of CCL2 in the nasal secretions (7.6ng ml–1) where pathogenesis of AOM commences were significantly higher (P = 0.007) compared with the NW when the children were healthy (2.9ng ml–1); the levels in the MEF at the site of infection were ~30-fold higher (228ng ml–1) than those of nasal secretions at the onset of AOM.
Fig. 2.
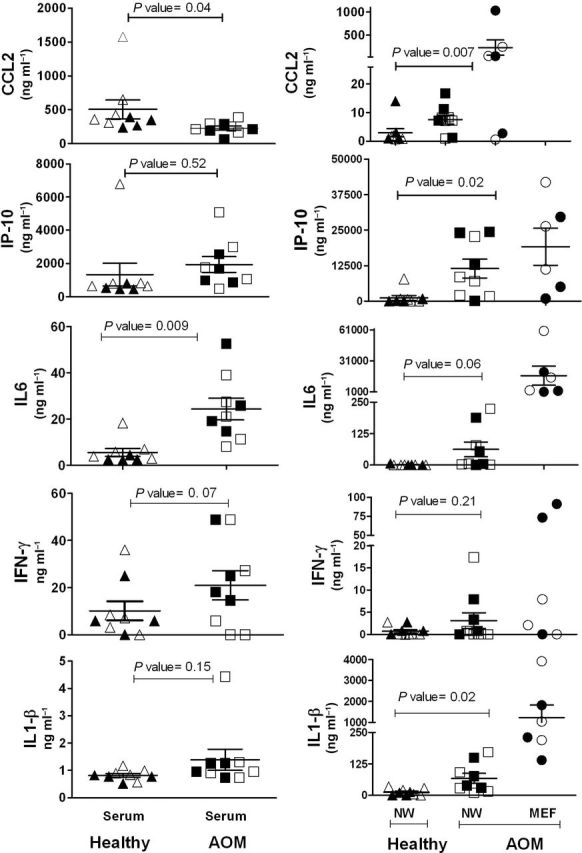
The cytokine levels of CCL2, IP-10, IL-6, IFN-γ and IL-1-β in the sera of healthy and NTHi-induced AOM children and mucosal samples (NW of healthy children and the NW and MEF of NTHi-induced AOM children). The amount shown is in nanograms per milliliter (ng ml–1) and the P values are calculated using the paired t-test. The bold filled symbols represent samples from children for whom transcriptome analysis was performed in the study.
Unlike the transcriptome results, the levels of IL-6 in serum samples of children at onset of AOM were significantly higher compared with the levels when the children were healthy, a result that was further reflected with even higher quantities of protein in the nasal secretions and MEF (Figure 2). No significant difference was observed in the concentrations of IP-10 and IFN-γ proteins during the healthy versus AOM stage in either serum or mucosal samples. The amounts of IP-10 and IL1-β were also the same during the healthy and AOM stages in serum samples but significantly higher in NW during AOM compared with the healthy state. The amounts of IL-6 and IL1-β were >100-fold higher at the site of infection (MEF) compared with serum samples of children with AOM.
Discussion
Number of genes mobilized during AOM due to NTHi in young children.
To our knowledge, this is the first report on the transcriptome profile in PBMCs of children at onset of AOM due to NTHi infection. NTHi infection was associated with activation and suppression of both innate and adaptive immune responses. The percentage of genes differentially expressed (4.8%) is consistent with that in other reports (5, 6). In our recent study regarding Spn-induced AOM in children, 6.2% of genes showed >2-fold change compared with the healthy state (2).
Genes associated with antibacterial activity.
The LPS in the outer membrane of Gram-negative bacteria elicits strong immune responses in animals. In this study, we found 9% (9/100) of the differentially regulated genes involved LPS, but most were down-regulated. The observed down-regulation of the universal antibacterial gene DEFB1 is not consistent with the observations of Lee et al. (7), who used an in vitro radial assay to show that both DEFB1 and DEFB2 had bactericidal/bacteriostatic activity against NTHi. However, human β-defensin gene regulation may be variable in different cell types (8, 9). In addition, the current study also showed that most of the regulated genes classified in the antibacterial group have a function related to responses unique to the Gram-negative bacterial outer membrane envelope, such as AZU1. Miller et al. (10) measured AZU1 protein in 47 middle ear effusions of children and found that the concentration of AZU1 in NTHi antigen-positive middle ear effusions were significantly higher than in either S. pneumoniae antigen-positive or NTHi antigen-negative middle ear effusions.
Genes associated with cytokines
Most of the genes regulating cytokines, including IL-6, were down-regulated. IL-6 is a potent inducer of the acute phase response, plays an essential role in the final differentiation of B cells into immunoglobulin-secreting cells, is involved in lymphocyte and monocyte differentiation and positively regulates the JAK–STAT cascade and defense response to Gram-negative bacteria.
Genes associated with T-cell activation, migration and proliferation
It is known that pathogen recognition receptors, such as the TLRs, play a role in AOM susceptibility (11–13). Hirano et al. (14) found that TLR4 played an important role in relation to Th1 function for the optimal development of acquired immune responses to outer membrane proteins of NTHi administered intra-nasally in mice However, in human children, we did not find the differential regulation of TLR4. Instead, we found that TLR7 was significantly down-regulated. TLR7 plays an important role against viral infections (15) and all four of the studied children had concurrent upper respiratory viral infections as is typical for children who develop AOM. In an in vitro experiment, Sakai et al. (16) found that up-regulation of TLR7 expression in human airway epithelial cells occurred, but they also showed that NTHi could induce expression of deubiquitinase, which, in turn, acted as a negative regulator of NTHi-induced TLR7 expression (16).
Genes associated with the complement system
The role of the complement system in the pathogenesis of otitis media has been previously reported. Veltri and Sprinkle (17) found soluble immune complexes in the MEF, which included complement components. Shimizu et al. (18) demonstrated that complement factors were important in bactericidal activity in MEF. In the current study, the up-regulation of CFB and down-regulation of C1QB not only verified the importance of the complement system in immune defense during NTHi infection in young children but also indicated that the alternative pathway predominated. Stenfors and Räisänen (19) identified complement component C3-coated bacteria in MEF from children with AOM. Bernstein et al. (20) also found elevated levels of CFB in children with otitis media with effusions. In a chinchilla model, Figueira et al. (21) showed that complement was a key arm of host innate immunity against NTHi-induced otitis media and LPS sialylation had a more profound impact on the amount of alternative-pathway-mediated C3 binding when LPS was the main C3 target.
Comparison of differentially expressed genes in AOM due to NTHi and S. pneumoniae
Clinically, in children, NTHi is known to produce less evidence of local inflammation and systemic illness during AOM compared with S. pneumoniae (discussed in the following paragraphs); so finding less up-regulation of pro-inflammatory cytokines might be expected at onset of AOM due to NTHi. In a rat model of experimental AOM caused by NTHi and S. pneumoniae, otomicroscopically, AOM appeared 1 day after NTHi inoculation and 3 days after S. pneumoniae inoculation. NTHi-induced AOM was less severe than S. pneumoniae-induced AOM. IL-6 mRNA levels peaked at 6h post-inoculation for NTHi-induced AOM and at 1–3 days for S. pneumoniae-induced AOM (22). Therefore, the possibility exists that the transcriptome signature differences between NTHi and S. pneumoniae may be due to either a different severity of infection or a difference in peak immune response transcription in PBMCs.
Protein analysis
The differences between the transcriptome signature and the protein results in serum as well as nasal secretions and MEF are of interest. Only the levels of CCL2 in serum were lower in the AOM stage compared with the levels in the healthy stage of children, similar to the transcript levels of CCL2 from PBMCs. Otherwise, cytokine transcript levels in circulating PBMCs from the AOM stage and the healthy stage of children were not reflected in serum cytokine levels. Several explanations may be at play (1). It is known that the middle ear is devoid of immune cells during health and they arrive to the NW and MEF from the blood. The transcription of immune factors in the NW and MEF may involve those cells that have migrated to the site of infection in response to the microbe. Some of those cells may have been depleted from the periphery where we sampled cells for this study. [To evaluate this possibility, we sought to study the transcriptome from the cellular infiltrate in the NW and MEF of children but found the number of cells too small to allow analysis by microarray (2)]. The timing of the immune responses may differ between the PBMCs and the cells that migrate to the middle ear. Tong et al. (23) used a rat model of AOM to determine the induction of gene expression for pro-inflammatory mediators in response to NTHi. Their results suggest that transcription events for the immune response following AOM due to NTHi occur over a relatively prolonged time frame. A single sampling at onset of clinical illness from AOM due to NTHi captures only that moment in time and children may differ in the duration of early infection before otalgia occurs and medical care is sought (3). The possibility of post-transcriptional regulation of cytokine expression should be raised as this could lead to a lack of correlation between transcript levels measured by transcriptome/qRT–PCR and protein levels measured by Luminex (4). The discordance could be explained by the possibility that PBMCs are not the primary source of elevated serum or mucosal inflammatory cytokines in children during NTHi-induced AOM infection. Elevated cytokines may be derived from cytokine-producing cells other than PBMCs, such as epithelial cells, which undergo different alterations in cytokine expression in response to infection. In fact, our observation of higher cytokine expression in the mucosal secretions compared with PBMCs suggests the involvement of other cells producing cytokines during infection. Such discrepancy in the PBMC transcript levels versus serum protein levels has been previously described (24).
General conclusions, limitations of study results and future directions
In summary, we have demonstrated that NTHi induces relatively weaker up-regulation of host immune defense genes in PBMCs of young children with AOM compared with those observed in young children following AOM caused by S. pneumoniae. The important signatures for NTHi in children suggested that the balance of the immune response was toward suppression. About half of the differentially regulated genes associated with antibacterial activity and cell-mediated immune response were activated and half were suppressed. Further, 90% of the genes associated with a pro-inflammatory cytokine response were down-regulated. The genes associated with the classic complement pathway were down-regulated although alternative complement pathway genes were up-regulated. This study therefore demonstrates that NTHi infection is associated with a predominant down-regulation of genes associated with the immune response in PBMCs in young children, unlike the response observed with S. pneumoniae infection (2).
Limitations of our study include those inherent in clinical studies of gene regulation in humans, where genetic heterogeneity, timing of exposure to pathogens, dose and strain of the pathogen are involved. Even though we only studied four children, the power of our analysis is in the ability to study the transcriptome profile in the same child before and after infection, proven to be caused by a specific pathogen. By studying the same child with paired samples and restricting our analysis to those responses that were shared (albeit to varying degrees) among all four children, we avoided the difficulties in microarray analysis of the human host due to individual divergence. In addition, to avoid the technical error and subjectivity in microarray assay interpretation, the data were blinded and compiled by an outside laboratory (Phalanx Biotech). False discovery rate (FDR) or other statistical methods may be a more preferable way to pick up differentially expressed genes. However, with the data set we had, the FDR methodology was not successful. Even after controlling FDR at a pre-specified level (i.e., 5%), very few genes were selected. Because of the small sample size, we also tried a non-parametric method, the Wilcoxon two-sample test, to generate P values moderated with either FDR controlling or Benjamin–Hochberg adjustment, but this methodology also allowed selection of only a few genes. Despite the limitations of fold change, we contend that the results of fold differences have non-trivial biological implications.
We used PBMCs as the source of clinical material because blood is the reservoir for immune cells that migrate to the middle ear during AOM. Leukocytes isolated from the peripheral blood constitute an accessible source of clinically relevant information, from which a comprehensive molecular phenotype can be obtained by microarray analysis. Therefore, although further study with more samples will be needed to further confirm our results, the results of this challenging study strongly indicate the potential value of this research.
Future parallel comparisons of the gene expression patterns to different otopathogens during AOM in children might further specify the features of pathogen-specific genes which could facilitate the identification and the development of novel immune-modulating therapeutics. Further study of both peripheral blood and tissue-specific cells could find functional differences among immune cell subsets.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
National Institutes of Health grant R01DC08671 (to M.E.P.).
Supplementary Material
Acknowledgements
This study would not have been possible without the help and dedication of Dr Janet Casey at Legacy Pediatrics. We thank Dr Robert Osgood from the Rochester Institute of Technology for assistance with the RT–PCR experiments, the collaborating pediatricians from Long Pond Pediatrics, Genesis Pediatrics, Rainbow Pediatrics and Lewis Pediatrics; we also thank the parents who consented to this challenging study. K.L. contributed to the design and performance of the research, analyzed the data and wrote the first draft of the paper. L.C., PhD, contributed to analysis of the data. R.K. performed the protein analysis. M.E.P. contributed to the design of the research, analyzed data and finalized the writing of the paper.
References
- 1. Casey J. R., Adlowitz D. G., Pichichero M. E. 2010. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine Pediatr. Infect. Dis. J. 29: 304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu K., Chen L., Kaur R, Pichichero M. 2012. Transcriptome signature in young children with acute otitis media due to Streptococcus pneumoniae Microbes Infect. 14:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu K., Janet C, Michael P. 2010. Serum intercellular adhesion molecule 1 variations in young children during acute otitis media Clin. Vacc. Immunol. 17: 1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaur R., Adlowitz D. G., Casey J. R., Zeng M, Pichichero M. E. 2010. Simultaneous assay for four bacterial species including Alloiococcus otitidis using multiplex-PCR in children with culture negative acute otitis media Ped. Infect. Dis. J. 29: 741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenberger C. M., Scott M. G., Gold M. R., Hancock R. E., Finlay B. B. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression J. Immunol. 164: 5894 [DOI] [PubMed] [Google Scholar]
- 6. Ichikawa J. K., Norris A., Bangera M. G., et al. 2000. Interaction of Pseudomonas aeruginosa with epithelial cells: identification of differentially regulated genes by expression microarray analysis of human cDNAs. Proc. Natl. Acad. Sci. U.S.A. 97: 9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee H. Y., Andalibi A., Webster P., et al. 2004. Antimicrobial activity of innate immune molecules against Streptococcus pneumoniae, Moraxella catarrhalis and nontypeable Haemophilus influenzae . BMC Infect. Dis. 4: 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chung W. O., Dale B. A. 2004. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect. Immun. 72: 352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu A. Y., Destoumieux D., Wong A. V., et al. 2002. Human beta-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J. Invest. Dermatol. 118: 275 [DOI] [PubMed] [Google Scholar]
- 10. Miller M. B., Koltai P. J., Hetherington S. V. 1990. Bacterial antigens and neutrophil granule proteins in middle ear effusions. Arch. Otolaryngol. Head Neck Surg. 116: 335 [DOI] [PubMed] [Google Scholar]
- 11. Shuto T., Imasato A., Jono H., et al. 2002. Glucocorticoids synergistically enhance nontypeable Haemophilus influenzae-induced Toll-like receptor 2 expression via a negative cross-talk with p38 MAP kinase. J. Biol. Chem. 277: 17263 [DOI] [PubMed] [Google Scholar]
- 12. Imasato A., Desbois-Mouthon C., Han J., et al. 2002. Inhibition of p38 MAPK by glucocorticoids via induction of MAPK phosphatase-1 enhances nontypeable Haemophilus influenzae-induced expression of toll-like receptor 2. J. Biol. Chem. 277: 47444 [DOI] [PubMed] [Google Scholar]
- 13. Takeda K., Akira S. 2004. TLR signaling pathways. Semin. Immunol. 16: 3 [DOI] [PubMed] [Google Scholar]
- 14. Hirano T., Kodama S., Moriyama M., Kawano T., Suzuki M. 2009. The role of Toll-like receptor 4 in eliciting acquired immune responses against nontypeable Haemophilus influenzae following intranasal immunization with outer membrane protein. Int. J. Pediatr. Otorhinolaryngol. 73: 1657 [DOI] [PubMed] [Google Scholar]
- 15. Hemmi H., Kaisho T., Takeuchi O., et al. 2002. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 3: 196 [DOI] [PubMed] [Google Scholar]
- 16. Sakai A., Koga T., Lim J. H., et al. 2007. The bacterium, nontypeable Haemophilus influenzae, enhances host antiviral response by inducing Toll-like receptor 7 expression: evidence for negative regulation of host anti-viral response by CYLD. FEBS J. 274: 3655 [DOI] [PubMed] [Google Scholar]
- 17. Veltri R. W., Sprinkle P. M. 1976. Secretory otitis media. An immune complex disease. Ann. Otol. Rhinol. Laryngol. 85: 135 [DOI] [PubMed] [Google Scholar]
- 18. Shimizu T., Harada T., Majima Y., Sakakura Y. 1988. Bactericidal activity of middle ear effusion on a single isolate of non-typable Haemophilus influenzae . Int. J. Pediatr. Otorhinolaryngol. 16: 211 [DOI] [PubMed] [Google Scholar]
- 19. Stenfors L. E., Räisänen S. 1992. Opsonization of middle ear bacteria during chronic suppurative and secretory otitis media. Acta Otolaryngol. 112: 96 [DOI] [PubMed] [Google Scholar]
- 20. Bernstein J. M., Schenkein H. A., Genco R. J., Bartholomew W. R. 1978. Complement activity in middle ear effusions. Clin. Exp. Immunol. 33: 340 [PMC free article] [PubMed] [Google Scholar]
- 21. Figueira M. A., Ram S., Goldstein R., Hood D. W., Moxon E. R., Pelton S. I. 2007. Role of complement in defense of the middle ear revealed by restoring the virulence of nontypeable Haemophilus influenzae siaB mutants. Infect. Immun. 75: 325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Melhus A., Ryan A. F. 2000. Expression of cytokine genes during pneumococcal and nontypeable Haemophilus influenzae acute otitis media in the rat. Infect. Immun. 68: 4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tong H. H., Chen Y., Liu X., DeMaria T. F. 2008. Differential expression of cytokine genes and iNOS induced by nonviable nontypeable Haemophilus influenzae or its LOS mutants during acute otitis media in the rat. Int. J. Pediatr. Otorhinolaryngol. 72: 1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O’Rourke R. W., Kay T., Lyle E. A., et al. 2006. Alterations in peripheral blood lymphocyte cytokine expression in obesity. Clin. Exp. Immunol. 146: 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


